Abstract
Glioblastoma (GBM) has limited therapeutic options. DNA repair mechanisms contribute GBM cells to escape therapies and re-establish tumor growth. Multiple studies have shown that POLD2 plays a critical role in DNA replication, DNA repair and genomic stability. We demonstrate for the first time that POLD2 is highly expressed in human glioma specimens and that expression correlates with poor patient survival. siRNA or shRNA POLD2 inhibited GBM cell proliferation, cell cycle progression, invasiveness, sensitized GBM cells to chemo/radiation-induced cell death and reversed the cytoprotective effects of EGFR signaling. Conversely, forced POLD2 expression was found to induce GBM cell proliferation, colony formation, invasiveness and chemo/radiation resistance. POLD2 expression associated with stem-like cell subsets (CD133+ and SSEA-1+ cells) and positively correlated with So×2 expression in clinical specimens. Its expression was induced by So×2 and inhibited by the forced differentiation of GBM neurospheres. shRNA-POLD2 modestly inhibited GBM neurosphere-derived orthotopic xenografts growth, when combined with radiation, dramatically inhibited xenograft growth in a cooperative fashion. These novel findings identify POLD2 as a new potential therapeutic target for enhancing GBM response to current standard of care therapeutics.
Keywords: POLD2, cytoprotective oncogene, GBM stem-like cells, Chemo-/radiation Sensitizer
Introduction
The recurrence rate in GBM patients remains nearly 100% despite best current treatment consisting of surgical resection followed by ionizing radiation plus the DNA alkylating agent temozolomide. This points to the importance of DNA repair mechanisms in the ability of GBM cells to escape therapy and re-establish tumor growth. Novel therapeutic targets for sensitizing GBM to DNA-damaging radiation/chemotherapy are urgently needed.
Pol δ is a DNA polymerase closely associated with chromosomal DNA synthesis. Pol δ also participates in multiple DNA repair pathways such as base excision repair (BER), nucleotide excision repair (NER), and mismatch repair (MMR) [1, 2]. Polδ has an important 3’–5’ exonuclease activity, functions as an important gap-filling enzyme in the late post-excision stage of repair [3, 4], and serves to repair DNA lesions arising from exposure to mutagens [5]. Pol δ is also involved in DNA strand elongation during homologous recombination in yeast [6, 7]. Polδ consists of four subunits p125, p50, p66, and p12 that are encoded by the POLD1, POLD2, POLD3, POLD4 genes, respectively [8, 9]. POLD1/p125 is the catalytic subunit while POLD2/p50, POLD3/p66 and POLD4/p12 serve regulatory functions. There is a tight correlation between Polδ dysfunction, genome instability and carcinogenesis [10]. Depleting POLD1 or POLD3 results in the accumulation of DNA strand breaks, impaired S-phase progression and accumulation of chromosome abnormalities [11]. Decreased expression of POLD4 has been shown to be significantly associated with genomic instability in lung cancer [12]. Consistent with the central roles of Polδ in genome replication and repair, POLD1 and POLD2 are both essential for cell proliferation [13]. Several studies have shown that POLD1 mutation is a major cause of inherited cancer such as colorectal adenocarcinoma, endometrial carcinomas and breast cancer [13–16]. POLD2 serves as a scaffold for the assembly of Polδ by interacting simultaneously with all of the other three subunits [17]. Yeast two-hybrid assays have shown that only POLD2 interacts with translesion synthesis polymerase (Polη), indicating that POLD2 may play a unique role within the replication complex. Polη/POLD2 interaction contributes to DNA damage tolerance [17]. Emerging experimental and clinical evidence have shown that POLD2 is aberrently expressed in multiple cancers. In ovarian carcinomas, POLD2 was strongly up-regulated in poorly differentiated serous carcinomas (PDSC) when compared to control tissues and other examined histological subgroups of ovarian carcinomas [18]. POLD2 as a signature gene was significantly related with ovarian cancer patient survival [18, 19].
A multigene predictor model using TCGA glioblastoma data recently found that POLD2 is one of 7 landscape genes significantly associated with patient survival, suggesting that POLD2 may play a vital role in GBM pathogenesis and/or therapeutic resistance [20]. However, the expression and clinical significance of POLD2 and its function in DNA repair remain unknown in glioblastoma. In this study, we examined the expression, function and pre-clinical effects of POLD2 gain- and loss-of-functions in combination with DNA-damaging therapies in gliomas. Our data demonstrate that POLD2 is highly expressed in human GBM and GBM stem-like cells and that expression correlates with poor patient survival. Silencing POLD2 expression inhibited GBM cell growth and GBM stem-like cell self-renewal, and sensitized GBM cells to cell death induced by temozolomide and γ-radiation. Inhibition of POLD2 expression attenuated EGF-induced cell proliferation and the effects of EGF-induced cytoprotection against chemo-/γ-radiation-induced cell death. POLD2 inhibition in combination with ionizing radiation completely inhibited tumor growth in vivo. These findings identify POLD2 as a new potential therapeutic target for GBM.
Materials and Methods
Clinical samples
46 primary gliomas tissues (WHOII, astrocytoma n=9, oligodendroglioma n=4, mixed oligoastrocytoma n=2; WHOIII, anaplastic astrocytoma n=11, anaplastic oligodendroglioma n=3; WHOIV, GBM, n=17) and 7 control non-neoplastic brain tissues were collected from Xinqiao Hospital, Third Military Medical University, Chongqing. The gliomas were diagnosed and categorized by two neuropathologists according to the 2016 WHO Classification of Tumors of the Central Nervous System [21]. Informed consent was signed by all patients. The present study was approved by the Ethical Committee of Xinqiao Hospital, Third Military Medical University, Chongqing and Second Xiangya Hospital of Central South University, Changsha.
Patient dataset analysis
POLD2 expression (mRNA) was analyzed using GlioVis (https://gliovis.bioinfo.cnio.es, data exported September 2017) and the Cancer Genome Atlas (TCGA). The datasets were exported directly from GlioVis. Based on the status of MGMT methylation in TCGA GBM datasets, the survival analysis were carried out using the optimal cutoff as provided in http://gliovis.bioinfo.cnio.es/.
Vectors, transfections and transduction
All plasmid transfections were performed using Lipofectamine® 2000 Reagent (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. A172 and U87 cells were plated ( 4×105 cells /10 cm dish) and transfected with control siRNA or POLD2 siRNA mix at the final concentration of 100 nM (Ribobio, Guangzhou, China, POLD2 siRNA: target sequence 1: GTGGCAACCTACACCAACT; target sequence 2: GTGGAGGACTATTGCTTTG; target sequence 3: GACAGAACGTGAGTGACAT ). Cells were collected 48h later for further analysis. To generate lentivirus, 293FT cells were co-transfected with plasmids pLM-POLD2 (pLenti-GIII-CMV-GFP-2A-Puro, Abm, Richmond, Canada); or three pLKO-shRNA POLD2 and shRNA-control vectors (Johns Hopkins ChemCORE Facility); or pLM-mCitrine-Sox2 and control vector (Addgene) plus packaging mix plasmids (Open Biosystems, Waltham, MA, USA). Conditioned medium containing lentivirus was collected 48–72 hrs later and used to infect cells. Cells were collected 48–72 hrs after infection to assess transgene expression for mRNA or protein analysis. Three shRNA-POLD2 sequences were evaluated for POLD2 knockdown efficiency (sequence 1: CCCACTTGACACAGATAGGTT; sequence 2: CGAGTTTGATCCCACCAATTA; sequence 3: GCCAAATACCTCACCAAGAAA). Only sequence 3 was found to sufficiently knockdown POLD2 (>80%) and the sequence 3 was used for all shRNA-POLD2 experiments. Cells were cultured in puromycin (1μg/ml) to establish stable shRNA-POLD2 cell lines.
Cell culture and treatment
A172 and U87 cells were originally purchased from ATCC (Manassas, VA, USA). HEB normal glial cell line was purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). A172, U87 and HEB cells were cultured as adherent monolayers in DMEM containing 10% FBS and GBM-derived neurosphere lines GBM1A, GBM1B and GBM-KK were cultured as spheres in serum-free medium containing DMEM/F-12 (Invitrogen, Carlsbad, CA, USA), supplemented with 1% bovine serum albumin, 20 ng/ml epidermal growth factor and 10 ng/ml fibroblast growth factor as previously described [22–24]. Subconfluent cells (~50% confluence) were transduced with lentivirus expressing either shRNA-POLD2 or shRNA-Control for 24 hrs before exposure to EGF and/or TMZ (0.6% DMSO) or γ-radiation.
