Abstract
The present study examined the predictive values of amyloid PET, 18F-FDG PET, and nonimaging predictors (alone and in combination) for development of Alzheimer dementia (AD) in a large population of patients with mild cognitive impairment (MCI). Methods: The study included 319 patients with MCI from the Alzheimer Disease Neuroimaging Initiative database. In a derivation dataset (n = 159), the following Cox proportional-hazards models were constructed, each adjusted for age and sex: amyloid PET using 18F-florbetapir (pattern expression score of an amyloid-β AD conversion–related pattern, constructed by principle-components analysis); 18F-FDG PET (pattern expression score of a previously defined 18F-FDG–based AD conversion–related pattern, constructed by principle-components analysis); nonimaging (functional activities questionnaire, apolipoprotein E, and mini-mental state examination score); 18F-FDG PET + amyloid PET; amyloid PET + nonimaging; 18F-FDG PET + nonimaging; and amyloid PET + 18F-FDG PET + nonimaging. In a second step, the results of Cox regressions were applied to a validation dataset (n = 160) to stratify subjects according to the predicted conversion risk. Results: On the basis of the independent validation dataset, the 18F-FDG PET model yielded a significantly higher predictive value than the amyloid PET model. However, both were inferior to the nonimaging model and were significantly improved by the addition of nonimaging variables. The best prediction accuracy was reached by combining 18F-FDG PET, amyloid PET, and nonimaging variables. The combined model yielded 5-y free-of-conversion rates of 100%, 64%, and 24% for the low-, medium- and high-risk groups, respectively. Conclusion: 18F-FDG PET, amyloid PET, and nonimaging variables represent complementary predictors of conversion from MCI to AD. Especially in combination, they enable an accurate stratification of patients according to their conversion risks, which is of great interest for patient care and clinical trials.
Keywords: mild cognitive impairment, amyloid load, PCA, Cox model, 18F-florbetapir, 18F-FDG, PET
Amyloid PET using 18F-florbetapir and 18F-FDG PET are established biomarkers of amyloid-β (Aβ) pathology and neuronal injury in Alzheimer disease, respectively (1). Both modalities have shown variable performance when adopted to predict progression to Alzheimer dementia (AD) (2,3).
Stratifying mild cognitive impairment (MCI) subjects according to their conversion risk is of great interest for clinical practice and clinical trials (e.g., patient counseling, initiation of pharmacologic and nonpharmacologic treatments, and inclusion in trials). A recent study by our group evaluated 18F-FDG PET by voxelwise principle-components analysis (PCA) and validated a PCA-derived AD conversion–related pattern (ADCRP) that showed high accuracy in prediction of conversion from MCI to AD (4). This study was in contrast to other studies (5–7). The difference in the results can be explained by methodology (PCA vs. conventional 18F-FDG PET analysis) or patient populations (highly selected research vs. real-life clinical population (5)). Conversely, the aforementioned studies favor amyloid PET over 18F-FDG PET for predicting conversion (5,7).
In the present study, we extended our previous work by also including amyloid PET and explored the predictive values of 18F-FDG PET, amyloid PET, and nonimaging variables, alone and in combination, in their ability to stratify MCI patients according to their conversion risk. As in our previous study, we took advantage of the large patient dataset of the Alzheimer Disease Neuroimaging Initiative (ADNI) database, which enables proper validation by independent derivation and validation datasets. For a fair comparison of modalities and in additional to conventional volume-of-interest analyses (yielding continuous and binary measures of amyloid load), we also applied voxelwise PCA to the amyloid PET data to assess the Aβ-based ADCRP (Aβ-ADCRP).
MATERIALS AND METHODS
Cohort
The present data were obtained from the ADNI database (ClinicalTrials.gov identifier NCT00106899; further information on the ADNI project is available at www.adni-info.org). The study was approved by ADNI, and written informed consent was obtained by the ADNI from all subjects at the baseline visit (study inclusion) and before protocol-specific procedures were performed, according to the ADNI protocols. Of the 544 subjects with MCI (suspected incipient Alzheimer disease with subjective and objective memory deficits) used in our previous study (4), we included 319 patients for whom 18F-florbetapir PET at the baseline visit was also available. Participants were evaluated at baseline and at 6- to 12-mo intervals after initial evaluation for up to 10 y. The initial inclusion criteria were a diagnosis of MCI, a mini-mental state examination (MMSE) score of at least 24 points at the time of PET imaging, a minimal follow-up time of at least 6 mo, and no bidirectional change of diagnosis (MCI to AD, and back to MCI) within the follow-up time window. The subjects were dichotomized into MCI patients who converted to AD (MCI converters) and those who did not (MCI nonconverters).
