Highlights
-
•
We find that there are highly homology in HR region of SARS-CoV-2 and SARS-CoV.
-
•
We firstly used molecular dynamics method in designing antivirus peptide.
-
•
Our antivirus peptide can prevent virus membrane fuse with host cell.
-
•
Furthermore, we first calculate the binding energy of HR1 and HR2 in SARS-CoV-2 spike protein.
Keywords: Blocking peptide, Molecular dynamics, SARS-CoV-2, Spike protein
Abstract
An outbreak caused by 2019 novel coronavirus (2019-nCoV) was first identified in Wuhan City, Hubei Province, China. The new virus was later named SARS-CoV-2. The virus has affected tens of thousands of patients in the world. The infection of SARS-CoV-2 causes severe pneumonia and even death. It is urgently needed to find a therapeutic method to treat patients with SARS-CoV-2 infection. Studies showed that the surface spike (S) protein is essential for the coronavirus binding and entry of host cells. The heptad repeats 1 and 2 (HR1 and HR2) in the S protein play a decisive role in the fusion of the viral membrane with the host cell membrane. We predicted the HR1 and HR2 regions in S protein by sequence alignment. We simulated a computational model of HR1/2 regions and the fusion core. The binding energy of HR1 and HR2 of the fusion core was –33.4 kcal/mol. We then designed antivirus peptides by molecular dynamics simulation of the fusion core. The binding energy of HR2-based antiviral peptide to HR1 was −43.0 kcal/mol, which was stronger than the natural stage of the fusion core, suggesting that the predicted antiviral peptide can competitively bind with HR1 to prevent forming of the fusion core. The antiviral peptides can prevent SARS-CoV-2 membrane fusion and can potentially be used for the prevention and treatment of infections.
1. Introduction
Coronavirus is a positive-sense RNA enveloped virus with various hosts, including avian species and mammals, especially bats [1]. It is emerging as a global health threat, with three previously unknown deadly coronaviruses outbreaks in the past two decades. In November 2002, the first case of Severe Acute Respiratory Syndrome (SARS) occurred in Foshan, China. As of August 2003, 8069 cases had been reported globally, and 919 of them died. The Middle East Respiratory Syndrome Coronavirus (MERS-CoV) in 2012 is the second highly pathogenic coronavirus found in the 21st century. MERS-CoV has a high fatality rate, with up to 40% of the patients died [2]. The 2019 Novel Coronavirus (SARS-CoV-2) is first discovered in December 2019 and is affecting Millions of patients in all over the world.
Patients infected with SARS-CoV-2 develop varying degrees of symptoms, ranging from a fever or mild cough, to pneumonia, to more severe or even death [3]. Currently, the virus has a fatality rate of about 2%–4%. Though the fatality rate is lower than SARS and MERS, the SARS-CoV-2 is highly contagious. On February 28, the World Health Organization (WHO) raised the global risk assessment to very high. Therefore, the development of therapeutic strategies targets SARS-CoV-2 is urgently needed.
The detailed mechanism underlying SARS-CoV-2 entering the host cell is unclear. The spike protein (S protein) of SARS-CoV-2 consists of S1 and S2 subunits on the viral membrane, which is similar to SARS-CoV. The S1 subunit contains the receptor-binding domain (RBD) for binding the cellular receptor. Furin, a kind of cell surface protein which can cleaves S protein into S1 and S2 and enhance the ability of viral fusion with host cell membrane [4]. The S2 subunit, comprised of fusion peptide (FP), heptad repeat 1 (HR1), heptad repeat 2 (HR2), transmembrane domain (TM), and cytoplasmic domain fusion (CP), is responsible for mediating viral fusion and entry. The S2 subunit of SARS-CoV-2 is highly conserved and is 99% homologous to the S2 subunit of the two bat SARS-Like Coronaviruses (SL-CoV ZXC21 and ZC45) and the human SARS-Like Coronavirus. Recent research highlights that angiotensin-converting enzyme Ⅱ (ACE2), a known receptor for SARS, is the recognition receptor for SARS-CoV-2 [[5], [6], [7]]. Thus, the infection pathway of the SARS-CoV-2 should be similar to other known coronaviruses, such as SARS and MERS [8], warranting the use of peptidic inhibitors against S2 as an effective prevention and treatment strategy for fighting the SARS-CoV-2 [9].
