Abstract
The development of bacterial resistant strains is a global health concern. Designing antibiotics that limit the rise of pathogenic resistance is essential to efficiently treat pathogenic infections. Self-assembling amphiphilic molecules are an intriguing platform for the treatment of pathogens due to their ability to disrupt bacterial membranes and function as drug nanocarriers. We have designed cationic peptide amphiphiles (PAs) that can form micelles, nanofibers, and twisted ribbons with the aim of understanding antimicrobial activity at the supramolecular level. We have found that micelle-forming PAs possess excellent antimicrobial activity against various Gram-positive and Gram-negative pathogens, such as methicillin-resistant S. aureus and multidrug resistant K. pneumoniae with MICs ranging between 1–8 µg/mL when compared to nanofibers with MICs >32 µg/mL. The data suggest the antimicrobial activity of the PAs depends on their morphology, amino-acids sequence, the length of the alkyl tail, and the overall hydrophobicity of the PA. Scanning electron microscopy, confocal microscopy, and flow cytometry studies using MRSA and E. coli K12 strains showed that PAs increase cell membrane permeability, and disrupt the integrity of the pathogen’s membrane, leading to cell lysis and death. PAs are a promising platform to develop new antimicrobials that could work as nanocarriers to develop synergistic antibacterial therapies.
Keywords: peptide amphiphiles, cationic nanostructures, micelles, antimicrobials, supramolecular structure-activity relationships, self-assembly
Graphical Abstract
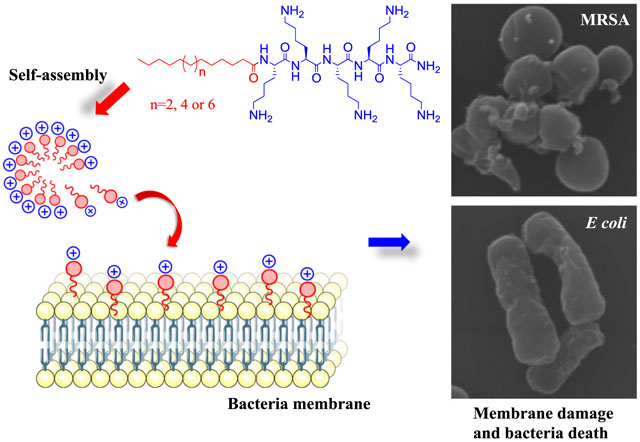
1. Introduction
The efficient treatment of infections has been aggravated by the unceasing proliferation of pathogenic resistance. The “nightmare bacteria” is a great health concern, and a cause of social issues for the general population.1–4 For example, drug resistant pathogens.1, 5 such as Staphylococci, Enterococci, Klebsiella pneumoniae, and Pseudomonas spp lead to prolonged illness and risk of death.6 In fact, in the United States, around 2 million people are infected each year by resistant pathogens causing over 20,000 annual deaths.3 These “super bugs” are a result of intrinsic resistance to antibiotics, and they acquire resistance via genetic mutations and gene transfer due to the overuse of current antibiotics in agriculture, human, and veterinary medicine.2, 7 Therefore, it is important to find new antimicrobial agents that are not affected by traditional mechanisms of bacterial resistance is essential.8
Antimicrobial peptides (AMPs) are endogenous molecules found in the innate immune system of humans and other organisms.5, 9 Unlike conventional antibiotics that target essential biosynthetic pathways, AMPs provide microbicidal activity by a mechanism of action associated with physical disruption of the pathogen’s membrane, which leads to cell death by leakage of cytoplasmic components.10, 11 The development of resistance against AMPs is greatly reduced,12, 13 because it is metabolically costly for the bacteria to mutate or repair membrane’s molecular constituents.14 Nevertheless, AMP-based antimicrobial agents usually suffer drawbacks, such as limited bioavailability and a short half-life due to proteolytic degradation. Self-assembling peptide amphiphiles (PAs) can mimic AMP activity while providing more resistance to proteolytic hydrolysis due to the formation of nanostructures (which may limit enzyme accessibility) and the acylation of their N-terminus.15 PA-based materials are also biocompatible, biodegradable, and possess diverse biological applications.16, 17 For example, Niu and co-workers reported lipopeptides with a broad-spectrum of activity against dangerous pathogens such as; multi-drug resistant S. epidermidis, MRSA, Vancomycin-resistant E. faecalis, and multi-drug resistant (MDR) P. aeruginosa.15 The authors postulated the length of the alkyl-chain tail to be key for the interaction with the membrane and found that unsaturated alkyl tails can decrease hemolytic activity, enhance selectivity, and increase resistance to proteolytic degradation.15 Shankar et al.9 reported the antimicrobial activity of lipopeptides assembled into nanofibers and nanoneedles, and hypothesized that different shapes may influence their biological activities. The authors reported that needle-shaped assemblies have high antimicrobial and hemolytic activity, while nanofibers displayed less hemolytic activity and toxicity.9
Other studies have indicated that the shape of the nanostructure plays an important role in antimicrobial activity suggesting that spherical nanostructures are more effective antibacterials than rod-like systems.9, 12, 18 Intrigued by those precedents, we sought to understand how the shape affects the antibacterial activity of PAs. We designed a series of cationic PAs of different morphologies and investigated the relationship between the shape, charge, and antimicrobial activity. In this report, we demonstrated that PA micelles are more effective antibacterials than nanofibers, showing a broad-spectrum of activity towards various bacteria and fungi, including clinically relevant pathogens such as: MRSA, K. pneumoniae, C. albicans, and C. neoformans. The observed supramolecular structure-activity relationships (SSAR) correlated with parameters such as charge and hydrophobicity with antimicrobial activity and cytotoxicity. Self-assembling PAs that damage the bacterial membrane show strong potential as antimicrobials and may be combined with traditional antibiotics to develop therapies with synergistic antibacterial action.
2. Experimental section
Materials.
Dichloromethane (DCM), N,N-dimethylformamide (DMF), and Octadecanoic acid were obtained from Fisher Chemical. The amino acids Fmoc-Lys(Boc)-OH and Fmoc-Orn(Boc)-OH were purchased from Creosalus (Louisville, KY), Fmoc-amino acids and N,N,N′,N′-Tetramethyl-O-(1H-benzotriazol-1-yl)uronium hexafluorophosphate (HBTU) were purchased from AAPPTec, P3 Biosystems or Novabiochem. Anthracene-9-carboxylic acid, Hexadecanoic and 2-n-Hexyldecanoic acid were obtained from Alfa Ceasar. Rink amide resin was purchased from CreoSaulus (Louisville, KY). N,N-diisopropylethylamine (DIPEA) was purchased from Oakwood Chemical and 4-methyl piperidine from Acros Organic. DMEM – high glucose (Hylcone), and Cosmic Calf Serum were obtained from Thermo Scientific, fetal bovine serum, 1% penicillin/streptomycin, and Medium 199 were obtained from Gibco Invitrogen. Hydrocortisone, Sodium Selenite, Apo-Transferrin bovine, Insulin, and Gentamicin were purchased from Sigma Aldrich and Recombinant human EGF from R&D systems. The 96-well flat-bottomed polystyrene microtiter plates (Nunclon Delta Surface) were from Thermo Scientific).
General Procedure for Synthesis and Characterization.
PAs were synthesized manually or using a Focus XC peptide synthesizer (AAPPTec) with standard Fmoc solid-phase peptide chemistry on a 0.28 mmol scale on Rink amide resin following literature reports.19–21 All coupling and deprotection steps were done on the Focus XC synthesizer using default settings. Mass spectrometry, to identify the PAs, was done in a SIEX 4800 MALDI TOF/TOF analyzer (Ontario, Canada). The products were purified using a preparative scale reverse phase HPLC (Agilent) with a C18 column as stationary phase of 5μm, 100 Å pore size and 150 × 21.1 mm (Phonomenex) with a gradient of ACN/H2O (containing 0.1 % of TFA). The eluted fractions containing the desired products and with a purity >95% were collected, while the organic solvent was removed. The pH of the water solution was adjusted to 7 before lyophilizing. The purity of the new PAs was confirmed by an analytical HPLC instrument using a C18 column at wavelength of 220 nm with a linear gradient of Acetronitrile (0.1% TFA) and water (0.1% TFA) from 5% to 95% for 30 min.
Self-Assembly Protocol.19, 21
All PA samples were self-assembled in water at 1 mg/mL and the pH adjusted to 7. The solutions were maintained at room temperature for 24 hours before testing. Additionally, PA 4 and PA 7 were self-assembled under various conditions and MIC was determined. Aged samples were prepared 3 days before testing and were self-assembled in water at a 2 mg/mL concentration. For annealing, PAs were self-assembled in water at 2 mg/mL, heated at 80°C for 3 hours and then cooled at room temperature. For HIFP, solutions were prepared at 2 mg/mL concentration in HFIP. HFIP was evaporated and PAs were reconstituted in water and the pH was adjusted to 7. Solutions were maintained at room temperature 24 hr before testing. In order to exchange TFA with HCl the PAs were dissolved in HCl (0.1 M) and were evaporated to remove TFA and then lyophilized. Solutions were prepared at 2 mg/mL, the pH was adjusted to 7, and solutions were maintained at room temperature for 24 hours before testing.
Circular Dichroism (CD).19, 21
The PAs were dissolved in MMilli-Q water (Milli-Q Integral water purification system, Billerica, MA) to 2 mM at pH 7. CD studies were done in a J-815 Jasco Circular Dichroism Spectrometer (Easton, MD) with a 0.1 mm quartz cuvette at 25 °C. The wavelengths were set at 190–300 nm with a scan speed of 100 nm/min. The response time was set at 2 sec, and the bandwidth was set to 1 nm. The water was used as a reference (blank) and five scans were recorded for each PA.
Transmission Electron Microscopy (TEM) Imaging.20, 21
The PAs were dissolved in ultrapure water to give a final concentration of 2 mM and aged for 24 hr before the experiments. Approximately 10 μL of sample was applied into a copper grid and allowed to absorb for 2–5 min, covered with a folded piece of filter paper like a tent. The excess PA was removed from the grid by inverting the forceps and touching only the edge of the grid to a clean piece of filter paper. Then, 1 drop of the negative stain (NanoVan) was added for 2 min. The excess stain was blotted off as before, and the PA was visualized at several magnifications.
Antibacterial Assay.22
Staphylococcus aureus USA 300 JE2, Staphylococcus aureus USA300 LAC, Escherichia coli K12 were provided by Dr. Bayles’ research lab and Pseudomonas aeruginosa PA01 was provided by Dr. Gus Wang’s lab (both at the Departament of Pathologhy and Microbiology of the University of Nebraska Medical Center – UNMC). The MICs of the PAs were studied using the broth microdilution method. Bacterial cultures were made by the direct colony suspension method to 1.5 × 108 colony forming unit CFU/ml and dilute for 2 mL into 40 mL of Muller Hinton Broth (MHB) to a final concentration of ~105 CFU/mL. A stock solution of each PAs was prepared in ultrapure water at 1 mg/ml concentration and pH was adjusted to 7. Then, serial dilutions were made in MHB, in Cellstar 96-well microtiter plates (Greiner, Bio-One). Each well was inoculated with 10 μL of bacterial cultures. The plates were incubated statically for 16–24 h at 37°C. The lowest concentration of PA that prevented bacterial growth was considered the MIC. The O.D value was set at 600 nm and was recorded with an AccuSkan, MultiSkan FC (Thermo Fisher Scientific). Vancomycin and Gentamicin were used as positive controls and media was used as negative control. The assay was performed in triplicate.
Isothermal Titration Microcalorimetry (ITC).
