Abstract
Extracellular vesicles (EVs) are small membrane-bound organelles naturally released from cells and potentially function as vehicles of intercellular communication. Cells release numerous sub-species of EVs, including exosomes and microvesicles, which are formed via distinct cellular pathways and molecular machineries and contain specific proteins, RNAs and lipids. Accumulating evidence indicates that the repertoire of molecules packaged into EVs is shaped by both the physiological state of the cell and the EV biogenesis pathway involved. Although these observations intimate that precisely regulated pathways sort molecules into EVs, the underlying molecular mechanisms that direct molecules for secretion remain poorly defined. Recently, with the advancement of mass spectrometry, next-generation sequencing techniques and molecular biology tools, several mechanisms contributing to EV cargo selection are beginning to be unraveled. This review examines strategies employed to reveal how specific proteins, RNAs and lipids are directed for secretion via EVs.
Keywords: Cargo, protein, RNA or lipid species incorporated in to extracellular vesicles, EXOmotif, short RNA sequence that targets microRNAs for secretion in extracellular vesicles
1. Introduction
Extracellular vesicles (EVs) are a heterogenous collection of small membrane-bound organelles released from cells into the extracellular environment. Originally described as a mechanism to selectively eliminate proteins, nucleic acids and lipids from cells, EVs are now viewed as an important mode of intercellular communication that impacts diverse physiological and pathological processes [1–4]. EVs are naturally secreted by all cell types and these vesicles are comprised of heterogenous, yet overalapping, subpopulations in terms of physical size, density and even molecular composition. These physical similarities have made it challenging to discriminate the biogenesis pathways and functional roles of specific EV subpopulations. Nevertheless, extensive biochemical and microscopic characterization over the last two decades has revealed that EVs can broadly be divided into two classes, exosomes and microvesicles, which are distinguished by their modes of biogenesis [1, 2, 4].
The term exosome was initially used to describe small EVs, ranging from approximately 50 nm to 200 nm in diameter, released from reticulocytes during differentiation into erythrocytes [5, 6]. Seminal studies in this system revealed exosomes arise from the endo-lysosomal trafficking pathway during the formation of multivesicular bodies (MVBs) and are released extracellularly when MVBs fuse with the plasma membrane. Fundamentally, exosome biogenesis involves cellular machinery that drives intraluminal budding from the limiting membrane of MVBs [1, 4]. Upon membrane scission, the invaginated portions of membrane and cytoplasmic material are sequestered into intraluminal vesicles (ILVs) that fill MVB cisternae and serve as exosome precursors. Regulated exocytosis of MVBs then releases these ILVs with their contents into the extracellular space as exosomes. Importantly, early studies also revealed that EV biogenesis distinctly occurs at the plasma membrane [4, 7–9]. This second class of EVs, termed microvesicles, range in size between 50nm to 1,000nm, and are formed by pathways that drive outward budding or membrane shedding from the surface of the cell. Microvesicles were actually first observed as tiny particles released from platelets, and for many years, dismissed as irrelevant cellular dust [10]. Although the physical similarities of exosomes and microvesicles has hindered characterization, accumulating evidence indicates these two EV classes encapsulate distinct molecular repertoires and likely play different biological functions [1, 4].
Accumulating evidence supports that EV biogenesis is a finely tuned process and that the molecular composition and rates of production of exosomes and microvesicles vary greatly depending on cell-type and physiological state [1, 4]. Moreover, profiling of EVs reveals that exosomes and microvesicles are enriched in specific lipids, RNAs and proteins, yet largely devoid of others[1, 4]. Together, these observations highlight the existence of a tightly regulated machinery that directs the sorting of molecules into EVs for extracellular secretion (Fig. 1A–F). EV lipidomic analyses demonstrate membranes of exosomes and microvesicles contain high concentrations of cholesterol, sphingomyelin and ceramide, which may reflect roles for these lipid species in EV biogenesis and cargo sorting [11, 12]. Exosome membranes are also enriched in numerous tetraspanins including CD63, CD9 and CD81, and adhesion molecules such as integrins and intercellular adhesion molecule −1 (ICAM-1) [13, 14], whereas microvesicles contain high levels of select transmembrane proteins including the metastatic integrins ITGA6, ITGA4, matrix metalloprotease MT1-MMP and an oncogenic form of the epidermal growth factor receptor (EGFRvIII) [15–17]. Although luminal proteins vary amongst EV subpopulations, they frequently include machinery involved in EV biogenesis such as endosomal sorting complexes required for transport (ESCRT) components and accessory proteins, syntenin, ARF6, as well as various Rab GTPases, annexins, chaperones, enzymes and RNA-binding proteins (RBPs) [1, 4]. Finally, considerable evidence supports that select small RNA species including microRNA (miRNA), transfer RNA (tRNA), vault RNAs, and small nucleolar RNA (snoRNA) are packaged into EVs, presumably through mechanisms that are functionally intertwined with RBP loading [18–25].
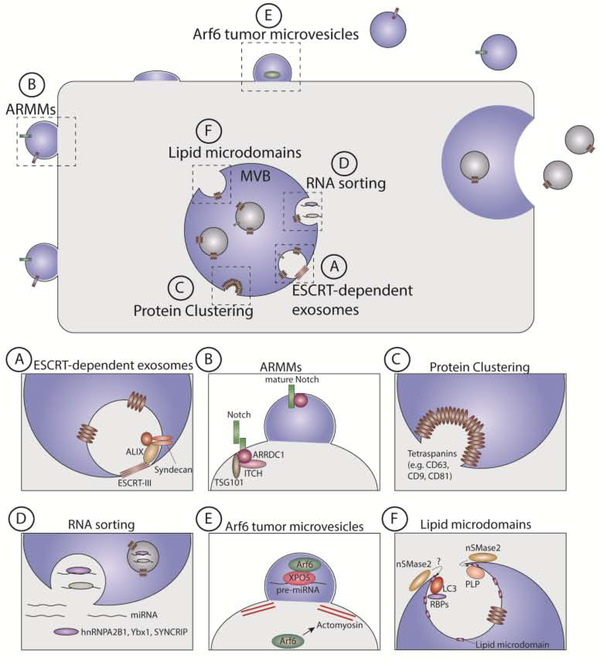
Figure 1. Mechanisms that specify cargo loading into extracellular vesicles.
(A) ALIX and Endosomal Sorting Complexes Required for Transport (ESCRT)-III components direct the loading of Syntenin and syndecan into exosomes formed at multivesicular bodies (MVBs). (B) Arrestin domain containing protein 1 (ARRDC1) controls the biogenesis and cargo sorting of ARRDC1-mediated microvesicles (ARMMs) at the plasma membrane. A complex comprising ARRDC1, TSG101 and Nedd4 family E3 ligases such as ITCH coordinate the packaging and secretion of the NOTCH2 receptor in ARMMs. (C) The clustering of proteins such as tetraspanins (e.g. CD63, CD9, CD81) at the MVB and plasma membrane is sufficient to drive their segregation into extracellular vesicles (EVs). The biophysical mechanisms regulating clustering-dependent EV biogenesis and cargo loading remain an exciting area for future investigation. (D) YBX1, SYNCRIP and sumoylated hnRNPA2B1 control the sorting of miRNAs into EVs. (E) Arf6 regulation of actomyosin contractility drives the formation of tumor microvesicles that contain Exportin-5 (XPO5) and pre-miRNAs. (F) Neutral Sphingomyelinase 2 (nSMase2)-dependent production of ceramide promotes the formation of lipid microdomains containing myelin proteolipid protein (PLP), which subsequently bud or are sorted into EVs formed from the limiting membrane of MVBs. Sorting of the autophagy protein MAP1LC3B (LC3) and associated RNA-binding proteins (RBPs) into EVs also appears to require nSMase2 activity.
The unique repertoire of proteins, RNAs and lipids packaged and secreted in EVs represents a molecular code that not only contains information about the state of a cell at the time of secretion but also is postulated to reprogram adjacent cells and tissues during both normal physiology and disease [1, 4]. To decrypt this code, and potentially leverage EVs to diagnose and treat disease, we require more detailed insight into the mechanisms that direct the loading of molecules into EVs in specific physiological contexts. Although the pathways that control EV cargo selection and packaging remain poorly understood, recent advances have begun to shed light on some of the molecular mechanisms that specify proteins for secretion in EVs. This review examines emerging methods and technologies being employed to decipher how diverse molecules are specifically incorporated into EVs.
2. Approaches and Methods to Dissect Mechanisms of EV Loading
Since the discovery of EVs several decades ago, a major focus of the field has been to uncover the specialized mechanisms controlling the sorting of molecules into EVs for secretion outside of cells. This process has turned out to be far more complicated than anyone imagined. Much of this complexity stems from the fact that EVs are released as a heterogenous mixture vesicles with overlapping size, similar morphology, and variable composition, making it difficult to isolate specific subpopulations and assess changes in cargo loading under various experimental conditions [1, 4]. In addition, the pleiotropic nature of a subset of components implicated in EV biogenesis and cargo sorting have made it challenging to establish cause and effect or interrogate these pathways on the scale required for functional assays [26]. However, recent advances in EV isolation and systems technologies such as mass spectrometry, next generation sequencing, and nanoscale flow cytometry, coupled with emerging cell and molecular biology tools, have helped to reveal mechanisms that contribute to EV loading and secretion. Below, we summarize the methods and technologies employed to uncover how specific (2.1) proteins, (2.2) RNAs and (2.3) lipids are incorporated into EVs.
2.1. Defining Mechanisms that Specify Proteins for Secretion in EVs
To date, the best characterized EV cargoes are EV-associated proteins. This has in part driven by widespread utilization of mass spectrometry for proteomic profiling of EVs. Although these proteomic profiling approaches have been critical for the identification of proteins packaged into EVs, they provide limited information about the mechanisms that contribute to the packaging of individual targets. To overcome these limitations, researchers have begun to employ orthogonal systems approaches in addition to classical gain-or loss-of-function strategies to reveal the machinery involved in EV packaging.
2.1.1. Identification of EV Cargo Proteins via Mass Spectrometry and Profiling Approaches
The first step to deciphering how specific proteins are packaged into EVs involves determining the repertoire of protein cargoes that are secreted in EVs. Nearly two decades ago, the first EV proteomics dataset was published and reported the identification of roughly 10 proteins found to be enriched in the EVs of dendritic cells [27]. By comparison, current liquid chromatography–electrospray ionization tandem mass spectrometry (LC/ESI-MS/MS) approaches allow routine identification and quantification of hundreds to thousands of proteins in EVs from cell culture media or body fluids [28]. These advances have revealed the EV proteome to be enriched in components of the ESCRT machinery, tetraspanin membrane proteins (CD63, CD9, and CD81), adhesion proteins (integrins, ICAM) and chaperones [1, 3, 4]. However, the sensitivity of modern mass spectrometry techniques has also uncovered significant variability in portions of the EV proteome between cell-types, experimental conditions and EV isolation strategies, highlighting the heterogeneous nature of EVs and the presence of extracellular protein contaminants that can obscure the EV proteome.
