SUMMARY
Although intramuscular (i.m.) administration is the most commonly used route for licensed vaccines, subcutaneous (s.c.) delivery is being explored for several new vaccines under development. Here, we use rhesus macaques, physiologically relevant to humans, to identify the anatomical compartments and early immune processes engaged in the response to immunization via the two routes. Administration of fluorescently labeled HIV-1 envelope glycoprotein trimers displayed on liposomes enables visualization of targeted cells and tissues. Both s.c. and i.m. routes induce efficient immune cell infiltration, activation, and antigen uptake, functions that are tightly restricted to the skin and muscle, respectively. Antigen is also transported to different lymph nodes depending on route. However, these early differences do not translate into significant differences in the magnitude or quality of antigen-specific cellular and humoral responses over time. Thus, although some distinct immunological differences are noted, the choice of route may instead be motivated by clinical practicality.
Graphical Abstract
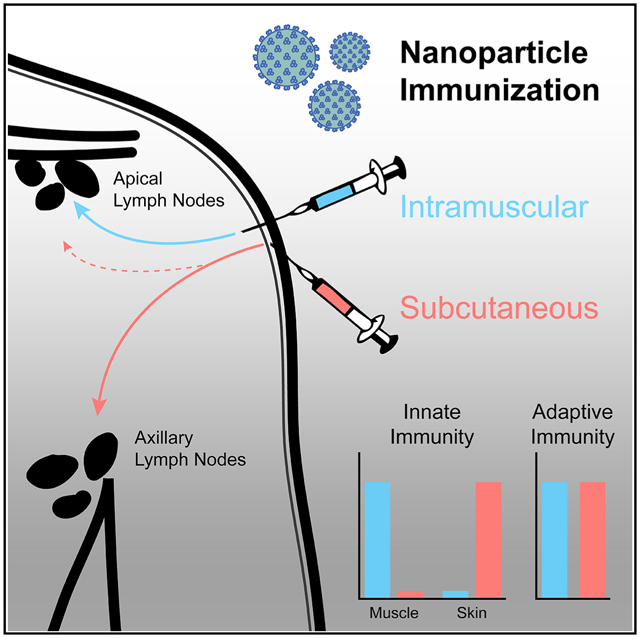
In Brief
Route of immunization, especially intramuscular versus subcutaneous administration, is often debated. Ols et al. use a rhesus macaque model to determine the tissues targeted by a nanoparticle vaccine administered by either route. The authors demonstrate that tissue dissemination is route dependent, but innate and adaptive immune responses develop comparably.
INTRODUCTION
The majority of licensed vaccines are administered by intramuscular (i.m.) injection, but some are approved for subcutaneous (s.c.) or intradermal (i.d.) use. Intramuscular administration is often preferred because it is easy to perform and generally well tolerated, with a low risk for adverse reactions at the site of injection. However, vaccine delivery to the skin as a highly immunocompetent site compared with the muscle has long been considered a strategy to amplify vaccine responses. Administration of the yellow fever virus vaccine or influenza vaccines into the skin compared with i.m. injection results in enhanced responses in healthy individuals and, importantly, also in non- or low responders (Roukens et al., 2012). Intradermal immunization was also shown to allow antigen dose reduction without loss of efficacy. We recently found that i.d. administration of an mRNA vaccine resulted in more efficient activation of antigen-presenting cells (APCs) at the site of injection compared with i.m. vaccination and was accompanied by transiently higher levels of vaccine-specific T cell responses and antibody (Ab) titers (Liang et al., 2017a; Lindgren et al., 2017).
Similar to i.d. delivery, s.c. immunization is thought to result in better targeting of immune cells and improved vaccine responses compared with i.m. administration. However, several clinical trials have revealed no significant differences between s.c. and i.m. vaccination in generating humoral immune responses to hepatitis B (Wahl and Hermodsson, 1987); hepatitis A (Fisch et al., 1996); herpes zoster virus (Diez-Domingo et al., 2015); influenza (Cook et al., 2006); diphtheria toxin (Mark et al., 1999); measles, mumps, rubella, and varicella (Gillet et al., 2009; Knuf et al., 2010); and tick-borne encephalitis virus (Hopf et al., 2016). These data, in combination with adverse events such as reactogenicity at site of injection, which were more frequent in s.c. immunized individuals in a majority of clinical trials, have motivated i.m. vaccination as the desired route of delivery.
The development of new vaccine platforms on the basis of nanoparticle structures over the past decade, such as polymer particles, liposomes, and self-assembling protein nanoparticles, has reignited the debate on route of administration. Many of these platforms have been designed to be given in the skin to better target APCs and efficiently drain to lymph nodes (LNs) (Moyer et al., 2016; Trevaskis et al., 2015). Importantly, testing of new vaccines typically begins in small animal models, usually in mice, which have different anatomy from humans and in which i.m. administration cannot be comprehensively evaluated. Therefore, studies of different routes of vaccine administration in small animals are insufficient to guide choices regarding immunization route in humans. A better understanding of immunological mechanisms involved in immunization of the skin versus the muscle in primates such as rhesus macaques (RMs) is therefore critical. RMs resemble humans to a greater degree than rodents in their anatomy, LN drainage, immune cell subsets, and immune receptor expression (Thompson and Loré, 2017).
We have previously used RMs to explore vaccine trafficking after i.m. immunization using HIV-1 envelope glycoprotein (Env) as the model antigen (Liang et al., 2017b). In the present study, we extend this to define and compare the initial events leading to vaccine responses after s.c. versus i.m. administration of another Env-based vaccine on the basis of well-ordered HIV-1 Env trimers covalently coupled to synthetic liposomes (Ingale et al., 2016; Martinez-Murillo et al., 2017; Pauthner et al., 2017) administered with or without Matrix-M adjuvant. To track the uptake and dissemination of the vaccine after injection, we fluorescently labeled the Env trimers and the liposomes with separate fluorescent dyes. We analyzed the site of injection and identified the LNs in which adaptive immune responses were primed following immunization.
RESULTS
Subcutaneous and Intramuscular Administration Target the Skin and Muscle, Respectively
To perform a systematic comparison of early immune functions after s.c. or i.m. administration, we fluorescently labeled HIV-1 Env clade C 1086 native flexibly linked (NFL) trimers (Guenaga et al., 2017) with Alexa Fluor 680 and covalently coupled them to synthetic liposomes labeled with TopFluor Cholesterol to enable in vivo tracking (Figure S1A). The trimer antigenicity and stability were confirmed to be intact after labeling (Figures S1B–S1D). Naive RMs (n = 3) received injections at six different sites simultaneously so that s.c. and i.m. administration of antigen, or PBS as control, could be compared in the same animal (Figures S1E and S1F). This also allowed multiple data collection while limiting the number of animals used for experimentation. We analyzed several tissues after 24 h because we have previously observed high levels of antigen uptake and local innate immune activity at this time point (Liang et al., 2017a, 2017b). Env:liposome uptake was detected in the muscle after i.m. injection, but not in the skin over the muscle injection site (Figures 1A and 1B). In contrast, s.c. immunization showed only a few Env:-liposome+ cells in the muscle but large numbers in the skin, indicating targeting of different tissue compartments depending on the route. The most abundant Env+ cells at the site of injection were neutrophils and monocytes with both routes (Figure 1C). However, s.c. administration targeted a more diverse set of cells than i.m., likely since the skin contains multiple populations of resident APCs. With both routes, we detected cells that were only liposome+ and not Env+, which could be a consequence of labeling instability and accumulation of the liposome dye in the cells, as this could be observed in vitro (Figure S1G).
Figure 1. Vaccine Uptake Is Restricted to the Site of Injection and Targets Distinct Anatomical LNs.
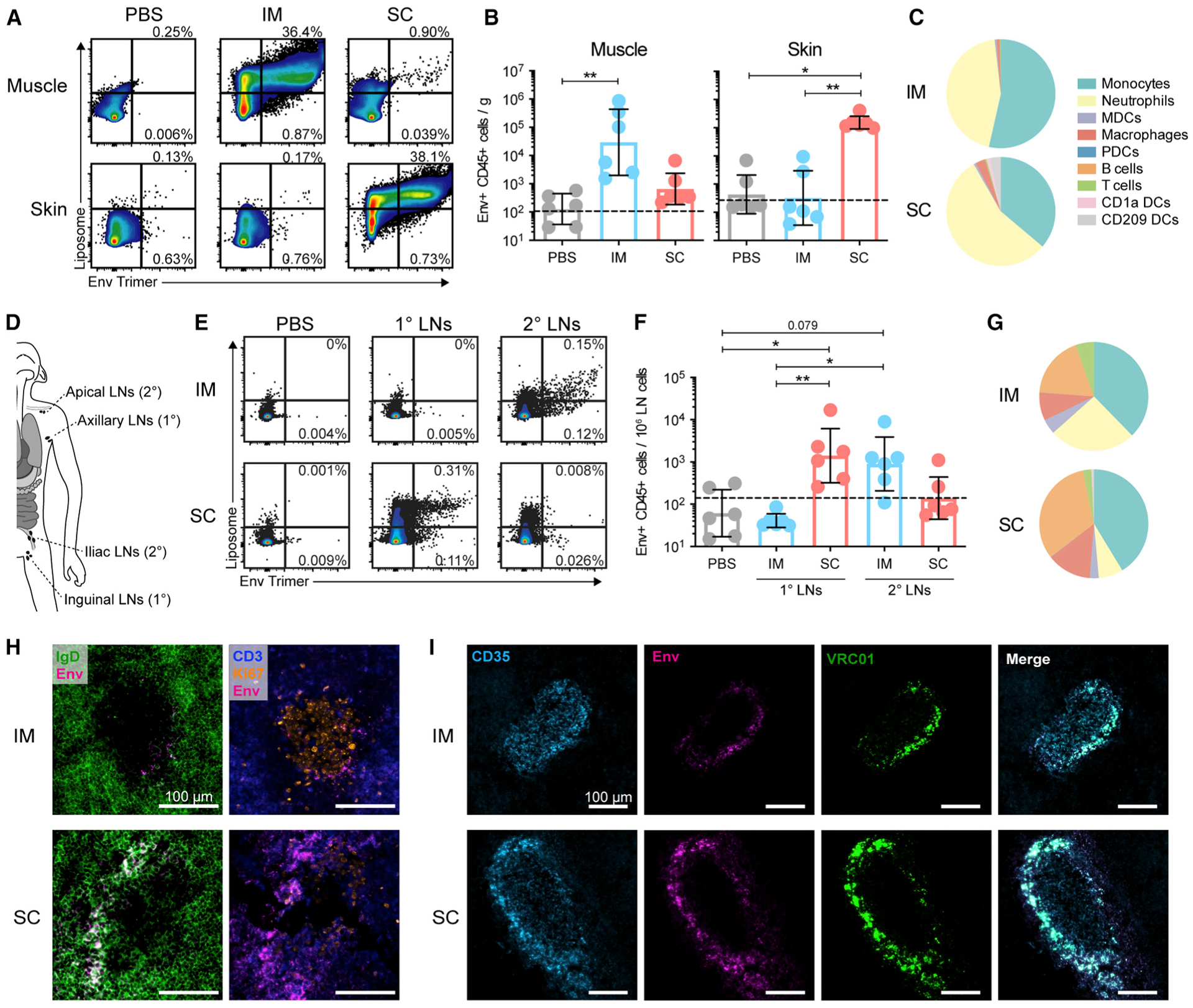
(A) Flow cytometry gating of Env:liposome signals at the site of injection, gated on CD45+ cells. (B) Quantification of Env+ CD45+ cells per gram of muscle or skin tissue. (C) Proportions of Env+ CD45+ cell subsets in the muscle and skin after i.m. and s.c. injection, respectively. (D) Schematic of LN clusters analyzed and their classification as 1° (axillary/inguinal) or 2° (apical/iliac) LNs on the basis of proximity to the injection site (deltoid/quad). (E) Flow cytometry gating of Env:liposome signals in LNs, presented as in (A). (F) Quantification of Env+ CD45+ cells in LNs. (G) Proportions of Env+ CD45+ cell subsets in the draining LNs (sum of 1° and 2° LNs). (H) Representative images of Env localization in LNs stained for CD3 (blue), IgD (green), Env-AF680 (magenta), and Ki67 (orange). (I) Representative images of Env signal verification with VRC01 antibody. LNs stained for CD35 (cyan), Env-AF680 (magenta), and VRC01 (green).
