Abstract
Advances in the field of T cell memory, including the discovery of tissue residency, continue to add to the list of defined T cell subsets. Here, we briefly review the role of resident memory T cells (TRM) in protective immunity, and propose that they exhibit developmental and migrational plasticity. We discuss T cell classification, the concept of cell type versus ‘subset’, and the difficulty of inferring developmental relationships between cells occupying malleable differentiation states. We propose that popular subset strategies do not perfectly define boundaries of developmental potential. We integrate TRM into a ‘terrace’ model that classifies memory T cells along a continuous axis of decreasing developmental potential. This model also segregates cells on the basis of migration properties, although different migration properties are viewed as parallel differentiation states that may be permissive to change.
Keywords: T cell, tissue resident, Trm, T cell differentiation
Introduction
“We must be clear that when it comes to atoms, language can be used only as in poetry. The poet, too, is not nearly so concerned with describing facts as with creating images and establishing mental connections”
-Niels Bohr
Primary T cell responses are “inside out”
The adaptive immune system must balance massive clonal diversity in the naïve lymphocyte repertoire with surveillance efficiency. T cells are tactile, and probe for antigen on the surface of cells. To increase the expediency by which very rare (naïve) T clones detect cognate antigen, they limit their surveillance to secondary lymphoid organs (SLOs) rather than explore the entire organism. Recognition of antigen within SLOs (typically internal sites that drain body surfaces) results in priming and activation of small populations of naïve T cells. Primed naïve T cells undergo massive clonal expansion. These now-activated T cells radiate out to solid organs and barrier sites where they survey the tissue, recognize infected cells, and execute effector functions that contribute to the restoration of homeostasis. This could be considered an inside (originating in draining LNs) out (then migrating to peripheral tissues) immune response (Figure 1A).
Figure 1. The anatomic topology of primary and recall T cell responses.
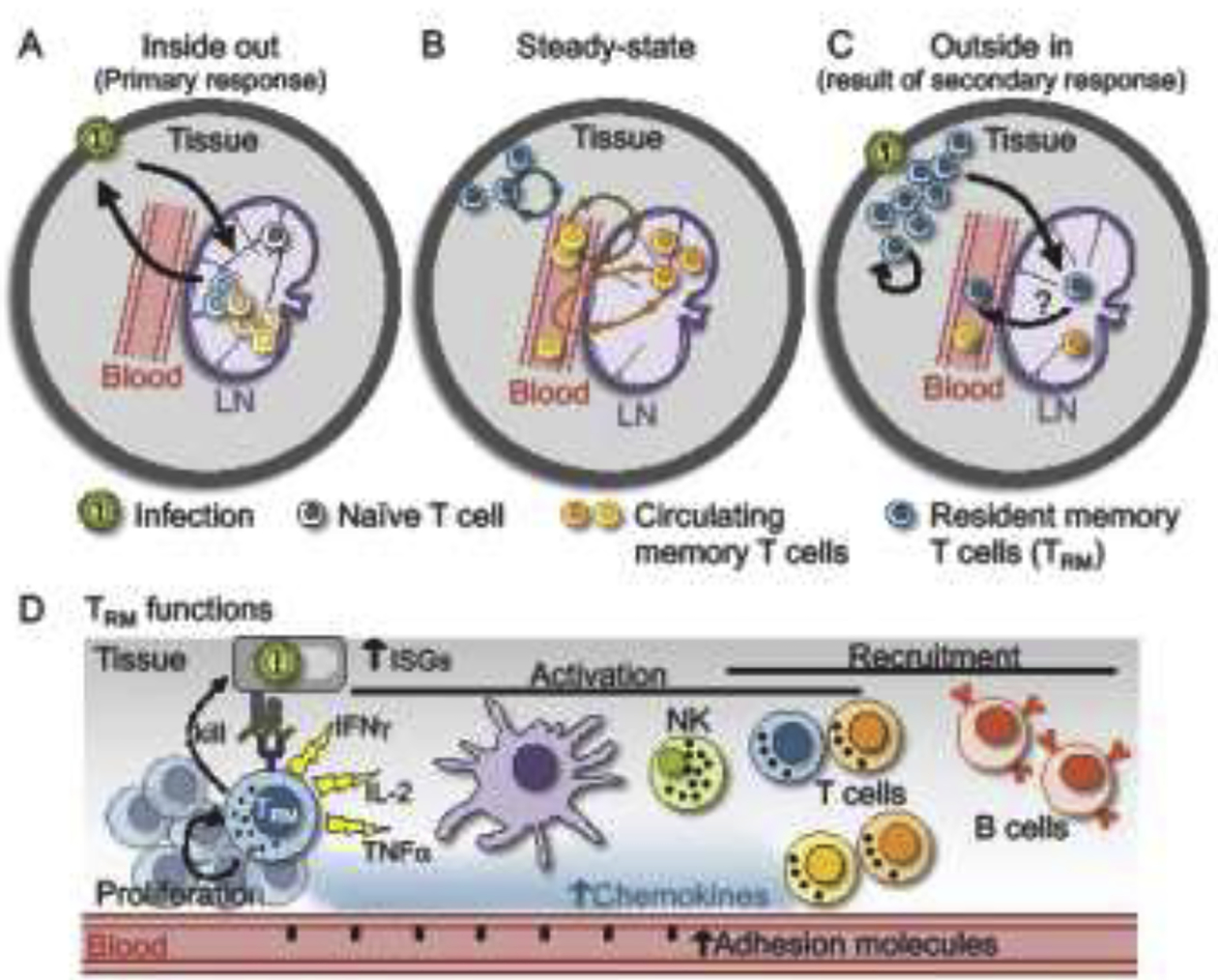
A) During a primary infection, antigen drains from peripheral tissues to draining lymph nodes (LN) which activates naïve T cells specific for that pathogen. Cells expand in lymphoid tissue and migrate out to peripheral tissues to control the infection (which could be referred to as an inside-out immune response). B) At steady-state, memory T cells patrol for reinfection and are heterogeneous, consisting of resident cells that are absent from circulation (TRM), cells that circulate through blood and lymph, and cells that recirculate though tissue using blood and lymph as conduits. C) Following secondary exposure to antigen, TRM can proliferate, give rise to an expanded local population of long-lived progeny, redistribute to draining LNs where they may remain resident, and possibly rejoin the circulation. TRM redistribution from the periphery to LNs and the circulating pool could be referred to as an outside-in immune response. Not pictured: Reactivation of LN circulating memory T cells (i.e., TCM) recapitulates an inside-out immune response. D) Upon sensing cognate antigen within tissue, TRM reactivate and alert the local immune system of a reinfection through chemokine and cytokine production. This leads to upregulation of interferon stimulated genes (ISGs), maturation of DCs, activation of T cells and NK cells, adhesion molecule upregulation and recruitment of CD8+ T cells and B cells. TRM also proliferate and upregulate cytotoxic molecules, likely contributing to killing of infected cells.
