Abstract
Grass carp reovirus (GCRV), the genus Aquareovirus in family Reoviridae, is viewed as the most pathogenic aquareovirus. To understand the molecular mechanism of how aquareovirus initiates productive infection, the roles of endosome and microtubule in cell entry of GCRV are investigated by using quantum dots (QDs)-tracking in combination with biochemical approaches. We found that GCRV infection and viral protein synthesis were significantly inhibited by pretreating host cells with endosome acidification inhibitors NH4Cl, chloroquine and bafilomycin A1 (Bafi). Confocal images indicated that GCRV particles could colocalize with Rab5, Rab7 and lysosomes in host cells. Further ultrastructural examination validated that viral particle was found in late endosomes. Moreover, disruption of microtubules with nocodazole clearly blocked GCRV entry, while no inhibitory effects were observed with cytochalasin D treated cells in viral infection, hinting that intracellular transportation of endocytic uptake in GCRV infected cells is via microtubules but not actin filament. Notably, viral particles were observed to transport along microtubules by using QD-labeled GCRV. Altogether, our results suggest that GCRV can use endosomes and microtubules to initiate productive infection.
Electronic supplementary material
The online version of this article (10.1007/s12250-019-00178-1) contains supplementary material, which is available to authorized users.
Keywords: Aquareovirus, Cell entry, Quantum dot, Endosome, Microtubule
Introduction
Aquareoviruses, members of the family Reoviridae, have been isolated from a wide variety of aquatic organisms in both sea and freshwater origins. The pathogens are nonenveloped viruses, and can cause infection in aquatic organisms including bony fish, shellfish, and crustaceans worldwide (King et al. 2011). Although some of aquareoviruses are isolated from breeding aquatics with minor symptoms, several members of this group are important pathogens. Such as grass carp reovirus (GCRV), which is able to cause fatal hemorrhagic disease of grass carp and also can induce cell–cell fusion and produce characteristic cytopathic effect (CPE) in its permissive cells (Fang et al. 1989; Ke et al. 1990; Guo et al. 2013). GCRV is recognized to be the most pathogenic amongst isolated aquareoviruses (Rangel et al. 1999; Jaafar et al. 2008). Previous studies have been demonstrated that core proteins (VP1–VP4, and VP6) of GCRV are mainly responsible for viral replication, while outer capsid proteins VP5 and VP7 play critical role in viral entry and infection (Fang et al. 2005; Li and Fang 2013). More than twenty GCRV strains have been reported so far, and the whole genome sequences have been completed from ten of the GCRV isolates. Based on the phylogenetic relationship of core protein VP6, three groups were classified, with representative isolates GCRV-873 (group I), GCRVHZ08 (group II), and HGDRV (group III) (wang et al. 2012; Rao and Su 2015).
Nonenveloped viruses are unable to take advantage of membrane fusion to enter cells, and instead, must disrupt lipid membrane to form pores for initiating efficient cell entry. In general, reoviruses are activated for entry and infection by undergoing a series of programmed proteolytic cleavages to generate infectious subvirion particles (ISVPs) for productive infection (Sturzenbecker et al. 1987; Fang et al.2008). In addition, release of the myristoylated protein μ1 in MRV or VP5 in aquareovirus at N-terminus is necessary for reovirus membrane penetration at an early stage of viral infection (Chandran et al. 2002). Recent study indicated that the autocleavage of outer capsid protein VP5 of aquareovirus is required for efficient infection and its N-terminal myristoylated short peptide played key role for penetrating cell membrane and promoting infectivity (Chen et al. 2018). Upon internalization, viruses are usually delivered to endosomal compartments and transported along actin filaments or microtubules (Doherty and McMahon 2009; Mainou et al. 2013). Like other non-enveloped viruses, uncoating in MRV or aquareovirus results in the removal of outer-capsid protein σ3/VP7 and conformational rearrangement of outer-capsid protein μ1/VP5, which allows endosomal membrane penetration and release of transcriptionally active viral core particles into the cytoplasm (Li and Fang 2013; Suomalainen and Greber 2013). The detailed event of how GCRV initiates its infection in host cells is largely unknown.
Inorganic semiconductor nanocrystal quantum dots (QDs) possess remarkable photostability and brightness, which have been shown to be powerful tool for single particle with long-term imaging of viral infection in live cells (Bruchez et al. 1998; Medintz et al. 2005). Previously, we have revealed that the caveolae/raft-mediated endocytosis is required for aquareovirus entry by using QD-labeled GCRV in combination with biochemical analyses (Zhang et al. 2017, 2018). In this study, we further identified that the infectivity of GCRV was suppressed by endosome acidification inhibitors, indicating that GCRV infection requires a low pH intracellular compartment. Moreover, GCRV infection was obviously prohibited by disrupting microtubules with the specific drug nocodazole. This result provides basis for further revealing molecular pathogenesis of aquareovirus.
Materials and Methods
Cell and Virus
The Ctenopharyngodon idellus kidney (CIK) cell line was used in this study for the propagation of GCRV. CIK cells were grown at 28 °C in Eagle’s minimum essential medium (MEM, Invitrogen) including 2 mmol/L l-glutamine supplemented with 10% fetal bovine serum (MEM-10). The GCRV-873, the type strain of GCRV, was isolated from fresh aquaculture fishery in Shaoyang county, Hunan province (Ke et al.1990). The virus cultivated in Eagle’s MEM supplemented to contain 2% fetal bovine serum (MEM-2) was grown in monolayers of CIK cells as previously described (Fang et al. 1989; Ke et al. 1990).
Antibodies and Inhibitors
The anti-biotin mouse monoclonal antibody was obtained from Santa Cruz Biotechnology. The alkaline-phosphatase-coupled goat anti-mouse IgG or goat anti-rabbit IgG was the product of Sigma-Aldrich, and peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG was purchased from Thermo Pierce. The Alexa Fluor 488 donkey anti-rabbit IgG (H + L) was purchased from Invitrogen Co. (Invitrogen, Carlsbad). The mouse and rabbit polyclonal antibodies (pAbs) against proteins VP3, VP5 and VP7 of GCRV were generated in our laboratory as reported previously (Yan et al. 2015a; Zhang et al.2018). Pharmacological inhibitors chloroquine (CQ), NH4Cl, cytochalasin D (Cyto D), bafilomycin A1 (Bafi), and nocodazole (Noco) were purchased from Sigma-Aldrich.
