Abstract
Aphids live in symbiosis with a variety of bacteria, including the obligate symbiont Buchnera aphidicola and diverse facultative symbionts. The symbiotic associations for one aphid species, especially for polyphagous species, often differ across populations. In the present study, by using high-throughput 16S rRNA sequencing, we surveyed in detail the microbiota in natural populations of the cotton aphid Aphis gossypii in China and assessed differences in bacterial diversity with respect to host plant and geography. The microbial community of A. gossypii was dominated by a few heritable symbionts. Arsenophonus was the most dominant secondary symbiont, and Spiroplasma was detected for the first time. Statistical tests and ordination analyses showed that host plants rather than geography seemed to have shaped the associated symbiont composition. Special symbiont communities inhabited the Cucurbitaceae-feeding populations, which supported the ecological specialization of A. gossypii on cucurbits from the viewpoint of symbiotic bacteria. Correlation analysis suggested antagonistic interactions between Buchnera and coexisting secondary symbionts and more complicated interactions between different secondary symbionts. Our findings lend further support to an important role of the host plant in structuring symbiont communities of polyphagous aphids and will improve our understanding of the interactions among phytophagous insects, symbionts, and environments.
Electronic supplementary material
The online version of this article (10.1007/s00248-019-01435-2) contains supplementary material, which is available to authorized users.
Keywords: Polyphagous species, Arsenophonus, Host specialization, Symbiont–symbiont interactions
Introduction
Aphids are well known for their symbiotic associations with bacteria. Almost all aphid species harbor the primary endosymbiont Buchnera aphidicola, which inhabits specialized bacteriocytes and provides aphids with important nutrients for their growth and reproduction [1–4]. Buchnera is strictly maternally inherited [5, 6] and has undergone parallel diversification with its aphid hosts [7–14].
Aphids also host multiple secondary (or facultative) bacterial symbionts that are generally not essential for their survival or reproduction. Some are commonly studied, such as Arsenophonus, Fukatsuia symbiotica, Hamiltonella defensa, Regiella insecticola, Rickettsiella viridis, and Serratia symbiotica from the Gammaproteobacteria; Rickettsia and Wolbachia from the Alphaproteobacteria; and Spiroplasma from the Mollicutes [15–21]. These secondary symbionts reside in bacteriocytes, sheath cells, or hemocoel [22] and are transmitted maternally and horizontally [18, 23, 24]. They have environmentally dependent effects on host aphids, including increasing heat tolerance [25–29], protecting against parasitic wasps [30–34] and fungal pathogens [29, 35–37], influencing aphid fitness on host plants [38–40], modifying body color [41], and affecting aphid reproduction [42, 43]. More details are reviewed in Oliver et al. [44], Zytynska and Weisser [45], and Guo et al. [46]. In addition, several facultative symbionts seem to have established co-obligate associations along with Buchnera in certain aphid species, such as Serratia symbiotica, Erwinia haradaeae, Fukatsuia symbiotica, Hamiltonella defensa, and Sodalis in some Lachninae species [21, 47–51] and Wolbachia in Pentalonia nigronervosa [52, 53].
The associations between microbial symbionts and aphids are quite different in different aphid species. Symbionts carried by one aphid species also often vary across populations. There seems to be a widespread pattern in polyphagous aphids that the populations feeding on different host plants differ in their symbiont communities [54–56]. Most studies have focused on the pea aphid Acyrthosiphon pisum, which consists of at least eleven biotypes adapted to specific host plants [57]. Facultative symbionts in the pea aphid exhibit large variation across plant-adapted populations in terms of composition and prevalence [35, 54, 58–61]. Links between particular symbionts and plants have been observed, such as associations between Hamiltonella defensa and alfalfa and Regiella insecticola and clover. A nonrandom distribution of bacterial symbionts across host plants has also been reported in other polyphagous aphid species such as Aphis craccivora [55, 62] and the oligophagous aphids Phylloxera notabilis [63] and Aphis citricidus [64]. Nevertheless, several studies have highlighted the role of geography in structuring the community of aphids’ bacterial partners. Tsuchida et al. [65] revealed characteristic geographical distribution patterns of secondary symbionts that infected Acyrthosiphon pisum in Japan, particularly for Regiella. Jones et al. [66] found that the symbiont communities of Aphis gossypii and Pentalonia caladii varied across aphid populations from different Hawaiian islands. Some studies also indicated correlations between aphid symbionts and other factors, including developmental stage of aphids [67, 68], rearing condition [69], plant species richness [70], and season [71].
The cotton aphid, Aphis gossypii Glover, is a cosmopolitan insect pest causing serious economic losses in agriculture. It feeds on many important crops, including cotton, cucurbits, citrus, eggplant, peppers, potato, and flowering ornamental plants such as Hibiscus [72]. Several studies have been conducted on bacterial communities. Najar-Rodríguez et al. [73] and Jones et al. [66] investigated the microbial diversity of natural aphid populations from Japan and Australia and from Hawaii, respectively, and highlighted the effect of geography on bacterial profiles. Zhao et al. [74] also found distinct bacterial community structures from different geographic populations feeding on Bt cotton in northern China. A. gossypii is currently controlled primarily by insecticides, which have been reported to influence the bacterial communities associated with aphids [75, 76]. In addition, by utilizing quantitative PCR, Ayoubi et al. [77] uncovered development-associated dynamics in the abundance of symbionts within A. gossypii.
Although geography has been proposed to have a role in structuring the bacterial communities of A. gossypii, samples used in previous studies were restricted to a few plants. A detailed and deep exploration of the microbiota in natural populations of A. gossypii is still lacking. In this study, using Illumina sequencing of 16S rRNA gene, we characterized the microbial communities of A. gossypii collected from diverse plants and different regions in China, assessed differences in bacterial community according to host plant and geography, and discussed the interactions between symbionts.
Material and Methods
Sample Collection and DNA Extraction
A total of 110 samples of Aphis gossypii feeding on plants belonging to 25 families were collected from 23 regions of China (Table S1). Specimens from the same colony were preserved in 75% and 100% ethanol for making voucher slides and DNA extraction, respectively. The slide-mounted specimens were identified based on the external morphology. All voucher specimens and samples were deposited in the National Zoological Museum of China, Institute of Zoology, Chinese Academy of Sciences, Beijing, China.
A single adult was chosen from each sample for DNA extraction. To remove microbial contaminants from the body surface, each aphid individual was washed with 70% ethanol for 5 min and then rinsed with sterile ultrapure water once for 5 min and four times for 1 min. DNA was extracted from the whole body of a single individual using DNeasy Blood & Tissue Kit (QIAGEN, Hilden, Germany) following the manufacturer’s protocol. A blank sample of sterile ultrapure water was also processed through the same extraction protocol to serve as a negative control during the DNA extraction. The standard cytochrome oxidase subunit I (COI) barcodes were amplified by universal primers (LCO1490: 5′-GGTCAACAAATCATAAAGATATTGG-3′; HCO2198: 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) [78] to test the quality of DNA extracts, to verify the aphid species identification, and to detect contamination from parasitoid wasps.
PCR Amplification, Library Preparation, and Sequencing
DNA was amplified using the universal primers of the V3–V4 region of 16S rRNA gene (338F: 5′-ACTCCTACGGGAGGCAGCA-3′; 806R: 5′-GGACTACHVGGGTWTCTAAT-3′). The first polymerase chain reaction (PCR) was carried out in a 50-μL volume containing 1.5 μL (10 μM) of each primer, 0.4 U Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA, USA), 10 μL 5× Q5 Reaction Buffer (New England Biolabs), 10 μL 5× Q5 High GC Enhancer (New England Biolabs), 1 μL dNTPs (New England Biolabs), and 40–60 ng DNA extract. The reaction conditions were as follows: initial denaturation at 95 °C for 5 min, followed by 15 cycles of 95 °C for 1 min, 50 °C for 1 min, 72 °C for 1 min, and final elongation at 72 °C for 7 min. The PCR products were purified using VAHTS™ DNA Clean Beads (Vazyme Biotech, Nanjing, China). In the next step, 10 μL of the purified product was ligated to adapter and sample barcode in a 40-μL volume containing 1 μL (10 μM) of each fusion primer and 20 μL of 2× Phusion High-Fidelity PCR Master Mix (New England Biolabs). The second PCR conditions were as follows: 98 °C for 30 s, 10 cycles of 98 °C for 10 s, 65 °C for 30 s, and 72 °C for 30 s, followed by a final extension at 72 °C for 5 min. Negative amplification controls (sterile ultrapure water) were also included in all PCR reactions. The final PCR products were recovered using 1.8% agarose gel electrophoresis, purified with VAHTS™ DNA Clean Beads (Vazyme Biotech) and then quantified by NanoDrop 2000 (Thermo Scientific, Wilmington, DE, USA). All positive PCR products were mixed at a mass ratio of 1:1. Finally, the library pool was submitted to an Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) for paired-end sequencing. The raw reads have been deposited in the NCBI Sequence Read Archive (SRA) database under BioProject accession number PRJNA543947.
