Highlights
-
•
Deep learning method enables rapid identification of COVID-19 on chest CT exams.
-
•
Deep learning method mitigates the radiologists’ workload in COVID-19 screening.
-
•
It is essential to validate the deep learning method in large-scale dataset.
Abbreviations: COVID-19, Coronavirus disease 2019; RT-PCR, Reverse-transcriptase–polymerase-chain-reaction; CT, Computed tomography; GGO, Ground glass opacities; AI, Artificial intelligence; AUC, Area under the receiver-operating characteristics curve; RHWU, Renmin Hospital of Wuhan University; 1st HCMU, The First Hospital of China Medical University; BYH, Beijing Youan Hospital; ROC, Receiver-operating characteristics
Keywords: Coronavirus disease 2019, Deep learning, Multi-view model, Computed tomography
Abstract
Purpose
To develop a deep learning-based method to assist radiologists to fast and accurately identify patients with COVID-19 by CT images.
Methods
We retrospectively collected chest CT images of 495 patients from three hospitals in China. 495 datasets were randomly divided into 395 cases (80%, 294 of COVID-19, 101 of other pneumonia) of the training set, 50 cases (10%, 37 of COVID-19, 13 of other pneumonia) of the validation set and 50 cases (10%, 37 of COVID-19, 13 of other pneumonia) of the testing set. We trained a multi-view fusion model using deep learning network to screen patients with COVID-19 using CT images with the maximum lung regions in axial, coronal and sagittal views. The performance of the proposed model was evaluated by both the validation and testing sets.
Results
The multi-view deep learning fusion model achieved the area under the receiver-operating characteristics curve (AUC) of 0.732, accuracy of 0.700, sensitivity of 0.730 and specificity of 0.615 in validation set. In the testing set, we can achieve AUC, accuracy, sensitivity and specificity of 0.819, 0.760, 0.811 and 0.615 respectively.
Conclusions
Based on deep learning method, the proposed diagnosis model trained on multi-view images of chest CT images showed great potential to improve the efficacy of diagnosis and mitigate the heavy workload of radiologists for the initial screening of COVID-19 pneumonia.
1. Introduction
In December 2019, a pneumonia associated with coronavirus disease 2019 (COVID-19) pneumonia emerged in Wuhan, Hubei Province, China. COVID-19 can be transmitted through airborne, surface of objects, and especially respiratory droplets and close contacting with the infected ones [1]. Until March 15th, 2020, COVID-19 has caused 81,048 infection cases and 3204 deaths in China. It has caused 75,333 infection cases and 2624 deaths outside China [2].
Currently, the real-time reverse-transcriptase–polymerase-chain-reaction (RT-PCR) test is widely used as the gold standard for confirming COVID-19 pneumonia. However, it has the nature of time consuming and high false negative [3]. Thus, it is urgent to find an effective and accurate method to screen COVID-19. Computed tomography (CT) imaging is a critical tool in the initial screening of COVID-19 pneumonia. Shi et al. studied the CT image data of 81 COVID-19 pneumonia patients and found that in the Chest CT scan, COVID-19 pneumonia often presented bilateral and subpleural ground glass opacities (GGO) [4]. Ai et al. showed that the diagnostic results of CT images were highly consistent with RT-PCR assays for the diagnosis of COVID-19 pneumonia. They also found that CT had a very high sensitivity for the diagnosis of COVID-19 pneumonia [5]. Although CT images have shown great potential for diagnosing COVID-19 pneumonia, currently, it is needed to manually identify radiographic features of COVID-19 pneumonia from all the thinner layer CT images (average 300 layers per patient) by trained radiologists. This will significantly increase the workload of radiologists, and could delay the diagnosis and following treatment.
The artificial intelligence (AI) methods, especially the deep learning methods, have showed its advanced performance in medical applications. In some detailed classification, segmentation and detection problem, deep learning can achieve human-level accuracy, such as in pulmonary nodules diagnosis [6], breast cancer diagnosis [7], retina image segmentation [8], etc. In clinical applications of lung diseases, Coudray et al. studied the use of pathological images of lung cancer tissues to predict lung cancer subtypes and gene mutations [9]. They trained the deep learning network inceptionV3 to establish prediction models, and achieved good performance in independent validation sets. Wang et al. used CT image data from lung regions to predict EGFR gene mutations using an end-to-end deep learning network, and achieved the area under the receiver-operating characteristics curve (AUC) of 0.81 in the validation set of 241 cases [10]. Hence, the deep learning-based methods can be used to assist radiologists to improve the diagnostic efficiency and accuracy in the diagnosis of COVID-19 pneumonia. In this study, we aimed to train a diagnostic model based on deep learning network on the chest CT images to screen the patients with COVID-19 pneumonia.
2. Materials and methods
2.1. Datasets
This study was approved by the institutional review board of Renmin Hospital of Wuhan University (RHWU), The First Hospital of China Medical University (1st HCMU) and Beijing Youan Hospital (BYH) in China. Written informed consent was waived by institutional review board for retrospective study.
