Abstract
The objective of this study was to investigate the effect of EF24, an NF-κB-inhibitor, on the expression of negative regulators in IL-1R pathway, namely ST2 and SIGIRR. Murine JAWS II dendritic cells (DC) were challenged with lipopolysaccharide (LPS, 100 ng/ml) for 4 h, followed by treatment with 10 μM EF24 for 1 h. ST2 and SIGIRR expression was monitored by qRT-PCR and immunoblotting. ST2L and MyD88 interaction was studied by co-immunoprecipitation, and IL-33, a ST2L ligand, was assayed by ELISA. Activation of transcription factor SP1 was examined by confocal microscopy, immunoblotting, and EMSA. The effect of EF24 on accumulation of ubiquitinated proteins in DCs and proteolysis of fluorogenic peptides by purified proteasome was studied. We found that EF24 upregulated the expression of ST2 and SIGIRR and decreased the interaction of the membrane-bound ST2 (ST2L) with MyD88, and significantly reduced IL-33 levels in LPS-stimulated DCs. Simultaneously it increased the activation of transcription factor SP1and restored the basal level of ubiquitinated proteins in LPS-stimulated DCs. Moreover, EF24 inhibited trypsin- and chymotrypsin-like activity of proteasome by directly interacting with 26S proteasome. The results suggest that EF24 activates endogenous anti-inflammatory arm of IL-1R signaling, most likely by stabilizing SP1 against proteasomal degradation.
Keywords: Lipopolysaccharide, Inflammation, Dendritic cells, IL-1R, EF24, ST2, SIGIRR
1. Introduction
Innate arm of immune response is responsible for local and systemic homeostasis after tissue injury. The inflammatory response associated with this process is meant for the protection of affected tissue from the primary insult [1], but uncontrolled or deregulated activation of inflammatory and immune responses can be detrimental to the host and cause inflammatory disorders of chronic and lethal consequences. The manifestation of this inflammatory reaction is preceded by recruitment of immune cells, such as neutrophils, monocytes and dendritic cells (DCs). These immune cells express special cell surface receptors mainly belonging to the interleukin-1 receptor (IL-1R) superfamily. Upon stimulation, the IL-1Rs recruit adaptor proteins which initiate a molecular cascade culminating into the secretion of effector inflammatory principles such as interleukin (IL)-6 and tumor necrosis factor (TNF)-α [2, 3].
IL-1R superfamily is characterized by the presence of a conserved intracellular Toll/IL-1R (TIR) domain [4, 5]. In addition to the receptors for IL-1, this superfamily is populated by IL-18 receptors, Toll-like receptors, and an orphan receptor called Suppression of Tumorigenicity 2 (ST2) [6]. The TLR group exhibits extracellular leucine-rich repeats, whereas IL-1R1 and ST2 groups contain extracellular immunoglobulin (Ig) domains. The third group of IL-1R superfamily is occupied by cytosolic adaptor proteins such as myeloid differentiation primary response 88 (MyD88), Toll-IL-1R (TIR) domain-containing adapter-inducing interferon-β (TRIF), and Toll-like receptor adaptor molecule (TRAM). A relatively recent IL-1R member is single immunoglobulin IL-1R-related (SIGIRR) which differs from other members in having only one Ig domain in its extracellular region [7].
The common signaling pathway is tightly regulated at different levels by pro-inflammatory or anti-inflammatory role affected by these IL-1R members. For instance, the expression of decoy receptor IL-1R2 binds the pro-inflammatory ligands, making them unavailable for transducing pro-inflammatory signaling via IL-1R1. Similarly, ST2 and SIGIRR are purported as two negative regulators of pro-inflammatory IL-1R signaling. They are fringe regulatory molecules belonging to orphan class of receptors. ST2 can be membrane-bound (ST2L) or soluble form (sST2). Binding of ST2L with interleukin-33 (IL-33) activates T helper 2 (TH2) response [8]. It has also been suggested that induced expression of ST2L can sequester MyD88, making latter unavailable for other MyD88-dependent pro-inflammatory IL-1R pathways [6]. In addition, abundance of sST2 purportedly prevents IL-33 from binding to ST2L and activating TH2 response [8]. On the other hand, SIGIRR-mediated negative regulation of inflammation follows two entirely different mechanisms. Immunoglobulin domain of SIGIRR interferes with heterodimerization of IL-1R to its accessory proteins, whereas its intracellular TIR domain inhibits IRAK-TRAF6 interaction, thus inhibiting the activation of nuclear factor-kappa B (NF-κB) [9].
To date, no small anti-inflammatory molecule has been shown to modulate ST2 and SIGIRR expression. The primary objective of this study was to investigate the effect of EF24, 3,5-bis(2-fluorobenzylidene)piperidin-4-one, on the expression of ST2, and SIGIRR in lipopolysaccharide (LPS)-stimulated DCs. DCs are the most potent antigen presenting cells, and compared to macrophages, DCs are better able to ‘preserve’ antigenic information because of their low destructive capacity [10]. LPS-stimulated DCs serve as an in vitro model of sterile inflammation by mimicking systemic inflammation associated with hemorrhagic shock without sepsis. In our previous study we have demonstrated that EF24 interferes with NF-κB signaling in DCs, resulting in inhibition of their maturation and expression of major histocompatibility class (MHC) II molecules [11]. Putatively, EF24 inhibits the β isoform of inhibitor of κB kinase (IKKβ) [11, 12]. Here, we report that EF24 increases the expression of SIGIRR and ST2, negative regulators of IL-1R/TLR signaling, in LPS-stimulated DCs.
2. Results
We used immortalized C57BL/6 mouse-derived JAWS II DCs to elucidate the modulation of ST2 and SIGIRR by EF24. As described in our previous reports, JAWS II cells showed typical immunophenotypic and morphologic characteristics of DCs when cultured in presence of GM-CSF [11, 17]. This included the appearance of dendrites and typical cytokine profile upon stimulation with LPS [11, 17].