During coronavirus infection, the FP of S2 subunit is inserted into the target cell membrane, exposing the HR1 and HR2 domain and allowing the HR1 and HR2 domain to form a 6-helix bundle that brings the cellular and viral lipid bilayers into proximity, initiating membrane fusion process. During the membrane fusion process, the S protein has three conformational states: a prefusion native state, a pre-hairpin intermediate state, and a stable post-fusion hairpin state. Before binding with the receptor, the S protein exit in prefusion native state. When S protein bind with the receptor, the S1 subunit will be degraded and expose the S2 unit. The S protein is in the pre-hairpin intermediate state. Then three pairs of HR1 and HR2 form a six-helix bundle structure, and the fusion core takes shape [10]. Currently, the S protein is in the stable post-fusion hairpin state. When the blocking peptide bind to HR1, which can prevent S2 conformationally change to the post-fusion hairpin state will inhibit membrane fusion and consequently virus entry into cells.
Based on the HR2 sequences, we designed a peptidic inhibitor that specifically binds to the HR1 domain of SARS-CoV-2 and prevents the S2 subunit from forming the pre-hairpin conformation, thus potentially blocking SARS-CoV-2 infection.
2. Methods
2.1. Protein structure modeling
Homologous modeling was used for protein structure modeling. The sequence data of S protein was downloaded from NCBI (GenBank: QHQ82464.1). The SWISS-MODEL(https://swissmodel.expasy.org/) was used to search the template structure of the S protein [[11], [12], [13]]. Structure 6 acd.1.C was selected as the template structure of SARS-CoV-2 S protein (GMQE is 0.77, QSQE is 0.83, Identity is 66.01). After modeling, the structure was downloaded in PDB format.
2.2. HR region identification
The S protein of SARS-CoV-2 with the HR1 and HR2 region of SARS-CoV and MERS-CoV were aligned by ClustalW (Gap Opening Penalty is 10, Gap Extension Penalty is 0.2).
2.3. The fusion core modeling
When the sequences of HR1 (NQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQ) and HR2 (GINASVVNIQKEIDRLNEVAKNLNESLIDL) were obtained, we used a linker with 22 amino acids (LVPRGSGGSGGSGGLEVLFQGP) to connect the HR1 and HR2 to form the fusion core. The linker is flexible enough to enable HR2 bind with HR1 spontaneously and it had been proved that can work successfully in the SARS-CoV and MERS-CoV fusion core [14,15]. The structure of the fusion core was obtained by modeling from the protein sequence. SWISS-MODEL was used to search the template structure of the fusion core; the structure 1wnc.1.B from SWISS-MODEL modeling was selected as the template of the fusion core. When the modeling process completed, the result was downloaded in PDB format. All the structure figures were generated by PyMOL (https://pymol.org/2/).
2.4. Connecting the HR2 domain to the HR1 domain by molecular docking technology
We first separated the HR2 domain form the HR1/HR2 complex and saved it as a PDB file. Then GRAMM-X (http://vakser.compbio.ku.edu/resources/gramm/grammx/) was used to dock the HR2 and HR2-based peptide to fusion core [16,17]. We used the Umbrella Sampling method in GROMACS software to calculate the binding energy [18]. To simulate the system, we select the GROMACS96 53A6 force field and put the protein into a rectangular box with the simple point charge (SPC) water [19,20]. NaCl was added into the testing system with the concentration 0.1 M, including neutralizing counterions. We set the pulling force was along the z-axis. All pulling simulation was conducted with the GROMACS software 2020 version [21]. We select 170 windows, and each window performs 0.1 ns for a total simulation time of 17 ns utilized for umbrella sampling. The weight histogram analysis method (WHAM) was used to analyze the result [22].