In (A)–(G), geometric mean and gSD is displayed. Data points represent individual tissue samples. n = 6 per group. Dashed line represents the limit of detection. See methods for calculation. *p < 0.05 and **p < 0.01. In (H) and (I), representative images of n = 3 LNs per group are shown. Image brightness was increased to allow visualization. See also Figures S1 and S2.
In line with the Env uptake in specific tissues, we observed robust immune cell infiltration after Env:liposome administration compared with PBS (Figures S2A–S2C). Again, s.c. immunization induced cell infiltration restricted to the skin, while i.m. administration exclusively showed infiltration to the muscle. The cell recruitment was induced mainly by the adjuvant Matrix-M, although the liposomes alone also induced some cell infiltration (Figures S2D–S2G).
Intramuscular and Subcutaneous Administration Drain to Anatomically Distinct LNs
The transport of vaccine antigen to the local LNs is crucial for priming of T and B cell responses (Liang et al., 2017b). We and others have shown, using both flow cytometry and positron emission tomography (PET)/computed tomography (CT), that vaccine transport after i.m. injection is restricted to the local LNs and is not disseminated systemically (Liang et al., 2017a, 2017b; Lindsay et al., 2019). To identify the LNs targeted by immunization, LNs were classified as the primary (1°; axillary or inguinal) or secondary (2°; apical or iliac) draining LNs on the basis of their proximity to the injection site (Figure 1D). Subcutaneous administration was found to predominantly target the 1° LNs, with lower Env:liposome signal observed in the 2° LNs. In contrast, i.m. immunization almost exclusively drained to the 2° LNs (Figures 1E, 1F, and S2H). This is likely the result of how the lymphatics drain the skin compared with the muscle, with deeper injections targeting more internally localized LNs. Env+ neutrophils and monocytes, but also B cells, were the most abundant cells in the draining LNs (Figures 1G and S2I). In fact, assessment of Env distribution by imaging of LN cryosections demonstrated that Env localized primarily within B cell follicles and was associated with follicular dendritic cells (FDCs) (Figure 1H). This was confirmed by amplifying the endogenous Env signal with an anti-Env Ab (VRC01) (Figures 1I and S2J). As found by flow cytometry, Env+ neutrophils were also detected in LN cryosections, but mainly in the para-cortex of the LNs (Figure S2K). Collectively, these data may indicate that Env+ neutrophils and APCs migrate from the site of injection, while Env+ B cells capture antigen that disseminated into the LNs.
Comparable Adaptive Immune Responses to HIV-1 Env after Intramuscular and Subcutaneous Immunization
To assess if the distinct tissue targeting between s.c. and i.m. administration would influence adaptive Env-specific responses, we immunized naive RMs four times with unlabeled Env:liposomes formulated with Matrix-M adjuvant (n = 5 per group) (Figures 2A and S1B–S1D). Env-specific IgG titers in plasma reached peak levels after the third immunization, and no significant differences were observed between the groups (Figure 2B). Neutralization of the tier 1 pseudoviruses was also comparable between the groups (Figure 2C), while neutralization of the autologous tier 2 1086 pseudovirus was detected in only some i.m. immunized animals, although this finding was not statistically significant. Furthermore, no significant differences were observed in other measured adaptive immune parameters, including IgG avidity, IgA titers, memory B cells in blood, plasma cells in the bone marrow, and CD4 T cells in circulation (Figures 2D–2H and S2L). These data demonstrate that the differences in tissue compartments targeted by i.m. and s.c. administration did not translate into significant differences in the vaccine-specific adaptive immune responses.
Figure 2. Adaptive Immune Responses to HIV-1 Env Are Comparable.
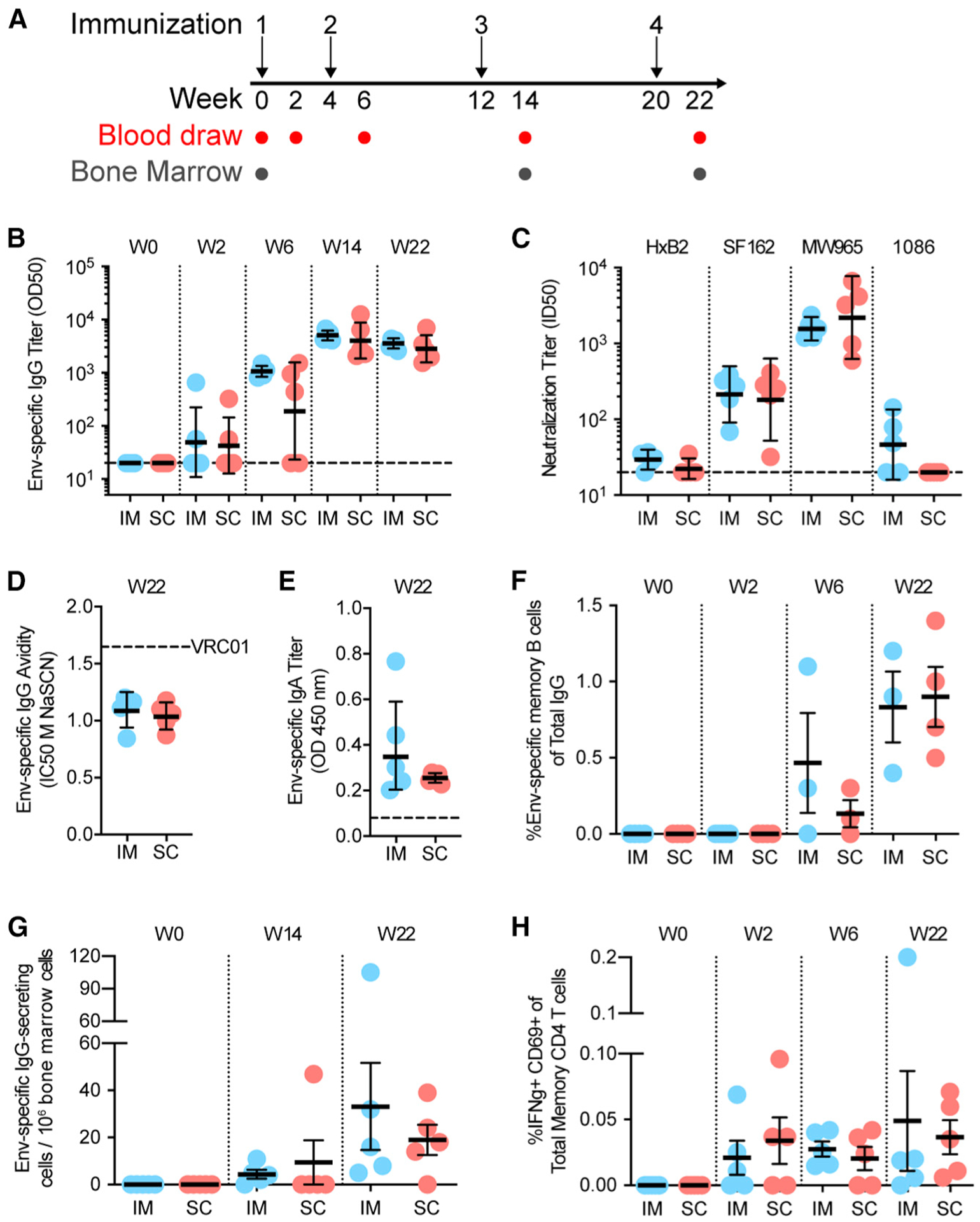
(A) Schematic of i.m. and s.c. immunization and sampling schedule. (B) Anti-1086 Env IgG OD50 binding titers measured using ELISA. (C) Tier 1 (H×B2, SF162, MW965) and autologous tier 2 (1086) neutralization at week 22. (D) Env-specific IgG avidity as measured using a chaotropic wash ELISA using NaSCN. Mean of three independent experiments is displayed. (E) Env-specific IgA titers in plasma measured using ELISA. Max OD of 20-fold plasma dilution is displayed. (F) Env-specific memory B cell responses in blood measured using ELISpot. (G) Env-specific plasma cells in bone marrow measured using ELISpot. (H) Env-specific CD4+ memory T cell responses in blood measured by intracellular cytokine recall assay.