E.coli ATCC 29425 and S. aureus ATCC 25923 were used for ITC experiments. The calorimetric measurements were conducted on a TAM III microcalorimetric system with a stainless-steel sample cell (1 mL volume) at 298.15 ± 0.01 K. The reference cell was filled with 765 μL of buffer and the sample cell was initially loaded with 600 μL bacteria solution at 1.5 ×108 CFU/mL, and then the PAs were injected into the sample cell (10 μL) per injection via a Hamilton syringe (500 μL) using a 612 Thermometric Lund pump until the addition was completed. Solutions of PA 4 and PA 7 at 25 μg/mL for titration into E. coli and S aureus; PA 12 at 180 μg/mL for titration into E. coli and 45 μg/mL for titration into S aureus; and PA 15 at 500 μg/mL for titration into E. coli and 170 μg/mL for titration into S aureus. The interval between two injections (12 minutes) was long enough to reach equilibrium. The solution in the sample cell was stirred at 90 rpm with a gold propeller. The ITC curves were fitted with NanoAnalyze software in an ITC package (supplied by TA Instruments Inc.).
Scanning Electron Microscopy (SEM) Characterization:
E. coli K12 and S. aureus JE2 USA 300 (MRSA) bacterial strain were grown in Muller Hinton Broth and incubated at 37°C. The resultant mid-log phase cultures were diluted to a final concentration of 1.5 ×108 CFU/mL (0.5 McFarland). The bacteria were treated with PAs at twice the MIC value and incubated for 1 h at 37°C. A control was prepared with no PA added. After PA addition, the bacteria were washed three times with HyClone™ Dulbecco’s Phosphate Buffered Saline solution (GE Healthcare Life Science) and the samples fixed with 2.0% (v/v) glutaraldehyde and 2% (v/v) of paraformaldehyde in phosphate buffer (0.1 M). Samples for SEM imaging were fixed by immersion in a solution of 2% glutaraldehyde, 2% paraformaldehyde in Sorenson’s phosphate buffer (0.1 M, pH 6.2) for a minimum of 24 h at 4°C. Samples were then placed on glass chips coated with 0.1% Poly-L Lysine and allowed to adhere for 30 minutes. Chips were then washed three times with phosphate buffer to clear excess fixative. Samples were post-fixed in a 1% aqueous solution of osmium tetroxide for 30 minutes to aid in conductivity. Subsequently, samples were dehydrated in a graded ethanol series (50, 70, 90, 95, 100%). Following dehydration, samples were critical point dried and attached to aluminum SEM stubs with double-sided carbon tape. Silver paste was applied to increase conductivity. The following day, samples were coated with ~50nm Gold-Palladium alloy in a Hummer VI Sputter Coater (Anatech USA) and imaged at 30kV in a FEI Quanta 200 SEM operating in high vacuum mode.
Live/Dead Assay for Bacterial Cultures.
S. aureus JE2 USA 300 was grown to the mid log phase in MHB at 37°C under constant shaking at 150 rpm, washed twice and diluted to a concentration of 106 CFU/mL in a saline solution (0.85%). The bacteria were treated with PA 2, PA 5 and PA 10 at their MIC values and a fixed Propidium Iodide (PI) concentration of 10 μg/mL for 30 minutes at 37°C. The cells were immediately washed and resuspended in saline 0.85%. As control we used cultures with PI, no PI and treated with EtOH 70%. Readings were obtained with a flow cytometry at a wavelength of 488 nm (excitation).
Confocal Laser Scanning Microscopy.
S. aureus JE2 USA 300 was grown to the mid log phase in MHB at 37°C under shaking at 150 rpm, washed twice, centrifuged for 5 min at 2000g, and diluted to 1 × 108 CFU/mL in PBS. The bacteria were incubated with PA 2, PA 3, PA 5 and PA 10 at twice MIC values for 60 minutes at 37°C. Cells were washed with PBS, and incubated in dark with PI (10 μg/mL final concentration) for 15 min at 37°C. Immediately the cells were washed and re-suspended in PBS. Slides were prepared using antifade reagent with DAPI. Untreated control without the addition of PAs (negative control) and EtOH 70% (positive control) were performed following the exact same procedure for the treated samples. Images were captured using a confocal laser-scanning microscope (Zeiss 710) and the data were analyzed using Zen 2010 software.
Inner Membrane Permeabilization Assay.
Inner membrane permeabilization was determined by measuring the L-galactosidase release into the medium using o-Nitrophenyl-β-D-galactopyranoside (ONPG) as a substrate.23 E. coli K12 was grown (logarithmic phase) in LB medium supplemented with 2% lactose, washed and resuspended in PBS to 106 CFU/ml. Bacterial suspension (90 μL) was added into each well of a 96-well followed by 10 μL ONPG S(30 mM) and serial dilutions of PAs (100 μL) in PBS. Polymyxin B (PMB), a potent membrane-active peptide, was used as a reference compound. The o-nitrophenol was quantified by measuring absorbance at 415 nm at various time intervals at 37°C. Wells containing bacteria suspension, ONPG and PBS were used as control. The experiment was performed in triplicate.24
Cytotoxicity assay.22
Immortalized Human Keratinocyte (HaCaT) cell line was donated by Dr. David Oupicky Lab (Center for Drug Delivery and Nanomedicine – UNMC) and Immortalized Human Colon Epithelial Cell (HCEC) line was donated by Dr. Robert Lewis Lab (Eppley Cancer Institute – UNMC). The HaCaT cells were cultured in DMEM – high glucose, with 10% fetal bovine serum, and 1% penicillin/streptomycin. HCEC cells were cultured in DMEM – high glucose without sodium bicarbonate and sodium pyruvate, with Medium 199 (Invitrogen), Cosmic Calf Serum, Hydrocortisone, Sodium Selenite, Apo-Transferrin bovine, Insulin, Gentamicin, and Recombinant human EGF. The assays were carried out in sterile 96-well flat-bottomed polystyrene microtiter plates. Plates contained 100 μL of a cell suspension in each well (5000 cells/well) and were pre-incubated for 24 hours at 37 °C in a humidified environment with 5% CO2. The samples to be tested were 2-fold diluted and 10 μL of each concentration were added to test plates in triplicate to get final concentrations ranging from 150 to 4,68 μg/mL. The test plate containing the compounds and the control (cells in culture medium) was incubated for 24 h. The plates were further incubated with 50 μL MTT solution (0.5 mg/ml) at 37 °C for 2 hrs. The top medium was then removed and 100 μL DMSO was added to each well to dissolve the formazan crystals.25 The absorbance of the solution was determined at 600 nm using a multi-well plate reader AccuSkan, MultiSkan FC (Thermo Fisher Scientific).
In Vivo Antibacterial Studies.
Galleria mellonella were purchased from Knutson’s live bait (Michigan, USA) and maintained on wood chips in the dark until used. The experiments were performed according to the protocol of Ramarao et al 2012.26 Briefly, larvae (2–3 cm long and 200–300 mg) were selected 24 hr before infection and placed into an empty pedri dish. For a lethal dose, 1.5 × 108 CFU/mL was used for S. aureus JE2 USA 300. Bacterial culture were centrifuged (5000 xg for 5 min), washed twice and re-suspended in PBS buffer. 10 μL of bacteria suspension were injected on the first right proleg of the larvae using a microsyringe (LS, Innovative Labor system). After 2 hr, stock solutions of PA 2 and PA 5 were diluted in PBS to 300 μg/mL and 10 μL of solutions (to obtain 15 mg/kg body weight) and were injected into different prolegs and incubated at 37°C. Insects were observed for the production of pigmentation and time of death was monitored over 96 hours. Larvae unresponsive to touch and showing the black pigmentation were considered dead. All tests in G. mellonella were perfomed in triplicate (n=10 larvae per each replicate). As a control, we used G. mellonella infected but untreated (no drug), or injected with 30 % DMSO in PBS. For statistical testing, survival data were analyzed using the Kaplan-Meier method. All statistical analyses were performed using Sigma Plot, version 5.04 (Systat Software Inc. (SSI), in San Jose, California, USA).
3. Results and discussion
PAs Design and Characterization
All PAs were synthesized by standard Fmoc Solid-Phase Peptide Synthesis (SPPS), using rink amide resin, and their identity was confirmed by mass spectroscopy as described in the Experimental Section. The PA sequences and the physicochemical and morphological characteristics are listed in Table 1. The structures, morphological characteristics and purities of the PAs are shown on the SI (Figures S1–S3, Table S1).
Table 1:
Physicochemical and morphological characteristics of PAs.
| PAs | Sequence | Retention timea (min) | LogPb | Morphology by TEMc | Zeta potential (mV)c |
|---|---|---|---|---|---|
| PA 1 | C12K5 | 10.95 | 1.93 | Micelles | 34.66 ± 3.21 |
| PA 2 | C14K5 | 12.49 | 2.81 | Micelles | 34.50 ± 2.10 |
| PA 3 | C16K5 | 14.26 | 3.70 | Micelles | 39.50 ± 4.41 |
| PA 4 | C18K5 | 15.78 | 4.58 | Micelles | 31.46 ± 1.75 |
| PA 5 | C20K5 | 17.27 | 5.47 | Micelles | 41.43 ± 1.38 |
| PA 6 | 2-Hexyldec-K5 | 12.66 | 3.63 | Wormlike Micelles | 21.83 ± 0.47 |
| PA 7 | C18K-K5 | 16.45 | 4.24 | Micelles | 45.43 ± 2.58 |
| PA 8 | (C8)2K-K5 | 12.36 | 11.51 | Micelles | 18.83 ± 1.69 |
| PA 9 | (C18)2K-K5 | 27.78 | 16.92 | Short Fibers | 41.40 ± 1.05 |
| PA 10 | AnthraceneK-K5 | 8.60 | 1.27 | Micelles | 38.33 ± 1.77 |
| PA 11 | C18O5 | 15.62 | 2.37 | Micelles | 39.80 ± 1.55 |
| PA 12 | C18R5 | 9.51 | 3.54 | Micelles | 14.10 ± 1.21 |
| PA 13 | C10F4K5 | 16.29 | 6.02 | Twisted Ribbons | 40.63 ± 1.95 |
| PA 14 | C16F3K3 | 21.27 | 7.31 | Twisted Ribbons | 44.03 ± 1.25 |
| PA 15 | C18V3K5 | 18.62 | 6.26 | Nanofibers | 16.56 ± 2.55 |
| PA 16 | C16(VK)4 | 16.84 | 6.28 | Twisted Ribbons | 40.56 ± 2.31 |
| PA 17 | C16V3A3K3 | 19.15 | 4.83 | Nanofibers | 33.76 ± 0.63 |
| PA 18 | C18K31-peptoid | 16.35 | 4.56 | Micelles | 36.26 ± 1.46 |
Lysine (K), Ornithine (O), Arginine (R), Phenylalanine (F), Valine (V) and Alanine (A), Caprylic acid (C8), Decanoic acid (C10), Dodecanoic acid (C12), Myristic acid (C14), Hexadecanoic acid (C16), Octadecanoic acid (C18), Eicosanoic acid (C20).
Linear gradient of Acetronitrile (0.1% TFA) and water (0.1% TFA) from 5% to 95% in 30 min.
Calculated from Molecular Operating Environment (MOE).
Aqueous solution of 2 mmolar concentration in pH 7.
PAs lacking amino acids that favored ß-sheet interactions self-assembled into micelles (PA 1–5, PA 7–8, PA 10–12, and PA 18). Meanwhile, PAs that displayed strong intermolecular H-bonds formed nanofibers (PA 9, PA 15 and PA 17) (Table S1). The transmission electron (TEM) micrographs of some selected PAs are shown in Figure 1 (the remaining TEM micrographs can be found on the SI S4). PA 13 (C10F4K5) and PA 14 (C16F3K5) formed twisted ribbons due to π-π stacking as shown by Pashuck et al (2010).27 A previous report by Stupp and associates demonstrated that PAs containing Valine (V) and Glutamic acid (E) dimer sequences of various lengths also lead to twisted ribbons.28 They showed that flat β-sheet nanostructures tend to twist because the length of the peptide sequence suppresses lateral growth favoring the transformation to cylindrical morphology. Thus, as expected PA 16, with sequence C16(VK)4, presented that morphology.
