Abstract
The objective of the study was to elucidate the microRNA (miRNA) profile of an enriched human corneal epithelial stem cell (CESC) population in comparison to differentiated central corneal epithelial cells (CCECs) by small RNA sequencing. The CESCs were enriched by differential enzymatic treatment to isolate the basal limbal epithelial cells followed by laser capture microdissection of cells with nucleus to cytoplasm ratio ≥0.7, from donor tissues. Small RNA sequencing was carried out using Illumina NextSeq. 500 platform and the validation of differentially expressed miRNAs by quantitative real-time PCR (qPCR) and locked nucleic acid miRNA in-situ hybridization (LNA-ISH). The sequencing identified 62 miRNAs in CESCs and 611 in CCECs. Six miRNAs: hsa-miR-21-5p, 3168, 143-3p, 10a-5p, 150-5p and 1910-5p were found to be significantly upregulated in enriched CESCs, which was further confirmed by qPCR and LNA-ISH. The expression of hsa-miR-143-3p was exclusive to clusters of limbal basal epithelial cells. The targets of the upregulated miRNAs were predicted to be associated with signaling pathways -Wnt, PI3K-AKT, MAPK and pathways that regulate pluripotency of stem cells, cell migration, growth and proliferation. Further studies are essential to elucidate their functional role in maintenance of stemness. The findings of the study also hypothesize the inherent potential of hsa-miR-143-3p to serve as a biomarker for identifying CESCs.
Subject terms: Adult stem cells, Non-coding RNAs
Introduction
The anterior surface of the human eye is defined by the tear film and cornea. The cornea acts as the natural barrier by preventing the underlying delicate structures from harmful radiation and potent infections1. The terminally differentiated cells in the superficial layer of the human corneal epithelium are normally shed and the cell loss is replenished by the progeny of the corneal epithelial stem cells (CESCs) that reside in the basal epithelial layer of the limbus to maintain tissue homeostasis. The CESCs give rise to cells that can proliferate, differentiate and migrate centripetally2,3. These adult stem cells possess slow cycling4, label retaining cell property5 and has the ability to form holoclones6,7. Loss or dysfunction of the CESCs results in conjunctivalization, vascularization and loss of vision, a condition termed as limbal stem cell deficiency8,9.
Even though it is well accepted that the limbus is the site of stem cells for corneal epithelial homeostasis10–12, the regulatory mechanism governing the maintenance of these CESCs is still not clear. MicroRNAs (miRNAs) are non-coding RNAs approximately 22-nucleotide-long and they act as key players in regulating gene expression by RNAi machinery. In the last decade, the importance of miRNAs as a potential epigenetic regulator of stem cell potency, proliferation, differentiation and survival in embryonic stem cells13,14, induced pluripotent stem cells15,16 and adult tissue resident stem cells like human skin/hair follicle17 has been reported.
It has been demonstrated that only 25% of the isolated limbal basal epithelial cells are stem cells based on the analysis of two parameters, high levels of p63 expression and greater nucleus to cytoplasmic area7. It is crucial to make use of a highly enriched CESCs in order to identify the profile of miRNA specific to adult tissue resident stem cells. Therefore, in this study we have made use of the strategy we have developed earlier7,18 to enrich CESCs to the extent of 80% and this enriched population are known to express stem cell markers ΔNp63α and ABCG218. It was thus possible to identify a specific profile of miRNA, significantly up-regulated in CESCs compared to differentiated CCECs19. Accordingly, the major findings of the study will serve as a platform to decipher the unresolved questions relating to the miRNA regulation of CESCs.
Methods
Samples
Human donor tissues were handled in accordance with the tenets of the Declaration of Helsinki and the study was approved by Institutional Ethics Committee Aravind Medical Research Foundation (RES2013038BAS). Informed consent was obtained for all donor eyes including the minors from the legally authorized representative - either the donor’s parents or family through the Eye Banks of Aravind Eye Care System (Madurai, Coimbatore, Pondicherry and Tirunelveli). The donor globes not suitable for transplantation (procured within 8 hours of death and received within 24 hours for research) and limbal rims obtained after corneal transplantation (received within 10 days of storage in Cornisol media at 4 °C in sterile condition) were included in this study. Inclusion criteria were donor age less than 70 years (range: 11 to 70 years), non-vascularized, with no history of diabetes and ocular infection. The donor globes were observed under stereo binocular microscope and those with intact limbus with radial ridges of palisades of Vogt were used for the study.
Enrichment of corneal epithelial stem cells
The corneal region together with adjacent sclera was dissected out from the donor globes and the corneal endothelium was removed using a sterile cotton tip20,21. Central cornea was punched out using 8 mm trephine and the central corneal epithelial cells (CCECs) were isolated following the protocol of Arpitha et al.20. Briefly, the corneal epithelial sheet was separated by dispase II (2 mg/ml) treatment and the individual cells were obtained after 0.25% trypsin treatment. For enrichment of the CESCs, a two step-protocol was followed18. The limbal basal epithelial cells (LBECs) were isolated from limbal tissues by differential enzymatic treatment – 0.25% trypsin followed by 2 mg/ml dispase II treatment7. The cytosmears of isolated LBECs on membrane slide were stained with Giemsa. The cells were focused with 20X magnification and the image was projected on the computer monitor connected to an automated microscope. The area of nucleus and cytoplasm of each cell and the nucleus to cytoplasmic (N/C) ratio was then calculated using PALM Robo Software 4.3 SP2. The corneal epithelial stem cells (CESCs) with N/C ratio >0.7 were selectively marked and cut using Positioning and Ablation Laser MicroBeam (PALM) and collected on the adhesive cap of the collection tube which positioned to overhang the area under focus. Catapulted cells collected in the adhesive cap can be checked by moving it directly over the objective lens at any point of time during the collection. The enriched CESCs in the adhesive cap of the collection tube were then incubated for 30 minutes at 42 °C in 15 µL of extraction buffer (ARCTURUS PicoPure RNA Isolation Kit) and collected to the bottom of the tube from the cap by a brief centrifugation. The collected cell lysate was stored at −80 °C until RNA isolation18,22.