3. Results
3.1. SARS-CoV-2 fusion core modeling
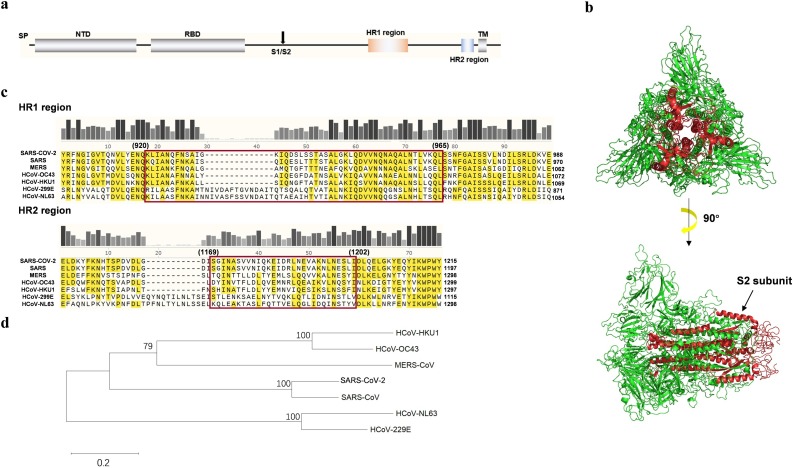
The full-length amino-acid sequence of SARS-CoV-2 S protein was accessed from the NCBI database (GenBank: QHQ82464.1). By multiple sequence alignment with SARS-CoV and MERS-CoV, the positions of HR1 (Y904 to E988) and HR2 (E1151 to Y1215) regions were identified on the S protein (Fig. 1 a). SWISS-MODEL was used to model the S protein of SARS-CoV-2. The 3-dimension model of S protein with the highest Global Model Quality Estimate (GMQE) number was shown in Fig. 1b. The cleavage position which separate S1 and S2 subunit is at R685 and the S2 subunit forms helices inside the intact S protein [4]. When the furin cut the S protein from the cleavage site (R685/S686), the S2 subunit is exposed and forms the fusion core.
Fig. 1.
Sequence alignment of coronavirus HRs and SARS-CoV-2 fusion core formation. (a) A sketch map of the SARS-CoV-2 Spike protein (S protein). The function structure and potential domains are labeled on the figure. The site indicated by an arrow is the cleavage site between S1 and S2. (b) The S protein structure simulated by SWISS-MODEL. S2 subunit in the center of the complex is indicated by red color. (c) Sequence alignment of HR regions among SARS-CoV-2, SARS-CoV, MERS-CoV, HCoV-OC43, HCoV-HKU1, HCoV-299E and HCoV-NL63. The HR domains are indicated by the red box. The HR2 sequences are identical between SARS-CoV-2 and SARS-CoV. (d) Phylogenetic trees constructed by maximum likelihood method from the S proteins of the seven coronaviruses that can infect humans. SP, signal peptide; NTD, N-terminal domain; RBD, receptor binding domain; TM, transmembrane domain; HR, heptad repeats; S1/S2, enzyme cutting site. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The full sequences of S protein from seven coronaviruses that can infect human were multiple aligned by ClustalW (Fig.1c). Based on the phylogenetic tree analysis (Fig. 1d), HCoV-299E and HCoV-NL63 have less homology relationship with other viruses. The full length of S protein of SARS-CoV-2 shared 73% and 26% sequence identity with SARS-CoV and MERS-CoV, respectively. There is only 42.7% sequence similarity between the SARS S1 subunit and the SARS-CoV-2 S1 subunit. The low similarity of the full-length S protein and S1 subunit between SARS and SARS-CoV-2 indicates the high evolution of beta-coronavirus, an RNA virus.
Although the S1 subunit in coronavirus is not conserved, the HR regions in the S2 subunit of SARS-CoV-2 were 93% similar to that of SARS-CoV (Fig. 1c). The highly conserved HR regions suggest that the cell entry mechanism of SARS-CoV-2 is similar to that of SARS. In the HR region of the seven coronaviruses, short amino-acid sequences is highly conserved, such as AIS and RLD in HR1 and KWPW in HR2. These highly conserved amino-acid short sequences may be the functional module of the HR region that mediating formation of fusion core. After multiple sequence alignment, the homologous HR1 (from Y904 to E965) and HR2 (from E1151 to Y1215) sequences of SARS-CoV-2 were obtained. Using LearnColi-VMF and referring to the fusion core construction strategy of SARS-CoV and MERS-CoV, HR1 and HR2 were screened in more detail to obtain sequences of HR1 (from Q920 to L965) and HR2 (from I1169 to I1202) (Fig. 1c) [14,15,23,24]. Then the HR region of SARS-CoV-2 is connected by linker with 22 amino, which is flexible and long enough to allow the HR1 and HR2 region to interact spontaneously to form a peptide of 102 amino acids in length [24].