Sequence Processing and Analyses
Paired-end reads were assembled using FLASH v1.2.11 [79]. The merged tags with an average quality score lower than 20 in a 50-bp sliding window were trimmed using Trimmomatic v0.33 [80]. The remaining tags shorter than 300 bp were also removed. High-quality clean tags were then obtained after removing chimeras with UCHIME v8.1 [81]. The denoised sequences were clustered into operational taxonomic units (OTUs) at 97% sequence similarity by the UCLUST module from QIIME [82]. Taxonomy was assigned to all OTUs by searching against the Silva databases [83] using the RDP classifier within QIIME [84]. The OTUs were then filtered with a threshold value of 0.005% of all sequences [85], except for the OTUs that could be classified. Finally, an OTU table containing the number of sequences per sample and taxonomic information was generated.
Statistical Analyses
Alpha diversity indices (i.e., Shannon and Simpson diversity indices) for each sample were calculated using the diversity function in the R package vegan [86]. The relative abundance of each bacterial genus was estimated by normalizing the number of sequences assigned to each genus against the total number of sequences obtained for a given sample using the decostand function of vegan. To better investigate the symbiont and secondary symbiont communities, all OTUs assigned to the reported symbionts of aphids were screened out from the OTU table, and the relative abundance of each symbiont was calculated.
All samples of A. gossypii were grouped according to geographic region and host plant (Table S2). First, we compared the alpha diversity indices of symbiont communities from different groups. Shannon and Simpson diversity indices for each group (sample size ≥ 3) were calculated with vegan. The alpha diversity data were found to deviate from normality (P < 0.05, Shapiro–Wilk test); therefore, we performed the non-parametric Kruskal–Wallis test to check for significant differences across all groups and conducted the non-parametric Wilcoxon tests for pairwise group comparisons.
Next, we investigated the patterns of beta diversity to address the relative importance of geography and host plant on symbiont and secondary symbiont communities. Beta diversity, i.e., the variation of symbiont community composition among differently grouped samples, was quantified using Bray–Curtis dissimilarity which considered the presence/absence and relative abundance of the individual symbiont. The Bray–Curtis distance was calculated between each pair of samples using the vegdist function in vegan. In the analyses of symbiont community, to reduce the influence of the most abundant Buchnera, the relative abundance data were logarithmically transformed with the decostand function of vegan. For the grouping scheme of geographic region, we assessed variation in community composition across all 23 groups, across ten groups with a sample size ≥ 3 (Zhejiang 1, 2, and 3 were treated as one group), and across three groups colonizing Rhamnaceae (sample size ≥ 3) (Beijing, Heilongjiang, and Liaoning 2). For the grouping scheme of host plant, we assessed variation in community composition among all 25 groups, among eleven groups with a sample size ≥ 3, among six groups with a sample size ≥ 5, and among eight groups from Beijing (sample size ≥ 3) (Asteraceae, Buxaceae, Crassulaceae, Cucurbitaceae, Lamiaceae, Malvaceae, Rhamnaceae, and Verbenaceae).
Principal component analysis (PCA) was firstly performed on the relative abundance matrix using the prcomp function in the R package stats to visualize variation among different groups in symbiont and secondary symbiont community compositions. PCA reduces the dimension of multivariate data and interprets such data diagrammatically. The resulting ordination was plotted with the R package ggbiplot [87]. Then, we used unconstrained and constrained ordination methods to visualize the Bray–Curtis dissimilarity. For the unconstrained ordination approach, we performed nonmetric multidimensional scaling (NMDS) on the Bray–Curtis distance matrix using the metaMDS function in vegan and presented two-dimensional plots by the R package ggplot2 [88]. NMDS is found to always produce better ordinations than PCA [89]. For the constrained ordination approach, constrained principal coordinate analysis (cPCoA) was performed on the Bray–Curtis distance matrix using capscale and anova.cca functions in vegan and the resulting ordination was visualized by ggplot2. These ordination techniques are useful in representing community variation in response to environmental factors, such as geography and host plant in this study. In an ordination, samples that are close are more similar to one another than those that are far apart.
Based on the Bray–Curtis distance matrices, differences in symbiont and secondary symbiont community structures were also statistically analyzed with analysis of similarities (ANOSIM) and permutational multivariate analysis of variance (PERMANOVA). These two analyses are both resemblance-based permutation methods widely used in ecology and PERMANOVA is generally more powerful than ANOSIM to detect changes in community composition [90]. ANOSIM and PERMANOVA were applied using the anosim function and adonis function in vegan, respectively, and P values were obtained using 999 permutations. To further identify which symbionts were driving the differences in secondary symbiont community, we carried out the analysis of variance (ANOVA) tests in STAMP v2.1.3 [91] based on the relative abundances of each secondary symbiont from groups with a sample size ≥ 3. Pairwise group comparisons of the average relative abundances were then conducted using the post hoc Scheffé test, where the Bonferroni-adjusted P values were used to control the false discovery rate.
Furthermore, to test the effect of geographic distances among sampling sites in structuring the symbiont and secondary symbiont communities, the Pearson correlation coefficient between geographic distance matrix and Bray–Curtis distance matrix was calculated using Mantel test in vegan. The geographic distance matrix was generated using the Geographic Distance Matrix Generator v1.2.3 [92]. Mantel test allows to look for the correlation between two distance matrices. The null hypothesis that inter-point distances in these two matrices are not correlated was tested through a permutation procedure.
Finally, to explore potential interactions among different symbionts associated with A. gossypii, the Spearman correlation coefficients (ρ) between symbionts were calculated based on their relative abundances using the cor function in stats and were visualized in a heatmap with the R package corrplot [93].
Results
The Bacterial Diversity of Aphis gossypii
Overall Bacterial Diversity
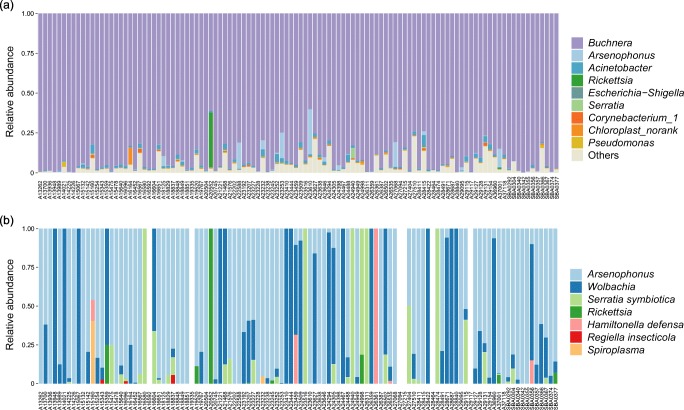
After quality control, we obtained 3,867,639 16S rRNA gene sequences (35,160 reads per sample). A total of 1524 OTUs were identified at 97% similarity and were assigned into 39 phyla (Proteobacteria, 96.73% of total sequences), 104 classes (Gammaproteobacteria, 94.96%), 180 orders (Enterobacteriales, 93.48%), 310 families (Enterobacteriaceae, 93.48%), and 630 genera (Table S3). The bacterial community of A. gossypii was dominated by the primary endosymbiont Buchnera aphidicola (average relative abundance across all samples 91.79%), followed by the secondary symbiont Arsenophonus (1.11%) and the bacteria Acinetobacter (0.99%) (Fig. 1a, Table S3).
Fig. 1.
Barplots of bacterial communities (a) and secondary symbiont communities (b) associated with Aphis gossypii across all samples
Symbiont Diversity
The alpha diversity of the symbiont community was very low (mean Shannon index = 0.063, mean Simpson index = 0.969) (Table S4). A total of eight aphid symbionts were detected in the cotton aphid. All samples harbored the primary endosymbiont Buchnera aphidicola. Along with Arsenophonus, they were also infected with Rickettsia (average relative abundance across all samples 0.32%), Serratia symbiotica (0.07%), Wolbachia (0.04%), Hamiltonella defensa (< 0.005%), Regiella insecticola (< 0.005%), and Spiroplasma (< 0.005%) (Fig. 1b, Table 1). Within the secondary symbiont community, the most prevalent bacteria were Arsenophonus (infection frequency 82/110), followed by Wolbachia (68/110), and Serratia symbiotica (32/110). Hamiltonella defensa, Regiella insecticola, and Spiroplasma were found to be low in both infection rate and abundance (Table 1). Many samples (69/110) were infected by at least two secondary symbionts in various combinations (Table 2). Co-infection with Arsenophonus and Wolbachia was the most common type (30/110), followed by multiple infections with Arsenophonus, Serratia symbiotica, and Wolbachia (13/110).