Small RNA sequencing and data analysis
The total RNA was extracted from pooled CESCs (2046 cells) and CCECs (8.55 × 105 cells) using ARCTURUS PicoPure RNA Isolation Kit from two donor tissues (Age: 60 and 62 years) for small RNA sequencing. Total RNA was used for small RNA library construction using TruSeq Small RNA Library Prep Kit (Illumina, California, USA) following manufacturer’s protocol. Libraries were quantified using Qubit dsDNA HS Assay Kit (Invitrogen, California, USA) and validated in bioanalyzer using High Sensitivity DNA Kit (Agilent, California, USA). Based on the library QC report, the library generated was suitable for sequencing on Illumina. For CCECs, library distribution was in the range of 120bp-180bp and for CESCs the range was 120bp-160bp indicating the presence of small RNA inserts. The effective insert length was in the range of ~10bp- 40 bp; with combined size of adapters flanking the library being ~120 bp. Cluster generation and sequencing was carried out using NextSeq. 500 High-Output v2 Kit (Illumina, Inc.) using 75 cycles chemistry at the Genotypic Technology Pvt Ltd (Bangalore, India).
First, the quality assessment of raw deep sequencing data was performed with the FastQC tool. Adapter and low-quality reads were discarded from raw sequencing data using our Perl script, allowing no mismatches for adapter identification. Further, the data were uploaded into Oasis 2.0 online software23,24 for the miRNA analysis. Briefly, the data with the read size (15–32 nt) and low abundance reads (<5 reads) were discarded and remaining data were aligned to Homo sapiens hg19 genome reference. For differential expression (DE) analysis, NOISeq R software package were applied on the TMM normalized reads (R version 3.5.1). The variable expression of miRNAs between two cell types was considered significant when the fold change was 2 or greater and the probability score was more than 0.9. The results were represented in the volcano plot constructed by R version 3.5.125 based on the NOISeq scores: the M-value and D-value.
Quantitative real time PCR
For confirmation of the sequencing data, quantitative real time PCR was carried out using (i) CESCs from 33 pairs of limbal rims (cells from 11 pairs were pooled as one sample to get around 3500 cells/pool, n = 3) and CCECs from three pairs of corneal button and (ii) LBECs and CCECs from limbal rims and corneal buttons respectively, were pooled to obtain a minimum of 1 × 106 cells each (pool 1: 7 pairs; pool 2: 7 pairs; pool 3: 6 pairs of tissues), but after ensuring that there was no bias of age. The RNA concentration was estimated using Qubit RNA HS Assay Kit (Invitrogen, California, USA) in Qubit2.0. fluorometer. RNA (15 ng) from CESCs (n = 3) and CCECs (n = 3) and 1 µg of RNA from LBECs (n = 3) and CCECs (n = 3) was reverse transcribed using miScript II RT Kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol. Due to the low quantity of RNA (15 ng) obtained, the reverse transcribed cDNA from CESCs was pre-amplified using miScript PreAMP PCR Kit (Qiagen, Hilden, Germany) for 12 cycles (Denaturation: 30 seconds at 94 °C and annealing/ extension: 3 minutes at 60 °C) preceded by initial activation: 15 minutes at 95 °C. Hence the same protocol was followed for CCECs for comparison. The qPCR amplification was performed for 40 cycles using 2X miScript SYBR Green PCR Master Mix, 10X miScript Universal Primer and miRNA specific miScript Primer Assays (Supplementary Table S1 for list of miRNA Primer Assays used and Table S2 for custom designed miRNA primer sequence). Experiment was done in triplicates and signals were normalized to small nucleolar RNA U6 (RNU6B) which was run in parallel as reference miRNA. Relative miRNA expression was calculated using comparative threshold cycle (Ct) method (2-ΔΔCt) and the result was represented as mean ± SEM. Statistical analysis of qPCR data was carried out using Mann-Whitney U test and miRNAs with fold change >2 and p value <0.05 were considered significant.
LNA in-situ hybridization
For validation of differential expression of miRNAs in its native environment, LNA in-situ hybridization was carried out following protocol of Obernosterer et al.26 with some modifications. Briefly, The corneoscleral region from the donor eye ball was separated and fixed with 4% paraformaldehyde (Sigma-Aldrich, Missouri, United States) for 30 minutes at room temperature and then the tissue was incubated overnight at 4 °C in 30% sucrose solution prepared in 1X phosphate buffered saline (PBS) of pH 7.4 (Ambion, California, United States). The tissue was embedded in optimal cutting temperature (OCT) compound (Leica, Wetzlar, Germany), and cryosections (10 μm) of limbal epithelium and corneal epithelium were taken on SuperFrost Plus slides (Thermofisher Scientific, Massachusetts, United States). The cryosections were fixed with ice cold acetone for 10 minutes at room temperature and then hybridized with biotinylated LNA miRNA probes (Eurogentec, Liège, Belgium; Supplementary Table S3 for list of LNA miRNA probes used) at 50 °C overnight by placing coverslip over the sections to prevent drying. U6 small nucleolar RNA probe and LNA scrambled microRNA probe were used as positive and negative control respectively. After hybridization, the slides were soaked in prewarmed 5X SSC Buffer (Ambion, California, United States) to remove the coverslips. The sections were incubated at 60 °C for 1 hour in 0.2X SSC buffer followed by blocking with 10% fetal bovine serum (Invitrogen, California, USA) in 0.1 M Tris (pH 7.5) and 0.15 M sodium chloride solution at room temperature for 1 hour. To detect the hybridized probes, the sections were incubated with Streptavidin-Fluorescein Isothiocyanate (FITC) Conjugate (BD Biosciences, New Jersey, United States) in 1:1000 in 5% bovine serum albumin (Sigma-Aldrich, Missouri, United States) at 25 °C in dark for 2 hours. The sections were then washed twice with 1X PBS for 5 minutes and mounted using fluorescent mounting media with propidium iodide (Vector Laboratories, California, United States). The images were acquired using confocal laser scanning microscope (Leica SP8, Germany).
miRNA Target prediction, Gene Ontology and Pathway Analysis
miRWalk (Version 3.0) and mirDIP (Version 4.1.1.6) were used to predict the target genes of miRNAs. Experimentally validated targets from miRWalk and which were targets with> 10 sources in mirDIP databases were filtered for the analysis. For pathway prediction, ClueGO package in Cytoscape 3.2.0 was used. KEGG pathway list was chosen with p-value ≤ 0.01. Further, the network of miRNAs, target genes and the associated pathways was constructed by Cytoscape 3.2.027.
