Abstract
So far, ascoviruses have only been identified from Lepidoptera host insects and their transmission vectors—endoparasitic wasps. Here, we reported the first finding of a complete novel ascovirus genome from a Diptera insect, Dasineura jujubifolia. Initially, sequence fragments with homology to ascoviruses were incidentally identified during metagenomic sequencing of the mitochondria of D. jujubifolia (Cecidomyiidae, Diptera) which is a major pest on Ziziphus jujuba. Then a full circular viral genome was assembled from the metagenomic data, which has an A+T percentage of 74% and contains 142,600 bp with 141 open reading frames (ORFs). Among the 141 ORFs, 37 were conserved in all sequenced ascoviruses (core genes) including proteins predicted to participate in DNA replication, gene transcription, protein modification, virus assembly, lipid metabolism and apoptosis. Multi-gene families including those encode for baculovirus repeated open reading frames (BROs), myristylated membrane proteins, RING/U-box E3 ubiquitin ligases, and ATP-binding cassette (ABC) transporters were found in the virus genome. Phylogenetic analysis showed that the newly identified virus belongs to genus Toursvirus of Ascoviridae, and is therefore named as Dasineura jujubifolia toursvirus 2 (DjTV-2a). The virus becomes the second reported species of the genus after Diadromus pulchellus toursvirus 1 (DpTV-1a). The genome arrangement of DjTV-2a is quite different from that of DpTV-1a, suggesting these two viruses separated in an early time of evolution. The results suggest that the ascoviruses may infect a much broader range of hosts than our previous knowledge, and shed lights on the evolution of ascoviruses and particularly on that of the toursviruses.
Electronic supplementary material
The online version of this article (10.1007/s12250-019-00177-2) contains supplementary material, which is available to authorized users.
Keywords: Dasineura jujubifolia toursvirus 2 (DjTV-2a), Ascovirus, Toursvirus
Introduction
Ascoviruses are a group of large DNA viruses which infect Lepidoptera insects and transmitted by endoparasitic wasps. Ascoviridae comprises two genera, the Ascovirus and Toursvirus based on genome organization and host range (Asgari et al. 2017). So far the International Committee on Taxonomy of Viruses (ICTV) has recognized three species including Spodoptera frugiperda ascovirus 1 (SfAV-1), Trichoplusia ni ascovirus 2 (TnAV-2), Heliothis virescens ascovirus 3 (HvAV-3) in Ascovirus and one specie Diadromus pulchellus toursvirus 1 (DpTV-1, previously also called DpAV-4) (Bigot et al. 2009) in Toursvirus (Asgari et al.2017). The genome organization of members of the Ascovirus is collinear, that of the member of Toursvirus is quite different (Asgari et al. 2017). The members of Ascovirus infect the insect family of Noctuidae, while DpTV-1 replicates in the insect family Yponomeutidae, as well as its ichneumonid vector (Asgari et al. 2017). In this manuscript, ascoviruses refer to the members of Ascoviridae including those belonging to Toursvirus. So far, 10 ascoviruses have been fully sequenced including 9 viruses belonging to Ascovirus and 1 virus belonging to Toursvirus, and they contain a double stranded circular DNA genome with length of about 110–200 kb (Asgari et al. 2017). Here we report the genome analysis of a new toursvirus incidentally identified during metagenomic sequencing of Dasineura jujubifolia Jiao and Bu (Diptera: Cecidomyiidae) (Jiao et al.2017). Following the traditional scheme of using the host’s name, we named this virus as Dasineura jujubifolia toursvirus 2 (DjTV-2a).
Dasineura jujubifolia is one of the major pests of Ziziphus jujuba Miller (Rhamnaceae) (Jiao et al.2017). The plantation of jujube is very popular in Shandong, Hebei, Shanxi, Sanxi, Henan and Xinjiang provinces in China and it makes significant contribution to the local economy. At present, D. jujubifolia is found widely distributed in various jujube areas in Xinjiang, China and it occurs four to five generations a year (Li et al.2010). Larvae of D. jujubifolia damage young shoots and new leaves by sucking juice from the leaves during the sprouting of jujube trees (Jiao et al.2017). The infestation causes severe damage to tender buds and the leaves are curled and finally dry off. These affect the photosynthesis of the jujube trees, and eventually lead to a decrease yield of fruits (Li et al.2010). To understand the molecular biology of D. jujubifolia, the mitochondria genome of the pest was sequenced by metagenomics. To our surprise, a substantial portion of the sequence matched to that of ascoviruses, and a full ascovirus genome was assembled from the metagenomic data.
Here we reported the genome analysis of this new toursvirus DjTV-2a from D. jujubifolia and compared it with other published ascovirus sequences. The results showed that it is a novel viral species belonging to Toursvirus and is distinctly related to DpTV-1a, the previous solo member of the genus. The results enrich our understanding of the molecular organization and evolution of ascoviruses.
