Abstract
Primary liver cancer (PLC) may be mainly classified as the following four types: hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma (ICC), hepatoblastoma (HB), and combined hepatocellular carcinoma and intrahepatic cholangiocarcinoma (cHCC-ICC). The majority of PLC develops in the background of tumor microenvironment, such as inflammatory microenvironments caused by viral hepatitis, alcoholic or nonalcoholic steatohepatitis, carbon tetrachloride (CCl4), 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC), and necroptosis-associated hepatic cytokine microenvironment caused by necroptosis of hepatocytes. However, the impact of different types of microenvironments on the phenotypes of PLC generated by distinct oncogenes is still unclear. In addition, the cell origin of different liver cancers have not been clarified, as far as we know. Recent researches show that mature hepatocytes retain phenotypic plasticity to differentiate into cholangiocytes. More importantly, our results initially demonstrated that HCC, ICC, and cHCC-ICC could originate from mature hepatocytes rather than liver progenitor cells (LPCs), hepatic stellate cells (HSCs) and cholangiocytes in AKT-driven, AKT/NICD-driven and AKT/CAT-driven mouse PLC models respectively by using hydrodynamic transfection methodology. Therefore, liver tumors originated from mature hepatocytes embody a wide spectrum of phenotypes from HCC to CC, possibly including cHCC-ICC and HB. However, the underlying mechanism determining the cancer phenotype of liver tumors has yet to be delineated. In this review, we will provide a summary of the possible mechanisms for directing the cancer phenotype of liver tumors (i.e., ICC, HCC, and cHCC-ICC) in terms of oncogenic driver genes and tumor microenvironment. Moreover, this study initially revealed the cell origin of different types of liver cancer.
Subject terms: Liver cancer, Liver cancer
Facts
Liver tumor phenotype is defined by a combination of driving oncogenes but also the types of tumor microenvironments.
Necroptosis-associated hepatic microenvironment facilitates formation of ICC, whereas apoptosis-associated hepatic microenvironment promotes formation of HCC.
HCC, ICC, and cHCC-ICC could originate from mature hepatocytes in mouse models by using hydrodynamic transfection methodology.
Open questions
In addition to mature hepatocytes, it is not clear whether liver cancer could originate from hepatic progenitor cells, hepatic stellate cells and bile duct cells.
It’s uncertain whether the formation of cHCC-ICC was jointly caused by necroptosis environment and apoptosis environment, which needs to be verified in the future.
In the course of chemoembolization therapy for patients with HCC, a phenotypic transition from HCC to ICC was observed. The possible mechanism may lie in the necroptosis-associated hepatic microenvironment caused by chemoembolization therapy, suggesting the cell environment may directly affect the choice of treatment methods.
Introduction
Primary liver cancer (PLC) is the fifth most prevalent cancer and the third common cause of cancer-related mortality worldwide1. PLC is insensitive to various treatments, which could be partly explained by its wide genetic variations, reflecting in the diverse phenotypes and histological characters. PLC may be mainly classified as the following four types: hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma (ICC), hepatoblastoma (HB), and combined HCC and intrahepatic cholangiocarcinoma (cHCC-ICC)2. cHCC-ICC, as an intermediate variant of PLC, has attracted more and more attention in recent years. cHCC-ICC is described as demonstrating histologic features of both hepatocellular and biliary epithelial differentiation3. It is a rare primary liver malignancy, accounting for 1–14.2% of cases. However, the cell origin of PLC is still controversial.
From morphological and pathological perspectives, HCC and ICC were previously considered to originate from hepatocytes and cholangiocytes, respectively4. In addition, some subtypes of HCC with fetal hepatoblasts features are thought to arise from hepatic progenitor cells, which may differentiate into hepatocytes and bile duct epithelial cells under certain stimuli5. Generally, the cell origin of PLC may be derived from the following four types of cells: hepatocytes, cholangiocytes, hepatoblasts, and liver stem/progenitor cells. However, the cell origin of PLC and the underlying mechanism for the phenotypic determination remains unclear. Recently, some well-established lineage-tracing mouse experiments have demonstrated that HCC or ICC originates from mature hepatocytes rather than liver progenitor cells (LPCs), hepatic stellate cells (HSCs), and cholangiocytes. For example, one study showed that ICC could originate from hepatocytes in mice when the PI3K-AKT and Notch pathways were coactivated6. On a similar note, Mu et al. demonstrated that hepatocytes represent the cell of origin for HCC in mice. Moreover, for the subtype of HCC with a progenitor signature, it does reflect progenitor origin, but dedifferentiation of hepatocyte-derived tumor cells7. Therefore, liver tumors originated from mature hepatocytes consist of a wide spectrum of phenotypes from HCC to CC, possibly encompassing cHCC-ICC and HB4.
In this review, we will summarize the potential mechanisms for determining the cancer phenotype of hepatocyte-derived mouse liver tumors, including ICC, HCC, and cHCC-ICC, in terms of oncogenic driver genes and tumor microenvironment by combining our previous work and the latest research progress. More importantly, it may help us screen of innovative therapeutic approaches against this deadly malignancy in the future.
Regulatory molecules and tumor microenvironment that commit ICC formation
Previous studies have shown that ICC may originate from the cells lining the bile ducts, biliary duct cells (BDCs) or liver stem/progenitor cells8. Nevertheless, recent studies have demonstrated that mature hepatocytes possess a capacity for cholangiocytes transdifferentiation under certain conditions9. For example, Nishikawa et al.10 found that cultured hepatocytes expressed several bile duct markers including cytokeratin (CK) 19 in a three-dimensional organoid culture system, which containing insulin and epidermal growth factor. Likewise, Michalopoulos et al.11 showed that hepatocytes can transdifferentiate into BDCs and help repair the damaged biliary epithelium when its proliferative capacity is being compromised. Moreover, a recent study also showed that mature hepatocytes exhibited the bile duct-like phenotype after chronic liver injury both in vivo and in vitro9. In addition, the notion that cell origin of ICC is mature hepatocytes was subsequently confirmed in another chemically induced ICC mouse model12, as well as a study by electroporating oncogenic transposon plasmids into the left liver lobe of mice13. Recent studies have shown a significant difference between the primary BDCs and the hepatocytes transdifferentiated BDCs. Morphologically, these hepatocytes transdifferentiated BDCs are not mature cholangiocytes with reserve for hepatocyte differentiation. Functionally, hepatocyte-derived ductules are not conducive to bile drainage. Importantly, a recent study demonstrated that these hepatocytes transdifferentiated BDCs are transcriptionally distinct from the primary BDCs as shown by RNA-sequencing analysis and ultrastructural analysis. Interestingly, these hepatocytes transdifferentiated BDCs keep their origin memory and could revert back to hepatocytes upon cessation of injury, which reflecting an adaptive injury escape mechanism14. Mechanistically, TGFβ signaling has been identified associated with the formation of the transdifferentiated BDCs from hepatocytes.
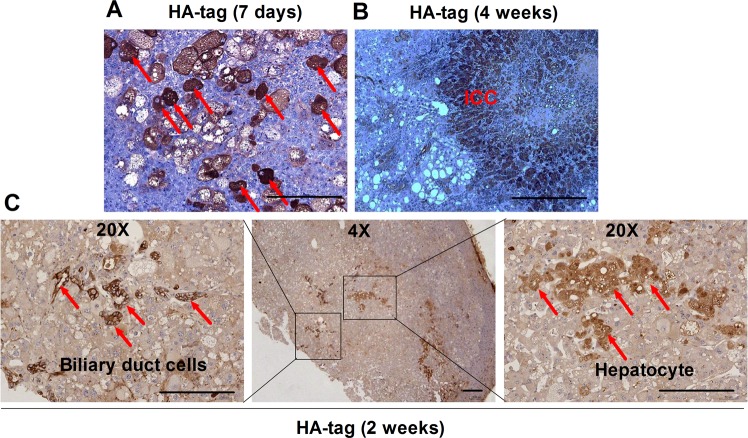
In our previous research, we applied hydrodynamic tail vein injection of hemagglutinin (HA) tagged AKT and NICD plasmid (AKT/NICD), along with Sleeping Beauty (SB) plasmids into BALB/c mice (6–8 weeks) to initiate ICC development. After 7 days, some scattered HA-tag strongly positive hepatocytes were detected in AKT/NICD-injected livers (Fig. 1a). After 4 weeks, immunohistochemical results showed that HA-tag protein was also expressed in ICC, indicating that ICC could originate from these HA-tag positive hepatocytes (Fig. 1b) After 2 weeks, we found that some HA-stained hepatocytes and BDCs appeared at the same time, which further proved the hepatocytes transdifferentiation (Fig. 1c). Accordingly, cellular reprogramming- transition from hepatocyte towards a more ICC-like phenotype might be induced by genetic and cellular alterations occurring during tumorigenesis. Based on the latest research, mechanism underlying hepatocyte-derived ICC formation can be summarized as the following aspects (Fig. 2).