Statistical analysis
Mann-Whitney U test was performed to determine the statistical significance between the two experimental groups and p < 0.05 was considered statistically significant. STATA 14.0 (Texas, USA) statistical software was used for analysis. Non-parametric test, Mann-Whitney U was chosen since the qPCR data followed non-gaussian distribution based on the Shapiro-Wilk normality test.
Results
Characteristics of small RNA sequencing and differentially expressed miRNAs between CESCs and CCECs
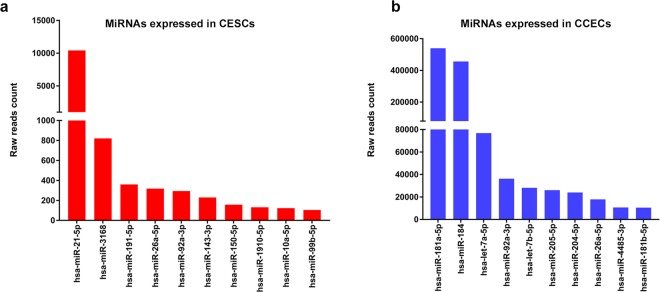
The entire quantity of total RNA isolated from enriched CESCs (pooled cells from two pairs of donor tissues) was used for library preparation as the concentration was lesser than the detection limit in the Qubit analysis. For CCECs, 10 ng of total RNA (RIN 2.8) was used. The sequencing generated more than 11 million reads in CESCs and CCECs. After quality assessment and refinement, the high quality reads were mapped to human hg 19 genome. A total of 38 miRNAs in CESCs and 301 miRNAs in CCECs were detected. Further, differential expression (DE) analysis using NOISeq after normalization were performed on all detected miRNAs. A total of 127 DE miRNAs were identified in CESCs compared to CCECs with fold change >±2. Among them 29 miRNAs were upregulated and 64 miRNAs were downregulated. The fold change values of the miRNAs were summarized as volcano plot in Fig. 1. The top ten miRNAs that were highly expressed in CESCs (Fig. 2a) and CCECs (Fig. 2b) based on raw reads counts were represented graphically. Based on fold difference between CESCs and CCECs and raw reads counts, fourteen differentially expressed miRNAs and two miRNAs without significant difference in expression were selected for validation by qPCR expression analysis (Table 1).
Figure 1.
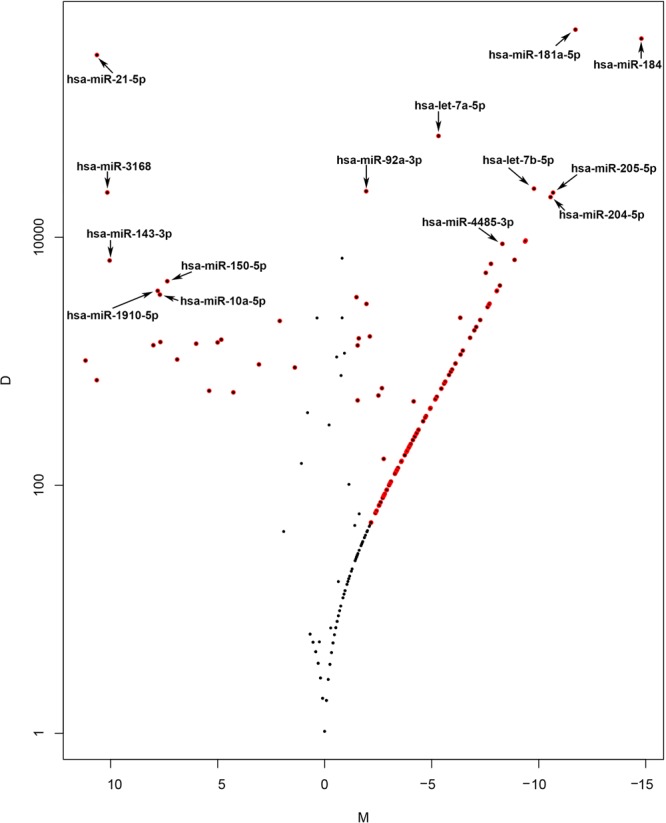
Volcano plot showing differentially expressed miRNAs in CESCs and CCECs. M-D values in noise are represented by black dots, and miRNAs with p-value ≤ 0.9 by red dots. M –value (Mean fold change): log2(x1/x2). D –value (Difference in expression levels): |x1-x2 | . where x1 represents the expression level in CESCs and x2 in CCECs.
Figure 2.
MicroRNAs with high read counts in corneal epithelial stem cells and central corneal epithelial cells by small RNA sequencing. (a) Bar diagram showing the top ten miRNAs expressed in enriched CESCs based on raw read counts. (b) Bar diagram showing the top ten miRNAs expressed in CCECs based on raw read counts.
Table 1.
Differentially expressed miRNAs CESCs vs. CCECs validated by qPCR.
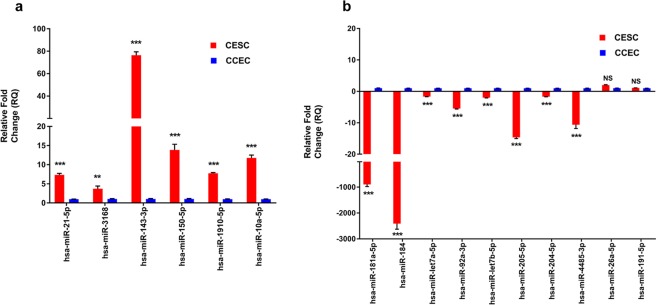
| miRNA | NGS fold change | qPCR fold change |
|---|---|---|
| hsa-miR-21-5p | 11 | 7.34 ± 0.38 |
| hsa-miR-3168 | 10 | 3.71 ± 0.72 |
| hsa-miR-143-3p | 10 | 76.44 ± 3.07 |
| hsa-miR-150-5p | 7 | 13.86 ± 1.49 |
| hsa-miR-1910-5p | 8 | 7.76 ± 0.17 |
| hsa-miR-10a-5p | 8 | 11.76 ± 0.75 |
| hsa-miR-181a-5p | −12 | −894 ± 83.22 |
| hsa-miR-184 | −15 | −2409 ± 214.9 |
| hsa-let-7a-5p | −5 | −1.66 ± 0.062 |
| hsa-let-7b-5p | −10 | −1.98 ± 0.11 |
| hsa-miR-205-5p | −11 | −14.64 ± 0.39 |
| hsa-miR-204-5p | −11 | −2.18 ± 0.08 |
| hsa-miR-4485-3p | −9 | −10.63 ± 1.21 |
| hsa-miR-92a-3p | −2 | −5.47 ± 0.16 |
| hsa-miR-191-5p | NS | 1.10 ± 0.02 |
| hsa-miR-26a-5p | NS | 1.99 ± 0.11 |
NS-not significant.
