Abstract
ATG8 family proteins are evolutionary conserved ubiquitin-like modifiers, which become attached to the headgroup of the membrane lipid phosphatidylethanolamine in a process referred to as lipidation. This reaction is carried out analogous to the conjugation of ubiquitin to its target proteins, involving the E1-like ATG7, the E2-like ATG3 and the E3-like ATG12–ATG5–ATG16 complex, which determines the site of lipidation. ATG8 lipidation is a hallmark of autophagy where these proteins are involved in autophagosome formation, the fusion of autophagosomes with lysosomes and cargo selection. However, it has become evident that ATG8 lipidation also occurs in processes that are not directly related to autophagy. Here we discuss recent insights into the targeting of ATG8 lipidation in autophagy and other pathways with special emphasis on the recruitment and activation of the E3-like complex.
Subject terms: Macroautophagy, Phagocytosis
Introduction
The Atg8 protein is a ubiquitin-like protein, which was identified in S. cerevisiae in the course of screens designed to discover genes required for the process of macroautophagy (hereafter referred to as autophagy) or the related cytoplasm-to-vacuole targeting (Cvt) pathway1–3. Subsequent sequence analysis revealed homology of Atg8 to the mammalian LC3 and GABARAP proteins4–6. In total the human genome contains seven functional ATG8 genes (LC3A, LC3B, LC3B2, LC3C, GABARAP, GABARAPL1, and GABARAPL2)7, of which LC3B2 appears to be expressed at very low levels8. The genome of many plants codes for even more ATG8 proteins. For example, Arabidopsis thaliana has 9 ATG8 genes9. Structural studies revealed that the core of the ATG8 structure consists of an ubiquitin-like fold10. However, unlike ubiquitin, which is conjugated to the lysine residues of target proteins via an isopeptide bond involving its C-terminal glycine residue, ATG8 proteins become attached to the amino headgroup of membrane lipids11. In vivo the main target of this conjugation, which is also referred to as lipidation, is phosphatidylethanolamine (PE), although at least in vitro phosphatidylserine can also serve as substrate12–14. Because the process of autophagy can be conveniently traced employing ATG8 proteins as markers, they have been widely used to monitor and study the pathway15,16. In this review, we will summarize how ATG8 proteins function in autophagy. It is becoming increasingly clear that there are roles for these proteins in pathways that are not strictly related to autophagy and we will also summarize the current knowledge about the association of ATG8s with these pathways. Finally, we will discuss how the ATG8 conjugation machinery is activated and targeted to the correct membrane with special emphasis on the E3-like ATG12–ATG5–ATG16 complex.
The function of ATG8 proteins in autophagy
Autophagy mediates the delivery of various cytoplasmic substances into lysosomes (the vacuole in yeast) for degradation. This is achieved by the sequestration of this material referred to as cargo within double membrane vesicles named autophagosomes (Fig. 1). Autophagosomes are generated de novo and first appear as small membrane structures called isolation membranes or phagophores. The isolation membranes capture the cargo as they grow. Subsequently, the isolation membranes close to give rise to autophagosomes, which eventually fuse with a lysosome wherein the inner membrane and the cargo are finally degraded17.
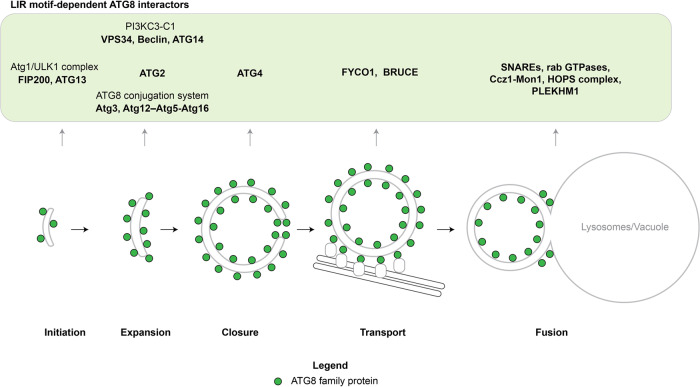
Fig. 1. LIR motif-dependent ATG8 interactions.
During the process of macroautophagy, ATG8 family proteins participate in autophagosome formation at different stages from initiation to the fusion with the lytic compartment. These interactions are typically mediated by LIR motifs in the target proteins. In the scheme, the ATG8 interactors found in yeast and humans are shown. The ATG8 protein family members are depicted in green.
The formation of autophagosomes depends on a number of highly conserved factors that are collectively referred to as the autophagy machinery and that can be subdivided into distinct functional modules. According to the guidelines, we will refer to the human proteins with all capital letters (ATG) and to the S. cerevisiae proteins with only the first letter in capital (Atg; Table 1). In particular, these are (1) the ULK1/2 kinase complex composed of the ULK1/2 protein kinase, the FIP200 scaffold protein, ATG13 and ATG101, (2) vesicles containing the ATG9A protein, (3) the class III phosphatidylinositol 3-phosphate kinase complex 1 (PI3KC3-C1) composed of the VPS34 lipid kinase, VPS15, Beclin and ATG14, (4) the WIPIs and ATG2, and (5) the ATG8 conjugation system including the ATG12–ATG5–ATG16L1 complex18–20. Humans have six ATG8 proteins that are expressed at considerable levels and that can be subdivided into the LC3 (LC3A, LC3B, LC3C) and GABARAP (GABARAP, GABARAPL1, GABARAPL2) families. In their entirety we will refer to them as ATG8 proteins unless a particular family member is mentioned.
Table 1.
Atg/ATG nomenclature.
| Species Protein group | S. cerevisiae (Atg) | H. sapiens (ATG) |
|---|---|---|
| Protein kinase complex | Atg1, Atg13, Atg11/Atg17, Atg29, Atg31 | ULK1/2, ATG13, FIP200, ATG101 |
| Vesicles | Atg9 | ATG9A |
| Lipid kinase complex | Vps34, Vps15, Atg6, Atg14 | VPS34, VPS15, Beclin, ATG14 |
| PI3P sensors and lipid transfer | Atg21, Atg18, Hsv2, Atg2 | WIPI1, WIPI2, WIPI3, WIPI4, ATG2 |
| Ubiquitin-like conjugation machineries | Atg7, Atg10, Atg5, Atg12, Atg16, Atg3, Atg8, Atg4 | ATG7, ATG10, ATG5, ATG12, ATG16L1, ATG3, LC3s/GABARAPs, ATG4A/B/C/D |
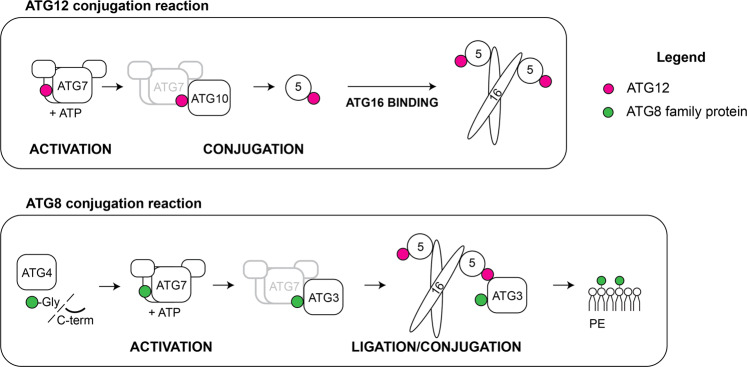
The lipidation of ATG8 proteins occurs analogous to the conjugation of ubiquitin to target proteins (Fig. 2)11,21. First the C-terminus of the ATG8 proteins becomes proteolytically cleaved by ATG4 family proteases to expose a glycine residue22. ATG8 is subsequently transferred to a cysteine residue in the E1-like ATG7 under the consumption of ATP. From there ATG8 is transferred to the E2-like ATG3, which mediates the attachment of ATG8 via its C-terminal glycine to the headgroup of PE. This last step is promoted by the ATG12–ATG5–ATG16 complex, which acts in an E3-like manner and which is itself the product of a ubiquitin-like conjugation reaction (Fig. 2)12,23,24. To form the complex, the ubiquitin-like ATG12 is activated by ATG7, transferred to the E2-like ATG10 and from there to an internal lysine residue of ATG5. The ATG12–ATG5 conjugate subsequently associates with the dimeric coiled-coil protein ATG1625–27. Although in vitro ATG8 conjugation can occur in the absence of the E3-like complex, its presence vastly accelerates the reaction and it is essential for ATG8 lipidation in cells12,23,25–27. The attachment of ATG8 proteins to PE is reversible as the ATG4 proteases can remove them from the membrane28,29.
