Abstract
Vibrio alginolyticus is a major cause of Vibriosis in farmed marine aquatic animals and has caused large economic losses to the Asian aquaculture industry in recent years. Therefore, it is necessary to control V. alginolyticus effectively. The virulence mechanism of V. alginolyticus, the Type III secretion system (T3SS), is closely related to its pathogenicity. In this study, the T3SS gene tyeA was cloned from V. alginolyticus wild-type strain HY9901 and the results showed that the deduced amino acid sequence of V. alginolyticus tyeA shared 75–83% homology with other Vibrio spp. The mutant strain HY9901ΔtyeA was constructed by Overlap-PCR and homologous recombination techniques. The HY9901ΔtyeA mutant exhibited an attenuated swarming phenotype and an ~40-fold reduction in virulence to zebrafish. However, the HY9901ΔtyeA mutant showed no difference in growth, biofilm formation and ECPase activity. Antibiotic susceptibility test was observed that wild and mutant strains were extremely susceptible to Amikacin, Minocycline, Gentamicin, Cefperazone; and resistant to oxacillin, clindamycin, ceftazidime. In contrast wild strains are sensitive to tetracycline, chloramphenicol, kanamycin, doxycycline, while mutant strains are resistant to them. qRT-PCR was employed to analyze the transcription levels of T3SS-related genes, the results showed that compared with HY9901 wild type, ΔtyeA had increased expression of vscL, vscK, vscO, vopS, vopN, vscN, and hop. Following vaccination with the mutant strain, zebrafish had significantly higher survival than controls following infection with the wild-type HY9901 (71.2% relative percent survival; RPS). Analysis of immune gene expression by qPCR showed that vaccination with HY9901ΔtyeA increased the expression of IgM, IL-1β, IL-6, and TNF-α in zebrafish. This study provides evidence of protective efficacy of a live attenuated vaccine targeting the T3SS of V. alginolyticus which may be facilitated by up-regulated pro-inflammatory and immunoglobulin-related genes.
Keywords: Vibrio alginolyticus, live attenuated vaccine, type III secretory system, tyeA, regulator
Introduction
Vibrio alginolyticus, a Gram-negative motile rod bacterium, is widely found in seawater and estuaries in various regions of the world (Thompson et al., 2004; Jones et al., 2013) V. alginolyticus is highly pathogenic in marine animals (Ben Kahla-Nakbi et al., 2009; Jun et al., 2015) and is associated with diseases of fish, shellfish, shrimp, and coral (Chang et al., 2008; Sadok et al., 2013). It is also a potential zoonotic pathogenic bacteria and incurs great economic losses to the aquaculture industry with raised pressures for the industry to develop controls in recent years. Zoonotic infections can result from human food poisoning causing diarrhea and other symptoms (Li et al., 2009; Sganga et al., 2009). Therefore, it is of great importance to understand the pathogenic mechanisms of V. alginolyticus in order to develop an effective vaccine.
Type III secretory system (T3SS) is a highly conserved secretory system in several Gram-negative bacteria such as Yersinia spp., Salmonella spp., Vibrio cholerae, Pseudomonas spp. (Anand et al., 2003; Masato et al., 2014; Khavong and Lorena, 2016; Dorothea et al., 2017). When V. alginolyticus infects the host, the T3SS can secrete virulence proteins outside the cell or on the surface of the host cell, or even directly inject virulent proteins into the host cell, resulting in cell death (Pallen et al., 2005). Although the T3SS mechanism is usually conserved in Gram-negative pathogens, the functions of its regulator vary widely. Comparative genome analysis showed that the T3SS of V. alginolyticus was similar to that of Vibrio parahaemolyticus (Martinez-Argudo and Blocker, 2010), but little is known about the regulator TyeA of V. alginolyticus T3SS.
Until now, there was no related research on tyeA gene in Vibrio alginolyticus, and its function was still being explored. It has been found that TyeA is a regulator of T3SS and involved in regulation of effector proteins expression (Sundberg and Forsberg, 2003). Numerous studies have shown that the YopN-TyeA heterodimer plays a particularly important role in regulating the secretion of effector proteins in Yersinia (Iriarte et al., 1998; Cheng and Schneewind, 2000; Schubot et al., 2004; Ferracci et al., 2005). In order to understand the function of TyeA in T3SS, we first constructed a mutant of tyeA gene, studied the biology and pathogenicity of the HY9901ΔtyeA strain and the transcription levels of T3SS-related genes were then analyzed by qRT-PCR. In addition, we found that HY9901ΔtyeA mutant can be used as an effective live vaccine against Vibrio alginolyticus in zebrafish and induces the expression of zebrafish pro-inflammatory and immunoglobulin-related genes.
Materials and Methods
Bacterial Strains, Plasmid, and Experimental Fish
The bacterial strains, plasmids, and zebrafish used in this study are listed in Table 1. V. alginolyticus HY9901 was isolated from diseased red snapper (Lutjanus sanguineus) (Cai et al., 2007) in Zhanjiang Port, Guangdong Province. E. coli DH5 α, E. coli MC1061 (λ pir) (Rubirés et al., 1997), S17-1 (λ Pir) (Simon et al., 2019), and suicide plasmid pRE112 (Edwards et al., 1998) were preserved in our laboratory. Healthy zebrafish (Danio rerio) were purchased from Zhanjiang Aquatic Market, 3–4 cm long and ~0.2 g in weight. The zebrafish were tested by bacteriological recovery tests and kept in seawater in a circulation system at 28°C for 2 weeks before the experiment.
Table 1.
Bacterial strains, plasmids, and experimental fish used in this study.
| Strains, plasmids | Relevant characteristics | Source |
|---|---|---|
| V.alginolyticus HY9901 | Wild type, isolated from diseased Lutjanus sanguineus off the Southern China coast | (Cai et al., 2007) |
| E. coli DH5α | supE44 ΔlacU169 (ϕ80 lacZDM15) hsdR17 recA1 gyrA96 thi-1 relA1 | TakaRa |
| MC1061 (λpir) | lacY1 galK2 ara-14 xyl-5 supE44 λpir | (Rubirés et al., 1997) |
| pRE112 | pGP704 suicide plasmid, pir dependent, oriT, oriV, sacB, C mr | (Edwards et al., 1998) |
| S17-1 (λpir) | T prSmrrecA thi pro hsdR-M+RP4:2-Tc: Mu: K m T n7 λpir | (Simon et al., 2019) |
| S17-1-pRE-ΔtyeA | S17-1 containing plasmid of pRE-ΔtyeA, C mr | This study |
| pMD18-T | Cloning vector, Ampr | TakaRa |
| Zebrafish | experimental fish, purchased from Zhanjiang Aquatic Market | Zhanjiang Aquatic Market |
Reagents and Primers
TIANamp Bacteria DNA Kit (Beijing, Tiangen Biotech Co., Ltd.); Easy PureTMQuick GelExtraction Kit and Easy PureTM Plasmid MiniPrepKit (Beijing, TransGen Biotech Co., Ltd.); pMD18-T vector, ExTaqDNA polymerase, Prime START MHS DNA polymerase, KpnI, SacI, and T4DNA ligase were all purchased from TaKaRa (Japan). The primers were synthesized by Sangon Biotech Co., Ltd.
Cloning and Sequencing of the tyeA Gene From V. alginolyticus HY9901
A pair of primers tyeAP1-F/tyeAP1-R were designed as shown in Table 2 according to the V. alginolyticus gene sequence (GenBank Number: GU074526.1). PCR was performed in a Thermocycler (Bio-Rad, CA, USA) under the following optimized amplification conditions: an initial denaturation at 95°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s. Five microlitre of each amplicon was examined on a 1% agarose gel stained with ethidium bromide. The PCR product was recovered from the agarose gel to ligate into the pMD18-T vector and transformed into E. coli DH5α (Table 1). The inserted fragment was sequenced by Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). Similarity analyses of the determined nucleotide sequences and deduced amino acid sequences were performed by BLAST programs (http://blast.ncbi.nlm.nih.gov /Blast.cgi) and aligned using the program Clustal-X (version 1.81). Protein analysis was conducted with ExPASy tools (http://expasy.org/tools/). Location of the domain was predicted using the InterProScan program (http://www.ebi.ac.uk/Tools/pfa/iprscan/).
Table 2.