In order to obtain reliable EV proteomics data, researchers have begun to employ more rigorous EV isolation procedures. For many years, EVs were isolated through standard differential centrifugation protocols that precipitate all EV subpopulations together along with contaminating proteins aggregates and other material [29, 30]. Many current EV isolation workflows employ high-resolution density gradient fractionation and/or immunoaffinity capture strategies to purify EV subpopulations from heterogenous extracellular pools [13, 14]. Furthermore, approaches such as size exclusion chromatography, ultrafiltration, asymmetric-flow field-flow fractionation (AF4) and tangential flow filtration (TFF) have emerged as ways to isolate specific EV populations and separate them from non-EV protein contaminants [31–34]. Together, these isolation strategies, when coupled with EV proteomics, have led to better definition of the global EV proteome and revealed proteins enriched in specific EV populations. For example, annexin A1 was previously viewed as a universal marker of EVs, but advanced purification techniques indicate that it is specifically enriched in microvesicles shed from the plasma membrane [13]. Nevertheless, the increased rigor of these approaches come at a price. These methodologies are generally costlier and less efficient than traditional protocols with respect to time and yield. Ultimately, this may impact the amount of material that can be processed and the proteomic coverage of EV populations.
New protein labelling approaches for mass spectrometry have also helped to more precisely define the EV proteome. Until relatively recently, quantitative measurements of EV proteins predominately relied upon label free strategies such as peak area determination and spectral counts [28, 35]; quantitative approaches based upon stable isotope stable isotope labelling in cell culture (SILAC) can only be used in vitro and the sheer volume of media required for most EV preparations render SILAC largely impractical for EV proteomics. However, new post-digestion peptide labelling reagents including tandem mass tags (TMT) and isotope tags for relative and absolute quantitation (iTRAQ) enable the precise quantification of proteins in EVs from basically any source, including human plasma EVs, and can be multiplexed to allow 8–10 different samples to be analyzed in a single experiment [35–37]. Recent studies employing TMT mass spectrometry have helped to define the proteome of breast cancer exosomes and highlight the utility of this approach [38].
Mass spectrometry-based proteomics can also be employed to dissect the topology of proteins found within membranes [39]. By combining proteomics with proteinase treatment and biotin labelling, the Lötvall group was able to map surface exposed peptides of transmembrane proteins found within EVs [40]. This clever approach surprisingly found that a number of transmembrane proteins commonly detected in EVs including SCAMP3 appeared to have a reversed topology. This study reinforces that mass spectrometry can also be leveraged to interrogate structural aspects of EV biology and may also serve to highlight proteins sorted into EVs via unconventional pathways and/or discriminate non-classical EV subpopulations.
Although most current strategies to discriminate and isolate EV subpopulations rely upon criteria including subcellular origin, size, sedimentation properties, floatation density and/or select protein markers, it is important to note that these definitions are largely operational and likely underestimate the actual diversity of EV subtypes and biogenesis pathways [41]. It is also unclear whether the cargoes of EV subpopulations are completely distinct or constitute partly overlapping protein combinations. Hence, it may be virtually impossible to employ these approaches to purify and define the molecular composition of each EV subpopulation via proteomic profiling. To overcome this potential issue and more precisely define EV subtypes researchers are developing methods to scrutinize EVs at the level of individual vesicles. For example, the single EV analysis (SEA) platform which employs a microfluidic system to cyclically capture and immune-stain single EVs on-chip, has emerged as a promising multiplexable technique to profile the content of individual EVs within complex mixtures [42]. Moreover, advances in flow cytometry technology have now made it possible to characterize the distribution of a limited set of EV cargoes and select biophysical parameters at single EV resolution [16, 43]. Undoubtedly, the integration of these high resolution EV analysis techniques with proteomic data from advanced isolation strategies will provide a powerful means to define subpopulations, functional relationships between EV proteins, and the mechanisms that underlie cargo packaging and release.
2.1.2. Protein-Protein Interaction Screens for the Identification of Sorting Pathways
Although mass spectrometry-based profiling has identified several hundred to possibly thousands of proteins secreted via EVs, the physical and functional relationship between most EV proteins remains largely unclear. Nevertheless, one can predict that protein-protein interactions between cargo and the packaging machinery delivers these proteins to EVs prior to secretion. Indeed, bioinformatic analyses reveal that the EV proteome consists of a dense network of physically and functionally interconnected proteins [44, 45]. Protein interaction maps from these analyses also highlight that many EV cargoes tend to interact with discrete node proteins with relatively few other network connections, potentially pointing to common machinery (node proteins) involved in the regulation of entire clusters of cargo [45, 46]. Collectively, these bioinformatic data indicate that strategies aimed at identifying protein interaction partners amongst EV cargoes may be a practical approach to uncover mechanisms involved in cargo sorting and EV biogenesis.
Classical protein interaction discovery strategies including yeast two hybrid assays have been amongst the most successful for identifying physical connections between EV proteins and uncovering machinery linked to cargo loading. For example, yeast two hybrid assays helped to implicate the ESCRT-III associated protein ALIX in the loading and biogenesis of syndecan and CD63-positive exosomes [47]. Employing the syndecan and CD63 adaptor Syntenin-1 as bait, Baietti and colleagues identified ALIX as potential Syntenin-1 interactor. Recognizing that ALIX, Syntenin-1 and CD63 are highly enriched in exosomes and that ALIX has roles in MVBs biogenesis and virus budding [48, 49], this group then examined the functional relationship between these proteins in EV biogenesis and cargo loading. These studies revealed that genetic depletion of ALIX and Syntenin-1 dramatically impairs the packaging and secretion of syndecans and CD63 in exosomes (Fig. 1A) [47]. Remarkably, this exosome secretion pathway was also found to be coordinated by the cargo itself; siRNAs targeting syndecan and CD63 impaired the loading of ALIX and Syntenin-1 into EVs and caused an overall reduction in exosome secretion [47, 50]. Therefore, molecular interactions between ALIX, Syntenin-1 and syndecan drives the segregation these proteins into EVs.
Yeast two hybrid interaction screens also helped to reveal mechanisms involved in the biogenesis and packaging of a subpopulation of EVs termed arrestin domain containing protein 1 mediated microvesicles (ARMMs) [51]. In particular, screens employing arrestin domain containing protein 1 (ARRDC1) as bait identified the ESCRT-I component TSG101 as an interaction partner. TSG101, like ALIX, is highly enriched in EVs and implicated in the budding of human immunodeficiency virus (HIV) [48, 49]. Ectopic expression of ARRDC1 induces dramatic relocalization of TSG101 to the plasma membrane where it facilitates the formation of ARRDC1-positive plasma membrane microvesicles through a mechanism that also requires VPS4, an ESCRT-III component, and the NEDD4 E3 ubiquitin ligase WWP2 [51]. Proteomic profiling has recently revealed that ARMMs are enriched in components of the NOTCH pathway including the receptor NOTCH2 and ITCH, an E3 ubiquitin ligase related to WWP2 (Fig. 1B) [52]. Furthermore, ARRDC1 and ITCH were found to biochemically interact, and siRNA-mediated depletion of ITCH impairs the packaging and secretion of NOTCH2 in ARMMs [52]. Collectively, these examples illustrate that EV cargo and sorting machinery such as ESCRT proteins are often packaged into EVs together as complexes. Hence, approaches focused on defining the biochemical relationship between the proteins contained within EVs can yield important insights into EV loading pathways.
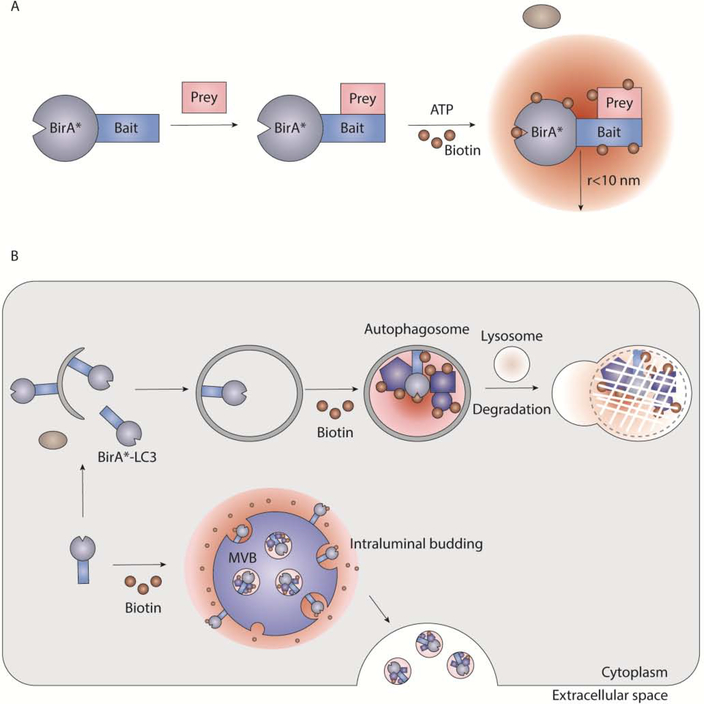
Novel biochemical strategies to identify protein interactions may also have utility for isolating the machinery and/or cargo of EV biogenesis pathways. Recently, our group developed a proximity-dependent biotinylation strategy (BioID) [53, 54] to identify protein targets of secretory autophagy, an emerging secretion pathway controlled by the autophagy machinery [55]. In this strategy, the E. coli BirA biotin ligase (BirA*) was fused to MAP1LC3B (LC3), a protein critical for cargo binding within the autophagy pathway, to identify targets of secretory autophagy biotinylated by this recombinant probe, thereby enabling their purification from conditioned media (Fig. 2A,B). Indeed, cells expressing the BirA*-LC3 recombinant probe released numerous biotinylated proteins into the conditioned media, which were purified on streptavidin after depletion of the free biotin from media via precipitation-based protocols. Moreover, this strategy was combined with SILAC to perform quantitative proteomics of the BirA*-LC3-labelled secretome in comparison to BirA* alone. Proteomic data from these studies notably revealed that the BirA*-LC3-labelled secretome was enriched in proteins previously found to be packaged into EVs, including many RBPs [55]. These results broached that LC3 and the autophagy machinery control the loading and secretion of proteins in EVs. Further analyses demonstrated that LC3 is enriched within EVs and directly mediates the loading of interacting proteins such as RBPs through mechanisms that require the autophagy machinery that conjugates LC3 to lipid at cellular membranes as well as neutral sphingomyelinase 2 (nSMase2) (Fig 1F) [55]. However, genetic evidence indicates that LC3-dependent EV loading and secretion is independent of classical autophagy. In addition to uncovering a new role for LC3 in sorting cargo into EVs, this study also demonstrates that proximity-dependent biotinylation can be utilized to identify protein interaction networks that contribute to the loading of EV cargo. The development of methods to induce proximity-dependent biotinylation in vivo raises the possibility that researchers may be able to study EV cargo loading and secretion during developmental processes or specific pathophysiological conditions [56–58].
