Abstract
Cerebrospinal fluid (CSF) Aβ42 and tau protein levels are established diagnostic biomarkers of Alzheimer’s disease (AD). However, their inadequacy to represent clinical efficacy in drug trials indicates the need for new biomarkers. Sequential window acquisition of all theoretical fragment ion spectra (SWATH)-based mass spectrometry (MS) is an advanced proteomic tool for large-scale, high-quality quantification. In this study, SWATH-MS showed that VGF, chromogranin-A, secretogranin-1, and opioid-binding protein/cell adhesion molecule were significantly decreased in 42 AD patients compared to 39 controls, whereas 14-3-3ζ was increased (FDR < 0.05). In addition, 16 other proteins showed substantial changes (FDR < 0.2). The expressions of the top 21 analytes were closely interconnected, but were poorly correlated with CSF Aβ42, tTau, and pTau181 levels. Logistic regression analysis and data mining were used to establish the best algorithm for AD, which created novel biomarker panels with high diagnostic value (AUC = 0.889 and 0.924) and a strong correlation with clinical severity (all p < 0.001). Targeted proteomics was used to validate their usefulness in a different cohort (n = 36) that included patients with other brain disorders (all p < 0.05). This study provides a list of proteins (and combinations thereof) that could serve as new AD biomarkers.
Subject terms: Alzheimer's disease, Dementia
Introduction
Cerebrospinal fluid (CSF) levels of Aβ1–42 (Aβ42), total Tau (tTau), and phosphorylated Tau181 (pTau181) are diagnostic markers of Alzheimer’s disease (AD)1,2. These biomarkers are currently used to confirm the core pathology of AD, i.e., Aβ and tau pathology, in deceased patients while they are alive3, and are also monitored in AD therapeutic trials4. However, accumulating evidence has demonstrated a lack of utility of these markers after clinical onset of AD due to early concentration plateaus5 and poor reflection of the clinical benefits of therapeutics that target them6. CSF proteins related to synaptic function, neurodegeneration, inflammation, and Aβ metabolism have recently been suggested as supplementary biomarkers7; however, they have not yet been clinically adopted.
Advances in proteomic techniques have allowed the characterisation and quantification of several CSF proteins related to AD8. Targeted mass spectrometric methods, such as multiple reaction monitoring (MRM) and parallel reaction monitoring (PRM), are powerful tools for detailed quantification and have been used frequently in AD biomarker research9,10. However, these approaches are hypothesis-driven and are therefore useful only for measuring preselected proteins, minimising the likelihood of discovering novel biomarkers11. In contrast, the unbiased proteomic approach allows for the discovery of innovative biomarkers and development of new biological hypotheses of disease. However, the possibility that changes in proteins present in low quantities will remain undetected, and that different molecules with similar mass spectrometry (MS) signatures may not be distinguishable, are significant obstacles to the application of this technique in biomarker research9,11. Sequential window acquisition of all theoretical fragment ion spectra (SWATH)-based MS is a powerful and advanced proteomic technology that permits more precise identification of disease-specific changes in large protein pools. This tool can qualify and quantify proteins reliably and reproducibly at a large scale with deep proteome coverage9,12,13. In this study, we conducted a SWATH-based proteomic analysis to explore novel CSF biomarkers related to AD, and to determine whether proteomic changes can provide new insight into AD pathophysiology. Furthermore, we aimed to build new biomarker panels that can supplement Aβ- or tau-centred conventional biomarkers. To decrease the possibility of pathologies other than AD and age-related effects affecting our results, we recruited patients with early onset AD and age-matched controls after confirming the clinical diagnosis using CSF AD biomarkers14. We also screened for genetic mutations in AβPP, PSEN1, and PSEN2, and explored the impact of genetic variants on CSF proteomic analyte expression.
Results
AD-dependent changes in the CSF quantitative proteomic profile
The AD (n = 42) and control (n = 39) groups had similar demographic characteristics but significantly different clinical features, based on the Mini-Mental State Examination (MMSE), Clinical Dementia Rating Scale (CDR), CDR sum-of-boxes (SOB) scores, number of APOE ε4 alleles, medial temporal atrophy grades, and CSF Aβ42, tTau, and pTau181 levels (Table 1).
Table 1.
Baseline data for the subjects included in this study.
| Control (n = 39) | AD (n = 42) | p-value | |
|---|---|---|---|
| Sex (M:F) | 10: 29 | 14: 28 | 0.476 |
| Age at sampling (yo) | 58.9 ± 6.3 | 60.3 ± 5.7 | 0.309 |
| Education (y) | 10.2 ± 3.2 | 10.2 ± 4.0 | 0.962 |
| Duration of illness (y) | — | 2.0 ± 1.2 | |
| MMSE | 28.3 ± 1.6 | 18.9 ± 6.4 | <0.001 |
| CDR | 0 ± 0 | 1.1 ± 0.8 | <0.001 |
| CDR-SOB | 0 ± 0.1 | 5.5 ± 5.3 | <0.001 |
| APOE ε4 carriers (%) | 12.8 | 45.2 | 0.001 |
| MTA visual grade61 | 0.1 ± 0.4 | 2.2 ± 0.9 | <0.001 |
| CSF Aβ42 (pg/mL) | 704.2 ± 141.4 | 348.4 ± 88.5 | <0.001 |
| CSF tTau (pg/mL) | 207.7 ± 55.3 | 637.8 ± 301.8 | <0.001 |
| CSF pTau181 (pg/mL) | 42.2 ± 12.6 | 78.3 ± 20.1 | <0.001 |
Values are shown in mean ± standard deviation.
p-values are determined through either independent t-test or Chi-squared tests depending on the character of the variables.
Abbreviations: AD, Alzheimer’s disease; APOE, apolipoprotein E; CDR-SOB, clinical dementia rating scale sum-of-boxes; CSF, cerebrospinal fluid; F, female; M, male; MTA, medial temporal atrophy determined by Schelten’s criteria in severe side61; MMSE, mini-mental state examination; y, years; yo, year-old.