Preparation of Quantum Dot Labeled GCRV
Quantum dot labeling of GCRV were performed as described elsewhere (Zhang et al. 2017, 2018). In brief, the purified virions were first incubated with sulfosuccinimidyl-6-biotinamidohexanoate (sulfo-NHS-LC-biotin, Thermo Scientific) at 28 °C for 2 h to obtain biotinylated GCRVs (bio-GCRVs), and then were separated from Unbound biotin by using ZebTM spin desalting column (Thermo Scientific) according to the manufacturer’s instruction. After biotinylation, the purified bio-GCRVs were incubated with streptavidin-conjugated QDs (SA-QDs, 625 nm, Invitrogen) for 3 h at room temperature. The further purification of QDs labeled GCRV were performed by using sucrose density gradient centrifugation (SDGC) as described previously (Fang et al. 2005).
Real-Time qPCR Analysis
The real-time quantitative PCR (qPCR) was performed following previous reported methods (Yan L et al. 2015). The virus genome was amplified by qPCR for detecting the expression level of minor core protein VP2. The primers used for the real-time qPCRs are as the following: the forward primer 5′-TACGCCTACACCTTACTTCAA-3′ and reverse primer 5′-CGGTTCGGTCCACTCTATT-3′ (114 bp). In brief, total RNAs were extracted from the infected CIK cells at 24 hpi by using RNA extraction kit (Omega) according to kit instruction. Then qPCR was performed on a real-time thermo cycler (BIO-RAD) with the CFX 96 software. Each reaction was performed in triplicate and β-actin was analyzed as a control for each experiment. The relative expression ratios were calculated by a mathematical model and Graphpad Prism V5.01 was used for relative quantification.
Infection and One-Step Growth Curve
One-step growth curve was performed in CIK cells as described previously (Furlong et al. 1988; Maginnis et al.2008; Zhang et al. 2013). Initially, cells were infected with QD-bio-GCRV, bio-GCRV and GCRV at an MOI of 5, and then the viral infected cell supernatant was taken out at 8, 12, 24, 36, 48, 60, 72, 84, and 96 h postinfection (hpi) respectively. All collected samples were stored at − 80 °C for further titer determination by immunofluorescence assay (IFA) using antibody against protein VP5. Briefly, the collected supernatant from virus-infected cells was serially diluted in serum-free MEM and then added into the CIK cells for 48 h cultivation at 28 °C. After washing twice with PBS, the cells were fixed with 4% paraformaldehyde and processed for immunofluorescence staining. The number of immunofluorescent focus was counted under fluorescence microscopy. The viral titer was determined by examining the mean number of immunofluorescent focus units (IFU) of virus presented in three random microscope fields, and three independent experiments were performed.
Infection assays were performed by determining the plaque formation unit (PFU) in the presence of the inhibitors according to previously described method (Furlong et al. 1988; Yan L et al. 2015). In brief, CIK cells were grown in 24-well tissue culture plates before the day of carrying out inhibitor treatment experiment. The cultured cells were pretreated with all the indicated drugs for 1 h at 28 °C. And then, cells were inoculated with GCRV at an MOI of 1 in the drug-containing medium at 4 °C for 1 h. The infected cells were washed with PBS to remove un-adsorped virions, and then were maintained in MEM-2 in the presence of the inhibitors at 28 °C for 48 h. Finally, the cells were fixed with 4% paraformaldehyde and stained with methylrosaniline chloride solution.
Western Blot (WB) Analysis
WB was used for viral entry and infection analysis. CIK cells were plated in 6-well microplates (Corning) at a density of 2 × 106 cells for cultivation. After treating with inhibitors, cells were then inoculated with GCRV at an MOI of 5 in the presence of drugs for 1 h at 4 °C. Infected CIK cells were washed twice with PBS and incubated with fresh MEM-2 medium plus inhibitors at 28 °C for 24 h. In each experiment, cells were treated with drugs at indicated concentration for 60 min before or after virus addition and maintained during the infection. All the cells were harvested and washed twice with PBS, then lysed in RIPA buffer (Beyotime Institute). Cell lysates were fractionated by SDS-PAGE and further analyzed by WB with specific primary antibodies against VP3, VP5 and VP7 of GCRV. Alkaline-phosphatase-coupled goat anti-rabbit IgG or goat anti-mouse IgG were used as secondary antibodies. β-actin was detected simultaneously as a loading control.
Transmission Electron Microscopy (TEM) for Ultrastructural Examination
Ultrastructural examinations were performed as described previously (Fan et al.2010; Zhang et al. 2018). In brief, GCRV particles were added to the CIK cells with the serum-free MEM at 4 °C for 20 min to allow binding to cell surface receptors. After removing the supernatants, the infected cells were incubated with fresh MEM-2 at 28 °C for 30 min. The collected cell samples were fixed with 2% glutaraldehyde for preparing ultrathin serial sections. Ultrathin sections were obtained using a Leica EM UC7 (Leica Microsystems, Vienna, Austria) and mounted on copper grids. All the grids were examined by using an FEI G20 Tecnai transmission electron microscope (FEI, Tecnai G20 TWIN).
Immunofluorescence Microscopy
CIK cells (1 × 105 per well) were seeded and cultured in glass-bottomed petri dishes overnight at 28 °C, then infected with QD-bio-GCRVs. Meanwhile, QD and GCRV + QD were served as controls. Following 20 min postinfection (mpi), all the cells were fixed with 4% paraformaldehyde at room temperature for 30 min and permeabilized with 0.2% Triton X-100 for 15 min. After blocking with BSA, cells were washed three times with PBS and then incubated with an antibody against protein VP5 or VP1. Alexa 488-labeled donkey anti-mouse IgG (H + L) antibody (Invitrogen) was used as second antibody, and hoechst 33342 (Invitrogen) was used to stain the nuclei. Imaging was processed with a PerkinElmer UltraView VOX system using Nikon Ti microscope.
Inhibitors Blocking Assay
To analyze GCRV uptake, CIK cells were pretreated with all the listed inhibitors. Cells were incubated with the indicated inhibitors prior to exposure to the QD-bio-GCRV. After pretreatment, cells were washed with PBS and incubated with QD-bio-GCRV at an MOI of 100 at 28 °C for 1 h. Then the cells were incubated in MEM-2 in the presence of inhibitor for 3 h. Image acquisition was performed under the laser confocal microscope (Nikon). To quantify the effect of a drug treatment, the location of QD-bio-GCRV in infected cells with drugs or mock treatment was evaluated by determining the mean number of cells showed virus internalization. Three independent experiments were performed, and cells were selected in ten random microscope fields at ×60 magnification in each experiment.
For investigating the entry route of the QD-bio-GCRV, cells were pretreated with different concentrations of inhibitors for 1 h prior to incubation with QD-labeled virus. After pretreatment, cells were washed with PBS and inoculated with QD-bio-GCRV at an MOI of 5 for 1 h. Following adsorption, infected cells were washed three times with PBS and incubated in MEM-2 in the presence of inhibitors. And then, the cell lysates and supernatants of GCRV-infected cells were collected at 24 hpi for WB assays and viral titration, respectively.