In (B)–(E), geometric mean and gSD are displayed. In (F)–(H), mean and SEM are displayed. In (B)–(H), data points represent individual animals. n= 5 per group. In (F), n = 3–5 per group. In (B)–(H), no statistically significant differences. See also Figures S1 and S2.
Pre-existing Immunity Alters Vaccine Trafficking Dynamics
Because the vast majority of vaccines are given as a regimen with multiple immunizations, we assessed if the pre-existing immunity evident in a boost would alter the pattern observed with the different routes. By administering labeled Env:liposomes to animals with high levels of anti-Env Ab titers (here referred to as high-titer animals; n = 3) (Figure 3A), we compared the uptake and distribution of antigen to the naive animals in Figure 1. The high-titer animals showed a trend toward enhanced antigen uptake compared with naive animals, which was most prominent after i.m. administration (Figure 3B). The enhanced uptake may be attributed to immune complex formation, as in vitro exposure of primary cells to Env in the presence of plasma from the high-titer animals increased uptake (Figure 3C). The representation of Env+ cell subsets was to a large degree similar in naive versus high-titer animals (Figures S3A–S3F).
Figure 3. Pre-existing Immunity Alters Vaccine Trafficking Dynamics.
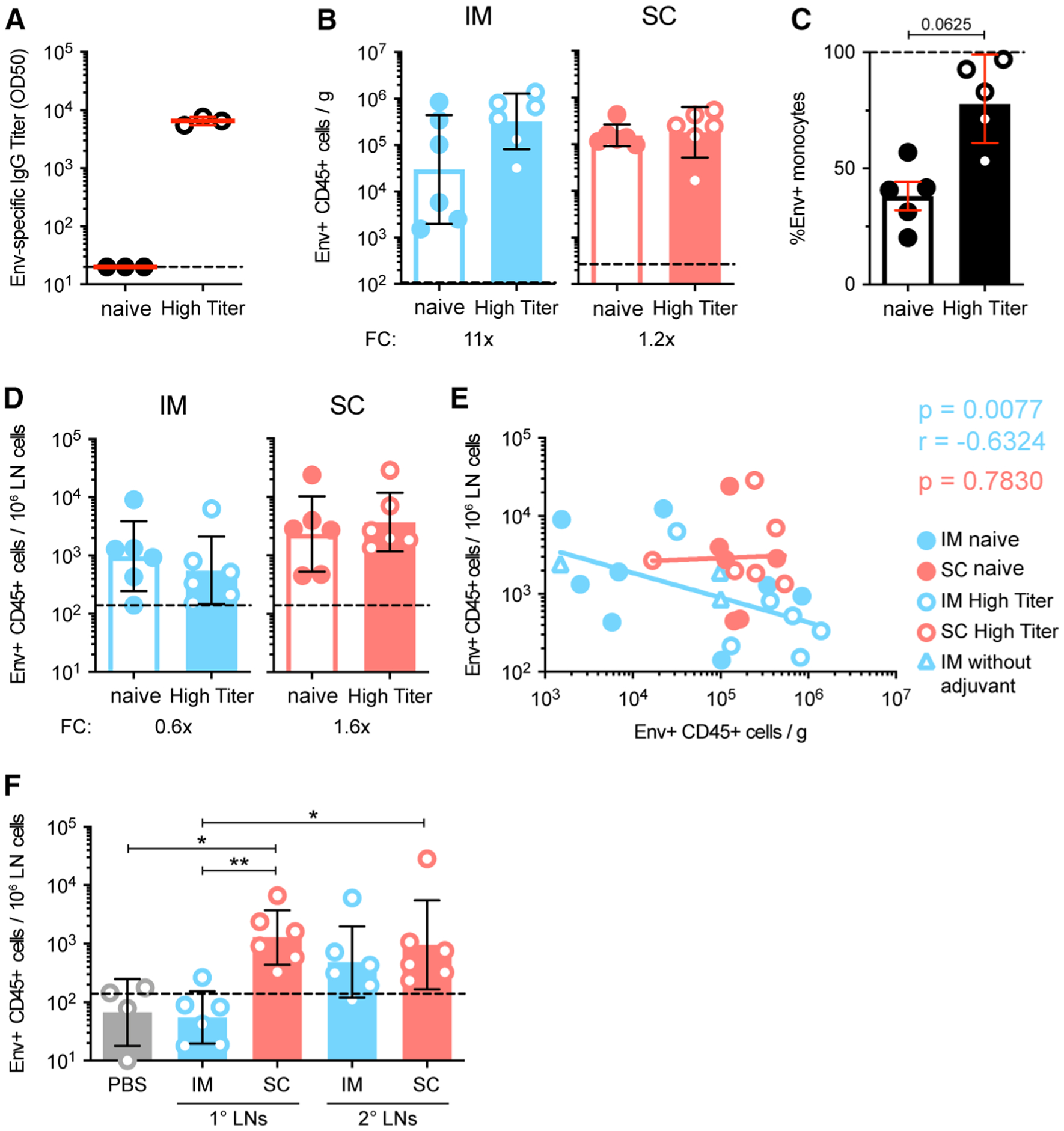
(A) Anti-1086 Env IgG OD50 binding titers on day of immunization in naive and high-titer animals measured using ELISA. (B) Quantification of Env+ CD45+ cells per gram of muscle or skin tissue of naive and high-titer animals. (C) Env:liposome uptake by isolated human monocytes in vitro with plasma from naive or high-titer animals. Two independent experiments; n = 5 human donors. (D) Quantification of Env+ CD45+ cells in the draining LNs of naive and high-titer animals. (E) Spearman correlation of Env+ CD45+ cells at the site of injection and in the draining LNs. (F) Quantification of Env+ CD45+ cells in 1° and 2° LNs of high-titer animals.
In (A)–(F), geometric mean and gSD are displayed. FC, fold change. Naive animal data are the same as displayed in Figure 1. Data points represent individual tissue samples. n = 6 per group. Dashed line represents the limit of detection. See methods for calculation. *p < 0.05 and **p < 0.01. See also Figure S3.
In the LNs, there was a trend toward decreased Env+ cell numbers for i.m. administration compared with naive animals (Figure 3D). In fact, when taking all data into account, there was a significant negative correlation between numbers of Env+ cells in the LNs and Env+ cells at the site of injection after i.m. administration, whereas no such pattern was found with s.c. (i.m., p = 0.0077, r = −0.6324; s.c., p = 0.7830; Figure 3E). Nonetheless, the preferential transport of Env to different LNs depending on the administration route remained in the high-titer animals (Figure 3F). Also, little systemic dissemination of the vaccine was observed in both naive and high-titer animals (Figure S3G).
Priming of T and B Cell Responses Occurs in the Local Draining LNs
With the consistent observation of differential targeting of LNs between the s.c. and i.m. routes, we assessed if the priming of adaptive responses also occurred in different LNs. We found that Env+ APCs, regardless of route, showed a higher cell differentiation profile than their counterparts with no Env signal (Figures S4A and S4B), and this activation was independent of the adjuvant (Figure S4C). This indicates that uptake of the Env:liposome complex is associated with cell activation. In line with this, we found that Env-specific T cells developed in the vaccine-draining LNs and not in the non-draining mesenteric LNs, regardless of i.m. or s.c. immunization (Figures 4A and 4B). Although the high-titer animals showed overall higher responses, both the high-titer and naive animals showed substantial proliferation of Env-specific T cells in their respective LNs. This suggests that antigen presentation and priming/reactivation of T cells occurs rapidly after immunization and exclusively in these LNs.
Figure 4. Priming of Adaptive Responses is Restricted to the Local Vaccine-Draining LNs.
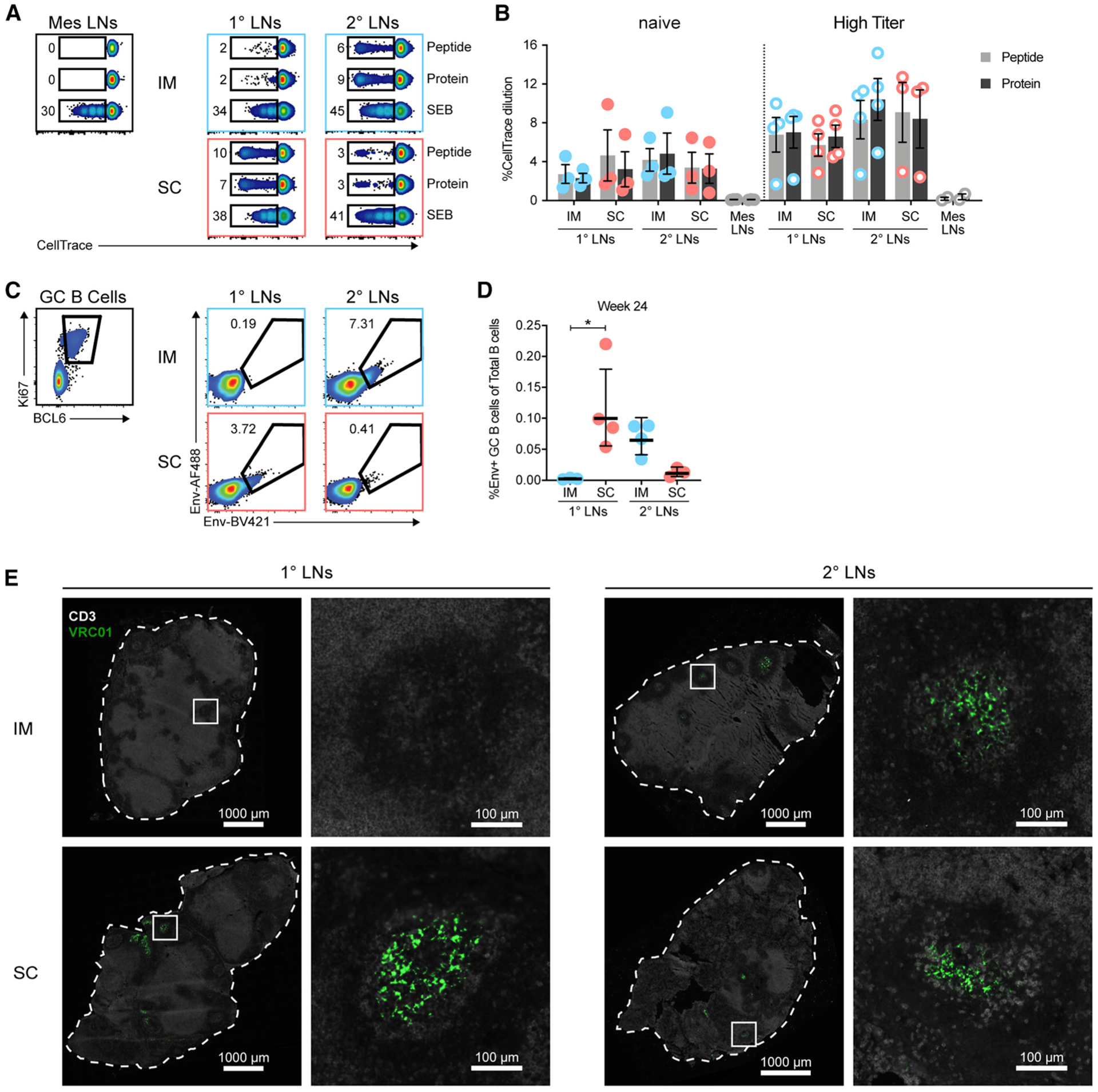
(A and B) T cell proliferation of LN cell suspensions from 24 h after immunization as measured using CellTrace dilution on day 5. (A) Representative flow cytometry plots of proliferating T cells in LNs of naive animals. LN samples are concatenated by condition. (B) Plotted is the percentage of CellTrace dilution in live CD3+ T cells. (C–E) LNs obtained from 30 days post-immunization four (week 24) of RMs from Figure 2. (C) GC B cells identified by expression of Ki67 and BCL6 from CD20+ CD3− cells. Env specificity was interrogated with dual-labeled probes. (D) Plotted is the percentage of Env-specific GC B cells of total CD20+ B cells. (E) Representative images of unlabeled Env localization in LNs stained for CD3 (white) and VRC01 (green). n = 4 LNs per group. Image brightness was increased to allow visualization.