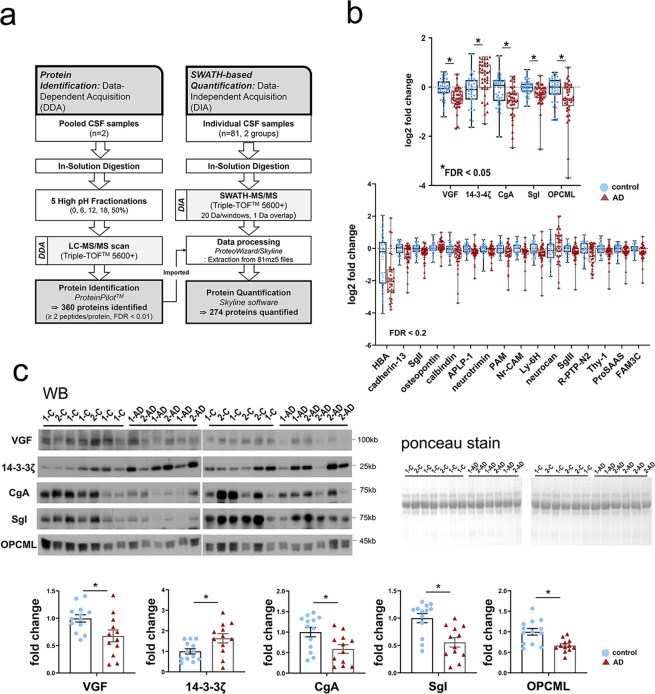
Spectral libraries for the reference map were established through fractional analysis of two pooled CSF samples using liquid chromatography–tandem MS (LC–MS/MS)-based DDA. A total of 360 CSF proteins were identified by two or more peptides per protein at an accuracy of FDR < 0.01. On the entire sample (n = 81), we then performed high-resolution proteomic analysis using SWATH-based MS to identify all fragmented compounds in a systematic and unbiased manner, via DIA (Fig. 1a). A total of 274 proteins were quantified through SWATH-MS proteomics across all samples (see Supplementary Table S1). Statistical analyses revealed significant differences between the AD and control groups in the expression of five proteins with FDR < 0.05 and in the expression of another 16 proteins with FDR < 0.2 after we corrected for multiple comparisons using the FDR method15,16 (Fig. 1b; see Supplementary Table S2).
Figure 1.
The results of SWATH–MS proteomic analysis and western blot validation. (a) Data-dependent acquisition (DDA) used pooled cerebrospinal fluid (CSF) samples to generate a CSF proteome library (left). Data-independent acquisition (DIA) was used to import DDA results to obtain the SWATH spectrum and quantify CSF proteins (right). (b) Box plots with scatter plots of log2 fold-change levels of CSF analytes revealing significant changes between AD and controls at *FDR < 0.05 (upper) and FDR < 0.2 (lower) on a t test with FDR correction. Box plots show the median and interquartile range; whiskers represent maximum and minimum values. (c) Western blot (WB) images and densitometric analyses show significant changes in indicated proteins in AD versus controls. The protein density in each lane was measured and normalized to total protein levels measured using Ponceau S staining of the corresponding polyvinylidene difluoride (PVDF) membrane. *p < 0.05 based on t test. Abbreviations: AD, Alzheimer’s disease; APLP-1, amyloid-like protein 1; C, control; CgA, chromogranin-A; SgI, secretogranin-1; FAM3C, protein FAM3C; FDR, false discovery rate; HBA, hemoglobin subunit alpha; Ly-6H, lymphocyte antigen 6 H; neurocan, neurocan core protein; Nr-CAM, neuronal cell adhesion molecule; OPCML, opioid-binding protein/cell adhesion molecule; PAM, peptidyl-glycine alpha-amidating monooxygenase; R-PTP-N2, receptor-type tyrosine-protein phosphatase N2; SgII, secretogranin-2; SgIII, secretogranin-3; Thy-1, Thy-1 membrane glycoprotein; WB, western blot; 14-3-3ζ, 14-3-3 protein zeta/delta; 1-, sample from the first cohort; 2-, sample from the second cohort.
Validation using western blot
We validated the AD-related changes in the top five candidate biomarkers using western blots of 25 CSF samples. Of these, five AD and eight control samples were from the cohort on which SWATH analysis was performed (the first cohort), whereas seven AD and five control CSF samples were from a different cohort (the second cohort). Consistent with the results of SWATH-MS, the protein expression of neurosecretory protein VGF, chromogranin-A (CgA), secretogranin-1 (SgI), and opioid-binding protein/cell adhesion (OPCML) decreased in AD samples, whereas that of 14-3-3 protein zeta/delta (14-3-3ζ) increased (Fig. 1c).
Exploration of genetic impact on protein expression levels
Next, we explored whether rare genetic variants contributed to differential expression among the 21 analytes differentially expressed in patients with AD (FDR < 0.2). The frequency of variants with minor allele frequency (MAF) < 0.01 was determined via whole-exome sequencing, performed on all AD subjects (see Supplementary Fig. S1). Only rs377747918 in the NCAN gene had much higher frequency (7.1%; 3 of 42 patients) in AD subjects than in the general population (<1%). However, there was no difference in the fold-change levels of neurocan core protein measured through rs377747918 (1.0 ± 0.6 vs. 1.4 ± 0.9, p = 0.479). Therefore, the genetic variant did not appear to affect CSF protein expression levels in our subjects.