Fig. 1. ICC could originate from hepatocytes.
a Immunohistochemical staining (IHC) showed that some scattered HA-tag strongly positive hepatocytes were detected in AKT/NICD-injected livers after 7 days. b IHC results showed that HA-tag protein was also expressed in ICC tissues after 4 weeks. c IHC results showed that some HA-stained hepatocytes and biliary duct cells appeared after 2 weeks. ICC: intrahepatic cholangiocarcinoma (scale bars, 50 μm).
Fig. 2. Schematic representation of regulatory molecules and tumor microenvironment that commit hepatocyte-derived ICC formation.
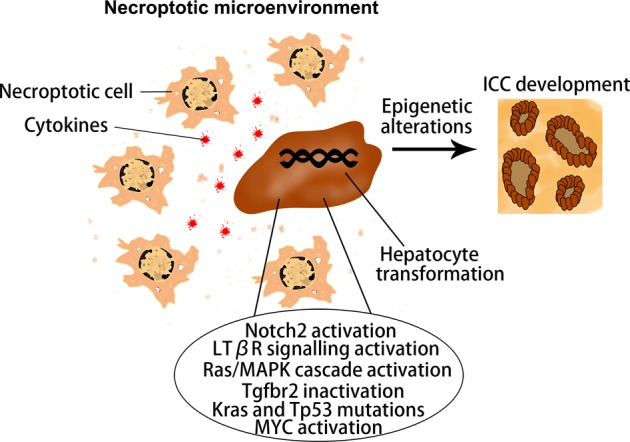
ICC: intrahepatic cholangiocarcinoma.
Regulatory molecules that commit ICC formation
Notch signaling pathway
Accumulating evidence suggests that the canonical Notch cascade controls hepatocyte-derived ICC formation in mice12,14. Notch is a highly conservative signaling pathway that regulates cell proliferation and differentiation and plays an important role in embryonic development and in cell fate determination15. By using well-established lineage-tracing mouse models, Yanger et al.16 report that the activation of Notch is sufficient to reprogram hepatocytes into biliary epithelial cells under injury conditions that provoke a biliary response. Likewise, Fan et al.6 and Sekiya et al.12 also show that ICC can originate from fully differentiated hepatocytes by using a mouse model of hepatocyte fate tracing. However, which Notch receptor is responsible for hepatocyte-derived ICC formation appears to be more important. Most recently, a new study reveals that Notch2, rather than Notch1, controls hepatocyte-derived ICC formation in mice. In this study, Chen et al. established a murine hepatocyte-derived ICC model by co-expression of AKT and Yap plasmids in mice liver14. They demonstrated that deletion of Notch2 skews AKT/Yap-induced ICC pathology towards a more hepatocellular adenoma-like phenotype. However, deletion of Notch1 in tumor cells does not affect the histological type14. Therefore, endogenous Notch signaling is required for hepatocyte-derived ICC. This finding suggested that Notch2 could serve as a target for treatment of this deadly disease, which has great impact on clinical practice in the foreseeable future.
Kras and Tp53 mutations
The recent study of Hill et al.17 indicates Kras and Tp53 mutations facilitate formation of hepatocyte-derived ICC in the context of chronic liver injury. By using Alb-Cre;KrasLSL-G12D;Tp53f/f transgenic mice that targeting Kras and Tp53 mutations to the mouse liver, Hill et al.17 demonstrated that selective induction of Kras and Tp53 mutations in mature hepatocytes in the setting of liver injury, such as DDC-induced chronic inflammation (3,5-diethoxycarbonyl-1,4-dihydrocollidine), could drive rapid progression of ICC17. More importantly, Tp53 has been identified as a key regulator in enabling hepatocyte-derived ICC in this context. Indeed, Tp53 has been shown to control plasticity in a number of different cellular contexts18 and thus Tp53 mutations may facilitate such transdifferentiation events that are implicated in hepatocyte-derived ICC pathogenesis19.
Tgfbr2
Another study reported that Tgfbr2 (TGF-β receptor II) restricts hepatocyte-derived ICC20. TGF-β pathway is closely related to the development of hepatic fibrosis both in mice and patients21. It is noteworthy that recent exon sequencing revealed a high frequency of mutations in Smad4, a key downstream mediator of TGF-β signals, in human cholangiocarcinoma22. Most recently, a new study reveals that hepatocyte-specific deletion of Tgfbr2 and PTEN mediated by AAV8-TBG-Cre promoted hepatocyte-derived ICC formation and reduced survival of mice20. Mechanistically, deletion of Tgfbr2 promotes the proliferation of cholangiocyte rather than hepatocytes, suggesting the pivotal role of epithelial Tgfbr2 in restricting cholangiocyte proliferation20. Although targeting TGF-β may be clinically effective for liver fibrosis, this approach may increase the risk of ICC, which needs to be paid enough attention in clinic.
c-Myc
The recent study of Hill et al.23 indicates c-Myc is required for hepatocyte-derived ICC in AKT/Fbxw7ΔF mice. The ubiquitin ligase F-box and WD repeat domain-containing 7 (FBXW7) plays an anti-cancer role in many cancers, such as HCC, colorectal cancer and gastric cancer24. It can lead to the degradation of several oncogenes, such as c-MYC and YAP25. Wang et al.23 generated a ICC mouse model by co-expression of Fbxw7ΔF (a dominant negative form of Fbxw7) and AKT plasmids in mice livers. Using lineage tracing technology, they confirmed that ICC lesions induced by AKT/Fbxw7ΔF derived from hepatocytes. Interestingly, selected deletion of c-Myc, as for the downstream targets of FBXW7, completely suppresses hepatocyte-derived ICC formation in AKT/Fbxw7ΔF mice23. Furthermore, in human ICC specimens, the expression level of Fbxw7 was negatively correlated with the transcription activity of c-myc34. Therefore, c-Myc could serve as a therapeutic target for ICC treatment, especially with respect to patients with low FBXW7 expression.
Ras/MAPK cascade
Ras/MAPK cascade may influence the formation of hepatocyte-derived ICC by promoting cell proliferation and regulating tumor microenvironment26. Previous studies have shown that Ras/MAPK pathway is significantly activated in human ICC27. In a recent study, Wang et al. generated a hepatocyte-derived ICC mouse model by hydrodynamic tail vein injection of AKT and YapS127A plasmids in mice livers26. They found that inhibition of Ras/MAPK cascade significantly delayed the progression of AKT/YapS127A-induced ICC. On the one hand, Ras/MAPK cascade can significantly promote the proliferation of cholangiocarcinoma cells. On the other hand, this cascade can recruit activated hepatic stellate cells (AHSC) and create hypoxic microenvironment in tumor tissues, which is key features of human ICC28. Because MEK is a key player in Ras/MAPK pathway, MEK inhibitors may be a therapeutic option for ICC in future clinical trials.
LTβR signaling
The recent study of Scarzello et al.29 indicates LTβR signaling accelerates formation of hepatocyte-derived ICC in AKT/β-catenin and AKT/NICD mouse models. LTβR is a member of the tumor necrosis factor (TNF) superfamily of receptors30 and implicated in the initiation of liver cancer31. AKT/CAT-induced tumors display multiple pathological features, including lipogenic hepatic foci, HB/HCC-like nodules and ICC-like lesions32, among which the first two types of pathological features are most common, while ICC-like lesions are relatively rare. However, when using LTβR agonists, more ICC-like tumors were observed in AKT/β-catenin mouse model, suggesting LTβR signaling skews AKT/β-catenin pathology towards a more ICC-like phenotype29. In addition, a role for LTβR signaling in promoting the progression of ICC was further confirmed using AKT/NICD-initiated ICC model. In preclinical and clinical research study of liver cancer, combination therapies are being widely explored and are attracting more attention increasingly. Immune agents blocking the activity of LTβR in combination with other drugs, such as Akt or β-catenin pathway inhibitor, may achieve better therapeutic effect in ICC.