Figure 1.
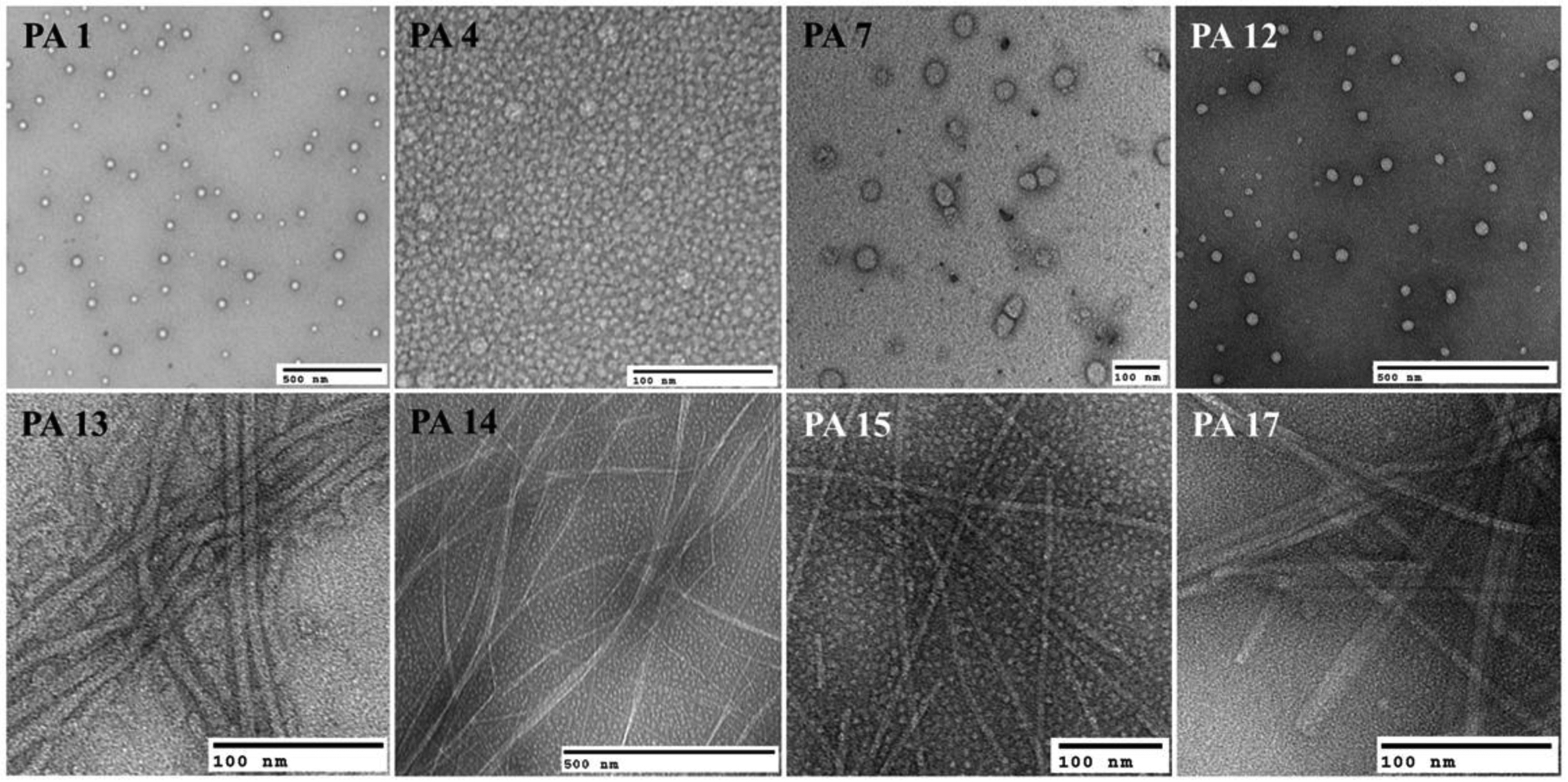
TEM micrographs of various PAs. Micelles (PA 1, PA 4, PA 7 and PA 12), nanofibers (PA 15 and PA 17) and twisted ribbons (PA 13 and PA 14).
The secondary structure of the PAs was assessed using Circular Dichroism (CD) (SI S3). Fiber-like assemblies, PA 9 and PA 13–17, exhibited a characteristic β-sheet CD signal with negative bands ~ 218 nm and positive bands ~ 196 nm. Meanwhile, micelle-forming structures showed the expected random coil signal.16 Hydrodynamic sizes of the micelles were evaluated by dynamic light scattering using a Zetasizer Nano ZS (Malvern Instruments) at 25°C. The PA 1, PA 4, PA 7 and PA 12 presented hydrodynamic radii of ~ 190 nm, 166 nm, 137 nm and 99 nm, respectively (the particle size distribution can be found in the SI S5).
Antimicrobial Activity
The antimicrobial activity of the PAs was evaluated against bacteria, as well as the fungi Candida albicans and Cryptococcus neoformans (Table 2). To our surprise, neither the nanofibers nor the twisted ribbons possessed significant activity against the bacteria at 32 μg/mL regardless of their positive charge. Additionally, the micelle forming PA 1 (C12K5) did not show antimicrobial activity against the tested microorganisms. However, increasing the hydrophobic tail length of the PAs: C14K5, C16K5, C18K5 and C20K5 lead to potent and broad-spectrum antimicrobials in vitro. PA 4 was effective against MRSA, E. coli and C. neoformans with a minimal inhibitory concentration (MIC) of 1 μg/mL. PA 4 was also active against P. aeruginosa (MIC = 4 μg/mL) and K. pneumoniae (MIC = 16 μg/mL). PA 5 showed potent activity against Gram-positive bacteria and fungi with MIC of 1 μg/mL, 4 μg/mL and ≤ 0.25 μg/mL against MRSA, C. albicans and C. neoformans, respectively. However, it did not possess significant activity against Gram-negative bacteria. These results indicate that, in addition to supramolecular shape, the length of the alkyl tail plays an important role in antimicrobial activity. This can be seen in Table 1, with the antimicrobials PA 2–5 (logP = 2.81, 3.70, 4.58, 5.47, respectively) being more hydrophobic than the inactive PA 1 (logP = 1.93). We believe this relates to the ease of insertion within the bacteria membrane;13 however, antimicrobial activity linked to an optimal hydrophilic lipid ratio should not be discarded.
Table 2:
Antibacterial activity of the PA (Antimicrobial PAs micelles are indicated with green color).
| MIC (μg/mL) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gram-positive | Gram-negative | Fungi | |||||||||
| PAs | Saa | Sab | Sac | Ecd | Ece | Paf | Pag | Kph | Abi | Caj | Cnk |
| PA 1 | >32 | >32 | >32 | ≥ 32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| PA 2 | 32 | ne | 8 | 32 | 16 | ne | >32 | >32 | >32 | >32 | 16 |
| PA 3 | 16 | ne | 2 | 8 | 2 | ne | 8 | 32 | 32 | 32 | 1 |
| PA 4 | 8 | 8 | 1 | 4 | 1 | 4 | 4 | 16 | 16 | 32 | 1 |
| PA 5 | 16 | 16 | 1 | 16 | 8 | 32 | >32 | >32 | >32 | 4 | ≤ 0.25 |
| PA 6 | 16 | 16 | >32 | 16 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| PA 7 | 8 | 8 | 1 | 8 | 8 | 16 | 8 | 32 | 16 | 2 | ≤ 0.25 |
| PA 8 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| PA 9 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| PA 10 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| PA 11 | 8 | 16 | 1 | 16 | 2 | 16 | 4 | 16 | 16 | 4 | ≤ 0.25 |
| PA 12 | 16 | 32 | >32 | ≥ 32 | >32 | >32 | >32 | >32 | >32 | >32 | 16 |
| PA 13 | 32 | 32 | 1 | >32 | 8 | >32 | >32 | >32 | 4 | >32 | 0.5 |
| PA 14 | >32 | >32 | 32 | >32 | >32 | ne | >32 | >32 | >32 | 8 | 32 |
| PA 15 | >32 | >32 | 16 | >32 | >32 | ne | >32 | >32 | >32 | >32 | ≤ 0.25 |
| PA 16 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| PA 17 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 |
| PA 18 | 8 | 8 | 2 | 16 | 4 | ne | 8 | 8 | 16 | 16 | 0.5 |
| Van | 2 | 2 | ne | ne | ne | ne | ne | ne | ne | ne | ne |
| Gen | ne | ne | ne | 0.25 | ne | 0.25 | ne | ne | ne | ne | ne |
Staphylococcus aureus JE2 MRSA;
Staphylococcus aureus LAC MRSA;
Staphylococcus aureus ATCC 43300 MRSA;
Escherichia coli K12;
Escherichia coli ATCC 25922;
Psedomonas aeruginosa PA01;
Psedomonas aeruginosa ATCC 27853;
Klebsiella pneumoniae ATCC 700603;
Acinetobacter baumannii ATCC 19606;
Candida albicans ATCC 90028 and
Cryptococcus neoformans var. grubii H99 ATCC 20882. ne: not evaluated. All PAs assemblies tested were prepared in water at pH 7.
The “reversed” PA 7 (C18K-K5) with intermediate hydrophobicity (PA 5 >> PA 7 >> PA 4) and spherical morphology was effective against MRSA (MIC = 1 μg/mL), E. coli and P. aeruginosa (MIC = 8 μg/mL), C. albicans (MIC = 2 μg/mL), and C. neoformans (≤ 0.25 μg/mL). Given this compound showed equal or lower potency than PA 4 against the tested bacteria, but a higher Zeta potential, we theorize that the lipophilic ratio may be more important for activity than the charge of the system. Changes in the hydrophobic tail, as can be seen in PA 8–10, resulted in lower activity, indicating that the presence of two alkyl chains or an aromatic chain did not result in antibacterial effect.13 The octadecanoyl analogue PA 11 (C18O5) was active against MRSA (MIC = 1 μg/mL), E. coli (MIC = 2 μg/mL), P. aeruginosa (MIC = 4 μg/mL), C. albicans (MIC = 4 μg/mL), and C. neoformans (≤ 0.25 μg/mL), slightly less active than PA 4. This may be related to the projection of positive charges from the peptide core (ornithine has one less methylene unit than lysine), which may affect the electrostatic interactions with the membranes. Perhaps the most perplexing results were seen with the arginine-PA 12, which was inactive against most of the pathogens. Arginine-based PAs had been described as an effective antibacterial agent29 that are able to kill pathogens at low MBC.30 For example, the lipopeptide C14R3 was reported as anti-staphylococcal with anti-biofilm activity against various resistant strains of S. aureus equivalent to that of daptomycin and vancomycin.31 However, the compound was not effective against P. aeruginosa and K. pneumoniae. The pKa values of lysine and arginine (~10.5 and ~12.8, respectively) suggest a positive charge at physiological pH, which is an essential characteristic for damaging membranes. However, molecular dynamics models demonstrated that arginine is the only amino acid residue that remains protonated inside the membranes.32 Thus, the data again suggest that overall hydrophobicity (PA 4 > PA 12) plays a more significant role in the antimicrobial activity than charge.