2.1. EF24 enhances the expression of ST2
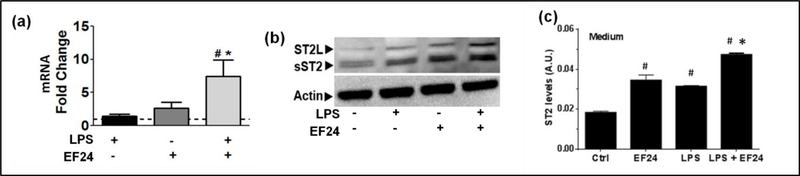
As shown in Figure 1a, EF24 upregulated total ST2 mRNA expression in LPS-stimulated DCs, but not in unstimulated DCs. Immunobloting revealed that both LPS alone and EF24 alone substantially increased the expression of sST2 and ST2L proteins (Figure 1b). However, EF24 treatment of LPS-stimulate cells showed large increase in sST2 and ST2L protein expression (Figure 1b). To further substantiate the effect of EF24 on sST2 expression in DCs, we next determined the concentration of ST2 in the cell culture medium by ELISA (Figure 1c), assuming that ST2 detected in medium would most likely be the soluble form sST2. EF24 treatment alone also significantly increased the content of ST2 in medium which was in line with the observation in immunoblots that EF24 increases ST2 protein expression in unstimulated cells. Compared to the basal levels, LPS alone also significantly increased ST2 in medium, but EF24 treatment further enhanced ST2 in medium of LPS-treated DCs.
Figure 1.
EF24 upregulates the expression of ST2. (a) ST2 mRNA expression (fold changes as compared to control) in JAWS II DCs. The cells were treated with LPS and EF24 as described in the material and methods. (b) A representative immunoblot showing the expression of ST2L and sST2 in DCs treated with LPS and EF24. (c) Effect of EF24 on sST2 in medium of LPS-stimulated JAWS II DCs. ELISA was performed in triplicate on samples from three separate sets of experiments. The values were normalized with protein concentrations in cell lysates. # p value < 0.05 versus Ctrl and * p value < 0.05 versus LPS treatment.
2.2. EF24 reduces interaction of ST2 with MyD88 and suppresses the expression of ST2L ligand IL-33
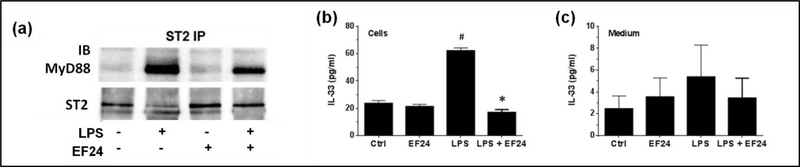
The TH2 inflammatory signaling of membrane-bound ST2 (ST2L) occurs via recruitment of a common intracellular adaptor protein MyD88 [6]. To test the hypothesis that EF24 treatment suppresses the downstream MyD88-dependent signaling of ST2L, we immunoprecipitated ST2 and probed the immunoprecipitate with MyD88 antibody. As shown in Figure 2a, large amounts of MyD88 co-immunoprecipitated with ST2 in LPS-stimulated DCs, however the treatment with EF24 substantially reduced the level of MyD88 bound to ST2. EF24 treatment by itself had no effect on this interaction in unstimulated DCs. Interestingly in LPS-treated cells, there was a corresponding decrease in the ST2L-specific immunoreactive band (Figure 2a, lower row), perhaps because of its interaction with MyD88. These results suggest that EF24 treatment reduces ST2L-MyD88 interaction.
Figure 2.
EF24 reduces ST2L-MyD88 interaction and suppresses IL-33 expression. (a) Co-immunoprecipitation of MyD88 with ST2 from whole cell lysates treated with EF24 and LPS. The blot shown is a representative of three separate experiments. (b) Effect of EF24 on IL-33 expression in LPS-stimulated JAWS II DCs. (c) Effect of EF24 on IL-33 levels in medium of LPS-stimulated JAWS II DCs. The ELISA was performed in triplicate on samples from three separate sets of experiments. The values were normalized with protein concentrations in cell lysates. # p value < 0.05 versus Ctrl and * p value < 0.05 versus LPS treatment.
MyD88 is recruited to ST2L upon interaction of IL-33 with ST2L. Therefore, we estimated the intracellular and extracellular concentration of IL-33 (Figure 2b). We found that the concentration of IL-33 in cell lysate was significantly increased by LPS, but this LPS-induced intracellular IL-33 was reduced to basal levels by EF24 treatment. The trend was similar in the medium, but the data variability precluded any statistically valid conclusions about extracellular IL-33 levels (Figure 2c).
2.3. EF24 treatment upregulates SIGIRR
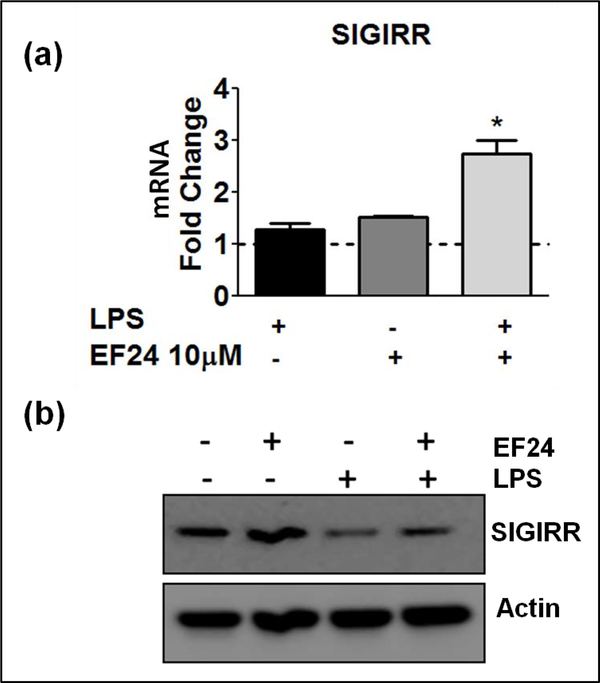
SIGIRR is another intracellular mediator belonging to the IL-1R family that has been shown to negatively regulate pro-inflammatory IL-1R1 signaling [9]. In our in vitro model of LPS-stimulated myeloid DCs, we found that EF24 treatment increased SIGIRR expression at both mRNA and protein levels (Figure 3a and 3b, respectively). LPS by itself reduced the protein levels of SIGIRR, but had no effect on its mRNA levels. EF24 alone had negligible effect on basal levels of SIGIRR protein.
Figure 3.
EF24 upregulates the expression of SIGIRR. (a) SIGIRR mRNA expression (fold changes as compared to control) in JAWS II DCs. The cells were treated with LPS and EF24 as described in the material and methods. # p value < 0.05 versus control and * p value < 0.05 versus LPS treatment. (b) A representative immunoblot of SIGIRR expression in cells treated with EF24 and LPS.