Protein co-expression network and gene set enrichment analysis
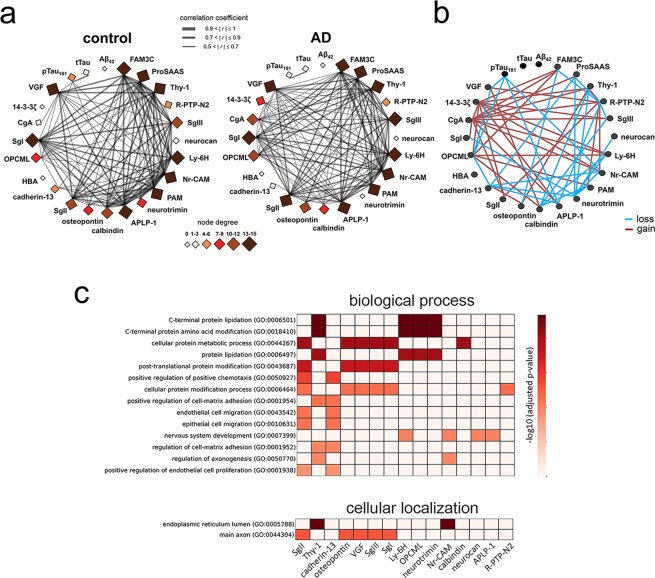
To obtain system-level insight into the altered CSF proteomic profile in AD patients, co-expression analysis was performed on the 21 top proteins with FDR < 0.2 and on AD diagnostic biomarkers, including CSF Aβ42, tTau, and pTau181 proteins (see Fig. 2a and Supplementary Table S3). Several proteins were strongly co-expressed; more than half of these had ≥ 12 close interconnections (edges on the network) in the AD group. The network changed distinctly with AD diagnosis (Fig. 2b). The largest changes were seen in the 14-3-3ζ node (gain of nine edges with AD diagnosis), CgA (gain of eleven edges with AD), and neurotrimin (loss of eight edges with AD). None of the examined proteins exhibited a significant correlation with CSF Aβ42 or tTau levels, but the following proteins had co-expression relationships with pTau181 in the control group: VGF, OPCML, APLP-1, and Nr-CAM. These findings suggest that AD-associated changes in the CSF proteome among our subjects were independent of Aβ pathology, but linked to tau pathology.
Figure 2.
Co-expression network and gene-based enrichment analysis of CSF analytes. (a) Correlation maps demonstrate the interrelationships between fold-change levels of 21 SWATH–mass spectrometry (MS) proteins and CSF levels of Aβ42, tTau, and pTau181 (node), as determined by Pearson’s correlation followed by the Bonferroni correction to correct for multiple comparisons. Edge thickness is proportional to the strength of the correlation. (b) Gains (red) and losses (blue) of protein-to-protein interrelationships in the AD group compared to the control group are revealed by comparison of the left and right network maps in (a). (c) Gene-based enrichment analysis results for the top 21 CSF analytes; Gene Ontology (GO) biological processes (upper) and GO-cellular components (lower) are presented in order of adjusted significance, which we determined by genetic association using the EnrichR tool (http://amp.pharm.mssm.edu/Enrichr). Abbreviations: AD, Alzheimer’s disease; APLP-1, amyloid-like protein 1; CgA, chromogranin-A; SgI, secretogranin-1; FAM3C, protein FAM3C; HBA, hemoglobin subunit alpha; Ly-6H, lymphocyte antigen 6 H; neurocan, neurocan core protein; Nr-CAM, neuronal cell adhesion molecule; OPCML, opioid-binding protein/cell adhesion molecule; PAM, peptidyl-glycine alpha-amidating monooxygenase; R-PTP-N2, receptor-type tyrosine-protein phosphatase N2; SgII, secretogranin-2; SgIII, secretogranin-3; Thy-1, Thy-1 membrane glycoprotein; 14-3-3ζ, 14-3-3 protein zeta/delta.
The results of Gene Ontology (GO) enrichment analysis using the gene sets of the top 21 proteins demonstrated that post-translational modifications, including in C-terminal amino acids, lipidation, and cellular protein metabolic processes, were related to CSF proteomic changes (Fig. 2c). To a lesser extent, chemotaxis, cell migration, cell-matrix adhesion, and axonogenesis were enriched biological processes. The main functional locations of these proteins were the lumen of the endoplasmic reticulum and the main axon.
Clinical utility of CSF analytes
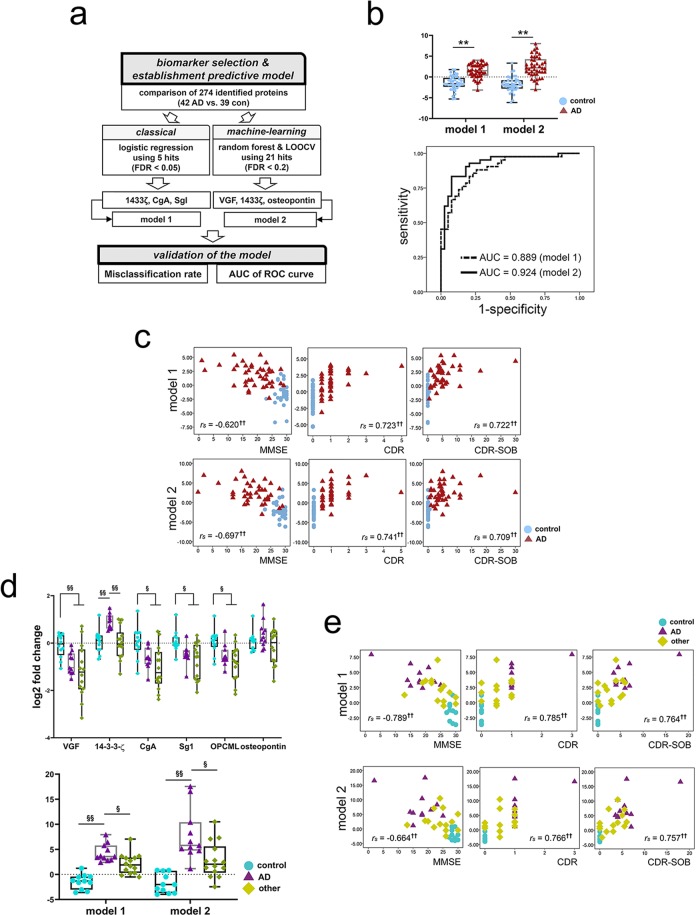
CSF expression of the top five analytes did not differ according to the presence of the APOE ε4 allele (p ≥ 0.05 overall). The fold-change in the top five CSF proteins according to AD diagnosis remained significant after we adjusted for age, education, sex, and the APOE ε4 allele covariates in multivariate analyses (B = −4.489, p = 0.001 for VGF; B = 1.339, p = 0.021 for 14-3-3ζ; B = −2.326, p = 0.015 for CgA; B = −3.981, p = 0.015 for SgI; B = −1.818, p = 0.046 for OPCML). The ability of these proteins to distinguish between AD and controls was fair, with 0.678–0.752 area under the curve (AUC) in receiver operating characteristic (ROC) analysis (see Supplementary Fig. S2). To construct a more robust model, we performed a backward stepwise logistic regression analysis. We determined that 14-3-3ζ, CgA, and SgI were significant variables and that a regression equation for their combined value (3.639 + 3.530 × 14-3-3ζ − 3.384 × CgA − 5.222 × SgI: model 1) enhanced the diagnostic accuracy of the biomarkers (AUC = 0.889). Next, because the data mining approach had been better than logistic regression for establishing a diagnostic algorithm, we conducted a random forest analysis with cross-validation using leave-one-out cross-validation (LOOCV) with more analytes (21 proteins with FDR < 0.2; Fig. 3a)17,18. Each protein was ranked according to importance score and further validated through LOOCV (see Supplementary Fig. S2). The panel of 14-3-3ζ, osteopontin, and VGF was most effective at distinguishing AD patients from controls. Their combined value through the regression equation (− 1.49 + 2.70 × 14-3-3ζ + 7.02 × osteopontin − 10.01 × VGF: model 2) contributed greatly to an AD diagnosis (AUC = 0.924; Fig. 3b), with a lower misclassification rate than logistic regression (see Supplementary Fig. S3). Because osteopontin was newly recruited into the biomarker algorithm using a data-mining approach, its AD-related change in CSF samples from the first and second cohorts was validated with western blot (n = 13). The results differed significantly between AD and control samples (p < 0.05, Supplementary Fig. S4).