The order of hydrophobicity for PAs with active systems showing in bold is PA 9 > PA 14 > PA 17 > PA 15 > PA 5 > PA 16 > PA 7 > PA 18 > PA 13 > PA 4 > PA 11 > PA 3 > PA 6 > PA 2 > PA 8 > PA 1 > PA 12 > PA 10, which suggests a hydrophobic range for activity. Intriguingly, the only two inactive PAs (PA 16 and PA 13) inside this range self-assemble into twisted ribbons, validating our observation of shape-dependent activity. In addition, the most effective PAs presented Zeta potential greater than + 31.46 mV and all the inactive PAs with high zeta values or > + 10 mV are either nanofibers or very hydrophobic/hydrophilic. Nanoparticles with zeta potential greater than +30 mV are considered cationic while values between – 10 mV and + 10 mV are approximately neutral.33 Positively charged nanoparticle are associated with cell membrane disruption and cytotoxicity, due to the fact that bacteria membranes are slightly negative.18, 33, 34 Thus, despite shape appearing to be the main driver of antibacterial activity, the relative potency of the supramolecular structures is strongly dependent on other factors such as charge and hydrophobicity. This is not a surpise, because cationic charges and amphiphilic character have been identified as essential characteristics of AMPs.12, 13,35
A recent investigation by Liang and co-workers has shown that antibacterial nanoparticles possess structure-dependent activity, supporting our observations.12 Further, Zoe et al36 has found that positively charged micelles target the negatively charged outer membrane of E. coli by electrostatic interactions destroying the cellular barrier function. The micelles then disintegrate the cell’s inner membrane through hydrophobic interactions between the hydrophobic tails of the PAs and the lipid component of the bacterial membrane, and these interactions lead to subsequent cytoplasmic leakage and cell death.36
In order to evaluate the effect of sample processing on antimicrobial activity, PA 4 and PA 7 were self-assembled under different conditions: i) aging for 3 days, ii) annealing, iii) using hexafluoroisopropanol (HFIP) to dissolve PA aggregates before assembly in water, iv) by exchanging the counter ion (TFA/HCl). The samples were tested against MRSA and E. coli K12 with no observed changes in antimicrobial activity (SI Table S2).
The minimum bactericidal concentration (MBC), dosage at which > 99.9% bacteria are killed, was determined for selected PAs against MRSA and E. coli. The MBC is reduced sharply from PA 12 to PA 4 against MRSA. PA 4 showed MBC at 16 μg/mL, corresponding to two-fold more than MIC while PA 12 did not show MBC up to 256 μg/mL. Similar results were obtained against E. coli, PA 4 and PA 12 killed > 99.9% bacteria at 8 μg/mL and 128 μg/mL, respectively. Thus, lysine-based PAs were more effective at killing MRSA and E. coli than arginine-based PAs.
Biofilm formation, a virulence factor associated with many bacterial infections, can significantly increase bacterial resistance to antibiotics. AMPs have shown potent anti-biofilm activity against multi-drug resistant bacteria strains at low concentrations and synergism with antibiotics.37 We investigated the efficacy of PA 4 micelles to inhibit the production of MRSA and E coli biofilms using a crystal violet staining assay.38 For the biofilm formation inhibition assays, initial S. aureus JE2 and E coli K12 inoculums were added in two-fold dilutions with concentration ranging from 16 to 4 μg/mL and 4 to 1 μg/mL for PA 4 and vancomycin or gentamicin, respectively. PA 4 reduced MRSA biofilm formation up to 68.80 ± 3.62 % at MIC while Vancomycin caused a reduction up to 92.80 ± 0.68 % (SI S6). At 2 × MIC, PA 4 was able to inhibit the adhesion of biofilms causing more than 90% reduction. PA 4 was less effective at inhibiting E coli biofilms. PA 4 did not reduce biofilms formation up to 16 ug/mL, which corresponds to 4 × MIC and caused a reduction of 39.5% at 32 ug/mL while Gentamicin caused a reduction of about 54. 3% at 2 ug/mL (SI S6).
Isothermal Titration Calorimetry (ITC)
We used isothermal titration calorimetry (ITC) to observe thermodynamic changes in the binding process of PA micelles and nanofibers to S. aureus and E. coli. Figure 2 shows the variation of observed enthalpy (ΔHobs) when PAs were titrated into S. aureus and E. coli solutions. The ITC curves show that micelles (PA 4, PA 7 and PA 12) possess a strong binding affinity with bacteria membranes, demonstrating exothermic values. With the addition of PAs, the ΔHobs values changed from larger exothermic to zero, reaching the saturation of interaction of the PAs with S. aureus membrane. The same result was observed for E. coli. However, a nanofiber (PA 15) showed a weak binding affinity with the bacterial membranes, displaying a line with small curvature. The electrostatic binding of positively charged PAs with negatively charged bacteria was highly associated with the observed exothermic effect. PA 4 and PA 7 (zeta potential values of 31.46 ± 1.75 and 45.43 ± 2.58 mV, respectively) show greater negative enthalpy change values to both S. aureus and E. coli. PA 12 (zeta potential value of 14.10 ± 1.21 mV) has a weak exothermic effect in E. coli, but it has a strong exothermic effect with S. aureus, indicating that the arginine group has stronger affinity with S. aureus. PA 15 (zeta potential value of 16.56 ± 2.55 mV) has the weakest interaction enthalpy values with S. aureus and E. coli.
Figure 2:
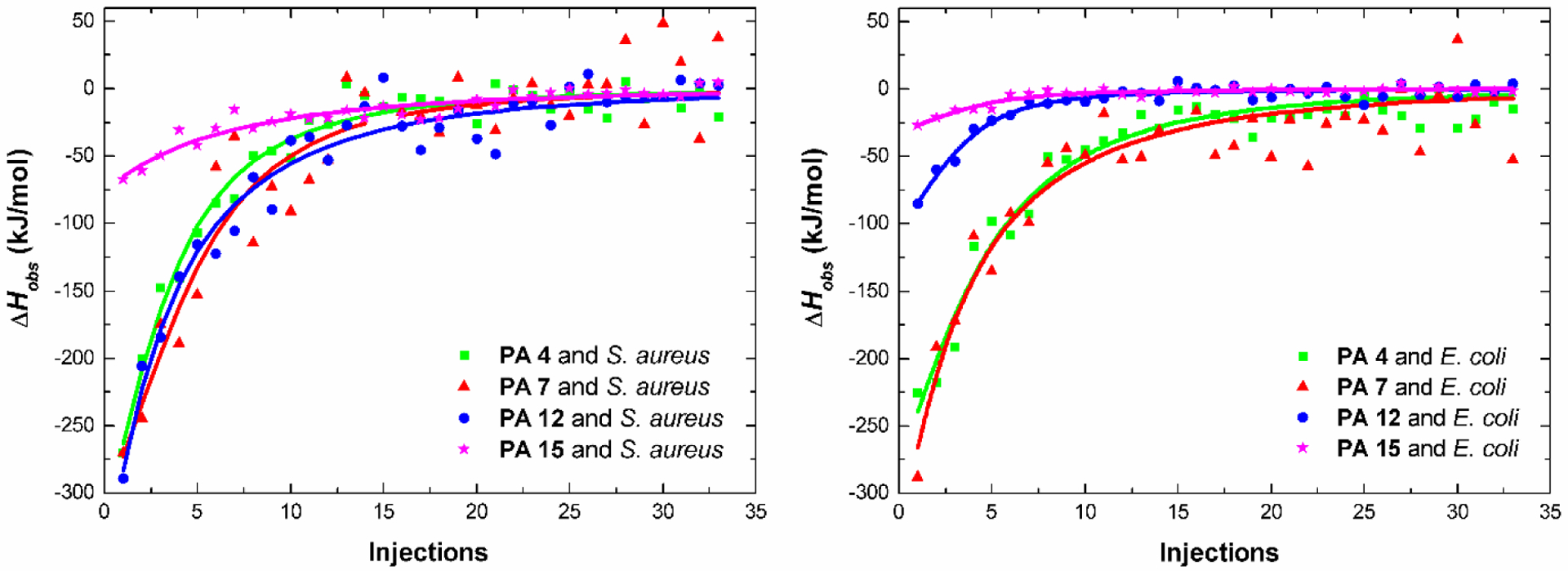
Variation of observed enthalpy changes (ΔHobs) in the binding process of S. aureus and E. coli bacteria membrane with micelles (PA 4, PA 7 and PA 12) and nanofibers (PA 15).
The ITC curves were analyzed by independent binding model as described in the SI. The binding constant (Kb) of PA decreased in the following order: KPA2 ˃ KPA5 ˃ KPA10 ˃ KPA13 for S aureus and KPA5 ˃ KPA2 ˃ KPA10 ˃ KPA13 for E. coli, respectively (Table 3). These results suggest that PA micelles possess better antimicrobial activity than nanofibers, because they are able to bind and interact more efficiently with the bacterial membrane. The micelle PA 4 antibacterial activity is 8-fold more potent than nanofiber PA 15 against S. aureus and 32-fold more potent against E. coli. Despite the impact that the hydrophobicity and cationic charge has on the biological activity of PAs, the most apparent difference in biological activity was observed when the morphology of nanostructures was modified. Both PA 4 (C18K5) and PA 15 (C18V3K5) possess the same overall charge but differ in their amino acid composition; PA 15 contains valine, which leads to nanofibers formation. The weaker intermolecular van der Walls interactions (when compared to hydrogen bonding) make micelles less stable than nanofibers.39 We hypothesize that the strength of intramolecular cohesion stabilizes the supramolecular structures and diminishes interaction with membrane. Recent studies suggest that intermolecular cohesion determines the ability of a supramolecular assembly to disrupt the membrane of cancer cells.40
Table 3:
Thermodynamic parameters of the binding of PAs with S. aureus and E. coli K12. Binding constant (Kb) and binding number (N) derived from ITC curves. (PAs self-assembled into micelles are indicated with green color).
| S. aureus | E. coli | |||
|---|---|---|---|---|
| PAs | N (×106) | Kb (×105) | N (×106) | Kb (×105) |
| PA 4 | 7.97 ± 1.10 | 4.15 ± 0.42 | 1.32 ± 0.25 | 6.08 ± 0.83 |
| PA 7 | 3.19 ± 0.59 | 2.87 ± 0.15 | 2.03 ± 0.11 | 8.06 ± 0.67 |
| PA 12 | 41.51 ± 2.24 | 1.97 ± 0.14 | 0.97 ± 0.26 | 2.00 ± 0.11 |
| PA 15 | 119.08 ± 5.37 | 0.57 ± 0.03 | 0.21 ± 0.03 | 0.23 ± 0.01 |
PA amphiphile micelles action on bacteria membrane:
To get visual insights into the mechanism of action of the PA micelles, the morphological changes of S. aureus JE2 and E. coli K12 exposed to micelles at 2 × MIC were observed by TEM and scanning electron microscopy (SEM). It was observed that after treatment with PA 4 and PA 7 membrane damage took place, with bacterial debris and burst bacterial membranes seen by TEM (SI S7) and SEM (Figure 3). S. aureus showed protruding bumps, cytoplasmic leakage and deep holes after treatment (1 hr) with PAs while E. coli presented deep craters and severe membrane deformations. The morphological changes indicate the PAs kill bacteria by disrupting their membranes (although other mechanism may also take place).