Materials and Methods
Sample Collection, Sequencing and Bioinformatics Analysis
The D. Jujubifolia larvae were collected from the jujube plantation of Tarim University, Xinjiang Province, China at the latitude of N 40°32′27.74″ and the longitude of E 81°18′8.41″. The mitochondria of D. jujubifolia was isolated from D. jujubifolia larvae using the Cytoplasmic and Mitochondrial Protein Extraction Kit (Sangon Biotech, Shanghai, China) and the DNA was extracted by using the Universal Genomic DNA Kit (Cwbio, Beijing, China). The mitochondrial DNA was sent to Sangon Biotech for sequencing using the MiSeq PE300 system. The generated paired-end reads were treated by cutadapt software (https://pypi.org/project/cutadapt/1.2.1/) to trim adaptors. The reads were performed by prinseq for quality filtering (Schmieder and Edwards 2011) and assembled to contigs with SPAdes software (http://cab.spbu.ru/software/spades/) (Bankevich et al.2012). All contigs were blasted with GenBank database. The ORFs were predicted by Softberry FGENESV program (http://www.softberry.com/berry.phtml?topic=virus0&group=programs&subgroup=gfindv), based on ATG as start codon and greater than 50 amino acid (aa).
The available genome sequences of ascoviruses were downloaded from GenBank database, including SfAV-1a (NC_008361) (Bideshi et al.2006), TnAV-6a (previously named as TnAV-2c, NC_008518) (Wang et al.2006), TnAV-6b (KY434117) (Liu et al.2018), HvAV-3e, (NC_009233) (Asgari et al.2007), HvAV-3f (KJ755191) (Wei et al.2014), HvAV-3g (JX491653)(Huang et al.2012), HvAV-3h (KU170628) (Huang et al.2017), HvAV-3i (MF781070) (Chen et al.2018), HvAV-3j (LC332918) (Arai et al.2018) and DpTV-1a (previously named as DpAV-4a, CU469068) (Bigot et al.2009). BLASTP (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome) was used for gene annotation and comparison. Tandem Repeats Finder (Benson 1999) and MIROPEATS (Parsons 1995) program were used to find repetitive regions. The annotated DjTV-2a genome sequence was submitted to GenBank with accession number: MK867691.
Phylogenetic Analysis
Phylogenetic analysis was performed using the amino acid (aa) sequences of DNA polymerase from DjTV-2a and the other complete sequenced ascoviruses, and with homologs from invertebrate iridescent virus 6 (IIV-6) (Jakob et al.2001) and Wiseana iridescent virus (WIV) (Wong et al.2011) as the outgroups. The sequences were aligned using MUSCLE with default parameters of MEGA6.0 (Tamura et al.2013). The alignment was used to generate phylogenetic trees using maximum likelihood with bootstrap 1000.
Results
General Organization of DjTV-2a Genome
As mentioned above, during the metagenomics sequencing of the mitochondrial DNA of D. jujubifolia, one of the assembled contigs was found to be a genome of an ascovirus and assigned as DjTV-2a. The genome was a circular double-stranded DNA with the length of 142,600 bp and was covered 4400 times by 2,122,516 sequence reads. The genome size of DjTV-2a was within the range of ascoviruses (from the 119,343 bp of DpTV-1a to the 199,721 bp of HvAV-3g). Among the 11 complete sequenced ascoviruses, DjTV-2a had the highest (74%) A+T content (Table 1).
Table 1.
Summary of the complete sequenced ascovirus genomes.
| Virus | Accession | Length (bp) | ORF no. | A + T (%) | Reference |
|---|---|---|---|---|---|
| SfAV-1a | NC_008361 | 156,922 | 123 | 50.7 | Bideshi et al. (2006) |
| HvAV-3e | NC_009233 | 186,262 | 180 | 54.1 | Asgari et al. (2007) |
| HvAV-3f | KJ755191 | 198,157 | 190 | 54 | Wei et al. (2014) |
| HvAV-3g | JX491653 | 199,721 | 194 | 54.2 | Huang et al. (2012) |
| HvAV-3h | KU170628 | 190,519 | 185 | 54.5 | Huang et al. (2017) |
| HvAV-3i | MF781070 | 185,650 | 181 | 54.6 | Chen et al. (2018) |
| HvAV-3j | LC332918 | 191,718 | 189 | 54.4 | Arai et al.(2018) |
| TnAV-6a | NC_008518 | 174,059 | 164 | 64.8 | Wang et al. (2006) |
| TnAV-6b | KY434117 | 185,664 | 178 | 64.6 | Liu et al. (2018) |
| DpTV-1a | CU469068 | 119,343 | 119 | 50.3 | Bigot et al. (2009) |
| DjTV-2a | MK867691 | 142,600 | 141 | 74 | This study |
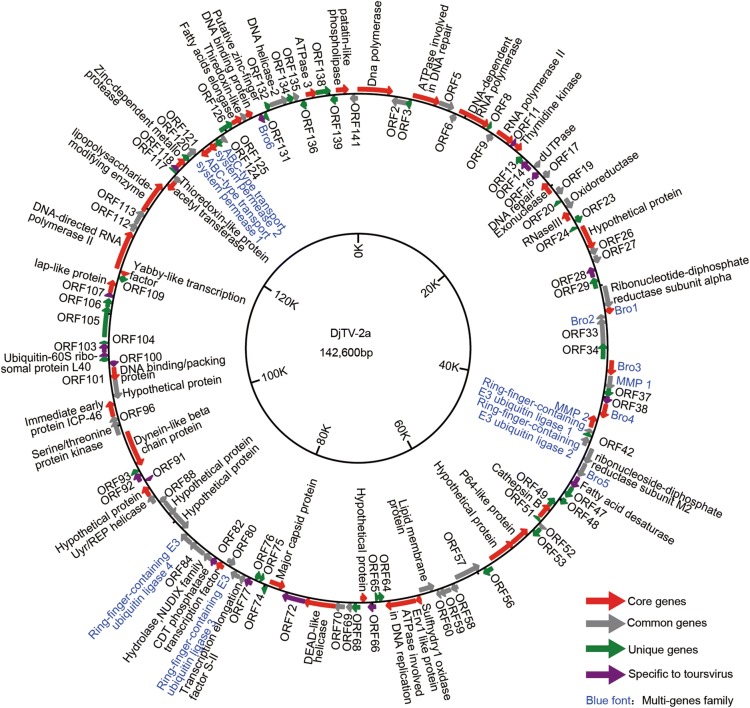