Table 1.
Infection prevalence and average relative abundance of symbionts across all samples of Aphis gossypii
| Symbiont | Infection frequency | Relative abundance (%) |
|---|---|---|
| Buchnera aphidicola | 110/110 | 91.79 |
| Arsenophonus | 82/110 | 1.11 |
| Wolbachia | 68/110 | 0.04 |
| Serratia symbiotica | 32/110 | 0.07 |
| Rickettsia | 9/110 | 0.32 |
| Hamiltonella defensa | 6/110 | < 0.005 |
| Regiella insecticola | 4/110 | < 0.005 |
| Spiroplasma | 2/110 | < 0.005 |
Table 2.
Infection pattern of secondary symbionts within Aphis gossypii
| Infection pattern | Secondary symbiont | Infection frequency |
|---|---|---|
| No infection | 6/110 | |
| Single infection | Arsenophonus | 19/110 |
| Wolbachia | 11/110 | |
| Serratia symbiotica | 4/110 | |
| Hamiltonella defensa | 1/110 | |
| Co-infection | Arsenophonus–Wolbachia | 30/110 |
| Arsenophonus–Serratia | 6/110 | |
| Serratia–Wolbachia | 3/110 | |
| Arsenophonus–Rickettsia | 2/110 | |
| Arsenophonus–Spiroplasma | 1/110 | |
| Serratia–Rickettsia | 1/110 | |
| Wolbachia–Rickettsia | 1/110 | |
| Multiple infections | Arsenophonus–Serratia–Wolbachia | 13/110 |
| Arsenophonus–Hamiltonella–Wolbachia | 3/110 | |
| Arsenophonus–Rickettsia–Wolbachia | 2/110 | |
| Arsenophonus–Hamiltonella–Spiroplasma | 1/110 | |
| Arsenophonus–Regiella–Serratia | 1/110 | |
| Arsenophonus–Regiella–Wolbachia | 1/110 | |
| Serratia–Rickettsia–Wolbachia | 1/110 | |
| Arsenophonus–Regiella–Serratia–Wolbachia | 1/110 | |
| Arsenophonus–Hamiltonella–Rickettsia–Serratia–Wolbachia | 1/110 | |
| Arsenophonus–Regiella–Rickettsia–Serratia–Wolbachia | 1/110 |
Symbiont Communities from Different Geographic Regions and Host Plants
Comparison of Alpha Diversity
No significant difference was detected among the alpha diversity indices of aphid symbionts from different geographic regions (P = 0.710 for Shannon index, P = 0.770 for Simpson index, Kruskal–Wallis test; P = 0.216–0.978 for Shannon index, P = 0.295–1.000 for Simpson index, Wilcoxon test). However, the Kruskal–Wallis test revealed statistical differences in the populations occupying different host plants (P = 0.006 < 0.01 for Shannon and Simpson indices). The symbionts within cotton aphid samples feeding on Buxaceae and Cucurbitaceae showed significantly higher and lower alpha diversities than samples on other plants, respectively (Buxaceae: P = 0.039 < 0.05 for Shannon index, P = 0.035 < 0.05 for Simpson index, Wilcoxon test; Cucurbitaceae: P = 0.002 < 0.01 for Shannon index, P = 0.015 < 0.05 for Simpson index, Wilcoxon test).
Pattern of Beta Diversity
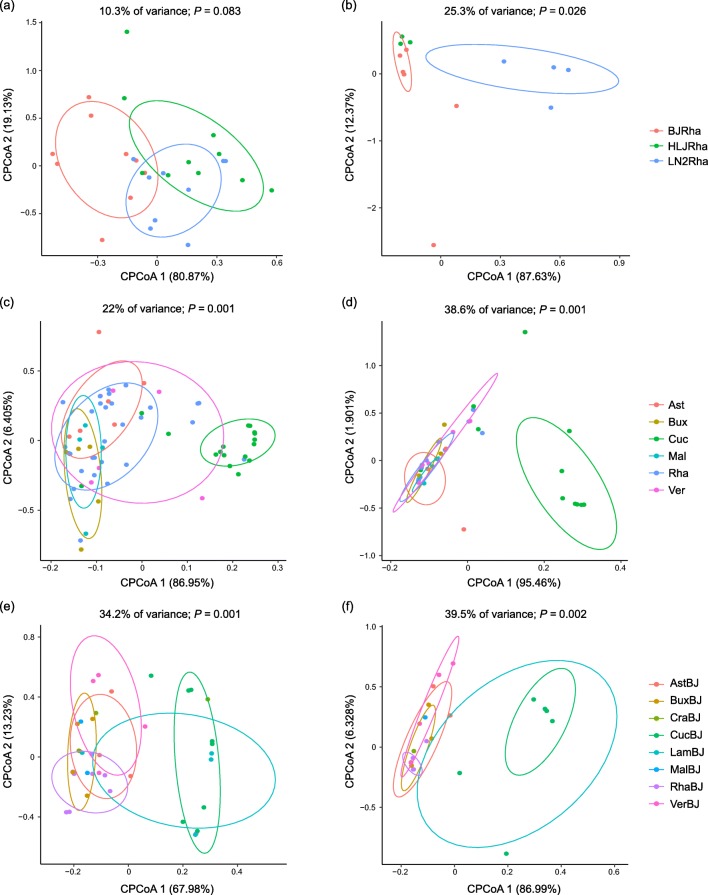
In the PCA, NMDS, and cPCoA analyses, no distinct clustering of symbiont composition for each geographic population was revealed (figures not shown), except for the cPCoA ordination of three groups feeding on Rhamnaceae (sample size ≥ 3), which showed that the samples from the same geographic region tended to cluster together and separate from others (Fig. 2a). ANOSIM tests found no significant differences across geographic regions (Table 3), whereas PERMANOVA tests detected statistical differences among 23 geographic populations (R2 = 0.265, P < 0.05) and across three groups feeding on Rhamnaceae (≥ 3 samples) (R2 = 0.170, P < 0.05). In the Mantel test, no significant correlation between Bray–Curtis dissimilarity and geographic distance could be observed (r = − 0.030, P = 0.681). For the symbiont communities associated with aphid populations occupying different host plants, although the PCA and NMDS ordinations did not show significant structuring patterns (figures not shown), both ANOSIM and PERMANOVA uncovered a strong effect of host plant on symbiont composition (P < 0.01, Table 3). Moreover, in the cPCoA analyses of six groups with a sample size ≥ 5 and eight groups from Beijing (sample size ≥ 3), the symbiont communities within aphids colonizing Cucurbitaceae tended to form a separate cluster (22–34.2% of variance, P = 0.001 < 0.01, Figs. 2c, e).
Fig. 2.
Constrained principal coordinate analyses (cPCoA) of Bray–Curtis distances of symbiont communities (a, c, e) and secondary symbiont communities (b, d, f) from three geographic groups feeding on Rhamnaceae (sample size ≥ 3) (a, b), six host plant groups with a sample size ≥5 (c, d), and eight host plant groups from Beijing (sample size ≥ 3) (e, f). See Table S2 for abbreviations
Table 3.
ANOSIM and PERMANOVA results for symbiont and secondary symbiont communities from different groups
| Group | Symbiont community | Secondary symbiont community | |||
|---|---|---|---|---|---|
| ANOSIM (R, P) | PERMANOVA (R2, P) | ANOSIM (R, P) | PERMANOVA (R2, P) | ||
| Geographic region | All 23 groups | 0.053, 0.249 | 0.265, 0.049 | 0.040, 0.223 | 0.256, 0.113 |
| 10 groups (sample size ≥3) | 0.002, 0.451 | 0.102, 0.386 | −0.006, 0.487 | 0.132, 0.189 | |
| 3 groups (on Rhamnaceae, sample size ≥3) | 0.091, 0.051 | 0.170, 0.034 | 0.010, 0.324 | 0.099, 0.223 | |
| Host plant | All 25 groups | 0.291, 0.001 | 0.433, 0.001 | 0.279, 0.002 | 0.491, 0.001 |
| 11 groups (sample size ≥3) | 0.258, 0.001 | 0.319, 0.001 | 0.280, 0.001 | 0.429, 0.001 | |
| 6 groups (sample size ≥5) | 0.239, 0.001 | 0.300, 0.001 | 0.281, 0.001 | 0.451, 0.001 | |
| 8 groups (from Beijing, sample size ≥3) | 0.278, 0.001 | 0.433, 0.001 | 0.304, 0.008 | 0.367, 0.006 | |
Significant P values (P < 0.05) are in italics.