Plasmids and Transfections
Plasmids pEGFP-actin, pcDNA3.1-GFP-MAP4, pEGFP-Rab5, pEGFP-Rab7 were kindly provided by Prof. Hanzhong Wang, Wuhan Institute of Virology, CAS. The transfection experiments were performed as described previously (Shao et al. 2013). Briefly, monolayers of CIK cells were transfected with indicated plasmid DNA mixed with Lipofectamine LTX PLUS reagent (Invitrogen) according to the manufacturer’s instruction. At 24 h posttransfection (hpt), cells were subjected to fluorescence microscopy observation for further entry assays.
Live Microscopy Experiments
In order to track the single-virus particle movement during entry into live cells, cells were grown on glass-bottom culture dishes for overnight cultivation. The nuclei of the CIK cells were pre-stained with hoechst 33342. Then the purified QD-bio-GCRVs were added to the cultured cells for dynamic tracking at 28 °C, and the real-time entry of the virus was imaged using Velocity software (PerkinElmer).
To visualize synchronized entry of labeled virus, the QD-bio-GCRVs were added to the drug-pretreated or the transfected cells and allowed to binding at 4 °C for 30 min in the presence of inhibitors. After washing with PBS, the infected cells were placed at 28 °C in the presence of inhibitors for real-time tracking. QDs were excited with 561 nm lasers. Serial images were recorded using the UltraView VOX (PerkinElmer) confocal system with an inverted microscope (Nikon).
Statistical Analysis
The statistical significance was determined using the unpaired Student’s t test. P values were interpreted as *P < 0.05; **P < 0.01. All graphs represent means and standard deviations of normalized data points for triplicate samples from each of three independent experiments.
Results
Infection and Real Time Tracking Assays of QD Labeled GCRV Particles
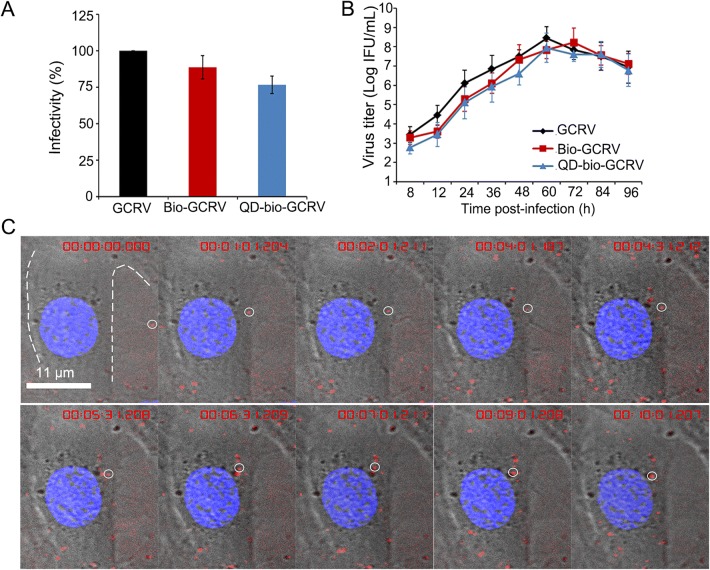
We previously identified that GCRV can use caveolae/raft-mediated endocytosis for cell entry (Zhang et al. 2018). To understand whether endosome and cytoskeleton are involved in GCRV productive entry, we obtained QD-labeled GCRV for live-virus tracking by using our previously established method of specific surface labeling of GCRV with QDs (Zhang et al. 2017). To evaluate whether the QD-labeled GCRV remains its original biological activities, the relative infectivity of QD-bio-GCRV was evaluated by qPCR analysis at 24 hpi, and the mRNAs encoding core protein VP2 (RNA-dependent RNA polymerase, RdRp) were detected to assess the expression levels of the RdRp. It appeared that the relative expression level of VP2 with modified GCRV in infected cells remained at 87% after biotinylation and at 72% after the conjunction with QDs (Fig. 1A). Further, the influence of the labeling on viral infectivity was also examined by one-step growth curve using IFA. As shown in Fig. 1B, QD-bio-GCRV exhibited similar growth kinetics as that of unmodified virus. Although there was a slight decrease in viral titer with QD-bio-GCRV by comparing to unmodified virus in the early phase of infection, both of them reached a maximum titer at 60 hpi, suggesting that the QD-modified GCRVs still retained relative high infectivity. This result indicated that the viral infectivity of QD-bio-GCRV is not significantly affected by the QD conjunction.
Fig. 1.
Infection and real time tracking assays of QD labeled GCRV particles. A Viral infectivity of QD-bio-GCRV, Bio-GCRV and GCRV was calculated by qPCR analysis at 24 hpi. B One-step growth curves of QD-bio-GCRV, Bio-GCRV and GCRV in CIK cells. Cells were infected with the indicated virus at an MOI of 5, and non-adsorbed viruses were removed at 1 hpi. The infection was allowed to proceed at 28 °C and the supernatant was taken at the indicated time. Titers were determined by IFU. The data shown represent the mean values and standard deviations from three independent experiments. C Snapshots of QD-labeled GCRV entering into live cell. QD-bio-GCRVs were added to the adherent cells directly for dynamic tracking at 28 °C. In situ real-time images of QD-bio-GCRVs in a live cell without synchronization of any kind. Single-particle of QD-labeled GCRV is indicated with white circle. Red fluorescence in the cytoplasm indicates internalized virus.
To test whether the QD-bio-GCRV could facilitate virus tracking during entry into host cells, the QD-bio-GCRVs were inoculated with CIK cells and imaged by confocal microscope for in situ real-time dynamic tracking. Snapshots clearly showed that a single-particle of QD-bio-GCRV attached to the cell membrane during very early stage of infection, and was soon internalized into the cytoplasm (Fig. 1C and Supplementary Movie S1). Altogether, the results strongly indicated that QD-bio-GCRVs could be used for virus imaging, and were real time tracking-capable in live cells.