Neutral comet assays
Neutral comet assays were performed using the Trevigen Neutral Comet Assay Kit according to the manufacturer’s instructions (Trevigen). Briefly, cells were mixed with melted LM Agarose (Trevigen). Neutral electrophoresis was conducted with 1X Neutral Electrophoresis Buffer at 21 V for 1 h at 4°C in the Comet Assay Electrophoresis System (Trevigen). The DNA was stained with SYBR Green I (Trevigen) and observed by fluorescence microscopy. Comet images were analyzed using CASP software (CASP, Wroclaw, Poland) [25]. Data are presented as the mean±SEM for 3 biological replicates with more than 40 cells analyzed per replicate.
Immunoblotting, Immunohistochemistry and Immunofluorescence
Western blotting was performed using a quantitative Western blot system (LICOR Bioscience, Lincoln, NE, USA) as previously described [23, 26]. The primary antibodies are listed in Supplementary Table 1. For immunofluorescence, A172 cells were grown on coverslips and GBM1A neurospheres were collected by cytospin onto glass slides. Cells were fixed with 4% paraformaldehyde and immunostained with γH2AX antibody (Cell Signaling) according to manufacturers’ protocols, followed by secondary antibody. Secondary antibodies were conjugated with Cy3. Coverslips were treated with Vectashield antifade solution containing 4′6-diamidino-2-phenylindole (Vector Laboratories). Tumor cell proliferation and apoptotic cells were assessed by Ki-67 and cleaved caspase-3 immunohistochemistry, respectively. Frozen tumor sections (7–8 μm thick) were immunostained with primary antibodies against Ki-67 and cleaved caspase-3 (Cell Signaling, Danvers, MA, USA) as previously described [22, 27, 28]. Apoptotic and cell proliferation indices were determined by computer-assisted quantification of the number of positively stained cells per microscopic field as previously described [22, 29].
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted from tissues and cell lines using RNeasy Mini Kit (Qiagen) following the manufacturer’s protocols. After reverse transcription reactions using the MuLV Reverse Transcriptase and Oligo (dT) primers from Applied Biosystems (ThermoFisher,Grand Island, NY, USA), qPCR was performed using Power SYBR green PCR kit (Applied Biosystems, ThermoFisher) with an iQ5 Detection System (Bio-Rad, Hercules, CA, USA). Gene expression was normalized to the 18S reference control. Forward and reverse primers for POLD2 are 5´- TCCAAATGAGACCCTTCCTG −3ánd 5´- CCACACAGCACTTCTCCTCA −3´, and for 18s are 5’- ACAGGATTGACAGATTGATAGCTC −3’ and 5’- CAAATCGCTCCACCAACTAAGAA-3’.
Cell proliferation, colony formation and neurosphere formation assay
Cell counting kit-8 (CCK-8) assay was conducted following the manufacturer’s protocols [28, 30]. Cells were transfected with POLD2 siRNA or control siRNA; alternatively, cells were transduced with lentivirus expressing shRNA-POLD2 or shRNA-control, POLD2 or control. For neurosphere formation, GBM1A cells expressing shRNA-POLD2 or shRNA-Control were dissociated into single cells and cultured in ultra-low attachment flasks (2.5 ×104 cells per ml). After 10 days, GBM neurospheres were embedded in 1% agarose and stained with 0.1% Wright stain solution. The numbers of spheres were quantified using computer-assisted image analysis. For colony formation assays, A172 cells expressing transgenic POLD2 or isogenic control or A172 cells expressing shRNA-POLD2 or shRNA-control were plated (500 cells/well) and cultured for 12 d. Cells were then fixed with 4% paraformaldehyde for 10 min, and stained with 0.1% crystal violet. Cells were destained by washing with water and plates were air-dried. Colonies of ≥50 cells were counted.
Cell invasion assay
The effects of POLD2 expression on cell invasiveness were determined using a transwell invasion assay as previously described [31]. Briefly, cells were transfected with POLD2 siRNA mix (or control siRNA) or transduced with lentivirus expressing shRNA-POLD2 (or shRNA-control) or POLD2 (or control) for 48 hrs. Transfected cells (1×105) were resuspended in 300 μL 0.1% FBS medium and placed in the upper chambers of the wells, 600μL 10% FBS medium were placed in the lower chambers. After incubation for 6 hrs at 37°C in 5% CO2, the cells on the upper membrane surface were mechanically removed. Cells that had migrated to the lower side of the membrane were fixed and stained with 0.1% crystal violet or DAPI. Migrated cells were counted under a microscope in five randomly chosen fields and photographs were taken.
Chromatin immunoprecipitation (ChIP)-PCR
So×2 binding sites 2kb upstream of the POLD2 translation start site were identified using the PROMO-algorithm. Chromatin immunoprecipitation was performed using the MAGnify Chromatin Immunoprecipitation System (Life Technologies Corporation) [24]. Briefly, DNA from 1×107 GBM1A neurospheres expressing Sox2 was crosslinked using 4% paraformaldehyde, and chromatin was isolated and fragmented by sonication. DNA fragments (~300 bp) were incubated with anti-So×2 (Cell Signaling) or IgG antibody overnight at 4 °C. Precipitation of DNA fragments complexed with So×2 at the POLD2 promoter was quantified using qRT–PCR. Forward and reverse primer sequences for So×2 Binding region −1 are 5´- AGATCGCACCACTGCACTC −3ánd 5´- CTACTTCCCTGGGCCTGTTT −3´; region-2 are 5’- CCAAAGTGGGAGACAGCACT −3’ and 5’- ACAGCTGCACACCAGCTTC −3’; region-3 are CTTGGGCCTTCTTTCTGTTG-3’ and 5’- CAGCCTCCTTGGTGAAGC −3.
Cell cycle analysis
Cell cycles were analyzed by flow cytometry on a FACSCalibur (Becton-Dickinson, Mountain View, CA) [32]. Cells were transfected with POLD2 siRNA mix or control siRNA for 48 hrs. Cells were subsequently trypsinized, dissociated and fixed with 75% ethanol at 4°C for 30 min. Cells were then incubated with DNase-free RNase at 37°C for 30 min followed by propidium iodide (100ng/ml) for 1 h at 37°C. The percentage of cells at each cell-cycle phase (G1/G0, S and G2/M) was analyzed using CellQuest software (Becton-Dickinson).
Tumor implantation and animal treatments in vivo
The effects of POLD2 on in vivo tumor growth were tested in an intracranial GBM xenograft model as previously described [26, 27, 32]. 0.5×104 GBM1A cells expressing shRNA-POLD2 or shRNA-control were stereotactically implanted into the right caudate/putamen of SCID immunodeficient mice (N=18/each group). After 6 weeks, the animals implanted with GBM1A cells expressing shRNA-control or shRNA-POLD2 were divided into two groups and treated with or without radiation (300cGy once per week) for three weeks. We irradiated animals using the small animal radiation research platform [33]. The animals were sacrificed 1 week following the last radiation dose by perfusion with 4% paraformaldehyde. Brains were removed, sectioned and stained with hematoxylin/eosin. Tumor sizes were quantified by measuring maximum tumor cross-sectional area on hematoxylin and eosin–stained brain coronal sections using computer-assisted image analysis (MCID software) and then applying the formula Volume = (square root of maximum cross-sectional area) [32, 34]. All animal procedures were approved by the Johns Hopkins Institutional Animal Care and Use Committee. Activated caspase-3 and Ki-67 immunohistochemistry were conducted using anti-cleaved caspase-3 (Cell Signaling Technology, Beverly, MA) and anti Ki-67 antibodies (Dako Corp., Carpinteria, CA), respectively, as previously described [35]. Apoptotic and cell proliferation indices were determined by computer-assisted quantification of the number of positively stained cells per microscopic field as previously described [22, 29].
Statistical analysis
Data are presented as mean ± standard error of mean (SEM). Significance of differences was determined by the GraphPad Prism (GraphPad Software, La Jolla, CA). Means were compared using analysis of one-way ANOVA. Post-hoc tests included either Student’s t-test, Dunnet’s test or Tukey test as indicated. Statistical significance was defined with a P-value of less than 0.05.