Figure 3.
Analysis of the clinical utility of SWATH analytes. (a) The process of establishing CSF biomarker panels for AD through two different approaches: logistic regression (model 1) and random forest with leave-one-out cross-validation (LOOCV) (model 2). (b) A comparison of the values of model 1 and model 2 for the AD group vs. the control group (upper). The area under the curve in receiver operating characteristic (ROC) analyses of model 1 and model 2 for the diagnosis of Alzheimer’s disease. **p < 0.01 based on t test. (c) Correlations between CSF biomarker panel values and cognitive function (Mini-Mental State Examination, MMSE) and dementia severity profiles (Clinical Dementia Rating Scale [CDR], CDR sum-of-boxes [SOB] scores). rs: Spearman’s rank correlation coefficient, †p < 0.05 and ††p < 0.01 based on Spearman’s rank correlation analysis. (d) A comparison of the values of six quantified proteins (log2 fold-change levels of each protein, upper) and CSF panels (equation values, lower) in three diagnostic groups from the second cohort. §p < 0.05 and §§p < 0.01 based on the Kruskal–Wallis test followed by pairwise post-hoc analysis with significance levels adjusted for the number of comparisons. (e) Correlations between CSF biomarker panel values and clinical profiles (MMSE, CDR, CDR-SOB). rs: Spearman’s rank correlation coefficient, †p < 0.05 and ††p < 0.01 based on Spearman’s rank correlation analysis. Abbreviations: AD, Alzheimer’s disease; AUC, area under the curve; CDR-SOB, clinical dementia rating scale sum-of-boxes; CgA, chromogranin-A; FDR, false discovery rate; SgI, secretogranin-1; CON, control; MMSE, mini-mental state examination; OPCML, opioid-binding protein/cell adhesion molecule; 14-3-3ζ, 14-3-3 protein zeta/delta.
Spearman’s rank correlation analysis showed that both models correlated strongly with cognitive performance (rs = − 0.620, p < 0.001, for model 1 and rs = − 0.697, p < 0.001, for model 2 with MMSE) and dementia severity (rs = 0.723, p < 0.001, for model 1 and rs = 0.741, p < 0.001, for model 2 with CDR; rs = 0.722, p < 0.001, for model 1 and rs = 0.709, p < 0.001, for model 2 with CDR-SOB; Fig. 3c). The models performed better than any individual biomarker (see Supplementary Fig. S5).
Validation using targeted PRM-MS in a different cohort including subjects with other brain disorders
We further validated candidate CSF biomarkers using high-resolution targeted PRM-MS in a different cohort (the second cohort) composed of subjects with other brain disorders (n = 14) in addition to AD (n = 11) and age-matched controls (n = 11; see Supplementary Table S4). The top five SWATH-MS analytes and osteopontin levels were quantified (see Supplementary Table S5), and their utility in combined algorithms was tested. Both CSF panels efficiently differentiated AD from other diseases and from controls (all p < 0.05; Fig. 3d). In addition, the panels had a strong persistent correlation with clinical profiles (Fig. 3e).
Discussion
In the current study, we adopted strict diagnostic criteria to minimise age- and mutation-related confounding effects, and used unbiased high-resolution proteomic analysis to derive a new AD-related CSF protein profile and co-expression network. We also proposed a list of candidate AD biomarkers, demonstrating their utility in terms of AD diagnosis and their ability to reflect clinical severity.
Among the top five proteomic hits, VGF, CgA, and SgI are neurosecretory granin proteins. VGF is involved in the regulation of neurite growth, neurogenesis, and synaptic plasticity in the brain19; its synthesis is highly regulated by neurotrophic factors20,21. CgA and SgI are critical in the formation of secretory vesicles, and are involved in vasodilation, anti-apoptosis, microglial activation, neurotransmitter release, and synaptic function22. Altered CSF levels in VGF, CgA, and SgI have been suggested to represent synaptic loss and neuronal degeneration in AD patients23–25. Similarly, we identified significant correlations between the expression levels of these proteins and metrics of cognitive deterioration and dementia severity. However, the direction of change in the markers in our AD patients was contradictory to those reported in some previous studies25. This discrepancy might be due to dynamic changes in neurosecretory granin proteins at various stages of AD. A transient rise in the CSF levels of these synaptic proteins may occur as an early event in AD, followed by continual decreases along with disease progression26. This finding is supported by previous biological studies showing that a pathway compensating for synaptic damage increased the sizes of vesicles and synapses during the early stage of AD27.
Changes in the levels of these synapse-related proteins are not unique to AD; they have also been detected in other neurological disorders, such as multiple sclerosis28, schizophrenia29, Parkinson’s disease30, and amyotrophic lateral sclerosis (ALS)31,32. Consistent with these findings, we also found diminished CSF levels of VGF, CgA, and Sg1 in subjects with other brain disorders (e.g., frontotemporal dementia, Parkinson’s disease, and cerebrovascular disease). However, an optimal combination of biomarkers identified through multivariate analysis improved the accuracy of classification of AD versus other brain disorders.