A total of 141 ORFs with a methionine start codon and a minimum protein size of 50 aa were identified in the DjTV-2a genome (Fig. 1). Following the tradition of other genomic studies of ascoviruses, the A of the ATG start codon of the DNA polymerase was assigned position 1 for the DjTV-2a genome. There were 86 ORFs in forward orientation whereas 55 were in the reverse orientation (Fig. 1, Supplementary Table S1). The ORFs covered 90% genome with an average density of one gene per 910 bp. The longest length of intergenic region (IGR) is 431 bp (between ORF104 and ORF105), and the shortest IGR is between ORF33 and ORF34 that the two genes overlap by 30 bp. Except for these two genes, there are also some small overlaps between ORF2 and ORF3, ORF70 and DEAD-like helicase, ORF98 and DNA binding/packing protein, and ORF138 and ORF139.
Fig. 1.
Circular map and gene organization of the DjTV-2a genome. ORFs are indicated by arrows. The direction of the arrows indicates the orientation of gene transcription. The colors represent gene types: red for core genes (present in all ascoviruses), grey for common gene (present in other viruses), green for unique genes (only in DjTV-2a), purple for gene specific to Toursvirus, Blue font for multigene family.
BlastP searches revealed that among 141 ORFs, 38 were unique to DjTV-2a (Fig. 1, green), while the other 103 ORFs had homologues in the NCBI protein databases. Among these 103 ORFs, 18 had similarities only with DpTV-1a thus are considered to be specific to the genus Toursvirus (Fig. 1, purple). Thirty-seven ORFs present in all 11 sequenced ascoviruses and they are assigned as core genes for Ascoviridae (Fig. 1, red). For the rest 48 ORFs, 41 had similarities with some ascoviruses, while 7 had similarities with viruses other than ascoviruses (Fig. 1, grey). Apart from ascoviruses, DjTV-2a is also close to iridoviruses that it shares 61 genes with IIV-6 and 48 genes with WIV (Supplementary Table S1).
Core Genes
Among the 37 core proteins conserved in all sequenced ascoviruses (Table 2), seven ORFs are likely involved in DNA replication and repairing including DNA polymerase (ORF1), ATPase involved in DNA repair (ORF4), thymidine kinase (ORF12), DNA repair exonuclease (ORF18), ATPase involved in DNA replication (ORF63), DEAD-like helicase (ORF71), and ATPase3 involved in DNA metabolism (OR137). Six ORFs are predicted to participate in RNA transcription, including DNA-dependent RNA polymerase (ORF7), RNA polymerase II subunits (ORF10 and ORF111), RNase III (ORF22), CDT phosphatase transcription factor (ORF81), and yabby-like transcription factor (ORF110).
Table 2.
Classification of the 37 genes found in all sequenced ascoviruses.
| Classification | Gene |
|---|---|
| DNA replication and repairing (7) | DNA polymerase (orf1*), ATPase involved in DNA repair (orf4), thymidine kinase (orf12*), DNA repair exonuclease (orf18), ATPase involved in DNA replication (orf63*), DEAD-like helicase (orf71*), ATPase3 (orf137*) |
| RNA transcription (6) | DNA-dependent RNA polymerase (orf7*), RNA polymerase II(orf10*), RNase III (orf22*), CDT phosphatase transcription factor (orf81*), yabby-like transcription factor (orf110), DNA-directed RNA polymerase II (orf111*) |
| Structure proteins (3) | P64-like protein (orf54), major capsid protein (orf73*), DNA binding/packing protein (orf99*) |
| Protein modification (3) | hypothetical protein (orf25), hypothetical protein (orf55), sulfhydry1 oxidase Erv1 like protein (orf62) |
| Lipid metabolism (3) | acetyl transferase (orf115), fatty acid elongase (orf127), patatin-like phospholipase (orf140) |
| Membrane transport (2) | ABC-type transport system permease 1 (orf122), ABC-type transport system permease 2 (orf123) |
| Apoptosis (2) | cathepsin B (orf50), iap-like protein (orf108) |
| Others (11) | bro1 (orf31), bro3 (orf35), bro4 (orf39), myristylated membrane protein-like protein 2 (orf40), orf67, orf90, dynein-like β chain (orf94*), immediate early protein ICP-46 (orf97*), lipopolysaccharide-modifying enzyme (orf114), zinc-dependent metalloprotease (orf119), putative zinc-finger DNA binding protein (orf129) |
*The asterisk-labeled proteins are common in iridoviruses and ascoviruses.