We retrospectively collected high-resolution CT images of 495 patients with pneumonia, including 368 patients with laboratory-confirmed COVID-19 pneumonia and 127 patients with other pneumonia (47 bacterial pneumonia, 67 other viral pneumonia, 11 mycoplasma pneumonia and 2 fungal pneumonia). The dataset was collected from three hospitals in China, including RHWU (265 COVID-19 and 35 other pneumonia), 1 st HCMU (103 COVID-19, 46 other pneumonia) and BYH (46 other pneumonia).
The dataset and demographic characteristics of patients are detailed in Table 1 . The chest examinations were performed with a CT scanner (Optima CT680, GE Medical Systems, USA;) in RHWU. The CT protocol was as follows: tube voltage, 120 kVp; tube current, 249 mA; and mAs, 108.7. The images were acquired at lung (window width, 1500 HU; window level, -750 HU) settings. For patients in the 1st HCMU, the examinations were performed with a CT scanner (Discovery, CT750 HD, GE Medical Systems, USA). The CT parameter was set as: tube voltage, 120 kVp; tube current, 219 mA; and mAs, 95.6. The images were acquired at mediastinal (window width, 250 HU; window level, 40 HU) settings. LightSpeed VCT (GE Medical Systems, USA) scanner was used for BYH with the CT protocol: tube voltage, 120 kVp; tube current, 392 mA; and mAs, 238.9. The images were acquired at lung (window width, 1500 HU; window level, -750 HU) settings. All the images were standardized by mapping the pixel values in a range from 0 to 255 and reconstructed with a slice thickness of 0.625 mm or 5 mm.
Table 1.
Datasets and demographic characteristics of patients with COVID-19 pneumonia and other pneumonia.
| Training set (n = 395) | Validation set (n = 50) | Testing set (n = 50) | ||||
|---|---|---|---|---|---|---|
| Overall | COVID-19 | No COVID-19 | COVID-19 | No COVID-19 | COVID-19 | No COVID-19 |
| Age | ||||||
| < 60 years | 203(16 - 59) | 64 (3 - 59) | 26 (1 - 58) | 8 (17 - 48) | 19 (22 - 56) | 8 (7 - 58) |
| ≥ 60 years | 91 (60 - 87) | 37 (60 - 89) | 11 (61 - 78) | 5 (62 - 84) | 18 (60 - 80) | 5 (60 - 83) |
| Gender | ||||||
| Male | 140 | 70 | 17 | 4 | 18 | 6 |
| Female | 154 | 31 | 20 | 9 | 19 | 7 |
| Renmin Hospital of Wuhan University, Wuhan, China | ||||||
| Age | ||||||
| < 60 years | 151 (23 - 59) | 19 (18 - 59) | 22 (29 - 58) | 4 (17 - 43) | 18 (22 - 56) | 1 (7 - 7) |
| ≥ 60 years | 54 (60 - 87) | 8 (60 - 87) | 6 (64 - 72) | 2 (65 - 84) | 14 (61 - 80) | 1 (63 - 63) |
| Gender | ||||||
| Male | 91 | 19 | 13 | 0 | 15 | 1 |
| Female | 114 | 8 | 15 | 6 | 17 | 1 |
| The First Hospital of China Medical University, Shenyang, China | ||||||
| Age | ||||||
| < 60 years | 52 (16 - 59) | 20 (19 - 59) | 4 (1 - 58) | 1 (48 - 48) | 1 (22 - 22) | 5 (24 - 58) |
| ≥ 60 years | 37 (60 - 85) | 16 (60 - 84) | 5 (61 - 78) | 1 (75 - 75) | 4 (60 - 66) | 3 (62 - 83) |
| Gender | ||||||
| Male | 49 | 24 | 4 | 1 | 3 | 3 |
| Female | 40 | 12 | 5 | 1 | 2 | 5 |
| Beijing Youan Hospital, Beijing, China | ||||||
| Age | ||||||
| < 60 years | 25 (3 - 59) | 3 (17 - 47) | 2 (37 - 56) | |||
| ≥ 60 years | 13 (60 - 89) | 2 (62 - 65) | 1 (60-60) | |||
| Gender | ||||||
| Male | 27 | 3 | 2 | |||
| Female | 11 | 2 | 1 | |||
Categorical data of age were presented as counts (range).
2.2. CT image pre-processing
For each set of chest CT images, the lung region in each axial CT slices was firstly segmented using threshold segmentation and morphological optimization algorithms. The lung regions in coronal and sagittal CT slices were segmented accordingly. Subsequently, for each patient of chest CT images, the slice with the most pixels in the segmented lung area from each of the axial, coronal and sagittal views was selected as the inputs of the deep learning network. Finally, the three-view CT images of 495 patients were randomly divided into 395 cases (80%, 294 of COVID-19, 101 of other pneumonia) of the training set, 50 cases (10%, 37 of COVID-19, 13 of other pneumonia) of the validation set and 50 cases (10%, 37 of COVID-19, 13 of other pneumonia) of the testing set. Each image (512 × 512 pixels) was resized into 256 × 256 pixels for deep learning network training, validation and testing.