Co-expression network analysis showed that the 14-3-3ζ protein had many co-expression relationships with other significant proteins in AD patients, the biological roles of which are known to be mainly related to synapse and neurite outgrowth33–39. The 14-3-3ζ protein had a unique power to discriminate between AD and other brain disorders (Fig. 3d). This common biomarker was incorporated into two biomarker panels that were obtained through different statistical approaches. The 14-3-3 family of proteins includes ubiquitous scaffolding proteins in the brain that regulate various signalling pathways by recognising phosphoserine or phosphothreonine40. In particular, the 14-3-3ζ protein plays a role as an effector during tau protein phosphorylation41 and regulates tau aggregation42. Enriched biological processes linked to AD-related proteomic changes in this study included post-translational protein modification, cell migration, and axonogenesis. Together, these results suggest that increased 14-3-3ζ protein levels in the brain43 enhance synaptic degeneration in AD patients via effects on tau pathology, which may be reflected in the expression changes observed in CSF synaptic proteins. However, the direct relationship between significant CSF protein changes and AD pathophysiology needs additional biological study, as we only evaluated proteins released into the CSF from brain tissue. Osteopontin was unique in that it was part of a diagnostic panel generated by the random forest method despite the fact that its fold-change in AD versus controls was moderate (FDR < 0.2). Osteopontin, a secreted glyco-phosphoprotein with a role in cell-matrix interactions and innate immunity, increases in AD patients during the acute phase of disease progression44. However, specificity to AD is unlikely given the results of our validation process and a previous report describing an increase in CSF osteopontin levels in LBD and PD in addition to AD45.
The new biomarker panels are thought to be better than the established CSF biomarkers with respect to clinical correlations of AD, but not in discriminating AD from control samples based on previous findings from our14 and other46–50 research groups. The new biomarkers demonstrated abundant interconnections and some connections with CSF pTau181 levels, which contrast to no co-expression with the Aβ42 protein. This result demonstrates the potential value of our candidate biomarkers as indicators of Aβ-independent pathology. The recent failure of Aβ-targeting therapy (despite evidence of effects on an Aβ-related biomarker)51,52 means that there is a need for additional biomarkers. Ideally, new biomarkers of Aβ- or tau-independent brain pathology should correlate with clinical symptoms53. In this context, the proteins (and combinations thereof) identified through SWATH–MS analysis in this study could be useful biomarkers for AD. The biological pathways related to post-translational protein modification, cell migration and axonogenesis are thought to synergistically enhance the protein–protein interactions involved in synaptic degeneration and brain dysfunction. The characteristic CSF protein profile described herein provides new targets for future biological research aimed at developing diagnostic tools for management of AD.
Methods
Participants
This study followed local clinical research regulations under the approval of the ethical review board of Soonchunhyang University Bucheon Hospital (SCHBC_IRB_2012-124) and Ajou University Medical Center (AJIRB-BMR-SMP-18-545). The study adhered to the principles of the Declaration of Helsinki. All participants and their legal guardians (in cases of dementia) gave written informed consent. Four university hospitals in the capital area of South Korea participated in this project and prospectively collected samples from January 2014 to January 2017 that were used for this study. The clinical diagnosis of AD was based on the National Institute on Aging and Alzheimer’s Association diagnostic guidelines2, incorporating information from a comprehensive neuropsychological battery54, laboratory and neurological examinations, and neuroimaging, which were performed within 1 month before collecting the CSF. Adherence to the CSF criteria for AD was considered to include AD subjects who met the diagnostic cut-offs of our laboratory14. When pathogenic AD mutations of AβPP, PSEN1, and PSEN2 were identified, the subjects were excluded from the study. The age-matched control subjects had no history of a neurological disorder or systemic disorder that could potentially affect cognitive function, showed normal cognition on a neuropsychological test, and exhibited no abnormality on brain computed tomography or MRI. Moreover, CSF levels of Aβ42, tTau, and pTau181 in the normal range were mandatory in the control group. As blood contamination can affect CSF proteomic results, samples with >10/mm3 erythrocytes on a routine CSF analysis were excluded from the proteomic analysis in both groups. Clinical follow-up was performed at >6 months after the initial puncture to ensure the correct diagnosis. The second cohort, composed of subjects with various neurological disorders, was drawn from two university hospitals, and CSF samples were collected from January 2016 to June 2019. The basic requirements for participation were the same as those for the first cohort.
CSF collection and preparation for analysis
CSF was sampled and stored according to the established protocol for a biomarker study55. CSF analyses were performed at a biomarker core laboratory. All CSF samples were thawed immediately prior to analysis. Aβ42, tTau, and pTau181 protein levels were measured using the INNOTEST enzyme-linked immunosorbent assay kit (Fuijrebio Diagnostics, Ghent, Belgium).
SWATH-based MS
A DDA process with two pooled CSF samples from four individuals was executed to identify all fragmented compounds in a systematic and unbiased manner, and a CSF proteome library for SWATH–MS-based proteomic analysis was generated. Next we digested and prepared the CSF protein samples (n = 81) according to a previously described method for protein quantification by SWATH–MS12,56. The size of this study was comparable to a previous report on CSF57. A Triple-TOF 5600+ mass spectrometer (AB Sciex, Concord, ON, Canada) was used for all experiments. All spectra generated from DDA were searched using the ProteinPilot searching algorithm (SCIEX, Framingham, MA, USA) against a Uniprot human protein sequences database (UP000005640_9606_cRAP.fasta: total 21,159 protein entries) with the following search parameters: fully tryptic digestion; <50 ppm precursor ion tolerance; <0.5 Da fragment ion mass tolerance; fixed modifications for cysteine (+57 Da: carbamidomethylation); and biological modifications/artefacts, such as methionine oxidation (+16 Da). To reduce the false identification rate of proteins, we used a cut-off of two or more peptides as a qualification criterion, which permitted a peptide confidence level of >0.99. A CSF proteome spectral library was constructed using the Skyline software and the identified peptides58. All raw SWATH–MS data (WIFF files) were converted to the mz5 format using the ProteoWizard software, Version 3.0.6965 (http://proteowizard.sourceforge.net/). DIA data were extracted using the Skyline software, and DDA results were imported into Skyline with a cut-off criterion of 0.95. After extraction using the Skyline software, 274 proteins (1,006 peptides) were quantified in the 81 CSF samples. Two or more peptides were identified in all individual proteins, and 199 proteins had more than two peptides per protein. The areas under the peptide peaks for individual proteins were summed and used in expression analysis. Details of the SWATH–MS analysis are provided in Supplementary Information.