In (B) and (D), mean and SEM are displayed. Data points represent individual LN clusters. n = 3 or 4 LNs per group. *p < 0.05. See also Figure S4.
Similarly, analysis of 1° and 2° LNs obtained 30 days after the fourth immunization with unlabeled Env:liposomes revealed that Env-specific germinal centers were most prominent in the different LNs depending on route (Figures 4C and 4D). Localization of Env in these LNs was again detectable by VRC01 staining within the B cell follicles, most likely retained on FDCs (Figure 4E). These results reaffirm that priming of vaccine-specific adaptive immune responses is restricted to the local draining LNs and that this is determined by the route of vaccine administration. Nevertheless, either set of draining LNs is capable of inducing strong and comparable adaptive T cell and B cell responses to the vaccine antigen.
DISCUSSION
Subcutaneous administration has emerged as the proposed route of administration for several new vaccines under development, with the intention to increase immunogenicity. However, i.m. administration is to date the most commonly used route by far for licensed vaccines, and multiple clinical trials have demonstrated no difference in adaptive immune responses between s.c. and i.m. delivery (reviewed in Zhang et al., 2015). In this study, we aimed to dissect the innate immune events that precede the generation of vaccine-specific responses after s.c. and i.m. administration to improve the understanding of their mechanistic differences. Evaluation of the skin and muscle of the site of injection, as well as the draining LNs, revealed that distinct anatomical compartments were targeted, but with similar levels of vaccine antigen uptake and cell activation with the two administration routes. In addition, no statistically significant differences were observed in the vaccine-specific adaptive immune responses over a 22 week period with four immunizations.
Although s.c. immunization targeted cells of the skin, we speculate that this is not as efficient as with i.d. administration. Antigen dose-sparing effects have only been observed with i.d. immunization in clinical trials (reviewed in Herzog, 2014; Zhang et al., 2015). In mice, cell-associated trafficking by skin-resident dendritic cells (DCs) after i.d. administration was shown to be required for efficient priming of B and T cell responses (Levin et al., 2017), while these cells were dispensable after s.c. administration (Woodruff et al., 2014). This suggests that s.c. administration may involve less cell-mediated transport to the LNs. The larger number of Env+ B cells in the LNs draining s.c. injections compared with the i.m.-draining LNs found in the present study indicates that antigen trafficking with s.c. is very efficient in RMs too. On this note, the rates of lymphatic drainage after i.m. or s.c. injection were shown to be similar in an ovine cannulation model (Neeland et al., 2016).
Differential targeting of LNs by route of administration has been suggested by studies using Evans blue dye administration in RMs (Pauthner et al., 2017). However, Evans blue dye, with its high affinity for serum albumin (Yao et al., 2018), is most likely transported by different mechanisms than foreign vaccine antigens, for which cellular transport may constitute a large part. In addition, conclusions on antigen transport drawn from LN biopsies are dependent on the LNs collected and analyzed. Lymphatic drainage is complex, and there are numerous distinct LN clusters. We chose to collect axillary/inguinal LNs as the 1° draining LNs and apical/iliac LNs as the 2° on the basis of their proximity to the injection site. Similar lymphatic dissemination after i.m. vaccine administration was recently also shown by radio-labeling of a vaccine and whole-body PET/CT detection (Lindsay et al., 2019), a less biased approach although of lower resolution. Our findings highlight the importance of identifying and sampling the correct LNs depending on the route of administration when evaluating immune responses after vaccination.
Immune complexes have been proposed to facilitate antigen uptake and cell activation but also to lead to faster antigen clearance (reviewed in Lu et al., 2018). We hypothesize that the increased uptake we observed in the boost setting is a result of immune complex formation at the site of injection. This effect was most noticeable with i.m. delivery, perhaps because the muscle is highly vascularized and antibodies would have more access. However, it is not clear whether the increased uptake results in more degradation or accumulation of Env at the site of injection and therefore less Env would reach the LNs. Delayed draining kinetics could explain the lower levels of Env+ cells in the LNs, but this would require analyses of additional time points to confirm. Despite this, the elicited adaptive responses are comparable. Further studies are needed to delineate the role of immune complexes and phagocytes in a boost vaccination setting.
In conclusion, the anatomical differences of vaccine antigen delivery between i.m. and s.c. administration shown in this study provide important guidance for sampling and monitoring of immune responses in pre-clinical and clinical vaccine studies. In particular, caution should be taken with analysis of axillary/inguinal LNs after i.m. administration, as the immunological activity may be misleadingly low. We speculate that because the differences between i.m. and s.c. immunization of this type of vaccine candidate formulation were minimal, the choice of route should be motivated by safety, reproducibility, ease of administration, and practicality in clinical settings.
STAR★METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
This study did not generate new unique reagents. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Karin Loré (karin.lore@ki.se).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals and study design
This study was approved by the Local Ethical Committee on Animal Experiments. Thirteen Indian rhesus macaques, ten females and three males, of four to five years of age were housed in the Astrid Fagraeus laboratory at Karolinska Institutet according to the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care. All procedures were performed abiding to the provisions and general guidelines of the Swedish Board of Agriculture.
To follow the development of adaptive immune responses over time after vaccination, ten female macaques were split in two groups of five and administered unlabeled Env:liposomes in Matrix-M adjuvant four times at weeks 0, 4, 12, and 20 by intramuscular or subcutaneous injection. To analyze the early innate immune responses and track the fate of Env after immunization, labeled Env:liposomes in Matrix-M adjuvant were administered to three male macaques for prime/naive analyses and to three female macaques for boost/high titer analyses. To maximize data collection and minimize the use of animals for these tracking experiments, immunizations with labeled Env:liposomes were administered in multiple limbs per animal (Figure S1E). We have previously developed and optimized this model (Liang et al., 2015, 2017a, 2017b) to emphasize the ethical considerations of conducting terminal non-human primate studies. For these experiments, intramuscular administration of Env:liposomes in Matrix-M was performed in one deltoid and one quadricep while subcutaneous administration was performed in the contralateral deltoid and quadricep in the same animal for comparison. Phosphate-buffered saline (PBS) was administered to the calves. This way six data points were collected from each animal. Similarly, to study the contribution of the adjuvant, two female macaques were administered combinations of labeled Env:liposomes, Matrix-M adjuvant, and/or PBS at different sites.
Human blood
The collection and use of human samples were performed in accordance with the Helsinki declaration and approved by the institutional review board of ethics at the Karolinska Institutet, Stockholm, Sweden. Blood was collected from healthy human individuals after informed consent. The age and sex of the donors is unknown as they come from an anonymous sample bank.
METHOD DETAILS
Generation of clade C 1086 NFL trimer and liposomes
The cleavage-independent clade C 1086 NFL trimers were generated as previously described with specific modification (Guenaga et al., 2017). In brief, TD CC+ mutations were inserted into 1086 Env gp140. To covalently conjugate 1086 NFL trimers to liposomes, a free cysteine residue was genetically engineered, following a 16 amino acid liker as described previously (Bale et al., 2017). The 1086 NFL trimers were transiently expressed in 293F cells (Yang et al., 2018). Env proteins were harvested four days post transfection and purified by lectin affinity chromatography (Galanthus nivalis, Vector Labs) followed by size exclusion chromatography (SEC) on a Superdex 200 10/300 GL (GE Healthcare). The trimer peak was subjected to negative selection by the non-neutralizing mAb, F105, to remove disordered trimers. The flow-through from the F105 column, containing the well-ordered trimers, was resolved by a second SEC step.
In brief, the liposomes were comprised of DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine), cholesterol, and PE-MCC (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidomethyl)cyclohexane-carboxamide]) at the molar ratio of 50:34:16. The components were mixed in chloroform, in glass beaker and placed overnight in a desiccator under vacuum to yield a lipid film on the glass. The film was hydrated in PBS, pH 6.7, with vigorous shaking at 37°C followed by sonication for 20 to 30 s. The liposomes were extruded by sequentially passing them 14 to 15 times through a series of membrane filters (Whatman Nuclepore Track-Etch membranes) with pore sizes of 1.0, 0.8, 0.2, and 0.1 μm, respectively. The liposomes were incubated overnight with 1086 NFL trimers (900 mg protein per 300 mL of the liposomes) for covalent conjugation. The cysteine residues on the 1086 NFL trimers were reduced in 1.0 mM TCEP- PBS, pH 6.7, prior to coupling to liposomes. The trimer-conjugated liposomes were purified by a S200 size exclusion column to separate the trimer-coupled liposomes from unbound trimers. The amount of trimers conjugated to the liposomes was determined by a Bradford assay using a standard trimer curve generated with the Advanced Protein Assay reagent (Cytoskeleton Inc.) (Ingale et al., 2016).