Figure 3:
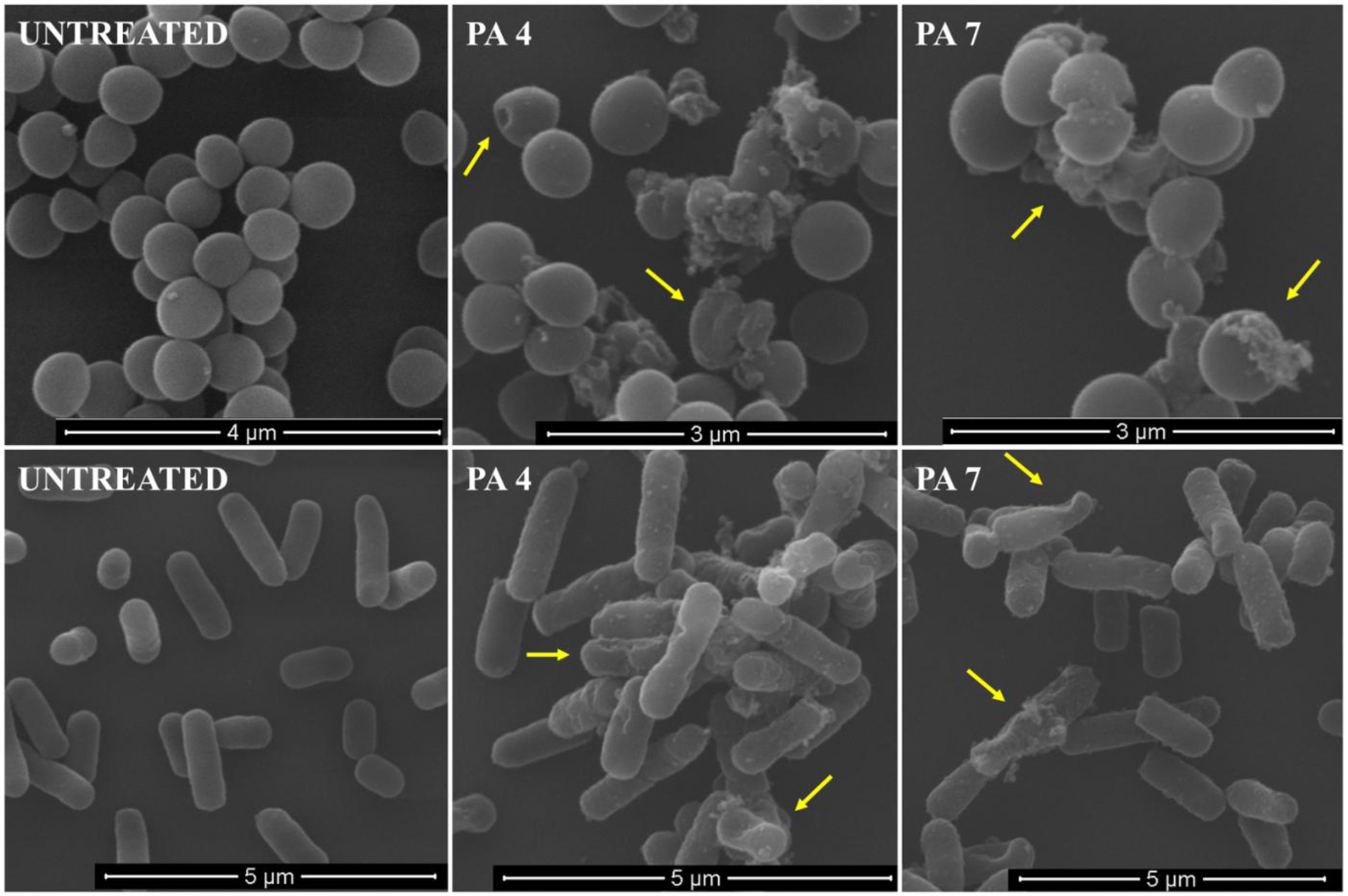
SEM micrographs of S. aureus JE2 MRSA (up) and E. coli K12 (down) cells treated with PA 4 and PA 7 at 2X MIC.
We further confirmed the ability of PAs to disrupt cell membrane using confocal microscopy by a double staining method with DAPI (4′,6-diamidino-2-phenylindole dihydrochloride) and PI (propidium iodide), as fluorophores. DAPI binds bacterial cells irrespective of their viability by intercalating with DNA giving a bright blue fluorescent complex. However, PI only crosses damaged cell membranes, intercalating with DNA forming a bright red fluorescent complex.15 After treatment with PA 4 and PA 7, cells showed PI fluorescence, suggesting that their action involves permeation of the microorganism’s membrane, similar to other AMPs (Figure 4 and S8).10
Figure 4:
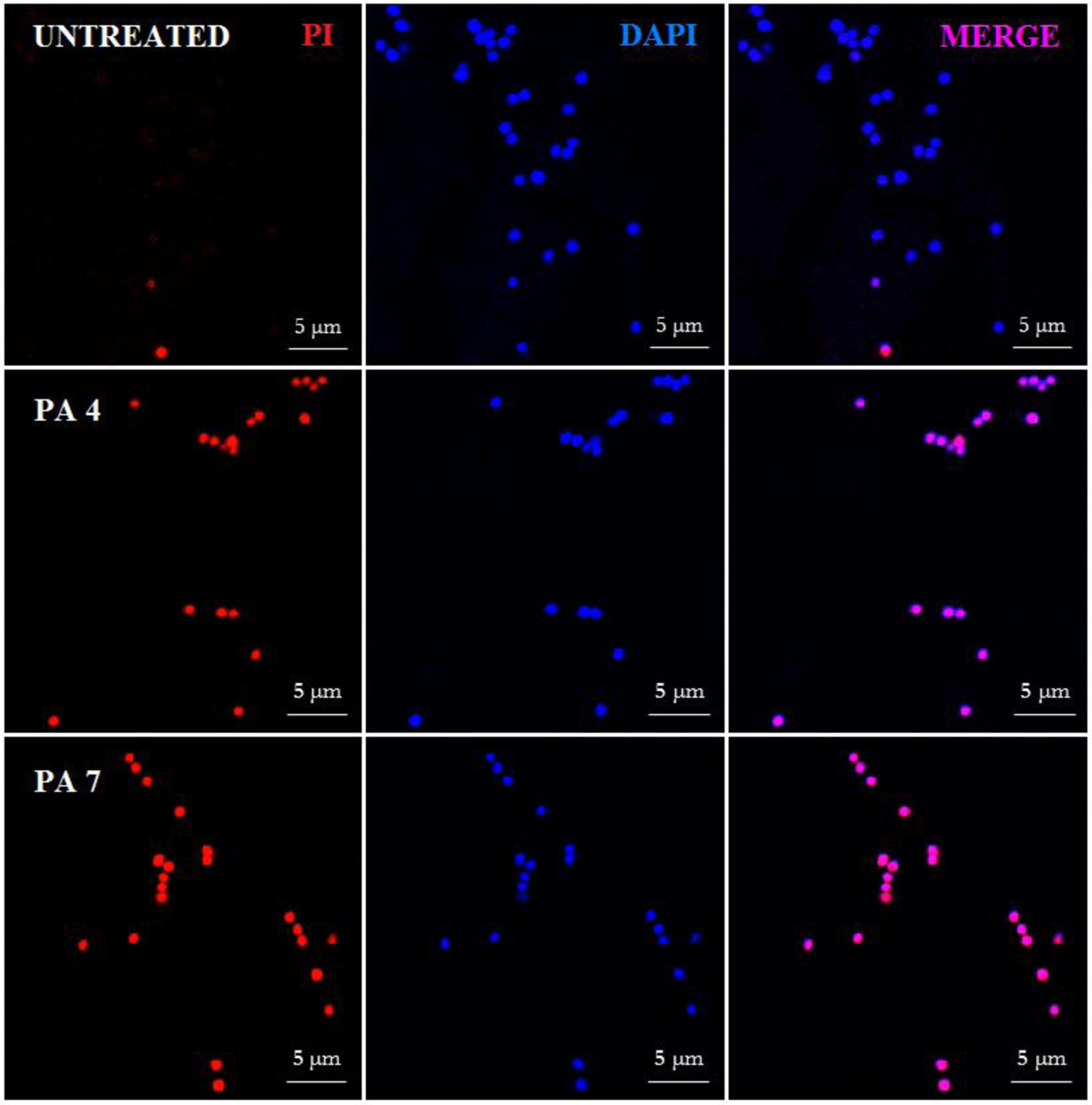
Confocal fluorescence microscopic images of Sa JE2 MRSA treated with PA 4 and PA 7 at twice MIC. Control was made with no PA addition.
The incorporation of PI in S. aureus and E. coli K12 treated with PAs was further evaluated by flow cytometry. The viability of S. aureus with no PA treatment was detected to be about 91%, suggesting intact bacterial cytoplasmic membranes (Figure 5). After treatment with PA 4 and PA 7, the cell viability decreased to 29.1% and 12.7% respectively, indicating cell death presumably due to permeabilized membranes. PA 4 and PA 7 were able to kill S. aureus at 1 × MIC after 30 minutes of treatment. However, the inactive PA 12 did not kill cells at 1 × MIC (Figure 5) and 2 × MIC (SI S9). The viability of S aureus after treatment with PA 12 at 1 × MIC was 92.7 %, this result is consistent with the MBC > 256 μg/mL, discussed previously.
Figure 5:
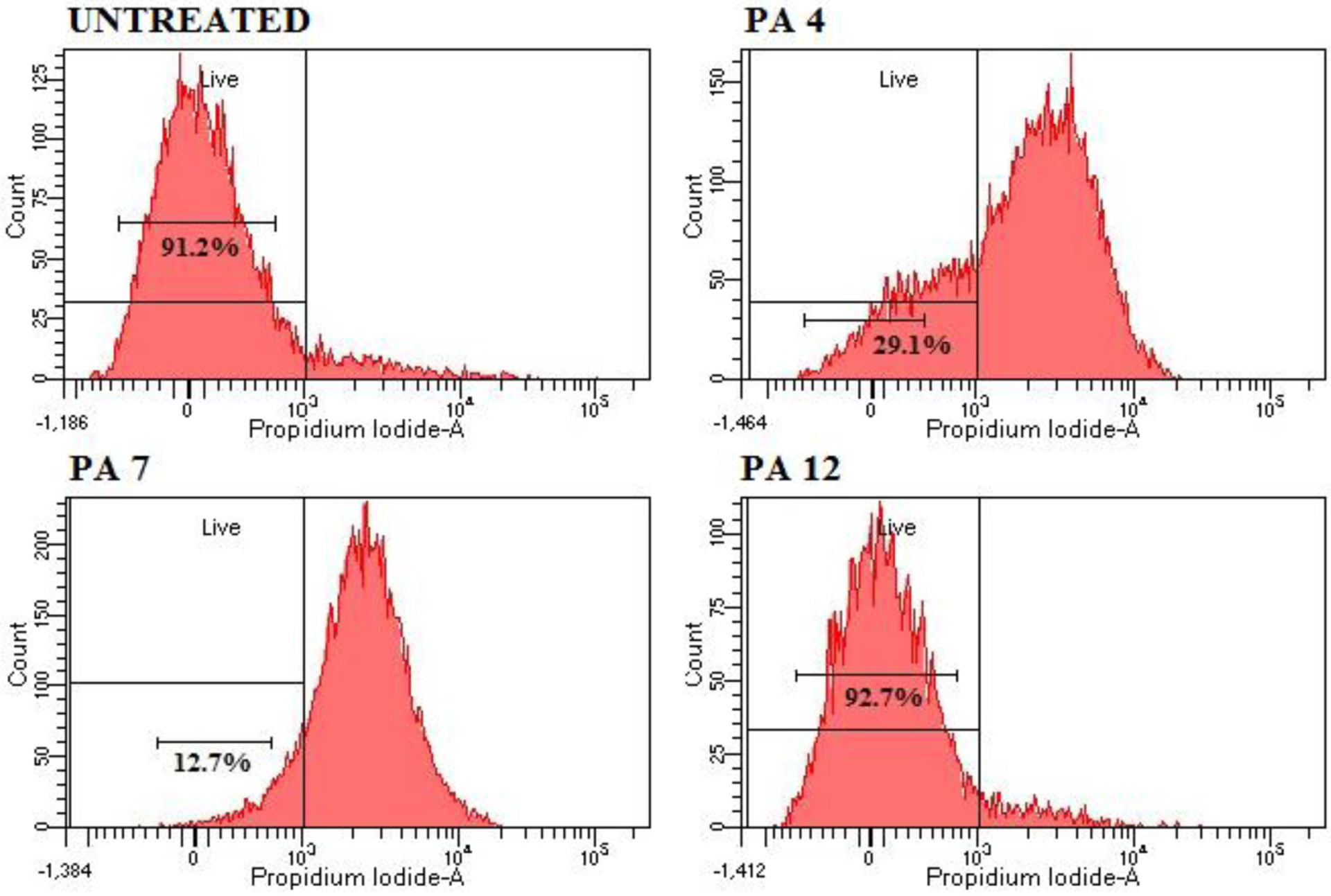
Flow cytometry analysis. Cell membrane disruption of S. aureus JE2 MRSA treated with PA 4 and PA 7 at MIC was determined by an increase in fluorescent intensity of PI. Negative control - No PA; PA 4 at 8 μg/mL; PA 7 at 8 μg/mL and PA 12 at 16 μg/mL. Treatment with EtOH 70% was used as positive control and the analysis can be found on the SI.