Evidence that resident memory T cells mediate “outside in” responses
Following resolution of infection, the host is left with a stable pool of memory T cells that patrol the body for reinfection. Such memory T cells are orders of magnitude more numerous than the naïve cells they differentiated from and can respond to antigen rapidly. Memory T cells represent a heterogeneous population of cells that circulate through blood and lymph [1,2], cells that extend this recirculation through non-lymphoid tissues (NLTs) [3–5], and cells that reside within NLT and, at steady-state, infrequently re-enter the circulation (Figure 1B). The latter are collectively referred to as resident memory T cells (TRM) [6,7]. TRM are remarkably abundant and broadly distributed throughout most of the body including internal organs, skin, and the mucosal barriers that form the most common sites of pathogen entry [8–11]. The positioning of TRM in frontline tissues allows them to be first responders in the event of reinfection.
TRM that encounter cognate antigen rapidly alert neighboring cells of a reinfection event, which has been termed a “sensing and alarm” function [12,13]. Within hours of antigen sensing, TRM alarm the immune system through the release of proinflammatory cytokines, establishing an antiviral state locally within the tissue, activating NK cells and T cells, promoting dendritic cell maturation and recruiting circulating innate and adaptive immune cells into the tissue (Figure 1D). These rapid TRM-mediated responses can accelerate protection against reinfection and in many cases are sufficient for protection without the contribution of circulating memory T cells [14–16]. TRM may also contribute to protection by killing infected cells directly, and are important for tumor immunosurveillance [17–19] as well as certain autoimmune [20–22], allergic, and inflammatory pathologies [23–25].
TRM arise from recently activated T cells that migrate to NLTs [26], and are thought to result from an inductive differentiation program that depends on tissue-derived developmental cues [27–30]. TRM differentiation is associated with downregulation of CD62L, CCR7, and KLF2 (a transcription factor that regulates recirculation) [31], expression of Hobit, Blimp1 [32], and/or Runx3 [33], coupled with cell surface expression of CD69 and sometimes CD103 (which may contribute to retention within tissues), although these are imperfect markers [8,26,34,35]. Indeed, a recent report referred to CD4+ ‘TRM’ that recirculated between blood and nonlymphoid tissues in the steady state (violating the definition of residence), yet expressed phenotypic signatures in common with TRM [36].
TRM have been reported to express markers, transcripts, and functions that are shared with effector and exhausted T cells, supporting one view that TRM are terminally differentiated cells; unable to mount recall responses that give rise to an amplified resident memory population, or differentiate into other subsets [37–39]. In contrast to this view, recent and ongoing work reveals that TRM are able to drive autonomous expansion of their population in response to antigen [38,40,41], can relocate to SLOs [42], and even stably re-enter circulation where they may transmogrify into memory T cells with different properties (R. Fonseca et al., submitted) [43]. This can be likened to an ‘outside-in’ topology; recall responses can be initiated at barrier sites (on the outside of the body) where TRM will proliferate, differentiate, migrate, and, after resolution of infection, may stably populate other tissue compartments including blood and lymphoid tissue (inside the body) (Figure 1C). In the event of systemic reactivation, we propose that those ‘Ex-TRM’ that have re-joined the circulating pool may remain poised for preferential homing back to previous sites of residence and reacquisition of a stable TRM program.
With advancing technologies, it has become apparent that there is considerable heterogeneity among memory T cells, including amongst those that share the property of residence [39,44,45]. This heterogeneity has encumbered the historical practice of delineating the diversity of memory into a few discrete and monolithic subsets. In this review, we will address what it means to be a subset, discuss common terminology through the lens of our emerging understanding of memory CD8+ T cell heterogeneity and plasticity, and propose where to integrate TRM into the broader lexicon and conceptualization of the T cell lineage.