Generation of fluorophore-labeled 1086 NFL trimer-conjugated liposomes
Fluor labeled liposomes were prepared similarly as described above with some modifications. Briefly, the fluorophore-labeled liposomes were comprised of DSPC, cholesterol, TopFluor cholesterol, and PE-MCC at the molar ratio of 50:32:2:16. The components were mixed and placed in the dark in a desiccator under vacuum to yield a lipid film. The film was hydrated in PBS, pH 6.7 and the liposomes were extruded by sequentially passing them across a series of membrane filters same as above. The TopFluor-labeled liposomes were incubated overnight with TCEP reduced 1086 NFL trimers for covalent conjugation. The trimer-conjugated liposomes were purified by passage through a S200 column by SEC to separate the trimer-coupled liposomes from unbound trimers. Next, the trimers on the TopFluor-labeled liposomes were labeled by Alexa Fluor® 680 (AF680) according to the manufacturer’s instructions (ThermoFisher Scientific). The double fluorophore-labeled trimer:liposomes were further purified by passage through a S200 size exclusion column. The amount of trimer conjugated to the liposomes was determined by a Bradford assay.
Immunogenicity immunizations and sample collection
Ten female RMs were allocated to two groups (n = 5/group) receiving either IM or SC administration of clade C 1086 NFL trimer-coupled liposomes (100 μg) formulated with Matrix-M adjuvant (75 μg; Novavax AB, Uppsala, Sweden). Immunizations were split between both quads (0.5 ml/injection) and animals were immunized at week 0, 4, 12, and 20. The animals were lightly sedated with ketamine at 10–15 mg/kg given intramuscularly (Ketaminol 100 mg/ml, Intervet, Sweden) during the immunizations, blood draws, and bone marrow aspirations. Bone marrow was sampled from the humerus as previously described (Spångberg et al., 2014). Mono-nuclear cells from peripheral blood (PBMCs) and bone marrow were obtained by standard density gradient centrifugation using Ficoll-Paque (GE Healthcare).
Tracking immunizations and terminal sample collection
For innate immune response studies, animals received two IM and two SC injections of Alexa Fluor 680-labeled Env trimer on TopFluor cholesterol-labeled liposomes (50 μg/site) formulated with Matrix-M adjuvant (37.5 μg/site) at different sites. PBS injections served as internal controls and were given either IM or SC. The final injection volumes were 0.5 mL and were administered on a marked injection site. Three male RMs were immunized for vaccine tracking in a naive setting and three female RMs from the immunogenicity experiment were immunized for the high titer setting. To assess the effect of the vaccine adjuvant and the Env:liposomes independently, two female RMs were immunized with either Env:liposomes in adjuvant, trimer:liposomes alone, adjuvant alone, or PBS. See Figure S2A for immunization schematic.
Tissue processing of tracking experiments
All tissues were sampled during necropsy and stored separately in RPMI1640 on ice, as previously described (Liang et al., 2017a). The skin and underlying muscle from marked injection sites was dissected for cell suspensions. Injection site tissues were weighed after removal of fat, connective tissue, and excess muscle or skin. Muscle and skin tissues were digested with 0.25 mg/ml Liberase TL (Roche) and 0.5 mg/ml DNase I (Sigma) at 37°C. Muscle was digested for 2 hours without agitation and skin for 1 hour with agitation (Liang et al., 2017a). R10 media (RPMI1640, 10% fetal calf serum (FCS), 1% L-glutamine, 1% penicillin/streptomycin) was used to quench enzyme activity and digestions were filtered through 70 mm cell strainers and washed with media. Samples were immediately stained for flow cytometry analysis upon completion of processing. LNs were mechanically disrupted using a plunger and 70 mm cell strainers. All individual LNs per LN cluster (axillary, apical, inguinal, external/common iliac, mesenteric) were pooled for analysis. Cell suspensions were washed and stained immediately.
Flow cytometry of tracking experiments
Cell suspensions representing approximately 2 g of injection site tissue or 5 million LN cells were stained for flow cytometry analysis. Briefly, Live/Dead fixable blue viability dye (Invitrogen) was used according to manufacturer’s protocol, FcR-blocking reagent (Miltenyi Biotec) was used, and a cocktail of fluorescent antibodies was added. Separate panels were used for analysis of muscle and skin as well as their respective draining LNs. The muscle panel included anti-human CD1c PE (AD5–8E7, Miltenyi), CD11c PE-Cy7 (3.9, Biolegend), CD66abce APC (TET2, Miltenyi), CCR7 PE-Dazzle594 (G043H7, Biolegend), CD3 APC-Cy7 (SP34–2, BD Biosciences), CD8 APC-Cy7 (RPA-T8, BD Biosciences), CD20 APC-Cy7 (L27, BD Biosciences), HLA-DR PE-Cy5.5 (Tu36, Invitrogen), CD14 BV570 (M5E2, Biolegend), CD123 BV510 (6H6, Biolegend), CD80 BV650 (L307.4, BD Biosciences), CD16 BV421 (3G8, Biolegend), and anti-NHP CD45 BV605 (D058–1283, BD Biosciences). The skin panel included anti-human CD1a PE (SK9, BD Biosciences), CD209 PerCP-Cy5.5 (DCN46, BD Biosciences), CD11c PE-Cy7 (3.9, Biolegend), CD66abce APC (TET2, Miltenyi), CCR7 PE-Dazzle594 (G043H7, Biolegend), CD3 APC-Cy7 (SP34–2, BD Biosciences), CD8 APC-Cy7 (RPA-T8, BD Biosciences), CD20 APC-Cy7 (L27, BD Biosciences), HLA-DR PE-Cy5.5 (Tu36, Invitrogen), CD14 BV570 (M5E2, Biolegend), CD123 BV510 (6H6, Biolegend), CD80 BV650 (L307.4, BD Biosciences), CD16 BV421 (3G8, Biolegend), and anti-NHP CD45 BV605 (D058–1283, BD Biosciences). Samples were spiked with AccuCount beads (Spherotech) and cell numbers were calculated according to the manufacturer’s protocol. At least 1 million events per sample were acquired on an LSRFortessa flow cytometer (BD) and data was analyzed using FlowJo v10 (FlowJo Inc).
In situ staining of LNs
Fresh LN biopsies were embedded in optimal cutting temperature (OCT) media and snap frozen with dry ice before storage at −80°C. Biopsies were cut into 8 μm-thick sections using a cryostat and mounted on superfrost plus glass slides (ThermoFisher Scientific). Sections were air-dried for 15 min before fixing with 2% PFA (Sigma) for 20 min. Tissues were blocked and permeabilized with 2% FCS in permwash buffer (tris-buffered saline containing 1% HEPES buffer (Sigma) and 0.1% saponin (Sigma)) for 30 min. BLOXALL reagent (Vector Laboratories) was additionally used, according to manufacturer’s protocol, for slides where tyramide signal amplification was employed. An avidin/biotin blocking kit (Vector Laboratories) was used for blocking of endogenous biotin. A combination of different antibodies was used for staining, including polyclonal rabbit anti-human CD3 (Dako), polyclonal goat anti-human IgD (Southern Biotech), mouse anti-human CD35 (E11, BD Biosciences), mouse anti-human Ki67 (B56, BD Biosciences), mouse anti-human neutrophil elastase (NP57, Dako), and biotinylated human anti-HIV-1 Env VRC01 (Wu et al., 2010) diluted in permwash buffer. The antibodies were added as a cocktail and incubated overnight at 4°C. Slides were washed with permwash solution three times and blocked with 1% donkey serum in permwash for 30 min. Biotinylated secondary antibodies and streptavidin-conjugated fluorophores were added sequentially for 30 min each, with additional avidin/biotin blocking performed between each secondary antibody and fluorophore pair. Secondary antibodies were all raised in donkey and included anti-rabbit, anti-goat, and anti-mouse (Jackson Immunoresearch). Streptavidin-conjugated fluorophores used included AF405, AF488, and AF555 (Invitrogen). For VRC01 staining of LNs, a Tyramide XX Biotin SuperBoost kit (Invitrogen) was used. Briefly, streptavidin-conjugated horseradish peroxidase (HRP) was added for 30 min at RT. After washing, tyramide XX biotin was added and the reaction was stopped using the kit’s stop solution after 7.5 min. Streptavidin-conjugated AF594 or AF488 (Invitrogen) was then added for 30 min. Some slides were also stained with 300nM DAPI (Invitrogen) for 10 min. After completion of staining, slides were washed with water, air-dried in the dark and mounted with Prolong Diamond anti-fade mounting media (Invitrogen) and 22×50 mm coverslips.
Images were captured using an automated confocal slide scanner (Pannoramic MIDI II FL, 3DHistech) utilizing a FLIR Grass-hopper3 camera equipped with a Zeiss 20x Plan-Apochromat 0.8NA objective, Lumencor SOLA SM light engine, and Pannoramic slide scanning software along with CaseViewer software.
In vitro vaccine experiments
Buffy coats from human blood donors were used to isolate PBMCs using standard density centrifugation or to isolate human monocytes with a Rosettesep human monocyte enrichment kit (STEMCELL Technologies) followed by standard density centrifugation. Isolated PBMCs were used for time-course experiments of Env:liposome signal by incubation of 1 million cells with 0.5 μg/mL labeled Env:liposomes in R10 media for 0, 1, 6, or 24 hours. Immune complex experiments were conducted using isolated human monocytes and RM plasma from a naive or a high titer animal. Briefly, for opsonization 0.1 μg/mL labeled Env:liposomes was incubated with 10% RM plasma in R10 media for 60 min at 37°C and was then added to 1 million monocytes and incubated for another 60 min at 37°C. After culture, cells were washed with PBS and stained with live/dead fixable blue viability dye (Invitrogen), FcR blocking reagent (Miltenyi Biotec), anti-human HLA-DR PE-Cy5.5 (Tu36, Life Technologies) and CD14 BV570 (M5E2, Biolegend). Cells were washed after staining and fixed with 1% PFA before acquisition on an LSRFortessa flow cytometer (BD Biosciences). Analysis was done using FlowJo v10 (FlowJo Inc.).