Results
Clinical gliomas express POLD2 and high levels of POLD2 are associated with poor survival.
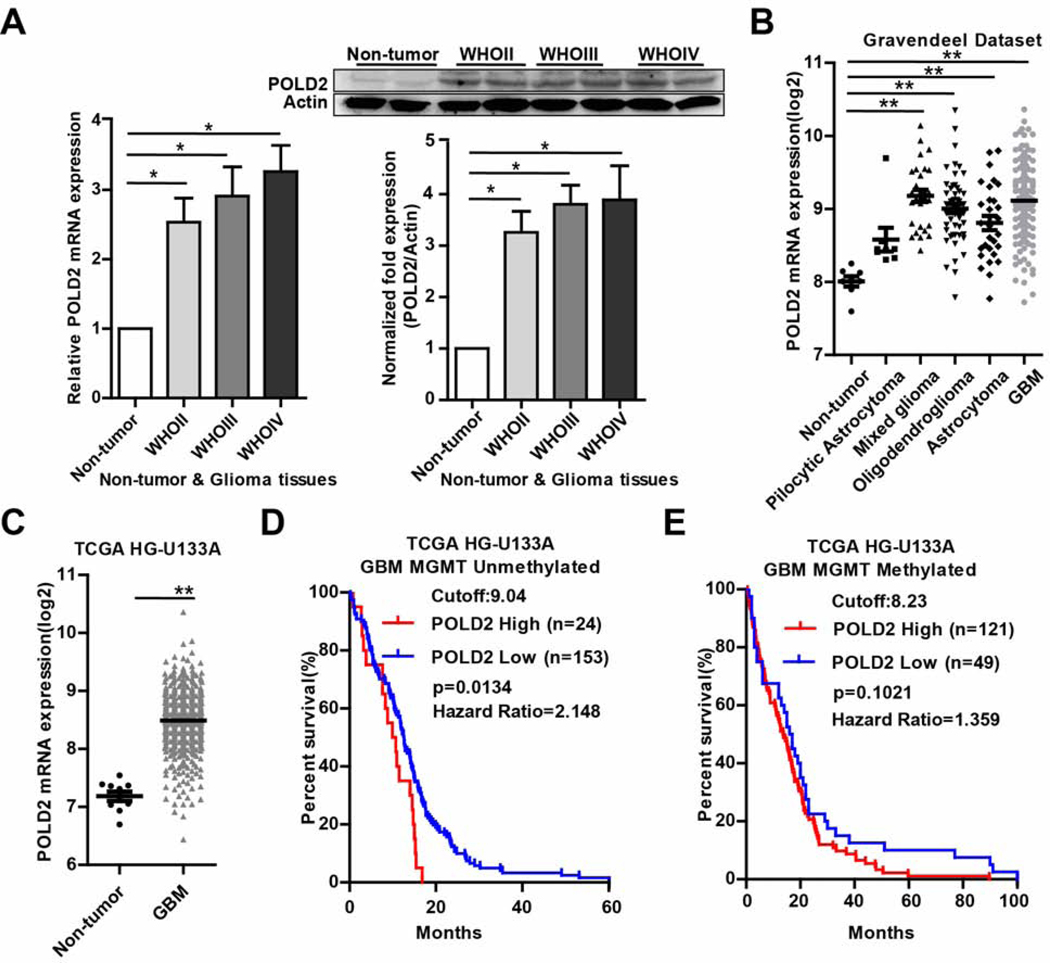
We analyzed POLD2 mRNA and protein expression in surgical glioma specimens. qRT-PCR revealed significantly elevated POLD2 expression (2.5–3.2 fold) in 46 glioma samples (WHOII, n=15; WHOIII, n=14; WHOIV, n=17) compared with non-tumor tissues (n=7) (Figure 1A, left panel). No significant differences were observed between the different glioma grades. Consistent with the qRT-PCR results, western blotting identified higher levels of POLD2 protein (3.2–3.8 fold) in glioma tissues compared with non-tumor tissues (Figure 1A, right panel). POLD2 expression was also analyzed in clinical specimens (https://gliovis.bioinfo.cnio.es). The Gravendeel dataset revealed significantly higher POLD2 expression in WHOII-IV glioma tissues with a trend of higher levels in WHOI tumors compared with non-tumor tissues (Figure 1B). Similarly, TCGA analysis showed higher POLD2 expression in GBM compared with non-tumor tissues (Figure 1C). Log-rank analysis of Kaplan-Meier survival curves in MGMT unmethylated GBM revealed a statistically significant association between high POLD2 expression and shorter survival (p=0.013) (Figure 1D) and a statistically insignificant trend (p=0.102) of shorter survival in MGMT methylated GBM patients (Figure 1E). These results suggest that POLD2 might play a role in therapeutic resistance especially in MGMT unmethylated GBMs that are currently the most therapeutically challenging.
Figure 1. Human glioma expresses POLD2 and high levels of POLD2 are associated with poor clinical outcome.
Total mRNA and protein were isolated from glioma surgical specimens (N=46) and non-neoplastic brain (N=7) and analyzed for POLD2 expression by qRT-PCR (A, left panel) and immunoblotting (A, right panel). POLD2 is highly expressed in WHOII-IV grade gliomas. The immunoblot show a representation of POLD2 protein in WHOII-IV grade gliomas. The bar graph represents immunoblot quantification by optical density (OD). Gravendeel and TCGA datasets show higher POLD2 expression in the WHOI-IV grade gliomas compared to normal tissues (B–C). TCGA dataset shows significantly poor survival for high POLD2-expressing patients in MGMT unmethylated GBM group. Survival data was retrieved from the GlioVis portal (http://gliovis.bioinfo.cnio.es/). Optimal expression cutoff was set using statistical algorithm by the GlioVis portal (D). *P < 0.05, **P < 0.01.