Sequences of primers used in this study.
| Primer name | Primer sequence(5′-3′) | Accession number |
|---|---|---|
| tyeA-for | GGAATCTAGACCTTGAGTCGATATCTCGACCATCGCGCAA | MN328351 |
| tyeA-int-rev | ATCTTCCCACGCTTCCTCTTCAACTTGATAAGCCATAATTCGTCC | |
| tyeA-int-for | GGACGAATTATGGCTTATCAAGTTGAAGAGGAAGCGTGGGAAGAT | |
| tyeA-rev | ACAGCTAGCGACGATATGTCAGGCCGGAGGTCATAGAGCT | |
| tyeA-up | CACATGAACTCGTTTCGGACTATT | |
| tyeA-down | TTTCTGGACGCAACAACTCTGA | |
| tyeAP1-F | ATGGCTTATCAAGTTTCTA | |
| tyeAP1-R | TCAATCCAACTCATCTTCC | |
| IL-6-F | GGTCAGACTGAATCGGAGCG | NM_001079833.1 |
| IL-6-R | CAGCCATGTGGCGAACG | |
| IL-6R-F | GCATGTGCTTAAAGTATCCTGGTC | NM_001114318.1 |
| IL-6R-R | TGCAAATTGTGGTCGGTATCTC | |
| IL-1β-F | TGGACTTCGCAGCACAAAATG | AY340959.1 |
| IL-1β-R | GTTCACTTCACGCTCTTGGATG | |
| IL-8-F | GTCGCTGCATTGAAACAGAA | XM_001342570.2 |
| IL-8-R | CTTAACCCATGGAGCAGAGG | |
| IgM-F | GTTCCTGACCAGTGCAGAGA | AF246193 |
| IgM-R | CCTGATCACCTCCAGCATAA | |
| TNF-α-F | TAGAACAACCCAGCAAAC | NC_007130.7 |
| TNF-α-R | ACCAGCGGTAAAGGCAAC | |
| rag-1-F | GAAGTATACCAGAAGCCTAAT | NC_007136.7 |
| rag-1-R | TTCCATTCATCCTCATCACA | |
| TLR5-F | GAAACATTCACCTGGCACA | NC_007131.7 |
| TLR5-R | CTACAACCAGCACCACCAGAATG | |
| c/ebpβ-F | GCCGTACCAGACTGCTCCGA | NC_007119.7 |
| c/ebpβ-R | AGCCGCTTCTTGCCTTTCCC | |
| β-actin-F | ATGGATGAGGAAATCGCTGCC | NM_131031.1 |
| β-actin-R | CTCCCTGATGTCTGGGTCGTC | |
| vscL-F | TACCACGGTGAGTGTAGTTC | ACY41051.1 |
| vscL-R | CGTAACCGACTTCAGGGA | |
| hop-F | CTTCGCTTTCGGTTTGCT | KX245315 |
| hop-R | AATACCATCCCACCCTGT | |
| vscO-F | GAGCTGGAAACATTAAGACA | ACY41065.1 |
| vscO-R | TTGCTGCAACTGAACGAA | |
| vscK-F | GGCGTTATCTCCCGTTCC | ACY41050.1 |
| vscK-R | CTCCGCCCACCATCAATA | |
| vopN-F | TGAACTCGTTTCGGACTA | ACY41067.1 |
| vopN-R | ACTTTCTGGACTCGCACT | |
| vscN-F | TAGGCGAAGAAGGAATGG | ACY41066.1 |
| vscN-R | GCGATAGAAGTGGCAACAA | |
| vopS-F | AGTTTTGGAAGTGTTAGCG | ACY41053.1 |
| vopS-R | ACATTGCCTCTGTCATCG | |
| 16S-F | TTGCGAGAGTGAGCGAATCC | NR_044825.2 |
| 16S-R | ATGGTGTGACGGGCGGTGTG |
Construction of In-frame Deletion Mutant of tyeA Gene
According to the method previously described Zujie et al. (2019), Overlap extension PCR was applied to generate an in-frame deletion of the tyeA gene on the V. alginolyticus wild-type HY9901 chromosome. The in-frame deletion of tyeA in the V. alginolyticus was generated according to the method of Rubirés et al. (1997). For the construction of ΔtyeA, two PCR fragments were generated from HY9901 genomic DNA. The first fragment was amplified using primers tyeA-for (contains a KpnI site at the 5′-end) and tyeA-int-rev, whereas primers tyeA-int-for and tyeA-rev (contains a SmaI site at the 5′-end) were used to amplify the second fragment. Both fragments contained a 20 bp overlapping sequence and were used as templates for the subsequent PCR procedure, which used primers tyeA-for and tyeA-rev. The PCR product was ligated into suicide vector pRE112 (Cmr) to generate pRE-ΔtyeA. This recombinant suicide plasmid was transformed into E. coli MC1061λpir and subsequently S17-1λpir. The single crossover mutants were obtained by conjugal transfer of the resulting plasmid into V. alginolyticus HY9901. Deletion mutants were screened on 10% sucrose TSA plates. Its presence was subsequently confirmed by PCR and sequencing using primers tyeA-up and tyeA-down.
Characterization of the ΔtyeA
Genetic Stability of Mutants HY9901ΔtyeA
HY9901ΔtyeA was inoculated onto a TSA plate and passaged blindly for 30 generations. In brief, a single colony was picked from Tryptic Soya agar (TSA) plates and cultured in Tryptic soya broth (TSB, HKM, Guangzhou, China) medium, shaking for 12 h, bacterial broth culture was streaked out on a TSA plate, and this process repeated 30 times. Its genetic stability was determined by the PCR method.
Growth Curve of Bacteria
The wild-type HY9901 strain and the ΔtyeA were cultured in TSB for 24 h. The HY9901 and HY9901 ΔtyeA were inoculated into TSB at the ratio of 1: 100 (OD600 = 0.5) at 28°C, with determination of OD600 every 2 h. This procedure was repeated three times per group.
Detection of Extracellular Protease Activity
According to the previously published method (Windle and Kelleher, 1997) wild-type strain HY9901 and HY9901ΔtyeA were coated on TSA plates coated with sterile cellophane, and cultured at 28°C for 24 h, washed with sterile PBS, centrifuged at 4°C for 30 min, and the supernatant filtered to obtain extracellular products. Inactivated sample (supernatant was boiled for 10 min) was used as a blank control.
Swarming Motility
Single colonies of wild-type strain HY9901 and HY9901ΔtyeA were selected by sterile toothpick and plated on TSA plates with agar content of 0.5%. Each group was incubated for 24 h at 28°C. The diameter of swarming circle was measured by Vernier calipers.
Detection of Biofilm Formation Ability Using Crystal Violet Ammonium Oxalate
With reference to the method described previously (Katharine and Watnick, 2003), wild strains HY9901 and HY9901 ΔtyeA (OD600 = 0.5) were transferred to a 96-well plate. Each well was inoculated with 200 μL of bacterium with 6 replicates per sample, the negative control was an inoculum of TSB only, and the culture temperature was 28°C. Samples were taken from the 96 well plate at 12, 24, 48, and 72 h, methanol fixed for 20 min, stained with Crystal violet ammonium oxalate dye for 15 min, rinsed with water and dried. Finally, 95% alcohol was added and incubated at room temperature for 30 min. OD570 was determined by Multimode Plate Reader(PerkinElmer EnSpire, EnSpire, Singapore) (OD600 = 1, bacterial concentration = 1 × 109 cfu/mL).
Detection of Biofilm Formation Ability Using
Laser Scanning Confocal Microscope (LSCM)
The wild strain HY9901 and the mutant strain HY9901ΔtyeA (OD600 = 0.5) were diluted 50-fold, added to a glass bottom culture dishes (spec: type 28.2 mm, class diameter 20 mm) (Wuxi NEST, Wuxi, China) and statically cultured in a 28°C biochemical incubator for 24 h, gently washed three times with physiological saline, and then combined with 10% SYTO9 green. Bacteria were incubated with fluorescent dyes in the dark for 20 min, washed three times with saline, mounted in 40% saline-glycerol and observed by confocal microscopy (Zeiss, LSM710, Germany). The excitation wavelength was 488 nm, scanned from the bottom to the top of the biofilm, Z-section was 1 μm apart, and biofilm parameters—biomass and maximum thickness were determined. Three samples were made for each strain and the average amount calculated.
Dose Response Challenge Test (LD50)
The injection concentrations used for the dose response of wild-type strain HY9901 and ΔtyeA were 104, 105, 106, 107, and 108 CFU/mL. A total of 330 fish were randomly divided into three groups (Table 3). The water temperature was adjusted to 28°C. The experiment was repeated three times. Five microlitre of bacterial solution was injected into fish by intramuscular injection. The control group was injected with equivalent volumes of PBS. Fish were monitored for 14 days or until no morbidities occurred. Specific mortality was determined by streaking head-kidney onto TSA. The LD50 of the mutant and wild strains was calculated by the Koch method (Reed and Muench, 1938).
Table 3.
Experiment of LD50.
| Concentration (CFU/mL) | HY9901 | Death rate (%) | ΔtyeA | Death rate (%) | Control (PBS) | Death rate (%) |
|---|---|---|---|---|---|---|
| 108 | 10 × 3 | 90 | 10 x 3 | 66.7 | – | – |
| 107 | 10 × 3 | 80 | 10 x 3 | 40 | – | – |
| 106 | 10 × 3 | 60 | 10 x 3 | 10 | – | – |
| 105 | 10 × 3 | 20 | 10 x 3 | 0 | – | – |
| 104 | 10 × 3 | 20 | 10 x 3 | 0 | – | – |
| 0(PBS) | – | – | – | – | 10 × 3 | 0 |
Each experiment involved 110 zebrafish with 10 fish per group in three replicate tanks and the experiment was repeated three times (total fish = 330).