Validation of differentially expressed miRNAs by qPCR
Validation of the sequencing data was carried out using RNA extracted from enriched CESCs and CCECs (after pooling cells from different donors) as specified under materials and methods. All six miRNAs hsa-miR-3168, hsa-miR-21-5p, hsa-miR-143-3p, hsa-miR-150-5p, hsa-miR-1910-5p and hsa-miR-10a-5p showed higher expression in CESCs with significant fold change (>2) compared to CCECs confirming the sequence data. Among them the magnitude of fold difference was much higher for hsa-miR-143-3p with 76.44 ± 3.07, while a ten-fold change was observed for hsa-miR-10a-5p and hsa-miR-150-5p and a five-fold change was observed for hsa-miR-21-5p and hsa-miR-1910-5p (Fig. 3a). The nine miRNAs that are identified to be down regulated in CESCs by sequencing, hsa-miR-184, hsa-miR-181a-5p, hsa-miR-92a-3p, hsa-miR-4485-3p, hsa-miR-205-5p, hsa-miR-99b-5p, hsa-miR-204-5p, hsa-let7a-5p and hsa-let7b-5p had reduced expression in CESCs compared to CCECs, thus validating the sequencing data. The top two miRNAs expressed in CCECs had fold change of 2409 ± 214.9 (hsa-miR-184) and 894 ± 83.22 (hsa-miR-181a-5p) compared to that of CESCs. Hsa-miR-26a-5p and hsa-miR-191-5p did not show significant difference in the expression pattern between the two populations (Fig. 3b).
Figure 3.
Quantitative Real time PCR validation of differentially expressed miRNAs. Relative miRNA expression (RQ) in (a) miRNAs highly expressed in CESCs in comparison to CCECs and (b) miRNAs highly expressed in CCECs in comparison to CESCs by qPCR using SYBR Green chemistry. Each sample (n = 3) was run in triplicate. The data were expressed as mean ± SEM and relative fold change of expression (RQ) was calculated by 2-∆∆CT method after normalization with RNU6B (Reference microRNA). (**P < 0.01; ***P < 0.001; NS P > 0.05; Mann–Whitney U test).
Localization of differentially expressed miRNAs by LNA in-situ hybridization
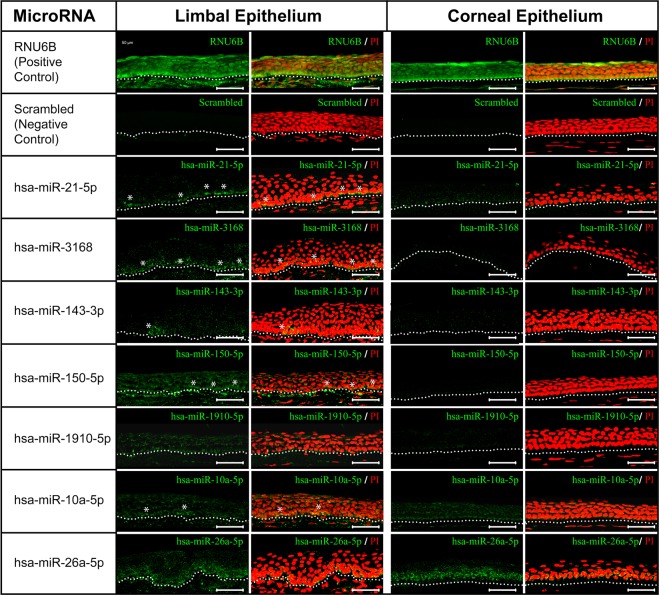
Expression of miRNAs hsa-miR-21-5p, hsa-miR-3168, hsa-miR-10a-5p, hsa-miR-1910-5p, hsa-miR-150-5p, hsa-miR-143-3p and hsa-miR-26a-5p was analyzed in cryosections of limbal epithelium (LE) and corneal epithelium (CE).
The differentially expressed miRNAs, hsa-miR-21-5p, hsa-miR-3168, hsa-miR-150-5p, hsa-miR-143-3p, hsa-miR-1910-5p and hsa-miR-10a-5p had higher expression in limbal basal epithelial cells compared to that of corneal basal epithelial cells. Interestingly the expression of hsa-miR-143-3p was exclusive to the clusters of cells in limbal basal epithelium. Among the limbal basal epithelial cells, higher expression of hsa-miR-21-5p, hsa-miR-3168, hsa-miR-150-5p and hsa-miR-10a-5p was observed in a few clusters, while a discontinuous positivity was observed for hsa-miR-1910-5p. The miRNA hsa-miR-26a-5p had equal expression in both limbal and corneal epithelial basal cells (Fig. 4).
Figure 4.
Locked nucleic acid in-situ hybridization of miRNAs highly expressed in CESCs. Expression of hsa-miR-21-5p, hsa-miR-3168 and hsa-miR-10a-5p (green) was higher in clusters of limbal basal epithelial cells compared to corneal epithelial cells, while the expression of hsa-miR-143–3p was restricted to clusters of cells in the limbal basal epithelium. Expression of hsa-miR-150-5p was evident in all layers of limbal epithelium however highly prominent in a few clusters in basal layer. Hsa-miR-1910-5p had discontinuous positivity in limbal basal epithelial layer, while hsa-miR-26a-5p had equal expression in the basal layer both limbal and corneal epithelium. Nuclei were stained with propidium iodide (PI, red). Positive control RNU6B was detected in all layers of epithelium both in limbus and cornea, whereas no signal was detected when hybridized with scrambled sequence. Asterisks represent the positivity in clusters. The dotted line demarcates the termination of the epithelium and beginning of the underlying stroma. Scale bar: 50 µm.