2.3. Multi-view fusion deep learning network architecture
We trained out deep learning network using the three-view images of maximum lung region. In previous deep learning-based studies on tumour diagnosis, it is believed that the largest slice of the tumour on CT images contains most of the information about the tumour [11]. Inspired by this, in this study, we selected the layer with the largest lung region as region of interest. As previous study of chest CT images has shown that images from multiple views can contribute to learning more useful information without introducing more redundant information [6]. To predict the classification of lung nodules by CT images, a multi-view deep learning model of lung nodules classification study showed good performance. Thus, we trained our deep learning network using the CT images from axial, coronal, and sagittal views of the lung regions for developing the diagnosis model to screen the COVID-19.
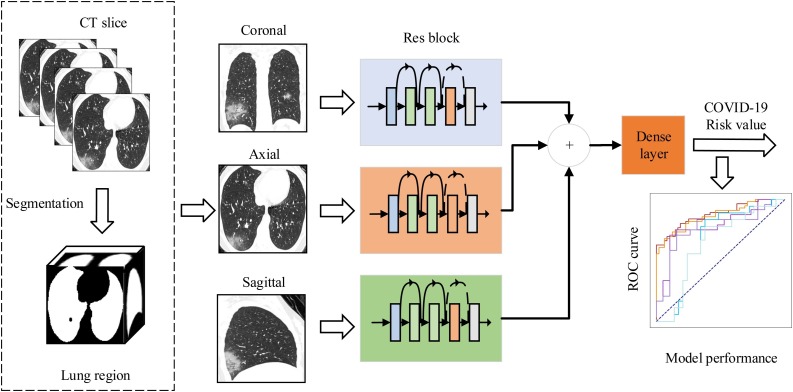
To discriminate the patients with COVID-19 pneumonia and other pneumonia, we developed a multi-view fusion model based on deep learning network as shown in Fig. 1 . Our model was constructed base on the modification of ResNet50 architecture [12]. We took the set of three-view images as the network input instead of the RGB three-channel image in typical ResNet50. We first trained our deep learning network using randomly selected 395 training set of three-view images. During the training phase, the validation set of three-view images were used to optimize the hyperparameters to achieve the best performance. The outputs of Res blocks were concatenated for each patient to facilitate input of Dense layer. The parameters of the deep network were updated through the training stage using an rmsprop optimizer with a learning rate of 1 × 10−5 and batchsize of 4.
Fig. 1.
The main framework of multi-view deep learning fusion model. We firstly extracted the lung region in CT slices using threshold segmentation method. Then, we trained our model based on the architecture of ResNet50. The inputs of the model are the corresponding CT images in axial, coronal, and sagittal views that selected from the maximum lung region selection. The three branch networks output feature maps that aggregated to feed into a fully connected dense layer. Finally, the layer outputs the risk value of COVID-19 pneumonia to evaluate the performance of the deep learning model.
2.4. Model implementation
The multi-view fusion deep learning network was implemented using Python (version, 3.6) language and based on the Keras (version, 2.1.6) framework. Deep learning network training and testing were performed using TITAN XP with server model. The performance indicators of the deep learning network are calculated in the R language (version, 3.5.1).
2.5. Performance evaluation
We evaluated the classification performance using AUC, accuracy, sensitivity, and specificity. AUC is an index to measure the performance of the classifier. The AUC is calculated according to the area under the ROC curve. Accuracy is calculated by (true positive samples + true negative samples) / (all samples). Sensitivity is calculated by (true positive samples) / (true positive samples + false negative samples). Specificity is calculated by (true negative samples) / (false positive samples + true negative samples). To evaluate the performance of multi-view fusion model, we trained a single-view, model using the axial images with the maximum lung region as input for comparison.
2.6. Statistical analysis
Delong test is used to test the differences between the AUC of single-view model and multi-view model. Mann-Whitney U test and Chi-square test are used to calculate the distribution differences in different sets of RHWU and 1st HCMU. P values <0.05 is considered indicative of statistical significance.