Tumor microenvironment that commits ICC formation
The so-called tumor microenvironment has been recognized as an important regulator in the initiation, development and treatment of various cancers. Recently, it has been found that necroptosis-associated hepatic cytokine microenvironment facilitates formation of ICC33. Tumor microenvironment is a complex environment for the survival and development of cancer cells, which mainly consists of cellular and non-cellular components34. Both components play a supporting role in the growth of tumors35. Very recently, it has been found that the microenvironment of cancer cells (especially the special form of cell death occurring in this environment) has a decisive influence on whether HCC or ICC occurs33. In necrotic apoptosis, a large number of cytokines are secreted from immune cells that are activated by damage-associated molecular patterns (DAMPs), which released from necroptosically dying hepatocytes36. While in apoptosis, vesicles are cleared by the immune system and there is no large amount of cytokine production in microenvironment37. Researchers found that hepatocytes with aberrantly activated oncogenes, if the cell death in their environment is caused by apoptosis, will give rise to HCC; on the other hand, if the cell death is caused by necroptosis, it will lead to ICC. These results were further validated in mouse models and human tissue samples33. Most importantly, the microenvironment formed by different apoptotic pathways had a great influence on two epigenetic regulators (Tbx3 and Prdm5), which are the key regulator in determining lineage commitment in liver cancer38. Interestingly, simultaneous Prdm5 overexpression and Tbx3 knockdown resulted in the development of ICC; however, Tbx3 overexpression combined with Prdm5 knockdown lead to the development of HCC33. There is evidence that inflammatory cytokines and infiltrated immune cells may play an important role in the formation of ICC because they may connect the bridge between the oncogenic driver genes and hepatic death. Seehawer et al.33found that some specific cytokines (e.g., Ccl4, Ccl8, Osm, Ccl6, Cxcl13, Pf4, and Aimp1) are secreted by immune cells, which are activated by DAMPs released from necroptosically dying hepatocytes33. These specific cytokines may act on hepatocytes together with aberrantly activated oncogenes and further led to ICC. However, they also found that the infiltrated immune cells (e.g., T cells, monocytes and (neutrophilic) granulocytes as well as B cells and antigen-presenting cells) were not obvious in the necroptosic microenvironment, suggesting the limited role of infiltrated immune cells in ICC formation33. In the future, in addition to the types of cancer, we should also study whether the tumor microenvironment directly affects the choice of treatments. In the course of chemoembolization therapy for patients with HCC, we found that some primary HCC could transform into ICC. The possible mechanism may lie in the necroptosis-associated hepatic cytokine microenvironment caused by chemoembolization therapy, which may promote a phenotypic transition from HCC to ICC. This may be one of the important reasons for drug resistance in patients with liver cancer.
Regulatory molecules and tumor microenvironment that may commit cHCC-ICC formation
cHCC-ICC is described as demonstrating histologic features of both hepatocellular and biliary epithelial differentiation. It is a rare primary liver malignancy, accounting for 1% to 14.2% of cases39. The diagnosis of cHCC-ICC is usually made at pathologic evaluation after either resection or transplantation and it is practically impossible to achieve an accurate, pre-operative diagnosis of cHCC-ICC with tumor markers or abdominal imaging40. Accordingly, the actual incidence may be higher due to frequent difficulty in accurate pathological assessment. Although little is known clinically about this type of malignancy, the data available indicate that it is aggressive and likely signify a unique subset of PLCs, which merits clinical distinction41. Recent lineage tracing experiments in mice have demonstrated that some subtypes of liver cancer, such as HCC and ICC, are derived from mature hepatocytes rather than from liver stem/progenitor cells7. Moreover, by analyzing systematic mutations, somatic copy number variations, and clonal analyses of human cHCC-ICC tissues, Moeini et al.42 and Wang et al.43 demonstrated that HCC and ICC components share a common cell of origin.
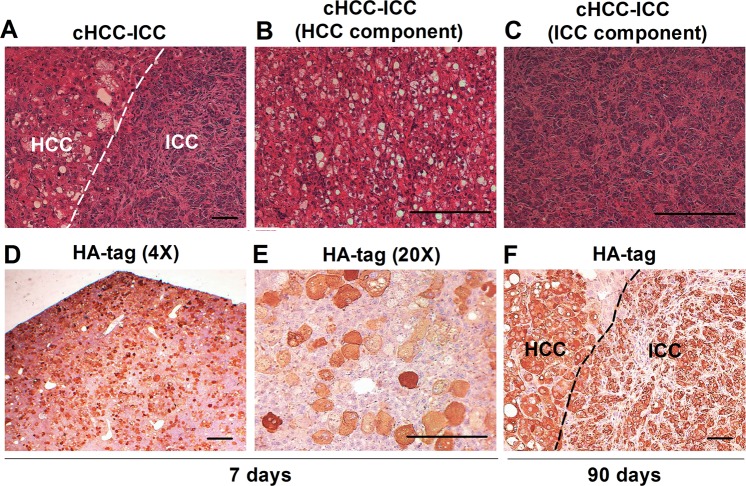
Consistently, our previous studies have shown that cHCC-ICC may originate from hepatocytes in AKT/CAT model. AKT/CAT-initiated tumors display multiple pathological characteristics, including lipogenic hepatic foci, HCC, ICC and cHCC-ICC. We found the moribund livers (5%) following 2 months of AKT and CAT injection displayed a pathological characteristics of cHCC-ICC containing both HCC and ICC two components (Fig. 3a–c). Some scattered HA-tag strongly positive hepatocytes were detected in AKT/CAT -injected livers after 7 days (Fig. 3d, e). Interestingly enough, after 90 days, IHC results showed that HA-tag protein was also expressed in both HCC and ICC two components of cHCC-ICC tumor tissues (Fig. 3f), indicating that HCC and ICC components might originate from these HA-tag positive hepatocytes.
Fig. 3. HCC and ICC components of cHCC-ICC could both originate from hepatocytes.
a–c The moribund livers (5%) following 2 months of AKT and CAT injection displayed a pathological characteristics of cHCC-ICC containing both HCC and ICC two components. d, e IHC showed that some scattered HA-tag strongly positive hepatocytes were detected in AKT/CAT -injected livers after 7 days. f IHC showed that HA-tag protein was also expressed in both HCC and ICC two components of cHCC-ICC tumor tissues after 90 days. HCC: hepatocellular carcinoma; ICC: intrahepatic cholangiocarcinoma; cHCC-ICC: combined hepatocellular carcinoma and intrahepatic cholangiocarcinoma; IHC: immunohistochemical staining (scale bars, 50 μm).
Therefore, liver tumors originated from mature hepatocytes consist of a wide spectrum of phenotypes from HCC to CC, possibly encompassing cHCC-ICC4. Based on the latest research, mechanism underlying hepatocyte-derived cHCC-ICC formation can be summarized as the following aspects (Fig. 4).
Fig. 4. Schematic representation of regulatory molecules and tumor microenvironment that may commit hepatocyte-derived cHCC-ICC formation.
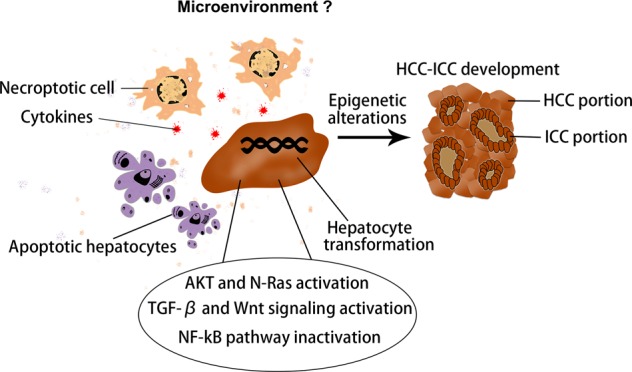
cHCC-ICC: combined hepatocellular carcinoma and intrahepatic cholangiocarcinoma.
TGF-β, Wnt, and Notch signaling
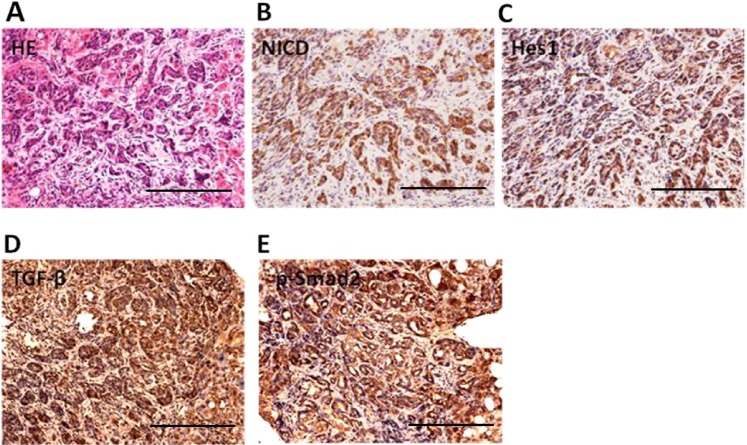
TGF-β, Wnt/β-catenin and Notch signalings were identified as the major signaling activated in human cHCC-ICC specimens41. Indeed, by using a genome-wide transcriptional analysis, Coulouarn et al.’s44 study showed that cHCC-ICC exhibited a gene signature characteristic of the activation of the Wnt/β-catenin pathway, which is closely related to the development of bile duct morphology. Interestingly, TGF-β signaling pathway has been reported to be activated in cHCC-ICC and could be attributed to the presence of the tumoral fibrous stroma with a cholangiocarcinoma-like gene expression trait45. Such results are in accord with a previous study published in Nature, suggesting that TGF-β signaling enhances the formation of the biliary system from hepatocytes through a transdifferentiation mechanism46. Therefore, TGF-β and Wnt/β-catenin pathway may be involved in the formation of ICC components in cHCC-ICC. In addition, mutations of TERT promoter and TP53, as well as substantial intratumoral heterogeneity, often appear in cHCC-ICC41. Consistently, our previous studies have shown that TGF-β and Notch signalings were activated in human cHCC-ICC, especially in the ICC components (Fig. 5).