Activation experiments were conducted using isolated human monocytes. Briefly, 0.5 μg/mL labeled Env:liposomes, labeled Env, unlabeled Env, 2.5 μg/mL TLR7/8 ligand (Invivogen), or media alone were added to 1 million monocytes and incubated for 24 hr at 37°C. After culture, cells were washed with PBS and stained with live/dead fixable blue viability dye (Invitrogen), FcR blocking reagent (Miltenyi Biotec), anti-human HLA-DR PE-Cy5.5 (Tu36, Life Technologies), CD14 BV570 (M5E2, Biolegend), CD11c PE-Cy7 (3.9, Biolegend), CCR7 PE-Dazzle594 (G043H7, Biolegend), and CD80 BV650 (L307.4, BD Biosciences). Cells were washed after staining and fixed with 1% PFA before acquisition on an LSRFortessa flow cytometer (BD Biosciences). Analysis was done using FlowJo v10 (FlowJo Inc.).
ELISA analysis of plasma samples
Env-specific IgG titers were measured by ELISA as previously described (Ingale et al., 2016). In brief, MaxiSorp 96-well plates (Nal-gene Nunc International) were coated overnight at 4°C with a mouse anti-His tag antibody (1.5 mg/ml; R&D Systems). The plates were blocked with PBS containing 2% milk for 1 hr at room temperature (RT) and then incubated with 1086 NFL trimers at 3 mg/ml for 1 hr at RT. The plates were subsequently incubated with plasma (5-fold serial dilutions starting at 1:20) for 1hr at RT. Env-specific IgG was detected by adding a secondary HRP conjugated anti-monkey IgG antibody (1:10,000; Nordic MUbio) and the signal was developed by addition of tetramethylbenzidine (TMB) substrate (Invitrogen). The addition of an equal volume of 1M H2SO4 stopped the reaction and the optical density (OD) was read at 450 nm and background was read at 550 nm. The plates were washed 6 times between each incubation step using PBS supplemented with 0.05% Tween 20. The half-max binding titers (OD50) for each sample was calculated by interpolation from mean OD50 values using the formula (ODmax-ODmin)/2).
A similar setup was used for detection of Env-specific IgA titers, but with the addition of a secondary HRP conjugated anti-monkey IgA antibody instead (1:1,000; Nordic MUbio). The IgA titers are reported as the max OD value obtained for the 1:20 plasma dilution.
Env-specific IgG avidity was measured using a chaotropic wash ELISA as previously described (Thompson et al., 2018), with some modifications. Plates were coated as described above. Plasma was normalized to an OD value of 1.5 and as a benchmark 0.25 μg/mL of VRC01 antibody was used. After sample incubation, plates were incubated with serial dilutions of sodium thiocyanate (NaSCN: 2, 1.75, 1.5, 1.25, 1, 0.75, 0.5, or 0 M) diluted in PBS for 10 min. The plates were then washed and developed as described above. The avidity of the plasma IgG is reported as IC50, which is the molar concentration of NaSCN needed to dissociate 50% of the plasma binding.
Pseudovirus neutralization assay
Ab neutralizing titers were assayed using a single round infectious HIV-1 Env pseudovirus assay using the TZM-bl target cells (Li et al., 2005). Serial dilutions of the plasma were assayed to determine the dilution that resulted in a 50% reduction in relative luciferace units (RLU). Neutralization dose-response curves were fit by non-linear regression using a 5-parameter hill slope equation using the R statistical software package. Neutralization capacities of the plasma were reported as ID50, which is the reciprocal of the plasma dilution producing 50% virus neutralization.
B cell ELISpot
To enumerate Env-specific plasma cells in bone marrow and memory B cells in blood enzyme-linked immunospot (ELISpot) assays were performed as previously described (Sundling et al., 2010). ELISpot plates (MAIPSWU10; Millipore) were coated with 10 μg/ml of goat anti-human IgG (Fcγ; Jackson ImmunoResearch). Dilution series of cells were transferred in duplicate and cultured overnight at 37°C. For bone marrow plasma cell enumeration, cells were plated directly without prior stimulation. For memory B cells in blood, cells were prestimulated for four days at 2 million cells/ml with 5 μg/ml CpG-B (ODN 2006; Invivogen), 10 μg/ml Pokeweed mitogen (PWM; Sigma-Aldrich), and 1:10,000 Protein A from Staphylococcus aureus Cowan strain (SAC; Sigma-Aldrich). Plates were washed with PBS-T, incubated with 0.25 μg/ml biotinylated goat anti-human IgG (Fcγ; Jackson ImmunoResearch Laboratories) for total IgG determination, 1 μg/ml biotinylated 1086 trimer for Env-specific determination, or 1 mg/ml biotinylated ovalbumin (OVA) in PBS-T. After another round of washing, streptavidin-conjugated alkaline phosphatase (Mabtech) diluted in PBS-T was added. BCIP/NBT substrate (Mabtech) was used to develop spots and counts were acquired with AID ELISpot reader (Autoimmun Diagnostika). Spots were background-subtracted using counts from OVA wells.
T cell stimulation and proliferation
To assess Env-specific T cell responses from the immunogenicity study, PBMCs were cultured at 1 million cells/ml in R10 alone (unstim), 1 μg/ml overlapping peptides (Douagi et al., 2010), or 10 μg/ml Env 1086 protein overnight. After 2 hours of stimulation, 10 μg/ml Brefeldin A (BFA; Invitrogen) was added to the cultures. After culture, cells were washed with PBS and stained with live/dead fixable blue viability dye (Invitrogen), anti-human CCR7 BV421 (G043H7, Biolegend), CD4 PE-Cy5.5 (S3.5, Invitrogen), CD8 BV570 (RPA-T8, Biolegend), and CD45RA PE-Cy5 (5H9, BD Biosciences). Cells were permeabilized using Cytofix/Cytoperm kit (BD Biosciences) and stained intracellularly for anti-human IL-21 AF647 (3A3-N2.1, BD Biosciences), IL-13 PE (JES10–5a2, Biolegend), TNF AF488 (MAb11, Biolegend), IL-2 BV605 (MQ1–17H12, BD Biosciences), IL-17A BV785 (BL168, Biolegend), CD69 ECD (TP1.55.3, Beckman Coulter), CD3 APC-Cy7 (SP34–2, BD Biosciences), and IFNg AF700 (B27, Biolegend). Cells were washed after staining and fixed with 1% PFA before acquisition on an LSRFortessa flow cytometer (BD Biosciences). Analysis was done using FlowJo v10 (FlowJo Inc.) and results were background subtracted using values from unstim cells.
Proliferation was used as the readout to assess priming of Env-specific T cells in LNs from the tracking animals. Briefly, LN cells were labeled with 0.5 μM CellTrace Violet (Invitrogen) at a cell concentration of 1 million/ml for 20 min at 37°C. Labeled cells were cultured for 5 days in R5 media (5% FCS) alone, 1 μg/ml overlapping peptides, 1 μg/ml Env 1086 protein, or 0.1 μg/ml staphylococcal enterotoxin B (SEB; Sigma). After culture, cells were washed with PBS and stained with live/dead fixable blue viability dye (Invitrogen), anti-human CD3 APC-Cy7 (SP34–2, BD Biosciences), CD4 PE-Cy5.5 (S3.5, Life Technologies) and CD8 BV570 (RPA-T8, Biolegend). Cells were washed after staining and fixed with 1% PFA before acquisition on an LSRFortessa flow cytometer (BD Biosciences). Analysis was done using FlowJo v10 (FlowJo Inc.).
GC B cell probing by flow cytometry
To assess Env-specific GC B cell responses, frozen LN cell suspensions were thawed and washed in R10, then stained with live/dead fixable blue viability dye (Invitrogen), tetramer Env probes in AF488 and BV421 for 30 min at 4°C. Cells were subsequently stained with anti-human CD20 BV570 (2H7, Biolegend), and CD3 APC-Cy7 (SP34–2, BD Biosciences) for an additional 20 min at 4°C. Cells were permeabilized using the transcription factor buffer set (BD Biosciences) and stained intracellularly for anti-human IgG BV786 (G18–145, BD Biosciences), BCL6 PE-Cy7 (K112–91, BD Biosciences), and Ki67 PE (B56, BD Biosciences). Tetramer Env probes were prepared by incubation of 4-fold molar excess of avi-tag biotinylated 1086 Env protein with either streptavidin-conjugated AF488 (Invitrogen) or streptavidin-conjugated BV421 (Biolegend).
QUANTIFICATION AND STATISTICAL ANALYSIS
No statistical methods were used to predetermine sample size. Statistical parameters including the exact value of n, the definition of center, dispersion, and precision measures are reported in the Figures and Figure Legends. Data were judged to be statistically significant when p < 0.05. In Figures, asterisks denote statistical significance as calculated using the two-tailed non-parametric Mann-Whitney U test for comparison of two groups or Kruskal-Wallis test with Dunn’s multiple comparisons correction when three or more groups were compared. Wilcoxon matched-pairs signed rank test was used for comparison of activation data of Env- and Env+ APCs. Non-parametric Spearman’s correlation was used to assess associations between measured parameters. (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001). Analyses were performed in GraphPad PRISM 8.
The lower limit of detection (LOD) for Env+ cells in tissues obtained from fluorescent vaccine tracking experiments was calculated by analysis of pre-vaccination blood samples, uninjected muscle or skin tissue, and mesenteric LNs. A LOD was calculated for Env+ CD45+ cells as well as each immune cell subset studied and is specific for the type of tissue analyzed. For immune cell infiltration, the theoretical LOD was calculated based on the assumption that at least a single CD45+ cell subset could be detected per 2.5 million events run. The average AccuCount bead event count (5,000 of 25,000 spiked beads) was then used to calculate the LOD to five cells per gram of tissue.