Figure 2. A proximity-dependent biotinylation strategy to identify targets of secretory autophagy.
(A) Diagram outlining the general principle of proximity-dependent biotinylation. A mutant version of the E. coli BirA biotin ligase (BirA*) is fused to a protein of interest (bait) and expressed within cells. Cell culture media supplemented with biotin triggers BirA* activity and biotinylation of prey within proximity of the bait protein. Biotinylated prey can be purified with streptavidin and identified via mass spectrometry. The radius (r) of BirA* biotin-labelling has been determined to be < 10 nm [133]. (B) To identify targets of secretory autophagy, the BirA* biotin ligase was fused to LC3, a protein recruited to autophagosomal membranes and involved with cargo recognition within the classical autophagy pathway. Cells expressing the BirA*-LC3 recombinant probe were treated with biotin and conditioned media from these samples was collected. EVs and soluble proteins in conditioned media were collected by precipitation and biotin-labelled targets were identified via mass spectrometry. This approach revealed that a pool of LC3 conjugated to the MVB limiting membrane specifies the loading of RBPs into EVs for secretion outside the cell.
2.1.3. Roles for Post-Translational Modifications in Protein Sorting
The ESCRT pathway was originally identified to drive the sorting of ubiquitin-conjugated membrane proteins into ILVs formed at a subset of MVBs that fuse with lysosomes targeting these membrane proteins for degradation [26]. Evidence implicating various ESCRT components in EV biogenesis and viral budding has prompted investigation into the role of post-translational modifications such as ubiquitin in specifying proteins for EV secretion. Although the overall contributions of post-translational modifications to EV cargo sorting still remain unclear, proteomic and cell biological approaches have revealed specific contexts in which modifications control the secretion of proteins in EVs.
Proteomic profiling has identified ubiquitin in EVs purified from diverse sources including cultured cells, human plasma, and urine [59]. Given the role of ubiquitin modification as a sorting signal for the ESCRT pathway during receptor turnover, these observations led to early hypotheses that ubiquitin earmarks proteins for packaging into EVs. However, studies examining the trafficking and secretion of major histocompatibility complex class II (MHCII), a membrane protein degraded via the ESCRT machinery in immature dendritic cells (DCs) and released in EVs from stimulated DCs, demonstrate that MHC-II is secreted independent of ubiquitin modification [60]. Additionally, genetic and biochemical data from a number of EV sorting paradigms suggest these processes may circumvent the ubiquitination requirements of classical ESCRT-dependent degradation pathway via direct recruitment of individual ESCRT components [47, 60–62]. These data support that ubiquitination is not a universal signal for delivering cargo to EVs.
Even though many EV proteins are packaged independent of ubiquitination, there are specific cell-types, physiological states and proteins where this post-translational modification appears important for EV loading. To comprehensively assess ubiquitination in murine myeloid-derived suppressor cells (MDSCs) EVs, Burke and colleagues combined antibody-based enrichment of di-glycl-lysine (KGlyGly) peptides, a characteristic tryptic product of ubiquitinated proteins, with mass spectrometry [63]. This study revealed that nearly 10% of the proteins in EVs from MDSCs were ubiquitinated, including numerous histones, Ezrin and HSP70. Proteomic analyses of EVs from human urine identified the KGlyGly ubiquitin signature in 15% of proteins and found that ~51% of modified proteins had K63-linked polyubquitin chains, a type of ubiquitin modification associated with lysosomal degradation, DNA repair and transcriptional regulation [64]. Interestingly, recent studies suggest that reduced Sirtuin-1 expression, as occurs in aging and many forms of cancer, promotes the secretion of exosomes enriched ubiquitinated cargo [65].
Select proteins may also require ubiquitin-modification for loading into EVs. Fas ligand (FasL) is an apoptosis inducing type II transmembrane protein that is secreted within EVs and also modified with ubiquitin [66, 67]. Notably, FasL mutants lacking lysine residues critical for ubiquitin modification display impaired trafficking to MVBs and EV-mediated secretion [67]. Macrophages infected with Mycobacterium tuberculosis also secrete EVs loaded with ubiquitinated proteins [68]. Interestingly, most of these modified EV cargoes are actually ubiquitinated mycobacterial proteins and pharmacological inhibition of ubiquitin E1-activating enzymes with PYR-41 or mutagenesis of the critical lysine residues in mycobacterial proteins was sufficient to impair their sorting into EVs. Overall, these examples strongly suggest ubiquitination influences the packaging of EVs but also emphasize the need for new tools and approaches to better define the relationship between ubiquitination and EV composition.
Proteomic and biochemical analyses have revealed that ubiquitination is not the only post-translational modification found on proteins within EVs. Recently, mass spectrometry identified the ubiquitin-like 3 (UBL3)/membrane-anchored Ub-fold protein (MUB) as a post-translational modification on diverse EV proteins and inhibition of UBL3 modification profoundly impaired the sorting of cargo into EVs [69]. In addition, Sanchez-Madrid and colleagues uncovered a role for small ubiquitin-related modifier −1 (SUMO-1) modification of heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNP A2B1) in targeting this protein and associated miRNAs for secretion in EVs (Fig. 1D; discussed below in section 2.2.1) [25]. This group has also recently revealed that interferon-1 (IFN-1) triggers TSG101 modification with the ubiquitin-like protein interferon-stimulated gene 15 (ISG15), triggering TSG101 degradation and impaired exosome secretion via mechanisms dependent on the lysosome or autophagy pathway [70]. The calcium-dependent phospholipid binding protein Annexin A2 is phosphorylated on Tyr23 and secreted in EVs in response to cellular calcium influx [71]. Interestingly, analyses of a phosphomimetic (Y23E) and phosphodefective (Y23A) mutants revealed that Annexin A2 phosphorylation is necessary and sufficient to drive its association with cholesterol-rich membrane microdomains that are segregated into EVs. Although the glycoproteome of EVs remains largely unexplored, 2-dimensional immunoblotting and mass spectrometry analyses have noted elevated levels of O-GlcNAcylated proteins in EVs from metastatic colorectal cancer cells [72, 73]. Moreover, O-GlcNAcylation of the neuronal protein crystallin αB is necessary for its localization to MVBs and incorporation into EVs [74]. Covalent lipid modification of a number of proteins has also been shown to facilitate their segregation into EVs [75, 76]. For example, the ESCRT accessory protein ALIX may be regulated by palmitoylation; furthermore, the LC3 conjugation machinery, which is involved in LC3 lipidation, is necessary for LC3 packaging and secretion via EVs [55, 77]. Finally, recent evidence also supports a role for limited proteolysis in the regulated sorting of select cargo into EVs. Caspase-8 mediated cleavage of lysyl-tRNA synthetase was found to expose a PDZ-binding motif in the carboxy-terminus of the protein which mediates the sorting and exosome-mediated secretion of the truncated isoform [78]. Collectively, these examples demonstrate that methods which scrutinize EV proteins for post-translational modifications including mass spectrometry and modification specific western analyses may reveal mechanisms that regulate the sorting of cargo into EVs.
2.1.4. The Role of Molecular Clustering in Specifying Proteins for EV Secretion
Although poorly understood, clustering of proteins and other molecules together can facilitate their segregation into EVs. This sorting mechanism was first noted for the transferrin receptor (TfR), which is secreted in EVs during reticulocyte maturation. Artificial clustering of TfR with the plant lectin concanavalin A, which binds to mannose-type N-glycans, or through anti-transferrin antibodies impairs TfR recycling and drives its robust secretion in EVs [79]. Subsequent studies revealed that artificial targeting of cytosolic proteins capable of oligomerizing to the plasma membrane was able to drive the budding of these proteins into microvesicles [80]. Together, these observations broached that clustering or high-order oligomerization at the plasma membrane is sufficient to segregate proteins into EVs for secretion outside of the cell.
Cargo clustering is implicated in sorting of numerous targets including tetraspanin family proteins (Fig. 1C). The clustering of premelanosome protein (PMEL) with CD63 at MVB membranes facilitates the ESCRT-independent sorting of PMEL into the ILVs of maturing melanosomes [81]. Moreover, trafficking studies in activated DCs demonstrate that MHC-II is targeted to CD9 enriched membrane microdomains that are sorted into EVs for extracellular release [60]. Single molecule tracking by total internal reflection fluorescence (TIRF) microscopy has revealed that CD9 cycles between tetraspanin-enriched microdomains (TEMs) and the remainder of the membrane [82]. In addition, biochemical analyses indicate these microdomains contain high levels of cholesterol, which together with tetraspanin proteins renders these structures highly resistant to physical and detergent-based disruption [83]. Structural analysis of CD81 has recently revealed that it contains a binding pocket for cholesterol and binding of this lipid may influence the interaction of partner proteins and clustering in membranes [84]. Given that tetraspanins can induce positive-membrane curvature [85], the interaction of numerous tetraspanins in microdomains may be sufficient to drive membrane budding and the segregation of associated proteins such as MHC-II into EVs. Similarly, protein clustering at cellular membranes as occurs with the assembly of the ESCRT machinery and formation of viral particles may provide the energy to drive membrane deformation and EV biogenesis [86, 87]. Although clustering-dependent EV biogenesis has been difficult to test, the recent development of tools enabling chemical or optogenetic control over protein oligomerization may provide the means to rigorously interrogate these models [88–92]. In addition, greater structural information about higher order clusters such as TEMs might enable the isolation of mutants with impaired oligomerization to assess their impact on EV biogenesis and sorting. Certainly, the role of protein clustering in EV loading and biogenesis remains an area that requires greater scrutiny.
2.2. Defining Mechanisms that Specify RNA for Secretion in EVs
Apart from proteins, EVs also contain RNA species that can be transferred into recipient cells to affect their function [1, 4]. High-throughput sequencing of EV-associated RNAs has identified diverse bio-types including messenger RNA (mRNA), long non-coding RNA (lncRNA), miRNA, tRNA, snoRNA, Y RNA, and Vault RNA [18, 20–24, 93]. Many studies have also revealed that the repertoire of RNAs encapsulated within EVs is distinct from the cells which they were derived, supporting that the incorporation of RNAs into EVs is a regulated process [21, 94–96]. Nevertheless, the importance of EV pathways in RNA secretion remains the subject of debate. This stems, in part, from observations that RNA species such miRNAs have been reported to be released at very low stoichiometric abundance in EVs [13, 23, 97]. Moreover, EV preparations subjected to high-resolution iodixanol density gradient ultracentrifugation showed that a significant proportion of miRNA species and other small non-coding RNAs do not fractionate with well-defined EV subtypes and may be associated with membrane-free ribonucleoprotein (RNP) complexes or other uncharacterized secretory structures [13]. It is also important to note that different RNA extraction methods and next-generation sequencing protocols are a source of bias that can contribute to discrepencies in the identification and quantification of RNA species in EV preparations [98, 99]. Irrespective of these issues, the RNAs secreted in EVs (or EV-associated fractions) are of immense interest because of their diagnostic potential and detailed understanding of the mechanisms that control the secretion of discrete species is critical to deciphering the physiological conditions associated with release. With the aid of emerging RNA sequencing and gene editing technologies, researchers have begun to uncover certain pathways involved in sorting various RNA species into EVs for secretion outside the cell.