Memory T cell subsetting: Developmental relationships
Our language concerning memory T cells often applies terms (and associated concepts) from developmental biology. Cellular differentiation is a progression of specification and determination events that result in discrete cell types that have reduced developmental potential. A cell lineage traces this differentiation history of a cell or tissue and ‘lineage commitment’ is the developmental point where a cell is irreversibly committed to becoming a certain cell type [46]. The T cell lineage, as with other cell types, ultimately begins with a single zygote, followed by iterative cell divisions and differentiation. Eventually, hematopoiesis gives rise to many lineage-committed cells of the immune system, including T cells. Thus, naïve T cells are the result of many cell divisions and differentiation steps (Figure 2). T cells themselves branch developmentally into clear lineages that do not typically transdifferentiate. Branches may include commitment to αβTCR vs γδTCR expression and/or single positive CD4 vs. CD8 expression. When mature naïve T cells recognize antigen, that likely also initiates an irreversible differentiation process coupled with division, that results in a large population of activated cells; many of which will die and some of which will further develop into memory cells. Important questions arise when we consider the possible branch points, or lack thereof, in this differentiation path following activation of naïve T cells. The way we conceptualize this process is intertwined with how we define subsets of cells and infer their developmental relationship.
Figure 2. Memory T cell lineage.
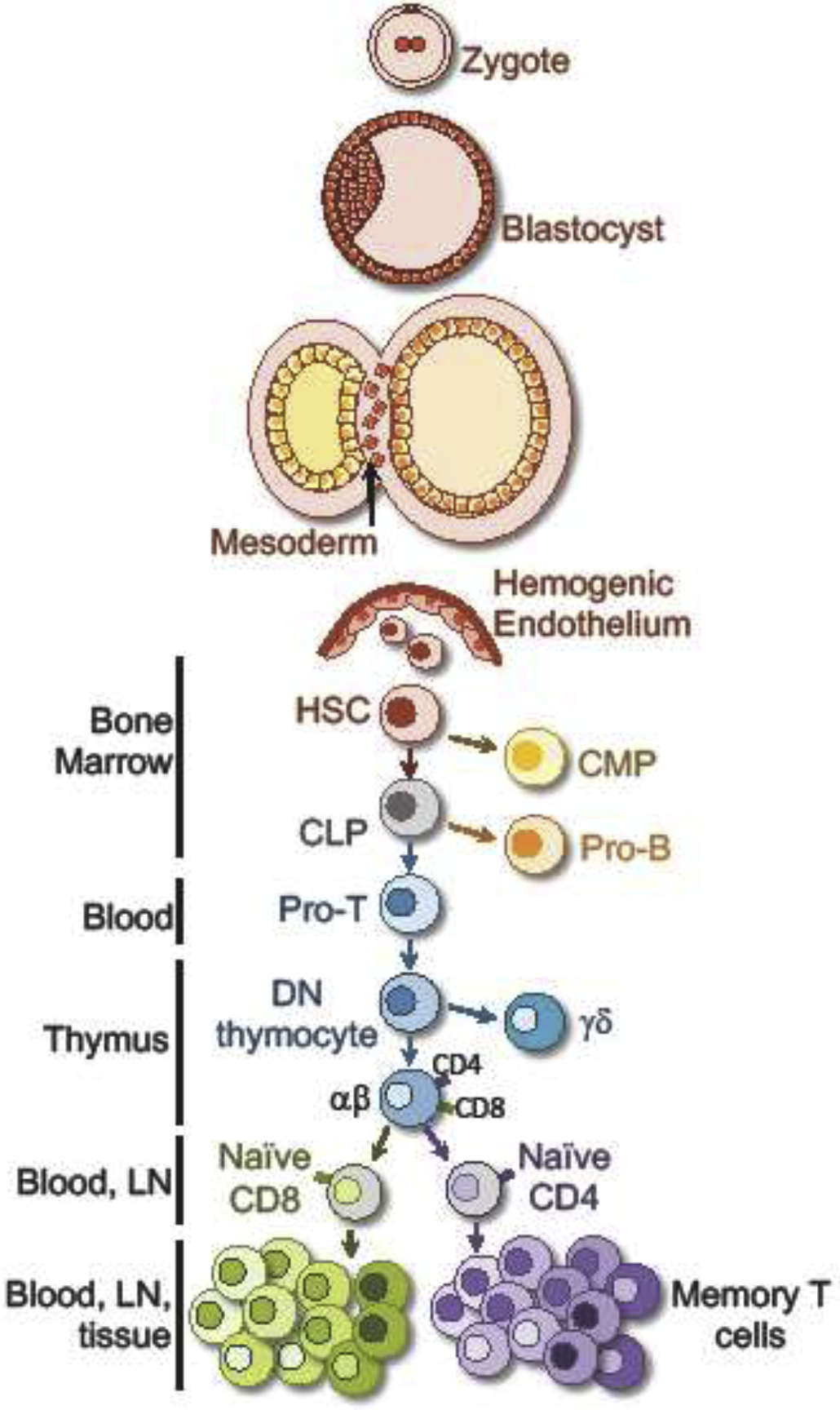
Memory T cells are a product of a series of lineage specification events that define cell type. These commitments become fixed under physiologic conditions and cells are unable to transdifferentiate or dedifferentiate (e.g. B cells do not become T cells). We argue that memory T cell subsets should not be viewed as a few discrete and distinct cell types that occupy clear steps in a linear differentiation path, but rather comprise a heterogeneous constellation of cells. Hematopoietic stem cell (HSC); Common myeloid progenitor (CMP); Common lymphoid progenitor (CL); Lymph node (LN).