POLD2 expression enhances GBM cell proliferation, clonogenicity and invasiveness
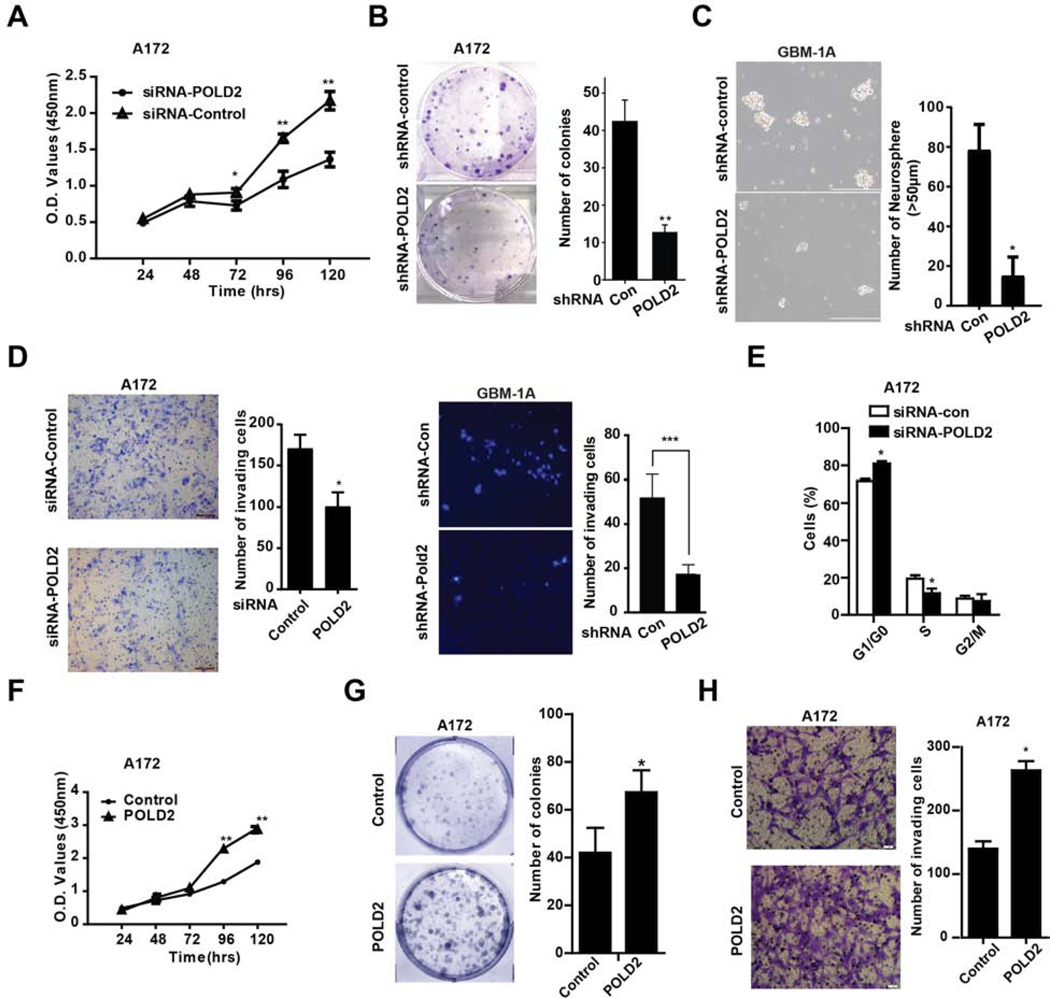
We assessed the effects of POLD2 expression on multiple parameters linked to glioma malignancy. POLD2 levels were significanty inhibited in GBM cells transfected with POLD2 siRNA mix or shRNA-POLD2, and elevated in cells transduced with lentivirus expressing full length POLD2 (Supplementary Figure 2–A,B,C). Following POLD2 knockdown, cell proliferation over 5 days was significantly reduced by 37 ± 2% in A172 cells and by 53 ± 1% in U87 cells, respectively, but did not significantly change cell proliferation in human normal glial cells (Figures 2A and Supplementary Figure 2D); colony formation was inhibited by 75.8 ± 9.1% in A172 cells (Figure 2B); and neurosphere formation was inhibited by 78.2 ± 2.1% and 50± 3.6% in GBM1A and GBM1B sphere cells, respectively (Figures 2C and Supplementary Figure 2E). Moreover, POLD2 inhibition significantly reduced the transwell invasion of A172 cells by 41 ± 2.9%, GBM1A spheres cells by 63.4±8.9% and U87 cells by 35 ± 2.5% (Figure 2D and Supplementary Figure 2F). POLD2 knockdown also increased the G0/G1 fraction from 72 ± 0.7% to 81 ± 0.7 % in A172 cells and from 76 ± 0.7% to 89 ± 1.6% in U87 cells and the percentage of S phase cells concurrently decreased from 20 ± 1% to 12 ± 1.4% in A172 cells and from 10 ± 1% to 6 ± 0.9% in U87 cells (Figures 2E and Supplementary Figure 2G). Conversely, forced POLD2 expression increased cell proliferation on the 4th day by 76 ± 1% (Figure 2F), colony formation by 60 ± 1.9% (Figure 2G) and the invasion of A172 cells by 88 ± 3.3% (Figure 2H).
Figure 2. Inhibition of endogenous POLD2 inhibits brain tumor cell proliferation, neurosphere formation, cell cycle progression and cell invasion.
A172 glioma cells were transfected with either POLD2 siRNA mix or control siRNA mix and assessed for cell proliferation by CCK-8 assay (A, n=6, ± SEM); A172 cells expressing shRNA-POLD2 or shRNA-control were plated (500 cells/well) and cultured for 12 d. Colonies of ≥50 cells were counted to estimate clonogenic potential (B); GBM1A spheres were transduced with lentivirus expressing shRNA-POLD2 or shRNA-control. Equal numbers of viable cells were cultured for 10 days and the number of spheres was quantified by computer-assisted image analysis (C); Cell invasions were measured by transwell invasion assay in A172 cells transfected with POLD2 siRNA or control siRNA (D, left panel) and GBM1A spheres transduced with lentivirus expressing shRNA-POLD2 or shRNA-control (D, right panel); Cell cycles were analyzed by propidium iodide flow cytometry in A172 cells transfected with POLD2 siRNA or control siRNA for 48 hrs (E). Controversly, cell proliferation (F), colony formation (G) and cell invasion (H) were analyzed in A172 cells transduced with lentivirus expressing transgenic POLD2 or isogenic control. *P<0.05, **P < 0.01
Inhibition of POLD2 expression sensitizes GBM cells to temozolomide and γ-radiation
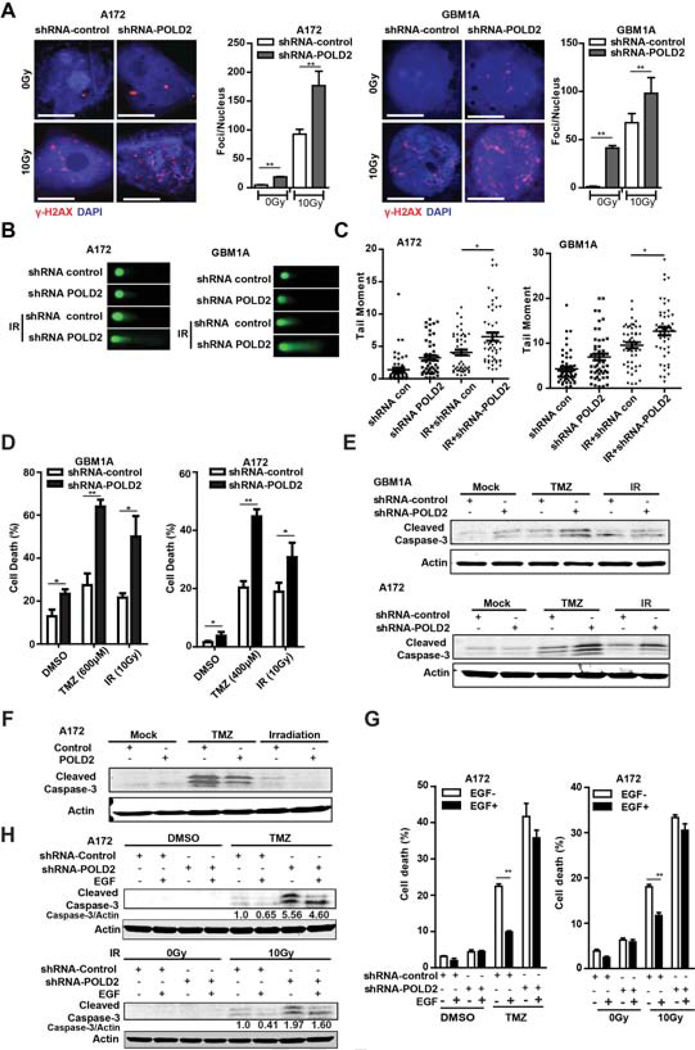
A172 cells and GBM1A neurospheres expressing shRNA POLD2 or shRNA control were treated +/− 10Gy radiation. POLD2 inhibition substantially increased DNA double-strand breaks (DSBs) in untreated control cells and in response to γ-radiation as determined by γ-H2AX immunofluorescence and western blot (Figure 3A and Supplementary Figure 3A–B). Comet assay showed that POLD2 knockdown also increased baseline DNA fragmentation in untreated cells and in response to 10 Gy γ-radiation (Figure 3B). Quantitative analysis confirmed more DNA damage occurred in shRNA POLD2 cells (Figure 3C). The effects of POLD2 inhibition on GBM cell death induced by either the alkylating agent temozolomide or γ-radiation were quantified by trypan blue assay. POLD2 inhibition significantly increased TMZ-induced A172 cell death from 20 ± 2% to 45 ± 2.5% and γ-radiation-induced A172 cell death from 19 ± 3% to 31 ± 5% (Figure 3D, right panel). Similar results were observed in patient-derived neurospheres (GBM1A) expressing shRNA-POLD2 or shRNA-Control (Figure 3D, left panel). Western blot analysis of cleaved caspase-3 showed that POLD2 knockdown increased the apoptosis response induced by either temozolomide or γ-radiation in A172 cells and in GBM1A neurospheres (Figure 3E). Conversely, forced POLD2 expression reduced temozolomide- and γ-radiation-induction of caspase-3 cleavage in A172 cells (Figure 3F), and decreased DNA double-strand breaks as determined by γ-H2AX immunofluorescence in GBM1A neurospheres in response to TMZ and γ-radiation (Supplementary Figure 3C).
Figure 3. Inhibition of POLD2 sensitizes glioma cells to chemo/radiation.