Fig. 5. IHC showed that TGF-β and Notch signalings were activated in the ICC components of human cHCC-ICC.
ICC: intrahepatic cholangiocarcinoma; cHCC-ICC: combined hepatocellular carcinoma and intrahepatic cholangiocarcinoma; IHC: immunohistochemical staining (scale bars, 50 μm).
NF-kB pathway
The function of NF-κB in liver cancer is still contradictory. Some studies have shown that the NF-κB pathway promotes inflammation-related cancer47, whereas inhibition of NF-κB activity in hepatocytes promotes the spontaneous formation of HCC, indicating that the NF-κB pathway function as tumor suppressor in hepatocytes48. A recent study, the first to analyze the role of NF-kB pathway in the progression of cHCC-ICC, indicates that block of NF-kB signaling skews c-Myc-driven HCC pathology towards a more cHCC-ICC-like phenotype49. It was well known that the tumor phenotype induced by c-Myc often manifests as HB50, however, ICC has not been reported previously. Importantly, after inhibition of NF-kB pathway, an additional tumor component resembling ICC was observed in this model, which was accompanied by MAPK activation, reflecting previous reports on the critical role of NF-kB pathway in cholangiocarcinoma51. Accumulating evidence suggests that liver cancer phenotype can be influenced by sequential oncogenic dysregulation and the inflammatory milieu29. Given NF-κB deletion led to an increase in infiltrating inflammatory cells52, it is reasonable that the chronic inflammatory environment caused by NF-κB ablation may modulate the phenotypic transition in this model.
AKT and N-Ras (N-Ras-V12, a persistently active form of N-Ras)
Activation of AKT and Ras pathways is often implicated in hepatocarcinogenesis. A previous study showed that overexpression of AKT and N-Ras in the mouse liver (AKT/Ras) by way of hydrodynamic gene transfer can accelerate both HCC and ICC development (i.e., cHCC-ICC), with ICC lesions accounting for about 10% of the total lesion area53. Mechanistically, mTORC1, FOXM1/SKP2, and c-Myc signaling cascades were found to be involved in the mediating AKT/N-Ras-induced hepatocarcinogenesis54. In addition, N-Ras-V12 oncogene was delivered to the livers of p19Arf-null or heterozygous mice to elicit tumor formation. The results showed that the tumor pathological type of this model was cHCC-ICC, further suggesting a key role of N-Ras-V12 in the development of cHCC-ICC55.
Tumor microenvironment that may commit cHCC-ICC formation
According to our knowledge, there is no report on the tumor microenvironment of cHCC-ICC so far. As mentioned earlier, necroptosis-associated hepatic cytokine microenvironment facilitates formation of hepatocyte-derived ICC, whereas apoptosis-associated hepatic cytokine microenvironment promotes formation of hepatocyte-HCC. Based on this, we speculate that hepatocytes with aberrantly activated oncogenes, if the cell death in their environment is jointly caused by necroptosis and apoptosis, will give rise to cHCC-ICC (Fig. 4). However, this viewpoint needs to be verified by experiments in the future.
Regulatory molecules and tumor microenvironment that commit HCC formation
Until recently, some well-established lineage-tracing mouse experiments have further demonstrated that HCC originates from mature hepatocytes rather than LPCs, hepatic stellate cells (HSCs) and biliary compartment both in genotoxic and genetic models7. In order to study the molecular mechanism of hepatocyte-derived HCC formation, various primary HCC mouse models were established (Table 1). For instance, mouse HCC induced by CCl4, diethylnitrosamine (DEN), or aristolochic acid was often accompanied with reactivation of a variety of fetal liver genes, such as Gpc3, Afp, Slpi, Spink3, and Abcd256–58. Moreover, various transgenic mouse models of HCC have been successfully generated by overexpression of oncogenes such as AKT, Myc, Bmi1, c-Met, Tgfa, E2F1, Ccnd1, Spry2Y55F, and HRASG12V, or genes that encode viral proteins, such as HbsAg, HBX, and SV40 T-Ag (Table 1)59–77. However, these transgenic mouse models have several limitations, such as high costs, time consuming and requiring high professional knowledge and skills. Hydrodynamic gene delivery is a new method that combines with the SB mediated somatic integration for long-term gene expression in mouse hepatocytes, which has been used in developing novel murine models for HCC (Table 1)4,6,23,32,33,49,54,55,78–87. Through this technique, Che et al.88 reveals a novel crosstalk between aberrant lipogenesis and cholesterol biosynthesis pathways in the progression of HCC. Shang et al.89 demonstrated that co-overexpression of focal adhesion kinase (FAK) and β-Catenin leads to HCC formation. Therefore, hydrodynamic transfection is a reliable method to induce liver tumor and can be used to study the role of genes with unknown functions in hepatocarcinogenesis.
Table 1.
The various mouse models of liver cancer.
| Genes | Tumor type | Mouse strains | Latency | Reference |
|---|---|---|---|---|
| Genetically engineered mouse models for liver cancer | ||||
| AAT | HCC | Transgenic mice using human alpha 1-antitrypsin M and Z genomic clones | 52–90 weeks | Geller et al.60 |
| NEMO−/− | HCC | NEMOΔhepa mice | 52 weeks | Beraza et al.61 |
| PTEN−/− | HCC | PTEN-deficient mice | 42–44 weeks | Watanabe et al.62 |
| PTEN−/− + GRP94−/− | cHCC-ICC | PTEN and GRP94 two liver-specific knockout mouse | 25 weeks | Chen et al.63 |
| HCV core | HCC | Transgenic for the HCV core gene | 80–105 weeks | Moriya et al.64 |
| TAK1−/− | HCC | Tak1Δhepa mice | 39 weeks | Inokuchi et al.65 |
| HBx | HCC | Transgenic mice expressed HBV-encoded gene products | 52–104 weeks | Chisari et al.66 |
| KRASG12D + HBx | HCC | Kras(G12D) and HBx double transgenic mice | 34 weeks | Ye et al.67 |
| c-myc | HB | c-myc single transgenic mice | 65–90 weeks | Thorgeirsson et al.68 |
| c-myc + EGF | HCC | Autocrine growth factor IgEGF and c-myc single transgenic mice | 12–18 weeks | Tönjes et al.69 |
| c-myc + E2F1 | HCC | c-Myc/E2F1 transgenic mouse | 26–39 weeks | Calvisi et al.70 |
| P53−/− + c-myc | HCC | c-Myc/p53KO mice | 21 weeks | Klocke et al.71 |
| P53−/− | HCC | P53Δhepa mice | 60 weeks | Katz et al.72 |
| EGF | HCC | Autocrine growth factor IgEGF transgenic mice | 24–36 weeks | Tönjes et al.69 |
| SV40 T-antigen | HCC | Mice expressing SV 40 early sequences | 20 weeks | Lou et al.73 |
| E2F-1 | HCC | E2f1 transgenic mice | 52 weeks | Lee et al.74 |
| APC−/− | HCC | APCΔhepa mice | 38 weeks | Colnot et al.75 |
| TGF- a | HCC | TGF-alpha transgenic mice | >52 weeks | Lee et al.76 |