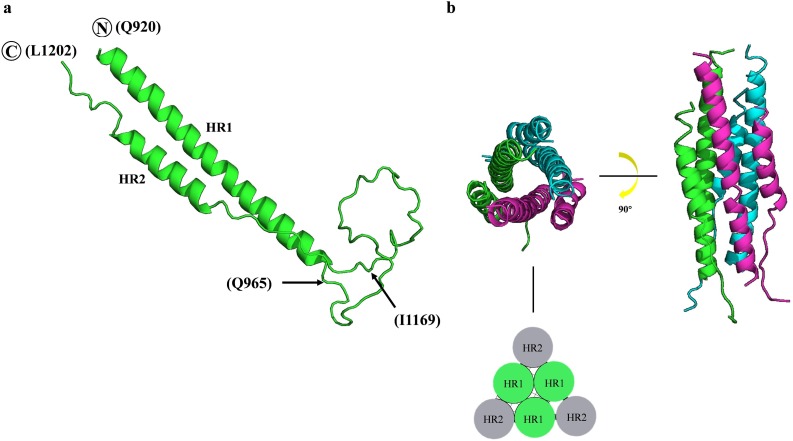
The fusion core was modeled by SWISS-MODEL in the homology modeling method and was visualized by PyMOL (Fig. 2 ). In the fusion core monomer (Fig. 2a), the N-terminal HR1 and C-terminal HR2 formed two helices structures, while the linker sequence maintained a linear structure. The SARS-CoV-2 HR1 form a terpolymer in the center and every HR2 contact with two HR1 (Fig. 2b).
Fig. 2.
The structure of the SARS-CoV-2 fusion core modeled by SWISS-MODEL. (a) The structure of the HR1/HR2 complex. The two helices in the molecule corresponded to the HR1 and HR2 domains. The HR1 region is from Q920 to Q965, and the HR2 region is from I1169 to L1202. The fragment between Q965 and I1169 is the linker. (b) The structure of the HR1/HR2 terpolymer. Three HR1/HR2 complexes are labeled by green, cyan and magenta. The left side is the top view of the terpolymer, and the side view of terpolymer is shown on the right side. The sketch map of fusion core is shown at the bottom. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Molecular interaction in the fusion core
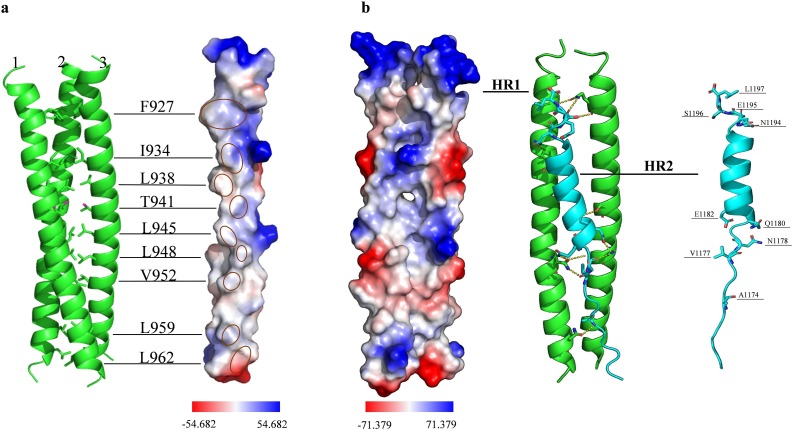
The HR1 and HR2 form helices inside the full-length S protein. HR residues with hydrophobic properties were concentrated in the center of the trimer (Fig. 3 a). A side groove between every two HR1 regions was exposed after furin digestion at the S1/S2 site (R685/S686). The sequence of HR1 peptide (HR1-P) is 919-NQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQ-965. The sequence of HR2 peptide (HR2-P) is 1171-GINASVVNIQKEIDRLNEVAKNLNESLIDL-1200. The HR2 helix can fold and bind into the side groove of two HR1 helices. HR2 connected with two HR1 fragments in the fusion core by hydrogen bonding and hydrophobic action (Fig. 3b). Nine residues in the ends of HR2 helix (A1174, V1177, N1178, Q1180, E1182, N1194, E1195, S1196 and L1197) interacted with the side groove of two HR1s and formed hydrogen bonds.
Fig. 3.
Detail amino acid interaction mediating SARS-CoV-2 fusion core formation. (a) Formation of the central coiled-coil. The three HR1 helices (green) are labeled 1, 2 and 3. The residues of HR1, which contributes to the coiled-coil formation, are shown as F927, I934, L938, T941, L945, L948, V952, L959, and L962. The vacuum electrostatics surface of single HR1 helix is shown on the right. The hydrophobic residues are labeled by red circles on the electrostatic surface. The redder the surface color, the greater the negative potential. The bluer the surface color, the greater the positive potential. (b) Binding of the HR2 to the side groove of two HR1 helices. The electrostatic surface of the side groove of two HR1 helices is shown on the left. HR2 (cyan) interaction with the side groove of two HR1 helices (green) is shown in the middle. HR2 binds to the side groove of two HR1 helices by eleven hydrogen bonds (dash lines). The residues of HR2 that form hydrogen bonds are shown in the right side. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To enhance the binding ability of HR2, we extended three amino acids at the beginning and end of the HR2-P sequence as the HR2-based antiviral peptide (HR2-anti-P: 1168-DISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL-1203) for further study.