Pathway and Gene Ontology analysis
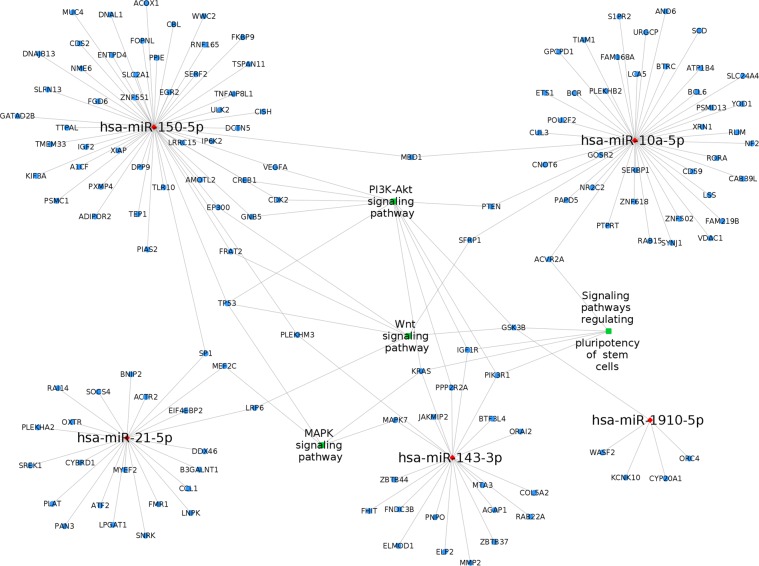
For the functional analysis, the targets of six DE miRNAs hsa-miR-3168, hsa-miR-10a-5p, hsa-miR-143-3p, hsa-miR-150-5p, hsa-miR-1910-5p, and hsa-miR-21-5p were predicted and their associated pathways and gene ontology were analyzed (Fig. S3). A total of 1055 targets were predicted for six miRNAs and they were enriched into 55 pathways with FDR < 0.01, are shown in Table S4. Further, stemness related pathways were visualized (Fig. 5). The target genes GSK3B (regulated by hsa-miR-1910-5p), KRAS (hsa-miR-143-3p), and PIK3R1 (hsa-miR-143-3p) were associated with PI3K-Akt signaling pathway and signaling pathways regulating pluripotency of stem cells. In addition, GSK3B was associated with Wnt signaling pathway, and KRAS with MAPK signaling pathway. Particularly in signaling pathway regulating the pluripotency of stem cells, hsa-miR-143-3p dysregulate the pathway through the target genes IGF1R, PIK3R1, KRAS; hsa-miR-1910-5p by GSK3B, and ACVR2A by hsa-miR-10a-5p.
Figure 5.
Network of differentially expressed miRNAs between CESCs and CCECs, their target genes and the stemness related pathways associated with them. Nodes red in colour are the miRNAs, blue are genes and green are their associated pathways.
Discussion
MicroRNA profiling of total human limbal epithelium28, human limbal basal epithelium29 and mouse limbal basal epithelium30,31 in comparison to central corneal epithelium revealed many differentially expressed miRNAs. In addition, reports are available on the role of miRNAs in differentiation28,32, transdifferentiation33, angiogenesis34–36, wound healing37,38, corneal transplant rejection39, corneal neovascularization40 and ocular infections41. But only a few reports are available on association of miRNAs in the regulation of stemness. The major challenge of profiling the miRNAs in CESCs was the low stem cell content (3-5%) in the total limbal epithelium. Hence an attempt was made to profile the miRNAs specific to adult stem cells of the limbus using a small number of highly enriched CESCs as close as eighty percent18 by small RNA sequencing platform. Since the yield of CESCs was minimum after enrichment, Arcturus picopure RNA isolation kit was used in this study for RNA isolation, to reduce the loss of short structured RNA with low GC during extraction from small number cells42.
Small RNA sequence data of CCECs was used as a quality control for the sequencing. Comparison of the data with published literature29,43 using total cornea or corneal epithelium (by NGS or microarray) identified a similar profile of miRNAs, thereby confirming the validity of the data. However, only 38 miRNAs were identified in CESC population and this could possibly due to the following concerns (i) low input RNA, that could not be quantified before library preparation ii) usage of LCM for enrichment process and iii) difference in method of isolation of CESCs and CCECs. In spite of this drawback, miRNAs identified to be highly expressed in the CESCs were confirmed to be specific by in-situ hybridization and qPCR.
Though the number of miRNAs detected in CESCs was only one tenth compared to CCECs, hsa-miR-3168, hsa-miR-21-5p, hsa-miR-143-3p, hsa-miR-150-5p, hsa-miR-1910-5p and hsa-miR-10a-5p had greater than seven folds increase in expression in the enriched CESCs indicating their specificity to stem cells. Among these six miRNAs, hsa-miR-316844 and hsa-miR-1910-5p45 were reported as novel miRNAs in human embryonic stem cells. Understanding their functional role in regulating adult tissue resident stem cells will be of significance. The remaining four miRNAs were reported to have a role in regulating other stem cells. Hsa-miR-21-5p suppresses marrow derived endothelial progenitor cells proliferation by activating TGFβ signaling through down regulation of WW domain-containing protein 1 (WWP1)46. Similarly, miR-10a down regulates proliferation of human cardiomyocyte progenitor cells by targeting GATA647. Hsa-miR-143-3p and hsa-miR-150-5p are known to be associated with self-renewal of mouse embryonic stem cells and human liver cancer stem cells respectively. MiR-143 promotes self-renewal by suppressing de novo methyltransferase gene DNMT3A48 and miR-150 inhibits cell proliferation by targeting C-MYB49.
Validation of the sequencing data by qPCR analysis revealed that fourteen differentially expressed miRNAs to have similar expression pattern (Fig. 3). In addition, all the differentially expressed miRNAs had several magnitudes of fold difference in CESCs than LBECs when compared to CCECs independently (Fig. S2). In addition, the up regulation of hsa-miR-3168 as well as the down regulation of hsa-let7a-5p and hsa-miR-204-5p was identified only with the enriched CESCs but not with LBECs. Thus, these observations highlight the importance of enrichment of CESCs and confirming the specificity of the identified miRNAs.
LNA in-situ hybridization revealed the location of the miRNAs in corneal and limbal tissue. Among the miRNAs that were highly expressed in CESCs, hsa-miR-3168, hsa-miR-21-5p, hsa-miR-150-5p and hsa-miR-1910-5p had higher expression in limbal basal epithelial layer compared to that of the corneal basal epithelial layer. Further, a few clusters of small cells had strong positivity (probably stem cells) compared to that of the other limbal basal epithelial cells. Likewise, the expression of hsa-miR-143-3p was confined only to clusters in limbal basal epithelial layer, indicating its very high specificity to stem cells.