A172 cells and GBM1A neurospheres expressing shRNA-POLD2 or shRNA-control were treated +/− γ-radiation (10Gy) and the phosphorylated histone variant H2AX (γH2AX) was determined by immunofluorescence (A); Comet assay demonstrated increased DNA fragmentation in response to POLD2 inhibition + γ-radiation (B); Quantitative analysis of comet assay tail moments showed increased DNA fragmentation in response to POLD2 inhibition + γ-radiation (C); A172 cells and GBM1A neurospheres expressing shRNA-POLD2 or shRNA-control were treated with TMZ or γ-radiation for 48h. The percentage of dead cells was determined by trypan blue staining (D); Cleaved caspase-3 was evaluated by immunoblotting in cells expressing shRNA-POLD2 or shRNA-control (E) and in cells expressing POLD2 or control (F) treated +/− TMZ or +/− γ-radiation; A172 cells were transduced with lentivirus expressing shRNA-POLD2 or shRNA-control for 24 hrs prior to stimulation with EGF for 1hr and then treated +/− TMZ or +/− γ-radiation for 48 hrs. Dead cells were measured by trypan blue staining (G) and cleaved caspase-3 was evaluated by immunoblot (H). *P < 0.05, **P < 0.01.
EGFR signaling has been shown to be involved in the cellular response to chemotherapy and radiotherapy through modulation of DNA repair mechanisms in GBM [36–38]. Gene expression analysis of clinical specimens reveals a positive correlation between POLD2 and EGFR in GBM specimens (supplementary Figure 4A). Moreover, we found that EGFR pathway activation induced POLD2 expression in a time-dependent manner (Supplementary Figure 4B). To determine if POLD2 mediates the effects of EGF on cytoprotection and cell proliferation, A172 cells expressing shRNA-POLD2 or shRNA-control were pretreated with EGF before treatment with TMZ or γ-radiation. The cell death and cell proliferation were measured by trypan blue staining, immunoblotting and CCK-8 assay. POLD2 inhibition attenuated the cytoprotective response to EGFR pathway activation. In shRNA-control-transfected cells treated with TMZ, EGF reduced cell death by 56% from 22 ± 0.7% to 10 ± 0.4%. In shRNA-POLD2transfected cells treated with TMZ, EGF reduced cell death by 14% from 42 ± 3.6% to 36 ± 2.1% (Figure 3G, left panel). In shRNA-control-transfected cells treated with γ-radiation, EGF reduced cell death by 35% from 18 ± 0.5% to 11.7 ± 0.7%. In shRNA-POLD2-transfected cells treated with γ-radiation, EGF reduced cell death by 8% from 33 ± 0.6% to 30 ± 1.4% (Figure 3G, right panel). Consistent with the trypan blue exclusion results, western blotting analysis showed that EGF treatment decreased caspase-3 cleavage by 35.1% in shRNA-con-transfected cells treated with TMZ and by 17.2% in shRNA-POLD2-transfected cells treated with TMZ. Similarly, EGF treatment decreased caspase-3 cleavage by 59.3% in shRNA-con-transfected cells treated with γ-radiation and by 18.6% in shRNA-POLD2-transfected cells treated with γ-radiation (Figure 3H). These findings demonstrated that POLD2 functions to protect cells from chemo/radiation-induced DNA damage and apoptosis.
POLD2 expression associates with GBM stem-like cells
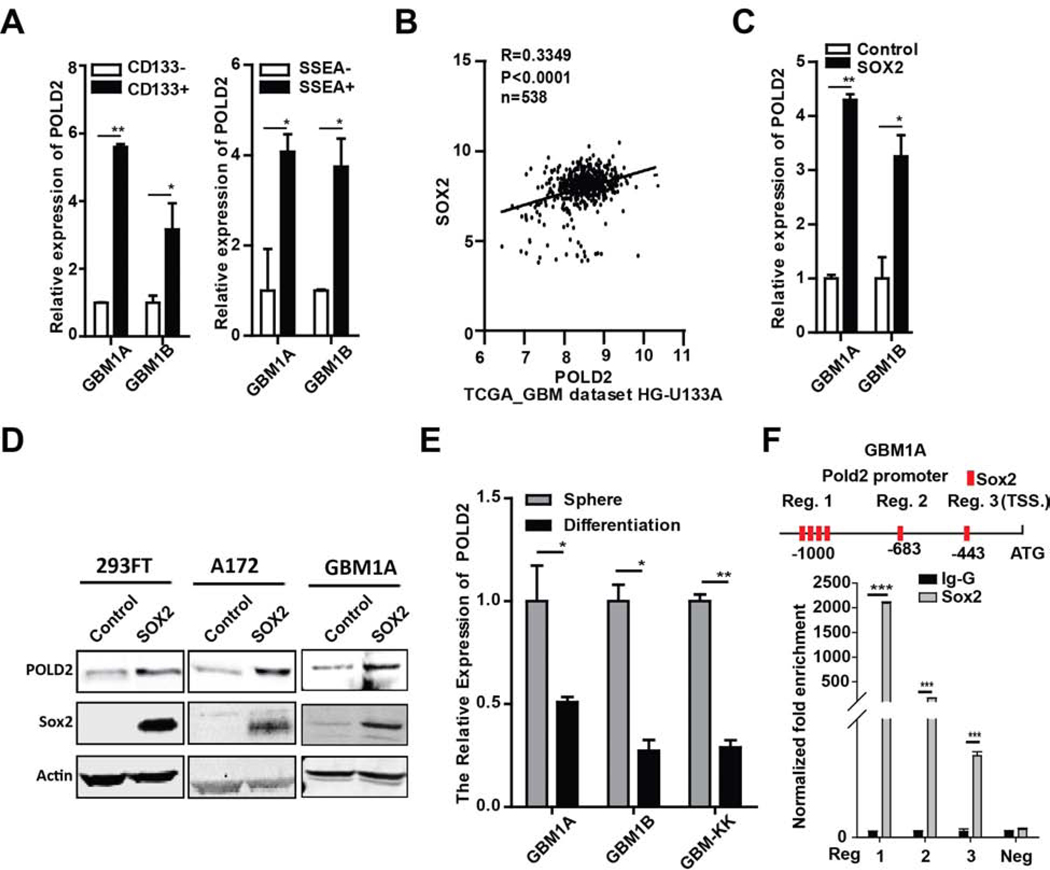
Our findings linking POLD2 expression to therapeutic resistance led us to ask if POLD2 expression associates with the GBM stem-like phenotype. We examined POLD2 expression in GBM cell fractions enriched for stem-like tumor-propagating cells. CD133+ and SSEA-1+ cell fractions from two distinct GBM neurosphere lines were separated from their prospective marker negative cells using flow cytometry [23, 26] and POLD2 expression was measured by qRT-PCR. POLD2 expression was 3.2–5.6 fold higher in CD133+ cells relative to CD133− cells and 3.7–4.1 fold higher in SSEA-1+ cells relative to SSEA-1- cells (Figure 4A). An analysis of TCGA gene expression data revealed a statistically significant correlation in the expression of POLD2 and SOX2, a marker and driver of the stem-like tumor-propagating phenotype (Figure 4B) [39, 40]. A functional relationship between Sox2 and POLD2 expression was confirmed using a complementary gain-of-function approach. Overexpressing Sox2 in 293FT, A172, GBM1A, and GBM1B cells significantly induced the expression of POLD2 mRNA and protein (Figure 4C–D). Conversely, forcing differentiation of GBM neurospheres, a condition that inhibits So×2 expression [23], significantly inhibited POLD2 expression (Figure 4E). To determine if So×2 regulates POLD2 expression through promoter binding in GBM spheres, the human POLD2 promoter sequence was found to contain several putative SO×2 binding sites (http://alggen.lsi.upc.es/) (Figure 4F, up panel). We used chromatin immunoprecipitation (ChIP) to determine if So×2 binds to the human POLD2 promoter. Anti-So×2 chromatin immunoprecipitation-PCR analysis showed enriched POLD2 promoter region 1 by ~ 2093-fold, promoter regions 2 and 3 by ~ 168 and ~ 13-fold, respectively, in GBM1A neurospheres expressing Sox2 isolate (Figure 4F, bottom panel). Taken together, these results demonstrate that POLD2 expression associates with GBM stem-like cells.
Figure 4. POLD2 is correlated with SOX2 and associated with the stem-like phenotype in GBM cells.