Structural Variation in Secondary Symbiont Communities
Geographical Variation in Community Structure
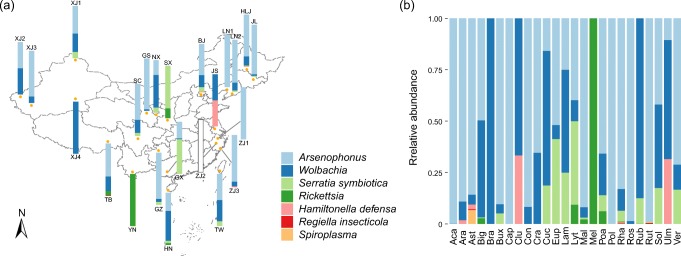
The barplot of secondary symbiont compositions of different geographic populations is shown in Fig. 3a. No recognizable clustering was observed in the ordination analyses (figures not shown), except for the cPCoA of three groups feeding on Rhamnaceae (sample size ≥ 3) (25.3% of variance, P = 0.026 < 0.05, Fig. 2b). Neither ANOSIM nor PERMANOVA revealed significant differences in secondary symbiont community among geographic populations (Table 3). Mantel test also found no significant correlation between Bray–Curtis dissimilarity and geographic distance (r = 0.011, P = 0.413).
Fig. 3.
Barplots of secondary symbiont communities from different geographic regions (a) and host plants (b). The bars display relative abundances of distinct secondary symbionts. See Table S2 for abbreviations
Community Variation with Respect to Host Plant
PCA did not exhibit good performance for the secondary symbionts from aphids exploiting different host plants (figures not shown). However, in the NMDS and cPCoA analyses of six groups with a sample size ≥ 5 and cPCoA of eight groups from Beijing (sample size ≥ 3), the communities within aphids feeding on Cucurbitaceae were clearly separated from other samples (cPCoA 38.6–39.5% of variance, P = 0.001–0.002 < 0.01, Figs. 2d, f). In addition, both ANOSIM and PERMANOVA revealed a significant effect of host plant on the secondary symbiont community (P < 0.01, Table 3). Among all detected secondary symbionts, the relative abundances of Arsenophonus and Wolbachia were found to significantly differ across different host plant groups (P < 0.01, ANOVA test, Figs. S1a, b). Arsenophonus was extremely dominant in the Acanthaceae, Caprifoliaceae, Polygonaceae, Rosaceae, and Rutaceae groups (average relative abundances across samples > 98%, Fig. 3b). However, it showed low abundance in the aphid samples feeding on Cucurbitaceae, Melastomataceae, and Ulmaceae (< 16%) and was not detected in the groups of Brassicaceae, Clusiaceae, and Rubiaceae (Fig. 3b). The post hoc Scheffé test also showed that the average relative abundance of Arsenophonus was significantly lower in the Cucurbitaceae group (P < 0.05, Fig. S1c). Wolbachia dominated in the communities from Brassicaceae, Clusiaceae, Cucurbitaceae, and Rubiaceae (> 65%, Fig. 3b). Its average relative abundance in the Cucurbitaceae group was significantly higher when tested using the post hoc Scheffé test (P < 0.05, Fig. S1d). However, Wolbachia accounted for much lower proportions (< 5%) in the communities from Acanthaceae, Asteraceae, Buxaceae, Melastomataceae, Rosaceae, and Rutaceae and was absent from the Caprifoliaceae and Polygonaceae groups (Fig. 3b).
The Correlations Between Different Symbionts
The Spearman correlation coefficients between symbionts were visualized as a heatmap (Fig. 4). Buchnera aphidicola seemed negatively correlated with secondary symbionts, particularly Arsenophonus (ρ = − 0.830, P < 0.01) and Wolbachia (ρ = − 0.211, P < 0.05) (Table S5). Both positive and negative correlations were observed between different secondary symbionts.
Fig. 4.
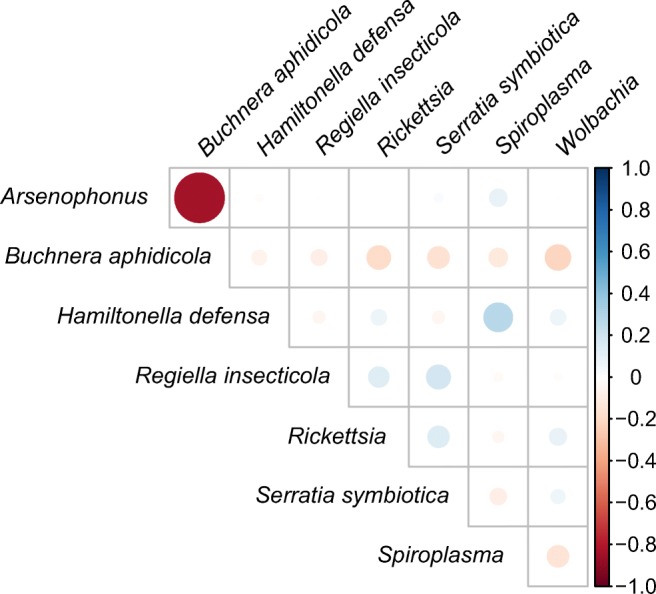
Heatmap of pairwise Spearman correlation coefficients of symbionts. Positive correlations are indicated as blue gradients from 0 to 1.0, and negative correlations are indicated as red gradients from 0 to − 1.0. The size of each circle is proportional to the significance level of the correlation coefficient
Discussion
Symbiont Diversity of Aphis gossypii
Our study revealed that the A. gossypii microbiota was dominated by a few bacterial taxa. Out of the top ten abundant genera, three were symbiotic bacteria, namely, Buchnera, Arsenophonus, and Rickettsia (Table S3). The third most abundant, Acinetobacter, has been reported in A. gossypii [73, 75, 76] and is common in insect gut communities [94, 95]. Many other bacteria detected here could be environmental or transient taxa.
As expected, Buchnera aphidicola was harbored by all aphid samples examined. It also showed the highest relative abundance in all samples. Considering the critical role of Buchnera in aphid survival and the long-term cospeciation of these two partners [8], the ubiquity and high abundance of Buchnera appear quite reasonable. Twelve OTUs belonging to Buchnera were identified. No phylogenetic concordance between Buchnera and their corresponding aphid hosts was found (data not shown). The presence of multiple phylotypes may therefore be correlated with mutation accumulation in the reduced Buchnera genomes, which seems to be caused by loss of DNA repair genes and fixation of slightly deleterious mutations through genetic drift [96, 97]. However, for phylotypes with extremely low abundance, the possibility that the mutations were artifacts introduced by PCR or sequencing errors cannot be ruled out.
Seven secondary symbionts were detected in this study, although their relative abundances were very low. Spiroplasma, which was not reported in A. gossypii previously, was identified in our sequencing data. Spiroplasma had extremely low relative abundance (< 0.005%) in two aphid samples, which may explain why it was not found before. The defensive symbiont Hamiltonella defensa was reported to infect all A. gossypii samples examined by Zhao et al. [74], Ayoubi et al. [77], and Zhang et al. [76]. However, in our study, it was just carried by six samples at very low abundance (< 0.005%). Our data showed that Arsenophonus was the predominant facultative symbiont, with the highest infection prevalence and abundance. It has been reported in previous studies of A. gossypii [66, 73–77]. Jousselin et al. [98] surveyed the distribution of Arsenophonus in aphids and revealed its high prevalence in the genus Aphis. Our results confirm their conclusion that Arsenophonus is a major bacterial partner of aphids. Most Arsenophonus in insects, including A. gossypii, was found to be associated with the lysogenic bacteriophage APSE [99], which is critical for Hamiltonella to confer protection against parasitoid wasps [100, 101]. In psyllids, the infection frequency of the APSE-bearing Arsenophonus has presented a positive correlation with parasitism, indicating a potential defensive role of Arsenophonus [102]. Therefore, we hypothesize that Arsenophonus in A. gossypii may play a similar role in providing resistance against parasitoids, especially in the case of rare Hamiltonella infection. Further experiments are required to determine the function of Arsenophonus in aphids.
Impact of Geography and Host Plant on Symbiont Community
Geography has been reported to influence the microbial profiles of the Japanese, Australian, Hawaiian, and Chinese populations of A. gossypii feeding on a limited number of plant species [66, 73, 74]. However, in the present study, it contributed little to the cotton aphid’s symbiont community structure. No significant differences in symbiont or secondary symbiont communities over space were detected in the ordination analyses or statistical tests, except for three geographic groups colonizing the same plant family Rhamnaceae. This result is consistent with previous studies in which only a few plant species were included and suggests that when the host plant is not taken into account (i.e., the same or very few plant species), geography has an influence on the symbiont composition of aphids. In addition, Mantel tests detected no significant correlation between the geographic distances among sampling sites and Bray–Curtis dissimilarities of symbiont or secondary symbiont communities, which suggested negligible effect of spatial distance on the symbiont community structure.