Weak Base NH4Cl Could Inhibit GCRV Infection
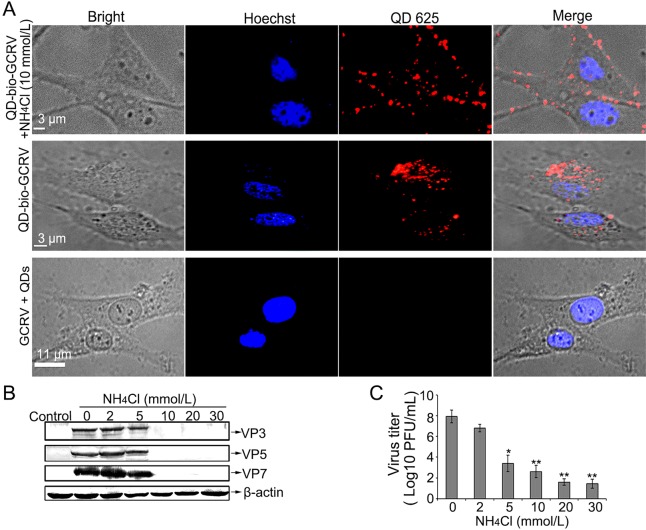
Previous studies indicated that MRV and aquareovirus infection can be inhibited by treating cells with inhibitors of acid-dependent lysosomal proteinases, such as the weak base NH4Cl (Chandran et al.1999; Yan S et al. 2015). To evaluate that the red fluorescent dots represent GCRV rather than free QDs, the viral infectivity was also examined by treating cells with inhibitor NH4Cl to prevent viral entry. As shown in Fig. 2A, most of the fluorescent dots of the QD-bio-GCRVs were located on the membrane of CIK cells pretreated with 10 mmol/L NH4Cl (Fig. 2A, upper panel). In contrast, such an inhibitory distribution of QD-bio-GCRV was not found in mock treated cells (Fig. 2A, middle panel). In addition, the strong red fluorescent signal was not detected in the cells incubated with unmodified GCRV and free QD (GCRV + QDs) (Fig. 2A, bottom panel), indicating that NH4Cl could efficiently inhibit QD-bio-GCRV internalization into cells. These results proved that QD-labeled GCRV can enter cells like native virus and GCRV entry could be inhibited by weak base NH4Cl.
Fig. 2.
Weak base NH4Cl could inhibit GCRV entry and infection. A Fluorescence distribution of QD-bio-GCRVs infected cells in the presence or absence of NH4Cl. Cells were incubated with or without 10 mmol/L NH4Cl prior to exposure to QD-bio-GCRV at an MOI of 100 at 28 °C. Imaging was performed at 3 hpi by the laser confocal microscope. B–C Viral proteins expression and virus yield assays in NH4Cl-treated CIK cells. Cells were pretreated with different drugs at the indicated concentrations and then infected with GCRVs in the presence of drugs. The expression level of viral proteins was analyzed by WB (B), and the progeny virus titer was determined by plaque assay (C). Three independent experiments, repeated three times for each sample, were performed. Asterisks denote a statistically significant change in virus titer compared to that in the absence of NH4Cl (*P < 0.05; **P < 0.01).
To further prove the effect of the agent NH4Cl on viral replication, infections then proceeded in the presence of different concentrations of NH4Cl (from 0 to 30 mmol/L) at 28 °C, then viral protein expression and infectious titers were determined at 24 hpi. As expected, the protein expression level and viral titers were reduced as concentration increased, which showed that NH4Cl could diminish GCRV infection in a dose-dependent manner (Fig. 2B, 2C), suggesting that infections with aquareovirus were blocked by NH4Cl. This result indicates that NH4Cl could inhibit GCRV infection.
GCRV Infection Requires Endosomal Acidification
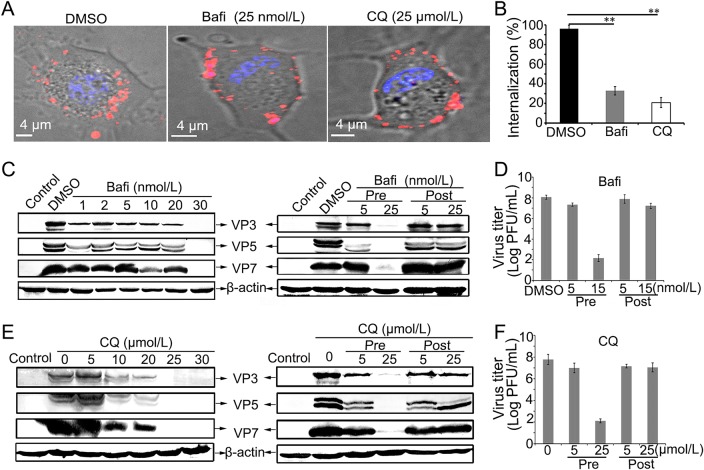
To initiate efficient infection, many viruses enter cells through cellular endosomal systems to make acidic environment (Forzan et al. 2007; Huang et al.2011; Mainou and Dermody 2012). Based on the aforementioned result that GCRV infection could be inhibited by weak base NH4Cl, we supposed that GCRV may utilize endocytic compartment during cell entry to initiate productive infection. To test this hypothesis, we next investigated whether GCRV infection needs the acidic environment of the endosomal compartment. We first treated CIK cells with different concentrations of Bafi and CQ, which are known to inhibit the acidification of endosomes and lysosomes (Sturzenbecker et al. 1987; Mainou and Dermody 2012). As shown in Fig. 3A, the GCRV infected CIK cells pre-treated with Bafi and CQ appeared a pattern that QD-bio-GCRV fluorescence signal rested on the cell membrane and failed to reach the cytosol, which was different from DMSO-treated cells. Further statistical analyses were performed to calculate the percent of the internalization. In the absence of drugs, almost all CIK cells were infected with the labeled GCRVs, with an internalization rate of 96.7% for DMSO. In the presence of endosome acidification inhibitor, the infection rate was significantly reduced, with an internalization rate of about 33% for Bafi, and 21% for CQ, respectively (Fig. 3B). Time-lapse images of the QD-bio-GCRV and CIK cells in the presence of CQ showed no obvious internalization in the visual field (see Supplementary Movie S2). These results demonstrate that both Bafi and CQ inhibit GCRV infection. To further confirm the inhibitory effect in virus replication cycle, we treated CIK cells with different concentrations of Bafi and CQ before and after viral infection, it showed that Bafi and CQ with high concentration could make a marked inhibitory effect on viral proteins synthesis (Fig. 3C, 3E, left panel) and progeny virus production (Fig. 3D, 3F) before virus adsorption to cells. However, when treated cells with high concentration Bafi and CQ after virus infection, no observed inhibition in both protein expression (Fig. 3C, 3E, right panel) and viral infectivity (Fig. 3D, 3F) could be detected, suggesting that the low endosomal pH is critical for GCRV entry, but not for replication.
Fig. 3.
The low-pH is essential for GCRV entry and replication in CIK cells. A–B Fluorescence profile and quantitative analysis of QD-bio-GCRVs in Bafi-, CQ-treated and untreated cells (DMSO). Bafi and CQ significantly reduced the internalization of QD-bio-GCRVs into the CIK cells. Cells were pretreated with indicated concentrations of inhibitors for 1 h and then incubated with QD-bio-GCRVs for 3 h in the presence of inhibitor. 150 cells were enumerated in each experiment, three independent experiments were carried out, error bars represent standard deviations (**P < 0.01). C–F Effects of inhibitors Bafi (C, D) and CQ (E, F) on GCRV infection in CIK cells. Cells were treated with drugs at different concentrations, or different times (before or after virus addition) at the indicated concentrations. Then the synthesis of viral proteins was detected by WB experiment, and viral production was examined by PFU assay.