| β-catenin(Dex3) + HRASG12V | HCC | Mouse strain containing a mutant beta-catenin allele of which exon 3 was sandwiched by loxP sequences [Catnb(lox(ex3))] | 8 weeks | Harada et al.77 |
| Application of the hydrodynamic transfection methodology to induce liver cancer | ||||
| myr-AKT | HCC | C57BL/6J, FVB/N | 6 months | Calvisi et al.78 |
| myr-AKT and DN90-b-catenin | HCC | C57BL/6J, FVB/N | 1 months | Calvisi DF et al.79 |
| myr-AKT and NRasV12 | cHCC-ICC | C57BL/6J, FVB/N | 1 months | Ho et al.54 |
| myr-AKT and NICD | ICC | C57BL/6J, FVB/N | 3 weeks | Fan et al.6 |
| c-Met and DN90-b-catenin | HCC | C57BL/6J, FVB/N | 3 months | Tward et al.80 |
| NRasV12 and DN90-b-catenin | HCC | C57BL/6J, FVB/N | 3 months | Lee et al.81 |
| NEMO (IKKγ) KO + c-Myc | cHCC-ICC | C57BL/6J, FVB/N | 45 days | He et al.49 |
| Myc and human NRASG12V | HCC | p19Arf−/− | 4 weeks | Seehawer et al.33 |
| mouse Myc and Akt1 | HCC | p19Arf−/− | 4 weeks | Seehawer et al.33 |
| FAK and DN90-b-catenin | HCC | 57BL/6J, FVB/N | 24 weeks | Shang et al.82 |
| myr-AKT and c-Myc | HCC | 57BL/6J, FVB/N | 8 weeks | Yamamoto et al.4 |
| myr-AKT/c-Myc/YAP | HCC | 57BL/6J, FVB/N | 3 weeks | Yamamoto et al.4 |
| myr-AKT and YAP | ICC | 57BL/6J, FVB/N | 6 weeks | Yamamoto et al.4 |
| c-Myc and YAP | HB | 57BL/6J, FVB/N | 16 weeks | Yamamoto et al.4 |
| NICD1 | ICC | 57BL/6J, FVB/N | 5 months | Fan et al.6 |
| HRasV12 and shP53 | Undifferentiated liver tumors | 57BL/6J, FVB/N | 1 week | Ju et al.83 |
| NRasV12 | cHCC-ICC | Ink4A/Arf−/− | 7 weeks | Carlson et al.55 |
| myr-AKT and Spry2Y55F | HCC | 57BL/6J, FVB/N | 4 months | Wang et al.84 |
| c-Myc and shp53 | HCC | 57BL/6J, FVB/N | 7 months | Ju et al.85 |
| AKT/Fbxw7ΔF | ICC | 57BL/6J, FVB/N | 10 weeks | Wang et al.23 |
| Nras-FAH and shP53 | HCC | Fah−/− | 10 weeks | Wangensteen et al.86 |
| Bmi1 and NRasV12 | HCC | 57BL/6J, FVB/N | 6 months | Xu et al.87 |
| Application of the chemical carcinogens to induce liver cancer | ||||
| Diethylnitrosamine (DEN) | HCC | 57BL/6J, FVB/N | 14 months | Ngo et al.57 |
| Aristolochic acid | HCC, cHCC-ICC | 57BL/6J, FVB/N | In a dose-dependent manner | Lu et al.58 |
Consistently, our previous studies have shown that HCC may originate from hepatocytes in AKT mouse model. AKT-initiated tumors were characterized by lipid rich droplets and high proliferation (Fig. 6c, d). Some scattered hepatocytes with strongly positive HA-tag were detected in AKT-injected livers after 7 days (Fig. 6a). IHC results showed that HA-tag protein was also expressed in HCC tumor tissues after 6 months (Fig. 6b), indicating that HCC might originate from these HA-tag positive hepatocytes. Based on the latest research, mechanism underlying hepatocyte-derived HCC formation can be summarized as the following aspects (Fig. 7).
Fig. 6. HCC could originate from hepatocytes.
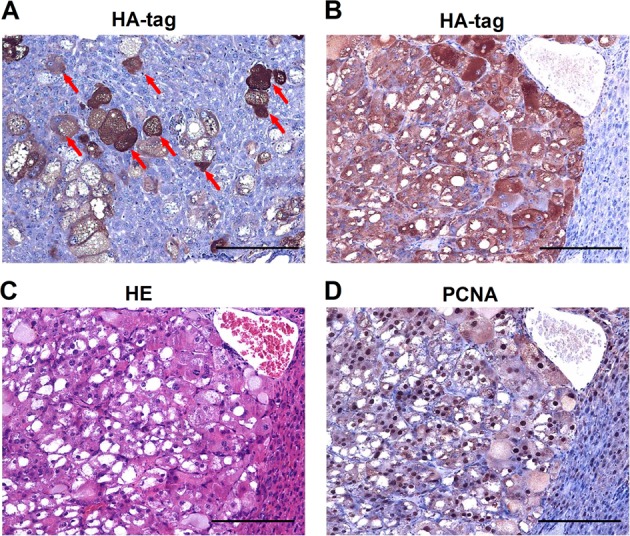
a IHC showed that some scattered HA-tag strongly positive hepatocytes were detected in AKT-injected livers after 7 days. b IHC results showed that HA-tag protein was also expressed in HCC tumor tissues after 6 months. c HE staining showed that AKT-initiated tumors were characterized by lipid rich droplets. d IHC results showed that PCNA protein was highly expressed in HCC tumor tissues after 6 months. HCC: hepatocellular carcinoma; IHC: immunohistochemical staining; HE: hematoxylin-eosin staining (scale bars, 50 μm).
Fig. 7. Schematic representation of regulatory molecules and tumor microenvironment that may commit hepatocyte-derived HCC formation.
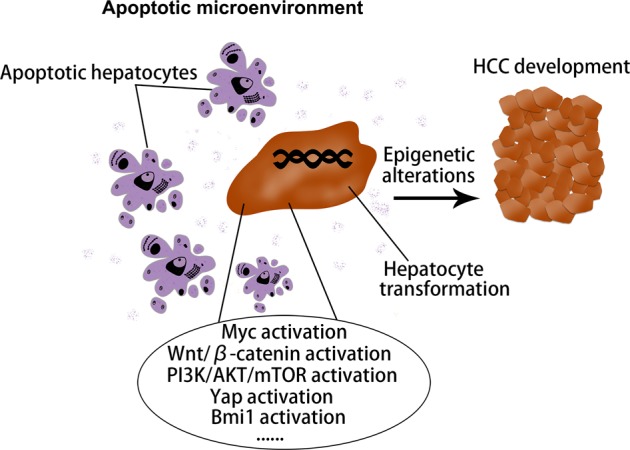
HCC: hepatocellular carcinoma.
Hepatocarcinogenesis due to the interaction of multiple genes
The occurrence of HCC is a complex process accompanied by the activation of multiple signaling pathways, which plays a synergistic role in the process of tumorigenesis90. Numerous studies have confirmed that PI3K/AKT/mTOR pathway and Wnt/β-catenin pathways play an important role in the development of HCC91. For instance, hydrodynamical codelivery of activated forms of AKT (pT3-EF1α-HA-myr-AKT) and β-catenin (pT3-EF1α-Δ90β-catenin, CAT) oncogenes into mouse livers using the SB transposon system efficiently and rapidly induces primary hepatic tumors. AKT/CAT-initiated tumors display multiple pathological characteristics, including early lipogenic hepatic foci and subsequent HB/HCC-like nodules, which is rich in lipids29. Importantly, this provides a good animal model for the study of steatosis-related liver cancer. In addition, the activated form of AKT was found to cooperate with activated Myc, Yap, NRasV12 or Spry2Y55F pathways to induce HCC formation in the mouse84. Hydrodynamical codelivery of the activated mutant of β-catenin and c-MET1 or NRasV1281 into mouse livers can also efficiently induce HCC over a short latency. Using the same method, Li et al.92 reported that the introduction of YAPS127A and PIK3CAH1047R (a constitutively active mutant of PI3K) induces liver cancers with many pathological features. Fan et al.93 found that Bmi1 is required for AKT/Ras -induced HCC development. Altogether, these results reflect the complex interaction of different oncogenes in hepatocyte-derived HCC formation.
A single gene sufficient for hepatocarcinogenesis
Activated PI3K/Akt/mTOR pathway is closely related to poor differentiation, early recurrence and poor prognosis of HCC94. Four weeks after hydrodynamical delivery of AKT plasmids, the livers are pale and greasy. Microscopically, hepatocytes were abundant with cytoplasmic lipid and characterized by the intermingled small ductular structures95. After 22-32 weeks of transfection, all AKT mice developed lethal liver cancer. In general, the livers of AKT mice were pale and enlarged. There were many tumor nodules on the surface. Microscopically, these tumor cells were characterized by increased cell volume and transparent cytoplasm due to fat accumulation95. This suggests that overexpression of AKT alone is sufficient to form liver cancer. For another example, MYC oncogene has been implicated in human liver cancer96. It was reported that MYC was over expressed in over 70% of viral or alcohol-related human HCC96. Hydrodynamic transfection of MYC caused lethal burden of liver cancer by 6–8 weeks post injection. Pathologically, MYC tumors are poorly differentiated and resemble human HBs with cancer stem cells-like properties97. All these studies demonstrate that a single gene, such as MYC or AKT, is sufficient for hepatocarcinogenesis, even if not combined with other oncogenes.