3. Results
3.1. Comparison between the single-view model and the multi-view model for COVID-19 classification
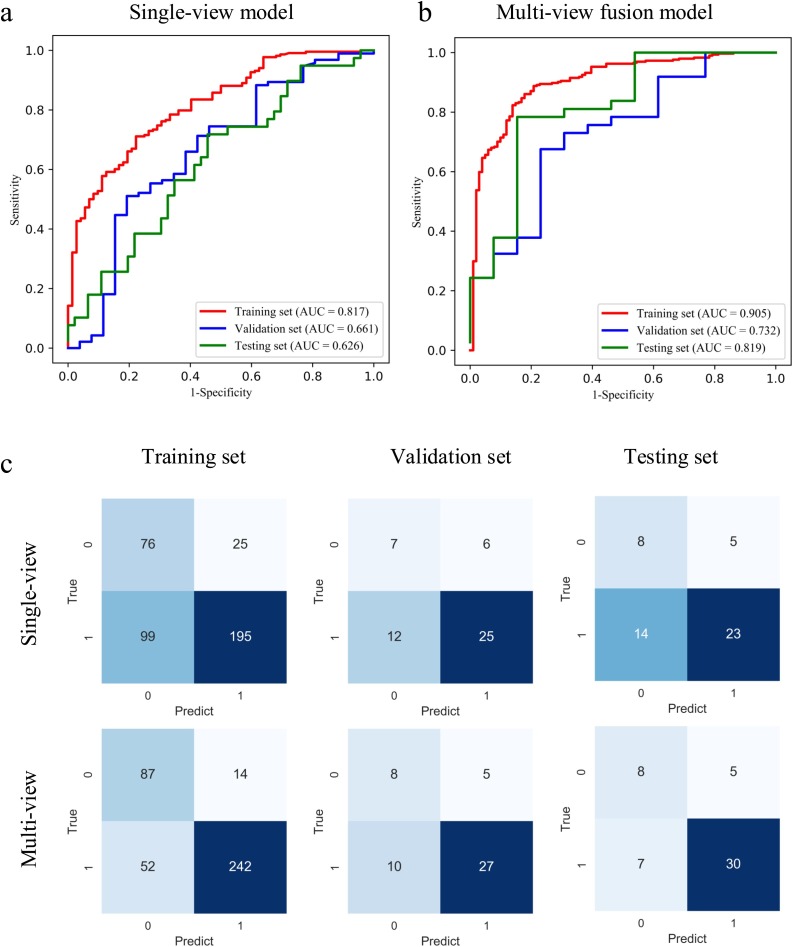
To illustrate the performance of the proposed model, we firstly evaluated the single-view model, in which only the axial-view CT images were used for training. We achieved AUC of 0.767, 0.642, and 0.634 in the training, validation, and testing set respectively. The accuracy was 0.640 and 0.620, sensitivity was 0.676 and 0.622 for the validation and testing sets. For the multi-view model, the AUC, accuracy, and sensitivity in the training set were 0.905 (p < 0.001), 0.833 and 0.823 respectively. The performance was comparable with the AUC of 0.732 (P = 0.423) and 0.819 (P = 0.08), accuracy of 0.700 and 0.760, and sensitivity of 0.730 and 0.811 for the validation and testing set. The receiver-operating characteristics (ROC) curve and the confusion matrix of the single-view and multi-view models in the training, validation, and testing set are shown in Fig. 2 . The performance comparison of the single-view and multi-view models is summarised in Table 2 . The results showed that we can achieve better performance in COVID-19 classification with multi-view model.
Fig. 2.
ROC curves of single-view (a) and multi-view (b) deep learning diagnosis model of COVID-19 pneumonia. The confusion matrix of two diagnosis models (c). 1 represents COVID-19 pneumonia, and 0 represents other pneumonia.
Table 2.
The performance of the single-view model and multi-view fusion model.
| Dataset | Model | AUC* | Accuracy* | Sensitivity* | Specificity* | P value |
|---|---|---|---|---|---|---|
| Training set | Single-view | 0.767 (0.718 - 0.816) |
0.686 (0.685 - 0.687) |
0.663 (0.609 - 0.717) |
0.752 (0.668 - 0.873) |
< 0.001 |
| Multi-view | 0.905 (0.871 - 0.939) |
0.833 (0.832 - 0.834) |
0.823 (0.780 - 0.867) |
0.861 (0.794 - 0.929) |
||
| Validation set | Single-view | 0.642 (0.465 - 0.820) |
0.640 (0.631 - 0.649) |
0.676 (0.525 - 0.827) |
0.538 (0.267 - 0.809) |
0.423 |
| Multi-view | 0.732 (0.569 - 0.895) |
0.700 (0.692 - 0.708) |
0.730 (0.587 - 0.873) |
0.615 (0.351 - 0.880) |
||
| Testing set | Single-view | 0.634 (0.469 - 0.799) |
0.620 (0.611 - 0.629) |
0.622 (0.465 - 0.778) |
0.615 (0.351 - 0.880) |
0.08 |
| Multi-view | 0.819 (0.673 - 0.965) |
0.760 (0.753 - 0.767) |
0.811 (0.685 - 0.937) |
0.615 (0.351 - 0.880) |
Note: Delong test is used to test the differences between the AUC of single-view model and multi-view model.
Quantitative data were presented as value (95% confidence interval).
3.2. Subgroup analysis of the multi-view model
To further verify the performance of the model, we conducted subgroup analysis for the two hospitals (RHWU and 1st HCMU) that both have COVID-19 and other pneumonia data in terms of gender and age.
Given the small datasets of subgroup for the validation and testing sets, we grouped them as one set for each hospital in subgroup analysis. Mann-Whitney U test and Chi-square test are used to calculate the distribution differences of age and gender groups in different sets for the two hospitals. The ROC curves of the gender and age groups are shown in Fig. 3, Fig. 4 , respectively.