The predicted targets of these miRNAs that were highly expressed in CESCs were identified to be associated with pathways regulating pluripotency of stem cells and other pathways including MAPK signaling, Ras signaling, Hippo signaling, ErbB signaling, Wnt signaling and PI3K-Akt signaling. Reports are available on the significance of Wnt50, PI3K/Akt51, MAPK52 signaling pathways in the regulation of stem cells. The direct targets of the miRNAs (highly expressed in CESCs) predicted to be involved in these signaling pathways are listed in Table 2. These targets are known to be associated with cell proliferation, differentiation, migration and apoptosis. Further studies are therefore essential to confirm the functional role of the identified miRNAs and their targets in the regulation of stemness in CESCs.
Table 2.
List of direct targets of selected miRNAs highly expressed in CESCs and their functional role.
| miRNA | Targets | Functional role | Reference |
|---|---|---|---|
| Hsa-miR-143-3p | MAPK7 | Inhibits adipocyte proliferation and enhance their differentiation | 58 |
| Inhibits proliferation, cell migration and invasion of breast cancer | 59 | ||
| KRAS | Suppresses colorectal cancer cell growth | 60 | |
| IGF1R | Regulates cell proliferation and apoptosis | 61 | |
| Regulation of myogenesis | 62 | ||
| PIK3R1 | Down regulation promotes propagation, migration, epithelial mesenchymal transition and stem cell phenotype of renal canal cells | 63 | |
| Hsa-miR-21-5p | MEF2C | Crucial for neuronal function | 64 |
| Hsa-miR-10a-5p | PTEN | Tumor suppressor | 65 |
| Hsa-miR-150-5p | VEGFA | Decrease migration, invasion and angiogenesis in Rheumatoid arthritis | 66 |
| CREB1 | A novel Wnt effector that may enhance epithelial mesenchymal transition of colorectal cancer cells | 67 | |
| CDK2 | Inhibits proliferation and tumorigenicity via retarding G1/S phase transition | 68 | |
| TP53 | Promotes proliferation of lung cancer cells | 69,70 |
One of the limitations of this study is the use of donor tissues. The RNA expression could have been altered due to the associated ischemia53 and the additional processing of the cells for enrichment of stem cells54. In spite of the low input of RNA for sequencing and low number of miRNAs identified in this study, confirmation of the high expression of miRNAs hsa-miR-3168, hsa-miR-21-5p, hsa-miR-143-3p, hsa-miR-150-5p, hsa-miR-1910-5p and hsa-miR-10a-5p in CESCs indicates their significant role in the regulation of stem cells. Transfection studies using hsa-miR-143-3p and hsa-miR-150-5p mimics in the primary limbal epithelial cells (n = 3) identified their increased colony forming potential, specifically their ability to form holoclones (based on colony morphology as defined by Barrandon and Green55), thereby indicating a strong regulatory influence of these miRNAs on maintenance of stemness56,57. Thus, the data generated serves as a platform to study the miRNAs associated with the maintenance of stemness. Further studies are essential to elucidate the functional role of these miRNAs in regulating the stemness in CESCs and the associated signaling pathways.
Supplementary information
Acknowledgements
The authors thank Department of Biotechnology for the financial support to the work, Council of Scientific & Industrial Research (CSIR) for providing fellowship to Lavanya Kalaimani, Ph.D student of Alagappa University, Karaikudi, Tamil Nadu, India. Mr. D. Saravanan, Manager, Rotary Aravind International Eye Bank, Madurai, India for his valuable help in sample collection for the study, Ms. R. Meenakshi, Aravind Communications, Aravind Eye Care System, India, for her help in preparing the figures, Mr. Mohammed Sithiq Uduman, Department of Biostatistics, Aravind Eye Care System, Madurai, India for helping us in the statistics.
Author contributions
K.L. work done, data analysis and interpretation, manuscript writing. D.B. study design, data analysis and interpretation, manuscript writing. S.U. data analysis and interpretation. A.V. data interpretation, proof reading. N.V.P. study design, data interpretation, proof reading. V.M. study design, data interpretation, proofreading. C.G.P. study design, obtained funding, data analysis and interpretation, manuscript writing.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-020-64273-0.
References
- 1.DelMonte DW, Kim T. Anatomy and physiology of the cornea. J Cataract Refract Surg. 2011;37:588–598. doi: 10.1016/j.jcrs.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 2.Le Q, Xu J, Deng SX. The diagnosis of limbal stem cell deficiency. Ocul Surf. 2018;16:58–69. doi: 10.1016/j.jtos.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rama P, et al. Limbal stem-cell therapy and long-term corneal regeneration. N. Engl. J. Med. 2010;363:147–155. doi: 10.1056/NEJMoa0905955. [DOI] [PubMed] [Google Scholar]
- 4.Cotsarelis G, Cheng SZ, Dong G, Sun TT, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: implications on epithelial stem cells. Cell. 1989;57:201–209. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 5.Arpitha P, Prajna NV, Srinivasan M, Muthukkaruppan V. A subset of human limbal epithelial cells with greater nucleus-to-cytoplasm ratio expressing high levels of p63 possesses slow-cycling property. Cornea. 2008;27:1164–1170. doi: 10.1097/ICO.0b013e3181814ce6. [DOI] [PubMed] [Google Scholar]
- 6.Pellegrini G, et al. Long-term restoration of damaged corneal surfaces with autologous cultivated corneal epithelium. Lancet. 1997;349:990–993. doi: 10.1016/S0140-6736(96)11188-0. [DOI] [PubMed] [Google Scholar]
- 7.Arpitha P, Prajna NV, Srinivasan M, Muthukkaruppan V. A method to isolate human limbal basal cells enriched for a subset of epithelial cells with a large nucleus/cytoplasm ratio expressing high levels of p63. Microsc. Res. Tech. 2008;71:469–476. doi: 10.1002/jemt.20573. [DOI] [PubMed] [Google Scholar]
- 8.Jhagta HS, Jain P. Limbal stem cell deficiency: A review. J. Clin. Ophthalmol. Res. 2015;3:71–75. doi: 10.4103/2320-3897.156582. [DOI] [Google Scholar]