The data were randomly split into derivation and validation datasets (Table 1). Age, sex, MMSE, functional activities questionnaire (FAQ) sum score, and median follow-up time did not differ significantly between the 2 datasets (P > 0.1). As would be expected, the prevalence of high-risk apolipoprotein E (APOE) genotypes (3/4 and 4/4) significantly differed between MCI converters and MCI nonconverters in each of the datasets (P < 0.01), without evidence of any interaction between subgroups and datasets (P > 0.1).
TABLE 1.
Clinical and Demographic Characteristics of Derivation and Validation Datasets
| Derivation dataset (n = 159) |
Validation dataset (n = 160) |
|||
| Characteristic | MCI converters (n = 41) | MCI nonconverters (n = 118) | MCI converters (n = 33) | MCI nonconverters (n = 127) |
| Age (y) | 72 ± 7 | 73 ± 8 | 73 ± 8 | 73 ± 8 |
| Sex (n) | ||||
| Male | 14 | 55 | 18 | 49 |
| Female | 27 | 63 | 16 | 111 |
| Mean FAQ ± SD | 2.4 ± 3.8 | 2.7 ± 3.7 | 2.3 ± 3.5 | 2.4 ± 3.8 |
| Mean MMSE ± SD | 27.8 ± 1.8 | 27.7 ± 1.8 | 28.1 ± 1.6 | 28.1 ± 1.5 |
| APOE ε4–positive rate | 78% | 49% | 90% | 40% |
| Amyloid-positive rate | 87% | 48% | 94% | 52% |
| Mean time to conversion (mo) | 41 | — | 37 | — |
| Follow-up time (mo) | ||||
| Median | 48 | 47 | ||
| 95% confidence interval | 36–51 | 35–52 | ||
PET Analysis
The PET acquisition details have been described in the study protocols of the ADNI project online. In the case of 18F-FDG PET, dynamic 3-dimensional scans with six 5-min frames were acquired 30 min after injection of 18F-FDG. All frames were motion-corrected to the first frame and added into a sum file. 18F-FDG PET scans were spatially normalized to an in-house 18F-FDG PET template in Montreal Neurological Institute space (8) and smoothed with an isotropic gaussian kernel of 12 mm in full width at half maximum. We assessed the pattern expression score (PES) of the previously validated ADCRP as described before (4).
In the case of 18F-florbetapir PET, dynamic 3-dimensional scans with four 5-min frames acquired 50–70 min after injection were used for analysis (details are provided in the ADNI acquisition protocols). Individual datasets were motion-corrected and summed to create single-image files, followed by spatial normalization to an in-house 18F-florbetapir PET template in Montreal Neurological Institute space, constructed of both amyloid-positive (n = 9) and amyloid-negative (n = 7) control scans from cognitively normal elderly people. Smoothing with an isotropic gaussian kernel of 12 mm in full width at half maximum was applied. For assessment of amyloid load with 18F-florbetapir PET, we performed voxelwise PCA on the combined group of MCI converters and nonconverters from the derivation dataset. To identify a significant pattern, the best combination of the principal components that account for maximal variability in the data was selected by a logistic regression analysis with group (MCI converters and MCI nonconverters) as the dependent variable and subject score as the independent variable (as previously described (9)). The obtained Aβ-ADCRP represents spatially covariant voxels associated with the conversion to AD, with each voxel being specifically weighted toward its relative contribution. For the derivation and the validation datasets, PES of Aβ-ADCRP was evaluated by a topographic profile–rating algorithm (10).
Additionally, we calculated the SUV ratio (SUVR) in regions with the highest β-amyloid burden in AD (Pittsburgh compound B volume-of-interest mask taken from a previous publication (11)) using the cerebellum as a reference region, yielding continuous SUVRs. As a common, clinically used measure, we also defined a binary amyloid status (0, amyloid-negative; 1, amyloid-positive) based on an SUVR cutoff of 1.3. All analyses were implemented in an in-house pipeline based on MATLAB (The MathWorks, Inc.) and Statistical Parametric Mapping (SPM12) (https://www.fil.ion.ucl.ac.uk/spm/).