Endosomes and Lysosomes are Required for GCRV Entry
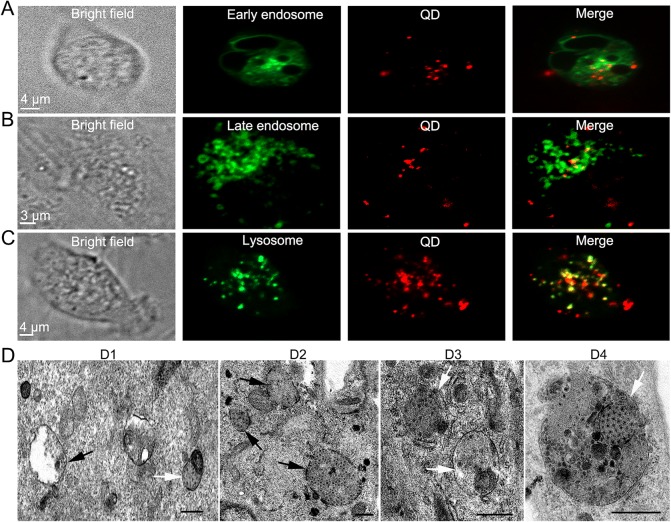
To determine whether GCRV entry was associated with the endosome and lysosome system, pEGFP-Rab5 and pEGFP-Rab7 were transfected into CIK cells to indicate early endosomes and late endosomes, and the LysoTracker (lysosome-specific dye) was used to stain the lysosomes. The transfected or stained cells were infected with GCRV, and examined by confocal immunofluorescence microscopy. As shown in Fig. 4A–4C, the colocalization between QD-bio-GCRVs and Rab5, Rab7 and lysosomes could be detected at 30 mpi, indicating that GCRV particles transport along early endosomes, late endosomes and lysosomes during internalization. To further prove the image results, the ultrastructural examinations using TEM were also conducted. It showed that GCRV particles were delivered to endocytic compartments and located in lysosomes during early entry (Fig. 4D1–4D4). All together, the results indicated that the endosome compartments and low pH are required for productive infection of GCRV.
Fig. 4.
Localization analysis of GCRV endocytic uptake. A–C QD-bio-GCRVs co-localize with Rab5, Rab7 and lysosomes during the early stage of infection in CIK cells. CIK cells expressing Rab5 (A) or Rab7 (B) were infected with QD-bio-GCRVs at 24 hpt, and then captured by confocal laser scanning microscope at 30 mpi. CIK cells treated with LysoTracker for 1 h (C) were also incubated with QD-bio-GCRVs for 30 min and then used for observation. D Ultrastructural analysis of GCRV internalization. CIK cells were infected with GCRV particles at an MOI of 500 at 4 °C for 30 min, then washed with PBS and fixed for TEM. Representative images reveal the organelles resembling endosomes and lysosomes containing GCRV particles in cells at 30 min (D1–2), and 60 min (D3–4) post-adsorption. Reovirus virions localized in structures that resemble endosomes and primary lysosomes are indicated by black arrows. Secondary lysosomes are indicated by white arrows. Scale bars, 500 nm.
GCRV Entry is Microtubule-Dependent and Actin-Independent
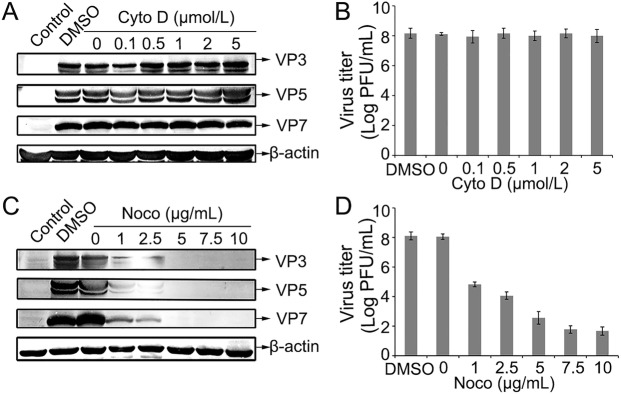
As is known, cytoskeleton plays critical roles during virus entry into cells. Upon internalization, many viruses are usually delivered to endosomal compartments and transported along actin filaments or microtubules (Doherty and McMahon 2009; Mainou et al. 2013). Cyto D is a cell-permeable inhibitor of actin polymerization by binding to F-actin polymer and preventing polymerization of actin monomers (Huang et al. 2011; Sánchez et al.2012). Noco can disrupt microtubules in cells by interfering with the polymerization of microtubules (Eilers et al. 1989; Mainou et al. 2013). To assess the effect of the inhibitors in GCRV infection, CIK cells were pretreated with various concentrations of Cyto D or Noco. It showed that the structural proteins (VP3, VP5 and VP7) synthesis and viral replication was not significantly affected by treating cells with Cyto D, even in the presence of very high concentrations of this drug (Fig. 5A, 5B), while the expression yields of the three structural proteins and viral titer were gradually decreased by pretreating cells with increased concentrations of Noco (Fig. 5C, 5D). These results indicate that actin is not essential in GCRV transportation, and microtubule may play an important role in the intracellular transport of virions.
Fig. 5.
GCRV entry is microtubule-dependent and actin-independent. A–D Effects of inhibitors Cyto D and Noco on GCRV infection in CIK cells. Cells were pretreated with different drugs at the indicated concentrations and then infected with GCRVs in the presence of drugs before being collected the cells. The supernatants of GCRV-infected cells were collected at the 24 h post-infection. The level of viral proteins of GCRV in the presence of Cyto D (A) or Noco (C) was analyzed by Western blot. The progeny virus titer of GCRV in the presence of Cyto D (B) or Noco (D) was determined by plaque assay.