Three proteins are putative virion structure proteins: P64-like protein (ORF54), major capsid protein (ORF73) (Long et al.2009) and DNA binding/packing protein (ORF99). ORF54 had high identity with P64 protein (ORF48) of SfAV-1a. Its theoretical isoelectric point is 9.85 and molecular weight (Mw) is about 67.4 kDa. P64 of SfAV-1a was recently identified as a large cationic protein that condenses viral genomic DNA for encapsidation (Bideshi et al. 2018). There are three core ORFs potentially participate in protein modification, including two hypothetical proteins with a PKc_like superfamily motif (ORF25, ORF55) and a sulfhydry1 oxidase Erv1 like protein (ORF62). ORF62 contains a conserved Erv1 domain with a CXXC motif which is likely to be a functional sulfhydryl oxidase participating in disulfide bond formation (White et al.2002; Bideshi et al.2006).
One of ascovirus infection characters is forming large vesicles containing virions, a process which should require proteins involved in membrane formation and transport. Indeed, three proteins related to lipid metabolism and two proteins involved in membrane transport were found to be conserved (Table 2). ORF115 is predicted to encode lysophospholipid acetyl transferase (LPLAT) which functions in de novo biosynthesis of cell membrane phospholipids (Cullinane et al.2005). Fatty acid elongase is involved in long chain fatty acid elongation system, its homologue (ORF127) is conserved in ascoviruses and also present in the fowlpox virus (Jakobsson et al.2006). ORF140 encodes a putative patatin-like phospholipase (PLP) which involved in lipid metabolism and turnover in mammals (Wilson and Knoll 2018). Two ABC-type transport system permeases (ORF122 and ORF123) were found in DjTV-2a. It has been suggested that the ascoviruses have the unique ability to manipulate the apoptotic pathway to facilitate the formation of virion-containing vesicles (Zaghloul et al. 2017). Two ORFs predicted to be involved in apoptosis are conserved in all ascoviruses (Table 2). In DjTV-2a, these two ORFs are orf50 encoding for caspase B and orf108 encoding for inhibitor of apoptosis (IAP)-like protein.
Eleven proteins with other or unknown function are also conserved among all sequenced ascoviruses (Table 2). Among the 37 ascovirus core genes, 14 are also conserved in iridoviruses including the genes involved in DNA replication, transcription, as well as structural proteins (Table 2, asterisk-labeled). This is in agreement with current understanding that ascoviruses are closely related to iridoviruses.
Multigene Families
One of important features was the presence of multigene families (MGFs) in the DjTV-2a genome. MGFs occur in large dsDNA viruses. TnAV-6a was found to encode 15 ORFs which could be divided into six gene families (Wang et al.2006). In DjTV-2a genome, there are 4 MGFs containing 14 ORFs (Table 3).
Table 3.
Multigene families in DjTV-2a and their copy numbers in other ascovirus genomes.
| Gene family | DjTV-2a | DpTV-1a | SfAV-1a | HvAV-3i | TnAV-6a |
|---|---|---|---|---|---|
| ABC-type transport system permeases | 2 | 2 | 0 | 3 | 6 |
| Bro | 6 | 13 | 3 | 25 | 3 |
| MMP | 2 | 1 | 1 | 1 | 1 |
| Ring-finger-containing E3 ubiquitin ligase | 4 | 0 | 3 | 2 | 0 |
As mentioned earlier, DjTV-2a encodes two putative ABC-type transport system permeases (ORF122 and ORF123). ABC (ATP-binding Cassette) transporters are a large superfamily of proteins found in living organisms including bacteria, archaea, and eukarya. These proteins typically transport a variety of compounds such as ions, sugars, amino acids, lipids, complex polysaccharides, peptides, etc., across cell membranes. ABC transporters have a core structure consisting of two transmembrane domains and two ABCs or nucleotide binding domains. In Ascoviridae, only Toursvirus contain two copies ORFs of ABC transporters, other ascoviruses only contain one gene (Table 3). The protein was identified as a component of the virions by MS/MS analysis in HvAV-3i (Chen et al.2019).