Statistical Analysis
For the derivation dataset, Cox proportional-hazards regressions were calculated using the “survival” package (12) in R (http://www.R-project.org/), each adjusted for age at baseline (years) and sex. As an initial step, we compared the 3 outcome measures of amyloid PET (PES of Aβ-ADCRP, continuous SUVR, and binary amyloid status) by Cox proportional-hazards regression and selected the most predictive measure for further analyses. Subsequently, the predictive accuracy for conversion from MCI to AD was tested for 18F-FDG PET (PES of ADCRP), amyloid PET (PES of Aβ-ADCRP), and nonimaging variables (FAQ, MMSE, and APOE ε4 genotype [positive or negative for the presence of at least 1 ε4 allele]) separately, both PES of ADCRP and PES of Aβ-ADCRP in combination with nonimaging variables, and finally all combined in the following models: amyloid PET (PES of Aβ-ADCRP); 18F-FDG PET (PES of ADCRP); nonimaging (FAQ, MMSE, and APOE); 18F-FDG PET + amyloid PET; amyloid PET + nonimaging; 18F-FDG PET + nonimaging; and amyloid PET + 18F-FDG PET + nonimaging. Continuous covariates were standardized by dividing the individual value by 2 times the SD of the respective variable to make the variables approximately equally scaled for appropriate comparison. Because conventional receiver-operating-characteristic analysis does not include time-to-event information, Harrell concordance was used instead to evaluate the goodness of the fit of the models.
In the validation dataset, the constructed Cox models were validated by means of offsetting the coefficients evaluated in the derivation dataset. For each model, the change in the Akaike information criterion (AIC) was computed using the model with the lowest AIC (i.e., best model) as a reference. Analysis of deviance was conducted for pairwise comparison between models.
Finally, the prognostic index (PI) (13) for conversion from MCI to AD was calculated for each subject using the regression coefficients gained by the respective models on the derivation dataset. Here, the PI is the sum of the product of the regression coefficients βi and predictor variables xi (with i being the index for the order of predictors in the model): PI = β1x1 +…+ βixi. The PI for each subject was calculated separately for each model. The validation dataset was stratified into 3 equally sized risk groups based on derived PI values (roughly reflecting the lowest, middle, and highest thirds of the total PI range for each of the 6 models). Separation between risk groups within different models was compared by Kaplan–Meier survival analysis. Interpretability of risk strata was limited after 60 mo by the small number of subjects with such long observation times. Thus, the display of the results (but not the statistical analysis) was restricted to a follow-up interval of 60 mo.
RESULTS
Aβ-ADCRP
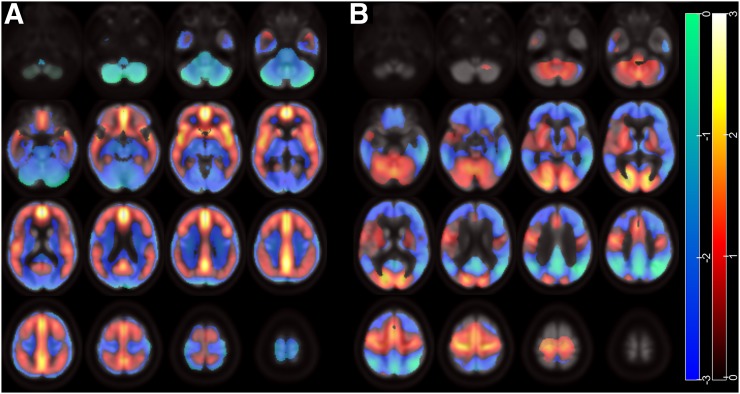
Scaled subprofile model PCA analysis identified 2 significant principal components (1 and 2) that accounted for a total of 37% variability in the data. The logistic regression model including these principal components yielded the highest significance and lowest AIC compared with principal component 1 or 2 alone; therefore, they were linearly combined to construct the Aβ-ADCRP (Fig. 1A), which allows for a highly significant separation between MCI converters and nonconverters (P = 2 × 10−12). The regions with the highest positive voxel loads (elevated amyloid load) include the posterior cingulate cortex and precuneus, the mesial frontal cortex, the insular region, the ventral striatum, and, to a slightly lesser extent, the lateral frontal, temporal, and parietal cortices.