CD133+/− and SSEA-1+/− cells were separated from GBM neurospheres by flow cytometry. CD133+ and SSEA-1+ cells express higher levels of POLD2 compared with CD133− and SSEA-1- cells as determined by normalized qRT-PCR (A); TCGA transcriptional dataset shows a positive correlation between Sox2 expression and POLD2 expression (B); POLD2 expression was quantified by qRT-PCR in GBM1A and GBM1B neurospheres expressing transgenic So×2 vs isogenic control (C); Immunoblotting showed elevated POLD2 expression in the cells expressing transgenic So×2 compared with cells expressing isogenic control (D); Forced differentiation inhibited POLD2 expression in patient-derived GBM1A, GBM1B and GBM-KK neurospheres (E); Ch-IP-PCR to measure binding of So×2 to the portion of the POLD2 promoter containing Sox2 binding sites in GBM1A cells expressing So×2 (F). *P < 0.05, **P < 0.01, ***P < 0.001.
POLD2 inhibition and ionizing radiation cooperatively inhibit GBM in vivo
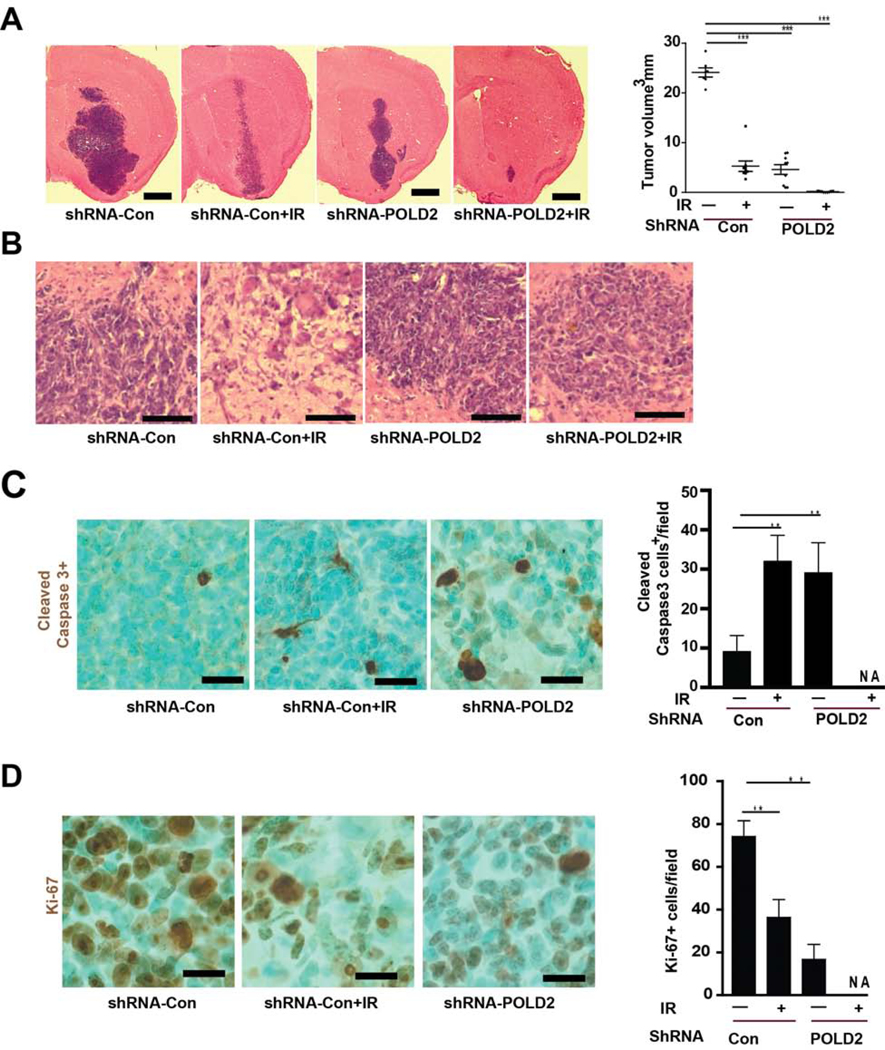
Our in vitro findings suggest that POLD2 knockdown and γ-radiation may have cooperative anti-tumor effects in vivo. To test this, pre-established orthotopic tumor xenografts derived from GBM neurospheres expressing shRNA-POLD2 (or shRNA-control) were treated +/− γ-radiation (300 cGy/week × 3 weeks) beginning on post-implantation week 6. Animals were sacrificed 7 days after completion of radiation, and tumor volumes were quantified from H&E-stained brain sections (Figure 5A). Radiation alone and POLD2-KD alone each inhibited tumor growth by ~ 80–85% (5 mm3 vs 24 mm3 and 4.6 mm3 vs 24 mm3, respectively). Combining radiation + POLD2-KD inhibited tumor growth by >99% (0.14 mm3 vs 24 mm3). Following radiation, an aggressive and invasive phenotype was observed at tumor edge derived from GBM1A expressing shRNA-control but not from GBM1A expressing shRNA-POLD2 (Figure 5B). Consistent with its effects in vitro, POLD2-KD increased the tumor apoptotic index (Figure 5C) and decreased the tumor proliferative index (Figure 5D) compared to both control tumors and tumors treated with radiation only as determined by immunohistochemistry for cleaved caspase3 and Ki-67, respectively. Tumors in the POLD2-KD + radiation group were not amenable to quantitative IHC analysis due to their exceedingly small sizes.
Figure 5. POLD2 expression inhibition and ionizing radiation cooperatively inhibit GBM growth, aggressive phenotypes and increase apoptosis in vivo.
Equal numbers of GBM1A neurosphere cells expressing shRNA-POLD2 or shRNA-control (5,000 cells/animal, shRNA-control: N=18, shRNA-POLD2: N=18) were injected into the right striatum of mice. At 6 weeks post-implantation, animals were treated with or without ionizing radiation (IR, 300 cGy/dose, once/per week) for three weeks. Animals were sacrificed one week following the last radiation treatment. Representative brain sections are shown (A, left panel). Tumor volumes were quantified by computer-assisted analysis of H&E-stained histological sections (A, right panel); H&E-stained representative photomicrographs at the tumor edge from GBM1A expressing shRNA-con, GBM1A expressing shRNA-con+IR, GBM1A expressing shRNA-POLD2, GBM1A expressing shRNA-POLD2+IR are shown (B); Tumor cell apoptosis was quantified by immunohistochemistry for cleaved caspase-3 (C) and tumor cell proliferation was quantified by Ki-67 imunohistochemistry (D). *P < 0.05, **P < 0.01.
Discussion
Several publications have shown the importance of human DNA polymerase (Pol δ) in DNA damage response (DDR) [1, 2, 41]. POLD2 as a subunit of the DNA polymerase Polδ exonuclease complex plays a crucial role in nucleotide excision repair (NER) [42]. However, the roles of POLD2 in chemo-/radiation therapy-induced DDR are unknown, especially in GBM. We now show for the first time that POLD2 expression is significantly higher in glioma compared with the non-tumor and link POLD2 up-regulation to multiple oncogenic mechanisms including cell proliferation, invasion, cell cycle progression and sensitivity to chemo-/radiation therapy. Inhibition of POLD2 expression significantly sensitized GBM cells to temozolomide and γ-radiation induced cell death in vitro and cooperated with radiation to substantially enhance GBM cell apoptosis and GBM xenograft growth inhibition in vivo. The capacity for POLD2 to mediate resistance to DNA-damaging therapeutics is likely due to its ability to complex with Pol ζ4 and thereby increases the efficiency of DNA synthesis [42]. Our findings are consistent with the recent findings of Givechian et al showing that POLD2 expression significantly associates with poor overall survival and cisplatin-based therapy resistance in bladder urothelial carcinoma [43].
Aberrant POLD2 expression has been detected in several neoplastic cell types but not previously in neoplastic stem cells [18, 20, 43]. We now show that POLD2 expression is enriched in GBM tumor-initiating SCs. POLD2 expression is shown to positively correlate with SO×2 expression in TCGA GBM datasets. Our findings demonstrate that SO×2 induces POLD2, that POLD2 silencing inhibits GBM SC self-renewal as spheres establish a mechanistic basis for this clinical association, and we provide evidence that POLD2 induces glioma malignancy, at least in part, by supporting the pool of GBM SCs. The capacity for POLD2 to support the neoplastic SC phenotype is particularly relevant in light of its role in chemo/radiation resistance. No other Pol δ subunits were found to be induced by So×2 in GBM neurospheres (Supplementary Figure 5), suggesting a unique oncogenic role for POLD2.