Compared with the limited impact of geography, the host plant appeared to have played a greater role in shaping the symbiotic bacterial community associated with A. gossypii. The alpha diversity of symbionts was found to be significantly different across aphid populations exploiting different plants. ANOSIM and PERMANOVA tests also revealed a strong effect of the host plant on both symbiont and secondary symbiont communities. These findings are consistent with previous studies that showed that the populations of polyphagous aphids colonizing different plants tended to harbor different symbiont communities (e.g., Acyrthosiphon pisum, Aphis craccivora, Aphis fabae, and Macrosiphum euphorbiae) [54–56].
It is worth noting that the Cucurbitaceae-feeding cotton aphids hosted unique symbiont communities. They showed lower alpha diversity and were clustered together and separated from other samples in some ordination analyses. The post hoc Scheffé tests revealed significantly low-abundance Arsenophonus but high-abundance Wolbachia within the Cucurbitaceae-feeding populations. Correlations between certain endosymbionts and host plants have been repeatedly reported in polyphagous aphids, especially in the extensively studied pea aphid Acyrthosiphon pisum [35, 54, 55, 59, 60, 62]. For instance, the clover-adapted biotype of pea aphid was found to be associated with Regiella insecticola around the world [35, 38, 58, 65], and Arsenophonus-bearing locust populations were reported in Aphis craccivora [55, 62]. A. gossypii is a typical polyphagous species with a very wide range of host plants. Genetic differentiation has been found to occur among its host-associated populations [103–105]. Both host plant transfer experiments [106, 107] and molecular studies [104, 105, 108] have confirmed the existence of a cucurbits-specialized host race in A. gossypii. The special symbiont communities within cucurbits-feeding populations support the ecological specialization of A. gossypii on Cucurbitaceae from the perspective of symbiotic bacteria. However, it is not clear whether the associated symbionts have played a substantive role in host plant specialization of A. gossypii. Some studies suggested that facultative symbionts had an important influence on the host plant use of aphids [38–40]; some, however, doubted the direct impact of facultative symbionts on the plant adaptation of aphids [109, 110]. Further works based on a more extensive sampling are needed to present a comprehensive landscape of microbiota in Cucurbitaceae-feeding cotton aphids. Assessments of fitness effects by particular facultative symbionts are also necessary to elucidate the exact role of endosymbionts in host specialization.
Symbiont–Symbiont Interactions
We conducted correlation analysis to assess the interactions between different symbionts. The correlation coefficients suggested antagonistic interactions between Buchnera aphidicola and secondary symbionts. In the pea aphid, Serratia symbiotica and Rickettsia have been reported to suppress the population density of Buchnera [111, 112]. Zhang et al. [76] also found a negative effect of Hamiltonella on the abundance of Buchnera in A. gossypii. These findings indicate competition between the primary and secondary symbionts for resources and survival niches within the same host aphid.
Multiple infections with secondary symbionts occurred commonly in our examined cotton aphids. Co-infections are often unstable [24, 113]. We hypothesize that such a high proportion of multiple infections may result from frequent horizontal transfers. A. gossypii is heteroecious holocyclic in China, alternating between primary host plants such as Punica, Hibiscus, and Rhamnus and various herbaceous secondary host plants [114]. The sexual phase [113] and migrations between different plants [115, 116] create opportunities for horizontal transfer of secondary symbionts among natural populations of A. gossypii. Co-infections may bring ecological benefits for the host aphids. Acyrthosiphon pisum co-infected with Hamiltonella–Serratia or Hamiltonella–Fukatsuia exhibited greater resistance to parasitoids [31, 32]. However, Polin et al. [117] reported that the Acyrthosiphon pisum strain co-infected with Rickettsiella viridis and Hamiltonella defensa was more exposed to ladybird predation than the singly Rickettsiella-infected strain. Ayoubi et al. [77] found that the Hamiltonella–Arsenophonus combination in A. gossypii conferred no resistance against parasitism by Aphidius matricariae. Therefore, the interactions between facultative symbionts seem very complicated, either synergistically or antagonistically, which was also indicated by the positive and negative Spearman correlation coefficients in our study.
Conclusions
Based on an extensive sampling from different plants and regions in China, we analyzed the diversity of symbiotic bacteria within Aphis gossypii using Illumina sequencing of 16S rRNA gene. The microbiota of A. gossypii was dominated by heritable symbionts, among which Buchnera aphidicola and Arsenophonus were the predominant symbiont and facultative symbiont, respectively. The symbiont diversity was found to vary with the host plant rather than geography, suggesting an important role of the host plant in shaping the bacterial community structure. The cucurbits-adapted aphid populations harbored unique symbiont communities, which provide a good model to explore the direct or indirect impacts of facultative symbionts on host specialization. Moreover, the interactions between coexisting symbionts within A. gossypii were revealed to be very complicated.
Electronic Supplementary Material
(DOCX 1.07 mb)
Acknowledgments
Thanks to all collectors for aphid sample collections. Thanks to Fendi Yang for making slides of voucher specimens.
Author Contributions
J.C. and G.Q. designed the project. G.Q. and L.J. identified voucher specimens. S.X. conducted molecular experiments and all analyses. J.C. assisted data analyses. S.X. and J.C. wrote the manuscript and all authors contributed to revisions.
Funding Information
The work was supported by the National Key R & D Program of China (No. 2016YFE0203100), the Strategic Priority Research Program A of the Chinese Academy of Sciences (No. XDA19050303), and the National Natural Sciences Foundation of China (Nos. 31620103916, 31430078).
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Contributor Information
Gexia Qiao, Email: qiaogx@ioz.ac.cn.
Jing Chen, Email: chenjing@ioz.ac.cn.
References
- 1.Buchner P. Endosymbiosis of animals with plant microorganisms. New York: Interscience Publishers; 1965. [Google Scholar]
- 2.Douglas AE. Requirement of pea aphids (Acyrthosiphon pisum) for their symbiotic bacteria. Entomol Exp Appl. 1992;65:195–198. doi: 10.1111/j.1570-7458.1992.tb01643.x. [DOI] [Google Scholar]
- 3.Douglas AE. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 4.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 5.Brough CN, Dixon AFG. Ultrastructural features of egg development in oviparae of the vetch aphid, Megoura viciae Buckton. Tissue Cell. 1990;22:51–63. doi: 10.1016/0040-8166(90)90089-R. [DOI] [PubMed] [Google Scholar]
- 6.Wegierek P, Michalik A, Wieczorek K, Kanturski M, Kobiałka M, Śliwa K, Szklarzewicz T. Buchnera aphidicola of the birch blister aphid, Hamamelistes betulinus (Horváth, 1896) (Insecta, Hemiptera, Aphididae: Hormaphidinae): molecular characterization, transmission between generations and its geographic significance. Acta Zool. 2017;98:412–421. doi: 10.1111/azo.12186. [DOI] [Google Scholar]
- 7.Munson MA, Baumann P, Clark MA, Baumann L, Moran NA, Voegtlin DJ, Campbell BC. Evidence for the establishment of aphid-eubacterium endosymbiosis in an ancestor of four aphid families. J Bacteriol. 1991;173:6321–6324. doi: 10.1128/jb.173.20.6321-6324.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc Lond B Biol Sci. 1993;253:167–171. doi: 10.1098/rspb.1993.0098. [DOI] [Google Scholar]
- 9.Baumann P, Moran NA, Baumann L. The evolution and genetics of aphid endosymbionts. Bioscience. 1997;47:12–20. doi: 10.2307/1313002. [DOI] [Google Scholar]