Baculovirus repeated open reading frame (BRO) was found in certain large dsDNA viruses (Bideshi et al.2003). DjTV-2a genome has 6 BRO homologues, including ORF31, ORF32, ORF35, ORF39, ORF45 and ORF130, which are designed as BRO1-6, respectively. Among them, BRO1, BRO3 and BRO4 are conserved in all ascoviruses (Table 3). In baculoviruses, a BRO protein of Bombyx mori nucleopolyhedrovirus (BmNPV Bro-A) was previously shown to interact with B. mori laminin, a component of the basal lamina (Kang et al. 2003). In AcMNPV, Ac-bro (Ac-2) gene had high expression levels in the midgut throughout the infection and was presumable to aid in disruption or reorganization of laminin surrounding the midgut, facilitating release of budded virus (Shrestha et al. 2018). Three to 29 bros were found in different genomes of ascoviruses, and their function remains to be elucidated.
There were two putative myristylated membrane proteins (MMPs) encoded by ORF36 and ORF40 in DjTV-2a genome, and these two proteins share 28% aa identity. MMP was identified as a viral protein of HvAV-3i by proteomics analysis (Chen et al.2019). MMP is a conserved protein in ascoviruses, however, only DjTV-2a contains two copies of MMP. MMP is also conserved in iridoviruses (Supplementary Table S1), while in poxvirus, MMP is involved in virus assembly (Ravanello and Hruby 1994).
Four potential RING/U-box E3 ubiquitin ligases were identified in DjTV-2a genome, including ORF41, ORF43, ORF79 and ORF85, with aa identities from 28% to 55%. RING/U-box proteins transfer ubiquitin from an E2-Ub conjugate to a target substrate for degradation. RING_UBox proteins have a variety of cellular functions, including oncogenesis, development, viral replication, signal transduction, cell cycle, and apoptosis. E3 ubiquitin ligases are not conserved in ascoviruses. SfAV contains three copies (Bideshi et al. 2006), HvAV contains 2 copies (Table 3), but DpTV-1a does not contain any copy (Table 3).
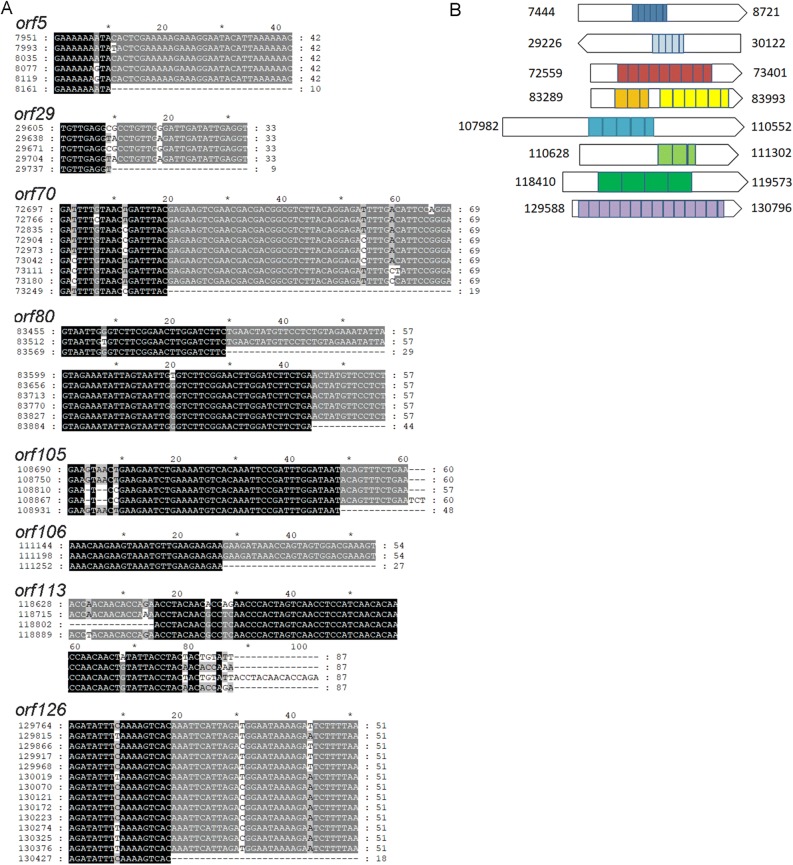
Genes with Intra-repeating Sequences
In DjTV-2a genome, there are 8 genes which have intra-repeating sequence (Fig. 2). These sequences repeat from 2 to 13 times within a gene, but there is no similarity between repeats from different genes. Except for those of orf5 and orf126, the other repeats had higher GC content than that of the average genome (26%). These repeats may be generated by sequence duplication during evolution. Interestingly, in other ascoviruses genome, there are also some genes which have intra-repeating sequence.
Fig. 2.
Genes with intra-repeating sequences. A The result shows intra-repeating sequences alignment. The numbers represent start position of repeat sequences and length. ORF80 has two parts. B It shows repeats sequence position in gene. The numbers indicate the start and end position of the ORFs.