2.4. EF24 treatment activates SP1/PU.1 and inhibits proteasome activity in LPS-stimulated DCs
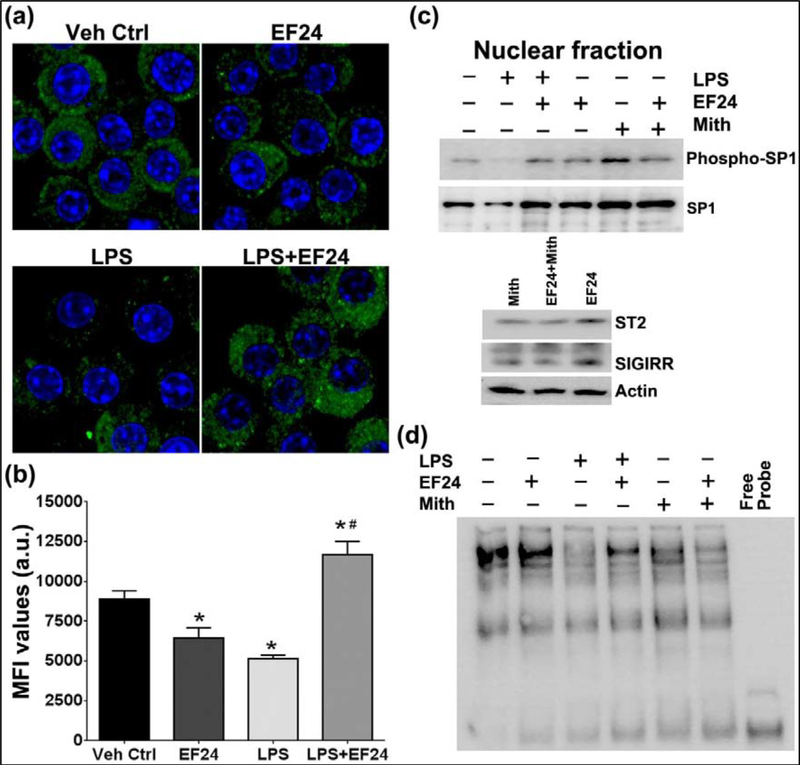
SP1/PU.1 is a putative transcriptional regulator of ST2 and SIGIRR expression. We monitored the activation of SP1 by labeling the LPS-stimulated DCs with primary antibody against the phosphorylated form of SP1 (Figure 4a–b). Confocal micrographs provided evidence that LPS treatment alone decreased the expression of phospho-SP1 and EF24 treatment induced the expression of phospho-SP1 in LPS-stimulated cells. These confocal observations were substantiated by immunoblotting results where the expression of phospho-SP1 was reduced by LPS stimulation, and subsequent EF24 treatment mitigated this LPS-effect (Figure 4c). Furthermore, EF24 was found to increase transcriptional activity of SP1 in DCs as determined by SP1 EMSA (Figure 4d). Whereas experimental evidence from expression-based techniques (confocal and immunoblotting) suggests that EF24 has no effect in unstimulated DCs, EMSA results suggest that EF24 induces DNA-binding of SP1 even in unstimulated DCs. This explains why EF24 treatment of unstimulated DCs moderately increased the expression of ST2 and SIGIRR (Figure 1 and Figure 3). When SP1 activation was blocked by a specific pharmacologic inhibitor mithramycin, EF24-induced expression of ST2 and SIGIRRR was abolished (Figure 4c, lower panel). Since mithramycin is a competitive inhibitor of SP1, which binds to GC-rich DNA sequences and displaces SP1 from its binding sites, expression levels of phospho-SP1 and SP1 were not affected by mithramycin (Figure 4c), but SP1 DNA-binding was reduced in EMSA (Figure 4d).
Figure 4.
EF24 treatment activates SP1 in LPS-stimulated DCs, but not in unstimulated DCs. (a) Confocal micrographs and (b) corresponding mean fluorescence intensity values for green fluorescence representing phosphorylated form of SP1. The nucleus was stained with Hoechst dye (blue). A representative set of images is shown. (c) Expression levels of phospho-SP1 and SP1 in the nuclear extract of dendritic cells (upper panel). Effect of mithramycin on EF24-induced ST2 and SIGIRR expression (lower panel). Mithramycin (Mith, 300 nM) was used as a control to specifically inhibit interaction of SP1 with its consensus DNA binding site. (d) Electrophoretic mobility shift assay showing induction of SP1activation by treatment of EF24 in LPS-stimulated DCs.
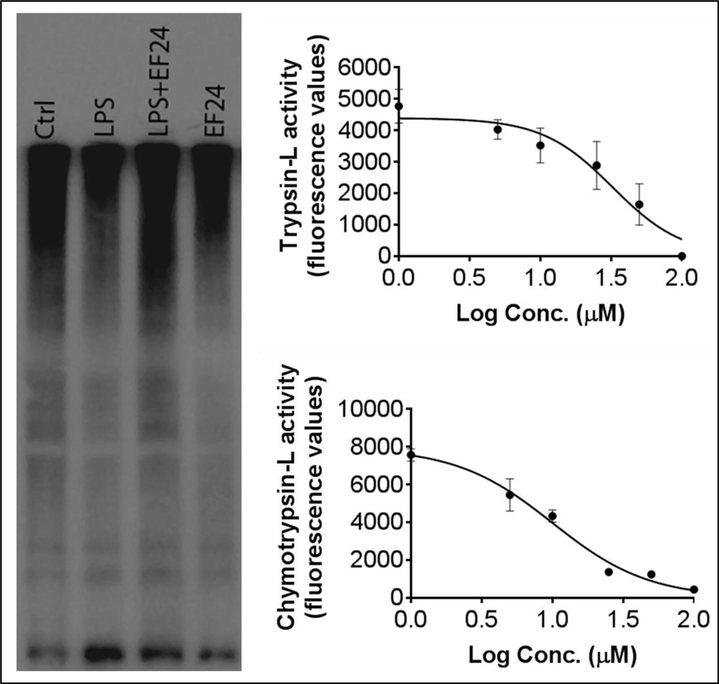
In addition to the phosphorylation-mediated regulation of activity, the expression of SP1 is also controlled by its degradation in proteasome-mediated proteolysis. We examined the status of proteasome activity in DCs by following the accumulation of ubiquitintated proteins (UbPr). As shown in Figure 5a, we found that LPS stimulation reduced the UbPr levels whereas EF24 treatment significantly restored the expression levels of UbPr in LPS-stimulated DCs. These results suggest that EF24 treatment can also increase the expression of SP1 by reducing its degradation by proteasome. We confirmed the inhibitory activity of EF24 on proteasome by assessing the direct effect of EF24 on the proteolytic activity of purified proteasome (Figure 5b). EF24 was found to significantly inhibit trypsin-like and chymotrypsin-like proteolytic activities of purified proteasome. The IC50 values of EF24 calculated from dose-response curves were 32.4 μM and 10.1 μM for trypsin-L and chymotrypsin-L activities (Figure 5b).
Figure 5.