2.2.1. Integrated Sequencing and Proteomics Strategies to Identify RNA Secretion Pathways
To date, the most common strategy to identify pathways that control RNA packaging into EVs has been to combine RNA sequencing or microarrays and mass spectrometry-based proteomics. This strategy derives from the logical assumption that RNAs are specified for secretion through their interactions with proteins that are sorted into EVs. Although mass spectrometry has identified diverse RBPs in EVs, limited material has made it difficult to determine if these proteins are bound to RNA and mediate the sorting of select RNA species. Hence, researchers have focused on identifying RNA sequence motifs or species that are enriched within EVs and then interrogate candidate RBPs via loss-of-function approaches for their requirement in the secretion of select RNAs.
Initial efforts to uncover pathways involved in RNA sorting focused on identifying sequence elements within secreted RNAs. Employing microarray analysis of activation-induced changes in the miRNA and mRNA profiles of primary T lymphoblasts and their exosomes, Villarroya-Beltri and colleagues identified the repertoire of RNAs enriched within exosomes (Fig. 1D) [25]. Bioinformatic analysis of this dataset revealed a short RNA sequence motif (EXOmotif) common to many of the miRNAs enriched within exosomes. Notably, mutagenesis of the EXOmotif within consensus miRNAs compromised their packaging in exosomes, whereas introduction of this motif into non-consensus miRNAs stimulated release, demonstrating that the EXOmotif is necessary and sufficient for targeting miRNAs into exosomes [25]. To identify potential regulators of this pathway, consensus EXOmotif RNAs and controls were conjugated to biotin and used to affinity purify interacting proteins from exosome lysates. Mass spectrometry analyses of these samples revealed that EXOmotif miRNA probes strongly enriched for the RBP hnRNPA2B1. Finally, classical loss of function and gain of function strategies including siRNA-mediated depletion and overexpression were employed to functionally support a role for hnRNPA2B1 in the sorting and secretion of EXOmotif miRNAs [25]. Together, these studies highlight that approaches aimed at identifying regulatory elements within secreted RNAs can reveal classes of RNAs that are secreted through common mechanisms. In fact, a very similar strategy was recently used to implicate SYNCRIP in the secretion of a different class of miRNAs (Fig. 1D) [100, 101].
Because numerous miRNAs and other RNA species within EVs lack obvious EXOmotifs, other pathways are also involved in specifying RNAs for secretion. Moreover, the issue of EV heterogeneity and crosstalk between pathways has the potential to confound analyses. The Schekman group developed a rigorous unbiased strategy to identify RNA species contained within CD63-positive EVs and dissect the mechanisms required for their loading [96]. In particular, they subjected EVs collected by differential centrifugation to density gradient floatation and then immunoaffinity purified CD63-positive exosomes from isolated fractions. This led to the identification of miR-223 as enriched in CD63-positive EVs, with most other miRNAs being of very low abundance. Employing an elegant cell free EV packaging assay that was optimized by studying the intraluminal budding of recombinant CD63-luciferase, they were able demonstrate miR-223 is selectively packaged into ILVs in vitro [96]. Mass spectrometry of proteins affinity purified with biotin-conjugated miR-223 identified a number of candidates potentially linked to this sorting pathway, including Y-box protein 1 (YBX1). Importantly, YBX1 was found to specifically bind miR-223 and classical complementation assays in YBX1 CRISPR knockout cells demonstrated that YBX1 is both necessary and sufficient for the secretion of miR-223 in CD63-positive exosomes (Fig. 1D) [96]. More recently, profiling studies of wild-type and YBX-1 knockout cells using thermostable group II intron reverse transcriptase sequencing (TGIRT-seq), which is able to read through modified RNA species, have implicated YBX-1 in the secretion of diverse small non-coding RNAs including tRNAs [23]. Although the underlying mechanisms that target YBX-1 (and bound RNA) to EVs remain unclear, YBX-1 has been shown to co-localize with cytoplasmic RNA granules including GW/processing-bodies (GW/P-bodies) and stress granules [102], two structures that associate with MVBs and endolysosomes [103–105]. Interestingly, the Argonaute (AGO) family of miRNA binding proteins are also enriched in GW/P-bodies and reported to be sorted into EVs [106, 107], suggesting a possible connection between RNA granules and RNP secretion in EVs. Overall, these studies emphasize the utility of combining classical biochemical techniques and modern systems approaches to uncover pathways that sort RNA into EVs.
EV profiling in the context of specific genetic manipulations has also helped to illuminate novel pathways involved in RNA loading. Recently, small RNA sequencing of microvesicles derived from tumor cells revealed a profound enrichment of pre-miRNAs, unprocessed forms of miRNAs [19]. A subset of tumor microvesicles (TMVs) are formed as a result of hyperactivated small GTPases including ARF6, which converge on the cellular contractile machinery and drive the fission of vesicles from the plasma membrane (Fig. 1E) [7, 108]. Notably, expression of constitutively active ARF6 (ARF6Q79L) was found to robustly stimulate the secretion of pre-miRNAs in TMVs, whereas dominant negative forms of this GTPase blocked the extracellular release of pre-miRNA species [19]. To uncover mechanisms involved in secretion of pre-miRNAs, D’Souza-Schorey and colleagues characterized the protein composition of TMVs. These studies revealed that TMVs, in addition to being enriched in ARF6, also contained high levels of Exportin-5, a nuclear export receptor that traffics pre-miRNAs to the cytoplasm for further processing. Subsequent analyses demonstrated that active ARF6 and Exportin-5 directly interact and this interaction is necessary for secretion of Exportin-5 bound pre-miRNAs (Fig. 1E) [19]. Biochemical and genetic data also implicated the guanine nucleotide exchange factor GRP1 and casein kinase 2 (CK2) in this RNA secretion pathway. This comprehensive study highlights how signaling pathways deregulated in cancer can affect RNA packaging and secretion in EVs and reinforce the value of gain-of-function and loss-of-function strategies for dissecting EV packaging pathways.
2.2.2. A Genetic Screen to Uncover Pathways that Target RNA for Secretion in EVs
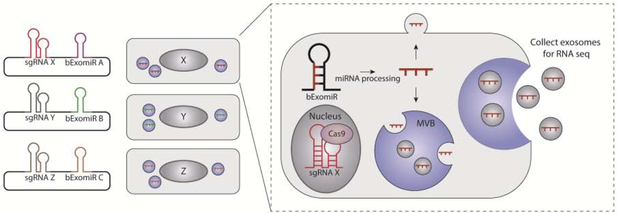
Although targeted genetic strategies have certainly yielded important insights into the mechanisms that specify RNAs for secretion, these approaches are most useful for characterizing pathways that have already been implicated in EV secretion. The unbiased interrogation of EV pathways to uncover mechanisms that regulate the sorting and secretion of RNA remains a fundamental challenge in the EV field. To tackle this problem, the Pfeffer group developed a clever CRISPR genome-wide screening strategy that identifies both genes and RNA sequence motifs that impact the secretion of miRNAs in EVs (Fig. 3) [109]. In this strategy, a library of artificially barcoded miRNAs containing the previously described EXOmotif (bEXOmiRs) was co-expressed with Cas9 and CRISPR single guide RNAs (sgRNAs) targeting each open reading frame such that each cell contained a unique bEXOmiR and sgRNA pair. Deep sequencing was then used to quantitively assess the levels of the barcoded miRNAs in EVs and identify genes affecting the packaging and secretion of RNA (Fig. 3). In addition to affirming the involvement of key EV players including Rab27a and neutral sphingomyelinase 2 (nSMase2/SMPD3), this screen also uncovered previously unrecognized roles for specific genes in MVB trafficking and exocytosis such as functions of the Wnt signaling pathway in EV release and regulation [109]. Moreover, this strategy may be adapted to specifically interrogate the secretion of other RNA species or proteins encapsulated within EVs. Certainly, genetic screens such as this will help to illuminate pathways and machinery, which to date, have been overlooked in the context of EV secretion.
Figure 3. A genome-wide CRISPR/Cas9 screen employing barcoded miRNAs (bEXOmiRs) to identify pathways involved in EV sorting and secretion (adapted from [109]).
F Oligonucleotides encoding single guide (sg) RNA-bEXOmiR pairs were synthesized and cloned into lentiviral expression vectors. Cells expressing Cas9 or deficient for Cas9 were transduced with lentiviral libraries at low multiplicity of infection to ensure that each cell expressed a unique sgRNA—bEXOmiR pair. Conditioned media from library expressing cells was collected after 48h and EVs were purified. The abundance of barcodes was assessed via deep sequencing from EV-extracted RNA and the levels quantified relative to Cas9-deficient cells. This approach identified an unrecognized role for the Wnt pathway in EV biogenesis.
2.3. Uncovering Mechanisms that Influence the Lipid Composition of EVs
Lipids are fundamental components of EV membranes and evidence indicates that select lipid species are enriched within EVs relative to cells. Indeed, studies have generally found that EVs are enriched in phosphatidylserine, cholesterol, sphingomyelin, ceramides, and phosphatidic acid, with the relative proportions of these lipids varying depending on the EV subpopulation, cell-type and experimental conditions [11]. Although these observations strongly support that specific lipid species are critical for EV biogenesis or function, the mechanisms incorporating these lipids into EVs have remained largely unclear. Recent studies have begun to address this gap in knowledge using various profiling and biochemical strategies.
Initial insight into the mechanisms that incorporate specific lipid species into EVs actually came from studies examining the trafficking and secretion of myelin proteolipid protein (PLP). In contrast to many other EV proteins, PLP is incorporated into EVs independent of the ESCRT machinery [12]. Lipidomic analyses of PLP-containing EVs via nano-ESI-MS/MS revealed profound enrichment of numerous lipids including ceramide. Ceramides are a class of lipids abundant within cell membranes, particularly membrane microdomains [110], and can be formed through a number of different routes including neutral sphingomyelinase 2 (nSMase2/SMPD3) driven hydrolysis of sphingomyelin [111]. Notably, pharmacological inhibitors or siRNAs targeting nSMase2 dramatically suppressed the biogenesis and secretion of EVs containing PLP, whereas purified bacterial sphingomyelinase was sufficient to stimulate the intraluminal budding of giant unilamellar vesicles containing sphingomyelin [12]. These data led to the model in which nSMase2 production of ceramides facilitates the formation of PLP-containing membrane microdomains that spontaneously bud to generate EVs (Fig. 1F). Although the secretion of numerous proteins and RNAs requires nSMase2 activity, wide variations in EV ceramide levels have led some to speculate this EV biogenesis pathway is restricted to specific cell types or conditions [11]. Ceramide may also affect EV composition through its metabolism into sphingosine 1-phosophate, which regulates cargo sorting through the sphingosine 1-phosphate receptor (S1PR). Therefore, EV lipidomics may reveal specific lipid species critical for the biogenesis of specific EV populations.