Fig. 2. The ATG12 and ATG8 ubiquitin-like conjugation systems.
Similar to the classical ubiquitination cascades, the ubiquitin-like proteins ATG12 and ATG8 undergo a series of enzymatic reactions. ATG8 is first modified by the ATG4 protease and, likewise for ATG12, is activated by the E1-like enzyme ATG7 with consumption of ATP (ACTIVATION). ATG8 and ATG12 are then conjugated to the target protein (ATG5) or lipid (Phosphatidylethanolamine, PE) by the cognate E2-like enzymes (ATG10 and ATG3) (CONJUGATION). The last step of ATG8 lipidation is stimulated by the E3-like ligase ATG12–ATG5–ATG16 complex (LIGATION), while the ATG12 conjugation system lacks a known enzyme for the E3-like ligase activity. The ATG12–ATG5 conjugate interacts with the coiled-coil protein ATG16 via the ATG5 subunit, and further assembles into a hexamer. Legend: the ATG8 protein family members are indicated in green and the ATG12 proteins are indicated in magenta.
The targeting and activation of ATG8 lipidation are genetically the most downstream events within the cascade of the core autophagy machinery depending on the presence of the upstream acting factors30,31. A crucial event is the recruitment of the E3-like complex. In autophagy the main mechanism for its recruitment is via the PI3KC3-C1-WIPI2 axis, where the PI3P generated by the PI3KC3-C1 recruits the WIPI2 proteins, which in turn recruit and activate the ATG12–ATG5–ATG16L1 complex and thus ATG8 lipidation32–34. Other mechanisms to recruit the E3 to the site of autophagosome biogenesis via the ULK1/2 kinase complex35–37, membrane binding23,38,39 and cargo receptor interaction40 do also exist and will be discussed in detail below.
The functions of the ATG8 proteins in autophagy are manifold and even though their lipidation is a downstream event they can play important roles in the recruitment of upstream components of the autophagy machinery via feedback loops41. Mechanistically, most of the many roles of ATG8 in autophagy can be attributed to their functions as binding platforms for other proteins42,43, in particular once they are attached to the membrane44. These interactions are generally, but not exclusively45,46, mediated by so-called LC3-interacting regions (LIRs, also referred to as ATG8-intercating motifs (AIMs) or LC3 recognition sequence (LRS))47–49. These LIR motifs, which are typically composed of two hydrophobic amino acids spaced by two random residues and preceded by negatively charged residues are located in unstructured regions of the ATG8-interacting proteins. While it is possible to predict LIR motifs with considerable confidence, the variations of these motifs necessitate their experimental confirmation50,51-53. The two hydrophobic residues within the LIR motifs bind to two hydrophobic pockets in the ATG8s54. The affinities of the LIR motifs for the ATG8 proteins are generally moderate with Kds in the low µM range55. However, due to the fact that the ATG8 proteins become highly concentrated on the nascent autophagosomal membrane in the course of their conjugation, these membranes efficiently attract proteins harboring LIR motifs44,56. Many core autophagy factors including the ULK1/Atg1 kinase, FIP200, ATG13, VPS34, Beclin, ATG14, ATG2, ATG3, ATG4 and the Atg12–Atg5–Atg16 complex contain LIR motifs57–64 (reviewed in52). The autophagy machinery thereby generates a positive feedback loop by first recruiting and activating the ATG8 lipidation machinery, which in turn serves to recruit further upstream factors (Fig. 2). However, the ATG4 proteins, which catalyze the reverse reaction i.e. ATG8 delipidation are also recruited to the membrane via their interaction with ATG862,63. Presumably the activity of the ATG4 proteins is inhibited by the activity of the ULK1/2 kinase before autophagosomes are completed65.
In addition to the core autophagy machinery, ATG8 proteins recruit factors to the completed autophagosome, which are important for the trafficking and fusion with lysosomes (Fig. 1)66,67. The ATG8 proteins of the GABARAP subfamily are particularly important in this respect in human cells68. Among the factors recruited are PLEKHM169, SNAREs70,71, FYCO172, BRUCE73, the HOPS complex74, the Mon1–Ccz1 complex75. In combination, these proteins and protein complexes mediate the recruitment of molecular motors that bring the completed autophagosomes in proximity to lysosomes, recruit and activate Rab GTPases, aid the tethering of the autophagosomes to the lysosomal membrane and recruit SNARE proteins that execute the final membrane fusion event (Fig. 1) (reviewed in66,67,76).
Apart from serving as docking platforms, ATG8 proteins have intrinsic membrane tethering and remodeling activities, which may aid the closure of isolation membranes to form autophagosomes and fusion with lysosomes. When concentrated on membranes, S. cerevisiae Atg8, C. elegans LGG-1 and LGG-2 as well as human LC3B, GABARAP, and GABARAPL2 efficiently tether small and giant unilamellar vesicles in vitro39,77–81. At least for Atg8 and LC3B, membrane fusion required high PE concentrations82. Yeast Atg8 was also reported to induce membrane curvature83. Furthermore, ATG8s were observed to mediate either hemifusion, a state where only the two contacting monolayers of the two opposing membranes fuse and thus no content mixing occurs84, or full fusion77–79. The membrane remodeling activities were attributed to the two N-terminal helices, which represent ATG8 unique extension of the ubiquitin fold and which are only loosely packed to the core of the globular proteins85. The loose packing of the N-terminus is particularly important for the fusogenic properties of the proteins79.
In summary, ATG8 proteins play a plethora of roles during the biogenesis of autophagosomes starting from early stages after autophagosome nucleation, via isolation membrane expansion, its closure, the transport of autophagosomes along microtubules to lysosomes until the final fusion event (Fig. 1). In addition, the ATG8 conjugation machinery is important to render the inner autophagosomal membrane an efficient target for lysosomal hydrolases86.
While autophagy was initially thought to be largely non-selective, it has become clear that the process can be highly specific with regard to the cargo that is captured by autophagosomes. This is particularly true for autophagosomes that are formed in the absence of starvation, when autophagy is induced by the presence of specific cargo and not by mTOR inactivation87. It is beyond the scope of this review to list all known selective autophagy pathways that degrade among other materials damaged mitochondria, bacterial pathogens and aggregated proteins88–91. The basis for selectivity in autophagy is at least in part mediated by the tethering of the cargo to the nascent autophagosomal membrane. This mechanism acts in conjunction with the local activation of autophagosome formation at the cargo40,92–97. Most cargoes do not directly bind the membrane but are linked to it by cargo receptors, which simultaneously bind the cargo and ATG8 proteins that decorate the membrane. A plethora of cargo receptors has been identified. The most extensively studied ones are the yeast Atg19 as well as the human p62/SQSTM1, NDP52, Optineurin and NBR1 proteins. These proteins are soluble and are recruited to the cargo on demand. There is also a growing number of adapter proteins that are embedded in the membrane of organelles, most notably the endoplasmic reticulum, and which have the ability to bind to ATG8 proteins. A comprehensive list of cargo receptors and their discussion is provided in the following reviews52,66,90. Many cargo receptors are dimeric or multimeric, and some of them have more than one LIR motif56,98–102, resulting in a high avidity interaction with ATG8 proteins when they are concentrated on the membrane44,56. ATG8-decorated membranes are therefore highly efficient recruiters of proteins, protein complexes and polymers that contain multiple LIR motifs.