Antibiotic Susceptibility
The susceptibility patterns of the HY9901 and HY9901ΔtyeA strains to 30 different antibiotics were determined according to the disc diffusion method using TSA (Bauer et al., 1966), and the diameters of the inhibition zones were measured using Vernier calipers. Resistant, intermediate and susceptible phenotype determinations were based on previously published susceptibility paper guidelines (HANGWEI, S1100, China). The strains were inoculated onto TSA plates and allowed to absorb onto agar for 10 min, and antibiotic discs were added after 24 h of incubation at 28°C (Cai et al., 2016).
Expression Analysis of T3SS-Related Genes
T3SS secretion was induced by Dulbecco's Modified Eagle Medium (DMEM) media, and strains HY9901 and HY9901 ΔtyeA were cultured for 12 h. The primers for T3SS related genes are shown in Table 2 (All primers were designed within the current study). The genes in this study were hop (Pang et al., 2018), vopN, vscN (Yonghong et al., 2016), vscO (Zhou et al., 2013), vopS, vscL, and vscK (Nguyen et al., 2000). 16S rRNA is used as an internal reference. According to the experimental method of Li et al. (2016), RNA was extracted, synthetic cDNA and real-time PCR were used to analyze the expression of T3SS related genes.
Preparation of Vaccine and Vaccination
The ΔtyeA immune concentration of 105 CFU/mL was selected by LD50 experiments, which has no mortality to zebrafish (Table 3). The mutant ΔtyeA was cultured in a shaking flask at 28°C for 18 h, washed and suspended in sterile artificial seawater, and the concentration of the bacterial solution was adjusted to 105CFU mL−1 by spectrophotometer. The experimental fish were randomly divided into two groups with 80 fish in each group. The water temperature was adjusted to 28°C. Each fish was injected with 5 μL ΔtyeA bacterial solution (105 CFU mL−1) by intramuscular injection and control fish were injected with 5 μL PBS per fish.
Determination of Vaccine Efficacy
Fish were challenged 28 days post vaccination. Thirty fish were randomly selected from each group (in triplicate) and given intramuscular injection of 5 μL V. alginolyticus the wild-type HY9901 (1 × 108 CFU mL−1). The water temperature was adjusted to 28°C and the number of moribund/mortalities was recorded for 14 days. Calculation of the relative percentage survival was performed at the end of the experiment (RPS (%) = (1-immunized group mortality / control group mortality) × 100%).
Immune Gene Expression of Zebrafish Induced by HY9901ΔtyeA Vaccine
Liver and spleen samples were taken from three fish from each group, respectively, 1 day before challenge. Immune-related genes expression levels were detected with real-time qPCR. Primers for IL6, IL6R, IL-1β, IL-8, IgM, TNF-α, rag-1, TLR5, and c/ebpβ are shown in Table 2, and β-actin was used as internal reference. The procedures of RNA extraction, cDNA synthesis, real-time qPCR for analysis of immune gene expression were described by Li et al. (2016).
Ethics Statement
All animal experiments were conducted strictly based on the recommendations in the “Guide for the Care and Use of Laboratory Animals” set by the National Institutes of Health. The animal protocols were approved by the Animal Ethics Committee of Guangdong Ocean University (Zhanjiang, China).
Biosecurity
The bacteria protocols were approved by the Biosecurity Committee of Guang dong Ocean University (Zhanjiang, China).
Statistical Analysis
The experimental data were analyzed by single factor analysis of variance (ANOVA) with SPSS17.0 software. **indicates extremely significant difference compared with the control group (p < 0. 01). *indicates significant difference compared with the control group (p < 0.05).
Results
Cloning of tyeA Gene and Construction of Mutant
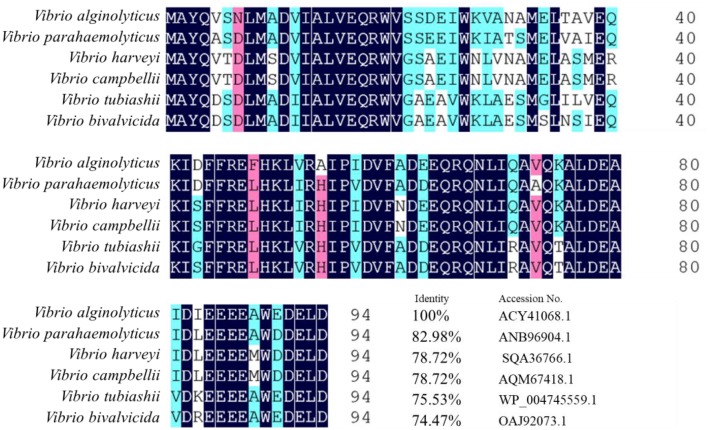
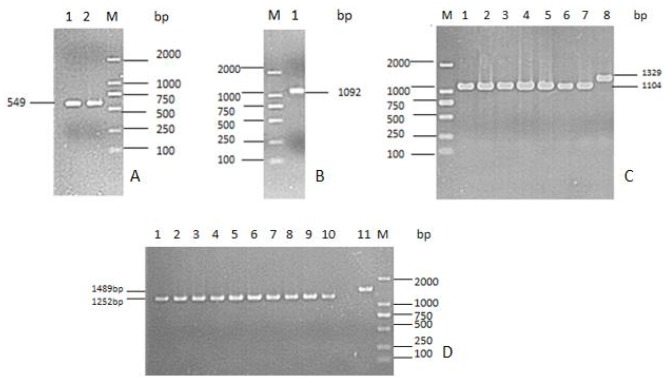
The tyeA gene consists of an open reading frame of 285 bp (Figure 1), encoding 95 amino acids (aa) with a predicted molecular weight of 10.98 kDa (tyeA accession no. MN328351). The deduced amino acid sequence analysis of TyeA show it has high homology of 75–83% with other Vibrio spp. (Figure 2), with the highest homology shared with V. parahaemolyticus (83%). An untagged in-frame deletion mutant HY9901ΔtyeA was constructed by over-lap PCR and forward and reverse screening methods. The mutant was confirmed by inabilityto grow on TSA supplemented with chloramphenicol, and verified by PCR by generating a fragment of ~1,092 bp. Using PCR the genome of HY9901ΔtyeA and wild-type strain HY9901 by primer tyeA-up/tyeA-down, a 1,252 bp fragment was obtained for HY9901ΔtyeA and fragment of 1,489 bp was obtained for HY9901 (Figure 3).
Figure 1.
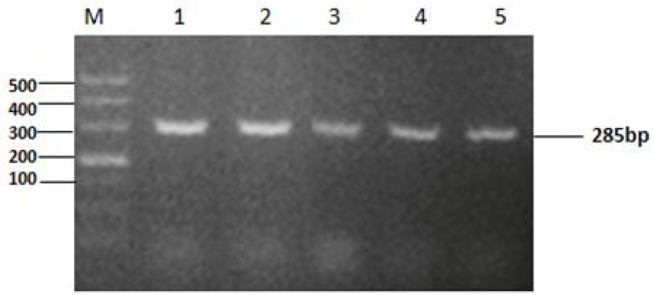
Cloning of tyeA gene. M: DL2000 marker. Lane 1-5:The 285bp fragment was amplified from genomic DNA of the wild-type strain HY9901 using primer pairs of tyeAP1-for/tyeAP1-rev (Lane 1, 2, 3, 4, 5:Tm = 56°C, 57°C, 58°C, 59°C, 60°C).
Figure 2.
Homology comparison of Vibrio parahaemolyticus HY9901 T3SS Regulator Protein TyeA. Vibrio alginolyticus T3SS regulatory protein TyeA Accession No.ACY41068.1; Vibrio parahaemolyticus T3SS regulatory protein TyeA Accession No.ANB96904.1; Vibrio harveyi T3SS regulatory protein TyeA Accession No.SQA36766.1; Vibrio campbellii T3SS regulatory protein TyeA Accession NO.AQM67418.1; Vibrio tubiashii T3SS regulatory protein TyeA Accession No.WP_004745559.1; Vibrio bivalvicida T3SS regulatory protein TyeA Accession No.OAJ92073.1.
Figure 3.
Construction and confirmation of the knockout mutant strain HY9901ΔtyeA. (A) M: DL2000 marker; Lane 1. The 549 bp upstream fragment amplified from genomic DNAs of the strain HY9901 using primer pairs of tyeA-for/tyeA-int-rev; Lane 2. The 543 bp downstream fragment amplified from genomic DNAs of the strain HY9901 using primer pairs of tyeA-int-for / tyeA-rev. (B) M: DL2000 marker; Lane 1. The 1,092 bp fragment amplified from genomic DNAs of HY9901ΔtyeA using primer pairs of tyeA-for/tyeA-rev. (C) M: DL2000 marker; Lane 1–7. The 1,104 bp fragment amplified from genomic DNAs of the HY9901ΔtyeA using primer pairs of tyeA-for/tyeA-rev; Lane 8. The 1,329 bp fragment amplified from genomic DNAs of the wild-type strain HY9901 using primer pairs of tyeA-for/tyeA-rev. (D) M: DL2000 marker; Lane 1–10. The 1,252 bp fragment amplified from genomic DNAs of HY9901ΔtyeA; Lane11. The 1,489 bp fragment amplified from genomic DNAs of the strain HY9901 using primer pairs of tyeA-up / tyeA-down.