Naïve T cells differentiate into ‘effector’ cells. Effector T cells have a somewhat ambiguous definition; sometimes being defined based on a functional property such as cytolysis or antiviral cytokine production, sometimes being defined as a cell that will soon die, and sometimes being defined as a cell that was ‘recently’ activated. The definition of memory is not particularly precise either, but usually invokes the idea that a significant period of time has elapsed since the naïve T cell was primed (although even this vague definition could be considered controversial). Superimposed on these vagaries, memory T cells are dissected into multiple discrete subsets, on the basis of phenotype, function, migration, perceived cell fate, or combinations of properties. There is obvious importance in being able to categorize cells into groups with shared properties, and subsetting imputes a division of labor among T cell varieties best suited to different desirable biological outcomes. Accordingly, vaccines that favor ‘central’ vs. ‘effector’ memory may have different efficacies that are contextual, rejuvenation of the ‘exhausted’ T cell subset may aid in clearing chronic infections or cancer [47,48], and CAR T cells might perform optimally if they comprised stem cell memory [49–52]. This has inspired a need to not only classify memory, but also to understand the developmental processes that contribute to the ontogeny of one subset or another.
But, how should we think about memory T cell subsets? To what extent does subsetting comprise an extension of irreversible developmental decisions or lineage commitments? Do some subsets give rise to others, but not the other way around? Or, could some subsets represent alternative differentiation states that are interconvertible? Are the qualitative axes by which subsets have been delineated robustly capturing differences in developmental relationships? Or rather, are they just highlighting functional differences of interest, without being founded on a more traditional developmental strategy for classification? Given recent advances in the field, has an opportunity been presented to restructure memory T cell classification, or to at least be more explicit about the definitions of each class and acknowledge when development principles are being eschewed in favor of (sometimes loosely defined) properties?
Criteria for subsetting and ontogeny models
There are a number of properties by which memory T cells have been Balkanized into subsets, and this process has carried implications, both semantic and conceptual. Firstly, subsets imply largely discrete homogenous entities (Figure 3A). Moreover, subset ontogeny has been diffracted through the prism of developmental biology, which imputes unidirectional lineage relationships and progressive loss of plasticity, and there has been a temptation to insert newly defined subsets into an existing lineage hierarchy. Confounding understanding, many subsets have been proposed, though not always clearly defined, with some gaining more traction than others. Often, a designation is based on a conveniently measurable cell-surface marker or transcription factor. These markers may imperfectly correlate with diverse and difficult-to-measure properties that relate more closely to function (e.g. what cytokines are made, trafficking patterns, proliferative capacity, developmental plasticity, etc.). Soon, the marker, and an associated subset name, becomes a synonym or substitute for the biology. In addition, the semantics (e.g. central vs. effector memory) can belie the actual properties (central memory T cells make effector cytokines while ‘effete’ might be a better label for some effector memory T cells) [53]. These problems are not unfamiliar to anyone working in the field of T cell memory and can be especially vexing to those in other fields [54,55].
Figure 3. Models of T cell differentiation.
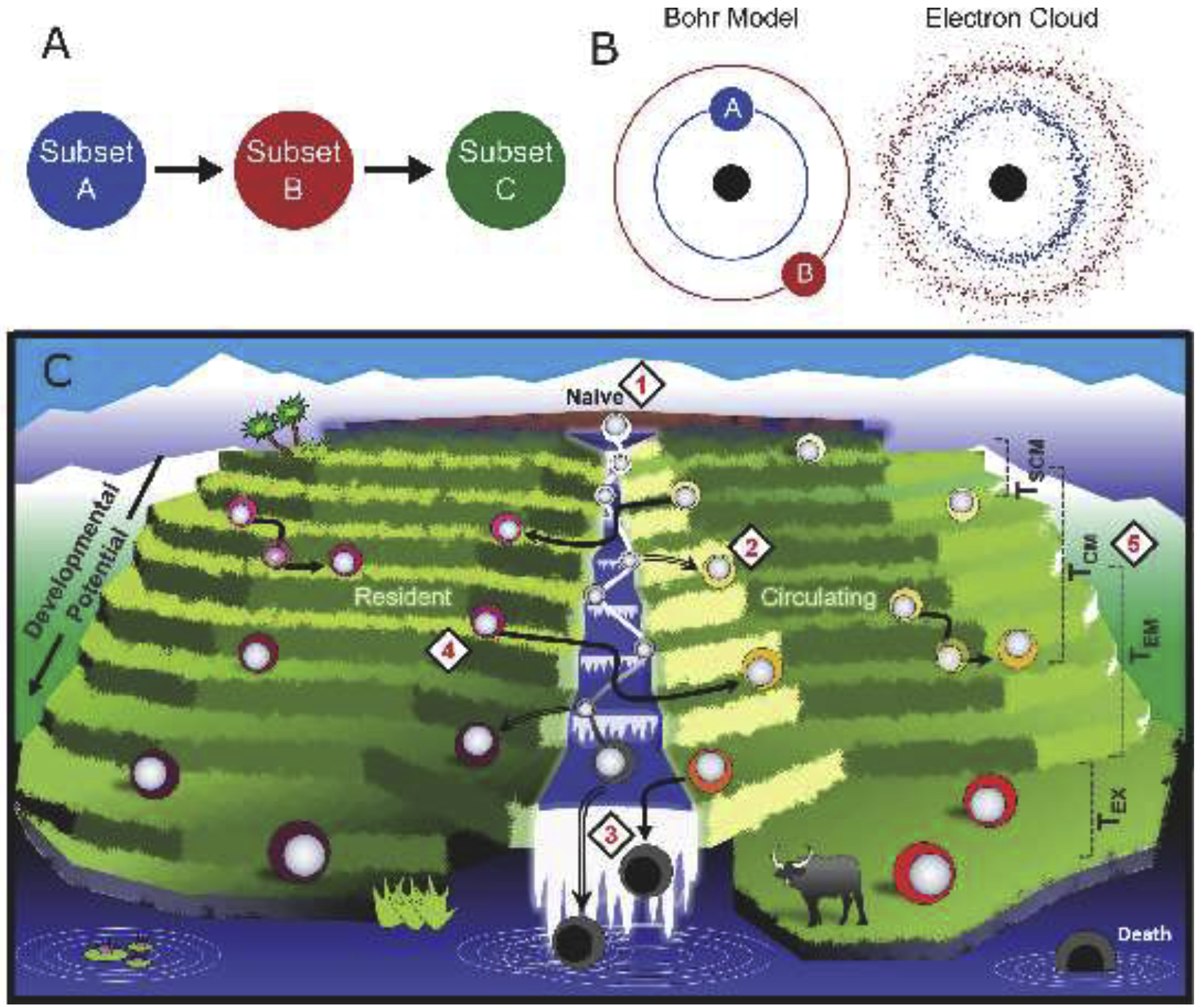
A) Defined subsets of memory T cells imply unidirectional differentiation between discrete cell types. B) Initial modeling of electron dynamics implied discrete orbits of movement (left), whereas the modern view highlights a probabilistic distribution, represented as an electron cloud (right). C) The ‘terrace’ model depicts naïve cell activation (1) and differentiation into memory T cells (2) along a continuum of developmental potential (depicted as terraces), though most activated T cells (depicted in the river) die (3). Migration status is used to further bifurcate memory T cells into a resident and circulating pool. This migration status is largely fixed, though may not be permanent for all cells (4). Subset nomenclature of circulating memory T cells is estimated, but does not reflect discrete cell types with stepwise loss of differentiation potential (5). The ‘terrace’ model is based on developmental potential and migration properties. Stem cell memory T cells (TSCM); Central memory T cells (TCM); Effector memory T cells (TEM); Exhausted T cell (TEX).