In addition, phosphatidic acid is enriched in the EVs released from some cell-types. One mechanism that generates phosphatidic acid is phospholipase D (PLD)-dependent hydrolysis of phosphtidylcholine [112]. Recently, the Zimmermann group performed a directed siRNA screen to identify ARF6 effectors that modulate the intraluminal budding of Syntenin into enlarged endosomes formed by ectopic expression of constitutively active Rab5 (Rab5Q79L)[113]. This screen identified PLD2 as a regulator of Syntenin intraluminal budding. Moreover, genetic or pharmacological inhibition of PLD2 impaired the secretion of EVs containing Syntenin, but not other EV subpopulations. Interestingly, a biosensor derived from the phosphatidic acid-binding domain of Spo20 (PABD-Spo20)[114] was found to specifically localize to the limiting membrane of Rab5Q79L endosomes, suggesting PLD2 likely drives the production of phosphatidic acid directly at the endosomal membrane [113]. This study highlights how localized production of lipid species can regulate EV biogenesis and sorting, as well as the utility of lipid sensors in defining the subcellular localization of lipid populations that regulate EV cargo sorting and secretion.
The unconventional phospholipid bis(monoacylglycero)phosphate (BMP), sometimes called lysobisphosphatidic acid (LBPA), is also critical for intraluminal budding at MVBs and may play a role in EV secretion [11]. Initial evidence implicating this lipid in ILV formation came from in vitro studies of liposomes reconstituted with BMP [115]. Remarkably, liposomes generated to mimic endosomes, harboring an acidic lumen with neutral external pH, that also contain BMP in their membranes spontaneously formed ILVs, whereas liposomes lacking BMP remained unilamellar. Moreover, this lipid-dependent intraluminal budding pathway was found to be regulated by ALIX, which binds to BMP directly, and is critical for endosome cholesterol homeostasis [115, 116]. Recent evidence indicates that endosomal cholesterol levels are an important regulator of endolysosome exocytosis through a pathway dependent on mucolipin-1 (MCOLN1) [117].
Given that membranes are composed of diverse lipids, perturbations to a single lipid species can sometimes dramatically alter membrane dynamics and trafficking to affect EV biogenesis. For example, the Llorente group recently revealed that inhibition of phosphatidylinositol 3,5-bisphosphate (PIP(3,5)P2) formation via genetic depletion or chemical inhibition of the phosphoinositide kinase PIKfyve dramatically stimulates the secretion of EVs [118]. Quantitative electron microscopy analysis of PIKfyve inhibited cells demonstrated a dramatic accumulation of MVBs and increased numbers of intraluminal vesicles per MVB, suggesting that PIKfyve production of PIP(3,5)P2 is important for MVB fusion with lysosomes. In support of this model, proteomic analysis of EVs from cells treated with PIKfyve inhibitors revealed a profound increase in the secretion of diverse autophagy proteins including LC3, NBR1 and p62/SQSTM1[118]. Although PIP(3,5)P2 is not a lipid species enriched in EVs, this study highlights the critical importance of cellular lipid metabolism and homeostasis in EV secretion pathways. Certainly, the functions of lipid species in EV biogenesis and sorting remains an understudied area. Presumably, the emergence of techniques to isolate specific subpopulations including immunoaffinity isolation and nanoFACs will enable researchers to scrutinize the role of lipids in distinct EV biogenesis pathways [119].
3. Conclusion and Future Perspectives
It is now clear that EVs contain a defined repertoire of proteins, RNAs and lipids that are incorporated through precisely regulated pathways. Importantly, these pathways are integrated with signaling networks that control growth and development as well as cellular stress responses. Hence, the set of molecules released in an EV represents information about the nature and physiological state of the cell from which it was derived. Although cells probably employ EVs as a mode of communication to coordinate cellular activities and maintain homeostasis, there is considerable excitement amongst researchers that this emerging intercellular this communication network can be leveraged to diagnose disease and deliver therapeutics.
Major efforts are currently underway to define the pathways and biomolecules that are secreted in EVs in order to develop a codex or key to decrypt this new language and interpret the messages being sent between cells in specific physiological contexts. In cancer, tumor-dervied EVs have been shown to play roles in immune evasion and metastatic progression. For example, melanoma and prostate cancer cells secrete EVs harboring programmed death-ligand 1 (PD-L1) on their surface, which suppresses the function of CD8 T cells and facilitates tumor growth [120, 121]. The repertoire of integrins on tumor-derived EVs has also been shown to impact their interaction with extracellular matrix molecules in distant tissues and can be employed to predict the risk of metastasis to specific organs in cancer patients [122]. In addition, the presence of circulating exosomes containing glypican-1, a putative biomarker of pancreatic cancer, can reliably detect pancreatic intraepithelial lesions before magnetic resonance imaging, highlighting the potential utility of EVs in non-invasive diagnostic tests to detect cancer at very early stages [123].
Accumulating evidence also implicates EVs in the pathogenesis of age-related neurodegenerative diseases including Parkinson’s disease, Alzheimer’s diseases, and amyotrophic lateral sclerosis (ALS) [124, 125]. These diseases are characterized by the harmful accumulation and aggregation of misfolded proteins within neurons, which may spread between cells via EV secretion pathways [125]. Indeed, genetic and pharmacological inhibition of EV secretion has been found to influence disease progression in a number of murine neurodegeneration models [126–128]. There is also the potential that EVs released during the early stages of neurodegeneration contain biomarkers that can be leveraged to more rapidly diagnose disease and perhaps even monitor the efficacy of therapeutic interventions.
Ultimately, with understanding of the mechanisms that contribute to cargo sorting it may be possible to engineer EVs to deliver therapeutics to treat various diseases. Emerging studies in this area show considerable promise; recent reports highlight the engineering of ARMMs and red blood cell EVs to package and deliver CRISPR-Cas9/guide RNA for the purposes of therapeutic genome editing [129, 130]. In fact, a number of biotechnology companies have developed EV-based therapeutics for the treatment of cancer, liver diseases and lysosomal storage disorders which will enter the first phase of clinical trials beginning in 2020 [131]. Research has also revealed that EVs from adult stem cells are capable of producing some of the same tissue regenerative effects as stem cell therapies [132], prompting plans to test these natural EVs in indications ranging from healing burns to cardiac repair.
Despite a host of exciting advances and potential applications for EVs, many questions regarding EV biogenesis and cargo capture remain unanswered. Is molecular clustering at the plasma membrane or endosome a primary mechanism that drives the segregation of cargo into EVs? How are RBPs such as hnRNP A2B1 and YBX1 and their associated RNAs incorporated into EVs? What are the functions of the different lipid classes enriched within EV subpopulations? How are EVs taken up by target cells? Advances in these areas will undoubtedly come through the development of new technologies and approaches, including those enabling the interrogation of EVs at the level of single vesicles. In addition, these technologies may serve as the basis for diagnostic platforms with the potential to catch diseases such as cancer at early stages when treatments are far more effective. Initially dismissed as “cellular dust”, EVs have become the subject of ever-increasing attention by the scientific community over the last decade; nevertheless, a rigorous understanding of the mechanisms underlying EV cargo selection remains crucial for unlocking their immense biological and therapeutic potential.
Highlights.
Extracellular vesicles (EVs) are enriched with specific proteins, RNAs and lipids indicating that molecules are loaded into EVs through regulated mechanisms.
The pathways that segregate molecules into EVs involve multiple different cellular machineries including ESCRT components, tetraspanins, autophagy proteins, GTPases, RBPs and lipid modifying enzymes, which drive the biogenesis and sorting of molecules into distinct EV subpopulations.
Emerging technologies, tools and approaches such as proximity-dependent biotinylation and CRISPR/Cas9-based genetic screens are beginning to reveal the complex mechanisms that specify molecules for secretion in different EV subtypes.
In order to leverage EVs to diagnose disease or deliver therapeutics there must be greater understanding of the mechanisms that package molecules into EVs in different physiological and pathophysiological contexts such as cancer and neurodegeneration.
Acknowledgements
We apologize to those researchers whose work we were unable to cite due to space limitations. We thank FuiBoon Kai (UCSF) for assistance in the preparation of figures. Fellowship support to AML includes a Banting Postdoctoral Fellowship from the Government of Canada (201409BPF-335868) and Cancer Research Society Scholarship for Next Generation of Scientists (22805). Grant support includes the NIH (CA201849, CA126792, CA201849, CA213775, AG05746 to JD) and the UCSF QB3 Calico Longevity Fellowship (to JD and AML).