Treating (30 min) E. coli with PA 4 and PA 7 at 1 and 2 × MIC showed 23.1%, 28.9%, 7% and 6.2 % dead cells (SI S10 –S11), respectively. As discussed previously, PA 4 displayed MBC of 8 μg/mL against E. coli after 3 hours incubations. The discrepancy in these results can be related to the time of incubation and indicate that PA 4 is able to kill S. aureus faster than E. coli. PA 12 did not kill E. coli at 1 and 2 × MIC (SI S11), consistent with an MBC of 128 μg/mL.
We further evaluated the ability of selected PAs to permeate the E. coli inner membrane by monitoring the hydrolysis of nitrophenyl-β-D-galactopyranoside (ONPG) by cytoplasmic β-galactosidase. Because ONPG cannot traverse the inner membrane, its hydrolysis to yellow o-nitrophenol suggests that permeabilization of the inner membrane and β-galactosidase escape have taken place.23, 24 Treatment of E. coli with PA 4 and PA 7 resulted in permeabilization of the inner membrane at 16 μg/mL, which are equivalent to 4 × and 2 × MIC, respectively, when compared with Polymyxin B (PMB) at 0.5 μg/mL. PMB, a drug known to disrupt E. coli’s membrane (IM).41 PA 12 did not produce permeabilization of the inner membrane at 32 μg/mL.
These results confirm that PAs micelles disrupt bacterial membrane more efficiently than nanofibers, killing bacteria with a mechanism of action associated with membrane disruption. A recent report has shown that the cellular internalization is strongly dependent on micelles’ stability.42 The more stable micellar structures enter cells by endocytosis disassembling inside while less stable micelles can intercalate into the membrane as monomers penetrating cells directly.42 Our current hypothesis is that the less stable micelles (when compared to nanofibers) work as a monomer reservoir; they disassemble leading to lipophilic tail intercalation into the membrane and subsequent bacterial death43 (Figure 7). Note the mechanism of action for Daptomycin involves disruption of the bacterial cell membrane by the insertion of a lipophilic chain into this barrier. This permits the flow of potassium ions out of the cell causing depolarisation, and consequently resulting in cell death. In addition, a study has revealed that this lipopeptide can self-assemble into spherical micelles raising the possibility of disruption of the cell membrane when it is disassembled at the bacterial membrane.43
Figure 7:
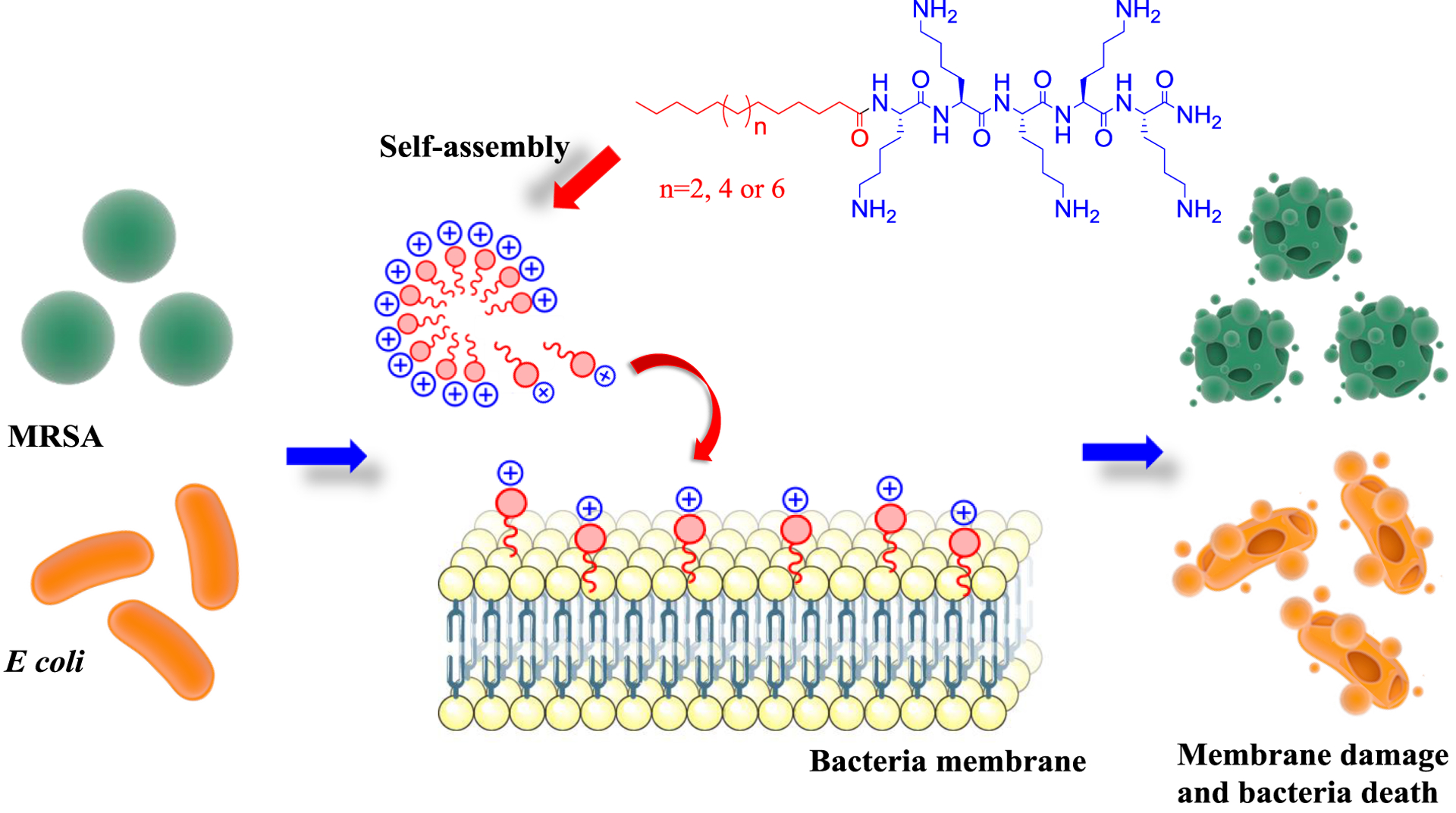
Proposed mechanism of action of PAs micelles.
Cytotoxicity and Hemolytic Activity
The cytotoxicity of the active PAs against immortalized human keratinocyte (HaCaT) and immortalized human colonic epithelial (HCEC) cells was assessed using the the MTT assay. In addition, we studied cell viability of human embryonic kidney cells (ATCC CRL-1573) using resazurin. The IC50 of selected systems was determined from the dose response curves of each PA. The hemolytic activity of active PAs was determined against human red blood cells and the HC10 and HC50 values (concentration at 10% and 50% haemolysis, respectively) calculated by curve fitting of the inhibition values vs. log(concentration) are summarized in the table 4.
Table 4:
Cytotoxicity of designed PA against HaCaT, HCEC and HEK-293 and hemolytic activity against Human red blood cells.
| IC50 (μg/mL) | HC50 (μg/mL) | |||
|---|---|---|---|---|
| PAs | HaCaTa | HCECb | HEK-293c | Human red blood cells |
| PA 2 | 21.42 | ne | >32 | 27.11 |
| PA 3 | 17.59 | ne | >32 | 8.31 |
| PA 4 | 52.0 | 28.3 | >32 | 22.3 |
| PA 5 | 35.9 | 22.5 | 20.8 | 4.9 |
| PA 8 | 40.4 | 19.3 | >32 | 8.1 |
| PA 11 | 37.1 | 18.7 | 20.3 | 16.1 |
| PA 12 | > 150 | > 150 | >32 | >32 |
| PA 13 | > 150 | 27.5 | >32 | >32 |
Immortalized Human Keratinocyte;
Immortalized Human Colonic Epithelial Cell;
Human embryonic kidney cells (ATCC CRL-1573).
HaCaT, HCEC cells were incubated with various PA concentration for 24 h and the cell viability was determined by MTT assay. Assays were performed in triplicate. Cytotoxicity against HEK-293 and Hemolytic Activity were performed by CO-ADD (The Community for Antimicrobial Drug Discovery), Australia.
Most of PAs showed IC50 values >32 μg/mL against HaCaT and HEK-293, except PA 5 and PA 11, which displayed toxicity against HEK-293 cells at 20.8 and 20.3 μg/mL respectively (Table 4). The PAs were found to be more toxic against HCEC; all of which showed values ˂ 32 μg/mL (except for PA 12). Also, PA 12 and PA 13 showed hemolytic activity with HC50 ˂ 32 μg/mL. As discussed previously, PA 1 (C12K5), PA 4 (C18K5) and PA 5 (C20K5) presented the same amino acid sequence with differing hydrophobic tail lengths. The PA 1 did not show antibacterial activity against the strains tested and did not show toxicity against HEK-293 cells and human red blood cells. PA 4 and PA 5 present similar antimicrobial activity but PA 5 exhibited lower IC50 values against mammalian cells. The hydrophobic tail and cationic charge are essential characteristics for antibacterial activity, but they also influence the cell viability; as lipophilicity increases, the hemolytic activity of the PAs also increases, compromising the selectivity of PAs. Shortening the tail length to 12 carbons resulted in a nontoxic PA but no antimicrobial activity. Other studies have reported a similar observation reducing the amphiphilic nature of the assemblies by decreasing the alkyl tail enhances cell viability.40 We expected that PA 2 (C14K5) and PA 3 (C16K5) would be less toxic for mammalian and red blood cells than PA 4, but the obtained data did not support our assumption. More studies need to be performed to find a proper balance of charge and hydrophobicity in order to design more selective PAs (Cytotoxicity/MIC).
In vivo studies
Galleria mellonella (wax moth larvae) has become a viable alternative to other animal models for the preliminary study of pathogenic bacteria. The wax moth larvae’s immune system shares structural and functional similarity to the vertebrate’s innate immune system.44 Furthermore, larvae are more cost-effective to establish, easier to maintain when compared with traditional mammalian model hosts, and they do not require ethical approval.45
First, we evaluated the toxicity of PA 4 and PA 7 at 30 mg/kg and 60 mg/kg towards the G. mellonella larvae (Figure 8 A). We found the PAs did not affect larval survival when compared with the PBS-treated control, and, therefore, they were deemed to be nontoxic at these doses. The effect of single doses of PA 4 and PA 7 (60 mg/kg body weight) 2 hours post infection with S. aureus JE2 MRSA (inoculum 3 × 108 CFU/mL) on the survival of G. mellonella larvae are shown in Figure 8 B and SI 12. Infected larvae treated with PA 4 and PA 7 showed a greater percentage of survival up to 48 hours when compared with untreated larvae, although a decrease was observed after the 48 hours. Experiments were performed to evaluate the efficacy of multiple doses, 2 hr post infection and 12 hr post first dose. Increase in survival was not observed when compared to a single dose effect (data not shown), which may be due to larvae trauma after multiple injections or metabolism of the PAs.