Phylogenetic Analysis
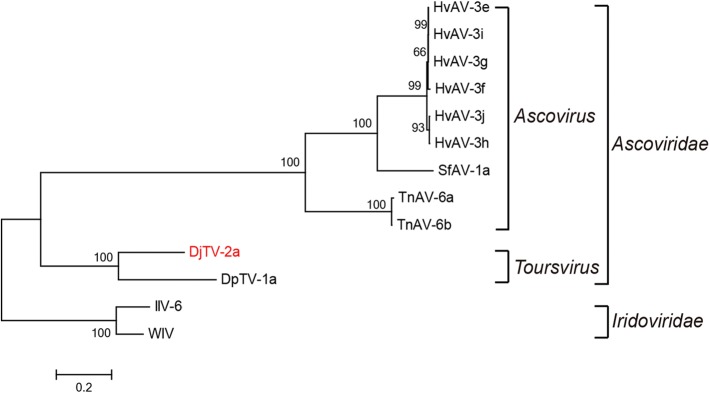
DNA polymerase is one of the best-studied enzymes in large dsDNA viruses. Phylogenetic analyses using DNA polymerases from different ascoviruses showed that the Ascoviridae could be clearly distinguished into two genera and DjTV-2a was close to DpTV-1a belonging to Toursvirus, the DNA polymerase of iridoviruses IIV-6 and WIV were used as outgroups (Fig. 3). Phylogenetic analyses also indicated that the separation of DjTV-2a and DpTV-1a appeared to be occurred much earlier than the separation of the current members of Ascovirus (Fig. 3).
Fig. 3.
Phylogenetic analyses of ascoviruses. Phylogenetic analysis was performed using the polymerase protein with maximum likelihood method and bootstrap value 1000 replicates. The numbers on the nodes indicate the bootstraps scores. DjTV-2a is indicated in red.
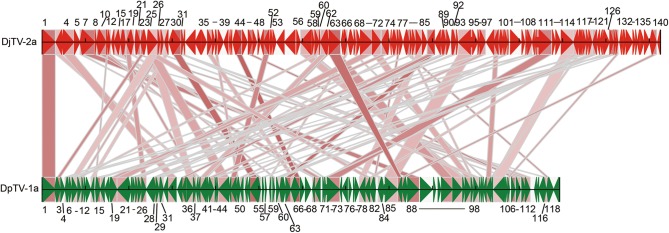
Comparison of DjTV-2a with DpTV-1a revealed 87 shared genes with an average aa identity of 39.7%. However, the gene order of DjTV-2a was quite different from that of DpTV-1a (Fig. 4). This again suggested that DjTV-2a was distinctly related to DpTV-1a and is a novel toursvirus.
Fig. 4.
Comparison of the genome structure of DjTV-2a and DpTV-1a. The genome of DjTV-2a was compared to that of DpTV-1a in ORF order and identity. The red arrows indicate DjTV-2a genes and green arrows indicate DpTV-1a genes. The direction of the arrows indicates the orientation of gene transcription. The homologue genes were with lined by lines with colors indicating the homology (the darker the color, the higher the identity). The number is orf order in genome (only those in forward direction were shown as a matter of convenience).
Discussion
Previously identified ascoviruses all infect Lepidoptera insects and are transmitted by endoparasitic wasps (Stasiak et al. 2005). The DjTV-2a genome, however, was found in D. jujubifolia belonging to Diptera, suggesting ascoviruses may distribute in a much wider range of insects than previously known. Among the previously identified ascoviruses, HvAV-3a and TnAV-2a have relative broad host spectra that they can replicate in different species of several noctuid genera (Bideshi et al. 2006). SfAV-1a has a narrow host range, only replicate in Spodoptera frugiperda species. DpTV-1a replicates in wasp vector (Diadromus pulchellus) and it transmitted to its lepidopteran host (Acrolepiopsis assectella), in which it replicates much more extensively (Bigot et al. 1997a, b; Stasiak et al. 2005). Since we did not have an isolate of the virus, the host range and transfer vector of DjTV-2a need to be identified in the future.
DjTV-2a is the second member of Toursvirus identified so far. Phylogeny and genome arrangement analyses showed that DjTV-2a is a distinct virus species related to DpTV-1a (Figs. 3, 4), suggesting these two viruses emerged from their common ancestor in an early time of evolution of tourviruses. DjTV-2a shares substantial genes with iridoviruses, supporting the hypothesis that Ascoviridae emerged recently from an invertebrate ancestor iridovirus lineage (Piégu et al. 2015). Interestingly, the viruses from Ascoviridae, Iridoviridae, Marseilleviridae shared 12 genes (Supplementary Table S1), including DNA-dependent RNA polymerase, RNA polymerase II, RNaseIII, ATPase involved in DNA metabolism, DEAD-like helicase, major capsid protein, Uyr/REP helicase, dynein-like beta chain protein, Immediate early protein ICP-46, DNA binding/packing protein, DNA-directed RNA polymerase II, and ATPase III, suggesting these viruses may use similar strategies for DNA replication and metabolism.
The identification of DjTV-2a raises numerous questions. For example, the virus genome was found in the field collected apparently healthy host, is this virus a commensal virus of the dipteran host? Are there similar viruses in other dipteran hosts? Is there an endoparasitic wasp associated with the virus for its transmission or is it transmitted vertically in the host insect, or both? So far, only the viral genome sequence of DjTV-2a is identified, isolation of the virus will be the next key step to further investigate these important questions. Nevertheless, the results of this study extended our knowledge of ascoviruses and opened a new area for future research.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the International Science and Technology Cooperation Program of Xinjiang Construction Corps (Grant No. 2017BC004) and the National Natural Science Foundation of China (Grant No. 31900154).