Western blot
Western blot analysis of CSF was performed with 12 AD and 13 control samples. As a loading control, the expression of the target proteins was normalized to total protein levels measured using Ponceau S staining. Further details are available in the Supplementary Information.
Targeted proteomics using PRM-MS
The sample preparation process was essentially the same as for the SWATH-MS analysis. The specifics of the process are described in the Supplementary Information. We targeted the top five proteins (FDR < 0.05) and osteopontin (FDR < 0.2) for PRM validation (see Supplementary Table S6). The latter was selected because of its contribution to the random forest model.
Statistical analysis
All statistical analyses were performed using R statistical computing software (version 3.6.1; R Foundation for Statistical Computing, Vienna, Austria). As the raw value of SWATH-MS analysis is spectral area, which relies on the number of peptides identified from a given protein, direct comparison between proteins is inadequate. Therefore, the protein level fold-changes were calculated relative to the mean of control group in all subjects within a protein to perform a statistical analysis59. The independent-samples t-test and the Mann–Whitney U test were used to analyse differences between the AD and controls, with the choice of test depending on whether data were normally distributed. The Kruskal–Wallis test was performed for three-group comparisons in the second cohort. The χ2 test was applied for categorical variables. The Benjamini–Hochberg FDR method was used to correct for multiple comparisons15,16. Binary logistic regression was used to conduct multivariate analysis with various covariates. Logistic regression analysis with backward stepwise selection or the random forest method with LOOCV was used to establish a diagnostic CSF algorithm. ROC curve analysis and LOOCV were performed to validate the diagnostic accuracy of the biomarkers. Correlation analyses were conducted using Spearman’s rank correlation and Pearson’s correlation tests to estimate the relationships between the CSF analytes and elements of the clinical profile and to build a co-expression network, respectively. The Bonferroni correction was used to adjust for errors in co-expression network analyses due to multiple comparisons.
Exome sequencing and calling of rare variants in gene sets of the SWATH analytes
Genomic DNA were extracted using the QIAamp® DNA blood midi kit (Qiagen, Valencia, CA, USA). Each exome library was prepared according to Agilent library preparation protocols (Agilent Human All Exon V5 + UTR kit; Agilent Technologies, Palo, Alto, CA, USA) and final libraries were then sequenced on an Illumina HiSeq. 2000 instrument (mean coverage × 200.3, 100-bp) by Macrogen (Seoul, Korea). To screen for effects of rare germline variants in 21 corresponding genes of CSF proteins with FDR < 0.2 on the expression changes in AD, the variants with MAF < 0.01 from the 1KG Database (East Asian) were checked against the open database of the Genome Aggregation Consortium (GnomAD; East Asian; http://gnomad.broadinstitute.org) and the Korean Reference Genome Database (KRGDB) from 1,722 Koreans (http://coda.nih.go.kr/coda/KRGDB/index.jsp). See the Supplementary Information for details.
Gene set enrichment analysis
The enrichment analysis was performed using the updated EnrichR tool (http://amp.pharm.mssm.edu/Enrichr) to identify overrepresented GO biological processes and cellular components of the gene sets from the CSF analytes60.
Supplementary information
Supplementary methods, Supplementary Figures.
Acknowledgements
This work was supported by Basic Science Research Program (NRF-2018R1A2B6009439 and 2019R1A5A2026045) and the Original Technology Research Program for Brain Science (NRF-2018M3C7A1056293) of the National Research Foundation of Korea (NRF) funded by the Korean government, MSIT, and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HI14C1942). We thank Prof. So Young Moon and Song Mi Han for their support during the revision.
Author contributions
S.A.P. and J.H.B. designed the experiments and major contribution in all the experiments. S.A.P., J.M.J. and B.P. conducted statistical analysis. J.M.J. performed bioinformatic analysis. J.S.P. and J.H.L. executed gene-protein association analysis. H.J.K., J.H.P., W.S.C., J.H.J., and S.H.C. performed a clinical association study. S.A.P., J.M.J., and J.H.B. wrote the manuscript. All authors reviewed the manuscript.
Data availability
The proteomic datasets generated during this study are included in this published article as supplementary information files. The availability of the genetic datasets that support the findings of this study are not publicly available. Data are however available from the authors upon reasonable request and permission of institutional review board.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-64461-y.