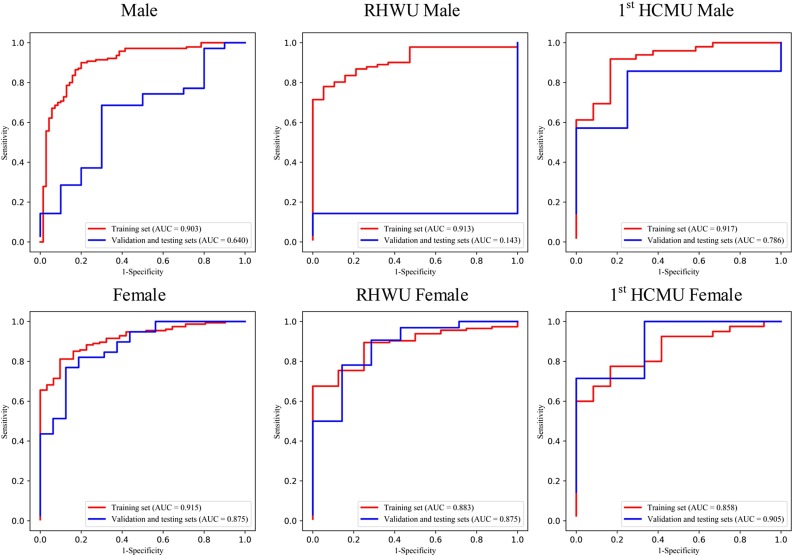
Fig. 3.
ROC curve of the gender group of multi-view deep learning diagnosis model of COVID-19 pneumonia.
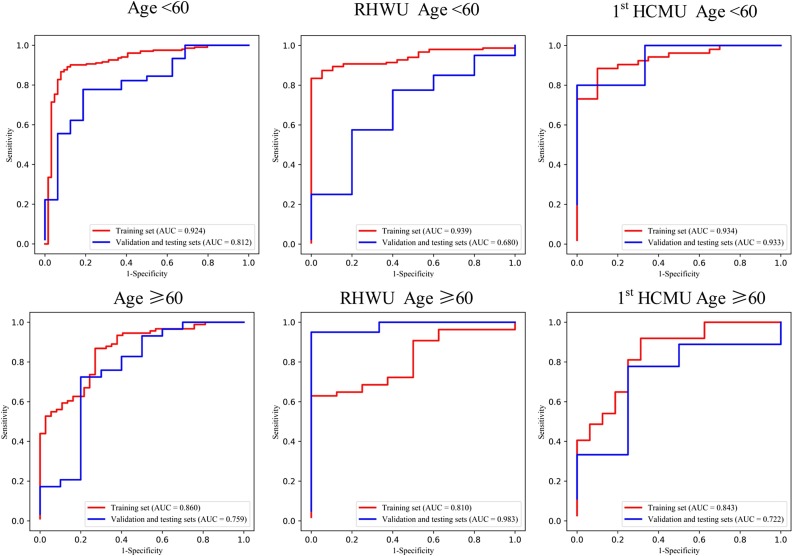
Fig. 4.
ROC curve of the age group of multi-view deep learning diagnosis model of COVID-19 pneumonia.
In age group analysis, the distribution between the two hospitals is significantly different on the training set (P < 0.001), but no significant different distribution is found on the validation and testing sets (P = 0.097). The performance is better in the group of age≥60 (AUC of 0.983) than in the group of age<60 (AUC of 0.680) on the validation and testing sets in for RHWU. Whereas the performance is opposite for 1st HCMU (AUC of 0.722 vs 0.933). In the gender group analysis, we achieved better performance on the validation and testing sets in the female group (AUC of 0.875 and 0.905) than in the male group (AUC of 0.143 and 0.786) for both RHWU and 1st HCMU. The performance of the subgroup analysis is detailed in Table 3 . It is noted that the statistical differences in both age and gender groups on the training set are partly due to the small dataset in different subgroups of the two hospitals.
Table 3.
The performance of multi-view fusion model in age and gender groups.
| subgroup | Overall |
RHWU |
1st HCMU |
P value |
||||
|---|---|---|---|---|---|---|---|---|
| Training set | Validation and testing sets | Training set | Validation and testing sets | Training set | Validation and testing sets | Training set | Validation and testing sets | |
| Age≥60 | 0.860 | 0.759 | 0.810 | 0.983 | 0.843 | 0.722 | < 0.001 | 0.097 |
| Age<60 | 0.924 | 0.812 | 0.939 | 0.680 | 0.934 | 0.933 | ||
| Male | 0.903 | 0.640 | 0.913 | 0.143 | 0.917 | 0.786 | 0.049 | 0.818 |
| Female | 0.915 | 0.875 | 0.883 | 0.875 | 0.858 | 0.905 | ||
Note: Mann-Whitney U test and Chi-square test are used to calculate the distribution differences of age and sex groups in different sets in the two hospitals.