Characterization of the tyeA
Genetic Stability of Mutants
After 30 generations of continuous passages of mutant ΔtyeA, the PCR genome of HY9901ΔtyeA and wild-type strain HY9901 using primers tyeA-up/tyeA-down, a fragment of 1,489 bp was obtained for HY9901, and fragment of 1,252 bp was obtained for HY9901ΔtyeA. This shows that the wild-type strain HY9901 has 237 bp more than the mutant ΔtyeA, and the mutant ΔtyeA has deleted the gene tyeA, which can stabilize the inheritance (Figure 4).
Figure 4.
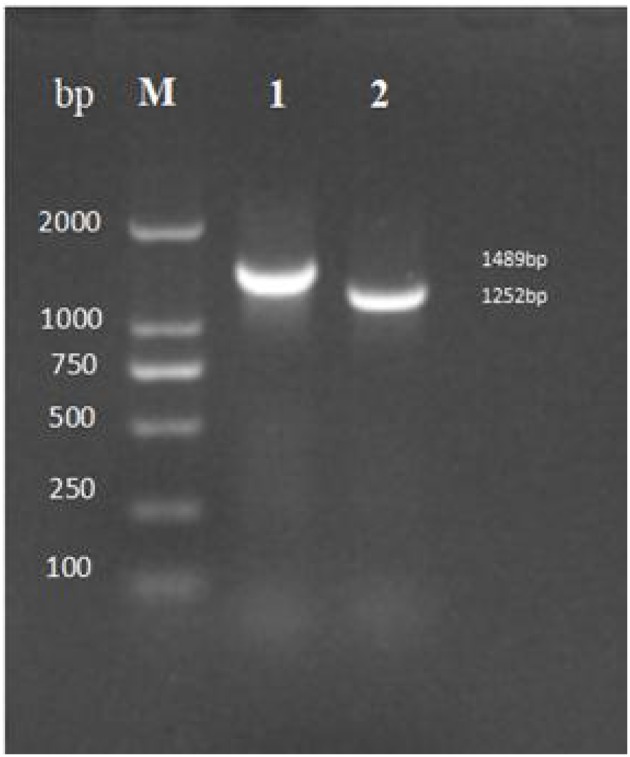
Genetic stability detection of HY9901ΔtyeA deletion mutant. M: DL2000 marker, Lane 1. A fragment of 1,489 bp is obtained for HY9901 using primer pairs of tyeA-up/tyeA-down. Lane 2. A fragment of 1,252 bp is obtained for HY9901ΔtyeA using primer pairs of tyeA-up/tyeA-down.
Comparison of Growth Rates of Wild Type and Mutant Bacteria
The growth rate of wild-type strain HY9901 and HY9901ΔtyeA was similar and not statistically significant (p > 0.05) (Figure 5), the deletion of tyeA gene has no effect on the growth of V. alginolyticus. The exponential growth phase of the two bacterial strains was from 0 to 4 h, with growth stationary at 18 h, OD600 ≈ 1.7.
Figure 5.
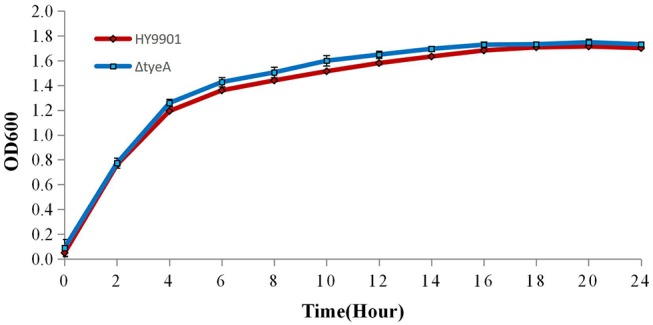
Growth rates of HY9901ΔtyeA and HY9901. Aliquots of cell culture were taken at various time points and measured for cell density at OD600.
Extracellular Protease Activity
Extracellular proteases, as metabolites of bacteria, play an important role as virulence factors in the process of infecting the host. Extracellular proteins have a variety of protease activities, including lecithin, amylase, lipase, and casein. The extracellular protease activity was not significantly different between HY9901ΔtyeA and the wild-type strain HY9901 (p > 0.05) (Table 4).
Table 4.
Comparison of biological characteristics between HY9901 and HY9901ΔtyeA.
| Characteristics | HY9901 | HY9901ΔtyeA |
|---|---|---|
| Activity of ECPase(A422)a | 1.03 ± 0.2 | 0.97 ± 0.2 |
| Biofilm thickness(μm)b | 60 ± 10 | 90 ± 20 |
| Swarming (mm)c | 34.9 ± 0.2 | 20.6 ± 0.4** |
| LD50(CFU mL−1)d | 5.8 × 105 | 2.6 × 107** |
Values are mean ± standard deviation for three trials. Significant differences between HY9901 and HY9901ΔtyeA indicated by asterisk.
p < 0.01.
Bacteria were incubated in TSB for 18 h at 28°C.
Bacteria were incubated in a glass bottom culture dish (NEST) for 24 h at 28°C.
Swarming diameters were measured after 24 h incubation on TSA containing 0.3% agar plates.
LD50were evaluated in healthy zebrafish with an average weight of 0.2 g.
Detection of Biofilm Formation Ability
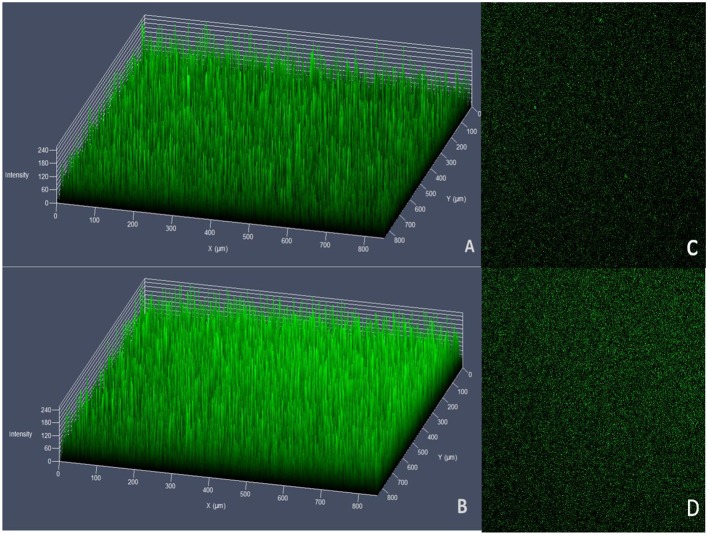
No significant differences were observed in biofilm formation at 24 h (p > 0.05) between wild strains and mutants of V. alginolyticus using the crystal violet ammonium oxalate assay (Figure 6) or by confocal microscopy (Figure 7 and Table 4). The biofilm fluctuates up and down within 72 h in crystal violet ammonium oxalate. The biofilm was thicker in HY9901 than in the mutant at 6 h (p < 0.05). After 12 h, the biofilm thickness was basically equal, and there was no significant change according to biological statistical analysis.
Figure 6.
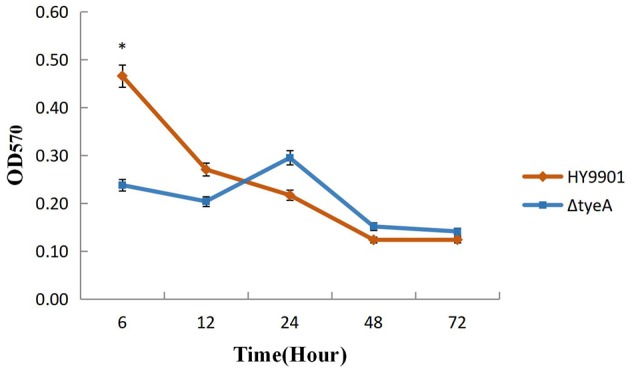
Measurement of biofilm by crystal violet ammonium oxalate. *indicates significant difference compared with the control group (p < 0.05). **indicates extremely significant difference compared with the control group (p < 0.01).
Figure 7.
Measurement of biofilm by LSCM. (A) HY9901 2.5d diagram. (B) HY9901ΔtyeA 2.5d diagram. (C) HY9901 2d diagram. (D) HY9901ΔtyeA 2d diagram; HY9901 Biofilm thickness:60 ± 10 μm, HY9901ΔtyeA Biofilm thickness:90 ± 20 μm.
Swarming Motility
The wild-type strain HY9901 and HY9901ΔtyeA were inoculated on the swarming plate, and the results were as follows: the swarming circle of wild strain was 34.9 ± 0.2 mm, mutant was 20.6 ± 0.4 mm (Table 4). The swarming circle diameter of HY9901ΔtyeA was significantly smaller than that of wild strain (p < 0.01), indicating that the swarming ability of HY9901ΔtyeA was significantly weakened.