It bears consideration whether immunology should borrow a page from physics. Niels Bohr elegantly visualized electrons as occupying discrete orbits, which advanced the field in its time and enabled further discoveries. However, the Bohr model had limitations, as many models do, and it was superseded by Erwin Schrodinger’s proposal that electrons fall within a probabilistic distribution that only approximate Bohr’s orbits, which was conceptualized as the ‘electron cloud’ (Figure 3B). T cell subsets may be fuzzy at the margins, or even represent a continuum of differentiation states upon which borders (and labels) have been roughly assigned.
We propose a model of T cell differentiation that is likened to terraced farming and based on developmental potential and migration (residence or circulation), while acknowledging that the current subset nomenclature may not be synonymous with these criteria (Figure 3C). Indeed, this model embraces the fact that simple subset designations will not capture discrete cell types. In the terrace model, naïve T cells can differentiate into diverse memory T cells that comprise a spectrum of developmental potentials. Unlike naïve T cells, memory T cells have passed through a stage of activation (symbolized as being in a river). Most activated T cells rapidly lose all developmental potential, and die (falling off the waterfall) without acquiring a stable memory differentiation state (occupying a flat terrace). It should be noted that activated T cells that will become memory cells may meet definitions of transiently being an ‘effector’ because they express anti-pathogen molecules and/or acquire epigenetic imprints of being competent to do so [56–58]. In this vernacular, however, being an ‘effector’ does not provide information about future differentiation potential, thus the word effector has been removed from the terrace model. As another point of emphasis, individual memory T cells exhibit a spectrum of developmental potential and are not clustered into a few discrete subsets. Therefore, existing T cell nomenclature only loosely captures the developmental relationship among circulating memory T cells (a few popular subsets have been depicted in Figure 3C while acknowledging some overlap in developmental potential). Moreover, the progression towards terminal T cell differentiation tends to correlate with impaired proliferative capacity and vulnerability to death.
This model is compatible with developmental biologist Conrad Waddington’s widely accepted creode, but rejects a common immunological interpretation that memory T cell subsets signify a few discrete cell types that are linearly organized by developmental potential and ontogeny, and dispenses with the notion that T cells with effector functions arise from terminally differentiated memory T cells [59]. This model also highlights migration because of its intrinsic relevance to understanding T cell biology given the primacy of location in relation to T cell antigen detection, metabolism, phenotype, and function. Thus, resident and circulating memory T cells exist on parallel descents of developmental potential. The property of residence or recirculation is not necessarily a fixed property, and TCR stimulus or inflammation may be a catalyst for change. Thus, developmental potential (irreversible), and migration status (amenable to change) form two continuous axes along which memory T cells may be placed.
Conclusions:
T cells are unusual somatic cells. They can remain quiescent for years, then undergo explosive proliferation and give rise to many progeny with diverse metabolisms, longevities, trafficking patterns, and functions. The growing application of single cell profiling accentuates memory T cell transcriptional, epigenetic, and posttranslational diversity. The establishment of resident memory T cells as a broadly distributed population united by a migration criterion has introduced an additional layer of complexity (and, yet another subset). TRM exhibit tissue-specific idiosyncrasies and even neighboring cells in the same organ may be dissimilar. The extent of memory T cell heterogeneity appears to be vast and may exceed the number of discrete subsets that would be practical to define in daily discourse, and this has made classification difficult. The subset labels most commonly applied (e.g., ‘effector’, ‘central’, ‘exhausted’, ‘resident’) intend to connote some useful demarcating qualities to organize the complexity.