DATA AND CODE AVAILABILITY
This study did not generate/analyze datasets or code.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-human CD1c PE (AD5-8E7) | Miltenyi | Cat#130-113-302, RRID:AB_2726081 |
| Mouse anti-human CD11c PE-Cy7 (3.9) | Biolegend | Cat#301608, RRID:AB_389351 |
| Mouse anti-human CD66abce APC (TET2) | Miltenyi | Cat#130-093-155, RRID:AB_871696 |
| Mouse anti-human CCR7 PE-Dazzle594 (G043H7) | Biolegend | Cat#353236, RRID:AB_2563641 |
| Mouse anti-human CD3 APC-Cy7 (SP34-2) | BD Biosciences | Cat#557757, RRID:AB_396863 |
| Mouse anti-human CD8 APC-Cy7 (RPA-T8) | BD Biosciences | Cat#557760, RRID:AB_396865 |
| Mouse anti-human CD20 APC-Cy7 (L27) | BD Biosciences | Cat#335829 |
| Mouse anti-human HLA-DR PE-Cy5.5 (Tu36) | Invitrogen | Cat#MHLDR18, RRID:AB_10372966 |
| Mouse anti-human CD14 BV570 (M5E2) | Biolegend | Cat#301832, RRID:AB_2563629 |
| Mouse anti-human CD123 BV510 (6H6) | Biolegend | Cat#306022, RRID:AB_2562068 |
| Mouse anti-human CD80 BV650 (L307.4) | BD Biosciences | Cat#564158, RRID:AB_2738630 |
| Mouse anti-human CD16 BV421 (3G8) | Biolegend | Cat#302038, RRID:AB_2561578 |
| Mouse anti-NHP CD45 BV605 (D058-1283) | BD Biosciences | Cat#564098, RRID:AB_2738590 |
| Mouse anti-human CD1a PE (SK9) | BD Biosciences | Cat#333167 |
| Mouse anti-human CD209 PerCP-Cy5.5 (DCN46) | BD Biosciences | Cat#558263, RRID:AB_647256 |
| Polyclonal rabbit anti-human CD3 | Dako | Cat#A0452, RRID:AB_2335677 |
| Polyclonal goat anti-human IgD FITC | Southern Biotech | Cat# 2030-02, RRID:AB_2795624 |
| Mouse anti-human CD35 (E11) | BD Biosciences | Cat#555451, RRID:AB_395844 |
| Mouse anti-human Ki67 (B56) | BD Biosciences | Cat#550609, RRID:AB_393778 |
| Mouse anti-human neutrophil elastase (NP57) | Dako | Cat#M075201-2 |
| Human anti-HIV-1 Env (VRC01), biotinylated | Produced in house Wu et al., 2010 | N/A |
| Polyclonal donkey anti-rabbit IgG, biotinylated | Jackson Immunoresearch | Cat#711-005-152, RRID:AB_2340585 |
| Polyclonal donkey anti-goat IgG, biotinylated | Jackson Immunoresearch | Cat#705-005-147, RRID:AB_2340385 |
| Polyclonal donkey anti-mouse IgG, biotinylated | Jackson Immunoresearch | Cat#715-005-150, RRID:AB_2340758 |
| Mouse anti-His tag (AD1.1.10) | R&D Systems | Cat#MAB050, RRID:AB_357353 |
| Polyclonal goat anti-monkey IgG antibody, HRP-conjugated | Nordic MUbio | Cat# GAMon/IgG(Fc)/PO |
| Polyclonal goat anti-monkey IgA antibody, HRP-conjugated | Nordic MUbio | Cat# GAMon/IgA(Fc)/PO |
| Polyclonal goat anti-human IgG (Fcγ-specific) | Jackson ImmunoResearch | Cat#109-005-008, RRID:AB_2337534 |
| Polyclonal goat anti-human IgG (Fcγ-specific), biotinylated | Jackson ImmunoResearch | Cat#109-065-008, RRID:AB_2337623 |
| Mouse anti-human CCR7 BV421 (G043H7) | Biolegend | Cat#353208, RRID:AB_11203894 |
| Mouse anti-human CD4 PE-Cy5.5 (S3.5) | Invitrogen | Cat#MHCD0418, RRID:AB_10376013 |
| Mouse anti-human CD8a BV570 (RPA-T8) | Biolegend | Cat#301038, RRID:AB_2563213 |
| Mouse anti-human CD45RA PE-Cy5 (5H9) | BD Biosciences | Cat#552888, RRID:AB_394517 |
| Mouse anti-human IL-21 AF647 (3A3-N2.1) | BD Biosciences | Cat#560493, RRID:AB_1645421 |
| Rat anti-human IL-13 PE (JES10-5a2) | Biolegend | Cat#501903, RRID:AB_315198 |
| Mouse anti-human TNF AF488 (MAb11) | Biolegend | Cat#502915, RRID:AB_493121 |
| Rat anti-human IL-2 BV605 (MQ1-17H12) | BD Biosciences | Cat#564165, RRID:AB_2738636 |
| Mouse anti-human IL-17A BV785 (BL168) | Biolegend | Cat#512338, RRID:AB_2566765 |
| Mouse anti-human CD69 ECD (TP1.55.3) | Beckman Coulter | Cat#6607110, RRID:AB_1575978 |
| Mouse anti-human IFN-gamma AF700 (B27) | Biolegend | Cat# 506516, RRID:AB_961351 |
| Mouse anti-human CD20 BV570 (2H7) | Biolegend | Cat#302332, RRID:AB_2563805 |
| Mouse anti-human IgG BV786 (G18-145) | BD Biosciences | Cat#564230, RRID:AB_2738684 |
| Mouse anti-human BCL6 PE-Cy7 (K112-91) | BD Biosciences | Cat#563582, RRID:AB_2738292 |
| Mouse anti-human Ki67 PE (B56) | BD Biosciences | Cat#556027, RRID:AB_2266296 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 1086 NFL TD CC+ trimer | Guenaga et al., 2017 | N/A |
| Galanthus nivalis lectin-agarose | Vector Laboratories | Cat#AL-1243 |
| DSPC | Avanti Polar Lipids | Cat#850365 |
| Cholesterol | Sigma | Cat#C3045 |
| PE-MCC | Avanti Polar Lipids | Cat#780200 |
| TopFluor Cholesterol | Avanti Polar Lipids | Cat#810255 |
| Streptavidin-conjugated Alexa Fluor 405 | Invitrogen | Cat#S32351 |
| Streptavidin-conjugated Alexa Fluor 488 | Invitrogen | Cat#S11223 |
| Streptavidin-conjugated Alexa Fluor 555 | Invitrogen | Cat#S32355 |
| Streptavidin-conjugated Alexa Fluor 594 | Invitrogen | Cat#S11227 |
| Streptavidin-conjugated BV421 | BioLegend | Cat#405225 |
| Streptavidin-ALP | Mabtech | Cat#3310-10-1000 |
| DAPI (4’,6-Diamidino-2-Phenylindole, Dilactate) | Invitrogen | Cat# D3571, RRID:AB_2307445 |
| ProLong Diamond Antifade Mountant | Invitrogen | Cat#P36965 |
| FcR Blocking Reagent, human | Miltenyi | Cat#130-059-901 |
| Tetramethylbenzidine (TMB) substrate | Invitrogen | Cat#002023 |
| BCIP/NBT substrate | Mabtech | Cat#3650-10 |
| HIV YU2 Env overlapping peptides | Douagi et al., 2010 | N/A |
| Brefeldin A | Invitrogen | Cat#B7450 |
| Staphylococcal enterotoxin B (SEB) | Sigma | Cat#S4881 |
| CpG class B (ODN 2006) | Invivogen | Cat#tlrl-2006 |
| Pokeweed mitogen | Sigma | Cat#L8777 |
| Protein A from Staphylococcus aureus Cowan strain | Sigma | Cat#P7155 |
| Liberase TL Research Grade | Sigma | Cat#5401020001 |
| Deoxyribonuclease I (DNase I) from bovine pancreas | Sigma | Cat#DN25 |
| Sodium thiocyanate (NaSCN) | Sigma | Cat#467871 |
| Critical Commercial Assays | ||
| Alexa Fluor 680 Protein Labeling Kit | Invitrogen | Cat#A20172 |
| LIVE/DEAD Fixable Blue viability dye | Invitrogen | Cat#L23105 |
| Avidin/Biotin Blocking kit | Vector Laboratories | Cat#SP-2001, RRID:AB_2336231 |
| BLOXALL Endogenous Peroxidase and Alkaline Phosphatase Blocking Solution | Vector Laboratories | Cat#SP-6000, RRID:AB_2336257 |
| Biotin XX Tyramide SuperBoost Kit, Streptavidin | Invitrogen | Cat#B40931 |
| Transcription Factor Buffer Set | BD Biosciences | Cat#562574 |
| Cytofix/Cytoperm Fixation/Permeabilization Solution kit | BD Biosciences | Cat#554714 |
| RosetteSep Human Monocyte Enrichment Cocktail | StemCell Technologies | Cat#15068 |
| CellTrace Violet Cell Proliferation Kit | Invitrogen | Cat#C34557 |
| AccuCount blank beads, 8.0-12.9 μm | Spherotech | Cat#ACBP-100-10 |
| Experimental Models: Cell Lines | ||
| Human: FreeStyle 293F | Invitrogen | Cat#R79007 |
| Human: TZM-bl | NIH AIDS Reagent Program | Cat#8129 |
| Experimental Models: Organisms/Strains | ||
| Indian-origin rhesus macaques (outbred) | PrimGen, PreLabs | N/A |
| Software and Algorithms | ||
| FlowJo v10 | FlowJo, LLC | RRID:SCR_008520, https://www.flowjo.com/solutions/flowjo/ |
| Prism v8 | GraphPad | RRID:SCR_002798, https://www.graphpad.com/scientific-software/prism/ |
| Caseviewer v2.3 | 3DHistech | RRID:SCR_017654, https://www.3dhistech.com/caseviewer |
| Other | ||
| Matrix-M | Novavax AB, Uppsala | N/A |
Highlights.