LD50 Determination
Zebrafish from quarantined stocks recognized as disease free (Xu et al., 2010) were used as models (Sullivan and Kim, 2008) to assess the virulence of the strain HY9901 and HY9901ΔtyeA. The results showed that the 50% lethal dose of HY9901ΔtyeA was 40 times higher than that of wild strain (Tables 3, 4). The symptoms of body surface hyperemia, abdominal redness and swelling and slow swimming were observed in the diseased fish. The results showed that the virulence of HY9901 ΔtyeA was significantly decreased when compared with the wild strain (p < 0.01).
Antibiotic Susceptibility
Both wild and mutant strains were extremely susceptible to amikacin, minocycline, gentamicin, Cefperazone; resistant to oxacillin, clindamycin, ceftazidime, penicillin, ampicillin, caebenicillin, cefazolin, ceftriaxone, cephradine, piperacillin. Wild strains were sensitive to tetracycline, chloramphenicol, kanamycin, doxycycline, while mutant strains were resistant to them (Table 5).
Table 5.
Drug sensitivity test results of the HY9901 and HY9901ΔtyeA.
| Antibiotic | Dose (μg) | Bacteriostatic circle diameter (mm) | |||
|---|---|---|---|---|---|
| HY9901 | Sensitivity | ΔtyeA | Sensitivity | ||
| Cefperazone | 75 | 0 | R | 0 | R |
| Oxacillin | 1 | 0 | R | 0 | R |
| Clindamycin | 2 | 0 | R | 0 | R |
| Ceftazidime | 30 | 0 | R | 0 | R |
| Penicillin | 10U | 0 | R | 0 | R |
| Ampicillin | 100 | 0 | R | 0 | R |
| Caebenicillin | 100 | 0 | R | 0 | R |
| Cefazolin | 30 | 8.0 ± 0.2 | R | 0 | R |
| Ceftriaxone | 30 | 9.3 ± 0.3 | R | 0 | R |
| Cephradine | 30 | 0 | R | 0 | R |
| Piperacillin | 100 | 0 | R | 0 | R |
| Cefuroxime | 30 | 0 | R | 0 | R |
| SMZ/TMP | 23.75/1.25 | 0 | R | 0 | R |
| Aboren | 30 | 0 | R | 0 | R |
| Vancomycin | 30 | 0 | R | 0 | R |
| Cephalexin | 30 | 0 | R | 0 | R |
| polymxinB | 200IU | 0 | R | 0 | R |
| Norfloxacin | 10 | 0 | R | 0 | R |
| Ofloxacin | 5 | 0 | R | 0 | R |
| Ciprofloxacin | 5 | 0 | R | 0 | R |
| Amikacin | 30 | 13.3 ± 0.3 | I | 10.0 ± 0.1 | I |
| Minocyline | 30 | 18.5 ± 0.2 | S | 14.3 ± 0.3 | I |
| Tetracyline | 30 | 13.5 ± 0.2 | I | 12.5 ± 0.2 | R |
| Gentamicin | 10 | 14.5 ± 0.1 | I | 12.5 ± 0.2 | I |
| Furazolidone | 300 | 10.5 ± 0.4 | R | 8.0 ± 0.1 | R |
| Chloramphenicol | 30 | 17.2 ± 0.3 | S | 0 | R |
| kanamycin | 30 | 14.1 ± 0.2 | I | 12.1 ± 0.2 | R |
| Erythromycin | 15 | 10.1 ± 0.2 | R | 0 | R |
| Doxycycline | 30 | 16.5 ± 0.3 | S | 10.6 ± 0.1 | R |
| Cefperazone | 30 | 14.2 ± 0.3 | I | 13.3 ± 0.3 | I |
S(susceptible) I(intermediate) R(resistance).
T3SS-related Gene Expression Analysis
QRT-PCR was employed to analyze the transcription levels of T3SS-related genes including vscL, vscK, vopN, vscO, vscN, vopS, and hop. The results showed that compared with HY9901 wild type, ΔtyeA had significantly increased expression of vscL, vscK, vscO, vopS (p < 0.05), vopN, vscN and hop (p < 0.01) (Figure 8).
Figure 8.
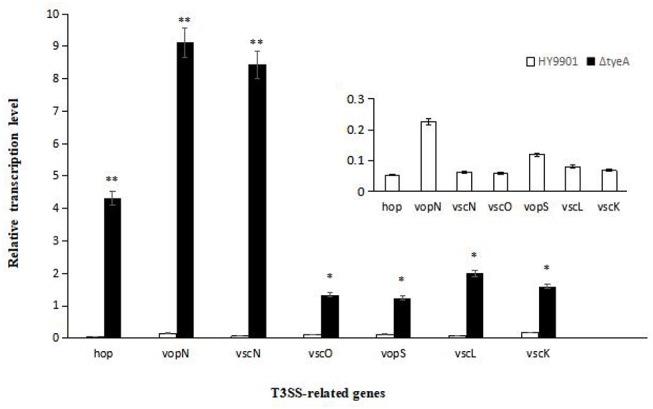
Expression of HY9901 and HY9901ΔtyeA T3SS-related genes induced by DMEM. ΔtyeA had significantly increased expression of vscL, vscK, vscO, vopS (p < 0.05), vopN, vscN, and hop (p < 0.01). * indicates significant difference compared with the control group (p < 0.05). ** indicates extremely significant difference compared with the control group (p < 0.01).
Vaccine Efficacy
Fish vaccinated with the mutant strain HY9901ΔtyeA were challenged 28 days post-vaccination with the wild strain. Within 14 days, the mortality rate in the control group injected with sterile PBS was 87%, the mortality rate in the injection immunization group was 25%, and the relative percentage survival was 71.2% (Figure 9).
Figure 9.
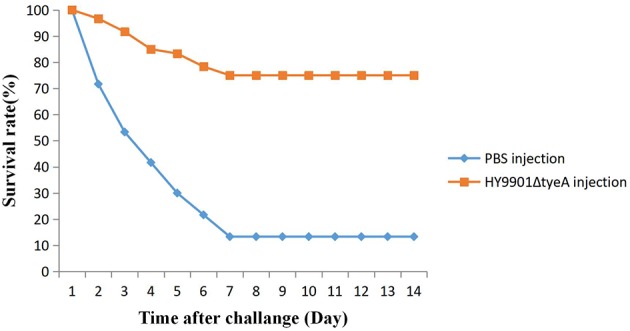
Survival in groups vaccinated with HY9901ΔtyeA and PBS following challenge with Vibrio alginolyticus HY9901.
Immune Gene Expression of Zebrafish Induced by HY9901ΔtyeA Vaccine
Detection of immune gene expression in zebrafish immunized with HY9901ΔtyeA live attenuated vaccine was assessed by qPCR to analyze the transcriptional levels of pro-inflammatory and immunoglobulin-related immune genes. The results showed that, the group vaccinated with the mutant strain HY9901ΔtyeA had significantly increased expression of IL6, IL6R, IL-1β, TNF-α, rag-1, TLR5, c/ebpβ genes in liver and IL6, IL8, IgM, TNF-α, rag-1 genes in spleen compared to control fish injected with PBS (p < 0.01) (Figure 10).
Figure 10.
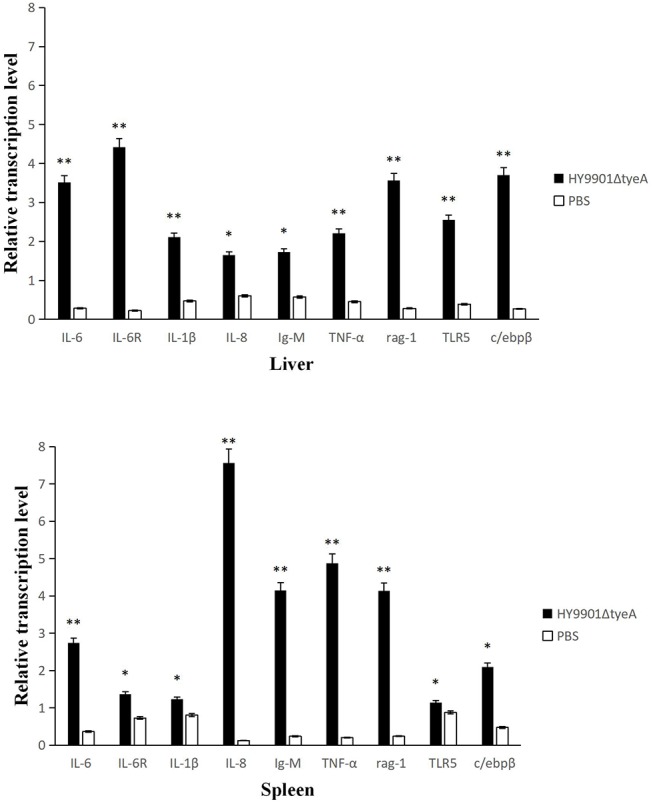
Comparative analysis of the expression of immune-related genes in liver and spleen of zebrafish given the live attenuated vaccine and unvaccinated zebrafish. The head kidney and spleen of grouper were sampled at 1 day before challenge, and the mRNA level of each immune-related gene was normalized to that of β-actin. Bars represent the mean relative expression of three biological replicates and error bars represent standard deviation.