The terrace model captures the essence of cellular differentiation and is based on a continuum of developmental potential. It deals with migrational heterogeneity, plasticity, and the issue that current subset naming conventions can be ambiguous or impermanent. One major shortcoming of the terrace model is that developmental potential and migration properties can be difficult to measure. However, it may provide a modern framework that embraces the complexity of T cell memory. Ultimately, a common language would help consolidate our current understanding of memory T cells, clarify gaps in knowledge, broaden the audience for our research, and accelerate therapeutic developments.
Highlights.
TRM reside in tissues without recirculating.
Memory T cells are more heterogenous than implied by a few discrete subset labels.
Popular subset labels do not perfectly define boundaries of developmental potential.
TRM are not terminally differentiated and exhibit plasticity.
A ‘terrace’ T cell differentiation model, based on developmental potential and migration, is proposed.
Acknowledgements
We would like to thank the members of the Masopust laboratory, particularly Andrew G. Soerens, and Vaiva Vezys for insightful discussions. The authors’ work has been funded by the National Institutes of Health – AI084913 (DM), DK114942 (SW), DE022732 (JMS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A: Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999, 401:708–12. [DOI] [PubMed] [Google Scholar]; • This seminal article proposes two subsets of memory T cells, distinguished by CCR7 expression, that have reciprocal functions and trafficking patterns.
- 2.Mueller SN, Gebhardt T, Carbone FR, Heath WR: Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol 2013, 31:137–61. [DOI] [PubMed] [Google Scholar]
- 3.Gerlach C, Moseman EA, Loughhead SM, Alvarez D, Zwijnenburg AJ, Waanders L, Garg R, de la Torre JC, von Andrian UH: The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity 2016, 45:1270–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This article provides evidence that peripheral memory T cells (TPM), identified by intermediate CX3CR1 expression, recirculate through nonlymphoid tissues.
- 4.Watanabe R, Gehad A, Yang C, Scott LL, Teague JE, Schlapbach C, Elco CP, Huang V, Matos TR, Kupper TS, et al. : Human skin is protected by four functionally and phenotypically discrete populations of resident and recirculating memory T cells. Sci Transl Med 2015, 7:279ra39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirai T, Zenke Y, Yang Y, Bartholin L, Beura LK, Masopust D, Kaplan DH: Keratinocyte-Mediated Activation of the Cytokine TGF-β Maintains Skin Recirculating Memory CD8+ T Cells. Immunity 2019, 50:1249–1261.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masopust D, Soerens AG: Tissue-Resident T Cells and Other Resident Leukocytes. Annu Rev Immunol 2019, 37:521–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szabo PA, Miron M, Farber DL: Location, location, location: Tissue resident memory T cells in mice and humans. Sci Immunol 2019, 4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steinert EM, Schenkel JM, Fraser KA, Beura LK, Manlove LS, Igyártó BZ, Southern PJ, Masopust D: Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell 2015, 161:737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gebhardt T, Wakim LM, Eidsmo L, Reading PC, Heath WR, Carbone FR: Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat Immunol 2009, 10:524–30. [DOI] [PubMed] [Google Scholar]
- 10.Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJC, Bickham KL, Lerner H, Goldstein M, Sykes M, et al. : Distribution and Compartmentalization of Human Circulating and Tissue-Resident Memory T Cell Subsets. Immunity 2013, 38:187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li N, van Unen V, Abdelaal T, Guo N, Kasatskaya SA, Ladell K, McLaren JE, Egorov ES, Izraelson M, Chuva de Sousa Lopes SM, et al. : Memory CD4+ T cells are generated in the human fetal intestine. Nat Immunol 2019, 20:301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schenkel JM, Fraser KA, Vezys V, Masopust D: Sensing and alarm function of resident memory CD8+ T cells. Nat Immunol 2013, 14:509–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ariotti S, Hogenbirk MA, Dijkgraaf FE, Visser LL, Hoekstra ME, Song J-Y, Jacobs H, Haanen JB, Schumacher TN, Ariotti S, et al. : T cell memory. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science 2014, 346:101–5. [DOI] [PubMed] [Google Scholar]
- 14.Schenkel JM, Fraser KA, Beura LK, Pauken KE, Vezys V, Masopust D: T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science 2014, 346:98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iijima N, Iwasaki A: T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science 2014, 346:93–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez-Ruiz D, Ng WY, Holz LE, Ma JZ, Zaid A, Wong YC, Lau LS, Mollard V, Cozijnsen A, Collins N, et al. : Liver-Resident Memory CD8+ T Cells Form a Front-Line Defense against Malaria Liver-Stage Infection. Immunity 2016, 45:889–902. [DOI] [PubMed] [Google Scholar]
- 17.Park SL, Buzzai A, Rautela J, Hor JL, Hochheiser K, Effern M, McBain N, Wagner T, Edwards J, McConville R, et al. : Tissue-resident memory CD8+ T cells promote melanoma-immune equilibrium in skin. Nature 2019, 565:366–371. [DOI] [PubMed] [Google Scholar]; •• This study provides evidence that TRM control tumor growth and are responsible for maintaining cancer-immune equilibrium.