Tumor microenvironment that commits HCC formation
Chronic liver inflammation has been implicated in tumorigenesis. Actually, most HCC develops in an inflammatory environment caused by viral hepatitis and alcoholic or nonalcoholic steatohepatitis98. Recent studies have shown that inflammation microenvironment can induce transformation of tumor types. For example, Matter et al.99 demonstrated that chronic liver inflammation caused by DDC (3,5-diethoxycarbonyl-1,4-dihydrocollidine) changed AKT/CAT-induced tumors pathology. AKT/CAT-induced tumors were steatotic and contained lipid droplets, whereas lipid content in tumors of AKT/CAT with DDC group was decreased significantly. Pathological types of AKT/CAT-induced liver cancer can be classified into three types: hepatocellular adenoma (HCA), HCC, and HB. In AKT/CAT group, the proportion of HCC was 5–25%, while in AKT/CAT with DDC group, the proportion of HCC was 5–50%, suggesting that chronic inflammation promotes the phenotypic transition from HCA to HCC. Likewise, chronic inflammation microenvironment induced by DDC can also reduce lipid droplets in AKT-NRASG12V tumors99. Altogether, this illustrates that driving oncogenes and tumor microenvironment jointly determined the hepatocyte-derived HCC formation.
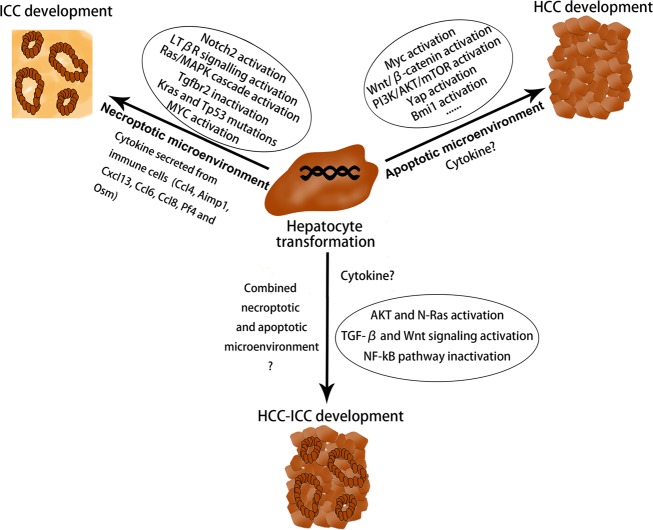
In summary, this review summarizes the possible mechanism of lineage determination in the development of PLC, including ICC, HCC, and cHCC-ICC (Fig. 8). We put forward the notion that the combined effects of oncogenic driver genes and tumor microenvironment decides the cancer phenotype of hepatocyte-derived mouse liver tumors. PLC always occurs inevitably in a variety of tumor microenvironments, in which different types of cell death such as necrosis, apoptosis or necroptosis occur. It is noteworthy that hepatocytes with aberrantly activated oncogenes will lead to ICC when cell death in their environment is caused by necroptosis with lots of cytokines production. In addition, various intracellular signaling cascades such as Notch2, MYC, Tgfbr2, and Ras/MAPK pathway in hepatocytes mediate the hepatocyte-derived ICC formation (Fig. 2). On the other hand, if the cell death in their environment is caused by apoptosis, hepatocytes with aberrantly activated oncogenes will give rise to HCC. It is well known that some classical cancer-related signalings such as MYC, Yap, Bmi1, Wnt/β-catenin, and PI3K/AKT/mTOR pathways were implicated in the hepatocyte-derived HCC formation (Fig. 7). cHCC-ICC is a rare primary liver malignancy and the incidence is increasing in the last twenty years, however, its pathogenesis is still poorly understood. Future work is needed to determine whether necroptosis, apoptosis or both occur in the tumor microenvironment that mediate the hepatocyte-derived cHCC-ICC formation (Fig. 4). In conclusion, the possible mechanism of lineage determination in the development of PLC has yet to be delineated. Deciphering the detailed roles of oncogenic driver genes and tumor microenvironment in PLC would certainly pave the way for the development of novel therapies.
Fig. 8. Oncogenic driver genes and tumor microenvironment determine the type of liver cancer.
HCC: hepatocellular carcinoma; ICC: intrahepatic cholangiocarcinoma; cHCC-ICC: combined hepatocellular carcinoma and intrahepatic cholangiocarcinoma.
Acknowledgements
The pT3-myr-AKT-HA, pT3-N90-beta-catenin and pT3-EF1a-NICD plasmids were kind gifts from Xin Chen and were obtained from Addgene (Cambridge, MA). pCMV/SB10 was a gift from Perry Hackett and were obtained from Addgene (Cambridge, MA). The authors thank Dr Xin Chen (UCSF, University of California, San Francisco campus) and Dr Perry Hackett (University of Minnesota) for sharing these plasmids. This work was supported by grants from National Natural Science Foundation of China (grant#81903075, 81670587) and Shanghai Excellent Youth training program (2018YQ62).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by Q. Chen
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Gang Wang, Qian Wang, Ning Liang, Hongyuan Xue
Contributor Information
Chaoxu Liu, Email: chaoxuliu@yahoo.com.
Zhangqian Chen, Email: Chenzq1@fmmu.edu.cn.
Xianli He, Email: hexianli999@126.com.
References
- 1.Shang N, et al. FAK is required for c-Met/beta-catenin-driven hepatocarcinogenesis. Hepatology. 2015;61:214–226. doi: 10.1002/hep.27402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connell LC, Harding JJ, Shia J, Abou-Alfa GK. Combined intrahepatic cholangiocarcinoma and hepatocellular carcinoma. Chin. Clin. Oncol. 2016;5:66. doi: 10.21037/cco.2016.10.02. [DOI] [PubMed] [Google Scholar]
- 3.Bergquist JR, et al. Mixed hepatocellular and cholangiocarcinoma: a rare tumor with a mix of parent phenotypic characteristics. HPB (Oxf.) 2016;18:886–892. doi: 10.1016/j.hpb.2016.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto M, et al. Oncogenic determination of a broad spectrum of phenotypes of hepatocyte-derived mouse liver tumors. Am. J. Pathol. 2017;187:2711–2725. doi: 10.1016/j.ajpath.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 5.Lee JS, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat. Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 6.Fan B, et al. Cholangiocarcinomas can originate from hepatocytes in mice. J. Clin. Invest. 2012;122:2911–2915. doi: 10.1172/JCI63212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mu X, et al. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. J. Clin. Invest. 2015;125:3891–3903. doi: 10.1172/JCI77995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marquardt JU, Andersen JB, Thorgeirsson SS. Functional and genetic deconstruction of the cellular origin in liver cancer. Nat. Rev. Cancer. 2015;15:653–667. doi: 10.1038/nrc4017. [DOI] [PubMed] [Google Scholar]
- 9.Nagahama Y, et al. Contributions of hepatocytes and bile ductular cells in ductular reactions and remodeling of the biliary system after chronic liver injury. Am. J. Pathol. 2014;184:3001–3012. doi: 10.1016/j.ajpath.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Nishikawa Y, et al. Transdifferentiation of mature rat hepatocytes into bile duct-like cells in vitro. Am. J. Pathol. 2005;166:1077–1088. doi: 10.1016/S0002-9440(10)62328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Michalopoulos GK, Barua L, Bowen WC. Transdifferentiation of rat hepatocytes into biliary cells after bile duct ligation and toxic biliary injury. Hepatology. 2005;41:535–544. doi: 10.1002/hep.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sekiya S, Suzuki A. Intrahepatic cholangiocarcinoma can arise from Notch-mediated conversion of hepatocytes. J. Clin. Invest. 2012;122:3914–3918. doi: 10.1172/JCI63065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gürlevik E, et al. Adjuvant gemcitabine therapy improves survival in a locally induced, R0-resectable model of metastatic intrahepatic cholangiocarcinoma. Hepatology. 2013;58:1031–1041. doi: 10.1002/hep.26468. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, et al. Notch2 controls hepatocyte-derived cholangiocarcinoma formation in mice. Oncogene. 2018;37:3229–3242. doi: 10.1038/s41388-018-0188-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang R, Engler A, Taylor V. Notch: an interactive player in neurogenesis and disease. Cell Tissue Res. 2018;371:73–89. doi: 10.1007/s00441-017-2641-9. [DOI] [PubMed] [Google Scholar]