Subcutaneous and intramuscular injection target the skin and the muscle, respectively
The immunization route affects the anatomical LN cluster that is targeted
Priming of adaptive immune responses occurs in the local vaccine-draining LNs
Comparable innate and adaptive immune responses with both immunization routes
ACKNOWLEDGMENTS
We wish to thank Dr. Mats Spångberg and the personnel at the Astrid Fagraeus laboratory at Karolinska Institutet for expert assistance and care of the non-human primates; Linda Stertman, Karin Lövgren Bengtsson, and Jenny Reimer at Novavax AB for sharing of Matrix-M adjuvant; Julia Campe, Fredrika Hellgren, Rubén Gimenez, Monika Àdori, and Néstor Vázquez Bernat for technical assistance with sample processing; and Sijy O’Dell and John Mascola for assistance with the pseudovirus neutralization assays and analysis. This work was supported by the Swedish Research Council (Vetenskapsrådet; K.L.), an instrument grant from Stiftelsen Olle Engkvist Byggmästare (188-0156; K.L.), an HIV Vaccine Research and Design (HIVRAD) grant from the National Institutes of Health (NIH) (P01 AI104722; L.Y., G.B.K.H., and R.T.W.), grant P01 AI124337 from the NIH (R.T.W.), the Scripps Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) (AI100663; L.Y. and R.T.W.), and funding from the International AIDS Vaccine Initiative (IAVI) (R.T.W.). The full list of donors to the IAVI can be found at https://www.iavi.org/. Also, this research was supported by intramural faculty salary grants from Karolinska Institutet (S.O., E.A.T., and A.L.) and a grant from the China Scholarship Council (A.L.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.02.111.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Bale S, Goebrecht G, Stano A, Wilson R, Ota T, Tran K, Ingale J, Zwick MB, and Wyatt RT (2017). Covalent linkage of HIV-1 trimers to synthetic liposomes elicits improved B cell and antibody responses. J. Virol 91, e00443–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook IF, Barr I, Hartel G, Pond D, and Hampson AW (2006). Reactogenicity and immunogenicity of an inactivated influenza vaccine administered by intramuscular or subcutaneous injection in elderly adults. Vaccine 24, 2395–2402. [DOI] [PubMed] [Google Scholar]
- Diez-Domingo J, Weinke T, Garcia de Lomas J, Meyer CU, Bertrand I, Eymin C, Thomas S, and Sadorge C (2015). Comparison of intramuscular and subcutaneous administration of a herpes zoster live-attenuated vaccine in adults aged R50 years: a randomised non-inferiority clinical trial. Vaccine 33, 789–795. [DOI] [PubMed] [Google Scholar]
- Douagi I, Forsell MNE, Sundling C, O’Dell S, Feng Y, Dosenovic P, Li Y, Seder R, Loré K, Mascola JR, et al. (2010). Influence of novel CD4 binding-defective HIV-1 envelope glycoprotein immunogens on neutralizing antibody and T-cell responses in nonhuman primates. J. Virol 84, 1683–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisch A, Cadilhac P, Vidor E, Prazuck T, Dublanchet A, and Lafaix C (1996). Immunogenicity and safety of a new inactivated hepatitis A vaccine: a clinical trial with comparison of administration route. Vaccine 14, 1132–1136. [DOI] [PubMed] [Google Scholar]
- Gillet Y, Habermehl P, Thomas S, Eymin C, and Fiquet A (2009). Immunogenicity and safety of concomitant administration of a measles, mumps and rubella vaccine (M-M-RvaxPro) and a varicella vaccine (VARIVAX) by intramuscular or subcutaneous routes at separate injection sites: a randomised clinical trial. BMC Med. 7, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenaga J, Garces F, de Val N, Stanfield RL, Dubrovskaya V, Higgins B, Carrette B, Ward AB, Wilson IA, and Wyatt RT (2017). Glycine substitution at helix-to-coil transitions facilitates the structural determination of a stabilized subtype C HIV envelope glycoprotein. Immunity 46, 792–803.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog C (2014). Influence of parenteral administration routes and additional factors on vaccine safety and immunogenicity: a review of recent literature. Expert Rev. Vaccines 13, 399–415. [DOI] [PubMed] [Google Scholar]
- Hopf S, Garner-Spitzer E, Hofer M, Kundi M, and Wiedermann U (2016). Comparable immune responsiveness but increased reactogenicity after subcutaneous versus intramuscular administration of tick borne encephalitis (TBE) vaccine. Vaccine 34, 2027–2034. [DOI] [PubMed] [Google Scholar]
- Ingale J, Stano A, Guenaga J, Sharma SK, Nemazee D, Zwick MB, and Wyatt RT (2016). High-density array of well-ordered HIV-1 spikes on synthetic liposomal nanoparticles efficiently activate B cells. Cell Rep. 15, 1986–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuf M, Zepp F, Meyer CU, Habermehl P, Maurer L, Burow HM, Behre U, Janssens M, Willems P, Bisanz H, et al. (2010). Safety, immunogenicity and immediate pain of intramuscular versus subcutaneous administration of a measles-mumps-rubella-varicella vaccine to children aged 11–21 months. Eur. J. Pediatr 169, 925–933. [DOI] [PubMed] [Google Scholar]
- Levin C, Bonduelle O, Nuttens C, Primard C, Verrier B, Boissonnas A, and Combadière B (2017). Critical role for skin-derived migratory DCs and Langerhans cells in TFH and GC responses after intradermal immunization. J. Invest. Dermatol 137, 1905–1913. [DOI] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al. (2005). Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol 79, 10108–10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Ploquin A, Hernández JD, Fausther-Bovendo H, Lindgren G, Stanley D, Martinez AS, Brenchley JM, Koup RA, Loré K, and Sullivan NJ (2015). Dissociation of skeletal muscle for flow cytometric characterization of immune cells in macaques. J. Immunol. Methods 425, 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Lindgren G, Lin A, Thompson EA, Ols S, Röhss J, John S, Hassett K, Yuzhakov O, Bahl K, et al. (2017a). Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol. Ther 25, 2635–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Lindgren G, Sandgren KJ, Thompson EA, Francica JR, Seu-bert A, De Gregorio E, Barnett S, O’Hagan DT, Sullivan NJ, et al. (2017b). Vaccine priming is restricted to draining lymph nodes and controlled by adjuvant-mediated antigen uptake. Sci. Transl. Med 9, eaal2094. [DOI] [PubMed] [Google Scholar]
- Lindgren G, Ols S, Liang F, Thompson EA, Lin A, Hellgren F, Bahl K, John S, Yuzhakov O, Hassett KJ, et al. (2017). Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+ PD-1+ CXCR3+ T follicular helper cells. Front. Immunol 8, 1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay KE, Bhosle SM, Zurla C, Beyersdorf J, Rogers KA, Vanover D, Xiao P, Araínga M, Shirreff LM, Pitard B, et al. (2019). Visualization of early events in mRNA vaccine delivery in non-human primates via PET-CT and near-infrared imaging. Nat. Biomed. Eng 3, 371–380. [DOI] [PubMed] [Google Scholar]
- Lu LL, Suscovich TJ, Fortune SM, and Alter G (2018). Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol 18, 46–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark A, Carlsson RM, and Granström M (1999). Subcutaneous versus intramuscular injection for booster DT vaccination of adolescents. Vaccine 17, 2067–2072. [DOI] [PubMed] [Google Scholar]
- Martinez-Murillo P, Tran K, Guenaga J, Lindgren G,Àdori M, Feng Y, Phad GE, Vázquez Bernat N, Bale S, Ingale J, et al. (2017). Particulate array of well-ordered HIV clade C Env trimers elicits neutralizing antibodies that display a unique V2 cap approach. Immunity 46, 804–817.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer TJ, Zmolek AC, and Irvine DJ (2016). Beyond antigens and adjuvants: formulating future vaccines. J. Clin. Invest 126, 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeland MR, Shi W, Collignon C, Taubenheim N, Meeusen ENT, Didierlaurent AM, and de Veer MJ (2016). The lymphatic immune response induced by the adjuvant AS01: a comparison of intramuscular and subcutaneous immunization routes. J. Immunol 197, 2704–2714. [DOI] [PubMed] [Google Scholar]
- Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, et al. (2017). Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 46, 1073–1088.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukens AHE, Gelinck LBS, and Visser LG (2012). Intradermal vaccination to protect against yellow fever and influenza. Curr. Top. Microbiol. Immunol 351, 159–179. [DOI] [PubMed] [Google Scholar]
- Spångberg M, Martinez P, Fredlund H, Karlsson Hedestam GB, and Sundling C (2014). A simple and safe technique for longitudinal bone marrow aspiration in cynomolgus and rhesus macaques. J. Immunol. Methods 408, 137–141. [DOI] [PubMed] [Google Scholar]
- Sundling C, Forsell MNE, O’Dell S, Feng Y, Chakrabarti B, Rao SS, Loré K, Mascola JR, Wyatt RT, Douagi I, and Karlsson Hedestam GB (2010). Soluble HIV-1 Env trimers in adjuvant elicit potent and diverse functional B cell responses in primates. J. Exp. Med 207, 2003–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EA, and Loré K (2017). Non-human primates as a model for understanding the mechanism of action of toll-like receptor-based vaccine adjuvants. Curr. Opin. Immunol 47, 1–7. [DOI] [PubMed] [Google Scholar]
- Thompson EA, Ols S, Miura K, Rausch K, Narum DL, Spångberg M, Juraska M, Wille-Reece U, Weiner A, Howard RF, et al. (2018). TLR-adjuvanted nanoparticle vaccines differentially influence the quality and longevity of responses to malaria antigen Pfs25. JCI Insight 3, 120692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis NL, Kaminskas LM, and Porter CJH (2015). From sewer to saviour - targeting the lymphatic system to promote drug exposure and activity. Nat. Rev. Drug Discov 14, 781–803. [DOI] [PubMed] [Google Scholar]
- Wahl M, and Hermodsson S (1987). Intradermal, subcutaneous or intramuscular administration of hepatitis B vaccine: side effects and antibody response. Scand. J. Infect. Dis 19, 617–621. [DOI] [PubMed] [Google Scholar]
- Woodruff MC, Heesters BA, Herndon CN, Groom JR, Thomas PG, Luster AD, Turley SJ, and Carroll MC (2014). Trans-nodal migration of resident dendritic cells into medullary interfollicular regions initiates immunity to influenza vaccine. J. Exp. Med 211, 1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang Z-YY, Li Y, Hogerkorp C-MM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. (2010). Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Sharma SK, Cottrell C, Guenaga J, Tran K, Wilson R, Behrens A-J, Crispin M, de Val N, and Wyatt RT (2018). Structure-guided redesign improves NFL HIV Env trimer integrity and identifies an inter-protomer disulfide permitting post-expression cleavage. Front. Immunol 9, 1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L, Xue X, Yu P, Ni Y, and Chen F (2018). Evans blue dye: a revisit of its applications in biomedicine. Contrast Media Mol. Imaging 2018, 7628037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang W, and Wang S (2015). Effect of vaccine administration modality on immunogenicity and efficacy. Expert Rev. Vaccines 14, 1509–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate/analyze datasets or code.