3.3. Calculation of the binding energy by the Umbrella Sampling method
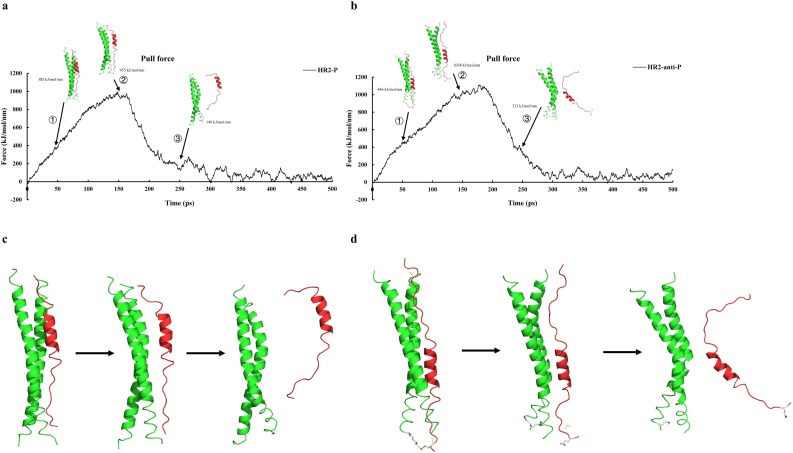
By molecular dynamics simulation the fusion core, an increasing force was applied to HR2-P or HR2-anti-P to pull away from the side groove of two HR1s. The pulling force indicates the binding stability of the fusion core. We simulated the process in 1 nanosecond (ns). Both HR2-P and HR2-anti-P disassociated from HR1s at about 250 ps (Fig. 4 a and b). When the simulation is around 150 ps, the pulling force will reach a peak and the complex began to separate. We set the umbrella sampling window as 0.2 nm and conducted umbrella sampling for frames 0–340. The time we simulated for each windows in Molecular Dynamics is 1 ns.
Fig. 4.
Dissociation curve of HR/HR2 complex with corresponding force vs. time. The first arrow means the point of maximum force, and the third arrow points to the dissociation of peptide HR2 or HR2-based peptide. The second arrow is the middle phase of the dissociation. Both HR2 (a) and HR2-anti-P (b) are dissociate at 250 ps. When HR2-anti-P is pulled out in the middle phase of the dissociation, it forms extra hydrogen bonds with HR1 (dash line). In contrast, when HR2 is pulled out in the middle phase of the dissociation, it does not form extra hydrogen bonds with HR1 (c).
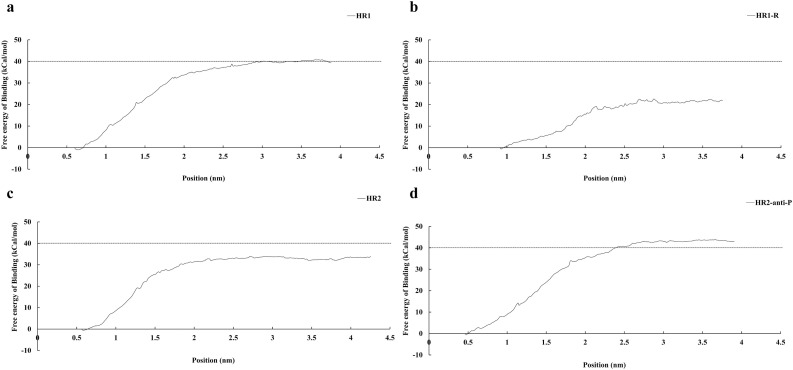
The binding energy of HR2-P to HR1s was −33.4 kcal/mol at 4.23 nm, while the binding energy of HR2-anti-P to HR1s was −43.0 kcal/mol at 3.89 nm (Fig. 5 c, d). When HR2-anti-P was pulled out of the HR1s, HR2-anti-P will formed extra hydrogen bond with HR1 in the middle phase at E1202 on HR2-anti-P and K921. The number of extra hydrogen bond exits from 7 ps to 99 ps. The extra hydrogen bonds can increase the binding energy because it can prevent HR2-anti-P from dissociating from the HR1 region. In contract, HR2-P did not form extra hydrogen bonds with HR1. In addition, the extra amino acid can form hydrogen bonds each other which can be helpful to stabilize the structure of HR2. The higher binding energy of HR2-anti-P suggests that HR2-anti-P can compete with HR2-P and block the binding of HR2-P to the side groove of two HR1s.