- 9.Haagdorens M, et al. Limbal Stem Cell Deficiency: Current Treatment Options and Emerging Therapies. Stem Cells Int. 2016;2016:9798374. doi: 10.1155/2016/9798374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tseng SC. Concept and application of limbal stem cells. Eye (Lond) 1989;3(Pt 2):141–157. doi: 10.1038/eye.1989.22. [DOI] [PubMed] [Google Scholar]
- 11.Pellegrini G, et al. Location and clonal analysis of stem cells and their differentiated progeny in the human ocular surface. J. Cell Biol. 1999;145:769–782. doi: 10.1083/jcb.145.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Priya CG, et al. Adult human buccal epithelial stem cells: identification, ex-vivo expansion, and transplantation for corneal surface reconstruction. Eye (Lond) 2011;25:1641–1649. doi: 10.1038/eye.2011.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/S1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 14.Suh M-R, et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Wilson KD, et al. MicroRNA profiling of human-induced pluripotent stem cells. Stem Cells Dev. 2009;18:749–758. doi: 10.1089/scd.2008.0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyoshi N, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Kasinathan JR, Namperumalsamy VP, Veerappan M, Chidambaranathan GP. A novel method for a high enrichment of human corneal epithelial stem cells for genomic analysis. Microsc. Res. Tech. 2016;79:1165–1172. doi: 10.1002/jemt.22771. [DOI] [PubMed] [Google Scholar]
- 19.Schermer A, Galvin S, Sun TT. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J CELL BIOL. 1986;103(1):49–62. doi: 10.1083/jcb.103.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arpitha P, Prajna NV, Srinivasan M, Muthukkaruppan V. High expression of p63 combined with a large N/C ratio defines a subset of human limbal epithelial cells: implications on epithelial stem cells. Invest. Ophthalmol. Vis. Sci. 2005;46:3631–3636. doi: 10.1167/iovs.05-0343. [DOI] [PubMed] [Google Scholar]
- 21.Espana EM, et al. Stromal niche controls the plasticity of limbal and corneal epithelial differentiation in a rabbit model of recombined tissue. Invest. Ophthalmol. Vis. Sci. 2003;44:5130–5135. doi: 10.1167/iovs.03-0584. [DOI] [PubMed] [Google Scholar]
- 22.Farris S, Wang Y, Ward JM, Dudek SM. Optimized Method for Robust Transcriptome Profiling of Minute Tissues Using Laser Capture Microdissection and Low-Input RNA-Seq. Front Mol Neurosci. 2017;10:185. doi: 10.3389/fnmol.2017.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capece V, et al. Oasis: online analysis of small RNA deep sequencing data. Bioinformatics. 2015;31:2205–2207. doi: 10.1093/bioinformatics/btv113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman R-U, et al. Oasis 2: improved online analysis of small RNA-seq data. BMC Bioinformatics. 2018;19:54. doi: 10.1186/s12859-018-2047-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Core Team, R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. (2018)
- 26.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- 27.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee SK-W, et al. MicroRNA-145 regulates human corneal epithelial differentiation. PLoS ONE. 2011;6:e21249. doi: 10.1371/journal.pone.0021249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teng Y, et al. Signature microRNAs in human cornea limbal epithelium. Funct. Integr. Genomics. 2015;15:277–294. doi: 10.1007/s10142-014-0417-9. [DOI] [PubMed] [Google Scholar]
- 30.Peng H, et al. microRNA-31/factor-inhibiting hypoxia-inducible factor 1 nexus regulates keratinocyte differentiation. Proc. Natl. Acad. Sci. USA. 2012;109:14030–14034. doi: 10.1073/pnas.1111292109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng H, et al. microRNA-103/107 Family Regulates Multiple Epithelial Stem Cell Characteristics. Stem Cells. 2015;33:1642–1656. doi: 10.1002/stem.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagosa S, et al. microRNA-184 Induces a Commitment Switch to Epidermal Differentiation. Stem Cell Reports. 2017;9:1991–2004. doi: 10.1016/j.stemcr.2017.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao X-Y, Xie H-T, Duan C-Y, Li J, Zhang M-C. Rat limbal niche cells can induce transdifferentiation of oral mucosal epithelial cells into corneal epithelial-like cells in vitro. Stem Cell Res Ther. 2018;9:256. doi: 10.1186/s13287-018-0996-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grimaldo S, et al. MicroRNA-184 Regulates Corneal Lymphangiogenesis. Invest. Ophthalmol. Vis. Sci. 2015;56:7209–7213. doi: 10.1167/iovs.15-17733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zong R, et al. Down-Regulation of MicroRNA-184 Is Associated With Corneal Neovascularization. Invest. Ophthalmol. Vis. Sci. 2016;57:1398–1407. doi: 10.1167/iovs.15-17417. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y, Zhang T, Ma X, Zou J. Subconjunctival injection of antagomir-21 alleviates corneal neovascularization in a mouse model of alkali-burned cornea. Oncotarget. 2017;8:11797–11808. doi: 10.18632/oncotarget.14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Funari VA, et al. Differentially expressed wound healing-related microRNAs in the human diabetic cornea. PLoS ONE. 2013;8:e84425. doi: 10.1371/journal.pone.0084425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.An J, et al. MicroRNA Expression Profile and the Role of miR-204 in Corneal Wound Healing. Invest. Ophthalmol. Vis. Sci. 2015;56:3673–3683. doi: 10.1167/iovs.15-16467. [DOI] [PubMed] [Google Scholar]
- 39.Wang T, Li F, Geng W, Ruan Q, Shi W. MicroRNA-122 ameliorates corneal allograft rejection through the downregulation of its target CPEB1. Cell Death Discov. 2017;3:17021. doi: 10.1038/cddiscovery.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukwaya A, Jensen L, Peebo B, Lagali N. MicroRNAs in the cornea: Role and implications for treatment of corneal neovascularization. Ocul Surf. 2019;17:400–411. doi: 10.1016/j.jtos.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 41.Xu, S. & Hazlett, L. D. MicroRNAs in Ocular Infection. Microorganisms 7, (2019). [DOI] [PMC free article] [PubMed]
- 42.Kim YK, Yeo J, Kim B, Ha M, Kim VN. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol. Cell. 2012;46(6):893–895. doi: 10.1016/j.molcel.2012.05.036. [DOI] [PubMed] [Google Scholar]