Effect of EF24 on proteasome activity. (a) Expression of ubiquitinated proteins in LPS-stimulated DCs. (b) Effect of EF24 on trypsin-like and chymotrypsin-L activity of purified 26S proteasome.
3. Discussion
Molecular signaling engendered by interaction of damage-associated molecular patterns and other putative ligands with IL-1R members is the earliest response in the inflammatory process post-injury [18]. Often, this protective response progresses into a vicious and self-damaging phenomenon which provides a pathophysiological basis of multiple organ dysfunction syndrome in systemic injuries such as hemorrhagic shock and trauma [19]. The two most important members of the IL-1R superfamily are TLR4 and IL-1R1. Whereas bacterial LPS is a putative ligand for TLR4, pro-inflammatory IL-1α and IL-1β cytokines are ligands for IL-1R1. EF24 treatment has been shown to suppress the expression of IL-1R1 and TLR4 in LPS-stimulated DCs [20]. This anti-inflammatory effect of EF24 is also duplicated in lung and liver intestinal tissues collected from a rat model of hemorrhagic shock [21–23]. Data suggests that these effects of EF24 are at transcriptional level controlled by NF-κB transcription factor [11, 12]. Herein, we provide evidence to hypothesize that anti-inflammatory effect of EF24 may also be contributed by the induction of endogenous anti-inflammatory molecules ST2 and SIGIRR. Even though they belong to the IL-1R superfamily, they are not known to directly interact with IL-1α or IL-1β. The effects of EF24 on the expression of ST2 and SIGIRR appear to be at transcriptional level, perhaps mediated by restoration of the basal induction of SP1 transcription factor. We have previously shown that EF24 does not alter the expression of two other negative regulators of inflammation, namely IL-1RA and IL-1R2 [20].
IL-1R pathway is characterized by in-built negative regulators such as ST2 and SIGIRR, the sole purpose of which appears to be that of preventing exaggerated inflammation. Upregulation of ST2 has been shown to play an important role in DC maturation, induction of tolerance, and T cell polarization [6, 24, 25]. The membrane-bound ST2L interacts with its ligand IL-33 resulting in the production of IL-10-dependent Th2 phenotype [26] and the development of tolerance against LPS [6]. On the other hand, a soluble form of ST2 (sST2) competitively attenuates Th2 response [8]. Macrophages from ST2-deficient mice produce enhanced levels of pro-inflammatory cytokines in response to LPS [6] and these mice display increased susceptibility to polymicrobial infection and impaired bacterial clearance [27]. It has been speculated that ST2 sequesters MyD88, making latter unavailable for other MyD88-dependent pro-inflammatory IL-1R pathways [6]. However, evidence is gathering against this simplistic explanation because it cannot explain several observations about antagonism of inflammation in ST2 knockout conditions. For instance, the severity of inflammation was found to be lower in the ST2 knockout mice than in wild type mice [28]. Secondly, binding of IL-33 to ST2 has been demonstrated as pro-inflammatory in nature, especially in the context of cardiovascular disease [8]. Therefore, a more realistic explanation appears to be based on the possibility that soluble form of ST2 sequesters IL-33 and thwarts it’s binding to the ST2L, preventing IL-33/ST2 signaling which is destined towards NF-κB activation. Soluble ST2 originates from alternative mRNA processing which has been shown to inhibit LPS stimulation of monocyte-derived dendritic cells [29]. Our results clearly show that EF24 treatment of LPS-stimulated DCs increases the secretion of soluble ST2 in medium (Fig. 1c). At the same time, the production of IL-33 was reduced in cells treated with EF24 (Fig. 2b). It is possible that the reduced interaction of ST2L and MyD88 (Fig. 2a) is a combined result of the suppression of IL-33 generation and its sST2-mediated sequestration. MyD88 is a common intracellular adaptor protein for multiple members of IL-1R family, and the downstream signaling of membrane-bound ST2 (ST2L) also occurs via MyD88 recruitment [6].
In addition to the effects on IL-33/ST2 axis, EF24 treatment increased SIGIRR expression in LPS-stimulated DCs (Fig. 3). Studies have shown that unlike ST2, SIGIRR (also known as TIR8) is expressed in DCs, but not in macrophages, and the mechanism of SIGIRR-mediated negative regulation of inflammation is also different [7, 30, 31]. According to Qin et al, SIGIRR inhibits IL-1R signaling by physically interacting with IL-1R1, TLR4, and MyD88 [9]. The Ig domain of SIGIRR interferes with the heterodimerization of IL-1R to its accessory proteins, whereas its intracellular TIR domain inhibits IRAK-TRAF6 interaction, thus inhibiting the activation of NF-κB [9]. It has been shown to constitutively interact with MYD88 and is required for dominant negative mutant MyD88 (MyD88lpr)-induced activation of DCs; MyD88lpr was unable to activate SIGIRR-deficient DCs, whereas restoration SIGIRR together with MyD88lpr was able to activate these DCs [32]. Mice lacking SIGIRR show hyper-responsiveness to LPS challenge and IL-1 administration, accompanied by enhanced activation of TLR/IL-1 pathways [7]. Also, a loss of function mutation in SIGIRR gene predisposes ischemic kidney to acute failure, whereas wild type SIGIRR prevents tissue injury by restraining the activation of intra-renal myeloid DCs [33]. Moreover, SIGIRR has also been shown to acts as a key local modulator of immune cell activation underlying kidney rejection and tolerance; Deletion of SIGIRR in the graft resulted in acute rejection of allo-transplanted kidney [34]. The post-transplant ischemia/reperfusion-induced inflammatory response was also more severe in SIGIRR(−/−) than in SIGIRR(+/+) grafts and was followed by expansion and maturation of resident DC precursors [34]. SIGIRR(−/−) DC precursors exhibited higher stimulatory activity and released more IL-6 after stimulation with a TLR4 ligand and TNF-α in vitro than SIGIRR(+/+) cells [34].
From above dispensation it is clear that increased sST2 and SIGIRR expression has anti-inflammatory consequences. To our knowledge no pharmacologic agent has been shown to increase the expression of these endogenous molecules, and this report is the first report in this respect. Although, the mechanism by which EF24 affects their expression is a matter of continued investigation, we found plausible role of SP1 transcription factor in this phenomenon. First, the expression of both ST2 and SIGIRR in LPS-stimulated cells was at mRNA level. SP1/PU.1 is a putative transcriptional regulator of ST2 and SIGIRR expression. Murine ST2 gene is mapped to chromosome 1, very tightly linked to the IL-1R1 locus, and contains two SP1-binding sites in the 5′ flanking region [35]. Similarly, Murine SIGIRR, mapped to chromosome 7 [36], also carries a putative SP1 binding site [37]. Like many other transcription factors, the activation and DNA binding of SP1 is also dependent on its phosphorylation status and nuclear translocation [38]. Here, we found that the nuclear translocation of phospho-SP1 is enhanced by EF24 treatment. This finding was confirmed when mithramycin, a selective SP1 inhibitor [39, 40], reduced the level of EF24-mediated nuclear translocation of SP1. Mithramicin treatment also abolished the increased expression of ST2 and SIGIRR by EF24 treatment. Based on these results we hypothesize that EF24 might be inducing ST2 and SIGIRR expression by restoring the activation state of SP1.