3.3. Visualization of the model on COVID-19 diagnosis
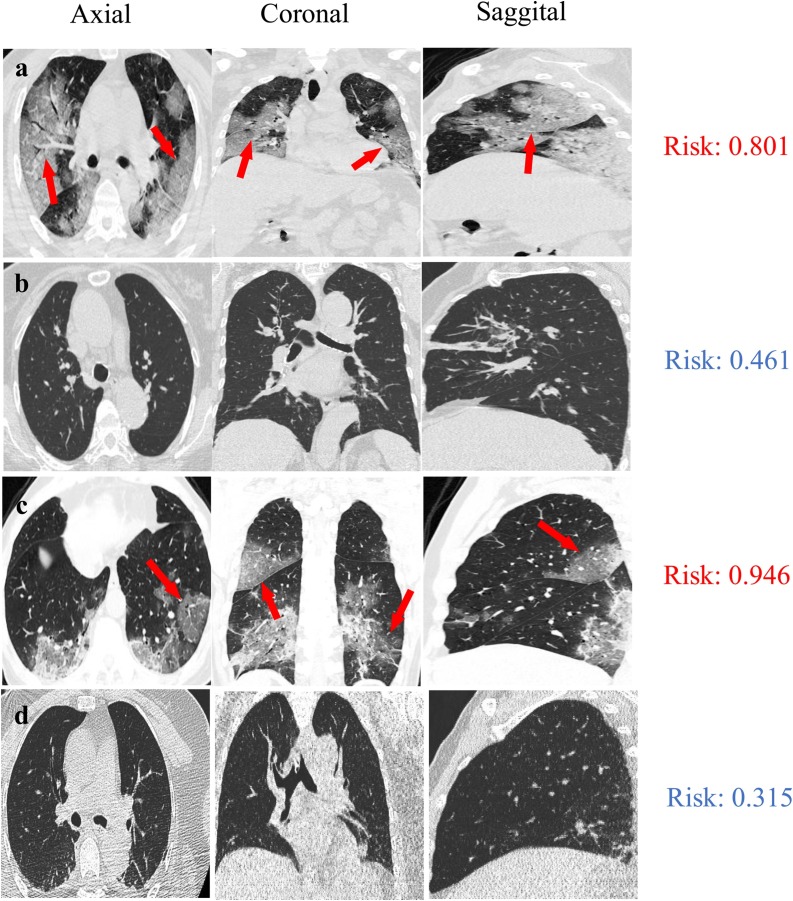
To visualize the diagnosis results, we presented the CT images of four patients in axial, coronal and sagittal views, including a 46-year-old male with COVID-19 pneumonia (Fig. 5 a), an 84-year-old female with bacterial pneumonia in validation set (Fig. 5b), a 62-year-old male with COVID-19 pneumonia (Fig. 5c) and a 52-year-old female with bacterial pneumonia in testing set (Fig. 5d). As previous reports on CT features for COVID-19 pneumonia [4,13,14], the characteristics of GGO were clearly seen in two classified COVID-19 patients (Fig. 5a, c, marked with red arrows). For these four patients, their risk scores with COVID-19 infections are 0.801, 0.461, 0.946, and 0.315 (range from 0 to 1), which were evaluated by the multi-view deep learning fusion model. In this study, we took the cut-off of the model as 0.653, according to Youden index. Normally, it takes more than ten minutes to examine the CT images of on patient and to identify the COVID-19 infected patient by a trained radiologist. In contrast, less than five seconds are taken to achieve the comparable screen result in COVID-19 pneumonia identification using multi-view deep learning model, which will mitigate over 100-fold workload of radiologist in the initial screening of patients with COVID-19 pneumonia.
Fig. 5.
Representative examples of pneumonia diagnosis for a 46-year-old male with COVID-19 pneumonia (a), an 84-year-old female with bacterial pneumonia in validation set (b), a 62-year-old male with COVID-19 pneumonia (c) and a 52-year-old female with bacterial pneumonia in testing set (d). The risk scores of these four patients with COVID-19 infections are 0.801, 0.461, 0.946, and 0.315 (range from 0 to 1), respectively, which assessed by the multi-view deep learning fusion model. The cut-off of the model is 0.653. The ground glass opacity in COVID-19 patients are marked with red arrows (a, c).
4. Discussion
As the COVID-19 has rapidly spread global-wide [2], it is very challenging for the radiologists to fast and accurately diagnose the pneumonias under tremendous workload. Artificial intelligence tools, more specifically, deep learning networks are the promising tools to assist radiologists in the initial screening of COVID-19. Eventually, it could mitigate the workload of radiologists, improve diagnosis efficiency and provide more timely and appropriate treatments for COVID-19 patients. In this study, we developed a multi-view deep learning fusion model to discriminate the patient with COVID-19 through CT images. Compared with the single-view model, the multi-view model used more information of CT images and showed better performance. For the single-view model, it is easy to be overfitting because there is less information to use. Moreover, if the training is conducted on the data of only one hospital, it is easy to fall into the local optimal solution, which is not necessarily effective to the other hospitals, so the performance of the model may fluctuate greatly among different hospitals.
To date, some efforts on COVID-19 diagnostic through AI approaches have been reported [[15], [16], [17], [18]]. Chen et al. [16] developed and validated deep learning diagnostic model of COVID-19 pneumonia using single view CT slices of 106 cases patients from single center. They achieved the accuracy, sensitivity and specificity of 0.989, 0.943 and 0.992, respectively, in the testing set. In another two centers study, Wang et al. [17] used 99 patients CT images data to identify COVID-19 pneumonia by transfer learning method. The AUC, accuracy, sensitivity and specificity of their model in the testing set were 0.780, 0.731, 0.670 and 0.760, respectively. It is important to note that most of patients with COVID-19 are more likely to have CT abnormalities characterized by bilateral and subpleural ground glass opacities (GGO), vascular thickening, spider web, and crazy-paving patterns [[19], [20], [21]], while patients with other pneumonia mainly show patchy or density increasing shadows [22]. The advanced AI-based algorithms can discriminate COVID-19 from other pneumonia by learning not only these typical CT image signs, but also some high-dimensional features, such as texture, wavelet information etc., which are unable processed by radiologists. Consequently, these deep learning-based studies showed significant effect on the initial screening of COVID-19 pneumonia through CT images. However, due to the small datasets and less than two centers were involved in these studies, the CT slices were taken as samples for developing deep learning model, resulting in inter-slice fusion and time cost issues. In addition, the generalization performance on multicentre data remains to be verified. Here, we utilized the information from the automatic segmentation of multi-view of CT images to train our deep learning network, which avoided the heavy workload of manually sketching the features in CT images of patients with pneumonia by the radiologists. Therefore, we could take one patient as a sample, which means that one patient only needs to be tested once. In this study, we established and verified the performance of the multi-view deep learning fusion model on 495 multi-center data. Our results further demonstrated that our model has the great potential to be used for the diagnosis of COVID-19 pneumonia.