Discussion
At present, the interaction mechanisms between some fish pathogens and hosts have been studied in depth. Based on these studies, the development of efficient new vaccines has become highly desirable in aquaculture (Pang et al., 2016). Attenuated live vaccines are one of the more efficient vaccines, especially compared to inactivated vaccines due to their greater activation of cellular immunity. Currently, the construction of attenuated strains by gene knockout is an important method to obtain candidate attenuated live vaccines.
In the current study, we knocked out the T3SS gene tyeA of V. alginolyticus, explored its biology and pathogenicity, and evaluated its effect as a live attenuated vaccine. Zhou et al. (2013) found that the mutant of T3SS chaperone escort vscO of V. alginolyticus showed an attenuated swarming ability and a 10-fold decrease in the virulence to fish. However, the ΔvscO mutant showed no difference in the biofilm formation and ECPase activity. The authors concluded that vscO is associated with the flagella of V. alginolyticus. The knockout of vscO gene results in the decline of group movement ability and thus reduced virulence (Zhou et al., 2013). Another mutant of V. alginolyticus, ΔsodB did not show any difference in growth when compared with wild type strains HY9901. However, the ΔsodB mutant resulted in the formation of biofilm, increased SOD activity and toxicity, increased ECPase activity and sensitivity to hydrogen peroxide, a decreased group movement ability and decreased adhesion to cytokine-induced killer (CIK) cells (Yanyan et al., 2019). Even though the mutant of T3SS effector Hop revealed an attenuated swarming phenotype and a 2,600-fold decrease in the virulence to grouper, the HY9901Δhop mutant showed no difference in morphology, growth, biofilm formation and ECPase activity (Pang et al., 2018). Similarly in the present study, there was no significant difference between HY9901ΔtyeA and wild type strains HY9901 in growth, biofilm formation and ECPase activity but the HY9901ΔtyeA mutant showed an attenuated swarming phenotype and a nearly 40-fold decrease in virulence to zebrafish. A polar flagellum and numerous lateral flagella contribute to the swimming and swarming motilities respectively, by which some bacteria access an appropriate niche inside the host after infection, as observed in V. parahaemolyticus (Enos-Berlage et al., 2005). Therefore, swarming ability is closely related to the virulence of some bacteria. Biofilm formation is a multicellular behavior through which bacteria colonize the surface of host tissues and tank surfaces and are thereby highly resistant to antibiotics and host immune defense mechanisms (Parsek and Singh, 2003; Verstraeten et al., 2008). Extracellular products (ECP) produced by bacteria include proteases, hemolysins and siderophores. As a metabolite of bacteria, extracellular proteases used as virulence factors by V. alginolyticus play an important role in the process of infection. Flagella assist in swimming and can help bacteria enter the appropriate niche in the host after Vibrio spp. infection. Motility is an important index used to judge the virulence of bacteria (Watnick et al., 2001). Numerous studies have shown that flagellin is essential for flagellum formation, trivial movement and symbiosis, and it also greatly affects the colonizing ability of Vibrio spp. (Millikan and Ruby, 2004). The results of this study showed that there was no significant difference in bacterial growth rate, biofilm, and extracellular enzyme activity between the tyeA knockout and the wild strain of V. alginolyticus, suggesting that TyeA may not affect these characteristics in V. alginolyticus. Nevertheless, this study found that HY9901ΔtyeA had significantly decreased swarming motility, which may be associated with bacterial flagellar movement. Therefore, the tyeA gene may have a positive regulation in the swarming motility of Vibrio alginolyticus, and its regulatory mechanism needs further investigation.
It is generally agreed that T3SS can be secreted by bacteria in vitro or by contacting host cells (Buttner and Bonas, 2002; Blondel Carlos et al., 2016). The secretory pathway of T3SS in vitro and in vivo is complex, sometimes the regulation is completed by a single regulatory protein, and sometimes the regulation is completed by multiple interacting regulatory proteins. For example, the transcriptional expression of the T3SS1 gene cluster of V. parahaemolyticus in DMEM can be regulated by the regulatory proteins ExsA and ExsD. Among them, ExsA is a positive transcriptional regulation protein and ExsD is a negative transcriptional regulation protein. It was hypothesize that ExsA might play a direct role in the T3SS1 promoter sequence (Zhou et al., 2008) based on the discovery that ExsA protein directly binds the upstream region of effector protein VP1668 and VP1687 in gel retardation assay (EMSA). Furthermore, the co-expression of recombinant proteins labeled with different antigens showed that: the combination of ExsD and ExsA blocks the expression of T3SS1; the combination of ExsC and ExsD releases ExsA, thus allowing the expression of T3SS1 (Zhou et al., 2010). According to Miller et al. (2016), in a bile salt environment, VttRA and VttRB of V.o cholerae can regulate the secretion of T3SS structural genes and some effector proteins. In terms of V. alginolyticus, ExsA and ExsC play a positive regulatory role on the effector proteins Va1686 and Va1687, ExsD, and ExsE play a negative regulatory role, ExsA and ExsC act as stimulus to the swarming of the side flagella of V. alginolyticus and on the contrary, ExsD and ExsE act as hindrance to the swarming of the side flagella (Liu et al., 2016). Interestingly, the results of fluorescence quantification and drug susceptibility tests in this study indicated that TyeA negatively regulated T3SS protein (vscL, vscK and vscO are apparatus proteins, while vopN, vscN, and hop are effector or regulatory proteins) and drug resistance genes. However, the systematic research of the network regulatory mechanism of various V. alginolyticus regulatory proteins on effector proteins in vitro and in vivo is still needed. TyeA, a regulator in T3SS, is widely studied in other strains (Sundberg and Forsberg, 2003; Schubot et al., 2004; Amer et al., 2016), and some progress has been made (Joseph and Plano, 2007; Plano and Schesser, 2013). However, its regulation mechanism in V. alginolyticus remains unknown. In order to further reveal the pathogenic mechanism of V. alginolyticus, so as to achieve the purpose of controlling vibriosis, the research on the regulatory mechanism of the regulatory gene tyeA in V. alginolyticus will be followed-up in future investigations.
Moreover, although zebrafish will not specifically be the target of the vaccine in the future, studies have shown that zebrafish are a good model for teleost immune responses (Streisinger et al., 1981). For example, Zhang et al. (2012) used zebrafish to investigate the immune response following administration of a live attenuated V. anguillarum vaccine. Hua and co-author's results showed that Th17 cells were activated following vaccination of zebrafish (Hua et al., 2013). At the same time, a large number of studies have shown that zebrafish can be infected by marine vibrios (O'toole et al., 2004; Rojo et al., 2007; Rodríguez et al., 2008). Therefore, it is feasible to use zebrafish to study the effectiveness of a marine Vibrio vaccine. In this study, genes associated with innate immunity or inflammatory factors: IL-1 β, IL6, IL8, and TNF- α were studied. Additionally the expression profiles of bacterial flagellum recognition factor TLR5, adaptive immune factors including rag-1, and immunoglobulin IgM were assessed, as well as blood corpuscle specific marker genes c/elbp. In summary, the results show that HY9901ΔtyeA can effectively induce the protective immune response of zebrafish associated with pro-inflammatory and immunoglobulin activity. V. alginolyticus has been resistant to most antibiotics according to the results of drug sensitivity testing in this study. Antibiotic resistance is closely linked to antibiotic abuse (Chee-Sanford et al., 2001; Burridge et al., 2010). In the future, the direction for the prevention and control of aquatic diseases such as Vibriosis is to develop Vibrio vaccine and thereby reduce the use of antibiotics, which has great significance for protecting the environment and developing aquaculture sustainably. In the current study, the relative percentage survival rate of zebrafish immunized with the vaccine candidate strain ΔtyeA was 71.2%, which indicated that the construction of the deletion strain could be used as one of the effective ways to develop a vaccine against this pathogen. It provides an experimental basis for the prevention and control of Vibrio spp. in aquaculture. Studies have shown that compared with inactivated vaccines, live attenuated vaccines have the advantages of long-lasting efficacy and inducing strong cellular immunity, which is in line with the current needs of aquaculture, however their use is not permitted under current regulations in Europe.
Conclusion
To sum up, we have successfully constructed the deletion strain of HY9901ΔtyeA. We found that the HY9901ΔtyeA strain exhibited decreased virulence to zebrafish, but the wild strain and the mutant strains were resistant to most antibiotics. The attenuated live vaccine was highly protective against V. alginolyticus and can induce protective immune responses in zebrafish. These results provide further evidence for the importance of T3SS in V. alginolyticus and provide reference for further study of this virulence factor. The immune effect and safety of the attenuated live vaccine will be tested and evaluated in aquaculture species in order to provide further theoretical basis and technical support for its application for the prevention and control of fish diseases caused by V. alginolyticus.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Accession No. MN328351.