There are three major mechanisms of SP1 regulation known in the literature. NF-κB and SP1 itself transcriptionally control the expression of SP1 gene, whereas the levels of SP1 protein are controlled by proteasome-mediated degradation [41–43]. Since previous studies have reported that LPS treatment induces large scale proteasome activation [44–46], we argued that the exaggerated proteasome activation in LPS-stimulated DCs plays a role in loss of SP1 protein. Global expression levels of UbPr are accepted as indicators of proteasome activity. We found that LPS reduced the UbPr level in DCs, suggesting that LPS induces proteasome activity; this effect of LPS was reversed by EF24 treatment (Fig. 5a). When EF24 was further investigated for direct proteasome-inhibiting activity in cell-free condition, EF24 was found to significantly reduce the trypsin-like and chymotrypsin-like proteolytic activity of purified 26S proteasome with moderate potency. Thus, one likely mechanism by which EF24 maintains basal level of SP1 activity in DCs is by interfering with LPS-induced proteasome activity.
4. Materials and Methods
4.1. Materials
Synthesis and chemical characterization of EF24 has been reported elsewhere [13]. A solution of EF24 in was prepared in water (< 0.1 endotoxin units/ml). Highly-purified lipopolysaccharide (LPS) of Escherichia coli O111: B4 was obtained from Calbiochem (Darmstadt, Germany). Antibodies for immunoblotting were obtained from Cell Signaling Technology (Danvers, MA), Santa Cruz Biotech (Dallas, TX), Abcam (Cambridge, MA), and Sigma-Aldrich (St. Louis, MO).
4.2. Cell culture and drug treatment
We employed an LPS-stimulated DCs model the characteristics of which have been described in our previous works [11, 14]. JAWS II DCs are immortalized and immature cells derived from C57BL/6 mice bone marrow (ATCC, Manassas, VA). The cells were maintained in α-modified minimum essential medium (αMMEM; Sigma, St Louis, MO) containing 20 % fetal bovine serum (FBS), L-glutamine (4 mM), penicillin (100 U/ml), streptomycin (100 μg/ml), gentamicin (50 μg/ml) and recombinant mouse granulocyte macrophage-colony stimulating factor (GM- CSF, 5 ng/ml; Peprotech, Rocky Hill, NJ) [14]. The cells were cultured in 6-well plates and treated with LPS (100 ng/ml) for 4 h, followed by EF24 (10 μM) for 1 h. Untreated cells and cells treated with 10 μM EF24 alone were taken as experimental controls.
4.3. Quantitative Real-time PCR
Total RNA was isolated from DCs using RNAeasy Mini Kit from Qiagen (Valencia, CA) and quantified on the basis of absorbance values at 260 nm. Conditions for reverse transcriptase reactions and PCR using SybrGreen II and Go Taq colorless master mix are described elsewhere [20]. The data were normalized using β-actin as the reference. Fold increase over control was calculated using the relative quantification method (2−ΔΔ Ct method). ST2 primers (Forward 5′- GAC AGT CTT TTC CAG GAG CA -3′ and Reverse 5′- GAG TTC AAC ATT CCC CTC CT -3′) and SIGIRR primers (Forward 5′- AGC TCC CAT TCC TTC ACT CT -3′ and Reverse 5′- TCG CTA TAG GAC ACG TAG GC -3′) were obtained from Real Time Primers (RTP, Elkins Park, PA).
4.4. Immunobloting
Whole cell lysates were prepared in a lysis buffer containing Tris-HCl (25 mM, pH 7.4), sodium dodecyl sulfate (SDS, 0.1%), Igepal (1%), ethylene diamine tetra acetic acid (2 mM), phenyl methyl sulfonyl fluoride (1 mM), sodium orthovanadate (0.2 mM), NaF (50 mM), NaCl (150 mM), leupeptin (1 μg/ml), and pepstatin (1 μg/ml). Approximately 25 μg of protein was fractionated on SDS-PAGE gels and the separated proteins were electro-transferred onto nitrocellulose membranes. After blocking non-specific sites with 5% skimmed milk in 0.4% Tween-20 in Tris-buffered saline (TBST), the membranes were incubated overnight at 4 °C with primary antibodies in TBST. The dilutions of primary antibodies against Phospho-SP1 (Abcam), SP1 (Abcam), SIGIRR (Santa Cruz Biotech) and mouse ST2 (Abcam) were made as recommended by the manufacturers. Afterwards, the membranes were washed and incubated with HRP-conjugated-anti-rabbit-IgG antibody (1:5,000 in TBST) for 2 h at room temperature. Protein bands were identified by SuperSignal West Femto detection reagent (Thermo Fisher Scientific, Rockford, IL). The membranes were also re-probed with anti-actin antibody (Sigma; 1:5,000 in TBST) after stripping them in a solution containing SDS (10%), Tris (0.5 M), and β-mercaptoethanol (35 μl/ml) at 60 °C for 45 min. The densitometric readings for immunoreactive protein bands were normalized with those for actin.
4.5. Confocal microscopy
For confocal detection of protein expression, DCs were seeded in 8-well chamber slides (25,000 cells per well). After treatment with LPS and EF24, the cells were fixed for 20 min with ice-cold 3.5% paraformaldehyde in phosphate-buffered saline (PBS). The fixed cells were incubated for 20 min on ice in αMMEM containing FBS (10%), saponin (0.05%) and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (10 mM) [15]. After washing with PBS containing 1% FBS and 0.05% saponin, the cells were blocked for 1 h at room temperature with 10% normal mouse serum (Sigma, St Louis MO). For probing SP1 activation, we used anti-SP1 (phospho T453) antibody (Abcam) at a dilution of 1:200. Following rigorous washing, the cells were incubated for 1 h with Alexa Fluor 488-labeled donkey anti-rabbit IgG antibody (10 μg/ml; Molecular Probes, Carlsbad, CA). Finally, 1 μg/ml Hoechst 33342 (Molecular Probes, Carlsbad, CA) dye was added to the cells. Confocal micrographs were obtained using Zeiss LSM-510META microscope (Oklahoma Medical Research Foundation, Oklahoma City, OK).