A certain degree of functional specialization among the human ATG8 proteins does exist21,103. For example, autophagosome formation and their degradation triggered by starvation and by PINK1/Parkin proceeds normally in HeLa cells lacking LC3 proteins, while GABARAP proteins are important for the fusion of autophagosomes with lysosomes68,104. This defect in fusion was attributed to reduced recruitment of PLEKHM1 to completed autophagosomes68. PLEKHM1 is a protein linking ATG8 positive membranes with Rab7 on lysosomes and the HOPS complex69. It was shown that its ATG8-interacting motif binds preferentially to GABARAP as opposed to LC3B via a GABARAP interaction motif (GIM), which is a variant of the canonical LIR motif105. In C. elegans, the GABARAP-like LGG-2 is more important for autophagosome formation and their fusion with lysosomes than the LC3-like LGG-174,79. Thus, current evidence points to a more important role of the GABARAP proteins in autophagy compared to the LC3 subfamily. However, the LC3 proteins may have crucial roles in selective autophagy pathways. For example, the degradation of p62 upon starvation-induced autophagy depends on the LC3 and GABARAP proteins68,106,107. LC3C is special among the ATG8 proteins as it binds to a noncanonical LIR motif termed CLIR for LC3C-interacting region, which is present in the autophagy receptor NDP52. LC3C and a functional CLIR motif were shown to be important for antibacterial autophagy108.
Given the many roles of the ATG8 proteins during autophagosome formation and cargo capture it is surprising that at least in some mammalian cells the presence of these proteins and their conjugation is not essential for autophagosome formation and the selective capture of mitochondria in PINK1/Parkin induced mitophagy68,86. This is in line with the fact that knockout mice lacking the ATG8 conjugation machinery are born viable109–112, while mice lacking components of the upstream machinery tend to have more severe phenotypes113. In contrast, in yeast Atg8 and its lipidation are essential for autophagosome formation16. How can the discrepancy between the finding that ATG8 proteins are defining features of autophagic membranes also in mammalian cells, that attract the autophagy machinery and cargo receptors be reconciled with their non-essentiality? First, the lipidation of ATG8 proteins is the most downstream step among the events occurring after initiation of autophagy30,31. Thus, autophagosomes are likely nucleated in the absence of ATG8 lipidation even in wild type cells. Redundant mechanisms for the recruitment of further autophagy factors during the expansion of isolation membranes must therefore exist in form of protein–lipid and protein–protein interactions. With regard to selective autophagy, ATG8s are indeed essential for the degradation of p62 upon starvation68. In PINK1/Parkin the induction of autophagosome formation occurs in vicinity of the mitochondria via the recruitment of FIP200 and the ULK1/2 complex by NDP5295. Therefore, in this and perhaps other forms of cargo-induced selective autophagy94,96,97 where the membrane is already in vicinity of the cargo from the beginning of the process the interaction of cargo receptors with ATG8 proteins might not be required to engulf the cargo. Instead, other proteins present on the cargo might link the cargo to the membrane. For example, cargo receptors bind components of the autophagy machinery, which in turn bind the membrane94–97,114. Regardless, the fact that the ATG8 proteins and their conjugation is not essential for autophagosome biogenesis and for the selection of certain cargoes does not mean that they are not important factors during these processes as most forms of macroautophagy are associated with ATG8 lipidation. It appears that the pathway is robust enough, such that it can proceed even in the absence of some core factors, albeit in a less efficient manner.
ATG8 conjugation to single membranes
For a long time, the process of ATG8 protein lipidation was considered a hallmark of autophagy that is exclusively associated with autophagosomes or their precursors. Indeed, many assays to follow the process of autophagy are based on the fact that these proteins are covalently coupled to these membranes and that a fraction of these proteins, which is present on the inner autophagosomal membrane is finally degraded within lysosomes115,116. However, it has become evident that ATG8 proteins are also present on membranes that are not associated with autophagosomes. The conjugation of ATG8 proteins to single membranes was first reported for a process called entosis, wherein one cell takes up another cell by a special form of phagocytosis, and for micropinocytosis117. Since then many endocytic events have been reported to be accompanied by ATG8 protein lipidation and they are commonly referred to as LC3-ascociated phagocytosis (LAP). ATG8 conjugation in LAP is independent of the ULK1/2 complex and ATG9 but requires the conjugation machinery including ATG5, ATG7 and ATG16, ROS production, and at least under certain conditions the activity of the PI3KC3 but not ATG14118–125. The function of ATG8 proteins on the endocytic organelles appears to be the acceleration of cargo degradation117. In addition to the plasma membrane-derived phagocytic and endocytic structures, ATG8 proteins have also been observed on endosomal and lysosomal membranes126,127. The distinction between endocytic and endo-lysosomal membranes is not always trivial as the membranes of endocytic vesicles and phagosomes fuse with endosomes and lysosomes. Agents that cause membrane damage are sufficient to elicit ATG8 lipidation on endo-lysosomal membranes suggesting that membrane defects are activators of the conjugation machinery126,127. The ROS produced during LAP may have a similar effect by oxidizing lipids and thus altering the membrane structure128. ATG8 protein conjugation has also been associated with the autophagic degradation of damaged endo-lysosomal structures by lysophagy129,130. Some of the ATG8 protein lipidation observed in lysophagy may actually correspond to their direct lipidation to the lysosomal membrane. Furthermore, it was demonstrated that ATG8 conjugation can also occur on the plasma membrane upon its damage by pathogen-derived factors and further that the presence of the lipidated ATG8 proteins aids plasma membrane repair131. ATG8 protein conjugation was also shown to be involved in various forms of non-conventional secretion including the release of exosomes and cytokines132–140.
A further non-autophagic function has been described for LC3C, which promotes COPII-dependent ER export via its interaction with TECPR2141. This is somewhat reminiscent for the intra-Golgi trafficking functions reported for GABARAP and GABARAPL2 (also known as GATE-16)6,142,143.
In summary, ATG8 proteins and their lipidation machinery have various functions that are not directly related to autophagy. Therefore, care needs to be taken when interpreting phenotypes of knockout studies and when following the production of lipidated ATG8 proteins as measures for autophagic activity.
Mechanisms regulating the targeting and activation of ATG8 lipidation
Given the many roles of the ATG8 proteins in various pathways, the question arises how their conjugation is targeted and activated at the right time and place. All conjugation events appear to be dependent on the ATG7/ATG3/ATG12–ATG5-ATG16 cascade and thus analogous to classical ubiquitination reactions requiring the activities of an E1, E2, and E3 (Fig. 1)21,144. The protein, which transfers ATG8 to PE is the E2-like ATG3. In vivo, yeast Atg3 has been localized together with Atg8 at the PAS and on the growing isolation membrane145. In vitro, the presence of ATG7 and ATG3 is sufficient for lipidation to occur on small unilamellar vesicles (SUVs) containing a high percentage of PE.
However, the activity of ATG3 is vastly stimulated by the ATG12–ATG5–ATG16 complex in vitro and all subunits of this protein complex are essential for detectable ATG8 conjugation in cells12. For the yeast conjugation system Atg16 is not required for the E3 activity of the complex in vitro, while for the mammalian system it is important, showing differences in the mechanism of action between the yeast and mammalian E3-like complex12,23,34,39.