Abbreviations
- EVs
extracellular vesicles
- MVB
multivesicular body
- ILV
intraluminal vesicle
- ICAM-1
intercellular adhesion molecule 1
- MT-MMP1
membrane-type matrix metalloproteinase-1
- EGFR
epidermal growth factor receptor
- ESCRT
endosomal sorting complexes required for transport
- ARF6
ADP Ribosylation Factor 6
- RBP, RNA
binding protein
- miRNA
microRNA
- tRNA
transfer RNA
- snoRNA
small nucleoloar RNA
- LC/ESI-MS/MS
liquid chromatography electrospray ionization tandem mass spectrometry
- AF4
asymmetric-flow field-flow fractionation
- TFF
tangential flow filtration
- SILAC
stable isotope labelling in cell culture
- TMT
tandem mass tags
- iTRAQ
isotope tags for relative and absolute quantitation
- SCAMP3
secretory carrier membrane protein 3
- SEA
single EV analysis
- nanoFACs
nano fluorescence associated cell sorting
- ALIX
ALG-2-Interacting Protein X
- ARMMs
arrestin domain contain protein 1 mediated microvesicles
- ARRDC1
arrestin domain containing protein
- TSG101
tumor susceptibility gene 101
- HIV
human immunodeficiency virus
- VPS4
vacuolar protein sorting 4
- NEDD4
Neural Precursor Cell Expressed, Developmentally Down-Regulated 4
- WWP2
WW Domain-Containing Protein 2
- LC3
MAP1LC3B
- MHC-II
major histocompatibility complex class II, DCs, dendritic cells
- MDSCs
myeloid derived suppressor cells
- KGlyGly
di-glycl-lysine
- HSP70
heat shock protein 70
- K63
lysine-63
- FasL
Fas ligand
- UBL3
ubiquitin-like 3
- MUB
membrane-anchored Ub-fold protein
- SUMO-1
small ubiquitin-like modifier 1
- hnRNPA2B1
heterogeneous nuclear ribonucleoprotein A2B1
- IFN-1
interferon-1
- ISG15
interferon-stimulated gene 15
- TfR
transferrin receptor
- PMEL
premelanosome protein
- TIRF
total internal reflection fluorescence
- TEM
tetraspanin-enriched microdomain
- mRNA
messenger RNA
- lncRNA
long non-coding RNA
- YBX1
Y-box protein 1
- TGIRT-seq
thermostable group II intron reverse transcriptase sequencing
- AGO
Argonaute
- TMVs
tumor microvesicles
- CK2
casein kinase 2
- GRP1
general receptor for phosphoinositides 1 associated scaffold protein
- CRISPR
clustered regularly interspaced short palindromic repeats
- bEXOmiR
barcoded miRNAs containing the EXOmotif
- nSMase2
neutral sphingomyelinase 2
- PLP
myelin proteolipid protein
- S1PR
sphingosine 1 phosphate receptor
- PLD
phospholipase D
- PABD-Spo20
phosphatidic acid-binding domain of Spo20
- PIP(3,5)P2
phosphatidylinositol 3,5-bisphosphate
- NBR1
neighbor of Brca1
- XPO5
Exportin-5
- BMP
bis(monoacylglycerol)phosphate
- LBPA
lysobisphosphatidic acid
- MCOLN1
mucolipin-1
- ALS
amyotrophic lateral sclerosis
Footnotes
Conflict of interest: AML has no potential conflicts of interest. JD is a Scientific Advisory Board Member for Vescor Therapeutics, LLC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mathieu M, Martin-Jaular L, Lavieu G, Thery C, Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication, Nat Cell Biol 21(1) (2019) 9–17. [DOI] [PubMed] [Google Scholar]
- [2].Meldolesi J, Exosomes and Ectosomes in Intercellular Communication, Curr Biol 28(8) (2018) R435–R444. [DOI] [PubMed] [Google Scholar]
- [3].Pegtel DM, Gould SJ, Exosomes, Annu Rev Biochem 88 (2019) 487–514. [DOI] [PubMed] [Google Scholar]
- [4].van Niel G, D’Angelo G, Raposo G, Shedding light on the cell biology of extracellular vesicles, Nat Rev Mol Cell Biol 19(4) (2018) 213–228. [DOI] [PubMed] [Google Scholar]
- [5].Harding C, Heuser J, Stahl P, Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes, J Cell Biol 97(2) (1983) 329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pan BT, Johnstone RM, Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor, Cell 33(3) (1983) 967–78. [DOI] [PubMed] [Google Scholar]
- [7].D’Souza-Schorey C, Clancy JW, Tumor-derived microvesicles: shedding light on novel microenvironment modulators and prospective cancer biomarkers, Genes Dev 26(12) (2012) 1287–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Dvorak HF, Quay SC, Orenstein NS, Dvorak AM, Hahn P, Bitzer AM, Carvalho AC, Tumor shedding and coagulation, Science 212(4497) (1981) 923–4. [DOI] [PubMed] [Google Scholar]
- [9].Poste G, Nicolson GL, Arrest and metastasis of blood-borne tumor cells are modified by fusion of plasma membrane vesicles from highly metastatic cells, Proc Natl Acad Sci U S A 77(1) (1980) 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wolf P, The nature and significance of platelet products in human plasma, Br J Haematol 13(3) (1967) 269–88. [DOI] [PubMed] [Google Scholar]
- [11].Skotland T, Hessvik NP, Sandvig K, Llorente A, Exosomal lipid composition and the role of ether lipids and phosphoinositides in exosome biology, J Lipid Res 60(1) (2019) 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M, Ceramide triggers budding of exosome vesicles into multivesicular endosomes, Science 319(5867) (2008) 1244–7. [DOI] [PubMed] [Google Scholar]
- [13].Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ, Reassessment of Exosome Composition, Cell 177(2) (2019) 428–445 e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Thery C, Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes, Proc Natl Acad Sci U S A 113(8) (2016) E968–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J, Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells, Nat Cell Biol 10(5) (2008) 619–24. [DOI] [PubMed] [Google Scholar]
- [16].Choi D, Montermini L, Jeong H, Sharma S, Meehan B, Rak J, Mapping Subpopulations of Cancer Cell-Derived Extracellular Vesicles and Particles by Nano-Flow Cytometry, ACS Nano 13(9) (2019) 10499–10511. [DOI] [PubMed] [Google Scholar]
- [17].Clancy JW, Sedgwick A, Rosse C, Muralidharan-Chari V, Raposo G, Method M, Chavrier P, D’Souza-Schorey C, Regulated delivery of molecular cargo to invasive tumour-derived microvesicles, Nat Commun 6 (2015) 6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bellingham SA, Coleman BM, Hill AF, Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells, Nucleic Acids Res 40(21) (2012) 10937–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Clancy JW, Zhang Y, Sheehan C, D’Souza-Schorey C, An ARF6-Exportin-5 axis delivers pre-miRNA cargo to tumour microvesicles, Nat Cell Biol 21(7) (2019) 856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kaur S, Abu-Shahba AG, Paananen RO, Hongisto H, Hiidenmaa H, Skottman H, Seppanen-Kaijansinkko R, Mannerstrom B, Small non-coding RNA landscape of extracellular vesicles from human stem cells, Sci Rep 8(1) (2018) 15503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nolte-’t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t Hoen PA, Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions, Nucleic Acids Res 40(18) (2012) 9272–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ogawa Y, Taketomi Y, Murakami M, Tsujimoto M, Yanoshita R, Small RNA transcriptomes of two types of exosomes in human whole saliva determined by next generation sequencing, Biol Pharm Bull 36(1) (2013) 66–75. [DOI] [PubMed] [Google Scholar]
- [23].Shurtleff MJ, Yao J, Qin Y, Nottingham RM, Temoche-Diaz MM, Schekman R, Lambowitz AM, Broad role for YBX1 in defining the small noncoding RNA composition of exosomes, Proc Natl Acad Sci U S A 114(43) (2017) E8987–E8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO, Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells, Nat Cell Biol 9(6) (2007) 654–9. [DOI] [PubMed] [Google Scholar]
- [25].Villarroya-Beltri C, Gutierrez-Vazquez C, Sanchez-Cabo F, Perez-Hernandez D, Vazquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sanchez-Madrid F, Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs, Nat Commun 4 (2013) 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hurley JH, ESCRTs are everywhere, EMBO J 34(19) (2015) 2398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Thery C, Regnault A, Garin J, Wolfers J, Zitvogel L, Ricciardi-Castagnoli P, Raposo G, Amigorena S, Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73, J Cell Biol 147(3) (1999) 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Pocsfalvi G, Stanly C, Vilasi A, Fiume I, Capasso G, Turiak L, Buzas EI, Vekey K, Mass spectrometry of extracellular vesicles, Mass Spectrom Rev 35(1) (2016) 3–21. [DOI] [PubMed] [Google Scholar]
- [29].Thery C, Amigorena S, Raposo G, Clayton A, Isolation and characterization of exosomes from cell culture supernatants and biological fluids, Curr Protoc Cell Biol Chapter 3 (2006) Unit 3 22. [DOI] [PubMed] [Google Scholar]
- [30].Shurtleff MJ, Temoche-Diaz MM, Schekman R, Extracellular Vesicles and Cancer: Caveat Lector, Annual Review of Cancer Biology 2 (2018) 395–411. [Google Scholar]
- [31].Busatto S, Vilanilam G, Ticer T, Lin WL, Dickson DW, Shapiro S, Bergese P, Wolfram J, Tangential Flow Filtration for Highly Efficient Concentration of Extracellular Vesicles from Large Volumes of Fluid, Cells 7(12) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lobb R, Moller A, Size Exclusion Chromatography: A Simple and Reliable Method for Exosome Purification, Methods Mol Biol 1660 (2017) 105–110. [DOI] [PubMed] [Google Scholar]
- [33].Oeyen E, Van Mol K, Baggerman G, Willems H, Boonen K, Rolfo C, Pauwels P, Jacobs A, Schildermans K, Cho WC, Mertens I, Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine, J Extracell Vesicles 7(1) (2018) 1490143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang H, Lyden D, Asymmetric-flow field-flow fractionation technology for exomere and small extracellular vesicle separation and characterization, Nat Protoc 14(4) (2019) 1027–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Schey KL, Luther JM, Rose KL, Proteomics characterization of exosome cargo, Methods 87 (2015) 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ, Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents, Mol Cell Proteomics 3(12) (2004) 1154–69. [DOI] [PubMed] [Google Scholar]
- [37].Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C, Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS, Anal Chem 75(8) (2003) 1895–904. [DOI] [PubMed] [Google Scholar]
- [38].Clark DJ, Fondrie WE, Liao Z, Hanson PI, Fulton A, Mao L, Yang AJ, Redefining the Breast Cancer Exosome Proteome by Tandem Mass Tag Quantitative Proteomics and Multivariate Cluster Analysis, Anal Chem 87(20) (2015) 10462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Calabrese AN, Radford SE, Mass spectrometry-enabled structural biology of membrane proteins, Methods 147 (2018) 187–205. [DOI] [PubMed] [Google Scholar]
- [40].Cvjetkovic A, Jang SC, Konecna B, Hoog JL, Sihlbom C, Lasser C, Lotvall J, Detailed Analysis of Protein Topology of Extracellular Vesicles-Evidence of Unconventional Membrane Protein Orientation, Sci Rep 6 (2016) 36338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Choi D, Lee TH, Spinelli C, Chennakrishnaiah S, D’Asti E, Rak J, Extracellular vesicle communication pathways as regulatory targets of oncogenic transformation, Semin Cell Dev Biol 67 (2017) 11–22. [DOI] [PubMed] [Google Scholar]