Author Contributions
HX and MY conceived and designed the experiments. HX performed the experiments. JW, ZH, GHH analysis the data. JW, FD, ZH, GHH, and MY wrote the manuscript. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interests.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
Footnotes
Jun Wang and Minglu Yang have contributed equally to the study.
Contributor Information
Minglu Yang, Email: ymlzkytd@163.com.
Zhihong Hu, Email: huzh@wh.iov.cn.
References
- Arai E, Ishii K, Ishii H, Sagawa S, Makiyama N, Mizutani T, Omatsu T, Katayama Y, Kunimi Y, Inoue MN, Nakai M. An ascovirus isolated from Spodoptera litura (Noctuidae: Lepidoptera) transmitted by the generalist endoparasitoid Meteorus pulchricornis (Braconidae: Hymenoptera) J Gen Virol. 2018;99:574–584. doi: 10.1099/jgv.0.001035. [DOI] [PubMed] [Google Scholar]
- Asgari S, Davis J, Wood D, Wilson P, McGrath A. Sequence and organization of the Heliothis virescens ascovirus genome. J Gen Virol. 2007;88:1120–1132. doi: 10.1099/vir.0.82651-0. [DOI] [PubMed] [Google Scholar]
- Asgari S, Bideshi DK, Bigot Y, Federici BA, Cheng XW, Consortium IR. ICTV Virus taxonomy profile: ascoviridae. J Gen Virol. 2017;98:4–5. doi: 10.1099/jgv.0.000677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bideshi DK, Renault S, Stasiak K, Federici BA, Bigot Y. Phylogenetic analysis and possible function of bro-like genes, a multigene family widespread among large double-stranded DNA viruses of invertebrates and bacteria. J Gen Virol. 2003;84:2531–2544. doi: 10.1099/vir.0.19256-0. [DOI] [PubMed] [Google Scholar]
- Bideshi DK, Demattei M-V, Rouleux-Bonnin F, Stasiak K, Tan Y, Bigot S, Bigot Y, Federici BA. Genomic sequence of Spodoptera frugiperda Ascovirus 1a, an enveloped, double-stranded DNA insect virus that manipulates apoptosis for viral reproduction. J Virol. 2006;80:11791–11805. doi: 10.1128/JVI.01639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bideshi DK, Spears T, Zaghloul HAH, Tan Y, Bigot Y, Federici BA. Ascovirus P64 Homologs: a novel family of large cationic proteins that condense viral genomic DNA for encapsidation. Biology (Basel) 2018;7:44. doi: 10.3390/biology7030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot Y, Rabouille A, Doury G, Sizaret PY, Delbost F, Hamelin MH, Periquet G. Biological and molecular features of the relationships between Diadromus pulchellus ascovirus, a parasitoid hymenopteran wasp (Diadromus pulchellus) and its lepidopteran host, Acrolepiopsis assectella. J Gen Virol. 1997;78:1149–1163. doi: 10.1099/0022-1317-78-5-1149. [DOI] [PubMed] [Google Scholar]
- Bigot Y, Rabouille A, Sizaret PY, Hamelin MH, Periquet G. Particle and genomic characteristics of a new member of the Ascoviridae: Diadromus pulchellus ascovirus. J Gen Virol. 1997;78(Pt 5):1139–1147. doi: 10.1099/0022-1317-78-5-1139. [DOI] [PubMed] [Google Scholar]
- Bigot Y, Renault S, Nicolas J, Moundras C, Demattei M-V, Samain S, Bideshi DK, Federici BA. Symbiotic virus at the evolutionary intersection of three types of large DNA viruses; iridoviruses, ascoviruses, and ichnoviruses. PLoS ONE. 2009;4:e6397. doi: 10.1371/journal.pone.0006397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZS, Hou DH, Cheng XW, Wang X, Huang GH. Genomic analysis of a novel isolate Heliothis virescens ascovirus 3i (HvAV-3i) and identification of ascoviral repeat ORFs (aros) Arch Virol. 2018;163:2849–2853. doi: 10.1007/s00705-018-3899-2. [DOI] [PubMed] [Google Scholar]
- Chen ZS, Cheng XW, Wang X, Hou DH, Huang GH. Proteomic analysis of the Heliothis virescens ascovirus 3i (HvAV-3i) virion. J Gen Virol. 2019;100:301–307. doi: 10.1099/jgv.0.001197. [DOI] [PubMed] [Google Scholar]
- Cullinane M, Baysse C, Morrissey JP, O’Gara F. Identification of two lysophosphatidic acid acyltransferase genes with overlapping function in Pseudomonas fluorescens. Microbiology. 2005;151:3071–3080. doi: 10.1099/mic.0.27958-0. [DOI] [PubMed] [Google Scholar]
- Huang GH, Wang YS, Wang X, Garretson TA, Dai LY, Zhang CX, Cheng XW. Genomic sequence of Heliothis virescens ascovirus 3 g isolated from Spodoptera exigua. J Virol. 2012;86:12467–12468. doi: 10.1128/JVI.02342-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GH, Hou DH, Wang M, Cheng XW, Hu Z. Genome analysis of Heliothis virescens ascovirus 3 h isolated from China. Virol Sin. 2017;32:147–154. doi: 10.1007/s12250-016-3929-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob NJ, Müller K, Bahr U, Darai G. Analysis of the first complete DNA sequence of an invertebrate iridovirus: coding strategy of the genome of Chilo iridescent virus. Virology. 2001;286:182–196. doi: 10.1006/viro.2001.0963. [DOI] [PubMed] [Google Scholar]
- Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45:237–249. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Jiao KL, Han PJ, Yang ML, Xiong RC, Wang YH, Bu WJ. A new species of gall midge (Diptera: Cecidomyiidae) attacking jujube, Ziziphus jujuba in China. Zootaxa. 2017;4247:487–493. doi: 10.11646/zootaxa.4247.4.10. [DOI] [PubMed] [Google Scholar]
- Kang WK, Imai N, Suzuki M, Iwanaga M, Matsumoto S, Zemskov EA. Interaction of Bombyx mori nucleopolyhedrovirus BRO-A and host cell protein laminin. Arch Virol. 2003;148:99–113. doi: 10.1007/s00705-002-0902-7. [DOI] [PubMed] [Google Scholar]
- Li L, Shataer A, Pan C, Cao Q. Study on Growth and Decline Law and Control of Dasineura datifolia Jiang in Aksy. J Xinjiang Agric Univ. 2010;33:36–39. [Google Scholar]
- Liu YY, Xian WF, Xue J, Wei YL, Cheng XW, Wang X. Complete genome sequence of a renamed isolate, Trichoplusia ni sscovirus 6b, from the United States. Genome Announc. 2018;6:e00148–e00218. doi: 10.1128/genomeA.00148-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long CM, Rohrmann GF, Merrill GF. The conserved baculovirus protein p33 (Ac92) is a flavin adenine dinucleotide-linked sulfhydryl oxidase. Virology. 2009;388:231–235. doi: 10.1016/j.virol.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Parsons JD. Miropeats: graphical DNA sequence comparisons. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- Piégu B, Asgari S, Bideshi D, Federici BA, Bigot Y. Evolutionary relationships of iridoviruses and divergence of ascoviruses from invertebrate iridoviruses in the superfamily Megavirales. Mol Phylogenetics Evol. 2015;84:44–52. doi: 10.1016/j.ympev.2014.12.013. [DOI] [PubMed] [Google Scholar]
- Ravanello MP, Hruby DE. Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virion assembly. J Virol. 1994;68:6401–6410. doi: 10.1128/JVI.68.10.6401-6410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha A, Bao K, Chen Y-R, Chen W, Wang P, Fei Z, Blissard GW. Global analysis of baculovirus Autographa californica multiple nucleopolyhedrovirus gene expression in the midgut of the Lepidopteran host Trichoplusia ni. J Virol. 2018;92:e01277–e01318. doi: 10.1128/JVI.01277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasiak K, Renault S, Federici BA, Bigot Y. Characteristics of pathogenic and mutualistic relationships of ascoviruses in field populations of parasitoid wasps. J Insect Physiol. 2005;51:103–115. doi: 10.1016/j.jinsphys.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Xue J, Seaborn CP, Arif BM, Cheng X-W. Sequence and organization of the Trichoplusia ni ascovirus 2c (Ascoviridae) genome. Virology. 2006;354:167–177. doi: 10.1016/j.virol.2006.06.029. [DOI] [PubMed] [Google Scholar]
- Wei YL, Hu J, Li SJ, Chen ZS, Cheng XW, Huang GH. Genome sequence and organization analysis of Heliothis virescens ascovirus 3f isolated from a Helicoverpa zea larva. J Invertebr Pathol. 2014;122:40–43. doi: 10.1016/j.jip.2014.08.003. [DOI] [PubMed] [Google Scholar]
- White CL, Senkevich TG, Moss B. Vaccinia virus G4L glutaredoxin is an essential intermediate of a cytoplasmic disulfide bond pathway required for virion assembly. J Virol. 2002;76:467–472. doi: 10.1128/JVI.76.2.467-472.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SK, Knoll LJ. Patatin-like phospholipases in microbial infections with emerging roles in fatty acid metabolism and immune regulation by Apicomplexa. Mol Microbiol. 2018;107:34–46. doi: 10.1111/mmi.13871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CK, Young VL, Kleffmann T, Ward VK. Genomic and proteomic analysis of invertebrate iridovirus type 9. J Virol. 2011;85:7900–7911. doi: 10.1128/JVI.00645-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaghloul HAH, Hice R, Arensburger P, Federici BA. Transcriptome analysis of the Spodoptera frugiperda ascovirus in vivo provides insights into how its apoptosis inhibitors and caspase promote increased synthesis of viral vesicles and virion progeny. J Virol. 2017;91:e00874–e00917. doi: 10.1128/JVI.00874-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.