Structural studies of the yeast and human conjugation machineries showed that ATG8 proteins are first bound by the extreme C-terminal domain (ECTD) of ATG7 through hydrophobic and aromatic residues that insert deeply into the two hydrophobic pockets of the ubiquitin-like fold, in a mode similar to that described for LIR motifs146. Subsequently, the substrates are transferred to the adenylation site (AD) of the ATG7 N-terminal domain containing the catalytic cysteine. ATG7 is a homodimer and binds the cognate E2s followed by conformational changes allowing the juxtaposition of the active site in the E2 with that of the E1. Thereby the substrate is transferred from ATG7 to ATG3146–148. The actual lipidation reaction is promoted by the ATG12–ATG5–ATG16 complex. The interaction of the complex and ATG3 is mediated by a patch in ATG12 and a loop in ATG3149. Details of the mechanisms of the E2 activation by the E3-like complex enzyme have been recently further elucidated. The so-called E123IR (E1-2-3 Interacting Region) in yeast Atg3 acts as an allosteric switch that initially interacts intra-molecularly locking the protein in an inactive state. Upon interaction with the Atg12–Atg5–Atg16 complex his inhibition is released allowing ATG8 attachment to PE150.
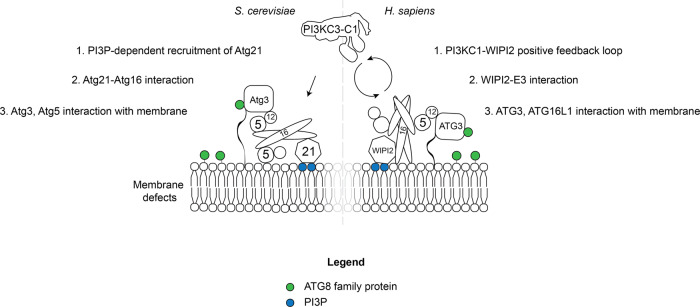
In order to correctly target ATG8 lipidation, the substrate i.e. the membrane has to be permissive for the activity of ATG3 and the ATG12–ATG5–ATG16 complex needs to be present. In autophagy, the main determinant for the recruitment of the E3 is the ATG16 subunit (Fig. 3)30,31,151. In mammalian cells ATG16L1 is bound by the WIPI2 proteins, which in turn are recruited to the membrane by PI3P produced by the PI3KC3-C132. In this manner, its recruitment is coupled to the activity of the lipid kinase complex. Additional targeting mechanisms also exists and likely work in conjunction with WIPI2 to robustly recruit and activate the E3 (Fig. 3)34. These mechanisms include the binding of ATG16L1 to FIP20035,36, which is a subunit of the ULK1/2 kinase complex152. In addition, ATG16L1 can directly bind to membranes that contain PI3P via two sites (Fig. 3)23,34,38. One site is located in the coiled-coil domain38 and the other site maps to a region C-terminal of this domain23. The extreme C-terminal WD domain of ATG16L1 is not required for its targeting in autophagy153. In combination, these targeting mechanisms increase the local concentration of the E3 and will thereby locally promote ATG8 lipidation. There may be additional activation mechanisms beyond the mere recruitment of the E3. For example, WIPI2 can directly activate the activity of the conjugation machinery (Fig. 3)34.
Fig. 3. Activation and localization of the ATG8 conjugation machinery in macroautophagy.
Schematic representation of the cascade that leads to ATG8 lipidation on the target membrane. In yeast and mammals PI3KC3-C1 activity is necessary to produce PI3P and to recruit PI3P-sensor proteins (Atg21 or WIPIs) to the pre-autophagosomal membrane. These proteins in turn bind the E3-like ligase via the ATG16 subunit and thereby recruit and activate the E3-like ligase activity. Correct membrane localization of the lipidation reaction is also mediated by the E2-like ATG3 that directly interacts with the ATG12 subunit of the E3 and its N-terminal amphipathic helix that inserts into the lipid bilayer (shown as black line). In yeast, ATG5 was shown to directly interact with poorly packed membranes, while in mammals ATG16L1 binds the membrane. Legend: the ATG8 protein family members are shown in green and the PI3P lipid is shown in blue.
In yeast, the situation is similar as the interaction of Atg16 with the WIPI2 homolog Atg21 is important for the recruitment of Atg12–Atg5–Atg16 complex to the site of autophagosome formation33. However, notable differences exist. Atg21 interacts with the coiled-coil domain of Atg16 whereas WIPI2 binds to a motif C-terminal of this domain in ATG16L132,33. The yeast the E3 can also bind to membranes but this interaction is mediated by the Atg5 subunit (Fig. 3)39. Furthermore, while Atg12–Atg5–Atg16 also interacts with the Atg1 kinase complex, the equivalent of the ULK1/2 complex, this interaction is mediated by the Atg12 subunit37.
Some specificity of the lipidation reaction may also be conferred by ATG3. Apart from the requirement for PE, membranes that display a high degree of membrane curvature, contain lipids with small headgroups or have membrane defects all facilitate ATG8 lipidation, likely by allowing insertion of amphipathic helix of ATG3 into the lipid bilayer39,154,155. Loosely, packed membranes containing a high percentage of lipids with unsaturated acyl chains appear to promote ATG8 conjugation at multiple levels34, consistent with their high abundance in autophagosomal membranes at least in yeast cells156.
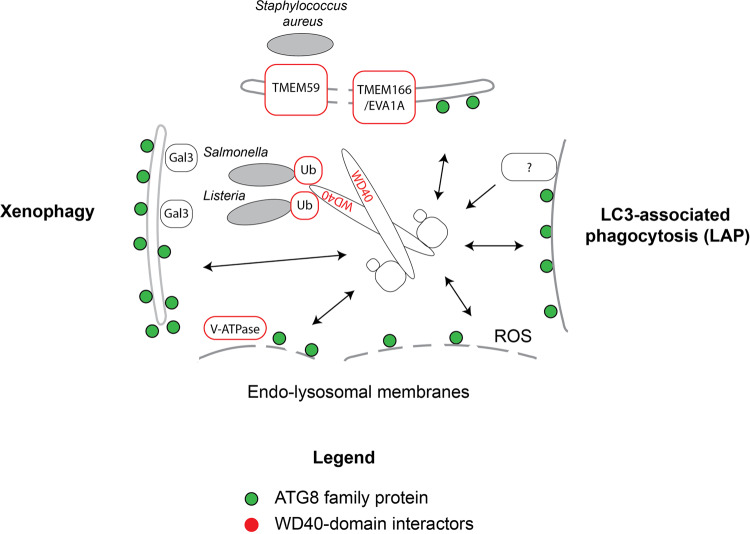
In contrast to the situation in autophagy, where a wealth of knowledge about the targeting of ATG8 lipidation exists, little is known about how this reaction is targeted in autophagy-independent events. The main difference is that the C-terminal WD40 domain of ATG16L1 is dispensable for ATG8 lipidation in autophagy but essential for ATG8 conjugation in LAP and on endosomes and lysosomes (Fig. 4)153,157. Potential recruiting factors of ATG16L1 via the WD40 domain are the TMEM166/EVA1A158, the TMEM59 protein159,160, the V-ATPase161 and ubiquitin, which is present of vacuoles containing bacterial pathogens162,163 (Fig. 4). In addition, it was shown that galectin-3 recruits ATG16L1 via TRIM16 to damaged lysosomes164,165. The C-terminal membrane binding site in ATG16L1 was also shown to promote its membrane recruitment and ATG8 lipidation induced by lysosomal damage23. It is conceivable that a combination of a proteinaceous recruitment factor in conjunction with hydrophobic defects such as caused by membrane damage and acyl chain oxidization is the most effective way to recruit the ATG12–ATG5–ATG16L1 complex and thus to elicit ATG8 lipidation. Perhaps, the lipidated ATG8 proteins first recruit factors for membrane repair and if this fails, cargo receptors and the autophagy machinery that mediate autophagic engulfment of the damaged membrane structure.