FIGURE 1.
ADCRPs of amyloid binding and glucose metabolism. (A) Aβ-ADCRP derived by PCA on 18F-florbetapir PET data. (B) ADCRP derived by PCA on 18F-FDG PET data. Voxels with negative region weights (coded as z score) are given in cool colors, and regions with positive region weights are depicted in warm colors. Data on neurologic orientation (i.e., left image side corresponds to patients’ left body side) are presented. Regions with increased amyloid load (A) and decreased metabolism (B) show positive and negative weights, respectively, whereas relatively spared regions are loaded with opposite weights due to data normalization.
For comparison, Figure 1B displays the previously defined 18F-FDG PET–based ADCRP (4), which showed prominent negative voxel loads (regional hypometabolism) in MCI converters compared with nonconverters in the temporoparietal cortex and in the posterior cingulate cortex and precuneus.
Derivation Dataset
The Cox proportional-hazards regression constructed with different measures of amyloid load based on 18F-florbetapir PET was penalized for multicollinearity among predictors and identified PES of Aβ-ADCRP to have a higher hazard ratio (3.1, P < 0.002) than continuous SUVR (hazard ratio, 1.7; P = 0.09) or binary amyloid status (hazard ratio, 1.4; P = 0.33). Therefore, PES of Aβ-ADCRP was used as a measure of amyloid load in all subsequent analyses.
Model characteristics and comparisons are summarized in Supplemental Table 1 (supplemental materials are available at http://jnm.snmjournals.org). Because we observed significant, although weak, correlations between PES of ADCRP and PES of Aβ-ADCRP (r = 0.33, P < 0.001), PES of ADCRP and MMSE, FAQ, and APOE (r = −0.22, P = 0.003; r = 0.24, P = 0.002; and r = 0.23, P = 0.003, respectively), and PES of Aβ-ADCRP and MMSE, FAQ, and APOE (r = −0.22, P = 0.005; r = 0.28, P < 0.001; and r = 0.47, P < 0.001, respectively), the models were computed using the ridge regression option to account for multicollinearity. Both the 18F-FDG PET and the amyloid PET models significantly predicted conversion to AD (both P < 0.001). The combined amyloid PET + 18F-FDG PET + nonimaging model showed the highest concordance (Harrell concordance, 0.87; P < 0.001) among constructed models.
Validation Dataset
The constructed Cox models were applied to the validation dataset (Table 2). Comparisons were done in 3 sequential steps, in which the most accurate model of the previous step served as the basis for more comprehensive models in subsequent steps (age and sex served as baseline variables). Change in AIC (ΔAIC) was calculated with reference to the model including both imaging and the nonimaging variables because this model yielded the lowest overall AIC (270.0).
TABLE 2.
Comparison of Cox Predictive Models: Validation Dataset
| Step | Model 1 | ΔAIC model 1 | Model 2 | ΔAIC model 2 | P* |
| 1 | Amyloid PET | 25.9 | Nonimaging | 19.7 | 0.001 |
| 18F-FDG PET | 23.9 | Nonimaging | 19.7 | 0.004 | |
| Amyloid PET | 25.9 | 18F-FDG PET | 23.9 | 5 × 10−4 | |
| 2 | Nonimaging | 19.7 | Amyloid PET + nonimaging | 8.9 | 1 × 10−4 |
| Nonimaging | 19.7 | 18F-FDG PET + nonimaging | 8.5 | 8 × 10−5 | |
| Amyloid PET + nonimaging | 8.9 | 18F-FDG PET + nonimaging | 8.5 | 0.008 | |
| 18F-FDG PET | 23.9 | 18F-FDG PET + amyloid PET | 10.7 | 2 × 10−5 | |
| Amyloid PET | 25.9 | 18F-FDG PET + amyloid PET | 10.7 | 9 × 10−6 | |
| 3 | 18F-FDG PET + amyloid PET | 10.7 | Amyloid PET + 18F-FDG PET + nonimaging | 0 | 1 × 10−4 |
| Amyloid PET + nonimaging | 8.9 | Amyloid PET + 18F-FDG PET + nonimaging | 0 | 2 × 10−4 | |
| 18F-FDG PET + nonimaging | 8.5 | Amyloid PET + 18F-FDG PET+ nonimaging | 0 | 4 × 10−4 |
Improvement in goodness of fit from models 1 to 2 (validation dataset).