- 10.Clark MA, Moran NA, Baumann P, Wernegreen JJ. Cospeciation between bacterial endosymbionts (Buchnera) and a recent radiation of aphids (Uroleucon) and pitfalls of testing for phylogenetic congruence. Evolution. 2000;54:517–525. doi: 10.1111/j.0014-3820.2000.tb00054.x. [DOI] [PubMed] [Google Scholar]
- 11.Jousselin E, Desdevises Y, Coeur d’acier A. Fine-scale cospeciation between Brachycaudus and Buchnera aphidicola: bacterial genome helps define species and evolutionary relationships in aphids. Proc R Soc Lond B Biol Sci. 2009;276:187–196. doi: 10.1098/rspb.2008.0679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L, Huang X, Zhang R, Jiang L, Qiao G. Phylogenetic congruence between Mollitrichosiphum (Aphididae: Greenideinae) and Buchnera indicates insect-bacteria parallel evolution. Syst Entomol. 2013;38:81–92. doi: 10.1111/j.1365-3113.2012.00647.x. [DOI] [Google Scholar]
- 13.Liu L, Li XY, Huang XL, Qiao GX. Evolutionary relationships of Pemphigus and allied genera (Hemiptera: Aphididae: Eriosomatinae) and their primary endosymbiont, Buchnera aphidicola. Insect Sci. 2014;21:301–312. doi: 10.1111/1744-7917.12113. [DOI] [PubMed] [Google Scholar]
- 14.Xu TT, Chen J, Jiang LY, Qiao GX. Historical and cospeciating associations between Cerataphidini aphids (Hemiptera: Aphididae: Hormaphidinae) and their primary endosymbiont Buchnera aphidicola. Zool J Linn Soc-Lond. 2018;182:604–613. doi: 10.1093/zoolinnean/zlx048. [DOI] [Google Scholar]
- 15.Chen DQ, Campbell BC, Purcell AH. A new Rickettsia from a herbivorous insect, the pea aphid Acyrthosiphon pisum (Harris) Curr Microbiol. 1996;33:123–128. doi: 10.1007/s002849900086. [DOI] [PubMed] [Google Scholar]
- 16.Jeyaprakash A, Hoy MA. Long PCR improves Wolbachia DNA amplification: wsp sequences found in 76% of sixty-three arthropod species. Insect Mol Biol. 2000;9:393–405. doi: 10.1046/j.1365-2583.2000.00203.x. [DOI] [PubMed] [Google Scholar]
- 17.Fukatsu T, Tsuchida T, Nikoh N, Koga R. Spiroplasma symbiont of the pea aphid, Acyrthosiphon pisum (Insecta: Homoptera) Appl Environ Microbiol. 2001;67:1284–1291. doi: 10.1128/AEM.67.3.1284-1291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russell JA, Latorre A, Sabater-Muñoz B, Moya A, Moran NA. Side-stepping secondary symbionts: widespread horizontal transfer across and beyond the Aphidoidea. Mol Ecol. 2003;12:1061–1075. doi: 10.1046/j.1365-294X.2003.01780.x. [DOI] [PubMed] [Google Scholar]
- 19.Moran NA, Russell JA, Koga R, Fukatsu T. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol. 2005;71:3302–3310. doi: 10.1128/Aem.71.6.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuchida T, Koga R, Fujiwara A, Fukatsu T (2014) Phenotypic effect of “Candidatus Rickettsiella viridis,” a facultative symbiont of the pea aphid (Acyrthosiphon pisum), and its interaction with a coexisting symbiont. Appl Environ Microbiol 80:525–533. 10.1128/Aem.03049-13 [DOI] [PMC free article] [PubMed]
- 21.Manzano-Marín A, Szabó G, Simon JC, Horn M, Latorre A. Happens in the best of subfamilies: establishment and repeated replacements of co-obligate secondary endosymbionts within Lachninae aphids. Environ Microbiol. 2017;19:393–408. doi: 10.1111/1462-2920.13633. [DOI] [PubMed] [Google Scholar]
- 22.Fukatsu T, Nikoh N, Kawai R, Koga R. The secondary endosymbiotic bacterium of the pea aphid Acyrthosiphon pisum (Insecta: Homoptera) Appl Environ Microbiol. 2000;66:2748–2758. doi: 10.1128/AEM.66.7.2748-2758.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen DQ, Purcell AH. Occurrence and transmission of facultative endosymbionts in aphids. Curr Microbiol. 1997;34:220–225. doi: 10.1007/s002849900172. [DOI] [PubMed] [Google Scholar]
- 24.Sandström JP, Russell JA, White JP, Moran NA. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol. 2001;10:217–228. doi: 10.1046/j.1365-294X.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- 25.Chen DQ, Montllor CB, Purcell AH. Fitness effects of two facultative endosymbiotic bacteria on the pea aphid, Acyrthosiphon pisum, and the blue alfalfa aphid, A. kondoi. Entomol Exp Appl. 2000;95:315–323. doi: 10.1046/j.1570-7458.2000.00670.x. [DOI] [Google Scholar]
- 26.Montllor CB, Maxmen A, Purcell AH. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. 2002;27:189–195. doi: 10.1046/j.1365-2311.2002.00393.x. [DOI] [Google Scholar]
- 27.Russell JA, Moran NA. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc R Soc Lond B Biol Sci. 2006;273:603–610. doi: 10.1098/rspb.2005.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burke G, Fiehn O, Moran N. Effects of facultative symbionts and heat stress on the metabolome of pea aphids. ISME J. 2010;4:242–252. doi: 10.1038/ismej.2009.114. [DOI] [PubMed] [Google Scholar]
- 29.Heyworth ER, Ferrari J. A facultative endosymbiont in aphids can provide diverse ecological benefits. J Evol Biol. 2015;28:1753–1760. doi: 10.1111/jeb.12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci U S A. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliver KM, Moran NA, Hunter MS. Costs and benefits of a superinfection of facultative symbionts in aphids. Proc R Soc Lond B Biol Sci. 2006;273:1273–1280. doi: 10.1098/rspb.2005.3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guay JF, Boudreault S, Michaud D, Cloutier C. Impact of environmental stress on aphid clonal resistance to parasitoids: role of Hamiltonella defensa bacterial symbiosis in association with a new facultative symbiont of the pea aphid. J Insect Physiol. 2009;55:919–926. doi: 10.1016/j.jinsphys.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Vorburger C, Gehrer L, Rodriguez P. A strain of the bacterial symbiont Regiella insecticola protects aphids against parasitoids. Biol Lett. 2010;6:109–111. doi: 10.1098/rsbl.2009.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frago E, Mala M, Weldegergis BT, Yang C, McLean A, Godfray HCJ, Gols R, Dicke M. Symbionts protect aphids from parasitic wasps by attenuating herbivore-induced plant volatiles. Nat Commun. 2017;8:1860. doi: 10.1038/s41467-017-01935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferrari J, Darby AC, Daniell TJ, Godfray HCJ, Douglas AE. Linking the bacterial community in pea aphids with host-plant use and natural enemy resistance. Ecol Entomol. 2004;29:60–65. doi: 10.1111/j.1365-2311.2004.00574.x. [DOI] [Google Scholar]
- 36.Scarborough CL, Ferrari J, Godfray HCJ. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781–1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- 37.Łukasik P, van Asch M, Guo H, Ferrari J, Godfray HCJ. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol Lett. 2013;16:214–218. doi: 10.1111/ele.12031. [DOI] [PubMed] [Google Scholar]
- 38.Leonardo TE, Muiru GT. Facultative symbionts are associated with host plant specialization in pea aphid populations. Proc R Soc Lond B Biol Sci. 2003;270:S209–S212. doi: 10.1098/rsbl.2003.0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuchida T, Koga R, Fukatsu T. Host plant specialization governed by facultative symbiont. Science. 2004;303:1989–1989. doi: 10.1126/science.1094611. [DOI] [PubMed] [Google Scholar]
- 40.Wagner SM, Martinez AJ, Ruan YM, Kim KL, Lenhart PA, Dehnel AC, Oliver KM, White JA. Facultative endosymbionts mediate dietary breadth in a polyphagous herbivore. Funct Ecol. 2015;29:1402–1410. doi: 10.1111/1365-2435.12459. [DOI] [Google Scholar]
- 41.Tsuchida T, Koga R, Horikawa M, Tsunoda T, Maoka T, Matsumoto S, Simon JC, Fukatsu T. Symbiotic bacterium modifies aphid body color. Science. 2010;330:1102–1104. doi: 10.1126/science.1195463. [DOI] [PubMed] [Google Scholar]
- 42.Leonardo TE, Mondor EB. Symbiont modifies host life-history traits that affect gene flow. Proc R Soc Lond B Biol Sci. 2006;273:1079–1084. doi: 10.1098/rspb.2005.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon JC, Boutin S, Tsuchida T, Koga R, Le Gallic JF, Frantz A, Outreman Y, Fukatsu T. Facultative symbiont infections affect aphid reproduction. PLoS One. 2011;6:e21831. doi: 10.1371/journal.pone.0021831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliver KM, Degnan PH, Burke GR, Moran NA. Facultative symbionts in aphids and the horizontal transfer of ecologically important traits. Annu Rev Entomol. 2010;55:247–266. doi: 10.1146/annurev-ento-112408-085305. [DOI] [PubMed] [Google Scholar]