- 16.Yanger K, et al. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill MA, et al. Kras and Tp53 mutations cause cholangiocyte- and hepatocyte-derived cholangiocarcinoma. Cancer Res. 2018;78:4445–4451. doi: 10.1158/0008-5472.CAN-17-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang H, et al. Cell cycle and p53 gate the direct conversion of human fibroblasts to dopaminergic neurons. Nat. Commun. 2015;6:10100. doi: 10.1038/ncomms10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llovet JM, et al. Hepatocellular carcinoma. Nat. Rev. Dis. Prim. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 20.Mu X, et al. Epithelial transforming growth factor-beta signaling does not contribute to liver fibrosis but protects mice from cholangiocarcinoma. Gastroenterology. 2016;150:720–733. doi: 10.1053/j.gastro.2015.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bataller R, Brenner DA. Liver fibrosis. J. Clin. Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan-On W, et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat. Genet. 2013;45:1474–1478. doi: 10.1038/ng.2806. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, et al. Loss of Fbxw7 synergizes with activated Akt signaling to promote c-Myc dependent cholangiocarcinogenesis. J. Hepatol. 2019;71:742–752. doi: 10.1016/j.jhep.2019.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeh CH, Bellon M, Nicot C. FBXW7: a critical tumor suppressor of human cancers. Mol. Cancer. 2018;17:115. doi: 10.1186/s12943-018-0857-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tu K, et al. Fbxw7 is an independent prognostic marker and induces apoptosis and growth arrest by regulating YAP abundance in hepatocellular carcinoma. Mol. Cancer. 2014;13:110. doi: 10.1186/1476-4598-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang XF, et al. Expression pattern of cancer-associated fibroblast and its clinical relevance in intrahepatic cholangiocarcinoma. Hum. Pathol. 2017;65:92–100. doi: 10.1016/j.humpath.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Dong MJ, et al. Efficacy of MEK inhibition in a K-Ras-driven cholangiocarcinoma preclinical model. Cell Death Dis. 2018;9:31. doi: 10.1038/s41419-017-0183-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sha M, et al. Isolation of cancer-associated fibroblasts and its promotion to the progression of intrahepatic cholangiocarcinoma. Cancer Med. 2018;7:4665–4677. doi: 10.1002/cam4.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scarzello AJ, et al. LTbetaR signalling preferentially accelerates oncogenic AKT-initiated liver tumours. Gut. 2016;65:1765–1775. doi: 10.1136/gutjnl-2014-308810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ware CF. Targeting lymphocyte activation through the lymphotoxin and LIGHT pathways. Immunol. Rev. 2008;223:186–201. doi: 10.1111/j.1600-065X.2008.00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haybaeck J, et al. A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell. 2009;16:295–308. doi: 10.1016/j.ccr.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stauffer JK, et al. Coactivation of AKT and beta-catenin in mice rapidly induces formation of lipogenic liver tumors. Cancer Res. 2011;71:2718–2727. doi: 10.1158/0008-5472.CAN-10-2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seehawer M, et al. Necroptosis microenvironment directs lineage commitment in liver cancer. Nature. 2018;562:69–75. doi: 10.1038/s41586-018-0519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lequeux A, et al. Impact of hypoxic tumor microenvironment and tumor cell plasticity on the expression of immune checkpoints. Cancer Lett. 2019;458:13–20. doi: 10.1016/j.canlet.2019.05.021. [DOI] [PubMed] [Google Scholar]
- 35.Lin CC, Korc M. Designer hydrogels: shedding light on the physical chemistry of the pancreatic cancer microenvironment. Cancer Lett. 2018;436:22–27. doi: 10.1016/j.canlet.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ou L, et al. The mechanisms of graphene-based materials-induced programmed cell death: a review of apoptosis, autophagy, and programmed necrosis. Int. J. Nanomed. 2017;12:6633–6646. doi: 10.2147/IJN.S140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elmore S. Apoptosis: a review of programmed cell death. Toxicol. Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki A, Sekiya S, Buscher D, Izpisúa Belmonte JC, Taniguchi H. Tbx3 controls the fate of hepatic progenitor cells in liver development by suppressing p19ARF expression. Development. 2008;135:1589–1595. doi: 10.1242/dev.016634. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Wang F, Kessinger A. Outcome of combined hepatocellular and cholangiocarcinoma of the liver. J. Oncol. 2010;8:2010. doi: 10.1155/2010/917356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panjala C, et al. The diagnostic conundrum and liver transplantation outcome for combined hepatocellular-cholangiocarcinoma. Am. J. Transpl. 2010;10:1263–1267. doi: 10.1111/j.1600-6143.2010.03062.x. [DOI] [PubMed] [Google Scholar]
- 41.Brunt E, et al. cHCC-CCA: consensus terminology for primary liver carcinomas with both hepatocytic and cholangiocytic differentation. Hepatology. 2018;68:113–126. doi: 10.1002/hep.29789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moeini A, et al. Mixed hepatocellular cholangiocarcinoma tumors: Cholangiolocellular carcinoma is a distinct molecular entity. J. Hepatol. 2017;66:952–961. doi: 10.1016/j.jhep.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Wang A, et al. Whole-exome sequencing reveals the origin and evolution of hepato-cholangiocarcinoma. Nat. Commun. 2018;9:894. doi: 10.1038/s41467-018-03276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coulouarn C, et al. Combined hepatocellular-cholangiocarcinomas exhibit progenitor features and activation of Wnt and TGFbeta signaling pathways. Carcinogenesis. 2012;33:1791–1796. doi: 10.1093/carcin/bgs208. [DOI] [PubMed] [Google Scholar]
- 45.Seok JY, et al. A fibrous stromal component in hepatocellular carcinoma reveals a cholangiocarcinoma-like gene expression trait and epithelial-mesenchymal transition. Hepatology. 2012;55:1776–1786. doi: 10.1002/hep.25570. [DOI] [PubMed] [Google Scholar]
- 46.Schaub JR, et al. De novo formation of the biliary system by TGFbeta-mediated hepatocyte transdifferentiation. Nature. 2018;557:247–251. doi: 10.1038/s41586-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pikarsky E, et al. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature. 2004;431:461–466. doi: 10.1038/nature02924. [DOI] [PubMed] [Google Scholar]
- 48.Sakurai T, Maeda S, Chang L, Karin M. Loss of hepatic NF-kappa B activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc. Natl Acad. Sci. USA. 2006;103:10544–10551. doi: 10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He J, et al. Block of NF-kB signaling accelerates MYC-driven hepatocellular carcinogenesis and modifies the tumor phenotype towards combined hepatocellular cholangiocarcinoma. Cancer Lett. 2019;458:113–122. doi: 10.1016/j.canlet.2019.05.023. [DOI] [PubMed] [Google Scholar]
- 50.Chow EK, Fan LL, Chen X, Bishop JM. Oncogene-specific formation of chemoresistant murine hepatic cancer stem cells. Hepatology. 2012;56:1331–1341. doi: 10.1002/hep.25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang C, et al. A systems biology perspective on cholangiocellular carcinoma development: focus on MAPK-signaling and the extracellular environment. J. Hepatol. 2009;50:1122–1131. doi: 10.1016/j.jhep.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 52.Luedde T, et al. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119–132. doi: 10.1016/j.ccr.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 53.Li L, et al. Differential requirement for de novo lipogenesis in cholangiocarcinoma and hepatocellular carcinoma of mice and humans. Hepatology. 2016;63:1900–1913. doi: 10.1002/hep.28508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho C, et al. AKT (v-akt murine thymoma viral oncogene homolog 1) and N-Ras (neuroblastoma ras viral oncogene homolog) coactivation in the mouse liver promotes rapid carcinogenesis by way of mTOR (mammalian target of rapamycin complex 1), FOXM1 (forkhead box M1)/SKP2, and c-Myc pathways. Hepatology. 2012;55:833–845. doi: 10.1002/hep.24736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Carlson CM, Frandsen JL, Kirchhof N, McIvor RS, Largaespada DA. Somatic integration of an oncogene-harboring Sleeping Beauty transposon models liver tumor development in the mouse. Proc. Natl Acad. Sci. USA. 2005;102:17059–17064. doi: 10.1073/pnas.0502974102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X, et al. Differential reactivation of fetal/neonatal genes in mouse liver tumors induced in cirrhotic and non-cirrhotic conditions. Cancer Sci. 2015;106:972–981. doi: 10.1111/cas.12700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ngo HKC, Kim DH, Cha YN, Na HK, Surh YJ. Nrf2 mutagenic activation drives hepatocarcinogenesis. Cancer Res. 2017;77:4797–4808. doi: 10.1158/0008-5472.CAN-16-3538. [DOI] [PubMed] [Google Scholar]
- 58.Lu ZN, et al. The mutational features of aristolochic acid-induced mouse and human liver cancers. Hepatology. 2020;71:929–942. doi: 10.1002/hep.30863. [DOI] [PubMed] [Google Scholar]
- 59.Lee JS, Grisham JW, Thorgeirsson SS. Comparative functional genomics for identifying models of human cancer. Carcinogenesis. 2005;26:1013–1020. doi: 10.1093/carcin/bgi030. [DOI] [PubMed] [Google Scholar]
- 60.Geller SA, et al. Hepatocarcinogenesis is the sequel to hepatitis in Z#2 alpha 1-antitrypsin transgenic mice: histopathological and DNA ploidy studies. Hepatology. 1994;19:389–397. doi: 10.1002/hep.1840190218. [DOI] [PubMed] [Google Scholar]