4.6. Enzyme-linked immunosorbent assay (ELISA)
The concentration of IL-33 and soluble ST2 in cultured DCs and media were measured by ELISA. The protocols for the treatment of JAWS II cells with LPS and EF24 were the same as described above. Cell-free culture medium as well as total cell lysates was collected. For IL-33, a commercially available mouse IL-33 ELISA Ready-SET go kit (eBioscience, San Diego, CA) was employed following the manufacturer’s instructions.
Mouse ST2 ELISA was set up in-house using rabbit anti-ST2 antibody (Santa Cruz Biotech). Briefly, an Immunolon 4 HBX plate (Thermo Electron Corporation, Milford, MA) was coated overnight at 4 °C with 50 μl of samples (and washed three times with 0.05% Tween-20 containing PBS (PBST). The wells were blocked with 3% bovine serum albumin (BSA) for 2 h, followed by washing with PBST. The wells were probed with 100 μl of 10 μg/ml rat anti-mouse ST2 antibody for 2 h. After washing thrice with PBST, 100 μl of HRP-conjugated goat anti-rabbit IgG antibody (Cell Signaling Technology) in blocking buffer (1:1,000) was added. The plate was allowed to incubate for 2 h at room temperature. The color was developed by adding 100 μl of 3,3′,5,5′-tetramethylbenzidine substrate solution (Sigma) and allowing the reaction for 30 min. The reaction was stopped by adding 50 μl of 0.2 N sulfuric acid and the absorbance was read at 450 nm.
4.7. Nuclear Extracts
For extraction of nuclear protein, we used a kit from Cayman chemical (Ann Arbor, MI). DCs were seeded at a density of 2 × 106 cells per well in a 6-well plate. Four hour after the treatment with LPS (100 ng/ml), the cells were treated with EF24 (10 μM) and mithramycin (Mith, 300 nM) for 1 hr followed by washing with ice-cold PBS. Nuclear fraction was isolated as per manufacturer’s protocol and protein concentration in the fractions was estimated by bicinchoninic acid protein assay (Pierce, Rockford, IL).
4.8. Electrophoretic Mobility Shift Assay for SP1
Electrophoretic Mobility Shift Assay (EMSA) was performed using EMSA assay kit (Signosis, Santa Clara, CA) following manufacturer’s instructions. Briefly, reaction mixture containing 5 μg nuclear extract, poly d(I-C), binding buffer, biotin labeled SP1 probe or cold probe (negative control) were incubate for 30 min at RT followed by running the samples on 6.5% non-denaturing polyacrylamide gel using Tris-borate buffer. After transfer, the protein-bound probe and free probe were immobilized with UV cross-linker followed by blocking and incubation with Streptavidin-HRP. Membrane was developed using Ultraquant image acquisition machine (Claremont, CA).
4.9. Ubiquitinated protein (UbPr) Expression in DCs and Proteasome Activity
Denaturing gel electrophoresis was conducted for the expression of UbPr in DC cell lysates. Immunoblotting method as described above was followed. Primary antibody against ubiquitin was purchased from Santa Cruz Biotech (Dallas, TX). For proteasome activity, we adapted a method described elsewhere [16]. Fluorogenic substrates for trypsin-like and chymotrypsin-like proteasome activities were Boc-Leu-Arg-Arg-AMC and Suc-Leu-Leu-Val-Tyr-AMC, respectively. Purified 26S proteasome preparation was obtained from Boston Biochem (Cambridge, MA). Approximately 100 ng of purified proteasome was incubated with 100 μl of assay buffer (50 mM Tris‐HCl, pH 7.5, 40 mM KCl, 1 mM dithiothreitol, 0.5 mg/ml bovine serum albumin) in presence of 2 mM ATP and 5 mM MgCl2. Fluorogenic substrates were added at 1 mM working concentration and the reaction was performed at 37 °C for 60 min. Over the reaction period, fluorescence readings were taken every 5 min at 380–440 nm. Boiled homogenate (to kill enzyme activity) and bortezomib (a known proteasome inhibitor) were used as assay controls. To examine the effect of EF24, proteasome preparations were allowed to interact with EF24 (1–100 μM, 10 min at room temperature) before the activity assay.
4.10. Data analysis
The data were analyzed by one-way analysis of variance (ANOVA) applying the Bonferroni post-test using Prism software (GraphPad, San Diego, CA, USA). Wherever applicable, the data are presented as mean ± standard error of mean (sem). For significance, p < 0.05 was the cutoff. The qRT-PCR data are expressed as fold changes compared to control.
5. Conclusions
We report for the first time that EF24 can enhance the expression of ST2 and SIGIRR which purportedly serve as endogenous negative regulators in the IL-1R pathway. So far the anti-inflammatory effects of EF24 have been solely speculated to be dependent on NF-κB inhibition. Although the direct evidence about the role of SP1 in resurrection of ST2 and SIGIRR is not available and the exact mechanism of SP1 modulation by EF24 is not known at present, these observations add to our recent reports that EF24 reduces the expression of pro-inflammatory IL-1R1 and TLR4, both in vitro as well as in vivo [11, 17, 20–23, 47]. Moreover, since proteasome inhibitors are known to inhibit NF-κB by reducing IκBα degradation [48, 49], direct inhibition of proteasome by EF24 also raises an investigational opportunity to further unravel mechanisms responsible for its NF-κB inhibition activity.
Acknowledgments
Funding for the work reported in this article was provided by the National Heart, Lung and Blood Institute (R01HL104286).