References
- 1.Shaw LM, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann. Neurol. 2009;65:403–413. doi: 10.1002/ana.21610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKhann GM, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7:263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jack CR, Jr., et al. NIA-AA Research Framework: toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018;14:535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blennow K. Biomarkers in Alzheimer’s disease drug development. Nat. Med. 2010;16:1218–1222. doi: 10.1038/nm.2221. [DOI] [PubMed] [Google Scholar]
- 5.Bateman RJ, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N. Engl. J. Med. 2012;367:795–804. doi: 10.1056/NEJMoa1202753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mattsson N, et al. Revolutionizing Alzheimer’s disease and clinical trials through biomarkers. Alzheimers Dement. (Amst.) 2015;1:412–419. doi: 10.1016/j.dadm.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Molinuevo JL, et al. Current state of Alzheimer’s fluid biomarkers. Acta Neuropathol. 2018;136:821–853. doi: 10.1007/s00401-018-1932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Portelius E, et al. Proteomic studies of cerebrospinal fluid biomarkers of Alzheimer’s disease: an update. Expert Rev. Proteomics. 2017;14:1007–1020. doi: 10.1080/14789450.2017.1384697. [DOI] [PubMed] [Google Scholar]
- 9.Meyer JG, Schilling B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert Rev. Proteomics. 2017;14:419–429. doi: 10.1080/14789450.2017.1322904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ludwig C, et al. Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol. Syst. Biol. 2018;14:e8126. doi: 10.15252/msb.20178126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chandramouli K, Qian PY. Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum. Genomics Proteomics. 2009;2009:239204. doi: 10.4061/2009/239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics. 2012;11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jylha A, et al. Comparison of iTRAQ and SWATH in a clinical study with multiple time points. Clin. Proteomics. 2018;15:24. doi: 10.1186/s12014-018-9201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park SA, et al. Cerebrospinal fluid biomarkers for the diagnosis of Alzheimer disease in South Korea. Alzheimer Dis. Assoc. Disord. 2017;31:13–18. doi: 10.1097/wad.0000000000000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995;57:289–300. [Google Scholar]
- 16.Diz AP, Carvajal-Rodriguez A, Skibinski DO. Multiple hypothesis testing in proteomics: a strategy for experimental work. Mol. Cell. Proteomics. 2011;10:M110.004374. doi: 10.1074/mcp.M110.004374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Touw WG, et al. Data mining in the Life Sciences with Random Forest: a walk in the park or lost in the jungle? Brief Bioinform. 2013;14:315–326. doi: 10.1093/bib/bbs034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu S, et al. Serum Protein-Based Profiles as Novel Biomarkers for the Diagnosis of Alzheimer’s Disease. Mol. Neurobiol. 2018;55:3999–4008. doi: 10.1007/s12035-017-0609-0. [DOI] [PubMed] [Google Scholar]
- 19.Alder J, et al. Brain-derived neurotrophic factor-induced gene expression reveals novel actions of VGF in hippocampal synaptic plasticity. J. Neurosci. 2003;23:10800–10808. doi: 10.1523/JNEUROSCI.23-34-10800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bonni A, Ginty DD, Dudek H, Greenberg ME. Serine 133-phosphorylated CREB induces transcription via a cooperative mechanism that may confer specificity to neurotrophin signals. Mol. Cell. Neurosci. 1995;6:168–183. doi: 10.1006/mcne.1995.1015. [DOI] [PubMed] [Google Scholar]
- 21.Possenti R, Di Rocco G, Nasi S, Levi A. Regulatory elements in the promoter region of vgf, a nerve growth factor-inducible gene. Proc. Natl. Acad. Sci. USA. 1992;89:3815–3819. doi: 10.1073/pnas.89.9.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartolomucci A, et al. The extended granin family: structure, function, and biomedical implications. Endocr. Rev. 2011;32:755–797. doi: 10.1210/er.2010-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perrin RJ, et al. Identification and validation of novel cerebrospinal fluid biomarkers for staging early Alzheimer’s disease. PLoS One. 2011;6:e16032. doi: 10.1371/journal.pone.0016032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrette O, et al. A panel of cerebrospinal fluid potential biomarkers for the diagnosis of Alzheimer’s disease. Proteomics. 2003;3:1486–1494. doi: 10.1002/pmic.200300470. [DOI] [PubMed] [Google Scholar]
- 25.Jahn H, et al. Peptide fingerprinting of Alzheimer’s disease in cerebrospinal fluid: identification and prospective evaluation of new synaptic biomarkers. PLoS One. 2011;6:e26540. doi: 10.1371/journal.pone.0026540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duits FH, et al. Synaptic proteins in CSF as potential novel biomarkers for prognosis in prodromal Alzheimer’s disease. Alzheimers Res. Ther. 2018;10:5. doi: 10.1186/s13195-017-0335-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merlo S, Spampinato SF, Sortino MA. Early compensatory responses against neuronal injury: a new therapeutic window of opportunity for Alzheimer’s disease? CNS Neurosci. Ther. 2019;25:5–13. doi: 10.1111/cns.13050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mattsson N, et al. Cerebrospinal fluid concentrations of peptides derived from chromogranin B and secretogranin II are decreased in multiple sclerosis. J. Neurochem. 2007;103:1932–1939. doi: 10.1111/j.1471-4159.2007.04985.x. [DOI] [PubMed] [Google Scholar]
- 29.Landen M, et al. Reduction of chromogranin A and B but not C in the cerebrospinal fluid in subjects with schizophrenia. Eur. Neuropsychopharmacol. 1999;9:311–315. doi: 10.1016/S0924-977X(98)00042-X. [DOI] [PubMed] [Google Scholar]
- 30.Cocco C, et al. Distribution of VGF peptides in the human cortex and their selective changes in Parkinson’s and Alzheimer’s diseases. J. Anat. 2010;217:683–693. doi: 10.1111/j.1469-7580.2010.01309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pasinetti GM, et al. Identification of potential CSF biomarkers in ALS. Neurology. 2006;66:1218–1222. doi: 10.1212/01.wnl.0000203129.82104.07. [DOI] [PubMed] [Google Scholar]
- 32.Kaiserova M, et al. Cerebrospinal fluid levels of chromogranin A and phosphorylated neurofilament heavy chain are elevated in amyotrophic lateral sclerosis. Acta Neurol. Scand. 2017;136:360–364. doi: 10.1111/ane.12735. [DOI] [PubMed] [Google Scholar]
- 33.Leifer D, Kowall NW. Thy-1 in hippocampus: normal anatomy and neuritic growth in Alzheimer’s disease. J. Neuropathol. Exp. Neurol. 1992;51:133–141. doi: 10.1097/00005072-199203000-00003. [DOI] [PubMed] [Google Scholar]