Fig. 4. ATG8 conjugation to single membranes in non-autophagic events.
Cartoon summarizing the recruitment of the ATG16L1 protein in ATG8 conjugation events that are not directly related to autophagy. The arrows indicate the recruitment of the ATG12–ATG5–ATG16L1 complex to single membranes. The factors recruiting the complex via the WD40 domain in ATG16L1 are highlighted in red.
In addition to the forward reaction catalyzed by the lipidation machinery, the reverse reaction in form of ATG4-mediated delipidation is a potential mechanism for the control of ATG8 protein density on the membrane. In fact, it was shown that in yeast inappropriately lipidated ATG8 is removed from membrane by Atg4166. Therefore, local activation of ATG8 lipidation in conjunction with inhibition of the ATG4-mediated reverse reaction may contribute to the control of the ATG8 densities on the membrane. This mechanism was already suggested during autophagosome biogenesis in yeast, where the Atg1 activity inhibits Atg465.
Conclusion
Considering the many factors that interact with ATG8 proteins in various pathway a perhaps obvious question is how the recruitment of the different factors is spatially and temporally regulated. For example, why are not all LIR containing autophagy factors recruited to the phagosomes during LAP and why are the cargo receptors and therefore the cargo attached to the inner but not the outer autophagosomal membrane? These questions are currently difficult to answer. Kinases that act together with antagonizing phosphatases are likely regulators. For example, TBK1 may locally increase the affinity of LIR motifs167 while phosphatases might quickly remove the phosphate. Furthermore, cargo receptors are frequently dimeric or even multimeric and will therefore select for surfaces with a high density of ATG8 proteins. They will also tend to have lower off-rates than monomeric ATG8 binders due to avidity effects. In contrast, monomeric proteins with higher affinities of their LIR motifs for ATG8 or higher on-rates might be preferentially recruited to membrane containing lower densities of ATG8. In addition, during autophagosome formation, the future outer membrane can make extensive and intimate contact with the ER and might therefore not be well accessible for cargo receptors168,169. Upon autophagosome completion a fraction of the ATG8 proteins is removed from the outer membrane, at least in yeast170. Therefore, the outer membrane may no longer be an attractive target for the receptors.
Since their original discovery the known roles of the ATG8 proteins have continuously expanded and more will certainly be discovered. This is reminiscent to ubiquitin itself, which was initially discovered as degradation tag in the ubiquitin-proteasome system and which is now implicated in virtually every complex cellular pathway171–173. ATG8 proteins are likely to keep surprising us with their cellular functions.
Acknowledgements
The authors thank Michael Lazarou for information about the expression of human ATG8 proteins. This work was supported by Human Frontiers Science Program RGP0026/2017 and ERC grant No. 646653.
Author contributions
S.M. and D.F. wrote the text and prepared the figures.
Conflict of interest
S.M is member of the scientific advisory board of Casma Therapeutics.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 2.Scott SV, et al. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc. Natl Acad. Sci. 1996;93:12304. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lang T, et al. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuznetsov SA, Gelfand VI. 18 kDa microtubule-associated protein: identification as a new light chain (LC-3) of microtubule-associated protein 1 (MAP-1) FEBS Lett. 1987;212:145–148. doi: 10.1016/0014-5793(87)81574-0. [DOI] [PubMed] [Google Scholar]
- 5.Legesse-Miller A, Sagiv Y, Porat A, Elazar Z. Isolation and characterization of a novel low molecular weight protein involved in intra-golgi traffic. J. Biol. Chem. 1998;273:3105–3109. doi: 10.1074/jbc.273.5.3105. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Bedford FK, Brandon NJ, Moss SJ, Olsen RW. GABAA-receptor-associated protein links GABAA receptors and the cytoskeleton. Nature. 1999;397:69–72. doi: 10.1038/16264. [DOI] [PubMed] [Google Scholar]
- 7.Shpilka T, Weidberg H, Pietrokovski S, Elazar Z. Atg8: an autophagy-related ubiquitin-like protein family. Genome Biol. 2011;12:226. doi: 10.1186/gb-2011-12-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai H, Inoue J, Kawano T, Inazawa J. A transcriptional variant of the LC3A gene is involved in autophagy and frequently inactivated in human cancers. Oncogene. 2012;31:4397–4408. doi: 10.1038/onc.2011.613. [DOI] [PubMed] [Google Scholar]
- 9.Kellner R, De la Concepcion JC, Maqbool A, Kamoun S, Dagdas YF. ATG8 expansion: a driver of selective autophagy diversification? Trends Plant Sci. 2017;22:204–214. doi: 10.1016/j.tplants.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Paz Y, Elazar Z, Fass D. Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J. Biol. Chem. 2000;275:25445–25450. doi: 10.1074/jbc.C000307200. [DOI] [PubMed] [Google Scholar]
- 11.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 12.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 13.Oh-oka K, Nakatogawa H, Ohsumi Y. Physiological pH and acidic phospholipids contribute to substrate specificity in lipidation of Atg8. J. Biol. Chem. 2008;283:21847–21852. doi: 10.1074/jbc.M801836200. [DOI] [PubMed] [Google Scholar]
- 14.Sou Y-s, Tanida I, Komatsu M, Ueno T, Kominami E. Phosphatidylserine in addition to phosphatidylethanolamine is an in vitro target of the mammalian Atg8 modifiers, LC3, GABARAP, and GATE-16. J. Biol. Chem. 2006;281:3017–3024. doi: 10.1074/jbc.M505888200. [DOI] [PubMed] [Google Scholar]
- 15.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirisako T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mercer, T. J., Gubas, A. & Tooze, S. A. A molecular perspective of mammalian autophagosome biogenesis. J. Biol. Chem.293, 5386–5395 (2018). [DOI] [PMC free article] [PubMed]
- 18.Bento CF, et al. Mammalian autophagy: how does it work? Annu. Rev. Biochem. 2016;85:685–713. doi: 10.1146/annurev-biochem-060815-014556. [DOI] [PubMed] [Google Scholar]
- 19.Hurley JH, Young LN. Mechanisms of autophagy Initiation. Annu. Rev. Biochem. 2017;86:225–244. doi: 10.1146/annurev-biochem-061516-044820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev. Cell Dev. Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 21.Mizushima N. The ATG conjugation systems in autophagy. Curr. Opin. Cell Biol. 2020;63:1–10. doi: 10.1016/j.ceb.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Kirisako T, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000;151:263–276. doi: 10.1083/jcb.151.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lystad AH, et al. Distinct functions of ATG16L1 isoforms in membrane binding and LC3B lipidation in autophagy-related processes. Nat. Cell Biol. 2019;21:372–383. doi: 10.1038/s41556-019-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizushima N, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 25.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J. Biol. Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 26.Mizushima N, et al. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 2003;116:1679. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 27.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p-Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanida I, et al. HsAtg4B/HsApg4B/autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phospholipid conjugates. J. Biol. Chem. 2004;279:36268–36276. doi: 10.1074/jbc.M401461200. [DOI] [PubMed] [Google Scholar]
- 29.Kauffman KJ, et al. Delipidation of mammalian Atg8-family proteins by each of the four ATG4 proteases. Autophagy. 2018;14:992–1010. doi: 10.1080/15548627.2018.1437341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 31.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6:764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dooley HC, et al. WIPI2 Links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12-5-16L1. Mol. Cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Juris L, et al. PI3P binding by Atg21 organises Atg8 lipidation. EMBO J. 2015;34:955. doi: 10.15252/embj.201488957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fracchiolla, D., Chang, C., Hurley, J. H. & Martens, S. A PI3K-WIPI2 positive feedback loop allosterically activates LC3 lipidation in autophagy. bioRxiv, 2019.2012.2018.880591, 10.1101/2019.12.18.880591 (2019). [DOI] [PMC free article] [PubMed]