ΔAIC is with reference to amyloid PET + 18F-FDG PET + nonimaging model that reached lowest AIC of all models (270.0).
Step 1
The 18F-FDG PET model (ΔAIC, 23.9) was significantly (P < 0.001) better than the amyloid PET model (ΔAIC, 25.9). However, the nonimaging model showed a significantly lower AIC (ΔAIC, 19.7) than either the 18F-FDG PET model (P < 0.005) or the amyloid PET model (P < 0.005).
Step 2
The 18F-FDG PET + nonimaging model (ΔAIC, 8.5) and the amyloid PET + nonimaging model (ΔAIC, 8.9) constituted significant improvements (P < 0.001) over the nonimaging model alone, with the former performing significantly better than the latter (P < 0.01).
Step 3
The complementary predictive value of the aforementioned variables is underlined by the combined amyloid PET + 18F-FDG PET + nonimaging model, which yielded the lowest AIC and was significantly superior to the 18F-FDG PET + nonimaging model (P < 0.001). For reasons of comprehensiveness, Table 2 also lists additional possible comparisons.
Conversion Analysis
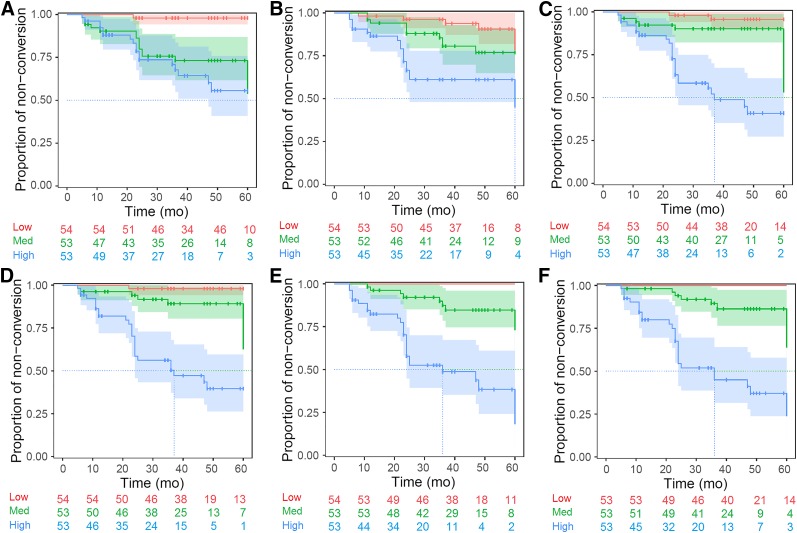
Each of the 6 constructed models was also applied to the validation dataset to calculate the individual PI for each subject and model. The resulting Kaplan–Meier plots are shown in Figure 2. Five-year free-of-conversion rates for the low-, medium- and high-risk groups and their comparisons are summarized in Table 3. The 18F-FDG PET and amyloid PET models showed significant (P < 0.05) strata separation only for the high-risk group and the low-risk group, respectively, whereas the nonimaging and the combined models showed significant separations between all 3 groups (P < 0.05) (Fig. 2). The strata separation was slightly better for the 18F-FDG PET + nonimaging model than for the amyloid PET + nonimaging model (Table 3; Fig. 2). However, the benefit of combining all the variables for risk group stratification was actually small based on Kaplan–Meier curves (Figs. 2E and 2F), although the amyloid PET + 18F-FDG PET + nonimaging model performed significantly better (P < 0.001) than the 18F-FDG PET + nonimaging model (Table 2).
FIGURE 2.
Kaplan–Meier curves of validation dataset. Risk strata using PI are based on amyloid PET model (A), 18F-FDG PET model (B), nonimaging model (C), amyloid PET + nonimaging model (D), 18F-FDG PET + nonimaging model (E), and amyloid PET + 18F-FDG PET + nonimaging model (F).
TABLE 3.