In future studies, a large, multicentre, and prospective dataset collection is necessary for training and validating AI models. More advanced AI algorithms also should be developed to achieve more efficient and accurate COVID-19 diagnosis. Due to the capability of disease quantification, AI-based method has great potential to predict the severity of COVID-19. Moreover, the deep learning-based model could be used for treatment decision-making in the patient with COVID-19. As the medical resources are seriously insufficient due to the outbreak of COVID-19 worldwide, fast diagnosis and accurate prediction of individualized prognosis of COVID-19 are crucial for the management of COVID-19. The use of AI combined with CT imaging as well as other clinical information, including epidemiology, nucleic acid detection results, clinical symptoms, and laboratory indicators, can avoid misdiagnosis and effectively improve the clinical effects.
This study has some limitations. First, we only took the slices of the maximum lung region in three views to build the training, validation, and testing set. More CT slices in different views could be added as input to improve the performance of the proposed model. Second, more CT images of COVID-19 patients from other hospitals throughout China, or even worldwide could be involved to validate the model. Finally, more detailed clinical information was unavailable for subgroup analysis, such as body mass index and the severity of COVID-19 infection. Nonetheless, the gender and age subgroup analyses in this study allowed a preliminary evaluation of the model.
In summary, we developed a multi-view fusion model to initial screening of COVID-19 pneumonia. This model achieved a better performance with single-view model and subgroup analysis. It showed great potential to improve the efficiency of diagnosis and mitigate the workload of radiologists. We envision that the multi-view deep learning-based tools can be used to assistant radiologist to fast and accurately identify COVID19 pneumonia.
Ethical standards
All human and animal studies have been approved by the appropriate ethics committee and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Informed consent
Written informed consent was waived by institutional review board for retrospective study.
CRediT authorship contribution statement
Xiangjun Wu: Methodology, Software, Formal analysis, Writing - original draft. Hui Hui: Methodology, Writing - review & editing, Conceptualization. Meng Niu: Investigation, Supervision. Liang Li: Investigation. Li Wang: Investigation. Bingxi He: Software. Xin Yang: Methodology. Li Li: Investigation. Hongjun Li: Investigation. Jie Tian: Conceptualization, Supervision. Yunfei Zha: Conceptualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no conflict of interest.
Acknowledgements
We thank all the patients involved in this study. We acknowledge Dr. Guoqing Xie and Dr. Peng Xiao for their work of data collecting in Wuhan. The authors would like to acknowledge the instrumental and technical support of multimodal biomedical imaging experimental platform, Institute of Automation, Chinese Academy of Sciences. This work was supported in part by the National Key Research and Development Program of China under Grant 2017YFA0700401, 2016YFC0103803, 2017YFA0205200, and 2019YFC0118100; Novel Coronavirus Pneumonia Emergency Key Project of Science and Technology of Hubei Province (No.2020FCA015); the National Natural Science Foundation of China under Grant 81671851, 81827808, 81527805, 81871332, and 81227901; the Strategic Priority Research Program under Grant XDB32030200 and the Scientific Instrument R&D Program under Grant YJKYYQ20170075 of the Chinese Academy of Sciences.
Contributor Information
Meng Niu, Email: niumeng@cmu.edu.cn.
Jie Tian, Email: jie.tian@ia.ac.cn.
Yunfei Zha, Email: zhayunfei999@126.com.
References
- 1.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W., Tsoi H.W., Lo S.K., Chan K.H., Poon V.K., Chan W.M., Ip J.D., Cai J.P., Cheng V.C., Chen H., Hui C.K., Yuen K.Y. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395(10223):514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO Coronavirus Disease (COVID-2019) Situation reports-54. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200314-sitrep-54-covid-19.pdf?sfvrsn=dcd46351_2 (Accessed March 15, 2020.