- 45.Zytynska SE, Weisser WW. The natural occurrence of secondary bacterial symbionts in aphids. Ecol Entomol. 2016;41:13–26. doi: 10.1111/een.12281. [DOI] [Google Scholar]
- 46.Guo J, Hatt S, He K, Chen J, Francis F, Wang Z. Nine facultative endosymbionts in aphids. A review. J Asia Pac Entomol. 2017;20:794–801. doi: 10.1016/j.aspen.2017.03.025. [DOI] [Google Scholar]
- 47.Lamelas A, Gosalbes MJ, Manzano-Marín A, Peretó J, Moya A, Latorre A. Serratia symbiotica from the aphid Cinara cedri: a missing link from facultative to obligate insect endosymbiont. PLoS Genet. 2011;7:e1002357. doi: 10.1371/journal.pgen.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manzano-Marín A, Latorre A. Settling down: the genome of Serratia symbiotica from the aphid Cinara tujafilina zooms in on the process of accommodation to a cooperative intracellular life. Genome Biol Evol. 2014;6:1683–1698. doi: 10.1093/gbe/evu133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manzano-Marín A, Simon JC, Latorre A. Reinventing the wheel and making it round again: evolutionary convergence in Buchnera-Serratia symbiotic consortia between the distantly related Lachninae aphids Tuberolachnus salignus and Cinara cedri. Genome Biol Evol. 2016;8:1440–1458. doi: 10.1093/gbe/evw085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Meseguer AS, Manzano-Marín A, Coeur d’acier A, Clamens AL, Godefroid M, Jousselin E. Buchnera has changed flatmate but the repeated replacement of co-obligate symbionts is not associated with the ecological expansions of their aphid hosts. Mol Ecol. 2017;26:2363–2378. doi: 10.1111/mec.13910. [DOI] [PubMed] [Google Scholar]
- 51.Manzano-Marín A, Coeur d’acier A, Clamens AL, Orvain C, Cruaud C, Barbe V, Jousselin E (2019) Serial horizontal transfer of vitamin-biosynthetic genes enables the establishment of new nutritional symbionts in aphids’ di-symbiotic systems. ISME J. 10.1038/s41396-019-0533-6 [DOI] [PMC free article] [PubMed]
- 52.De Clerck C, Fujiwara A, Joncour P, Léonard S, Félix ML, Francis F, Jijakli MH, Tsuchida T, Massart S. A metagenomic approach from aphid’s hemolymph sheds light on the potential roles of co-existing endosymbionts. Microbiome. 2015;3:63. doi: 10.1186/s40168-015-0130-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manzano-Marín A (2019) No evidence for Wolbachia as a nutritional co-obligate endosymbiont in the aphid Pentalonia nigronervosa. bioRxiv. 10.1101/609511 [DOI] [PMC free article] [PubMed]
- 54.Ferrari J, West JA, Via S, Godfray HCJ. Population genetic structure and secondary symbionts in host-associated populations of the pea aphid complex. Evolution. 2012;66:375–390. doi: 10.1111/j.1558-5646.2011.01436.x. [DOI] [PubMed] [Google Scholar]
- 55.Brady CM, Asplen MK, Desneux N, Heimpel GE, Hopper KR, Linnen CR, Oliver KM, Wulff JA, White JA. Worldwide populations of the aphid Aphis craccivora are infected with diverse facultative bacterial symbionts. Microb Ecol. 2014;67:195–204. doi: 10.1007/s00248-013-0314-0. [DOI] [PubMed] [Google Scholar]
- 56.Henry LM, Maiden MCJ, Ferrari J, Godfray HCJ. Insect life history and the evolution of bacterial mutualism. Ecol Lett. 2015;18:516–525. doi: 10.1111/ele.12425. [DOI] [PubMed] [Google Scholar]
- 57.Peccoud J, Ollivier A, Plantegenest M, Simon JC. A continuum of genetic divergence from sympatric host races to species in the pea aphid complex. Proc Natl Acad Sci U S A. 2009;106:7495–7500. doi: 10.1073/pnas.0811117106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simon JC, Carré S, Boutin M, Prunier-Leterme N, Sabater-Muñoz B, Latorre A, Bournoville R. Host-based divergence in populations of the pea aphid: insights from nuclear markers and the prevalence of facultative symbionts. Proc R Soc Lond B Biol Sci. 2003;270:1703–1712. doi: 10.1098/rspb.2003.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frantz A, Calcagno V, Mieuzet L, Plantegenest M, Simon JC. Complex trait differentiation between host-populations of the pea aphid Acyrthosiphon pisum (Harris): implications for the evolution of ecological specialisation. Biol J Linn Soc. 2009;97:718–727. doi: 10.1111/j.1095-8312.2009.01221.x. [DOI] [Google Scholar]
- 60.Henry LM, Peccoud J, Simon JC, Hadfield JD, Maiden MJC, Ferrari J, Godfray HCJ. Horizontally transmitted symbionts and host colonization of ecological niches. Curr Biol. 2013;23:1713–1717. doi: 10.1016/j.cub.2013.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gauthier JP, Outreman Y, Mieuzet L, Simon JC. Bacterial communities associated with host-adapted populations of pea aphids revealed by deep sequencing of 16S ribosomal DNA. PLoS One. 2015;10:e0120664. doi: 10.1371/journal.pone.0120664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brady CM, White JA. Cowpea aphid (Aphis craccivora) associated with different host plants has different facultative endosymbionts. Ecol Entomol. 2013;38:433–437. doi: 10.1111/een.12020. [DOI] [Google Scholar]
- 63.Medina RF, Nachappa P, Tamborindeguy C. Differences in bacterial diversity of host-associated populations of Phylloxera notabilis Pergande (Hemiptera: Phylloxeridae) in pecan and water hickory. J Evol Biol. 2011;24:761–771. doi: 10.1111/j.1420-9101.2010.02215.x. [DOI] [PubMed] [Google Scholar]
- 64.Guidolin AS, Cônsoli FL. Symbiont diversity of Aphis (Toxoptera) citricidus (Hemiptera: Aphididae) as influenced by host plants. Microb Ecol. 2017;73:201–210. doi: 10.1007/s00248-016-0892-8. [DOI] [PubMed] [Google Scholar]
- 65.Tsuchida T, Koga R, Shibao H, Matsumoto T, Fukatsu T. Diversity and geographic distribution of secondary endosymbiotic bacteria in natural populations of the pea aphid, Acyrthosiphon pisum. Mol Ecol. 2002;11:2123–2135. doi: 10.1046/j.1365-294X.2002.01606.x. [DOI] [PubMed] [Google Scholar]
- 66.Jones RT, Bressan A, Greenwell AM, Fierer N. Bacterial communities of two parthenogenetic aphid species cocolonizing two host plants across the Hawaiian Islands. Appl Environ Microbiol. 2011;77:8345–8349. doi: 10.1128/Aem.05974-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schmid M, Sieber R, Zimmermann YS, Vorburger C. Development, specificity and sublethal effects of symbiont-conferred resistance to parasitoids in aphids. Funct Ecol. 2012;26:207–215. doi: 10.1111/j.1365-2435.2011.01904.x. [DOI] [Google Scholar]
- 68.Simonet P, Duport G, Gaget K, Weiss-Gayet M, Colella S, Febvay G, Charles H, Vinuelas J, Heddi A, Calevro F. Direct flow cytometry measurements reveal a fine-tuning of symbiotic cell dynamics according to the host developmental needs in aphid symbiosis. Sci Rep. 2016;6:19967. doi: 10.1038/srep19967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bansal R, Mian MA, Michel AP. Microbiome diversity of Aphis glycines with extensive superinfection in native and invasive populations. Environ Microbiol Rep. 2014;6:57–69. doi: 10.1111/1758-2229.12108. [DOI] [PubMed] [Google Scholar]
- 70.Zytynska SE, Meyer ST, Sturm S, Ullmann W, Mehrparvar M, Weisser WW. Secondary bacterial symbiont community in aphids responds to plant diversity. Oecologia. 2016;180:735–747. doi: 10.1007/s00442-015-3488-y. [DOI] [PubMed] [Google Scholar]
- 71.Liu XD, Lei HX, Chen FF. Infection pattern and negative effects of a facultative endosymbiont on its insect host are environment-dependent. Sci Rep. 2019;9:4013. doi: 10.1038/s41598-019-40607-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Blackman RL, Eastop VF (2000) Aphids on the world’s crops: an identification and information guide. 2nd edn. John Wiley & Sons Ltd, Chichester