Separation of Risk Strata by Different Models in Validation Dataset
| 5-y free-of-conversion rate |
Pairwise log-rank P |
|||||
| Parameter | Low | Medium | High | Medium vs. high | Low vs. medium | Low vs. high |
| Amyloid PET | 98% | 54% | 55% | 0.331 | 1 × 10−4 | 1 × 10−5 |
| 18F-FDG PET | 79% | 76% | 45% | 0.022 | 0.109 | 6 × 10−4 |
| Nonimaging | 95% | 54% | 40% | 1 × 10−4 | 0.032 | 7 × 10−8 |
| 18F-FDG PET + amyloid PET | 97% | 70% | 33% | 1 × 10−4 | 0.008 | 2 × 10−8 |
| Amyloid PET + nonimaging | 97% | 63% | 39% | 1 × 10−5 | 0.015 | 8 × 10−9 |
| 18F-FDG PET + nonimaging | 100% | 74% | 19% | 8 × 10−6 | 0.003 | 1 × 10−10 |
| Amyloid PET + 18F-FDG PET + nonimaging | 100% | 64% | 24% | 4 × 10−6 | 0.002 | 5 × 10−11 |
DISCUSSION
In a large cohort of subjects, 18F-FDG PET and 18F-florbetapir PET in combination with voxel-based PCA and nonimaging variables predicted conversion from MCI to AD. Interestingly, 18F-FDG PET outperformed amyloid PET in prediction accuracy, and the nonimaging model (including APOE, FAQ, and MMSE) was superior to both imaging models. Still, the nonimaging model was improved by adding amyloid data and (even more so) 18F-FDG PET data, and the model including amyloid PET, 18F-FDG PET, and nonimaging variables yielded the highest prediction accuracy, underscoring their complementary value. The only single-component model that allowed for significant separation between all 3 risk groups was the nonimaging model, whereas the best separation between risk strata was achieved by combining predictor variables.
To improve comparability between 18F-FDG and amyloid PET, a sophisticated method based on voxelwise PCA was applied to 18F-florbetapir PET data. The obtained network topography is consistent with previously published typical regions of amyloid deposition in AD (11,14), although it revealed some regions with unexpectedly high weighting (e.g., insular region). Interestingly, we found that the PES of Aβ-ADCRP and the PES of the 18F-FDG PET–based ADCRP showed comparably high correlations with MMSE and FAQ in the present sample of MCI patients. Finally, we demonstrated that prediction of conversion based on Aβ-ADCRP was superior to conventional amyloid PET analyses (i.e., continuous SUVR in AD-typical regions and binary amyloid status). Taken together, this finding strongly supports future exploration and possible clinical use of the Aβ-ADCRP.
Our finding that amyloid PET predicts development of AD is in line with several studies (5,15,16), with Schreiber et al. (15) and Ben Bouallègue et al. (16) contemplating an overlapping ADNI cohort. Previous studies compared predictive values of amyloid PET and 18F-FDG PET in smaller patient samples: Brück et al. (17) reported similar predictive accuracies for 18F-FDG and amyloid PET, whereas Frings et al. (5) and Trzepacz et al. (7) reported amyloid PET to be a better predictor (18F-FDG PET being not even a significant predictor in the study of Frings et al. (5)). In the present study, 18F-FDG PET was slightly superior to amyloid PET in predicting conversion, as is in line with a study by Prestia et al. (18), who described 18F-FDG PET as the best predictor of progression from MCI to AD among various biomarkers, including Aβ42 in cerebrospinal fluid. Variable results may be explained by different methodologies or patient populations.
The combined set of nonimaging variables more accurately predicted the conversion from MCI to AD than either 18F-FDG or amyloid PET. This effect was driven by the particularly high predictive value of the FAQ (4), probably because the clinical decision on dementia is highly influenced by impairment of activities of daily living, which FAQ assesses. Moreover, the predictive accuracy of nonimaging variables was improved by adding 18F-FDG and amyloid PET, alone or in combination, underlining their complementary value.
Part of the validation dataset of the current study (n = 81) is a subset of the derivation dataset (n = 272) of our previous 18F-FDG PET study (4), in which the ADCRP was established. Nonetheless, exclusion of these subjects in the current validation dataset did not relevantly change the results. In turn, and in contrast to the validation dataset, the amyloid PET model performed slightly better than the 18F-FDG PET model on the derivation dataset (Supplemental Table 1), which, however, might well be explained by the fact that the Aβ-ADCRP was defined on this dataset.
With this large cohort of subjects with long follow-up times (median follow-up based on inverse Kaplan–Meier method, 48 mo [95% confidence interval, 35–52 mo]), we demonstrated the benefit of combining available imaging and nonimaging information into a single quantifiable PI of conversion for each subject. The combination of imaging and nonimaging variables gave the best predictive accuracy, which is similar to the study by Ben Bouallègue et al. (16). The separation between risk groups increases significantly when imaging variables (amyloid PET and, even more, 18F-FDG PET) are combined with nonimaging variables into a single model or when both imaging variables are combined together.