Real Time Tracking of Microtubule Dependent Pathway
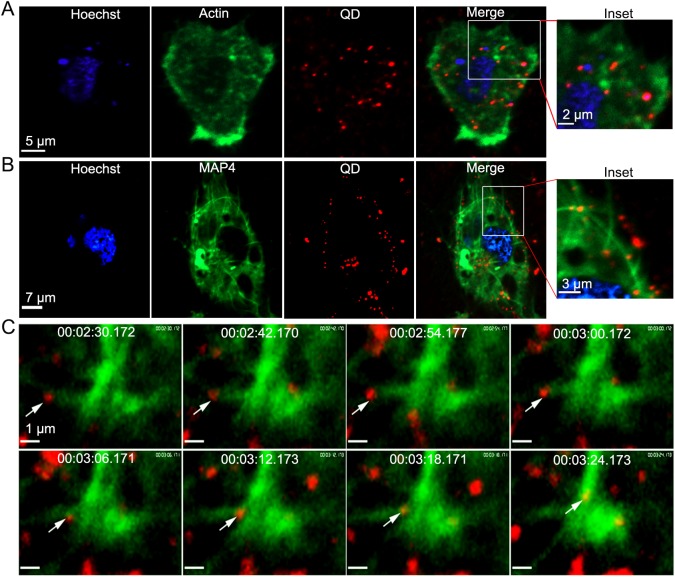
After finding the role of microtubule in viral transportation, it is important to capture interaction between GCRV and cytoskeleton microtubule in real time. To prove whether GCRV colocalize with actin or microtubule in live cells, EGFP-actin (GFP fused to the actin) and GFP-MAP4 (GFP fused to the microtubule-binding domain of mammalian microtubule-associated protein 4) were used to express in CIK cells to indicate actin filaments and microtubules, and then the cells were inoculated with GCRV. As shown in Fig. 6A, no obvious colocalization was observed between GCRV and actin on the CIK cells at 30 mpi. Under the same conditions, colocalization between GCRV particles and microtubules was clearly observed (Fig. 6B). Further real-time tracking recorded that GCRV particles were associated with the microtubules and moved along microtubules during infection (Fig. 6C, see Supplementary Movie S3), suggesting that actin may not be involved in GCRV internalization but microtubules may be crucial for intracellular transport of GCRV during entry.
Fig. 6.
Real time tracking of microtubule dependent pathway. A–B QD-bio-GCRVs co-localize with microtubules (B) but not with actin (A) during the early stage of infection. CIK cells were transfected with pEGFP-actin or pGFP-MAP4 prior to incubation with QD-bio-GCRVs. A magnification of the cell surface detail (box) showed many viral particles (red) colocalized with microtubule (green) near the cell membrane. And GCRV is not located in dot-like actin filaments. C Snapshots of a GCRV particle (white arrow) transporting along microtubules and being internalized into the cytoplasm. A movie depicting this process is also available (see Supplementary Movie S3). CIK cells were transfected with pGFP-MAP4 prior to incubation with QD-bio-GCRVs. Live cell images were captured using time-lapse confocal fluorescence microscopy after incubation with QD-bio-GCRVs for 30 min at 28 °C. The time that each snapshot was taken is indicated in white at the top of each frame.
Discussion
The efficient entry of viruses into host cells requires cellular endosome systems (Forzan et al. 2007; Huang et al.2011; Mainou and Dermody 2012). Previous studies on reovirus cell entry indicated that virion uncoating by endosomal proteases is an essential step of the viral life cycle, and exposure of virions in endocytic vacuoles leads to formation of ISVPs (Silverstein et al. 1972; Sturzenbecker et al. 1987), which showed the presence of both virions and ISVPs within endocytic vesicles at early times of infection (Maginnis et al. 2008; Schulz et al. 2012). The endosome compartment, which could provide acidic compartments, usually plays a vital role in virus entry into host cells. Accumulating evidences show that most enveloped and non-enveloped viruses depend on a low-pH step to trigger infection either by viral envelope fusion or virion uncoating within the endosome membrane via programmed proteases treatment and then release their genome into the cytosol (Sturzenbecker et al.1987; Forzan et al. 2007; Mainou and Dermody 2012).
In this study, using QD-labeled GCRV and combing biochemical assays, we first identified that weak base NH4Cl can strongly inhibit GCRV entry and infection. And we further confirmed that GCRV infection and viral protein synthesis were significantly inhibited by pretreating host cells with endosome acidification inhibitors CQ and Bafi A1. Given that the Rab GTPases coordinate vesicular transport and localize to specific organelles, it can serve as molecular markers to study endosome trafficking (Stenmark 2009). To clarify the above results, we also performed colocalization assays by transfection Rab5 and Rab7 into host cells and combined direct TEM observation. Confocal images indicated that GCRV particles colocalized with Rab5, Rab7 and lysosomes in host cells. But the overlapping in Rab5-marked early and Rab7-marked late endosomes is less than that of lysosomes. Since the spectral overlapping colocalization of fluorescently labeled GCRVs with tested endosome markers was recorded only at one indicated time point, this result is not surprising. Further ultrastructural examination validated that viral particle was found in late endosomes. The finding that QD-bio-GCRVs colocalized to early, late endosomes and lysosomes during entry into host cells is consistent with recent report that reovirus virions need to traffic to late endosomes and some particles are eventually delivered to lysosomes for proteolytic disassembly (Mainou and Dermody 2011, 2012; Patel et al. 2016). This result is also concordant with the reports that both clathrin- and caveolar-mediated pathways can traffic cellular cargoes to lysosomes (Doherty and McMahon 2009; Lopez and Arias 2010). These imaging results, combined with the data from our endosome inhibitor studies, confirm an important role of endosome in the early phases of GCRV infection. So we propose that GCRV enters host cells in a pH-dependent manner.
Following internalization, viruses encounter another issue of how to reach the replication site to initiate efficient infection. As such, viruses therefore need to use host cellular transport system for their trafficking (Doherty and McMahon 2009; Nicola et al. 2013). Studies revealed that many viruses delivered to endosomal compartments and transported to the cytosol or nucleus along actin filaments/microtubules during viral infection (Mabit et al. 2002; McDonald et al.2002). Our data indicated that no inhibitory effects were observed with Cyto D treated cells in viral internalization, while the inhibitor Noco significantly inhibited GCRV entry and viral structural protein synthesis during infection. Further confocal images showed that fluorescently labeled GCRVs strongly colocalized with microtubules in cells. Moreover, we observed that GCRV colocalized with and trafficked along microtubules by dynamic live-cell imaging and single-virus tracking, suggesting that GCRV involved in microtubule-based transport in infected cells. Similarly, the cytoplasmic movement of influenza virus has also been shown to rely on microtubules during different stages of infection (Liu et al. 2011). Orthoreoviruses including avian reovirus that cause infected cells to form syncytia and enters cells via caveolin-1, and unfusogenic MRVs also use microtubules trafficking route to enter cells (Salsman et al. 2005; Liu et al. 2008; Huang et al. 2011). This finding suggests that intracellular transportation of endocytic uptake via microtubules is a conserved cell entry mechanism used by different genera of reoviruses. The detailed molecular mechanism of how aquareovirus engages internalization and endosomal transport and interacts with host factors needs further systemic investigation.