- 18.Malik BT, Byrne KT, Vella JL, Zhang P, Shabaneh TB, Steinberg SM, Molodtsov AK, Bowers JS, Angeles CV., Paulos CM, et al. : Resident memory T cells in the skin mediate durable immunity to melanoma. Sci Immunol 2017, 2:eaam6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosato PC, Wijeyesinghe S, Stolley JM, Nelson CE, Davis RL, Manlove LS, Pennell CA, Blazar BR, Chen CC, Geller MA, et al. : Virus-specific memory T cells populate tumors and can be repurposed for tumor immunotherapy. Nat Commun 2019, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheuk S, Schlums H, Gallais Sérézal I, Martini E, Chiang SC, Marquardt N, Gibbs A, Detlofsson E, Introini A, Forkel M, et al. : CD49a Expression Defines Tissue-Resident CD8+ T Cells Poised for Cytotoxic Function in Human Skin. Immunity 2017, 46:287–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinbach K, Vincenti I, Egervari K, Kreutzfeldt M, van der Meer F, Page N, Klimek B, Rossitto-Borlat I, Di Liberto G, Muschaweckh A, et al. : Brain-resident memory T cells generated early in life predispose to autoimmune disease in mice. Sci Transl Med 2019, 11:1–14. [DOI] [PubMed] [Google Scholar]
- 22.Matos TR, O’Malley JT, Lowry EL, Hamm D, Kirsch IR, Robins HS, Kupper TS, Krueger JG, Clark RA: Clinically resolved psoriatic lesions contain psoriasis-specific IL-17-producing αβ T cell clones. J Clin Invest 2017, 127:4031–4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hondowicz BD, An D, Schenkel JM, Kim KS, Steach HR, Krishnamurty AT, Keitany GJ, Garza EN, Fraser KA, Moon JJ, et al. : Interleukin-2-Dependent Allergen-Specific Tissue-Resident Memory Cells Drive Asthma. Immunity 2016, 44:155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bošnjak B, Kazemi S, Altenburger LM, Mokrović G, Epstein MM: Th2-TRMs Maintain Life-Long Allergic Memory in Experimental Asthma in Mice. Front Immunol 2019, 10:840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zundler S, Becker E, Spocinska M, Slawik M, Parga-Vidal L, Stark R, Wiendl M, Atreya R, Rath T, Leppkes M, et al. : Hobit- and Blimp-1-driven CD4+ tissue-resident memory T cells control chronic intestinal inflammation. Nat Immunol 2019, 20:288–300. [DOI] [PubMed] [Google Scholar]
- 26.Masopust D, Choo D, Vezys V, Wherry EJ, Duraiswamy J, Akondy R, Wang J, Casey KA, Barber DL, Kawamura KS, et al. : Dynamic T cell migration program provides resident memory within intestinal epithelium. J Exp Med 2010, 207:553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackay LK, Rahimpour A, Ma JZ, Collins N, Stock AT, Hafon M-L, Vega-Ramos J, Lauzurica P, Mueller SN, Stefanovic T, et al. : The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat Immunol 2013, 14:1294–301. [DOI] [PubMed] [Google Scholar]
- 28.Casey KA, Fraser KA, Schenkel JM, Moran A, Abt MC, Beura LK, Lucas PJ, Artis D, Wherry EJ, Hogquist K, et al. : Antigen-independent differentiation and maintenance of effector-like resident memory T cells in tissues. J Immunol 2012, 188:4866–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson EA, Darrah PA, Foulds KE, Hoffer E, Caffrey-Carr A, Norenstedt S, Perbeck L, Seder RA, Kedl RM, Loré K: Monocytes Acquire the Ability to Prime Tissue-Resident T Cells via IL-10-Mediated TGF-β Release. Cell Rep 2019, 28:1127–1135.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wakim LM, Woodward-Davis A, Bevan MJ: Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc Natl Acad Sci U S A 2010, 107:17872–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skon CN, Lee J, Anderson KG, Masopust D, Hogquist KA, Jameson SC: Transcriptional downregulation of S1pr1 is required for the establishment of resident memory CD8+ T cells. Nat Immunol 2013, 14:1285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackay LK, Minnich M, Kragten NAM, Liao Y, Nota B, Seillet C, Zaid A, Man K, Preston S, Freestone D, et al. : Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science 2016, 352:459–63. [DOI] [PubMed] [Google Scholar]
- 33.Milner JJ, Toma C, Yu B, Zhang K, Omilusik K, Phan AT, Wang D, Getzler AJ, Nguyen T, Crotty S, et al. : Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature 2017, 552:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mackay LK, Braun A, Macleod BL, Collins N, Tebartz C, Bedoui S, Carbone FR, Gebhardt T: Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J Immunol 2015, 194:2059–63. [DOI] [PubMed] [Google Scholar]
- 35.Walsh DA, Borges da Silva H, Beura LK, Peng C, Hamilton SE, Masopust D, Jameson SC: The Functional Requirement for CD69 in Establishment of Resident Memory CD8+ T Cells Varies with Tissue Location. J Immunol 2019, 203:946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klicznik MM, Morawski PA, Höllbacher B, Varkhande SR, Motley SJ, Kuri-Cervantes L, Goodwin E, Rosenblum MD, Long SA, Brachtl G, et al. : Human CD4+CD103+ cutaneous resident memory T cells are found in the circulation of healthy individuals. Sci Immunol 2019, 4:eaav8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Farber DL, Yudanin NA, Restifo NP: Human memory T cells: generation, compartmentalization and homeostasis. Nat Rev Immunol 2014, 14:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park SL, Zaid A, Hor JL, Christo SN, Prier JE, Davies B, Alexandre YO, Gregory JL, Russell TA, Gebhardt T, et al. : Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat Immunol 2018, 19:183–191. [DOI] [PubMed] [Google Scholar]