Biomarkers of neurodegeneration derived from modalities other than 18F-FDG PET have also been shown to predict time to conversion from MCI to AD, especially MRI-based biomarkers (19) and cerebrospinal fluid total tau concentration (1). In this study, no comparison was performed against alternative neurodegenerative markers, and further studies are needed to compare the predictive powers of these biomarkers.
Although both 18F-FDG PET and amyloid PET were available for each ADNI patient analyzed in the present study, such is often not the case in clinical routine. We have shown that the combination of 18F-FDG PET and nonimaging variables is superior to the combination of amyloid PET and nonimaging variables. Furthermore, the risk stratification was fairly comparable between the 18F-FDG PET + nonimaging model, the 18F-FDG PET + amyloid PET model, and the model combining all 3 sets of variables, although this last model performed significantly better. Aside from lower costs and wider availability, an additional strength of 18F-FDG PET over amyloid PET may be the detection of neurodegenerative causes of MCI that are not associated with brain amyloidosis (20,21). Thus, further prospective studies on larger patient samples are warranted to define the predictive value and cost effectiveness of the present imaging and nonimaging variables (alone and in combination) in clinical routine.
CONCLUSION
18F-FDG PET, amyloid PET, and nonimaging variables represent complementary predictors of conversion from MCI to AD. The PES of the ADCRP (18F-FDG PET) yielded higher predictive accuracy than the PES of Aβ-ADCRP (18F-florbetapir PET). The combination of imaging and nonimaging variables enables accurate stratification of patients according to their conversion risk, which is of great interest for clinical practice and clinical trials.
DISCLOSURE
Data collection and sharing for this project were funded by the ADNI (National Institutes of Health grant U01 AG024904) and DOD ADNI (Department of Defense award W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC; Johnson & Johnson Pharmaceutical Research & Development LLC; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research provides funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (https://fnih.org/). The grantee organization is the Northern California Institute for Research and Education. No other potential conflict of interest relevant to this article was reported.
KEY POINTS
QUESTION: What are the predictive values of amyloid PET with 18F-florbetapir, 18F-FDG PET, and nonimaging predictors (APOE, FAQ, and MMSE) for development of AD in patients with MCI?
PERTINENT FINDINGS: In a large sample of patients with MCI (n = 319 from ADNI, split into a derivation and a validation dataset) and ADCRPs identified by PCA, we demonstrated that 18F-FDG PET, amyloid PET, and nonimaging variables represent complementary predictors of conversion from MCI to AD. 18F-FDG PET yielded higher predictive accuracy than amyloid PET (each alone and in combination with nonimaging variables). Using a PI and Kaplan–Meier analyses, we found that risk group separation was slightly better for the 18F-FDG PET + nonimaging model than for the amyloid PET + nonimaging model. The additional benefit of combining all the variables for risk group stratification was actually small, although the combination of all variables performed significantly better than the 18F-FDG PET + nonimaging model.
IMPLICATIONS FOR PATIENT CARE: PCA analyses of 18F-FDG and amyloid PET data and nonimaging variables represent complementary predictors for stratifying MCI subjects according to their conversion risk, which is of great interest for clinical practice and clinical trials (e.g., patient counseling, initiation of pharmacologic and nonpharmacologic treatments, and inclusion in trials).