- 73.Najar-Rodríguez AJ, McGraw EA, Mensah RK, Pittman GW, Walter GH. The microbial flora of Aphis gossypii: patterns across host plants and geographical space. J Invertebr Pathol. 2009;100:123–126. doi: 10.1016/j.jip.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 74.Zhao Y, Zhang S, Luo JY, Wang CY, Lv LM, Cui JJ. Bacterial communities of the cotton aphid Aphis gossypii associated with Bt cotton in northern China. Sci Rep. 2016;6:22958. doi: 10.1038/srep22958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang J, Pan Y, Zheng C, Gao X, Wei X, Xi J, Peng T, Shang Q. Rapid evolution of symbiotic bacteria populations in spirotetramat-resistant Aphis gossypii Glover revealed by pyrosequencing. Comp Biochem Physiol Part D Genomics Proteomics. 2016;20:151–158. doi: 10.1016/j.cbd.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 76.Zhang S, Luo J, Wang L, Zhang L, Zhu X, Jiang W, Cui J. Bacterial communities in natural versus pesticide-treated Aphis gossypii populations in North China. MicrobiologyOpen. 2019;8:e00652. doi: 10.1002/mbo3.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ayoubi A, Talebi AA, Fathipour Y, Mehrabadi M (2020) Coinfection of the secondary symbionts, Hamiltonella defensa and Arsenophonus sp. contribute to the performance of the major aphid pest, Aphis gossypii (Hemiptera: Aphididae). Insect Sci 27:86–98. 10.1111/1744-7917.12603 [DOI] [PubMed]
- 78.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- 79.Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 83.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/Aem.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2018) Package ‘vegan’, community ecology package. Version 2.5–2
- 87.Vincent QV (2011) ggbiplot: a ggplot2 based biplot. R package version 0.55. http://github.com/vqv/ggbiplot. Accessed 2 March 2019
- 88.Wickham H. ggplot2: elegant graphics for data analysis. 2. New York: Springer; 2016. [Google Scholar]
- 89.Fasham MJR. A comparison of nonmetric multidimensional scaling, principal components and reciprocal averaging for the ordination of simulated coenoclines, and coenoplanes. Ecology. 1977;58:551–561. doi: 10.2307/1939004. [DOI] [Google Scholar]
- 90.Anderson MJ, Walsh DCI. PERMANOVA, ANOSIM, and the Mantel test in the face of heterogeneous dispersions: what null hypothesis are you testing? Ecol Monogr. 2013;83:557–574. doi: 10.1890/12-2010.1. [DOI] [Google Scholar]
- 91.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ersts PJ (2019) Geographic Distance Matrix Generator v1.2.3. American Museum of Natural History, Center for Biodiversity and Conservation. http://biodiversityinformatics.amnh.org/open_source/gdmg. Accessed 14 August 2019
- 93.Wei T, Simko V, Levy M, Xie Y, Jin Y, Zemla J. Package ‘corrplot’. Statistician. 2017;56:316–324. [Google Scholar]
- 94.Geiger A, Fardeau ML, Grebaut P, Vatunga G, Josénando T, Herder S, Cuny G, Truc P, Ollivier B. First isolation of Enterobacter, Enterococcus, and Acinetobacter spp. as inhabitants of the tsetse fly (Glossina palpalis palpalis) midgut. Infect Genet Evol. 2009;9:1364–1370. doi: 10.1016/j.meegid.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 95.Priya NG, Ojha A, Kajla MK, Raj A, Rajagopal R. Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS One. 2012;7:e30768. doi: 10.1371/journal.pone.0030768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moran NA. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc Natl Acad Sci U S A. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 98.Jousselin E, Coeur d’acier A, Vanlerberghe-Masutti F, Duron O. Evolution and diversity of Arsenophonus endosymbionts in aphids. Mol Ecol. 2013;22:260–270. doi: 10.1111/mec.12092. [DOI] [PubMed] [Google Scholar]
- 99.Duron O. Arsenophonus insect symbionts are commonly infected with APSE, a bacteriophage involved in protective symbiosis. FEMS Microbiol Ecol. 2014;90:184–194. doi: 10.1111/1574-6941.12381. [DOI] [PubMed] [Google Scholar]
- 100.Moran NA, Degnan PH, Santos SR, Dunbar HE, Ochman H. The players in a mutualistic symbiosis: insects, bacteria, viruses, and virulence genes. Proc Natl Acad Sci U S A. 2005;102:16919–16926. doi: 10.1073/pnas.0507029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992–994. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hansen AK, Jeong G, Paine TD, Stouthamer R. Frequency of secondary symbiont infection in an invasive psyllid relates to parasitism pressure on a geographic scale in California. Appl Environ Microbiol. 2007;73:7531–7535. doi: 10.1128/AEM.01672-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Brévault T, Carletto J, Linderme D, Vanlerberghe-Masutti F. Genetic diversity of the cotton aphid Aphis gossypii in the unstable environment of a cotton growing area. Agric For Entomol. 2008;10:215–223. doi: 10.1111/j.1461-9563.2008.00377.x. [DOI] [Google Scholar]
- 104.Charaabi K, Carletto J, Chavigny P, Marrakchi M, Makni M, Vanlerberghe-Masutti F. Genotypic diversity of the cotton-melon aphid Aphis gossypii (Glover) in Tunisia is structured by host plants. Bull Entomol Res. 2008;98:333–341. doi: 10.1017/S0007485307005585. [DOI] [PubMed] [Google Scholar]
- 105.Carletto J, Lombaert E, Chavigny P, Brévault T, Lapchin L, Vanlerberghe-Masutti F. Ecological specialization of the aphid Aphis gossypii Glover on cultivated host plants. Mol Ecol. 2009;18:2198–2212. doi: 10.1111/j.1365-294X.2009.04190.x. [DOI] [PubMed] [Google Scholar]
- 106.Guldemond JA, Tigges WT, De Vrijer PWF. Host races of Aphis gossypii (Homoptera: Aphididae) on cucumber and chrysanthemum. Environ Entomol. 1994;23:1235–1240. doi: 10.1093/ee/23.5.1235. [DOI] [Google Scholar]
- 107.Wang L, Zhang S, Luo JY, Wang CY, Lv LM, Zhu XZ, Li CH, Cui JJ. Identification of Aphis gossypii Glover (Hemiptera: Aphididae) biotypes from different host plants in North China. PLoS One. 2016;11:e0146345. doi: 10.1371/journal.pone.0146345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Vanlerberghe-Masutti F, Chavigny P. Host-based genetic differentiation in the aphid Aphis gossypii Glover, evidenced from RAPD fingerprints. Mol Ecol. 1998;7:905–914. doi: 10.1046/j.1365-294x.1998.00421.x. [DOI] [Google Scholar]
- 109.Leonardo TE. Removal of a specialization-associated symbiont does not affect aphid fitness. Ecol Lett. 2004;7:461–468. doi: 10.1111/j.1461-0248.2004.00602.x. [DOI] [Google Scholar]
- 110.McLean AHC, van Asch M, Ferrari J, Godfray HCJ. Effects of bacterial secondary symbionts on host plant use in pea aphids. Proc R Soc Lond B Biol Sci. 2011;278:760–766. doi: 10.1098/rspb.2010.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Koga R, Tsuchida T, Fukatsu T. Changing partners in an obligate symbiosis: a facultative endosymbiont can compensate for loss of the essential endosymbiont Buchnera in an aphid. Proc R Soc Lond B Biol Sci. 2003;270:2543–2550. doi: 10.1098/rspb.2003.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sakurai M, Koga R, Tsuchida T, Meng XY, Fukatsu T. Rickettsia symbiont in the pea aphid Acyrthosiphon pisum: novel cellular tropism, effect on host fitness, and interaction with the essential symbiont Buchnera. Appl Environ Microbiol. 2005;71:4069–4075. doi: 10.1128/Aem.71.7.4069-4075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moran NA, Dunbar HE. Sexual acquisition of beneficial symbionts in aphids. Proc Natl Acad Sci U S A. 2006;103:12803–12806. doi: 10.1073/pnas.0605772103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang GX, Zhong TS. Economic insect fauna of China. Fasc. 25. Homoptera: Aphidinea, Part I. Beijing: Science Press; 1983. [Google Scholar]
- 115.Caspi-Fluger A, Inbar M, Mozes-Daube N, Katzir N, Portnoy V, Belausov E, Hunter MS, Zchori-Fein E. Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. Proc R Soc Lond B Biol Sci. 2012;279:1791–1796. doi: 10.1098/rspb.2011.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pons I, Renoz F, Noël C, Hance T. Circulation of the cultivable symbiont Serratia symbiotica in aphids is mediated by plants. Front Microbiol. 2019;10:764. doi: 10.3389/fmicb.2019.00764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Polin S, Le Gallic JF, Simon JC, Tsuchida T, Outreman Y. Conditional reduction of predation risk associated with a facultative symbiont in an insect. PLoS One. 2015;10:e0143728. doi: 10.1371/journal.pone.0143728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 1.07 mb)