Multiple mechanisms likely contribute to the up-regulation of POLD2 in astroglial malignancies. The gene for POLD2 resides on chromosome 7p13, which shows frequent gains in all WHO grades of astrocytoma [44]. POLD2 expression is down-regulated by the PTEN tumor suppressor [45], which is commonly lost and typically functions to repress oncogenic signaling pathways (e.g. Akt, receptor tyrosine kinase signaling) that are commonly hyperactive in malignant astrocytoma [45–48]. EGFR plays an important role in the cellular response to chemotherapy and radiotherapy through modulation of DNA repair mechanisms; as seen in previous studies, aberrant expression and activation of the epidermal growth factor receptor (EGFR) occur in up to 50% of GBM [49, 50]. TCGA transcriptional data reveals a positive correlation between POLD2 and EGFR in GBM specimens (supplementary Figure 4A). Our finding that POLD2 expression knock-down attenuates EGFR-dependent cell proliferation (supplementary Figure 4C) and cytoprotection establishes a direct oncogenic consequence for the link between POLD2, EGFR and potentially other PTEN-regulated signaling pathways. These, together with our novel findings, identify POLD2 as a potential therapeutic target in malignant glioma especially if used in conjunction with chemo/radiation therapy.
Supplementary Material
Highlights.
POLD2 is highly expressed in surgical glioma specimens and its expression correlates with poor patient survival.
POLD2 expression promotes GBM malignancy.
Inhibition of POLD2 expression sensitizes glioma cells to temozolomide and γ-radiation.
POLD2 expression is enriched in GBM stem-like cells.
POLD2-KD + γ-radiation cooperatively inhibit in vivo glioblastoma xenograft growth.
Acknowledgements
This work was supported by the scholarship from China Scholarship Council (CSC) and the United States NIH grants NS096754 and NS073611 (JL).
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Downey KM, Tan CK, So AG, DNA polymerase delta: a second eukaryotic DNA replicase, Bioessays, 12 (1990) 231–236. [DOI] [PubMed] [Google Scholar]
- [2].Kunkel TA, Burgers PM, Dividing the workload at a eukaryotic replication fork, Trends Cell Biol, 18 (2008) 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Overmeer RM, Gourdin AM, Giglia-Mari A, Kool H, Houtsmuller AB, Siegal G, Fousteri MI, Mullenders LH, Vermeulen W, Replication factor C recruits DNA polymerase delta to sites of nucleotide excision repair but is not required for PCNA recruitment, Mol Cell Biol, 30 (2010) 4828–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, Mullenders LH, Yamashita S, Fousteri MI, Lehmann AR, Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells, Mol Cell, 37 (2010) 714–727. [DOI] [PubMed] [Google Scholar]
- [5].Nicolas E, Golemis EA, Arora S, POLD1: Central mediator of DNA replication and repair, and implication in cancer and other pathologies, Gene, 590 (2016) 128–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Maloisel L, Fabre F, Gangloff S, DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension, Mol Cell Biol, 28 (2008) 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sebesta M, Burkovics P, Haracska L, Krejci L, Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities, DNA Repair (Amst), 10 (2011) 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Podust VN, Chang LS, Ott R, Dianov GL, Fanning E, Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme, J Biol Chem, 277 (2002) 3894–3901. [DOI] [PubMed] [Google Scholar]
- [9].Lee MY, Zhang S, Lin SH, Wang X, Darzynkiewicz Z, Zhang Z, Lee EY, The tail that wags the dog: p12, the smallest subunit of DNA polymerase delta, is degraded by ubiquitin ligases in response to DNA damage and during cell cycle progression, Cell Cycle, 13 (2014) 2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu L, Mo J, Rodriguez-Belmonte EM, Lee MY, Identification of a fourth subunit of mammalian DNA polymerase delta, J Biol Chem, 275 (2000) 18739–18744. [DOI] [PubMed] [Google Scholar]
- [11].Tumini E, Barroso S, Calero CP, Aguilera A, Roles of human POLD1 and POLD3 in genome stability, Sci Rep, 6 (2016) 38873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Huang QM, Tomida S, Masuda Y, Arima C, Cao K, Kasahara TA, Osada H, Yatabe Y, Akashi T, Kamiya K, Takahashi T, Suzuki M, Regulation of DNA polymerase POLD4 influences genomic instability in lung cancer, Cancer Res, 70 (2010) 8407–8416. [DOI] [PubMed] [Google Scholar]
- [13].Uchimura A, Hidaka Y, Hirabayashi T, Hirabayashi M, Yagi T, DNA polymerase delta is required for early mammalian embryogenesis, PLoS One, 4 (2009) e4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Valle L, Hernandez-Illan E, Bellido F, Aiza G, Castillejo A, Castillejo MI, Navarro M, Segui N, Vargas G, Guarinos C, Juarez M, Sanjuan X, Iglesias S, Alenda C, Egoavil C, Segura A, Juan MJ, Rodriguez-Soler M, Brunet J, Gonzalez S, Jover R, Lazaro C, Capella G, Pineda M, Soto JL, Blanco I, New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis, Hum Mol Genet, 23 (2014) 3506–3512. [DOI] [PubMed] [Google Scholar]
- [15].Chubb D, Broderick P, Frampton M, Kinnersley B, Sherborne A, Penegar S, Lloyd A, Ma YP, Dobbins SE, Houlston RS, Genetic diagnosis of high-penetrance susceptibility for colorectal cancer (CRC) is achievable for a high proportion of familial CRC by exome sequencing, J Clin Oncol, 33 (2015) 426–432. [DOI] [PubMed] [Google Scholar]
- [16].Bellido F, Pineda M, Aiza G, Valdes-Mas R, Navarro M, Puente DA, Pons T, Gonzalez S, Iglesias S, Darder E, Pinol V, Soto JL, Valencia A, Blanco I, Urioste M, Brunet J, Lazaro C, Capella G, Puente XS, Valle L, POLE and POLD1 mutations in 529 kindred with familial colorectal cancer and/or polyposis: review of reported cases and recommendations for genetic testing and surveillance, Genet Med, 18 (2016) 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Baldeck N, Janel-Bintz R, Wagner J, Tissier A, Fuchs RP, Burkovics P, Haracska L, Despras E, Bichara M, Chatton B, Cordonnier AM, FF483–484 motif of human Poleta mediates its interaction with the POLD2 subunit of Poldelta and contributes to DNA damage tolerance, Nucleic Acids Res, 43 (2015) 2116–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Elgaaen BV, Haug KB, Wang J, Olstad OK, Fortunati D, Onsrud M, Staff AC, Sauer T, Gautvik KM, POLD2 and KSP37 (FGFBP2) correlate strongly with histology, stage and outcome in ovarian carcinomas, PLoS One, 5 (2010) e13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Wang X, Wang SS, Zhou L, Yu L, Zhang LM, A network-pathway based module identification for predicting the prognosis of ovarian cancer patients, Journal of ovarian research, 9 (2016) 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bredel M, Scholtens DM, Harsh GR, Bredel C, Chandler JP, Renfrow JJ, Yadav AK, Vogel H, Scheck AC, Tibshirani R, Sikic BI, A network model of a cooperative genetic landscape in brain tumors, JAMA, 302 (2009) 261–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW, The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary, Acta Neuropathol, 131 (2016) 803–820. [DOI] [PubMed] [Google Scholar]