Ethics Statement
The animal study was reviewed and approved by Guangli Li and Guangdong Ocean University of ethics committee.
Author Contributions
SZ and HP designed the experiments. SZ, XT, and JL generated experimental data and wrote the manuscript. HP, RH, SM, and JJ conceived the work and critically reviewed the manuscript. All authors contributed extensively to the work presented in this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was funded by Shenzhen Science and Technology Project (JCYJ20170818111629778), the National Key Research and Development Program of China (2018YFD0900501), National Natural Science Foundation of China (No. 31402344, 31670129), Natural Science Foundation of Guangdong Province (No. 2017A030313174), Independent Project of Southern Marine Science and Engineering Guangdong Laboratory (Zhanjiang) (No. ZJW-2019-05), projects subsidized by special funds for science technology innovation and industrial development of Shenzhen Dapeng New District (No. PT202001-13).
References
- Amer A. A., Gurung Jyoti M., Costa Tiago R. D., Kristina R., Zavialov A. V., Forsberg A., et al. (2016). TyeA hydrophobic contacts required for regulating ysc-yop type, III secretion activity by yersinia pseudotuberculosis. Front. Cell. Infect. Microbiol. 6:66. 10.3389/fcimb.2016.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S., Tomoko K., Galán J. E. (2003). Synthesis and localization of the Salmonella SPI-1 type III secretion needle complex proteins PrgI and PrgJ. J. Bacteriol. 185, 3480–3883. 10.1128/JB.185.11.3480-3483.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45, 493–496. 10.1093/ajcp/45.4_ts.493 [DOI] [PubMed] [Google Scholar]
- Ben Kahla-Nakbi A., Chaieb K., Bakhrouf A. (2009). Investigation of several virulence properties among Vibrio alginolyticus strains isolated from diseased cultured fish in Tunisia. Dis. Aquat. Organ. 86, 21–28. 10.3354/dao02091 [DOI] [PubMed] [Google Scholar]
- Blondel Carlos J., Park Joseph S., Hubbard Troy P., Pacheco Alline R., Kuehl Carole J., Walsh Michael J., et al. (2016). CRISPR/Cas9 screens reveal requirements for host cell sulfation and fucosylation in bacterial type III secretion system-mediated cytotoxicity. Cell Host Microbe 20, 226–237. 10.1016/j.chom.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge L., Weis J. S., Cabello F., Pizarro J., Bostick K. (2010). Chemical use in salmon aquaculture: a review of current practices and possible environmental effects. Aquaculture 306, 7–23. 10.1016/j.aquaculture.2010.05.020 [DOI] [Google Scholar]
- Buttner D., Bonas U. (2002). Getting across–bacterial type III effector proteins on their way to the plant cell. EMBO J. 21, 5313–5322. 10.1093/emboj/cdf536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S. H., Wu Z. H., Jian J. C., Lu Y. S. (2007). Cloning and expression of gene encoding the thermostable direct hemolysin from Vibrio alginolyticus strain HY9901, the causative agent of vibriosis of crimson snapper (Lutjanus erythopterus). J. Appl. Microbiol. 103, 289–296. 10.1111/j.1365-2672.2006.03250.x [DOI] [PubMed] [Google Scholar]
- Cai X. H., Peng Y. H., Wang Z. C., Huang T., Xiong X. Y., Huang Y. C., et al. (2016). Characterization and identification of streptococci from golden pompano in China. Dis. Aquat. Organ. 119, 207–217. 10.3354/dao02998 [DOI] [PubMed] [Google Scholar]
- Chang C. C., Yeh M. S., Lin H. K., Cheng W. (2008). The effect of Vibrio alginolyticus infection on caspase-3 expression and activity in white shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 25, 672–678. 10.1016/j.fsi.2008.09.004 [DOI] [PubMed] [Google Scholar]
- Chee-Sanford J. C., Aminov R. I., Krapac I. J., Garrigues-Jeanjean N., Mackie R. I. (2001). Occurrence and diversity of tetracycline resistance genes in lagoons and groundwater underlying two swine production facilities. Appl. Environ. Microbiol. 67, 1494–1502. 10.1128/AEM.67.4.1494-1502.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L. W., Schneewind O. (2000). Yersinia enterocolitica TyeA, an intracellular regulator of the type III machinery, is required for specific targeting of YopE, YopH, YopM, and YopN into the cytosol of eukaryotic cells. J. Bacteriol. 182, 3183–3190. 10.1128/JB.182.11.3183-3190.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorothea R. A., Enrica B., Selma M., Da-Kang S., Xia L., Maria P., et al. (2017). Steps for shigella gatekeeper protein mxic function in hierarchical type III secretion regulation. J. Biol. Chem. 292, 1705–1723. 10.1074/jbc.M116.746826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R. A., Keller L. H., Schifferli D. M. (1998). Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207, 149–57. 10.1016/S0378-1119(97)00619-7 [DOI] [PubMed] [Google Scholar]
- Enos-Berlage J. L., Guvener Z. T., Keenan C. E., Mccarter L. L. (2005). Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol. Microbiol. 55, 1160–1182. 10.1111/j.1365-2958.2004.04453.x [DOI] [PubMed] [Google Scholar]
- Ferracci F., Schubot F. D., Waugh D. S., Plano G. V. (2005). Selection and characterization of Yersinia pestis YopN mutants that constitutively block Yop secretion. Mol. Microbiol. 57, 970–987. 10.1111/j.1365-2958.2005.04738.x [DOI] [PubMed] [Google Scholar]
- Hua Z., Chao F., Haizhen W., Minjun Y., Qin L., Qiyao W., et al. (2013). Transcriptome profiling reveals Th17-like immune responses induced in zebrafish bath-vaccinated with a live attenuated Vibrio anguillarum. PLoS ONE 8:e73871 10.1371/journal.pone.0073871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriarte M., Sory M. P., Boland A., Boyd A. P., Mills S. D., Lambermont I., et al. (1998). TyeA, a protein involved in control of Yop release and in translocation of Yersinia Yop effectors. EMBO J. 17, 1907–18. 10.1093/emboj/17.7.1907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones E. H., Feldman K. A., Palmer A., Butler E., Blythe D., Mitchell C. S. (2013). Vibrio infections and surveillance in Maryland, 2002-2008. Public Health Rep. 128, 537–45. 10.1177/003335491312800613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph S. S, Plano G. V. (2007). Identification of TyeA residues required to interact with YopN and to regulate Yop secretion. Adv. Exp. Med. Biol. 603, 235–345. 10.1007/978-0-387-72124-8_21 [DOI] [PubMed] [Google Scholar]
- Jun W., Yu-Hong S., Xue-Heng Z., Chang-Hong L., Ming-Yun L., Jiong C. (2015). Molecular characterization of an IL-1β gene from the large yellow croaker (Larimichthys crocea) and its effect on fish defense against Vibrio alginolyticus infection. Dongwuxue Yanjiu. 36, 133–141. 10.13918/j.issn.2095-8137.2015.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katharine K., Watnick I. P. (2003). Environmental determinants of Vibrio cholerae biofilm development. Appl. Environ. Microbiol. 69, 5079–88. 10.1128/AEM.69.9.5079-5088.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khavong P., Lorena N. (2016). Yersinia type III effectors perturb host innate immune responses. World J. Biol. Chem. 7, 1–13. 10.4331/wjbc.v7.i1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. X., Yao Z. J., Sun L. N., Hu W. J., Cao J. J., Lin W. X., et al. (2016). Proteomics analysis reveals a potential antibiotic cocktail therapy strategy for aeromonas hydrophila infection in biofilm. J. Proteome Res. 15, 1810–1820. 10.1021/acs.jproteome.5b01127 [DOI] [PubMed] [Google Scholar]
- Li X. C., Xiang Z. Y., Xu X. M., Yan W. H., Ma J. M. (2009). Endophthalmitis caused by vibrio alginolyticus. J. Clin. Microbiol. 47, 3379–3381. 10.1128/JCM.00722-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Lu S. Y., Orfe L. H., Ren C. H., Hu C. Q., Call D. R., et al. (2016). ExsE is a negative regulator for T3SS gene expression in vibrio alginolyticus. Front. Cell Infect. Microbiol. 6:177. 10.3389/fcimb.2016.00177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Argudo I., Blocker A. J. (2010). The Shigella T3SS needle transmits a signal for MxiC release, which controls secretion of effectors. Mol. Microbiol. 78, 1365–1378. 10.1111/j.1365-2958.2010.07413.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masato S., Olga D., Mekalanos J. J. (2014). Vibrio cholerae T3SS effector VopE modulates mitochondrial dynamics and innate immune signaling by targeting Miro GTPases. Cell Host Microbe 16, 581–591. 10.1016/j.chom.2014.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. A., Sofia M. K., Weaver J. W. A., Seward C. H., Dziejman M. (2016). Regulation by ToxR-like proteins converges on vttRB expression to control type 3 secretion system-dependent Caco2-BBE cytotoxicity in vibrio cholerae. J. Bacteriol. 198, 1675–1682. 10.1128/JB.00130-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millikan D. S., Ruby E. G. (2004). Vibrio fischeri flagellin a is essential for normal motility and for symbiotic competence during initial squid light organ colonization. J. Bacteriol. 186, 4315–4325. 10.1128/JB.186.13.4315-4325.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L., Paulsen I. T., Tchieu J., Hueck C. J., Saier M. H. (2000). Phylogenetic analyses of the constituents of Type III protein secretion systems. J. Mol. Microbiol. Biotechnol. 2, 125–44. [PubMed] [Google Scholar]