- 35.Gammoh N, Florey O, Overholtzer M, Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex–dependent and –independent autophagy. Nat. Struct. Mol. Biol. 2013;20:144–149. doi: 10.1038/nsmb.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimura T, et al. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep. 2013;14:284–291. doi: 10.1038/embor.2013.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harada K, et al. Two distinct mechanisms target the autophagy-related E3 complex to the pre-autophagosomal structure. eLife. 2019;8:e43088. doi: 10.7554/eLife.43088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudley LJ, et al. Intrinsic lipid binding activity of ATG16L1 supports efficient membrane anchoring and autophagy. EMBO J. 2019;38:e100554. doi: 10.15252/embj.2018100554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Romanov J, et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fracchiolla D, et al. Mechanism of cargo-directed Atg8 conjugation during selective autophagy. eLife. 2016;5:e18544. doi: 10.7554/eLife.18544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Padman BS, et al. LC3/GABARAPs drive ubiquitin-independent recruitment of Optineurin and NDP52 to amplify mitophagy. Nat. Commun. 2019;10:408. doi: 10.1038/s41467-019-08335-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wild P, McEwan DG, Dikic I. The LC3 interactome at a glance. J. Cell Sci. 2014;127:3. doi: 10.1242/jcs.140426. [DOI] [PubMed] [Google Scholar]
- 44.Wurzer B, et al. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. eLife. 2015;4:e08941. doi: 10.7554/eLife.08941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marshall RS, Hua Z, Mali S, McLoughlin F, Vierstra RD. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell. 2019;177:766–781.e724. doi: 10.1016/j.cell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krick R, et al. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J. Cell Biol. 2010;190:965–973. doi: 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pankiv S, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 48.Ichimura Y, et al. Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 2008;283:22847–22857. doi: 10.1074/jbc.M802182200. [DOI] [PubMed] [Google Scholar]
- 49.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 50.Xie Q, et al. hfAIM: A reliable bioinformatics approach for in silico genome-wide identification of autophagy-associated Atg8-interacting motifs in various organisms. Autophagy. 2016;12:876–887. doi: 10.1080/15548627.2016.1147668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Birgisdottir ÅB, Lamark T, Johansen T. The LIR motif—crucial for selective autophagy. J. Cell Sci. 2013;126:3237. doi: 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- 52.Johansen T, Lamark T. Selective autophagy: ATG8 family proteins, LIR motifs and cargo receptors. J. Mol. Biol. 2020;432:80–103. doi: 10.1016/j.jmb.2019.07.016. [DOI] [PubMed] [Google Scholar]
- 53.Kalvari I, et al. iLIR. Autophagy. 2014;10:913–925. doi: 10.4161/auto.28260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noda NN, et al. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 55.Rozenknop A, et al. Characterization of the Interaction of GABARAPL-1 with the LIR Motif of NBR1. J. Mol. Biol. 2011;410:477–487. doi: 10.1016/j.jmb.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 56.Sawa-Makarska, J.et al. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane–cargo apposition during selective autophagy. Nat. Cell Biol.16, 425–433, http://www.nature.com/ncb/journal/v16/n5/abs/ncb2935.html#supplementary-information (2014). [DOI] [PMC free article] [PubMed]
- 57.Bozic, M. et al. A conserved ATG2-GABARAP family interaction is critical for phagophore formation. EMBO Rep. e201948412, 10.15252/embr.201948412 (2020). [DOI] [PMC free article] [PubMed]
- 58.Alemu EA, et al. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements foR LC3-interacting region (LIR) motifs. J. Biol. Chem. 2012;287:39275–39290. doi: 10.1074/jbc.M112.378109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kraft C, et al. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 2012;31:3691–3703. doi: 10.1038/emboj.2012.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Birgisdottir ÅB, et al. Members of the autophagy class III phosphatidylinositol 3-kinase complex I interact with GABARAP and GABARAPL1 via LIR motifs. Autophagy. 2019;15:1333–1355. doi: 10.1080/15548627.2019.1581009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaufmann A, Beier V, Franquelim HenriG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469–481. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 62.Skytte Rasmussen M, et al. ATG4B contains a C-terminal LIR motif important for binding and efficient cleavage of mammalian orthologs of yeast Atg8. Autophagy. 2017;13:834–853. doi: 10.1080/15548627.2017.1287651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abreu S, et al. Conserved Atg8 recognition sites mediate Atg4 association with autophagosomal membranes and Atg8 deconjugation. EMBO Rep. 2017;18:765–780. doi: 10.15252/embr.201643146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sakoh-Nakatogawa M, Kirisako H, Nakatogawa H, Ohsumi Y. Localization of Atg3 to autophagy-related membranes and its enhancement by the Atg8-family interacting motif to promote expansion of the membranes. FEBS Lett. 2015;589:744–749. doi: 10.1016/j.febslet.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 65.Sanchez-Wandelmer J, et al. Atg4 proteolytic activity can be inhibited by Atg1 phosphorylation. Nat. Commun. 2017;8:295. doi: 10.1038/s41467-017-00302-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 2014;16:495–501. doi: 10.1038/ncb2979. [DOI] [PubMed] [Google Scholar]
- 67.Lőrincz, P. & Juhász, G. Autophagosome-lysosome fusion. J. Mol. Biol.10.1016/j.jmb.2019.10.028 (2019). [DOI] [PubMed]
- 68.Nguyen TN, et al. Atg8 family LC3/GABARAP proteins are crucial for autophagosome–lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol. 2016;215:857–874. doi: 10.1083/jcb.201607039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McEwan DavidG, et al. PLEKHM1 Regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 2015;57:39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 70.Gu Y, et al. Mammalian Atg8 proteins regulate lysosome and autolysosome biogenesis through SNAREs. EMBO J. 2019;38:e101994. doi: 10.15252/embj.2019101994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kumar S, et al. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J. Cell Biol. 2018;217:997–1013. doi: 10.1083/jcb.201708039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pankiv S, et al. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end–directed vesicle transport. J. Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ebner P, et al. The IAP family member BRUCE regulates autophagosome–lysosome fusion. Nat. Commun. 2018;9:599. doi: 10.1038/s41467-018-02823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Manil-Ségalen M, et al. The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev. Cell. 2014;28:43–55. doi: 10.1016/j.devcel.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 75.Gao J, Langemeyer L, Kümmel D, Reggiori F, Ungermann C. Molecular mechanism to target the endosomal Mon1-Ccz1 GEF complex to the pre-autophagosomal structure. eLife. 2018;7:e31145. doi: 10.7554/eLife.31145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kriegenburg F, Ungermann C, Reggiori F. Coordination of autophagosome-lysosome fusion by Atg8 family members. Curr. Biol. 2018;28:R512–R518. doi: 10.1016/j.cub.2018.02.034. [DOI] [PubMed] [Google Scholar]
- 77.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 78.Weidberg H, et al. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Developmental Cell. 2011;20:444–454. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 79.Wu F, et al. Structural basis of the differential function of the two C. elegans Atg8 homologs, LGG-1 and LGG-2, in autophagy. Mol. Cell. 2015;60:914–929. doi: 10.1016/j.molcel.2015.11.019. [DOI] [PubMed] [Google Scholar]
- 80.Landajuela A, et al. Lipid geometry and bilayer curvature modulate LC3/GABARAP-mediated model autophagosomal elongation. Biophysical J. 2016;110:411–422. doi: 10.1016/j.bpj.2015.11.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taniguchi, S., Toyoshima, M., Takamatsu, T. & Mima, J. Curvature-sensitive trans-assembly of human Atg8-family proteins in autophagy-related membrane tethering. bioRxiv, 870857, 10.1101/870857 (2020). [DOI] [PMC free article] [PubMed]