Figure 8:
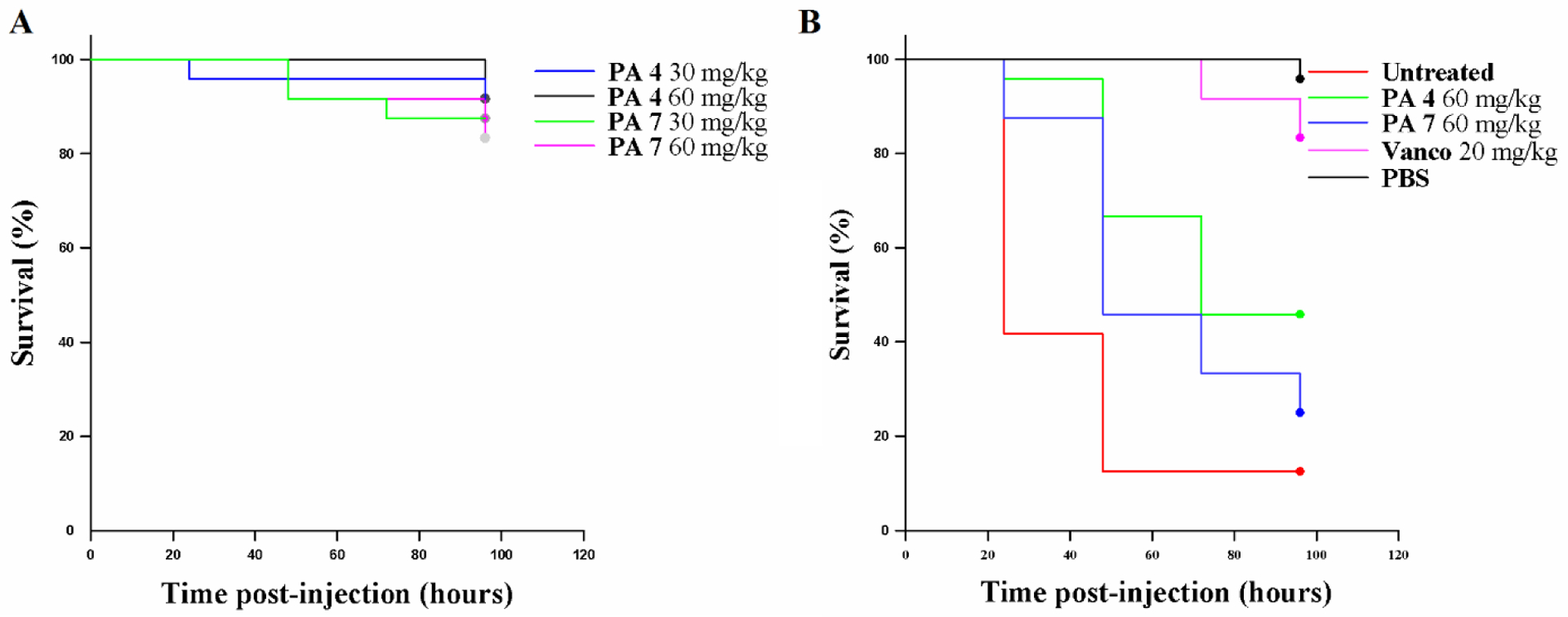
G. mellonella assays. Toxicity of selected PAs (A). MRSA infection of G. mellonella (B).
4. Conclusion
We have designed cationic PAs that self-assemble into supramolecular nanostructures such as micelles, nanofibers, and twisted ribbons. The micelles exhibited promising antimicrobial activity against a panel of Gram-positive and Gram-negative bacteria, as well as fungi with a mechanism of action associated with membrane damage. The results suggest that antibacterial activity is shape-dependent, related to hydrophobicity, and, to a lesser extent, Zeta potential. We hypothesize that intramolecular cohesion determines the antimicrobial activity because micelles are less stable than nanofibers,they may easily disassemble and insert in the bacteria membrane causing cell death. Thus, we believe the micelles work as a monomer reservoir. More studies are needed to find the proper balance between cationic charge and hydrophobicity to achieve efficient antimicrobial action, with low hemolytic activity and toxicity against mammalian cells. PAs micelles provide a great opportunity to develop new therapies in combination with other drugs to achieve synergistic antibacterial action as seen with other membrane disruptors such as pentamidine.46, 47
Supplementary Material
Figure 6:
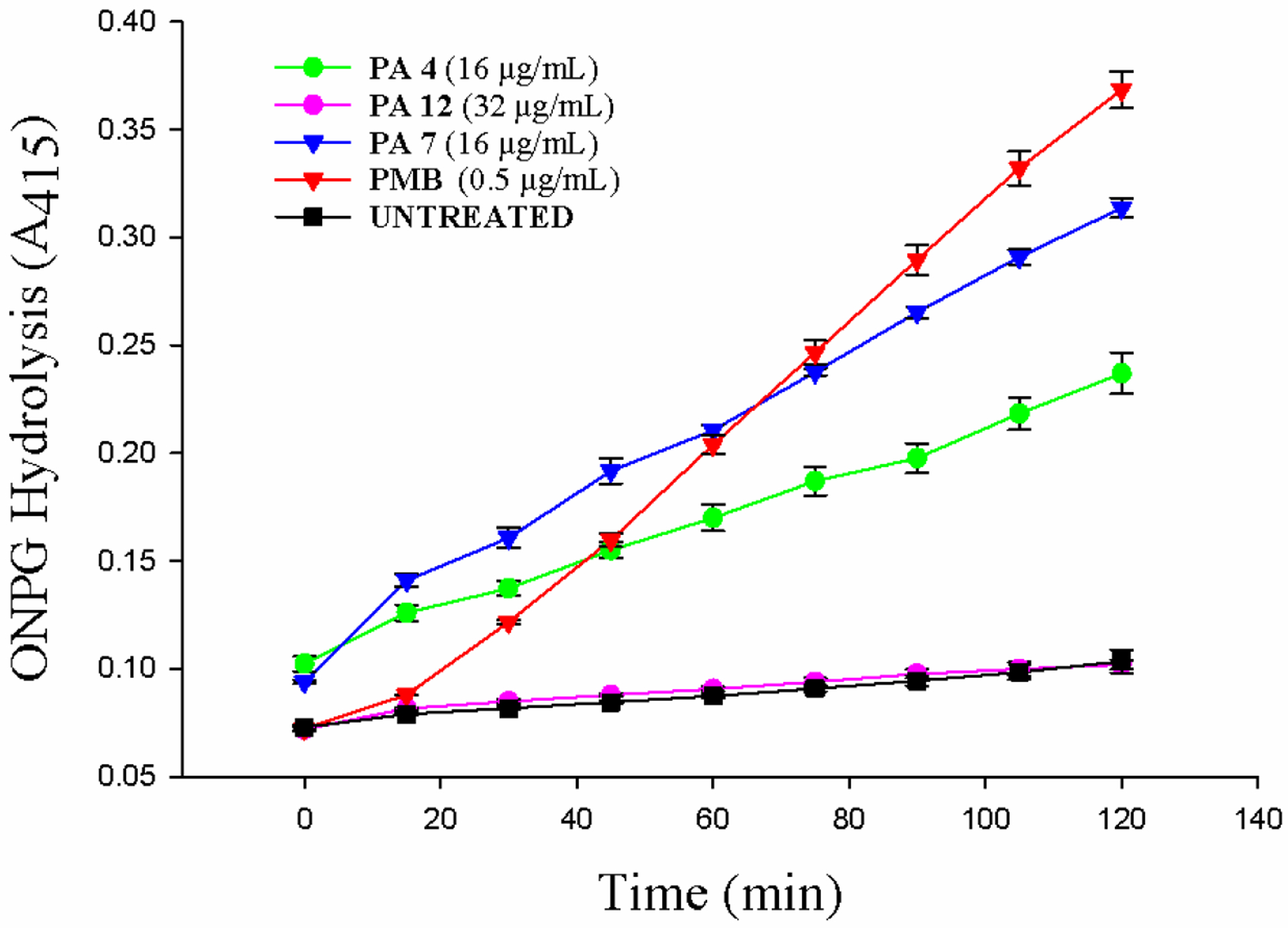
E. coli K12 inner membrane permeabilization. PA capability for permeabilizing E. coli internal membrane was evaluated by using ONPG substrate and treating bacteria with PA 4, PA 7 and PA 12 at 16, 16 and 32 μg/mL, respectively. PMB at 0.5 μg/mL was used as reference drug. The wells containing no test compound were used as control.
7. Acknowledgments
This work was supported by Start-up funds and NIH-COBRE (5P20GM103480- Nebraska Center for Nanomedicine). We would like to thank Mohamed A. Seleem and H. Andy Zhong in the Department of Chemistry at University of Nebraska Omaha for the logP calculations in MOE software. We thank the CO-ADD (The Community for Antimicrobial Drug Discovery), funded by the Welcome Trust (UK), at The University of Queensland (Australia) for doing antimicrobial tests against bacteria and fungi, cytotoxicity against HEC-293, and hemolytic assays in Human red blood cells. We would also like to express our gratitude to the Advanced Microscopy Core Facility at UNMC (Janice A. Taylor and James R. Talaska) confocal microscopy and the Flow Cytometry Research Facility at UNMC (Samantha Wall) for providing experimental assistance. The authors would like thank to Mehdi Bin Samad for providing some of the TEM images, and of the Electron Microscopy Core Facility (EMCF) at UNMC (Tom Bargar and Nicholas Conoan) for assistance. The EMCF is supported by the Nebraska Research Initiative (NRI) and the University of Nebraska Foundation, and the Office of the Vice Chancellor for Research.
5. Abbreviations used
- PAs
Peptide amphiphiles
- MRSA
methicillin-resistant S. aureus
- AMPs
Antimicrobial peptides
- HPLC
High Performance Liquid Chromatography
- TEM
Transmission Electron Microscopy
- ITC
Isothermal Titration Calorimetry
- CFU
colony forming units
- SEM
Scanning electron microscopy
- CD
Circular Dichroism
- MIC
minimal inhibitory concentration
- MBC
minimum bactericidal concentration
- ATCC
American Type Culture Collection
Footnotes
Supporting Information
PAs chemical structures; MALDI-TOF-MS spectra; HPLC chromatograms; CD spectra; nanostructures TEM micrographs; antibacterial activity in different self-assembling conditions; bacterial killing assay; inhibition of S aureus JE2 biofilms; experimental procedures for minimal inhibitory concentration, cytotoxicity and hemolysis from CO-ADD The University of Queensland (Australia); independent binding model for ITC analysis; additional TEM and SEM for S. aureus JE2 and E. coli K12; flow cytometry and in vivo antimicrobial assay pictures (PDF).
The authors declare that there are no conflicts of interest.