- 34.Liu L, Watanabe N, Akatsu H, Nishimura M. Neuronal expression of ILEI/FAM3C and its reduction in Alzheimer’s disease. Neuroscience. 2016;330:236–246. doi: 10.1016/j.neuroscience.2016.05.050. [DOI] [PubMed] [Google Scholar]
- 35.Pla V, et al. Secretory sorting receptors carboxypeptidase E and secretogranin III in amyloid beta-associated neural degeneration in Alzheimer’s disease. Brain Pathol. 2013;23:274–284. doi: 10.1111/j.1750-3639.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puddifoot CA, Wu M, Sung RJ, Joiner WJ. Ly6h regulates trafficking of alpha7 nicotinic acetylcholine receptors and nicotine-induced potentiation of glutamatergic signaling. J. Neurosci. 2015;35:3420–3430. doi: 10.1523/jneurosci.3630-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakurai T. The role of NrCAM in neural development and disorders-beyond a simple glue in the brain. Mol. Cell. Neurosci. 2012;49:351–363. doi: 10.1016/j.mcn.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Schilling S, et al. APLP1 Is a synaptic cell adhesion molecule, supporting maintenance of dendritic spines and basal synaptic transmission. J. Neurosci. 2017;37:5345–5365. doi: 10.1523/jneurosci.1875-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spellman DS, et al. Development and evaluation of a multiplexed mass spectrometry based assay for measuring candidate peptide biomarkers in Alzheimer’s Disease Neuroimaging Initiative (ADNI) CSF. Proteomics Clin. Appl. 2015;9:715–731. doi: 10.1002/prca.201400178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berg D, Holzmann C, Riess O. 14-3-3 proteins in the nervous system. Nat. Rev. Neurosci. 2003;4:752–762. doi: 10.1038/nrn1197. [DOI] [PubMed] [Google Scholar]
- 41.Hashiguchi M, Sobue K, Paudel HK. 14-3-3zeta is an effector of tau protein phosphorylation. J. Biol. Chem. 2000;275:25247–25254. doi: 10.1074/jbc.M003738200. [DOI] [PubMed] [Google Scholar]
- 42.Sadik G, et al. Phosphorylation of tau at Ser214 mediates its interaction with 14-3-3 protein: implications for the mechanism of tau aggregation. J. Neurochem. 2009;108:33–43. doi: 10.1111/j.1471-4159.2008.05716.x. [DOI] [PubMed] [Google Scholar]
- 43.Andreev VP, et al. Label-free quantitative LC-MS proteomics of Alzheimer’s disease and normally aged human brains. J. Proteome Res. 2012;11:3053–3067. doi: 10.1021/pr3001546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Y, et al. Elevated osteopontin levels in mild cognitive impairment and Alzheimer’s disease. Mediators Inflamm. 2013;2013:615745. doi: 10.1155/2013/615745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heywood WE, et al. Identification of novel CSF biomarkers for neurodegeneration and their validation by a high-throughput multiplexed targeted proteomic assay. Mol. Neurodegener. 2015;10:64. doi: 10.1186/s13024-015-0059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wallin AK, et al. CSF biomarkers for Alzheimer’s Disease: levels of beta-amyloid, tau, phosphorylated tau relate to clinical symptoms and survival. Dement. Geriatr. Cogn. Disord. 2006;21:131–138. doi: 10.1159/000090631. [DOI] [PubMed] [Google Scholar]
- 47.Mattsson N, et al. CSF biomarkers and incipient Alzheimer disease in patients with mild cognitive impairment. JAMA. 2009;302:385–393. doi: 10.1001/jama.2009.1064. [DOI] [PubMed] [Google Scholar]
- 48.Vemuri P, et al. MRI and CSF biomarkers in normal, MCI, and AD subjects: diagnostic discrimination and cognitive correlations. Neurology. 2009;73:287–293. doi: 10.1212/WNL.0b013e3181af79e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulder C, et al. Amyloid-beta(1-42), total tau, and phosphorylated tau as cerebrospinal fluid biomarkers for the diagnosis of Alzheimer disease. Clin. Chem. 2010;56:248–253. doi: 10.1373/clinchem.2009.130518. [DOI] [PubMed] [Google Scholar]
- 50.Jung, N.Y. et al. Comparison of diagnostic performances between cerebrospinal fluid biomarkers and amyloid PET in a clinical setting. J. Alzheimers Dis. 10.3233/JAD-191109, 10.3233/JAD-191109 (2020). [DOI] [PubMed]
- 51.Farlow M, et al. Safety and biomarker effects of solanezumab in patients with Alzheimer’s disease. Alzheimers Dement. 2012;8:261–271. doi: 10.1016/j.jalz.2011.09.224. [DOI] [PubMed] [Google Scholar]
- 52.Rinne JO, et al. 11C-PiB PET assessment of change in fibrillar amyloid-beta load in patients with Alzheimer’s disease treated with bapineuzumab: a phase 2, double-blind, placebo-controlled, ascending-dose study. Lancet Neurol. 2010;9:363–372. doi: 10.1016/s1474-4422(10)70043-0. [DOI] [PubMed] [Google Scholar]
- 53.Dhiman K, Blennow K, Zetterberg H, Martins RN, Gupta VB. Cerebrospinal fluid biomarkers for understanding multiple aspects of Alzheimer’s disease pathogenesis. Cell. Mol. Life Sci. 2019;76:1833–1863. doi: 10.1007/s00018-019-03040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park HK, et al. Cognitive profiles and neuropsychiatric symptoms in Korean early-onset Alzheimer’s disease patients: a CREDOS study. J. Alzheimers Dis. 2015;44:661–673. doi: 10.3233/jad-141011. [DOI] [PubMed] [Google Scholar]
- 55.Park SA, et al. A consensus in Korea regarding a protocol to reduce preanalytical sources of variability in the measurement of the cerebrospinal fluid biomarkers of Alzheimer’s disease. J. Clin. Neurol. 2015;11:132–141. doi: 10.3988/jcn.2015.11.2.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee WK, Baek JH, Ryoo SW, Yu YG. Swath-based comparative proteomic analysis of the Mycobacterium bovis BCG-Korea strain. Bull. Korean Chem. Soc. 2014;35:933–937. doi: 10.5012/bkcs.2014.35.3.933. [DOI] [Google Scholar]
- 57.Spreafico F, et al. Proteomic analysis of cerebrospinal fluid from children with central nervous system tumors identifies candidate proteins relating to tumor metastatic spread. Oncotarget. 2017;8:46177–46190. doi: 10.18632/oncotarget.17579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pursiheimo A, et al. Optimization of statistical methods impact on quantitative proteomics data. J Proteome Res. 2015;14:4118–4126. doi: 10.1021/acs.jproteome.5b00183. [DOI] [PubMed] [Google Scholar]
- 60.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scheltens P, et al. Atrophy of medial temporal lobes on MRI in “probable” Alzheimer’s disease and normal ageing: diagnostic value and neuropsychological correlates. J. Neurol. Neurosurg. Psychiatry. 1992;55:967–972. doi: 10.1136/jnnp.55.10.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods, Supplementary Figures.
Data Availability Statement
The proteomic datasets generated during this study are included in this published article as supplementary information files. The availability of the genetic datasets that support the findings of this study are not publicly available. Data are however available from the authors upon reasonable request and permission of institutional review board.