- 82.Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knorr RL, et al. Membrane morphology is actively transformed by covalent binding of the protein Atg8 to PE-Lipids. PLoS ONE. 2014;9:e115357. doi: 10.1371/journal.pone.0115357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martens S, McMahon H. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 2008;9:543–556. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 85.Kumeta H, et al. The NMR structure of the autophagy-related protein Atg8. J. Biomolecular NMR. 2010;47:237–241. doi: 10.1007/s10858-010-9420-1. [DOI] [PubMed] [Google Scholar]
- 86.Tsuboyama K, et al. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 2016;354:1036. doi: 10.1126/science.aaf6136. [DOI] [PubMed] [Google Scholar]
- 87.Zaffagnini G, Martens S. Mechanisms of selective autophagy. J. Mol. Biol. 2016;428:1714–1724. doi: 10.1016/j.jmb.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gomes LC, Dikic I. Autophagy in antimicrobial immunity. Mol. Cell. 2014;54:224–233. doi: 10.1016/j.molcel.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 89.Anding AL, Baehrecke EH. Cleaning house: selective autophagy of organelles. Developmental Cell. 2017;41:10–22. doi: 10.1016/j.devcel.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kirkin V, Rogov VV. A diversity of selective autophagy receptors determines the specificity of the autophagy pathway. Mol. Cell. 2019;76:268–285. doi: 10.1016/j.molcel.2019.09.005. [DOI] [PubMed] [Google Scholar]
- 91.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kamber RA, Shoemaker CJ, Denic V. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol. Cell. 2015;59:372–381. doi: 10.1016/j.molcel.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Turco, E., Fracchiolla, D. & Martens, S. Recruitment and activation of the ULK1/Atg1 kinase complex in selective autophagy. J. Mol. Biol.10.1016/j.jmb.2019.07.027 (2019). [DOI] [PMC free article] [PubMed]
- 94.Turco E, et al. FIP200 claw domain binding to p62 promotes autophagosome formation at ubiquitin condensates. Mol. Cell. 2019;74:330–346.e311. doi: 10.1016/j.molcel.2019.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Vargas JNS, et al. Spatiotemporal control of ULK1 activation by NDP52 and TBK1 during selective autophagy. Mol. Cell. 2019;74:347–362.e346. doi: 10.1016/j.molcel.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ravenhill BJ, et al. The cargo receptor NDP52 initiates selective autophagy by recruiting the ULK complex to cytosol-invading bacteria. Mol. Cell. 2019;74:320–329.e326. doi: 10.1016/j.molcel.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Smith MD, et al. CCPG1 Is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Developmental Cell. 2018;44:217–232.e211. doi: 10.1016/j.devcel.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abert C, Kontaxis G, Martens S. Accessory interaction motifs in the Atg19 cargo receptor enable strong binding to the clustered ubiquitin-related Atg8 protein. J. Biol. Chem. 2016;291:18799–18808. doi: 10.1074/jbc.M116.736892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kirkin V, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 100.Ciuffa R, et al. The selective autophagy receptor p62 forms a flexible filamentous helical scaffold. Cell Rep. 2015;11:748–758. doi: 10.1016/j.celrep.2015.03.062. [DOI] [PubMed] [Google Scholar]
- 101.Kim B-W, Beom Hong S, Hoe Kim J, Hoon Kwon D, Kyu Song H. Structural basis for recognition of autophagic receptor NDP52 by the sugar receptor galectin-8. Nat. Commun. 2013;4:1613. doi: 10.1038/ncomms2606. [DOI] [PubMed] [Google Scholar]
- 102.Li F, et al. Structural insights into the interaction and disease mechanism of neurodegenerative disease-associated optineurin and TBK1 proteins. Nat. Commun. 2016;7:12708. doi: 10.1038/ncomms12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Szalai P, et al. Autophagic bulk sequestration of cytosolic cargo is independent of LC3, but requires GABARAPs. Exp. Cell Res. 2015;333:21–38. doi: 10.1016/j.yexcr.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 104.Vaites LP, Paulo JA, Huttlin EL, Harper JW. Systematic analysis of human cells lacking ATG8 Proteins uncovers roles for GABARAPs and the CCZ1/MON1 regulator C18orf8/RMC1 in macroautophagic and selective autophagic flux. Mol. Cell. Biol. 2018;38:e00392–e00317. doi: 10.1128/MCB.00392-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rogov VV, et al. Structural and functional analysis of the GABARAP interaction motif (GIM) EMBO Rep. 2017;18:1382–1396. doi: 10.15252/embr.201643587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Weidberg H, et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Maruyama Y, et al. LC3B is indispensable for selective autophagy of p62 but not basal autophagy. Biochemical Biophysical Res. Commun. 2014;446:309–315. doi: 10.1016/j.bbrc.2014.02.093. [DOI] [PubMed] [Google Scholar]
- 108.von Muhlinen N, et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell. 2012;48:329–342. doi: 10.1016/j.molcel.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 111.Yoshii SaoriR, et al. Systemic analysis of Atg5-null mice rescued from neonatal lethality by transgenic ATG5 expression in neurons. Developmental Cell. 2016;39:116–130. doi: 10.1016/j.devcel.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 112.Sou Y-s, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 2008;19:4762–4775. doi: 10.1091/mbc.e08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kuma A, Komatsu M, Mizushima N. Autophagy-monitoring and autophagy-deficient mice. Autophagy. 2017;13:1619–1628. doi: 10.1080/15548627.2017.1343770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de la Ballina, L. R., Munson, M. J. & Simonsen, A. Lipids and lipid-binding proteins in selective autophagy. J. Mol. Biol.10.1016/j.jmb.2019.05.051 (2019). [DOI] [PubMed]
- 115.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat. Cell Biol. 2011;13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Heckmann BL, et al. LC3-associated endocytosis facilitates β-amyloid clearance and mitigates neurodegeneration in murine Alzheimer’s disease. Cell. 2019;178:536–551.e514. doi: 10.1016/j.cell.2019.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Heckmann BL, Boada-Romero E, Cunha LD, Magne J, Green DR. LC3-associated phagocytosis and inflammation. J. Mol. Biol. 2017;429:3561–3576. doi: 10.1016/j.jmb.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martinez J. LAP it up, fuzz ball: a short history of LC3-associated phagocytosis. Curr. Opin. Immunol. 2018;55:54–61. doi: 10.1016/j.coi.2018.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Heckmann BL, Green DR. LC3-associated phagocytosis at a glance. J. Cell Sci. 2019;132:jcs222984. doi: 10.1242/jcs.222984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Martinez J, et al. Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 2015;17:893–906. doi: 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 123.Martinez J, et al. Noncanonical autophagy inhibits the autoinflammatory, lupus-like response to dying cells. Nature. 2016;533:115–119. doi: 10.1038/nature17950. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 124.Martinez J, et al. Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells. Proc. Natl Acad. Sci. 2011;108:17396. doi: 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 125.Fazeli G, Stetter M, Lisack JN, Wehman AM. C. elegans blastomeres clear the corpse of the second polar body by LC3-associated phagocytosis. Cell Rep. 2018;23:2070–2082. doi: 10.1016/j.celrep.2018.04.043. [DOI] [PubMed] [Google Scholar]
- 126.Florey O, Gammoh N, Kim SE, Jiang X, Overholtzer M. V-ATPase and osmotic imbalances activate endolysosomal LC3 lipidation. Autophagy. 2015;11:88–99. doi: 10.4161/15548627.2014.984277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jacquin E, et al. Pharmacological modulators of autophagy activate a parallel noncanonical pathway driving unconventional LC3 lipidation. Autophagy. 2017;13:854–867. doi: 10.1080/15548627.2017.1287653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang C-S, et al. Autophagy protein rubicon mediates phagocytic NADPH oxidase activation in response to microbial infection or TLR stimulation. Cell Host Microbe. 2012;11:264–276. doi: 10.1016/j.chom.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hung Y-H, Chen LM-W, Yang J-Y, Yuan Yang W. Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat. Commun. 2013;4:2111. doi: 10.1038/ncomms3111. [DOI] [PubMed] [Google Scholar]