Conflicts of Interest: The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Abbreviations
- DCs
Dendritic cells
- EF24
3,5-bis(2-fluorobenzylidene)piperidin-4-one
- ELISA
Enzyme-linked immunosorbent assay
- EMSA
Electrophoretic mobility shift assay
- GM- CSF
Granulocyte macrophage-colony stimulating factor
- IC50
Concentration for 50% inhibition
- Ig
Immunoglobulin
- IKKβ
β isoform of inhibitor of κB kinase
- IL
Interleukin
- IL-1R
Interleukin-1 receptor
- LPS
Lipopolysaccharide
- qRT-PCR
Quantitative real-time polymerase chain reaction
- MHC
Major histocompatibility class
- MyD88
Myeloid differentiation primary response 88
- NF-κB
Nuclear factor-kappa B
- SP1
Specificity protein 1
- ST2
Suppression of Tumorigenicity 2
- sST2
Soluble ST2
- SIGIRR
Single Immunoglobulin IL-1R-related
- TH2
T helper 2
- TIR
Toll/IL-1R
- TLR
Toll-like receptor
- TNF
tumor necrosis factor
- TRIF
TRIF Toll–IL-1R (TIR) domain-containing adapter-inducing interferon-β
- UbPr
Ubiquitinated protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karin M; Lawrence T; Nizet V Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell 2006, 124, 823–835. [DOI] [PubMed] [Google Scholar]
- 2.Kobbe P; Lichte P; Schreiber H; Reiss LK; Uhlig S; Pape HC; Pfeifer R Inhalative IL-10 attenuates pulmonary inflammation following hemorrhagic shock without major alterations of the systemic inflammatory response. Mediators Inflamm 2012, 2012, 512974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavaillon JM Pro- versus anti-inflammatory cytokines: myth or reality. Cell Mol Biol (Noisy-le-grand) 2001, 47, 695–702. [PubMed] [Google Scholar]
- 4.Dinarello CA Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol 2009, 27, 519–550. [DOI] [PubMed] [Google Scholar]
- 5.Dunne A; O’Neill LA The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci STKE 2003, 2003, re3. [DOI] [PubMed] [Google Scholar]
- 6.Brint EK; Xu D; Liu H; Dunne A; McKenzie AN; O’Neill LA; Liew FY ST2 is an inhibitor of interleukin 1 receptor and Toll-like receptor 4 signaling and maintains endotoxin tolerance. Nat Immunol 2004, 5, 373–379. [DOI] [PubMed] [Google Scholar]
- 7.Wald D; Qin J; Zhao Z; Qian Y; Naramura M; Tian L; Towne J; Sims JE; Stark GR; Li X SIGIRR, a negative regulator of Toll-like receptor-interleukin 1 receptor signaling. Nat Immunol 2003, 4, 920–927. [DOI] [PubMed] [Google Scholar]
- 8.Kakkar R; Lee RT The IL-33/ST2 pathway: therapeutic target and novel biomarker. Nat Rev Drug Discov 2008, 7, 827–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin J; Qian Y; Yao J; Grace C; Li X SIGIRR inhibits interleukin-1 receptor- and toll-like receptor 4-mediated signaling through different mechanisms. J Biol Chem 2005, 280, 25233–25241. [DOI] [PubMed] [Google Scholar]
- 10.Savina A; Amigorena S Phagocytosis and antigen presentation in dendritic cells. Immunological Reviews 2007, 219, 143–156. [DOI] [PubMed] [Google Scholar]
- 11.Vilekar P; Awasthi S; Natarajan A; Anant S; Awasthi V EF24 suppresses maturation and inflammatory response in dendritic cells. Int Immunol 2012, 24, 455–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kasinski AL; Du Y; Thomas SL; Zhao J; Sun SY; Khuri FR; Wang CY; Shoji M; Sun A; Snyder JP; Liotta D; Fu H Inhibition of IkappaB kinase-nuclear factor-kappaB signaling pathway by 3,5-bis(2-flurobenzylidene)piperidin-4-one (EF24), a novel monoketone analog of curcumin. Mol Pharmacol 2008, 74, 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lagisetty P; Powell DR; Awasthi V Synthesis and structural determination of 3 3,5-bis(2-fluorobenzylidene)-4-piperidone analogs of curcumin. J Mol Str 2009, 936, 23–28. [Google Scholar]
- 14.Awasthi S; Awasthi V; Magee DM; Coalson JJ Efficacy of antigen 2/proline-rich antigen cDNA-transfected dendritic cells in immunization of mice against Coccidioides posadasii. J Immunol 2005, 175, 3900–3906. [DOI] [PubMed] [Google Scholar]
- 15.Inaba K; Turley S; Yamaide F; Iyoda T; Mahnke K; Inaba M; Pack M; Subklewe M; Sauter B; Sheff D; Albert M; Bhardwaj N; Mellman I; Steinman RM Efficient presentation of phagocytosed cellular fragments on the major histocompatibility complex class II products of dendritic cells. J Exp Med 1998, 188, 2163–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kisselev AF; Goldberg AL Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods Enzymol 2005, 398, 364–378. [DOI] [PubMed] [Google Scholar]
- 17.Vilekar P; Awasthi V; Lagisetty P; King C; Shankar N; Awasthi S In vivo trafficking and immunostimulatory potential of an intranasally-administered primary dendritic cell-based vaccine. BMC Immunol 2010, 11, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mollen KP; Anand RJ; Tsung A; Prince JM; Levy RM; Billiar TR Emerging paradigm: toll-like receptor 4-sentinel for the detection of tissue damage. Shock 2006, 26, 430–437. [DOI] [PubMed] [Google Scholar]
- 19.Bone RC Immunologic dissonance: a continuing evolution in our understanding of the systemic inflammatory response syndrome (SIRS) and the multiple organ dysfunction syndrome (MODS). Ann Intern Med 1996, 125, 680–687. [DOI] [PubMed] [Google Scholar]
- 20.Vilekar P; Rao G; Awasthi S; Awasthi V Diphenyldifluoroketone EF24 Suppresses Pro-inflammatory Interleukin-1 receptor 1 and Toll-like Receptor 4 in lipopolysaccharide-stimulated dendritic cells. J Inflamm (Lond) 2015, 12, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yadav VR; Sahoo K; Roberts PR; Awasthi V Pharmacologic Suppression of Inflammation by a Diphenyldifluoroketone, EF24, in a Rat Model of Fixed-Volume Hemorrhage Improves Survival. J Pharmacol Exp Ther 2013, 347, 346–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yadav VR; Vilekar P; Awasthi S; Awasthi V Hemorrhage-induced interleukin-1 receptor pathway in lung is suppressed by 3,5-bis(2-fluorobenzylidene)-4-piperidone in a rat model of hypovolemic shock. Artif Organs 2014, 38, 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadav VR; Hussain A; Xie J; Kosanke S; Awasthi V The salutary effects of diphenyldifluoroketone EF24 in liver of a rat hemorrhagic shock model. Scand J Trauma Resusc Emerg Med 2015, 23, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rank MA; Kobayashi T; Kozaki H; Bartemes KR; Squillace DL; Kita H IL-33-activated dendritic cells induce an atypical TH2-type response. J Allergy Clin Immunol 2009, 123, 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turnquist HR; Sumpter TL; Tsung A; Zahorchak AF; Nakao A; Nau GJ; Liew FY; Geller DA; Thomson AW IL-1beta-driven ST2L expression promotes maturation resistance in rapamycin-conditioned dendritic cells. J Immunol 2008, 181, 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller AM Role of IL-33 in inflammation and disease. J Inflamm (Lond) 2011, 8, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buckley JM; Liu JH; Li CH; Blankson S; Wu QD; Jiang Y; Redmond HP; Wang JH Increased susceptibility of ST2-deficient mice to polymicrobial sepsis is associated with an impaired bactericidal function. J Immunol, 187, 4293–4299. [DOI] [PubMed] [Google Scholar]