Based on the results presented in this study and combined our previous finding (Zhang et al. 2018), we proposed that caveolae/raft-mediated with low pH and microtubule-dependent endocytic uptake is one of the productive entry pathways for GCRV infection. As schematically illustrated in Fig. 7, GCRV is internalized by endocytosis involving caveolae/lipid raft-mediated endocytic route and microtubules dynamics in a low pH manner. To our knowledge, this is the first time to disclose that endosome systems and microtubule dependent endocytosis is an efficient infection route for GCRV entry. The evidences provided in this study broaden our understanding of the pathways required for productive GCRV infection and may foster development of broadly active antivirals that inhibit key steps in the entry process of aquareovirus.
Fig. 7.
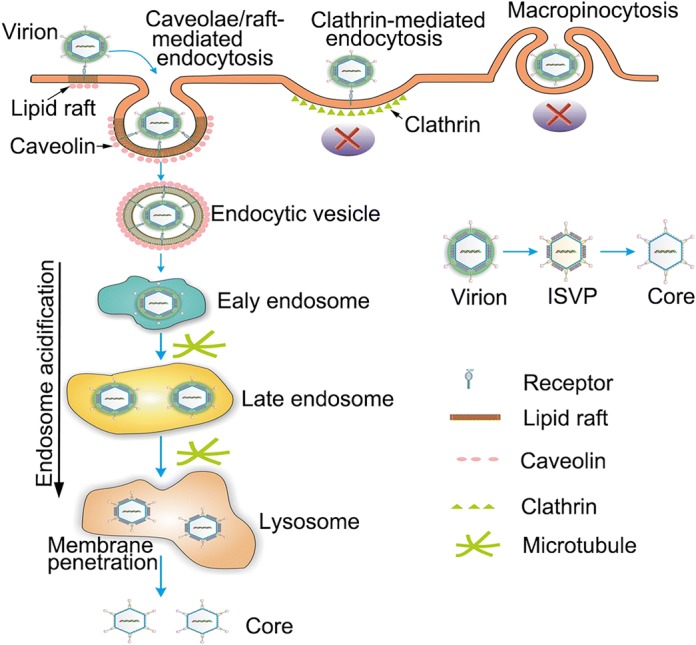
Model for GCRV productive entry into CIK cells. Following attachment to CIK cell surface, GCRV particle is endocytosed in a caveolae/raft-mediated and microtubule dependent manner to form endocytic vesicle, then transported into early, late endosomes and lysosome compartment for undergoing stepwise disassembly, and finally delivered the transcriptionally active core into the cytoplasm via outer capsid protein mediated membrane penetration.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Prof. Hanzhong Wang for providing plasmids encoding Rab5, Rab7, actin and MAP4; Ding Gao, Anna Du and Pei Zhang for ultrathin section preparation and EM observation. We also thank the support from “Center for Instrumental Analysis and Metrology, the Core Facility and Technical Support, Wuhan Institute of Virology”. This work is supported in part by grants from the National Natural Science Foundation of China (31672693, 31972838 and 31400139, 31372565).
Author Contributions
QF designed the experiments. FZ, GH, QC, ZR and QF carried out the experiments and analyzed the data. FZ and QF wrote the paper. All authors read and approved the final manuscript.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Animal and Human Rights Statement
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- Chandran K, Walker SB, Chen Y, Contreras CM, Schiff LA, Baker TS, Nibert ML. In vitro recoating of reovirus cores with baculovirus-expressed outer-capsid proteins μ1 and ς3. J Virol. 1999;73:3941–3950. doi: 10.1128/JVI.73.5.3941-3950.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Farsetta DL, Nibert ML. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein μ1 mediates membrane disruption. J Virol. 2002;76:9920–9933. doi: 10.1128/JVI.76.19.9920-9933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Guo H, Zhang F, Fang Q. N-terminal myristoylated VP5 is required for penetrating cell membrane and promoting infectivity in aquareoviruses. Virol Sin. 2018;33:287–290. doi: 10.1007/s12250-018-0036-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Eilers U, Klumperman J, Hauri H-P. Nocodazole, a microtubule-active drug, interferes with apical protein delivery in cultured intestinal epithelial cells (Caco-2) J Cell Biol. 1989;108:13–22. doi: 10.1083/jcb.108.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Shao L, Fang Q. Characterization of the nonstructural protein NS80 of grass carp reovirus. Arch Virol. 2010;155:1755–1763. doi: 10.1007/s00705-010-0753-6. [DOI] [PubMed] [Google Scholar]
- Fang Q, Ke L, Cai Y. Growth characterization and high titre culture of GCHV. Virol Sin. 1989;4:315–319. [Google Scholar]
- Fang Q, Shah S, Liang Y, Zhou H. 3D reconstruction and capsid protein characterization of grass carp reovirus. Sci China C Life Sci. 2005;48:593–600. doi: 10.1360/062004-105. [DOI] [PubMed] [Google Scholar]
- Fang Q, Seng E, Ding Q, Zhang L. Characterization of infectious particles of grass carp reovirus by treatment with proteases. Arch Virol. 2008;153:675–682. doi: 10.1007/s00705-008-0048-3. [DOI] [PubMed] [Google Scholar]
- Forzan M, Marsh M, Roy P. Bluetongue virus entry into cells. J Virol. 2007;81:4819–4827. doi: 10.1128/JVI.02284-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlong DB, Nibert M, Fields B. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol. 1988;62:246–256. doi: 10.1128/JVI.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Sun X, Yan L, Shao L, Fang Q. The NS16 protein of aquareovirus-C is a fusion-associated small transmembrane (FAST) protein, and its activity can be enhanced by the nonstructural protein NS26. Virus Res. 2013;171:129–137. doi: 10.1016/j.virusres.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Huang WR, Wang YC, Chi PI, Wang L, Wang CY, Lin CH, Liu HJ. Cell entry of avian reovirus follows a caveolin-1-mediated and dynamin-2-dependent endocytic pathway that requires activation of p38 mitogen-activated protein kinase (MAPK) and Src signaling pathways as well as microtubules and small GTPase Rab5 protein. J Biol Chem. 2011;286:30780–30794. doi: 10.1074/jbc.M111.257154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaafar FM, Goodwin AE, Belhouchet M, Merry G, Fang Q, Cantaloube J-F, Biagini P, de Micco P, Mertens PP, Attoui H. Complete characterisation of the American grass carp reovirus genome (genus Aquareovirus: family Reoviridae) reveals an evolutionary link between aquareoviruses and coltiviruses. Virology. 2008;373:310–321. doi: 10.1016/j.virol.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Ke L, Fang Q, Cai Y. Characteristics of a novel isolate of grass carp haemorrhagic virus. Acta Hydrobiol Sin. 1990;14:153–159. [Google Scholar]
- King AM, Lefkowitz E, Adams MJ, Carstens EB (2011) Virus taxonomy: 9th report of the international committee on taxonomy of viruses vol 9, Elsevier
- Li X, Fang Q. High-resolution 3D structures reveal the biological functions of reoviruses. Virol Sin. 2013;28:318–325. doi: 10.1007/s12250-013-3341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H-J, Lin P-Y, Wang L-R, Hsu H-Y, Liao M-H, Shih W-L. Activation of small GTPases RhoA and Rac1 is required for avian reovirus p10-induced syncytium formation. Mol Cell. 2008;26:396–403. [PubMed] [Google Scholar]