- 3.Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., Zeng B., Li Z., Li X., Li H. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020;126 doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi H., Han X., Jiang N., Cao Y., Alwalid O., Gu J., Fan Y., Zheng C. Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30086-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology. 2020:200642. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Y., Xia Y., Zhang J., Song Y., Feng D., Fulham M., Cai W. Knowledge-based collaborative deep learning for benign-malignant lung nodule classification on chest CT. IEEE Trans. Med. Imaging. 2019;38(4):991–1004. doi: 10.1109/TMI.2018.2876510. [DOI] [PubMed] [Google Scholar]
- 7.Braman N., Prasanna P., Whitney J., Singh S., Beig N., Etesami M., Bates D.D.B., Gallagher K., Bloch B.N., Vulchi M., Turk P., Bera K., Abraham J., Sikov W.M., Somlo G., Harris L.N., Gilmore H., Plecha D., Varadan V., Madabhushi A. Association of peritumoral radiomics with tumor biology and pathologic response to preoperative targeted therapy for HER2 (ERBB2)-Positive breast Cancer. JAMA Netw Open. 2019;2(4):e192561. doi: 10.1001/jamanetworkopen.2019.2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa P., Galdran A., Meyer M.I., Niemeijer M., Abramoff M., Mendonca A.M., Campilho A. End-to-End adversarial retinal image synthesis. IEEE Trans. Med. Imaging. 2018;37(3):781–791. doi: 10.1109/TMI.2017.2759102. [DOI] [PubMed] [Google Scholar]
- 9.Coudray N., Ocampo P.S., Sakellaropoulos T., Narula N., Snuderl M., Fenyo D., Moreira A.L., Razavian N., Tsirigos A. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat. Med. 2018;24(10):1559–1567. doi: 10.1038/s41591-018-0177-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang S., Shi J., Ye Z., Dong D., Yu D., Zhou M., Liu Y., Gevaert O., Wang K., Zhu Y., Zhou H., Liu Z., Tian J. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 2019;53(3) doi: 10.1183/13993003.00986-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen C., Liu Z., Guan M., Song J., Lian Y., Wang S., Tang Z., Dong D., Kong L., Wang M., Shi D., Tian J. 2D and 3D CT radiomics features prognostic performance comparison in non-small cell lung Cancer. Transl. Oncol. 2017;10(6):886–894. doi: 10.1016/j.tranon.2017.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He K.M., Zhang X.Y., Ren S.Q., Sun J. Deep Residual Learning for Image Recognition, 2016 Ieee Conference on Computer Vision and Pattern Recognition (Cvpr) 2016. pp. 770–778. [DOI] [Google Scholar]
- 13.Xu X., Yu C., Qu J., Zhang L., Jiang S., Huang D., Chen B., Zhang Z., Guan W., Ling Z., Jiang R., Hu T., Ding Y., Lin L., Gan Q., Luo L., Tang X., Liu J. Imaging and clinical features of patients with 2019 novel coronavirus SARS-CoV-2. Eur. J. Nucl. Med. Mol. Imaging. 2020 doi: 10.1007/s00259-020-04735-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., Du B., Li L.J., Zeng G., Yuen K.Y., Chen R.C., Tang C.L., Wang T., Chen P.Y., Xiang J., Li S.Y., Wang J.L., Liang Z.J., Peng Y.X., Wei L., Liu Y., Hu Y.H., Peng P., Wang J.M., Liu J.Y., Chen Z., Li G., Zheng Z.J., Qiu S.Q., Luo J., Ye C.J., Zhu S.Y., Zhong N.S. China medical treatment expert group for, clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fang M., He B., Li L., Dong D., Yang X., Li C., Meng L., Zhong L., Li H., Li H., Tian J. CT radiomics can help screen the coronavirus disease 2019 (COVID-19): a preliminary study. Sci. China Inf. Sci. 2020 doi: 10.1007/s11432-020-2849-3. [DOI] [Google Scholar]
- 16.Chen J., Wu L., Zhang J., Zhang L., Gong D., Zhao Y., Hu S., Wang Y., Hu X., Zheng B.Jm. Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: a prospective study. medRxiv. 2020 doi: 10.1038/s41598-020-76282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S., Kang B., Ma J., Zeng X., Xiao M., Guo J., Cai M., Yang J., Li Y., Meng X.Jm. A deep learning algorithm using CT images to screen for Corona Virus Disease (COVID-19) medRxiv. 2020 doi: 10.1007/s00330-021-07715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L., Qin L., Xu Z., Yin Y., Wang X., Kong B., Bai J., Lu Y., Fang Z., Song Q., Cao K., Liu D., Wang G., Xu Q., Fang X., Zhang S., Xia J., Xia J. Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology. 2020:200905. doi: 10.1148/radiol.2020200905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., Pan I., Shi L.-B., Wang D.-C., Mei J. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020 doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanne J.P. Chest CT findings in 2019 novel coronavirus (2019-nCoV) infections from Wuhan, China: key points for the radiologist. Radiology. 2020;295(1):16–17. doi: 10.1148/radiol.2020200241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai H.X., Hsieh B., Xiong Z., Halsey K., Choi J.W., Tran T.M.L., Pan I., Shi L.B., Wang D.C., Mei J., Jiang X.L., Zeng Q.H., Egglin T.K., Hu P.F., Agarwal S., Xie F., Li S., Healey T., Atalay M.K., Liao W.H. Performance of radiologists in differentiating COVID-19 from viral pneumonia on chest CT. Radiology. 2020:200823. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao D., Yao F., Wang L., Zheng L., Gao Y., Ye J., Guo F., Zhao H., Gao R. A comparative study on the clinical features of COVID-19 pneumonia to other pneumonias. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa247. [DOI] [PMC free article] [PubMed] [Google Scholar]