- [22].Oyinlade O, Wei S, Lal B, Laterra J, Zhu H, Goodwin CR, Wang S, Ma D, Wan J, Xia S, Targeting UDP-alpha-D-glucose 6-dehydrogenase inhibits glioblastoma growth and migration, Oncogene, 37 (2018) 2615–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Lopez-Bertoni H, Lal B, Li A, Caplan M, Guerrero-Cazares H, Eberhart CG, Quinones-Hinojosa A, Glas M, Scheffler B, Laterra J, Li Y, DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2, Oncogene, 34 (2015) 3994–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Lopez-Bertoni H, Lal B, Michelson N, Guerrero-Cazares H, Quinones-Hinojosa A, Li Y, Laterra J, Epigenetic modulation of a miR-296–5p:HMGA1 axis regulates Sox2 expression and glioblastoma stem cells, Oncogene, 35 (2016) 4903–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Konca K, Lankoff A, Banasik A, Lisowska H, Kuszewski T, Gozdz S, Koza Z, Wojcik A, A cross-platform public domain PC image-analysis program for the comet assay, Mutat Res, 534 (2003) 15–20. [DOI] [PubMed] [Google Scholar]
- [26].Li Y, Li A, Glas M, Lal B, Ying M, Sang Y, Xia S, Trageser D, Guerrero-Cazares H, Eberhart CG, Quinones-Hinojosa A, Scheffler B, Laterra J, c-Met signaling induces a reprogramming network and supports the glioblastoma stem-like phenotype, Proc Natl Acad Sci U S A, 108 (2011) 9951–9956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lal B, Goodwin CR, Sang Y, Foss CA, Cornet K, Muzamil S, Pomper MG, Kim J, Laterra J, EGFRvIII and c-Met pathway inhibitors synergize against PTEN-null/EGFRvIII+ glioblastoma xenografts, Mol Cancer Ther, 8 (2009) 1751–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xu QF, Pan YW, Li LC, Zhou Z, Huang QL, Pang JC, Zhu XP, Ren Y, Yang H, Ohgaki H, Lv SQ, MiR-22 is frequently downregulated in medulloblastomas and inhibits cell proliferation via the novel target PAPST1, Brain Pathol, 24 (2014) 568–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Abounader R, Ranganathan S, Lal B, Fielding K, Book A, Dietz H, Burger P, Laterra J, Reversion of human glioblastoma malignancy by U1 small nuclear RNA/ribozyme targeting of scatter factor/hepatocyte growth factor and c-met expression, J Natl Cancer Inst, 91 (1999) 15481556. [DOI] [PubMed] [Google Scholar]
- [30].Xu Q, Ahmed AK, Zhu Y, Wang K, Lv S, Li Y, Jiang Y, Oncogenic MicroRNA-20a is downregulated by the HIF-1alpha/c-MYC pathway in IDH1 R132H-mutant glioma, Biochem Biophys Res Commun, 499 (2018) 882–888. [DOI] [PubMed] [Google Scholar]
- [31].Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, Lopes B, Schiff D, Purow B, Abounader R, MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes, Cancer Res, 69 (2009) 7569–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Li Y, Lal B, Kwon S, Fan X, Saldanha U, Reznik TE, Kuchner EB, Eberhart C, Laterra J, Abounader R, The scatter factor/hepatocyte growth factor: c-met pathway in human embryonal central nervous system tumor malignancy, Cancer Res, 65 (2005) 9355–9362. [DOI] [PubMed] [Google Scholar]
- [33].Wong J, Armour E, Kazanzides P, Iordachita I, Tryggestad E, Deng H, Matinfar M, Kennedy C, Liu Z, Chan T, Gray O, Verhaegen F, McNutt T, Ford E, DeWeese TL, Highresolution, small animal radiation research platform with x-ray tomographic guidance capabilities, Int J Radiat Oncol Biol Phys, 71 (2008) 1591–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang Y, Farenholtz KE, Yang Y, Guessous F, Dipierro CG, Calvert VS, Deng J, Schiff D, Xin W, Lee JK, Purow B, Christensen J, Petricoin E, Abounader R, Hepatocyte growth factor sensitizes brain tumors to c-MET kinase inhibition, Clin Cancer Res, 19 (2013) 1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Goodwin CR, Rath P, Oyinlade O, Lopez H, Mughal S, Xia S, Li Y, Kaur H, Zhou X, Ahmed AK, Ho S, Olivi A, Lal B, Crizotinib and erlotinib inhibits growth of c-Met(+)/EGFRvIII(+) primary human glioblastoma xenografts, Clin Neurol Neurosurg, 171 (2018) 26–33. [DOI] [PubMed] [Google Scholar]
- [36].Chou RH, Wang YN, Hsieh YH, Li LY, Xia W, Chang WC, Chang LC, Cheng CC, Lai CC, Hsu JL, Chang WJ, Chiang SY, Lee HJ, Liao HW, Chuang PH, Chen HY, Wang HL, Kuo SC, Chen CH, Yu YL, Hung MC, EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4, Dev Cell, 30 (2014) 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Sharifi Z, Abdulkarim B, Meehan B, Rak J, Daniel P, Schmitt J, Lauzon N, Eppert K, Duncan HM, Petrecca K, Guiot MC, Jean-Claude B, Sabri S, Mechanisms and Antitumor Activity of a Binary EGFR/DNA-Targeting Strategy Overcomes Resistance of Glioblastoma Stem Cells to Temozolomide, Clin Cancer Res, (2019). [DOI] [PubMed] [Google Scholar]
- [38].Li L, Huang Y, Gao Y, Shi T, Xu Y, Li H, Hyytiainen M, Keski-Oja J, Jiang Q, Hu Y, Du Z, EGF/EGFR upregulates and cooperates with Netrin-4 to protect glioblastoma cells from DNA damage-induced senescence, BMC Cancer, 18 (2018) 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Li Y, Laterra J, Cancer stem cells: distinct entities or dynamically regulated phenotypes?, Cancer Res, 72 (2012) 576–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Takahashi K, Yamanaka S, Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors, Cell, 126 (2006) 663–676. [DOI] [PubMed] [Google Scholar]
- [41].Lee M, Wang X, Zhang S, Zhang Z, Lee EYC, Regulation and Modulation of Human DNA Polymerase delta Activity and Function, Genes (Basel), 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lee YS, Gregory MT, Yang W, Human Pol zeta purified with accessory subunits is active in translesion DNA synthesis and complements Pol eta in cisplatin bypass, Proc Natl Acad Sci U S A, 111 (2014) 2954–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Givechian KB, Garner C, Garban H, Rabizadeh S, Soon-Shiong P, CAD/POLD2 gene expression is associated with poor overall survival and chemoresistance in bladder urothelial carcinoma, Oncotarget, 9 (2018) 29743–29752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nishizaki T, Ozaki S, Harada K, Ito H, Arai H, Beppu T, Sasaki K, Investigation of genetic alterations associated with the grade of astrocytic tumor by comparative genomic hybridization, Genes Chromosomes Cancer, 21 (1998) 340–346. [DOI] [PubMed] [Google Scholar]
- [45].Matsushima-Nishiu M, Unoki M, Ono K, Tsunoda T, Minaguchi T, Kuramoto H, Nishida M, Satoh T, Tanaka T, Nakamura Y, Growth and gene expression profile analyses of endometrial cancer cells expressing exogenous PTEN, Cancer research, 61 (2001) 3741–3749. [PubMed] [Google Scholar]
- [46].Smith JS, Tachibana I, Passe SM, Huntley BK, Borell TJ, Iturria N, O’Fallon JR, Schaefer PL, Scheithauer BW, James CD, Buckner JC, Jenkins RB, PTEN mutation EGFR amplification, and outcome in patients with anaplastic astrocytoma and glioblastoma multiforme, J Natl Cancer Inst, 93 (2001) 1246–1256. [DOI] [PubMed] [Google Scholar]
- [47].Srividya MR, Thota B, Shailaja BC, Arivazhagan A, Thennarasu K, Chandramouli BA, Hegde AS, Santosh V, Homozygous 10q23/PTEN deletion and its impact on outcome in glioblastoma: a prospective translational study on a uniformly treated cohort of adult patients, Neuropathology, 31 (2011) 376–383. [DOI] [PubMed] [Google Scholar]
- [48].Carico C, Nuno M, Mukherjee D, Elramsisy A, Dantis J, Hu J, Rudnick J, Yu JS, Black KL, Bannykh SI, Patil CG, Loss of PTEN is not associated with poor survival in newly diagnosed glioblastoma patients of the temozolomide era, PLoS One, 7 (2012) e33684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA Jr., Hartigan J, Smith DR, Strausberg RL, Marie SK, Shinjo SM, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW, An integrated genomic analysis of human glioblastoma multiforme, Science, 321 (2008) 1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].An Z, Aksoy O, Zheng T, Fan QW, Weiss WA, Epidermal growth factor receptor and EGFRvIII in glioblastoma: signaling pathways and targeted therapies, Oncogene, 37 (2018) 1561–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.