- [42].Lee K, Fraser K, Ghaddar B, Yang K, Kim E, Balaj L, Chiocca EA, Breakefield XO, Lee H, Weissleder R, Multiplexed Profiling of Single Extracellular Vesicles, ACS Nano 12(1) (2018) 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Nolan JP, Duggan E, Analysis of Individual Extracellular Vesicles by Flow Cytometry, Methods Mol Biol 1678 (2018) 79–92. [DOI] [PubMed] [Google Scholar]
- [44].Choi DS, Kim DK, Kim YK, Gho YS, Proteomics, transcriptomics and lipidomics of exosomes and ectosomes, Proteomics 13(10–11) (2013) 1554–71. [DOI] [PubMed] [Google Scholar]
- [45].Choi DS, Yang JS, Choi EJ, Jang SC, Park S, Kim OY, Hwang D, Kim KP, Kim YK, Kim S, Gho YS, The protein interaction network of extracellular vesicles derived from human colorectal cancer cells, J Proteome Res 11(2) (2012) 1144–51. [DOI] [PubMed] [Google Scholar]
- [46].Choi DS, Kim DK, Kim YK, Gho YS, Proteomics of extracellular vesicles: Exosomes and ectosomes, Mass Spectrom Rev 34(4) (2015) 474–90. [DOI] [PubMed] [Google Scholar]
- [47].Baietti MF, Zhang Z, Mortier E, Melchior A, Degeest G, Geeraerts A, Ivarsson Y, Depoortere F, Coomans C, Vermeiren E, Zimmermann P, David G, Syndecan-syntenin-ALIX regulates the biogenesis of exosomes, Nat Cell Biol 14(7) (2012) 677–85. [DOI] [PubMed] [Google Scholar]
- [48].Mathivanan S, Fahner CJ, Reid GE, Simpson RJ, ExoCarta 2012: database of exosomal proteins, RNA and lipids, Nucleic Acids Res 40(Database issue) (2012) D1241–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Votteler J, Sundquist WI, Virus budding and the ESCRT pathway, Cell Host Microbe 14(3) (2013) 232–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Hurley JH, Odorizzi G, Get on the exosome bus with ALIX, Nat Cell Biol 14(7) (2012) 654–5. [DOI] [PubMed] [Google Scholar]
- [51].Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q, Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein, Proc Natl Acad Sci U S A 109(11) (2012) 4146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang Q, Lu Q, Plasma membrane-derived extracellular microvesicles mediate noncanonical intercellular NOTCH signaling, Nat Commun 8(1) (2017) 709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].May DG, Roux KJ, BioID: A Method to Generate a History of Protein Associations, Methods Mol Biol 2008 (2019) 83–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Roux KJ, Kim DI, Raida M, Burke B, A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells, J Cell Biol 196(6) (2012) 801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Leidal AM, Huang HH, Marsh T, Solvik T, Zhang D, Ye J, Kai F, Goldsmith J, Liu JY, Huang YH, Monkkonen T, Vlahakis A, Huang EJ, Goodarzi H, Yu L, W.A. P., Debnath J, The LC3 Conjugation Machinery Specifies the Loading of RNA-Binding Proteins into Extracellular Vesicles. Nat Cell Biol (2020) in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Spence EF, Dube S, Uezu A, Locke M, Soderblom EJ, Soderling SH, In vivo proximity proteomics of nascent synapses reveals a novel regulator of cytoskeleton-mediated synaptic maturation, Nat Commun 10(1) (2019) 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Uezu A, Kanak DJ, Bradshaw TW, Soderblom EJ, Catavero CM, Burette AC, Weinberg RJ, Soderling SH, Identification of an elaborate complex mediating postsynaptic inhibition, Science 353(6304) (2016) 1123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Uezu A, Soderling S, Identifying Synaptic Proteins by In Vivo BioID from Mouse Brain, Methods Mol Biol 2008 (2019) 107–119. [DOI] [PubMed] [Google Scholar]
- [59].Moreno-Gonzalo O, Fernandez-Delgado I, Sanchez-Madrid F, Post-translational add-ons mark the path in exosomal protein sorting, Cell Mol Life Sci 75(1) (2018) 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Buschow SI, Nolte-’t Hoen EN, van Niel G, Pols MS, ten Broeke T, Lauwen M, Ossendorp F, Melief CJ, Raposo G, Wubbolts R, Wauben MH, Stoorvogel W, MHC II in dendritic cells is targeted to lysosomes or T cell-induced exosomes via distinct multivesicular body pathways, Traffic 10(10) (2009) 1528–42. [DOI] [PubMed] [Google Scholar]
- [61].Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G, Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles, J Cell Sci 126(Pt 24) (2013) 5553–65. [DOI] [PubMed] [Google Scholar]
- [62].Geminard C, De Gassart A, Blanc L, Vidal M, Degradation of AP2 during reticulocyte maturation enhances binding of hsc70 and Alix to a common site on TFR for sorting into exosomes, Traffic 5(3) (2004) 181–93. [DOI] [PubMed] [Google Scholar]
- [63].Burke MC, Oei MS, Edwards NJ, Ostrand-Rosenberg S, Fenselau C, Ubiquitinated proteins in exosomes secreted by myeloid-derived suppressor cells, J Proteome Res 13(12) (2014) 5965–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Huebner AR, Cheng L, Somparn P, Knepper MA, Fenton RA, Pisitkun T, Deubiquitylation of Protein Cargo Is Not an Essential Step in Exosome Formation, Mol Cell Proteomics 15(5) (2016) 1556–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Latifkar A, Ling L, Hingorani A, Johansen E, Clement A, Zhang X, Hartman J, Fischbach C, Lin H, Cerione RA, Antonyak MA, Loss of Sirtuin 1 Alters the Secretome of Breast Cancer Cells by Impairing Lysosomal Integrity, Dev Cell 49(3) (2019) 393–408 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Frangsmyr L, Baranov V, Nagaeva O, Stendahl U, Kjellberg L, Mincheva-Nilsson L, Cytoplasmic microvesicular form of Fas ligand in human early placenta: switching the tissue immune privilege hypothesis from cellular to vesicular level, Mol Hum Reprod 11(1) (2005) 35–41. [DOI] [PubMed] [Google Scholar]
- [67].Zuccato E, Blott EJ, Holt O, Sigismund S, Shaw M, Bossi G, Griffiths GM, Sorting of Fas ligand to secretory lysosomes is regulated by mono-ubiquitylation and phosphorylation, J Cell Sci 120(Pt 1) (2007) 191–9. [DOI] [PubMed] [Google Scholar]
- [68].Smith VL, Jackson L, Schorey JS, Ubiquitination as a Mechanism To Transport Soluble Mycobacterial and Eukaryotic Proteins to Exosomes, J Immunol 195(6) (2015) 2722–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ageta H, Ageta-Ishihara N, Hitachi K, Karayel O, Onouchi T, Yamaguchi H, Kahyo T, Hatanaka K, Ikegami K, Yoshioka Y, Nakamura K, Kosaka N, Nakatani M, Uezumi A, Ide T, Tsutsumi Y, Sugimura H, Kinoshita M, Ochiya T, Mann M, Setou M, Tsuchida K, UBL3 modification influences protein sorting to small extracellular vesicles, Nat Commun 9(1) (2018) 3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernandez-Delgado I, Torralba D, Moreno-Gonzalo O, Baldanta S, Enrich C, Guerra S, Sanchez-Madrid F, ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins, Nat Commun 7 (2016) 13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Valapala M, Vishwanatha JK, Lipid raft endocytosis and exosomal transport facilitate extracellular trafficking of annexin A2, J Biol Chem 286(35) (2011) 30911–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chaiyawat P, Weeraphan C, Netsirisawan P, Chokchaichamnankit D, Srisomsap C, Svasti J, Champattanachai V, Elevated O-GlcNAcylation of Extracellular Vesicle Proteins Derived from Metastatic Colorectal Cancer Cells, Cancer Genomics Proteomics 13(5) (2016) 387–98. [PMC free article] [PubMed] [Google Scholar]
- [73].Phueaouan T, Chaiyawat P, Netsirisawan P, Chokchaichamnankit D, Punyarit P, Srisomsap C, Svasti J, Champattanachai V, Aberrant O-GlcNAc-modified proteins expressed in primary colorectal cancer, Oncol Rep 30(6) (2013) 2929–36. [DOI] [PubMed] [Google Scholar]
- [74].Kore RA, Abraham EC, Phosphorylation negatively regulates exosome mediated secretion of cryAB in glioma cells, Biochim Biophys Acta 1863(2) (2016) 368–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, Breakefield XO, Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters, Nat Commun 6 (2015) 7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Shen B, Wu N, Yang JM, Gould SJ, Protein targeting to exosomes/microvesicles by plasma membrane anchors, J Biol Chem 286(16) (2011) 14383–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Romancino DP, Buffa V, Caruso S, Ferrara I, Raccosta S, Notaro A, Campos Y, Noto R, Martorana V, Cupane A, Giallongo A, d’Azzo A, Manno M, Bongiovanni A, Palmitoylation is a post-translational modification of Alix regulating the membrane organization of exosome-like small extracellular vesicles, Biochim Biophys Acta Gen Subj 1862(12) (2018) 2879–2887. [DOI] [PubMed] [Google Scholar]
- [78].Kim SB, Kim HR, Park MC, Cho S, Goughnour PC, Han D, Yoon I, Kim Y, Kang T, Song E, Kim P, Choi H, Mun JY, Song C, Lee S, Jung HS, Kim S, Caspase-8 controls the secretion of inflammatory lysyl-tRNA synthetase in exosomes from cancer cells, J Cell Biol 216(7) (2017) 2201–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Vidal M, Mangeat P, Hoekstra D, Aggregation reroutes molecules from a recycling to a vesicle-mediated secretion pathway during reticulocyte maturation, J Cell Sci 110 ( Pt 16) (1997) 1867–77. [DOI] [PubMed] [Google Scholar]
- [80].Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ, Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes, PLoS Biol 5(6) (2007) e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G, The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis, Dev Cell 21(4) (2011) 708–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Espenel C, Margeat E, Dosset P, Arduise C, Le Grimellec C, Royer CA, Boucheix C, Rubinstein E, Milhiet PE, Single-molecule analysis of CD9 dynamics and partitioning reveals multiple modes of interaction in the tetraspanin web, J Cell Biol 182(4) (2008) 765–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Silvie O, Charrin S, Billard M, Franetich JF, Clark KL, van Gemert GJ, Sauerwein RW, Dautry F, Boucheix C, Mazier D, Rubinstein E, Cholesterol contributes to the organization of tetraspanin-enriched microdomains and to CD81-dependent infection by malaria sporozoites, J Cell Sci 119(Pt 10) (2006) 1992–2002. [DOI] [PubMed] [Google Scholar]
- [84].Zimmerman B, Kelly B, McMillan BJ, Seegar TCM, Dror RO, Kruse AC, Blacklow SC, Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket, Cell 167(4) (2016) 1041–1051 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Bari R, Guo Q, Xia B, Zhang YH, Giesert EE, Levy S, Zheng JJ, Zhang XA, Tetraspanins regulate the protrusive activities of cell membrane, Biochem Biophys Res Commun 415(4) (2011) 619–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Van Engelenburg SB, Shtengel G, Sengupta P, Waki K, Jarnik M, Ablan SD, Freed EO, Hess HF, Lippincott-Schwartz J, Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits, Science 343(6171) (2014) 653–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Henne WM, Stenmark H, Emr SD, Molecular mechanisms of the membrane sculpting ESCRT pathway, Cold Spring Harb Perspect Biol 5(9) (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Voss S, Klewer L, Wu YW, Chemically induced dimerization: reversible and spatiotemporal control of protein function in cells, Curr Opin Chem Biol 28 (2015) 194–201. [DOI] [PubMed] [Google Scholar]