Fig. 5.
The binding energies of different peptides with HR1. The binding energy of HR2 to HR1ΔGHR2 is −33.4 kcal/mol. The binding energy of HR2-anti-P to HR1ΔGHR2-anti-P is −43.0 kcal/mol, which is stronger than ΔGHR2. The binding energy of HR1 to HR1 ΔGHR1 is −40.3 cal/mol. And the revised HR1 binding energy ΔGHR1-R is −21.8kCal/mol. ΔGHR2-anti-P > ΔGHR1 > ΔGHR2 > ΔGHR1-R suggests that the HR2-anti-P can compete with HR2 to bind with HR1 during fusion core formation.
HR1:919-NQKLIANQFNSAIGKIQDSLSSTASALGKLQDVVNQNAQALNTLVKQ-965;
HR2: 1171-GINASVVNIQKEIDRLNEVAKNLNESLIDL-1200;
HR2-anti-P: 1168-DISGINASVVNIQKEIDRLNEVAKNLNESLIDLQEL-1203;
To test whether the HR1 can be use as blocking peptide, HR1-P was docking to the side groove of two HR1s (Fig. 5a). The binding energy of HR1 to the side groove of two HR1s is −40.3 kcal/mol, which is larger than that of HR2-P and HR2-anti-P. To explain why HR1-P has high binding energy, we made a guess that when HR1-P docked with two HR1s, they will form a HR1 trimer and the hydrophobic groups are wrapped in the center of the trimer just like in the S protein. Therefore, the binding energy is relatively high. If HR1-P is used as an antivirus peptide, HR1-P will bind to the side groove instead of forming trimer. Therefore, we docked the HR1-P to the HR1 trimer and calculated the binding energy again (Fig. 5b). By performing umbrella sampling again, the binding exergy of HR1-P is about −21.8 kcal/mol which proved that our conjecture is correct.
4. Discussion
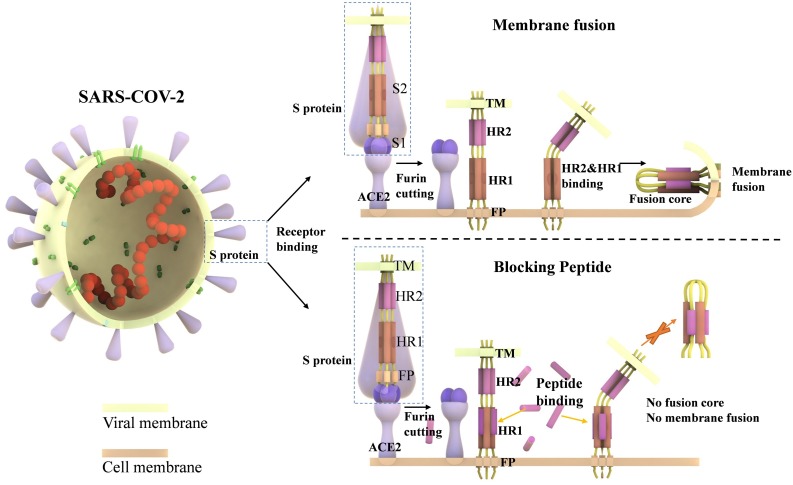
The existence of the HR region is a typical feature of class I enveloped viruses. Such a fusion core is also found in murine hepatitis virus (MHV) and SARS-CoV. The blocking peptides were used in the treatment of viral infections in vivo and in vitro, such as HIV, HCoV-229E, SARS-CoV and MERS-CoV [8,15,25]. In 1993, Wild C, Greenwell T, et al. synthesized a peptide based on the envelope glycoprotein of HIV. The anti-virus peptide has a significant effect of blocking the fusion of HIV-1 with host cells [25,26]. The fusion process is essential for virus entry into the host cells. When the S protein binds to the host receptor, the furin cut the S protein at the S1/S2 cleavage site. The HR1 and HR2 regions, which are located in the S2 subunit, are exposed and form the fusion core. The formation of fusion core induces the virus membrane fusion with the host cell membrane. The antivirus peptide can bind to HR1 more strongly and prevent HR2 from binding to HR1 to form the fusion core (Fig. 6 ).