- 43.Kulkarni M, et al. Genome-wide analysis suggests a differential microRNA signature associated with normal and diabetic human corneal limbus. Sci Rep. 2017;7:3448. doi: 10.1038/s41598-017-03449-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinton A, et al. sRNA-seq analysis of human embryonic stem cells and definitive endoderm reveals differentially expressed microRNAs and novel IsomiRs with distinct targets. Stem Cells. 2014;32:2360–2372. doi: 10.1002/stem.1739. [DOI] [PubMed] [Google Scholar]
- 45.Bar M, et al. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zuo K, et al. MiR-21 suppresses endothelial progenitor cell proliferation by activating the TGFβ signaling pathway via downregulation of WWP1. Int J Clin Exp Pathol. 2015;8:414–422. [PMC free article] [PubMed] [Google Scholar]
- 47.Liang D, et al. miR-10a regulates proliferation of human cardiomyocyte progenitor cells by targeting GATA6. PLoS ONE. 2014;9:e103097. doi: 10.1371/journal.pone.0103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao Y, et al. Vitamin C induces a pluripotent state in mouse embryonic stem cells by modulating microRNA expression. FEBS J. 2015;282:685–699. doi: 10.1111/febs.13173. [DOI] [PubMed] [Google Scholar]
- 49.Zhang J, et al. microRNA-150 inhibits human CD133-positive liver cancer stem cells through negative regulation of the transcription factor c-Myb. Int. J. Oncol. 2012;40:747–756. doi: 10.3892/ijo.2011.1242. [DOI] [PubMed] [Google Scholar]
- 50.Davidson KC, et al. Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc. Natl. Acad. Sci. USA. 2012;109:4485–4490. doi: 10.1073/pnas.1118777109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Romorini L, et al. AKT/GSK3β signaling pathway is critically involved in human pluripotent stem cell survival. Sci Rep. 2016;6:35660. doi: 10.1038/srep35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haston S, et al. MAPK pathway control of stem cell proliferation and differentiation in the embryonic pituitary provides insights into the pathogenesis of papillary craniopharyngioma. Development. 2017;144:2141–2152. doi: 10.1242/dev.150490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spruessel A, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. BioTechniques. 2004;36:1030–1037. doi: 10.2144/04366RR04. [DOI] [PubMed] [Google Scholar]
- 54.Bath C, Fink T, Vorum H, Hjortdal J, Zachar V. Technical brief: Optimized pipeline for isolation of high-quality RNA from corneal cell subpopulations. Mol. Vis. 2014;20:797–803. [PMC free article] [PubMed] [Google Scholar]
- 55.Barrandon, Y. & Green, H. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA 84, 2302–2306 (1987). [DOI] [PMC free article] [PubMed]
- 56.Priya CG, Kalaimani L, Bharanidharan D, Prajna V, Muthukkaruppan VR. Hsa-miR143-3p regulates human corneal epithelial stem cells. Invest. Ophthalmol. Vis. Sci. 2019;60(9):4825–4825. [Google Scholar]
- 57.Kalaimani, L. et al. Hsa‐miR‐150‐5p regulates human corneal epithelial stem cells through Wnt signaling. ACTA OPHTHALMOL, 97 (2019).
- 58.Zhang P, et al. MicroRNA-143a-3p modulates preadipocyte proliferation and differentiation by targeting MAPK7. Biomed. Pharmacother. 2018;108:531–539. doi: 10.1016/j.biopha.2018.09.080. [DOI] [PubMed] [Google Scholar]
- 59.Xia C, Yang Y, Kong F, Kong Q, Shan C. MiR-143-3p inhibits the proliferation, cell migration and invasion of human breast cancer cells by modulating the expression of MAPK7. Biochimie. 2018;147:98–104. doi: 10.1016/j.biochi.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 60.Chen X, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 61.Yang Z, Wang J, Pan Z, Zhang Y. miR-143-3p regulates cell proliferation and apoptosis by targeting IGF1R and IGFBP5 and regulating the Ras/p38 MAPK signaling pathway in rheumatoid arthritis. Exp Ther Med. 2018;15:3781–3790. doi: 10.3892/etm.2018.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soriano-Arroquia A, McCormick R, Molloy AP, McArdle A, Goljanek-Whysall K. Age-related changes in miR-143-3p:Igfbp5 interactions affect muscle regeneration. Aging Cell. 2016;15:361–369. doi: 10.1111/acel.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin Y, et al. PIK3R1 negatively regulates the epithelial-mesenchymal transition and stem-like phenotype of renal cancer cells through the AKT/GSK3β/CTNNB1 signaling pathway. Sci Rep. 2015;5:8997. doi: 10.1038/srep08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yelamanchili SV, Chaudhuri AD, Chen L-N, Xiong H, Fox HS. MicroRNA-21 dysregulates the expression of MEF2C in neurons in monkey and human SIV/HIV neurological disease. Cell Death Dis. 2010;1:e77. doi: 10.1038/cddis.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu T, et al. MiRNA-10a is upregulated in NSCLC and may promote cancer by targeting PTEN. Oncotarget. 2015;6:30239–30250. doi: 10.18632/oncotarget.4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen Z, Wang H, Xia Y, Yan F, Lu Y. Therapeutic Potential of Mesenchymal Cell-Derived miRNA-150-5p-Expressing Exosomes in Rheumatoid Arthritis Mediated by the Modulation of MMP14 and VEGF. J. Immunol. 2018;201:2472–2482. doi: 10.4049/jimmunol.1800304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guo Y-H, et al. Wnt/β-catenin pathway transactivates microRNA-150 that promotes EMT of colorectal cancer cells by suppressing CREB signaling. Oncotarget. 2016;7:42513–42526. doi: 10.18632/oncotarget.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li, X. et al. miR-150 inhibits proliferation and tumorigenicity via retarding G1/S phase transition in nasopharyngeal carcinoma. Int. J. Oncol. 10.3892/ijo.2017.3909 (2017). [DOI] [PMC free article] [PubMed]
- 69.Zhang N, Wei X, Xu L. miR-150 promotes the proliferation of lung cancer cells by targeting P53. FEBS Lett. 2013;587:2346–2351. doi: 10.1016/j.febslet.2013.05.059. [DOI] [PubMed] [Google Scholar]
- 70.Wang D-T, et al. miR-150, p53 protein and relevant miRNAs consist of a regulatory network in NSCLC tumorigenesis. Oncol. Rep. 2013;30:492–498. doi: 10.3892/or.2013.2453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.