- 28.Talabot-Ayer D; Martin P; Seemayer CA; Vigne S; Lamacchia C; Finckh A; Saiji E; Gabay C; Palmer G Immune-mediated experimental arthritis in IL-33 deficient mice. Cytokine, 69, 68–74. [DOI] [PubMed] [Google Scholar]
- 29.Nagata A; Takezako N; Tamemoto H; Ohto-Ozaki H; Ohta S; Tominaga S; Yanagisawa K Soluble ST2 protein inhibits LPS stimulation on monocyte-derived dendritic cells. Cell Mol Immunol, 9, 399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garlanda C; Riva F; Polentarutti N; Buracchi C; Sironi M; De Bortoli M; Muzio M; Bergottini R; Scanziani E; Vecchi A; Hirsch E; Mantovani A Intestinal inflammation in mice deficient in Tir8, an inhibitory member of the IL-1 receptor family. Proc Natl Acad Sci U S A 2004, 101, 3522–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polentarutti N; Rol GP; Muzio M; Bosisio D; Camnasio M; Riva F; Zoja C; Benigni A; Tomasoni S; Vecchi A; Garlanda C; Mantovani A Unique pattern of expression and inhibition of IL-1 signaling by the IL-1 receptor family member TIR8/SIGIRR. Eur Cytokine Netw 2003, 14, 211–218. [PubMed] [Google Scholar]
- 32.Drexler SK; Wales J; Andreakos E; Kong P; Davis A; Garlanda C; Mantovani A; Hussell T; Feldmann M; Foxwell BM Evidence for a DC-specific inhibitory mechanism that depends on MyD88 and SIGIRR. Scand J Immunol 2010, 71, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lech M; Avila-Ferrufino A; Allam R; Segerer S; Khandoga A; Krombach F; Garlanda C; Mantovani A; Anders HJ Resident dendritic cells prevent postischemic acute renal failure by help of single Ig IL-1 receptor-related protein. J Immunol 2009, 183, 4109–4118. [DOI] [PubMed] [Google Scholar]
- 34.Noris M; Cassis P; Azzollini N; Cavinato R; Cugini D; Casiraghi F; Aiello S; Solini S; Cassis L; Mister M; Todeschini M; Abbate M; Benigni A; Trionfini P; Tomasoni S; Mele C; Garlanda C; Polentarutti N; Mantovani A; Remuzzi G The Toll-IL-1R member Tir8/SIGIRR negatively regulates adaptive immunity against kidney grafts. J Immunol 2009, 183, 4249–4260. [DOI] [PubMed] [Google Scholar]
- 35.Tominaga S; Jenkins NA; Gilbert DJ; Copeland NG; Tetsuka T Molecular cloning of the murine ST2 gene. Characterization and chromosomal mapping. Biochim Biophys Acta 1991, 1090, 1–8. [DOI] [PubMed] [Google Scholar]
- 36.Thomassen E; Renshaw BR; Sims JE Identification and characterization of SIGIRR, a molecule representing a novel subtype of the IL-1R superfamily. Cytokine 1999, 11, 389–399. [DOI] [PubMed] [Google Scholar]
- 37.Kadota C; Ishihara S; Aziz MM; Rumi MA; Oshima N; Mishima Y; Moriyama I; Yuki T; Amano Y; Kinoshita Y Down-regulation of single immunoglobulin interleukin-1R-related molecule (SIGIRR)/TIR8 expression in intestinal epithelial cells during inflammation. Clin Exp Immunol 2010, 162, 348–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan NY; Khachigian LM Sp1 phosphorylation and its regulation of gene transcription. Mol Cell Biol 2009, 29, 2483–2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sleiman SF; Langley BC; Basso M; Berlin J; Xia L; Payappilly JB; Kharel MK; Guo H; Marsh JL; Thompson LM; Mahishi L; Ahuja P; MacLellan WR; Geschwind DH; Coppola G; Rohr J; Ratan RR Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. Journal of Neuroscience, 31, 6858–6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blume SW; Snyder RC; Ray R; Thomas S; Koller CA; Miller DM Mithramycin inhibits SP1 binding and selectively inhibits transcriptional activity of the dihydrofolate reductase gene in vitro and in vivo. J Clin Invest 1991, 88, 1613–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tapias A; Ciudad CJ; Roninson IB; Noe V Regulation of Sp1 by cell cycle related proteins. Cell Cycle 2008, 7, 2856–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirano F; Tanaka H; Hirano Y; Hiramoto M; Handa H; Makino I; Scheidereit C Functional interference of Sp1 and NF-kappaB through the same DNA binding site. Mol Cell Biol 1998, 18, 1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han I; Kudlow JE Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol Cell Biol 1997, 17, 2550–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reis J; Guan XQ; Kisselev AF; Papasian CJ; Qureshi AA; Morrison DC; Van Way CW 3rd; Vogel SN; Qureshi N LPS-induced formation of immunoproteasomes: TNF-alpha and nitric oxide production are regulated by altered composition of proteasome-active sites. Cell Biochem Biophys, 60, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qureshi N; Perera PY; Shen J; Zhang G; Lenschat A; Splitter G; Morrison DC; Vogel SN The proteasome as a lipopolysaccharide-binding protein in macrophages: differential effects of proteasome inhibition on lipopolysaccharide-induced signaling events. J Immunol 2003, 171, 1515–1525. [DOI] [PubMed] [Google Scholar]
- 46.Qureshi N; Morrison DC; Reis J Proteasome protease mediated regulation of cytokine induction and inflammation. Biochim Biophys Acta, 1823, 2087–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yadav VR; Hussain A; Sahoo K; Awasthi V Remediation of Hemorrhagic Shock-Induced Intestinal Barrier Dysfunction by Treatment with Diphenyldihaloketones EF24 and CLEFMA. J Pharmacol Exp Ther 2014, 351, 413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Traenckner EB; Wilk S; Baeuerle PA A proteasome inhibitor prevents activation of NF-kappa B and stabilizes a newly phosphorylated form of I kappa B-alpha that is still bound to NF-kappa B. EMBO J 1994, 13, 5433–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kanarek N; Ben-Neriah Y Regulation of NF-kappaB by ubiquitination and degradation of the IkappaBs. Immunol Rev 2012, 246, 77–94. [DOI] [PubMed] [Google Scholar]