- 39.Kumar BV, Ma W, Miron M, Granot T, Guyer RS, Carpenter DJ, Senda T, Sun X, Ho S-H, Lerner H, et al. : Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep 2017, 20:2921–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beura LK, Mitchell JS, Thompson EA, Schenkel JM, Mohammed J, Wijeyesinghe S, Fonseca R, Burbach BJ, Hickman HD, Vezys V, et al. : Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat Immunol 2018, 19:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakim LM, Waithman J, van Rooijen N, Heath WR, Carbone FR: Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science 2008, 319:198–202. [DOI] [PubMed] [Google Scholar]
- 42.Beura LK, Wijeyesinghe S, Thompson EA, Macchietto MG, Rosato PC, Pierson MJ, Schenkel JM, Mitchell JS, Vezys V, Fife BT, et al. : T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity 2018, 48:327–338.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masopust D, Vezys V, Wherry EJ, Barber DL, Ahmed R: Cutting Edge: Gut Microenvironment Promotes Differentiation of a Unique Memory CD8 T Cell Population. J Immunol 2006, 176:2079–2083. [DOI] [PubMed] [Google Scholar]
- 44.Savas P, Virassamy B, Ye C, Salim A, Mintoff CP, Caramia F, Salgado R, Byrne DJ, Teo ZL, Dushyanthen S, et al. : Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat Med 2018, 24:986–993. [DOI] [PubMed] [Google Scholar]
- 45.Clarke J, Panwar B, Madrigal A, Singh D, Gujar R, Wood O, Chee SJ, Eschweiler S, King EV, Awad AS, et al. : Single-cell transcriptomic analysis of tissue-resident memory T cells in human lung cancer. J Exp Med 2019, doi: 10.1084/jem.20190249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert SF: Developmental Biology. Sinauer Associates; 2000. [Google Scholar]
- 47.Im SJ, Hashimoto M, Gerner MY, Lee J, Kissick HT, Burger MC, Shan Q, Hale JS, Lee J, Nasti TH, et al. : Defining CD8(+) T cells that provide the proliferative burst after PD-1 therapy. Nature 2016, 537:417–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He R, Hou S, Liu C, Zhang A, Bai Q, Han M, Yang Y, Wei G, Shen T, Yang X, et al. : Follicular CXCR5-expressing CD8+ T cells curtail chronic viral infection. Nat Publ Gr 2016, 537:412–428. [DOI] [PubMed] [Google Scholar]
- 49.Cieri N, Oliveira G, Greco R, Forcato M, Taccioli C, Cianciotti B, Valtolina V, Noviello M, Vago L, Bondanza A, et al. : Generation of human memory stem T cells after haploidentical T-replete hematopoietic stem cell transplantation. Blood 2015, 125:2865–74. [DOI] [PubMed] [Google Scholar]
- 50.Xu Y, Zhang M, Ramos CA, Durett A, Liu E, Dakhova O, Liu H, Creighton CJ, Gee AP, Heslop HE, et al. : Closely related T-memory stem cells correlate with in vivo expansion of CAR.CD19-T cells and are preserved by IL-7 and IL-15. Blood 2014, 123:3750–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stemberger C, Graef P, Odendahl M, Albrecht J, Dössinger G, Anderl F, Buchholz VR, Gasteiger G, Schiemann M, Grigoleit GU, et al. : Lowest numbers of primary CD8(+) T cells can reconstitute protective immunity upon adoptive immunotherapy. Blood 2014, 124:628–37. [DOI] [PubMed] [Google Scholar]
- 52.Graef P, Buchholz VR, Stemberger C, Flossdorf M, Henkel L, Schiemann M, Drexler I, Höfer T, Riddell SR, Busch DH: Serial Transfer of Single-Cell-Derived Immunocompetence Reveals Stemness of CD8+ Central Memory T Cells. Immunity 2014, 41:116–126. [DOI] [PubMed] [Google Scholar]
- 53.Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R: Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat Immunol 2003, 4:225–234. [DOI] [PubMed] [Google Scholar]
- 54.Ahmed R, Bevan MJ, Reiner SL, Fearon DT: The precursors of memory: models and controversies. Nat Rev Immunol 2009, 9:662–668. [DOI] [PubMed] [Google Scholar]
- 55.Bluestone JA, Mackay CR, O’Shea JJ, Stockinger B: The functional plasticity of T cell subsets. Nat Rev Immunol 2009, 9:811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akondy RS, Fitch M, Edupuganti S, Yang S, Kissick HT, Li KW, Youngblood BA, Abdelsamed HA, McGuire DJ, Cohen KW, et al. : Origin and differentiation of human memory CD8 T cells after vaccination. Nature 2017, 552:362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Youngblood B, Hale JS, Kissick HT, Ahn E, Xu X, Wieland A, Araki K, West EE, Ghoneim HE, Fan Y, et al. : Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature 2017, 552:404–409. [DOI] [PMC free article] [PubMed] [Google Scholar]; • This article provides evidence that memory T cells retain an epigenetic imprint of T cell activation, including accessibility of genes associated with effector functions.
- 58.Bannard O, Kraman M, Fearon DT: Secondary replicative function of CD8+ T cells that had developed an effector phenotype. Science 2009, 323:505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Restifo NP, Gattinoni L: Lineage relationship of effector and memory T cells. Curr Opin Immunol 2013, 25:556–63. [DOI] [PMC free article] [PubMed] [Google Scholar]