- 61.Beraza N, et al. Hepatocyte-specific NEMO deletion promotes NK/NKT cell- and TRAIL-dependent liver damage. J. Exp. Med. 2009;206:1727–1737. doi: 10.1084/jem.20082152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe S, et al. Non-alcoholic steatohepatitis and hepatocellular carcinoma: lessons from hepatocyte-specific phosphatase and tensin homolog (PTEN)-deficient mice. J. Gastroenterol. Hepatol. 2007;22(Suppl 1):S96–S100. doi: 10.1111/j.1440-1746.2006.04665.x. [DOI] [PubMed] [Google Scholar]
- 63.Chen WT, et al. Liver-specific knockout of GRP94 in mice disrupts cell adhesion, activates liver progenitor cells, and accelerates liver tumorigenesis. Hepatology. 2014;59:947–957. doi: 10.1002/hep.26711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moriya K, et al. The core protein of hepatitis C virus induces hepatocellular carcinoma in transgenic mice. Nat. Med. 1998;4:1065–1067. doi: 10.1038/2053. [DOI] [PubMed] [Google Scholar]
- 65.Inokuchi S, et al. Disruption of TAK1 in hepatocytes causes hepatic injury, inflammation, fibrosis, and carcinogenesis. Proc. Natl Acad. Sci. USA. 2010;107:844–849. doi: 10.1073/pnas.0909781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chisari FV, et al. A transgenic mouse model of the chronic hepatitis B surface antigen carrier state. Science. 1985;230:1157–1160. doi: 10.1126/science.3865369. [DOI] [PubMed] [Google Scholar]
- 67.Ye H, et al. Synergistic function of Kras mutation and HBx in initiation and progression of hepatocellular carcinoma in mice. Oncogene. 2014;33:5133–5138. doi: 10.1038/onc.2013.468. [DOI] [PubMed] [Google Scholar]
- 68.Thorgeirsson SS, Santoni-Rugiu E. Transgenic mouse models in carcinogenesis: interaction of c-myc with transforming growth factor alpha and hepatocyte growth factor in hepatocarcinogenesis. Br. J. Clin. Pharmacol. 1996;42:43–52. doi: 10.1046/j.1365-2125.1996.03748.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tönjes RR, et al. Autocrine mitogen IgEGF cooperates with c-myc or with the Hcs locus during hepatocarcinogenesis in transgenic mice. Oncogene. 1995;10:765–768. [PubMed] [Google Scholar]
- 70.Calvisi DF, et al. Activation of the canonical Wnt/beta-catenin pathway confers growth advantages in c-Myc/E2F1 transgenic mouse model of liver cancer. J. Hepatol. 2005;42:842–849. doi: 10.1016/j.jhep.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 71.Klocke R, et al. Lack of p53 accelerates hepatocarcinogenesis in transgenic mice constitutively overexpressing c-myc in the liver. FASEB J. 2001;15:1404–1406. doi: 10.1096/fj.00-0487fje. [DOI] [PubMed] [Google Scholar]
- 72.Katz SF, et al. Disruption of Trp53 in livers of mice induces formation of carcinomas with bilineal differentiation. Gastroenterology. 2012;142:1229–1239 e3. doi: 10.1053/j.gastro.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 73.Lou DQ, et al. Conditional hepatocarcinogenesis in mice expressing SV 40 early sequences. Cancer Lett. 2005;229:107–114. doi: 10.1016/j.canlet.2004.12.032. [DOI] [PubMed] [Google Scholar]
- 74.Lee JS, et al. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat. Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- 75.Colnot S, et al. Liver-targeted disruption of Apc in mice activates beta-catenin signaling and leads to hepatocellular carcinomas. Proc. Natl Acad. Sci. USA. 2004;101:17216–17221. doi: 10.1073/pnas.0404761101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lee GH, Merlino G, Fausto N. Development of liver tumors in transforming growth factor alpha transgenic mice. Cancer Res. 1992;52:5162–5170. [PubMed] [Google Scholar]
- 77.Harada N, et al. Hepatocarcinogenesis in mice with beta-catenin and Ha-ras gene mutations. Cancer Res. 2004;64:48–54. doi: 10.1158/0008-5472.CAN-03-2123. [DOI] [PubMed] [Google Scholar]
- 78.Chen X, Calvisi DF. Hydrodynamic transfection for generation of novel mouse models for liver cancer research. Am. J. Pathol. 2014;184:912–923. doi: 10.1016/j.ajpath.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Calvisi DF, et al. Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology. 2011;140:1071–1083. doi: 10.1053/j.gastro.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tward AD, et al. Distinct pathways of genomic progression to benign and malignant tumors of the liver. Proc. Natl Acad. Sci. USA. 2007;104:14771–14776. doi: 10.1073/pnas.0706578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee SA, et al. Integration of genomic analysis and in vivo transfection to identify sprouty 2 as a candidate tumor suppressor in liver cancer. Hepatology. 2008;47:1200–1210. doi: 10.1002/hep.22169. [DOI] [PubMed] [Google Scholar]
- 82.Shang N, et al. Focal adhesion kinase and beta-catenin cooperate to induce hepatocellular carcinoma. Hepatology. 2019;70:1631–1645. doi: 10.1002/hep.30707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ju HL, et al. Investigation of oncogenic cooperation in simple liver-specific transgenic mouse models using noninvasive in vivo imaging. PLoS ONE. 2003;8:e59869. doi: 10.1371/journal.pone.0059869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang C, et al. Inactivation of Spry2 accelerates AKT-driven hepatocarcinogenesis via activation of MAPK and PKM2 pathways. J. Hepatol. 2012;57:577–583. doi: 10.1016/j.jhep.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ju HL, Han KH, Lee JD, Ro SW. Transgenic mouse models generated by hydrodynamic transfection for genetic studies of liver cancer and preclinical testing of anti-cancer therapy. Int J. Cancer. 2016;138:1601–1608. doi: 10.1002/ijc.29703. [DOI] [PubMed] [Google Scholar]
- 86.Wangensteen KJ, et al. A facile method for somatic, lifelong manipulation of multiple genes in the mouse liver. Hepatology. 2008;47:1714–1724. doi: 10.1002/hep.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu CR, et al. Bmi1 functions as an oncogene independent of Ink4A/Arf repression in hepatic carcinogenesis. Mol. Cancer Res. 2009;7:1937–1945. doi: 10.1158/1541-7786.MCR-09-0333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Che L, et al. Cholesterol biosynthesis supports the growth of hepatocarcinoma lesions depleted of fatty acid synthase in mice and humans. Gut. 2019;1:566–578. doi: 10.1136/gutjnl-2018-317581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shang N, et al. Focal adhesion kinase and beta-catenin cooperate to induce hepatocellular carcinoma. Hepatology. 2019;5:1631–1645. doi: 10.1002/hep.30707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen S, Cao Q, Wen W, Wang H. Targeted therapy for hepatocellular carcinoma: challenges and opportunities. Cancer Lett. 2019;460:1–9. doi: 10.1016/j.canlet.2019.114428. [DOI] [PubMed] [Google Scholar]
- 91.Tao J, et al. Activation of beta-catenin and Yap1 in human hepatoblastoma and induction of hepatocarcinogenesis in mice. Gastroenterology. 2014;147:690–701. doi: 10.1053/j.gastro.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li X, et al. Co-activation of PIK3CA and Yap promotes development of hepatocellular and cholangiocellular tumors in mouse and human liver. Oncotarget. 2015;6:10102–10115. doi: 10.18632/oncotarget.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fan L, et al. Bmi1 is required for hepatic progenitor cell expansion and liver tumor development. PLoS ONE. 2012;7:e46472. doi: 10.1371/journal.pone.0046472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou L, Huang Y, Li J, Wang Z. The mTOR pathway is associated with the poor prognosis of human hepatocellular carcinoma. Med. Oncol. 2010;27:255–261. doi: 10.1007/s12032-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 95.Li L, et al. Inactivation of fatty acid synthase impairs hepatocarcinogenesis driven by AKT in mice and humans. J. Hepatol. 2016;64:333–341. doi: 10.1016/j.jhep.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schlaeger C, et al. Etiology-dependent molecular mechanisms in human hepatocarcinogenesis. Hepatology. 2008;47:511–520. doi: 10.1002/hep.22033. [DOI] [PubMed] [Google Scholar]
- 97.Juric V, et al. Monocytes promote liver carcinogenesis in an oncogene-specific manner. J. Hepatol. 2016;64:881–890. doi: 10.1016/j.jhep.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.van der Windt DJ, et al. Neutrophil extracellular traps promote inflammation and development of hepatocellular carcinoma in nonalcoholic steatohepatitis. Hepatology. 2018;68:1347–1360. doi: 10.1002/hep.29914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Matter MS, et al. Oncogenic driver genes and the inflammatory microenvironment dictate liver tumor phenotype. Hepatology. 2016;63:1888–1899. doi: 10.1002/hep.28487. [DOI] [PMC free article] [PubMed] [Google Scholar]