- 130.Maejima I, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J. 2013;32:2336–2347. doi: 10.1038/emboj.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tan JMJ, et al. An ATG16L1-dependent pathway promotes plasma membrane repair and limits Listeria monocytogenes cell-to-cell spread. Nat. Microbiol. 2018;3:1472–1485. doi: 10.1038/s41564-018-0293-5. [DOI] [PubMed] [Google Scholar]
- 132.Leidal AM, et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 2020;22:187–199. doi: 10.1038/s41556-019-0450-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bel S, et al. Paneth cells secrete lysozyme via secretory autophagy during bacterial infection of the intestine. Science. 2017;357:1047. doi: 10.1126/science.aal4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.DeSelm CarlJ, et al. Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell. 2011;21:966–974. doi: 10.1016/j.devcel.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Guo H, et al. Atg5 disassociates the V1V0-ATPase to promote exosome production and tumor metastasis independent of canonical macroautophagy. Developmental Cell. 2017;43:716–730.e717. doi: 10.1016/j.devcel.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 136.Murrow L, Malhotra R, Debnath J. ATG12–ATG3 interacts with Alix to promote basal autophagic flux and late endosome function. Nat. Cell Biol. 2015;17:300–310. doi: 10.1038/ncb3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Dupont N, et al. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. J. Cell Biol. 2010;188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Manjithaya R, Anjard C, Loomis WF, Subramani S. Unconventional secretion of Pichia pastoris Acb1 is dependent on GRASP protein, peroxisomal functions, and autophagosome formation. J. Cell Biol. 2010;188:537–546. doi: 10.1083/jcb.200911149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Torisu T, et al. Autophagy regulates endothelial cell processing, maturation and secretion of von Willebrand factor. Nat. Med. 2013;19:1281–1287. doi: 10.1038/nm.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stadel D, et al. TECPR2 cooperates with LC3C to regulate COPII-dependent ER export. Mol. Cell. 2015;60:89–104. doi: 10.1016/j.molcel.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 142.Kittler JT, et al. The subcellular distribution of GABARAP and Its ability to interact with NSF suggest a role for this protein in the intracellular transport of GABAA receptors. Mol. Cell. Neurosci. 2001;18:13–25. doi: 10.1006/mcne.2001.1005. [DOI] [PubMed] [Google Scholar]
- 143.Sagiv Y, Legesse-Miller A, Porat A, Elazar Z. GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J. 2000;19:1494–1504. doi: 10.1093/emboj/19.7.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 145.Ngu M, Hirata E, Suzuki K. Visualization of Atg3 during autophagosome formation in Saccharomyces cerevisiae. J. Biol. Chem. 2015;290:8146–8153. doi: 10.1074/jbc.M114.626952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Noda NN, et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol. Cell. 2011;44:462–475. doi: 10.1016/j.molcel.2011.08.035. [DOI] [PubMed] [Google Scholar]
- 147.Kaiser SE, et al. Noncanonical E2 recruitment by the autophagy E1 revealed by Atg7-Atg3 and Atg7-Atg10 structures. Nat. Struct. Mol. Biol. 2012;19:1242–1249. doi: 10.1038/nsmb.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Yamada Y, et al. The crystal structure of Atg3, an autophagy-related ubiquitin carrier protein (E2) enzyme that mediates Atg8 lipidation. J. Biol. Chem. 2007;282:8036–8043. doi: 10.1074/jbc.M611473200. [DOI] [PubMed] [Google Scholar]
- 149.Metlagel Z, Otomo C, Takaesu G, Otomo T. Structural basis of ATG3 recognition by the autophagic ubiquitin-like protein ATG12. Proc. Natl Acad. Sci. 2013;110:18844. doi: 10.1073/pnas.1314755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zheng Y, et al. A switch element in the autophagy E2 Atg3 mediates allosteric regulation across the lipidation cascade. Nat. Commun. 2019;10:3600. doi: 10.1038/s41467-019-11435-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Fujita N, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell. 2008;19:2092–2100. doi: 10.1091/mbc.e07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008;181:497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fletcher K, et al. The WD40 domain of ATG16L1 is required for its non-canonical role in lipidation of LC3 at single membranes. EMBO J. 2018;37:e97840. doi: 10.15252/embj.201797840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nath S, et al. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat. Cell Biol. 2014;16:415–424. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hervás JH, et al. Human ATG3 binding to lipid bilayers: role of lipid geometry, and electric charge. Sci. Rep. 2017;7:15614. doi: 10.1038/s41598-017-15057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schütter, M., Giavalisco, P., Brodesser, S. & Graef, M. Local fatty acid channeling into phospholipid synthesis drives phagophore expansion during autophagy. Cell. 10.1016/j.cell.2019.12.005. [DOI] [PubMed]
- 157.Rai S, et al. The ATG5-binding and coiled coil domains of ATG16L1 maintain autophagy and tissue homeostasis in mice independently of the WD domain required for LC3-associated phagocytosis. Autophagy. 2019;15:599–612. doi: 10.1080/15548627.2018.1534507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hu J, et al. TMEM166/EVA1A interacts with ATG16L1 and induces autophagosome formation and cell death. Cell Death Dis. 2016;7:e2323–e2323. doi: 10.1038/cddis.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Boada-Romero E, et al. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J. 2013;32:566–582. doi: 10.1038/emboj.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Boada-Romero E, et al. The T300A Crohn’s disease risk polymorphism impairs function of the WD40 domain of ATG16L1. Nat. Commun. 2016;7:11821. doi: 10.1038/ncomms11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xu Y, et al. A bacterial effector reveals the V-ATPase-ATG16L1 axis that initiates xenophagy. Cells. 2019;178:552–566.e520. doi: 10.1016/j.cell.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 162.Fujita N, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J. Cell Biol. 2013;203:115–128. doi: 10.1083/jcb.201304188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ogawa M, et al. Streptococcus pneumoniae triggers hierarchical autophagy through reprogramming of LAPosome-like vesicles via NDP52-delocalization. Commun. Biol. 2020;3:25. doi: 10.1038/s42003-020-0753-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chauhan S, et al. TRIMs and galectins globally cooperate and TRIM16 and galectin-3 Co-direct autophagy in endomembrane damage homeostasis. Developmental Cell. 2016;39:13–27. doi: 10.1016/j.devcel.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Jia J, et al. Galectin-3 coordinates a cellular system for lysosomal repair and removal. Dev. Cell. 2020;52:69–87.e68. doi: 10.1016/j.devcel.2019.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nakatogawa H, Ishii J, Asai E, Ohsumi Y. Atg4 recycles inappropriately lipidated Atg8 to promote autophagosome biogenesis. Autophagy. 2012;8:177–186. doi: 10.4161/auto.8.2.18373. [DOI] [PubMed] [Google Scholar]
- 167.Wild P, et al. Phosphorylation of the autophagy receptor optineurin restricts salmonella growth. Science. 2011;333:228. doi: 10.1126/science.1205405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hayashi-Nishino M, et al. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat. Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 169.Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 170.Yu Z-Q, et al. Dual roles of Atg8−PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8:883–892. doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Goldstein G, et al. Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl Acad. Sci. USA. 1975;72:11–15. doi: 10.1073/pnas.72.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ciechanover A, Heller H, Elias S, Haas AL, Hershko A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl Acad. Sci. 1980;77:1365. doi: 10.1073/pnas.77.3.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hershko A, Ciechanover A, Heller H, Haas AL, Rose IA. Proposed role of ATP in protein breakdown: conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl Acad. Sci. 1980;77:1783. doi: 10.1073/pnas.77.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]