- Liu S-L, Zhang Z-L, Tian Z-Q, Zhao H-S, Liu H, Sun E-Z, Xiao GF, Zhang W, Wang H-Z, Pang D-W. Effectively and efficiently dissecting the infection of influenza virus by quantum-dot-based single-particle tracking. ACS Nano. 2011;6:141–150. doi: 10.1021/nn2031353. [DOI] [PubMed] [Google Scholar]
- Lopez S, Arias C. How viruses hijack endocytic machinery. Nat Educ. 2010;3:16–23. [Google Scholar]
- Mabit H, Nakano MY, Prank U, Saam B, Döhner K, Sodeik B, Greber UF. Intact microtubules support adenovirus and herpes simplex virus infections. J Virol. 2002;76:9962–9971. doi: 10.1128/JVI.76.19.9962-9971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maginnis MS, Mainou BA, Derdowski A, Johnson EM, Zent R, Dermody TS. NPXY motifs in the β1 integrin cytoplasmic tail are required for functional reovirus entry. J Virol. 2008;82:3181–3191. doi: 10.1128/JVI.01612-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainou BA, Dermody TS. SRC kinase mediates productive endocytic sorting of reovirus during cell entry. J Virol. 2011;85:3203–3213. doi: 10.1128/JVI.02056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainou BA, Dermody TS. Transport to late endosomes is required for efficient reovirus infection. J Virol. 2012;86:8346–8358. doi: 10.1128/JVI.00100-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainou BA, Zamora PF, Ashbrook AW, Dorset DC, Kim KS, Dermody TS. Reovirus cell entry requires functional microtubules. MBio. 2013;4:e00405–e00413. doi: 10.1128/mBio.00405-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater. 2005;4:435. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- Nicola AV, Aguilar HC, Mercer J, Ryckman B, Wiethoff CM. Virus entry by endocytosis. Adv Virol. 2013 doi: 10.1155/2013/469538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Mohl B-P, Roy P. Entry of bluetongue virus capsid requires the late endosome-specific lipid lysobisphosphatidic acid. J Biol Chem. 2016;291:12408–12419. doi: 10.1074/jbc.M115.700856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangel AA, Rockemann DD, Hetrick FM, Samal SK. Identification of grass carp haemorrhage virus as a new genogroup of aquareovirus. J General Virol. 1999;80:2399–2402. doi: 10.1099/0022-1317-80-9-2399. [DOI] [PubMed] [Google Scholar]
- Rao Y, Su J. Insights into the antiviral immunity against grass carp (Ctenopharyngodon idella) reovirus (GCRV) in grass carp. J Immunol Res. 2015 doi: 10.1155/2015/670437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salsman J, Top D, Boutilier J, Duncan R. Extensive syncytium formation mediated by the reovirus FAST proteins triggers apoptosis-induced membrane instability. J Virol. 2005;79:8090–8100. doi: 10.1128/JVI.79.13.8090-8100.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez EG, Quintas A, Pérez-Núñez D, Nogal M, Barroso S, Carrascosa ÁL, Revilla Y. African swine fever virus uses macropinocytosis to enter host cells. PLoS Pathogens. 2012;8:e1002754. doi: 10.1371/journal.ppat.1002754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz WL, Haj AK, Schiff LA. Reovirus uses multiple endocytic pathways for cell entry. J Virol. 2012;86:12665–12675. doi: 10.1128/JVI.01861-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Guo H, Yan L-M, Liu H, Fang Q. Aquareovirus NS80 recruits viral proteins to its inclusions, and its C-terminal domain is the primary driving force for viral inclusion formation. PLoS ONE. 2013;8:e55334. doi: 10.1371/journal.pone.0055334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein SC, Astell C, Levin DH, Schonberg M, Acs G. The mechanisms of reovirus uncoating and gene activation in vivo. Virology. 1972;47:797–806. doi: 10.1016/0042-6822(72)90571-5. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Sturzenbecker L, Nibert M, Furlong D, Fields B. Intracellular digestion of reovirus particles requires a low pH and is an essential step in the viral infectious cycle. J Virol. 1987;61:2351–2361. doi: 10.1128/JVI.61.8.2351-2361.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen M, Greber UF. Uncoating of non-enveloped viruses. Curr Opin Virol. 2013;3:27–33. doi: 10.1016/j.coviro.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Wang Q, Zeng W, Liu C, Zhang C, Wang Y, Shi C, Wu S. Complete genome sequence of a reovirus isolated from grass carp, indicating different genotypes of GCRV in China. J Virol. 2012;86:12466. doi: 10.1128/JVI.02333-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Zhang J, Guo H, Yan S, Chen Q, Zhang F, Fang Q. Aquareovirus NS80 initiates efficient viral replication by retaining core proteins within replication-associated viral inclusion bodies. PLoS ONE. 2015;10:e0126127. doi: 10.1371/journal.pone.0126127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Zhang J, Guo H, Yan L, Chen Q, Zhang F, Fang Q. VP5 autocleavage is required for efficient infection by in vitro-recoated aquareovirus particles. J Gen Virol. 2015;96:1795–1800. doi: 10.1099/vir.0.000116. [DOI] [PubMed] [Google Scholar]
- Zhang F, Zheng Z, Liu S-L, Lu W, Zhang Z, Zhang C, Zhou P, Zhang Y, Long G, He Z. Self-biotinylation and site-specific double labeling of baculovirus using quantum dots for single-virus in situ tracking. Biomaterials. 2013;34:7506–7518. doi: 10.1016/j.biomaterials.2013.06.030. [DOI] [PubMed] [Google Scholar]
- Zhang F, Yan S, Guo H, Chen Q, Fang Q. Characterization of viral entry and infection of quantum dot-labeled grass carp reovirus. Virol Sin. 2017;32:163–166. doi: 10.1007/s12250-016-3903-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Guo H, Zhang J, Chen Q, Fang Q. Identification of the caveolae/raft-mediated endocytosis as the primary entry pathway for aquareovirus. Virology. 2018;513:195–207. doi: 10.1016/j.virol.2017.09.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.