- [89].Stanton BZ, Chory EJ, Crabtree GR, Chemically induced proximity in biology and medicine, Science 359(6380) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Miyazaki Y, Mizumoto K, Dey G, Kudo T, Perrino J, Chen LC, Meyer T, Wandless TJ, A method to rapidly create protein aggregates in living cells, Nat Commun 7 (2016) 11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, Tucker CL, An optimized optogenetic clustering tool for probing protein interaction and function, Nat Commun 5 (2014)4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV, Optogenetic protein clustering and signaling activation in mammalian cells, Nat Methods 10(3) (2013) 249–52. [DOI] [PubMed] [Google Scholar]
- [93].Mateescu B, Kowal EJ, van Balkom BW, Bartel S, Bhattacharyya SN, Buzas EI, Buck AH, de Candia P, Chow FW, Das S, Driedonks TA, Fernandez-Messina L, Haderk F, Hill AF, Jones JC, Van Keuren-Jensen KR, Lai CP, Lasser C, Liegro ID, Lunavat TR, Lorenowicz MJ, Maas SL, Mager I, Mittelbrunn M, Momma S, Mukherjee K, Nawaz M, Pegtel DM, Pfaffl MW, Schiffelers RM, Tahara H, Thery C, Tosar JP, Wauben MH, Witwer KW, Nolte-’t Hoen EN, Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper, J Extracell Vesicles 6(1) (2017) 1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Chiou NT, Kageyama R, Ansel KM, Selective Export into Extracellular Vesicles and Function of tRNA Fragments during T Cell Activation, Cell Rep 25(12) (2018) 3356–3370 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F, Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells, Nat Commun 2 (2011) 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, Schekman R, Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction, Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M, Quantitative and stoichiometric analysis of the microRNA content of exosomes, Proc Natl Acad Sci U S A 111(41) (2014) 14888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Eldh M, Lotvall J, Malmhall C, Ekstrom K, Importance of RNA isolation methods for analysis of exosomal RNA: evaluation of different methods, Mol Immunol 50(4) (2012) 278–86. [DOI] [PubMed] [Google Scholar]
- [99].Srinivasan S, Yeri A, Cheah PS, Chung A, Danielson K, De Hoff P, Filant J, Laurent CD, Laurent LD, Magee R, Moeller C, Murthy VL, Nejad P, Paul A, Rigoutsos I, Rodosthenous R, Shah RV, Simonson B, To C, Wong D, Yan IK, Zhang X, Balaj L, Breakefield XO, Daaboul G, Gandhi R, Lapidus J, Londin E, Patel T, Raffai RL, Sood AK, Alexander RP, Das S, Laurent LC, Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation, Cell 177(2) (2019) 446–462 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Hobor F, Dallmann A, Ball NJ, Cicchini C, Battistelli C, Ogrodowicz RW, Christodoulou E, Martin SR, Castello A, Tripodi M, Taylor IA, Ramos A, A cryptic RNA-binding domain mediates Syncrip recognition and exosomal partitioning of miRNA targets, Nat Commun 9(1) (2018) 831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Santangelo L, Giurato G, Cicchini C, Montaldo C, Mancone C, Tarallo R, Battistelli C, Alonzi T, Weisz A, Tripodi M, The RNA-Binding Protein SYNCRIP Is a Component of the Hepatocyte Exosomal Machinery Controlling MicroRNA Sorting, Cell Rep 17(3) (2016) 799–808. [DOI] [PubMed] [Google Scholar]
- [102].Suresh PS, Tsutsumi R, Venkatesh T, YBX1 at the crossroads of non-coding transcriptome, exosomal, and cytoplasmic granular signaling, Eur J Cell Biol 97(3) (2018) 163–167. [DOI] [PubMed] [Google Scholar]
- [103].Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O, Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity, Nat Cell Biol 11(9) (2009) 1143–9. [DOI] [PubMed] [Google Scholar]
- [104].Lee YS, Pressman S, Andress AP, Kim K, White JL, Cassidy JJ, Li X, Lubell K, Lim DH, Cho IS, Nakahara K, Preall JB, Bellare P, Sontheimer EJ, Carthew RW, Silencing by small RNAs is linked to endosomal trafficking, Nat Cell Biol 11(9) (2009) 1150–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Liao YC, Fernandopulle MS, Wang G, Choi H, Hao L, Drerup CM, Patel R, Qamar S, Nixon-Abell J, Shen Y, Meadows W, Vendruscolo M, Knowles TPJ, Nelson M, Czekalska MA, Musteikyte G, Gachechiladze MA, Stephens CA, Pasolli HA, Forrest LR, St George-Hyslop P, Lippincott-Schwartz J, Ward ME, RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether, Cell 179(1) (2019) 147–164 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R, Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis, Cancer Cell 26(5) (2014) 707–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Standart N, Weil D, P-Bodies: Cytosolic Droplets for Coordinated mRNA Storage, Trends Genet 34(8) (2018) 612–626. [DOI] [PubMed] [Google Scholar]
- [108].Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D’Souza-Schorey C, ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles, Curr Biol 19(22) (2009) 1875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Lu A, Wawro P, Morgens DW, Portela F, Bassik MC, Pfeffer SR, Genome-wide interrogation of extracellular vesicle biology using barcoded miRNAs, Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Goni FM, Alonso A, Effects of ceramide and other simple sphingolipids on membrane lateral structure, Biochim Biophys Acta 1788(1) (2009) 169–77. [DOI] [PubMed] [Google Scholar]
- [111].Hannun YA, Obeid LM, Sphingolipids and their metabolism in physiology and disease, Nat Rev Mol Cell Biol 19(3) (2018) 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Frohman MA, The phospholipase D superfamily as therapeutic targets, Trends Pharmacol Sci 36(3) (2015) 137–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Ghossoub R, Lembo F, Rubio A, Gaillard CB, Bouchet J, Vitale N, Slavik J, Machala M, Zimmermann P, Syntenin-ALIX exosome biogenesis and budding into multivesicular bodies are controlled by ARF6 and PLD2, Nat Commun 5 (2014) 3477. [DOI] [PubMed] [Google Scholar]
- [114].Kassas N, Tryoen-Toth P, Corrotte M, Thahouly T, Bader MF, Grant NJ, Vitale N, Genetically encoded probes for phosphatidic acid, Methods Cell Biol 108 (2012) 445–59. [DOI] [PubMed] [Google Scholar]
- [115].Matsuo H, Chevallier J, Mayran N, Le Blanc I, Ferguson C, Faure J, Blanc NS, Matile S, Dubochet J, Sadoul R, Parton RG, Vilbois F, Gruenberg J, Role of LBPA and Alix in multivesicular liposome formation and endosome organization, Science 303(5657) (2004) 531–4. [DOI] [PubMed] [Google Scholar]
- [116].Chevallier J, Chamoun Z, Jiang G, Prestwich G, Sakai N, Matile S, Parton RG, Gruenberg J, Lysobisphosphatidic acid controls endosomal cholesterol levels, J Biol Chem 283(41) (2008) 27871–80. [DOI] [PubMed] [Google Scholar]
- [117].Vacca F, Vossio S, Mercier V, Moreau D, Johnson S, Scott CC, Montoya JP, Moniatte M, Gruenberg J, Cyclodextrin triggers MCOLN1-dependent endo-lysosome secretion in Niemann-Pick type C cells, J Lipid Res 60(4) (2019) 832–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hessvik NP, Overbye A, Brech A, Torgersen ML, Jakobsen IS, Sandvig K, Llorente A, PIKfyve inhibition increases exosome release and induces secretory autophagy, Cell Mol Life Sci 73(24) (2016) 4717–4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Pospichalova V, Svoboda J, Dave Z, Kotrbova A, Kaiser K, Klemova D, Ilkovics L, Hampl A, Crha I, Jandakova E, Minar L, Weinberger V, Bryja V, Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer, J Extracell Vesicles 4 (2015) 25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W, Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response, Nature 560(7718) (2018) 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, Montabana E, Lang UE, Fu Q, Fong L, Blelloch R, Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory, Cell 177(2) (2019) 414–427 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D, Tumour exosome integrins determine organotropic metastasis, Nature 527(7578) (2015) 329–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, Reissfelder C, Pilarsky C, Fraga MF, Piwnica-Worms D, Kalluri R, Glypican-1 identifies cancer exosomes and detects early pancreatic cancer, Nature 523(7559) (2015) 177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Hill AF, Extracellular Vesicles and Neurodegenerative Diseases, J Neurosci 39(47) (2019) 9269–9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Quek C, Hill AF, The role of extracellular vesicles in neurodegenerative diseases, Biochem Biophys Res Commun 483(4) (2017) 1178–1186. [DOI] [PubMed] [Google Scholar]
- [126].Asai H, Ikezu S, Tsunoda S, Medalla M, Luebke J, Haydar T, Wolozin B, Butovsky O, Kugler S, Ikezu T, Depletion of microglia and inhibition of exosome synthesis halt tau propagation, Nat Neurosci 18(11) (2015) 1584–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Dinkins MB, Enasko J, Hernandez C, Wang G, Kong J, Helwa I, Liu Y, Terry AV Jr., Bieberich E, Neutral Sphingomyelinase-2 Deficiency Ameliorates Alzheimer’s Disease Pathology and Improves Cognition in the 5XFAD Mouse, J Neurosci 36(33) (2016) 8653–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Iguchi Y, Eid L, Parent M, Soucy G, Bareil C, Riku Y, Kawai K, Takagi S, Yoshida M, Katsuno M, Sobue G, Julien JP, Exosome secretion is a key pathway for clearance of pathological TDP-43, Brain 139(Pt 12) (2016) 3187–3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Usman WM, Pham TC, Kwok YY, Vu LT, Ma V, Peng B, Chan YS, Wei L, Chin SM, Azad A, He AB, Leung AYH, Yang M, Shyh-Chang N, Cho WC, Shi J, Le MTN, Efficient RNA drug delivery using red blood cell extracellular vesicles, Nat Commun 9(1) (2018) 2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Wang Q, Yu J, Kadungure T, Beyene J, Zhang H, Lu Q, ARMMs as a versatile platform for intracellular delivery of macromolecules, Nat Commun 9(1) (2018) 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zipkin M, Exosome redux, Nat Biotechnol 37(12) (2019) 1395–1400. [DOI] [PubMed] [Google Scholar]
- [132].Riazifar M, Pone EJ, Lotvall J, Zhao W, Stem Cell Extracellular Vesicles: Extended Messages of Regeneration, Annu Rev Pharmacol Toxicol 57 (2017) 125–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ, Probing nuclear pore complex architecture with proximity-dependent biotinylation, Proc Natl Acad Sci U S A 111(24) (2014) E2453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]