Supplementary Material
Acknowledgments
Data used in preparation of this article were obtained from the ADNI database (http://adni.loni.usc.edu/). As such, the investigators within the ADNI contributed to the design and implementation of ADNI or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at https://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf. The study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
REFERENCES
- 1.Jack CR, Jr, Bennett DA, Blennow K, et al. NIA-AA research framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rice L, Bisdas S. The diagnostic value of FDG and amyloid PET in Alzheimer’s disease: a systematic review. Eur J Radiol. 2017;94:16–24. [DOI] [PubMed] [Google Scholar]
- 3.Cohen AD, Klunk WE. Early detection of Alzheimer’s disease using PiB and FDG PET. Neurobiol Dis. 2014;72:117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blazhenets G, Ma Y, Sorensen A, et al. Principal components analysis of brain metabolism predicts development of Alzheimer dementia. J Nucl Med. 2019;60:837–843. [DOI] [PubMed] [Google Scholar]
- 5.Frings L, Hellwig S, Bormann T, Spehl TS, Buchert R, Meyer PT. Amyloid load but not regional glucose metabolism predicts conversion to Alzheimer’s dementia in a memory clinic population. Eur J Nucl Med Mol Imaging. 2018;45:1442–1448. [DOI] [PubMed] [Google Scholar]
- 6.Grimmer T, Wutz C, Alexopoulos P, et al. Visual versus fully automated analyses of 18F-FDG and amyloid PET for prediction of dementia due to Alzheimer disease in mild cognitive impairment. J Nucl Med. 2016;57:204–207. [DOI] [PubMed] [Google Scholar]
- 7.Trzepacz PT, Yu P, Sun J, et al. Comparison of neuroimaging modalities for the prediction of conversion from mild cognitive impairment to Alzheimer’s dementia. Neurobiol Aging. 2014;35:143–151. [DOI] [PubMed] [Google Scholar]
- 8.Evans AC, Marrett S, Neelin P, et al. Anatomical mapping of functional activation in stereotactic coordinate space. Neuroimage. 1992;1:43–53. [DOI] [PubMed] [Google Scholar]
- 9.Spetsieris P, Ma Y, Peng S, et al. Identification of disease-related spatial covariance patterns using neuroimaging data. J Vis Exp. 2013;(76):50319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spetsieris PG, Eidelberg D. Scaled subprofile modeling of resting state imaging data in Parkinson’s disease: methodological issues. Neuroimage. 2011;54:2899–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frings L, Hellwig S, Spehl TS, et al. Asymmetries of amyloid-beta burden and neuronal dysfunction are positively correlated in Alzheimer’s disease. Brain. 2015;138:3089–3099. [DOI] [PubMed] [Google Scholar]
- 12.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. New York, NY: Springer; 2000:39–77. [Google Scholar]
- 13.Royston P, Altman DG. External validation of a Cox prognostic model: principles and methods. BMC Med Res Methodol. 2013;13:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jack CR, Jr, Barrio JR, Kepe V. Cerebral amyloid PET imaging in Alzheimer’s disease. Acta Neuropathol (Berl). 2013;126:643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber S, Landau SM, Fero A, Schreiber F, Jagust WJ; Alzheimer’s Disease Neuroimaging Initiative. Comparison of visual and quantitative florbetapir F 18 positron emission tomography analysis in predicting mild cognitive impairment outcomes. JAMA Neurol. 2015;72:1183–1190. [DOI] [PubMed] [Google Scholar]
- 16.Ben Bouallègue F, Mariano-Goulart D, Payoux P; Alzheimer’s Disease Neuroimaging Initiative. Joint assessment of quantitative 18F-florbetapir and 18F-FDG regional uptake using baseline data from the ADNI. J Alzheimers Dis. 2018;62:399–408. [DOI] [PubMed] [Google Scholar]
- 17.Brück A, Virta JR, Koivunen J, et al. [11C]PIB, [18F]FDG and MR imaging in patients with mild cognitive impairment. Eur J Nucl Med Mol Imaging. 2013;40:1567–1572. [DOI] [PubMed] [Google Scholar]
- 18.Prestia A, Caroli A, Wade SK, et al. Prediction of AD dementia by biomarkers following the NIA-AA and IWG diagnostic criteria in MCI patients from three European memory clinics. Alzheimers Dement. 2015;11:1191–1201. [DOI] [PubMed] [Google Scholar]
- 19.Jack CR, Jr, Wiste HJ, Vemuri P, et al. Brain beta-amyloid measures and magnetic resonance imaging atrophy both predict time-to-progression from mild cognitive impairment to Alzheimer’s disease. Brain. 2010;133:3336–3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohnen NI, Djang DS, Herholz K, Anzai Y, Minoshima S. Effectiveness and safety of 18F-FDG PET in the evaluation of dementia: a review of the recent literature. J Nucl Med. 2012;53:59–71. [DOI] [PubMed] [Google Scholar]
- 21.Cure S, Abrams K, Belger M, Dell’agnello G, Happich M. Systematic literature review and meta-analysis of diagnostic test accuracy in Alzheimer’s disease and other dementia using autopsy as standard of truth. J Alzheimers Dis. 2014;42:169–182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.