Fig. 6.
Mechanism of fusion core formation and blocking effect of anti-viral peptide. The S proteins are embedded in the viral membrane and are composed of the S1 and S2 subunits. The S1 subunit contains one receptor-binding domain (RBD). The S2 subunit mediates the virus/cell membrane fusion and the entry of the virus. RBD of the S1 subunit binds to the host ACE2 receptor when the SARS-CoV-2 contacts with the cell membrane. Furin cleaves the S protein into the S1 subunit and the S2 subunit. The fusion peptide (FP) of S2 is exposed and is inserted into the target cell membrane. Three HR1s and three HR2s combine to form the fusion core, pulling the viral membrane to fuse with the host cell membrane. The designed and computational-optimized anti-virus peptides can bind to HR1 more tightly, preventing the HR1s and HR2s from forming fusion core.
Targeting the viral fusion core with blocking peptides provides a new method for drug development. In 2004, Bosch et al. experimentally compared the ability of HR1 and HR2 peptides as inhibitors to block SARS-CoV infection in Vero cells [27]. They found that HR2-like peptidic inhibitor, but not HR1-like peptide inhibitor, dose-dependently inhibited viral infection. Our data also show a similar effect in SARS-CoV-2. TheΔGHR2 is −33.4 kcal/mol, stronger than ΔGHR1 −21.8 kcal/mol. Biochemical and electron microscopic analysis revealed that one HR1 binds three HR2 in an antiparallel pattern, thus may explain why HR1 derived peptide failed to inhibit viral infection. Thus, HR2-like peptide competitively inhibits the binding of the HR2 domain to the HR1 domain, thus blocking the formation of viral fusion core [14], while HR1-like peptide is less efficient in preventing HR1 and HR2 binding. These studies on SARS-CoV and our data on SARS-CoV-2 highlight that peptidic inhibitor based on the HR2 domain is a promising strategy to treat coronavirus infections.
Based on the genome sequences, it is expected that the SARS-CoV-2 shares high genetic similarity and a similar viral fusion core mechanism with SARS-CoV. Based on the HR2 region of SARS-CoV-2 and the use of biomolecular simulation, we have designed an HR2-based antivirus peptide with higher binding energy to HR1, thus can prevent the SARS-CoV-2 membrane fusion. When HR2-based peptide is pulled out and dissociates from HR1, more hydrogen bonds will form to prevent HR2 dissociation (Fig. 5d). Our findings provide a possible treatment for SARS-CoV-2 infection and also prepare for future clinical application research for SARS-CoV-2 therapy. We will further simulate the antivirus peptide with modification and test the blocking effect in cells and animal infections.
Funding
This work was supported by Shenzhen Science and Technology Innovation Commission grants (JCYJ20180305124812444 and KQJSCX20170728150303243).
CRediT authorship contribution statement
Rongsong Ling: Data curation, Software, Writing - original draft. Yarong Dai: Visualization, Writing - original draft. Boxuan Huang: Investigation, Writing - original draft. Wenjie Huang: Investigation, Writing - original draft. Jianfeng Yu: Methodology, Writing - review & editing. Xifeng Lu: Methodology, Writing - review & editing. Yizhou Jiang: Conceptualization, Funding acquisition, Writing - review & editing.
Contributor Information
Xifeng Lu, Email: x.lu@szu.edu.cn.
Yizhou Jiang, Email: jiangyz@szu.edu.cn.