8. References
- 1.Alekshun MN; Levy SB Molecular Mechanisms of Antibacterial Multidrug Resistance. Cell 2007, 128, 1037–1050. [DOI] [PubMed] [Google Scholar]
- 2.Blair JM; Webber MA; Baylay AJ; Ogbolu DO; Piddock LJ Molecular Mechanisms of Antibiotic Resistance. Nat. Rev. Microbiol 2015, 13, 42. [DOI] [PubMed] [Google Scholar]
- 3.CDC. Centers for Disease Control and Prevention. Antibiotic Resistance Threats in the United States; United States, 2013; p 114. [Google Scholar]
- 4.Taubes G The Bacteria Fight Back Science (Washington, DC, U. S.) 2008, 321, 356–361. [DOI] [PubMed] [Google Scholar]
- 5.Li J; Koh J-J; Liu S; Lakshminarayanan R; Verma CS; Beuerman RW Membrane Active Antimicrobial Peptides: Translating Mechanistic Insights to Design. Front. Neurosci.-Switz 2017, 11, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tenover FC Mechanisms of Antimicrobial Resistance in Bacteria. Am. J. Infect. Control 2006, 34, S3–S10. [DOI] [PubMed] [Google Scholar]
- 7.Odonkor ST; Addo KK Bacteria Resistance to Antibiotics: Recent Trends and Challenges. Int. J. Biol. Med. Res 2011, 2, 1204–1210. [Google Scholar]
- 8.Bush K; Courvalin P; Dantas G; Davies J; Eisenstein B; Huovinen P; Jacoby GA; Kishony R; Kreiswirth BN; Kutter E Tackling Antibiotic Resistance. Nat. Rev. Microbiol 2011, 9, 894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shankar SS; Benke SN; Nagendra N; Srivastava PL; Thulasiram HV; Gopi HN Self-Assembly to Function: Design, Synthesis, and Broad Spectrum Antimicrobial Properties of Short Hybrid E-Vinylogous Lipopeptides. J. Med. Chem 2013, 56, 8468–8474. [DOI] [PubMed] [Google Scholar]
- 10.Makovitzki A; Baram J; Shai Y Antimicrobial Lipopolypeptides Composed of Palmitoyl Di-and Tricationic Peptides: In Vitro and In Vivo Activities, Self-Assembly to Nanostructures, and a Plausible Mode of Action. Biochemistry 2008, 47, 10630–10636. [DOI] [PubMed] [Google Scholar]
- 11.Wimley WC Describing the Mechanism of Antimicrobial Peptide Action with the Interfacial Activity Model. ACS Chem. Biol 2010, 5, 905–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Y; Zheng W; Kuang L; Ma H; Liang H Hydrophilic phage-mimicking membrane active antimicrobials reveal nanostructure-dependent activity and selectivity. ACS Infect. Dis 2017, 3, 676–687. [DOI] [PubMed] [Google Scholar]
- 13.Ma Z; Yang J; Han J; Gao L; Liu H; Lu Z; Zhao H; Bie X Insights Into the Antimicrobial Activity and Cytotoxicity of Engineered α-Helical Peptide Amphiphiles. J. Med. Chem 2016, 59, 10946–10962. [DOI] [PubMed] [Google Scholar]
- 14.Ong ZY; Gao SJ; Yang YY Short Synthetic β-Sheet Forming Peptide Amphiphiles as Broad Spectrum Antimicrobials with Antibiofilm and Endotoxin Neutralizing Capabilities. Adv. Funct. Mater 2013, 23, 3682–3692. [Google Scholar]
- 15.Niu Y; Padhee S; Wu H; Bai G; Qiao Q; Hu Y; Harrington L; Burda WN; Shaw LN; Cao C Lipo-γ-AApeptides as a New Class of Potent and Broad-Spectrum Antimicrobial Agents. J. Med. Chem 2012, 55, 4003–4009. [DOI] [PubMed] [Google Scholar]
- 16.Dehsorkhi A; Castelletto V; Hamley IW Self-Assembling Amphiphilic Peptides. J. Pep. Sci 2014, 20, 453–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan T; Yu X; Shen B; Sun L Peptide Self-Assembled Nanostructures for Drug Delivery Applications. J. Nanomater 2017, 2017. [Google Scholar]
- 18.Liu L; Xu K; Wang H; Tan PJ; Fan W; Venkatraman SS; Li L; Yang Y-Y Self-Assembled Cationic Peptide Nanoparticles as an Efficient Antimicrobial Agent. Nat. Nanotechnol 2009, 4, 457. [DOI] [PubMed] [Google Scholar]
- 19.Conda-Sheridan M; Lee SS; Preslar AT; Stupp SI Esterase-Activated Release of Naproxen from Supramolecular Nanofibres. Chem. Commun 2014, 50, 13757–13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Samad MB; Maddeboina K; Rodrigues de Almeida N; Conda-Sheridan M Facile Protocol for the Synthesis of Self-Assembling Polyamine-based Peptide Amphiphiles (PPAs) and Related Biomaterials. J. Vis. Exp 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samad MB; Chhonker YS; Contreras JI; McCarthy A; McClanahan MM; Murry DJ; Conda-Sheridan M Developing Polyamine-Based Peptide Amphiphiles with Tunable Morphology and Physicochemical Properties. Macromol. Biosci 2017, 17. [DOI] [PubMed] [Google Scholar]
- 22.Krishnaiah M; de Almeida NR; Udumula V; Song Z; Chhonker YS; Abdelmoaty MM; do Nascimento VA; Murry DJ; Conda-Sheridan M Synthesis, Biological Evaluation, and Metabolic Stability of Phenazine Derivatives as Antibacterial Agents. Eur. J. Med. Chem 2018, 143, 936–947. [DOI] [PubMed] [Google Scholar]
- 23.Balakrishna R; Wood SJ; Nguyen TB; Miller KA; Kumar ES; Datta A; David SA Structural Correlates of Antibacterial and Membrane-Permeabilizing Activities in Acylpolyamines. Antimicrob. Agents Ch 2006, 50, 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim HR; Sugimoto Y; Aoki T Ovotransferrin Antimicrobial Peptide (OTAP-92) Kills Bacteria Through a Membrane Damage Mechanism. BBA-Gen. Subjects 2000, 1523, 196–205. [DOI] [PubMed] [Google Scholar]
- 25.Xu M; McCanna DJ; Sivak JG Use of the Viability Reagent PrestoBlue in Comparison with AlamarBlue and MTT to Assess the Viability of Human Corneal Epithelial Cells. J. Pharmacol. Tox. Met 2015, 71, 1–7. [DOI] [PubMed] [Google Scholar]
- 26.Ramarao N; Nielsen-Leroux C; Lereclus D The Insect Galleria Mellonella as a Powerful Infection Model to Investigate Bacterial Pathogenesis. J. Vis. Exp 2012, e4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pashuck ET; Stupp SI Direct Observation of Morphological Tranformation from Twisted Ribbons into Helical Ribbons. J. Am. Chem. Soc 2010, 132, 8819–8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moyer TJ; Cui H; Stupp SI Tuning Nanostructure Dimensions with Supramolecular Twisting. J. Phys. Chem. B 2012, 117, 4604–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan DI; Prenner EJ; Vogel HJ Tryptophan-and Arginine-Rich Antimicrobial Peptides: Structures and Mechanisms of Action. BBA-Biomembranes 2006, 1758, 1184–1202. [DOI] [PubMed] [Google Scholar]
- 30.Mi G; Shi D; Herchek W; Webster TJ Self-Assembled Arginine-Rich Peptides as Effective Antimicrobial Agents. J. Biomed. Mater. Res. A 2017, 105, 1046–1054. [DOI] [PubMed] [Google Scholar]
- 31.Mishra B; Lushnikova T; Wang G Small Lipopeptides Possess Anti-Biofilm Capability Comparable to Daptomycin and Vancomycin. RSC Adv. 2015, 5, 59758–59769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li L; Vorobyov I; Allen TW The Different Interactions of Lysine and Arginine Side Chains with Lipid Membranes. J. Phys. Chem. B 2013, 117, 11906–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clogston JD; Patri AK Zeta Potential Measurement Characterization of Nanoparticles Intended for Drug Delivery; 1rd ed; Humana Press, 2011, pp 63–70. In Springer. [DOI] [PubMed] [Google Scholar]
- 34.Wang H; Xu K; Liu L; Tan JP; Chen Y; Li Y; Fan W; Wei Z; Sheng J; Yang Y-Y The Efficacy of Self-Assembled Cationic Antimicrobial Peptide Nanoparticles Against Cryptococcus Neoformans for the Treatment of Meningitis. Biomaterials 2010, 31, 2874–2881. [DOI] [PubMed] [Google Scholar]
- 35.Travkova OG; Moehwald H; Brezesinski G The Interaction of Antimicrobial Peptides with Membranes. Adv. Colloid Interfac 2017, 247, 521–532. [DOI] [PubMed] [Google Scholar]
- 36.Zhou C; Wang F; Chen H; Li M; Qiao F; Liu Z; Hou Y; Wu C; Fan Y; Liu L Selective Antimicrobial Activities and Action Mechanism of Micelles Self-Assembled by Cationic Oligomeric Surfactants. ACS Appl. Mater. Inter 2016, 8, 4242–4249. [DOI] [PubMed] [Google Scholar]
- 37.Chung PY; Khanum R Antimicrobial Peptides as Potential Anti-Biofilm Agents Against Multidrug-Resistant Bacteria. J. Microbiol. Immunol 2017, 50, 405–410. [DOI] [PubMed] [Google Scholar]
- 38.Garrison AT; Bai F; Abouelhassan Y; Paciaroni NG; Jin S; Huigens III RW Bromophenazine Derivatives with Potent Inhibition, Dispersion and Eradication Activities Against Staphylococcus Aureus Biofilms. RSC Adv. 2015, 5, 1120–1124. [Google Scholar]
- 39.Cui H; Webber MJ; Stupp SI Self-Assembly of Peptide Amphiphiles: From Molecules to Nanostructures to Biomaterials. Biopolymers 2010, 94, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newcomb CJ; Sur S; Ortony JH; Lee O-S; Matson JB; Boekhoven J; Yu JM; Schatz GC; Stupp SI Cell Death Versus Cell Survival Instructed by Supramolecular Cohesion of Nanostructures. Nat. Commun 2014, 5, 3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanazawa K; Sato Y; Ohki K; Okimura K; Uchida Y; Shindo M; Sakura N Contribution of Each Amino Acid Residue in Polymyxin B3 to Antimicrobial and Lipopolysaccharide Binding Activity. Chem. Pharm. Bull 2009, 57, 240–244. [DOI] [PubMed] [Google Scholar]
- 42.Feiner-Gracia N; Buzhor M; Fuentes E; Pujals S; Amir RJ; Albertazzi L Micellar Stability in Biological Media Dictates Internalization in Living Cells. J. Am. Chem. Soc 2017, 139, 16677–16687. [DOI] [PubMed] [Google Scholar]
- 43.Kirkham S; Castelletto V; Hamley IW; Inoue K; Rambo R; Reza M; Ruokolainen J Self-Assembly of the Cyclic Lipopeptide Daptomycin: Spherical Micelle Formation Does not Depend on the Presence of Calcium Chloride. ChemPhysChem 2016, 17, 2118–2122. [DOI] [PubMed] [Google Scholar]
- 44.Pereira MF; Rossi CC; de Queiroz MV; Martins GF; Isaac C; Bosse JT; Li Y; Wren BW; Terra VS; Cuccui J Galleria Mellonella Is an Effective Model to Study Actinobacillus Pleuropneumoniae Infection. Microbiology 2015, 161, 387–400. [DOI] [PubMed] [Google Scholar]
- 45.Tsai CJ-Y; Loh JMS; Proft T Galleria Mellonella Infection Models for the Study of Bacterial Diseases and for Antimicrobial Drug Testing. Virulence 2016, 7, 214–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melander RJ; Melander C The Challenge of Overcoming Antibiotic Resistance: An Adjuvant Approach? ACS Infect. Dis 2017, 3, 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stokes JM; MacNair CR; Ilyas B; French S; Côté J-P; Bouwman C; Farha MA; Sieron AO; Whitfield C; Coombes BK; Brown ED Pentamidine Sensitizes Gram-Negative Pathogens to Antibiotics and Overcomes Acquired Colistin Resistance. Nat. Microbiol 2017, 2, 17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