- O'toole R., Hofsten J. V., Rosqvist R., Olsson P.-E., Wolf-Watz H. (2004). Visualisation of zebrafish infection by GFP-labelled Vibrio anguillarum. Microbial. Pathog. 37, 41–6. 10.1016/j.micpath.2004.03.001 [DOI] [PubMed] [Google Scholar]
- Pallen M. J., Beatson S. A., Bailey C. M. (2005). Bioinformatics analysis of the locus for enterocyte effacement provides novel insights into type-III secretion. BMC Microbiol. 5:9. 10.1186/1471-2180-5-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang H., Chen L., Hoare R., Huang Y., Zaohe W., Jian J. (2016). Identification of DLD, by immunoproteomic analysis and evaluation as a potential vaccine antigen against three Vibrio species in Epinephelus coioides. Vaccine 34, 1225–31. 10.1016/j.vaccine.2015.11.001 [DOI] [PubMed] [Google Scholar]
- Pang H. Y., Qiu M. S., Zhao J. M., Hoare R., Monaghan S. J., Song D. W., et al. (2018). Construction of a Vibrio alginolyticus hopPmaJ (hop) mutant and evaluation of its potential as a live attenuated vaccine in orange-spotted grouper (Epinephelus coioides). Fish Shellfish Immunol. 76, 93–100. 10.1016/j.fsi.2018.02.0 [DOI] [PubMed] [Google Scholar]
- Parsek M. R., Singh P. K. (2003). Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57, 677–701. 10.1146/annurev.micro.57.030502.090720 [DOI] [PubMed] [Google Scholar]
- Plano G. V., Schesser K. (2013). The Yersinia pestis type III secretion system: expression, assembly and role in the evasion of host defenses. Immunol. Res. 57, 237–245. 10.1007/s12026-013-8454-3 [DOI] [PubMed] [Google Scholar]
- Reed L. J., Muench H. (1938). A simple method of estimating fifty per cent endpoints12. Am. J. Epidemiol. 27, 493–497. 10.1093/oxfordjournals.aje.a118408 [DOI] [Google Scholar]
- Rodríguez I., Novoa B., Figueras A. (2008). Immune response of zebrafish (Danio rerio) against a newly isolated bacterial pathogen Aeromonas hydrophila. Fish Shellfish Immunol. 25, 239–49. 10.1016/j.fsi.2008.05.002 [DOI] [PubMed] [Google Scholar]
- Rojo I., De Ilarduya O. M., Estonba A., Pardo M. A. (2007). Innate immune gene expression in individual zebrafish after listonella anguillarum inoculation. Fish Shellfish Immunol. 23, 1285–1293. 10.1016/j.fsi.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Rubirés X., Saigi F., Piqué N, Climent N., Merino S., Albertí S., et al. (1997). A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J. Bacteriol. 179, 7581–7586. 10.1128/JB.179.23.7581-7586.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadok K., Mejdi S., Nourhen S., Amina B. (2013). Phenotypic characterization and RAPD fingerprinting of Vibrio parahaemolyticus and Vibrio alginolyticus isolated during Tunisian fish farm outbreaks. Folia Microbiol. 58, 17–26. 10.1007/s12223-012-0174-x [DOI] [PubMed] [Google Scholar]
- Schubot F. D., Jackson M. W., Penrose K. J., Cherry S., Tropea J. E., Plano G. V., et al. (2004). Three-dimensional structure of a macromolecular assembly that regulates type III secretion in yersinia pestis. J. Mol. Biol. 346, 1147–1161. 10.1016/j.jmb.2004.12.036 [DOI] [PubMed] [Google Scholar]
- Sganga G., Cozza V., Spanu T., Spada P. L., Fadda G. (2009). Global climate change and wound care: case study of an off-season Vibrio alginolyticus infection in a healthy man. Ostomy Wound Manage. 55, 60–62. [PubMed] [Google Scholar]
- Simon R., Priefer U., Pühler A. (2019). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotech. 1, 784–791. 10.1038/nbt1183-784 [DOI] [Google Scholar]
- Streisinger G., Walker C., Dower N., Knauber D., Singer F. (1981). Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 291, 293–296. 10.1038/291293a0 [DOI] [PubMed] [Google Scholar]
- Sullivan C., Kim C. H. (2008). Zebrafish as a model for infectious disease and immune function. Fish Shellfish Immunol. 25, 341–350. 10.1016/j.fsi.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Sundberg L., Forsberg A. (2003). TyeA of Yersinia pseudotuberculosis is involved in regulation of Yop expression and is required for polarized translocation of Yop effectors. Cell. Microbiol. 5, 187–202. 10.1046/j.1462-5822.2003.00267.x [DOI] [PubMed] [Google Scholar]
- Thompson F. L., Iida T., Swings J. (2004). Biodiversity of vibrios. Microbiol. Mol. Biol. Rev. 68, 403–431. 10.1128/MMBR.68.3.403-431.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten N., Braeken K., Debkumari B., Fauvart M., Fransaer J., Vermant J., et al. (2008). Living on a surface: swarming and biofilm formation. Trends Microbiol. 16, 496–506. 10.1016/j.tim.2008.07.004 [DOI] [PubMed] [Google Scholar]
- Watnick P. I., Lauriano C. M., Klose K. E., Croal L., Kolter R. (2001). The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39, 223–35. 10.1046/j.1365-2958.2001.02195.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windle H. J., Kelleher D. (1997). Identification and characterization of a metalloprotease activity from Helicobacter pylori. Infect. Immun. 65, 3132–3137. 10.1128/IAI.65.8.3132-3137.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Wang Y., Han Y., Chen J., Zhang X.-H. (2010). Mutation of a novel virulence-related gene mltD in Vibrio anguillarum enhances lethality in zebra fish. Res. Microbiol. 162, 144–150. 10.1016/j.resmic.2010.08.003 [DOI] [PubMed] [Google Scholar]
- Yanyan C., Fenglei W., Huanying P., Jufen T., Shuanghu C., Jichang J. (2019). Superoxide dismutase B (sodB), an important virulence factor of Vibrio alginolyticus, contributes to antioxidative stress and its potential application for live attenuated vaccine. Fish Shellfish Immunol. 89, 354–360. 10.1016/j.fsi.2019.03.061 [DOI] [PubMed] [Google Scholar]
- Yonghong Y., Huanying P., Zejun Z., Dawei S., Yu D., Jichang J., et al. (2016). Construction of live attenuated vaccine against Vibrio alginolyticus and the evaluation on its immunoprotection effect. Anim. Husbandry Feed Sci. 8, 85–93. 10.19578/j.cnki.ahfs.2016.02.007 [DOI] [Google Scholar]
- Zhang Z., Wu H., Xiao J., Wang Q., Liu Q., Zhang Y. (2012). Immune responses of zebrafish (Danio rerio) induced by bath-vaccination with a live attenuated Vibrio anguillarum vaccine candidate. Fish Shellfish Immunol. 33, 36–41. 10.1016/j.fsi.2012.03.031 [DOI] [PubMed] [Google Scholar]
- Zhou X., Konkel M. E., Call D. R. (2010). Regulation of type III secretion system 1 gene expression in Vibrio parahaemolyticus is dependent on interactions between ExsA, ExsC, and ExsD. Virulence 1, 260–272. 10.4161/viru.1.4.12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Shah D. H., Konkel M. E., Call D. R. (2008). Type III secretion system 1 genes in Vibrio parahaemolyticus are positively regulated by ExsA and negatively regulated by ExsD. Mol. Microbiol. 69, 747–764. 10.1111/j.1365-2958.2008.06326.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Pang H., Ding Y., Cai J., Huang Y., Jian J., et al. (2013). VscO, a putative T3SS chaperone escort of Vibrio alginolyticus, contributes to virulence in fish and is a target for vaccine development. Fish Shellfish Immunol. 35, 1523–31. 10.1016/j.fsi.2013.08.017 [DOI] [PubMed] [Google Scholar]
- Zujie Y., Zhuang G., Yuqian W., Wanxin L., Yuying F., Yuexu L., et al. (2019). Integrated succinylome and metabolomics profiling reveals crucial role of S-ribosylhomocysteine lyase in quorum sensing and metabolism of Aeromonas hydrophila. Mol. Cell. Proteomics 18, 200–215. 10.1074/mcp.RA118.001035 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated for this study can be found in the NCBI Accession No. MN328351.