References
- 1.Woo P.C.Y., Lau S.K.P., Huang Y., Yuen K.-Y. Coronavirus diversity, phylogeny and interspecies jumping. Exp. Biol. Med. (Maywood) 2009;234(10):1117–1127. doi: 10.3181/0903-MR-94. http://www.ncbi.nlm.nih.gov/pubmed/19546349 [Internet]. Oct [cited 2020 Feb 16], Available from: [DOI] [PubMed] [Google Scholar]
- 2.De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet [Internet] 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. https://isaric.tghn.org/protocols/ [cited 2020 Mar 1], Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. [Internet] 2020;176(February):104742. doi: 10.1016/j.antiviral.2020.104742. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W., Moore M.J., Vasllieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(November 6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020;29 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y., Guo Y., Pan Y., Zhao Z.J. Structure analysis of the receptor binding of 2019-nCoV. Biochem. Biophys. Res. Commun. [Internet] 2020 doi: 10.1016/j.bbrc.2020.02.071. http://www.ncbi.nlm.nih.gov/pubmed/32081428 Feb 17 [cited 2020 Mar 1]; Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia S., Xu W., Wang Q., Wang C., Hua C., Li W. Peptide-based membrane fusion inhibitors targeting HCOV-229E spike protein HR1 and HR2 domains. Int. J. Mol. Sci. 2018;19(February 2) doi: 10.3390/ijms19020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.JF-W Chan, Kok K.-H., Zhu Z., Hin C., KK-W To, Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from patients with acute respiratory disease in Wuhan, Hubei, China. Emerg. Microbes Infect. 2020;9 doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. [Internet] 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. Sep 29 [cited 2020 Feb 15]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guex N., Peitsch M.C., Schwede T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis. 2009;30(June SUPPL. 1) doi: 10.1002/elps.200900140. [DOI] [PubMed] [Google Scholar]
- 12.Bienert S., Waterhouse A., De Beer TAP Tauriello G., Studer G., Bordoli L. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017;45(January D1):D313–9. doi: 10.1093/nar/gkw1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(July W1):W296–303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu J., Xiao G., Xu Y., Yuan F., Zheng C., Liu Y. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem. Biophys. Res. Commun. 2004;319(June 1):283–288. doi: 10.1016/j.bbrc.2004.04.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao J., Lu G., Qi J., Li Y., Wu Y., Deng Y. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of middle east respiratory syndrome coronavirus. J. Virol. 2013;87(24):13134–13140. doi: 10.1128/JVI.02433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tovchigrechko A., Vakser I.A. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34(Web Server) doi: 10.1093/nar/gkl206. W310–4. [Internet]. 2006 Jul 1 [cited 2020 Feb 6], Available from: https://academic.oup.com/nar/article-lookup/doi/10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tovchigrechko A., Vakser I.A. Development and testing of an automated approach to protein docking. Proteins Struct. Funct. Bioinforma. [Internet] 2005;60(June 2):296–301. doi: 10.1002/prot.20573. 24 [cited 2020 Feb 6], Available from: [DOI] [PubMed] [Google Scholar]
- 18.Lemkul J.A., Bevan D.R. Assessing the stability of Alzheimer’s amyloid protofibrils using molecular dynamics. J. Phys. Chem. B [Internet] 2010;114(February 4):1652–1660. doi: 10.1021/jp9110794. 4 [cited 2020 Feb 6], Available from: [DOI] [PubMed] [Google Scholar]
- 19.Oostenbrink C., Villa A., Mark A.E., Van Gunsteren W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J. Comput. Chem. [Internet] 2004;25(13):1656–1676. doi: 10.1002/jcc.20090. Oct 1 [cited 2020 Feb 16], Available from: [DOI] [PubMed] [Google Scholar]
- 20.Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., Hermans J. Springer; Dordrecht: 1981. Interaction Models for Water in Relation to Protein Hydration; pp. 331–342. [Google Scholar]
- 21.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. [Internet] 2008;4(March 3):435–447. doi: 10.1021/ct700301q. 2 [cited 2020 Feb 16], Available from: [DOI] [PubMed] [Google Scholar]
- 22.Kumar S., Rosenberg J.M., Bouzida D., Swendsen R.H., Kollman P.A. THE weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. [Internet] 1992;13(October 8):1011–1021. doi: 10.1002/jcc.540130812. 1 [cited 2020 Feb 16], Available from: [DOI] [Google Scholar]
- 23.Singh M., Berger B., Kim P.S. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J. Mol. Biol. 1999;290(July 5):1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y., Lou Z., Liu Y., Pang H., Tien P., Gao G.F. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 2004;279(November 47):49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang S., Lin K., Strick N., Neurath A.R. HIV-1 inhibition by a peptide [3] Nature. 1993;365(6442):113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 26.Wild C., Greenwell T., Matthews T. A synthetic peptide from HIV-1 gp41 is a potent inhibitor of virus-mediated cell–cell fusion. AIDS Res. Hum. Retroviruses. 1993;9:1051–1053. doi: 10.1089/aid.1993.9.1051. [DOI] [PubMed] [Google Scholar]
- 27.Bosch B.J., Martina BEE Van Der Zee R., Lepault J., Haijema B.J., Versluis C. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U.S.A. 2004;101(June 22):8455–8460. doi: 10.1073/pnas.0400576101. [DOI] [PMC free article] [PubMed] [Google Scholar]