7-Iodo-5-aza-7-deazaguanosine represents a base-modified nucleoside with an entirely different recognition pattern as the parent purine compound. It shows an anti conformation at the glycosylic bond and an N conformation (O4′-endo) for the ribose moiety. Packing is controlled by hydrogen bonds and by a 7-iodo contact to 2′-oxygen of the ribose moiety.
Keywords: 7-iodo-5-aza-7-deazaguanosine, ribonucleoside, crystal structure, Hirshfeld surface analysis, base-pair prediction, crystal packing, all-purine RNA, pKa values
Abstract
The positional change of nitrogen-7 of the RNA constituent guanosine to the bridgehead position-5 leads to the base-modified nucleoside 5-aza-7-deazaguanosine. Contrary to guanosine, this molecule cannot form Hoogsteen base pairs and the Watson–Crick proton donor site N3—H becomes a proton-acceptor site. This causes changes in nucleobase recognition in nucleic acids and has been used to construct stable ‘all-purine’ DNA and DNA with silver-mediated base pairs. The present work reports the single-crystal X-ray structure of 7-iodo-5-aza-7-deazaguanosine, C10H12IN5O5 (1). The iodinated nucleoside shows an anti conformation at the glycosylic bond and an N conformation (O4′-endo) for the ribose moiety, with an antiperiplanar orientation of the 5′-hydroxy group. Crystal packing is controlled by interactions between nucleobase and sugar moieties. The 7-iodo substituent forms a contact to oxygen-2′ of the ribose moiety. Self-pairing of the nucleobases does not take place. A Hirshfeld surface analysis of 1 highlights the contacts of the nucleobase and sugar moiety (O—H⋯O and N—H⋯O). The concept of pK-value differences to evaluate base-pair stability was applied to purine–purine base pairing and stable base pairs were predicted for the construction of ‘all-purine’ RNA. Furthermore, the 7-iodo substituent of 1 was functionalized with benzofuran to detect motional constraints by fluorescence spectroscopy.
Introduction
The specific recognition of complementary nucleobases in the DNA coding system is based on the combination of purine and pyrimidine bases (Watson & Crick, 1953 ▸). Adenine pairs with thymine and guanine with cytosine. Nevertheless, an alternative coding system entirely composed of purines (‘all-purine’ DNA) was suggested by Wächtershäuser (1988 ▸). In this, purines glycosylated at nitrogen-9 (adenine and guanine) base pair with purines glycosylated at nitrogen-3 (motif I xanthine and motif II isoguanine, see Fig. 1 ▸; purine numbering is used throughout this article). The resulting purine–purine base pairs are isoelectronic and isogeometric to standard purine–pyrimidine Watson–Crick pairs. However, purines glycosylated at nitrogen-3 are unstable and difficult to handle (Leonard, 2020 ▸).
Figure 1.
Upper row: ‘all-purine’ pairs suggested by Wächtershäuser (1988 ▸). Lower row: purine–purine base pairing motifs of 5-aza-7-deazaguanine 2′-deoxyribonucleoside (2b) with antiparallel (aps) or parallel (ps) strand orientation (Seela et al., 2001 ▸). Notes: iGd = 2′-deoxyisoguanosine, dG = 2′-deoxyguanosine and R = β-d-2′-deoxyribofuranosyl.
Recent advances in the construction of ‘all-purine’ DNA used purine nucleosides glycosylated at nitrogen-9 (Seela et al., 1997 ▸, 2001 ▸; Seela & Melenewski, 1999 ▸; Seela & Rosemeyer, 2002 ▸). In this context, 5-aza-7-deazaguanine (imidazo[1,2-a]-1,3,5-triazine) 2′-deoxyribonucleoside (2b) has emerged as a promising purine analogue mimicking the pyrimidine site of a Watson–Crick base pair (Fig. 2 ▸). More specifically, 5-aza-7-deazaguanine is able to pair with isoguanine or guanine (Seela & Rosemeyer, 2002 ▸). The 5-aza-7-deazaguanine–isoguanine base pair forms stable ‘all-purine’ DNA with an antiparallel strand orientation (motif III, Fig. 1 ▸), whereas the 5-aza-7-deazaguanine–guanine pair leads to DNA with parallel strands (motif IV, Fig. 1 ▸). Furthermore, a configurational change at the anomeric centre from β-d to α-d resulted in heterochiral ‘all-purine’ DNA causing an additional change of the strand orientation (Seela et al., 2001 ▸). Size expansion of the double helix was studied with tricyclic purine bases (Winnacker & Kool, 2013 ▸).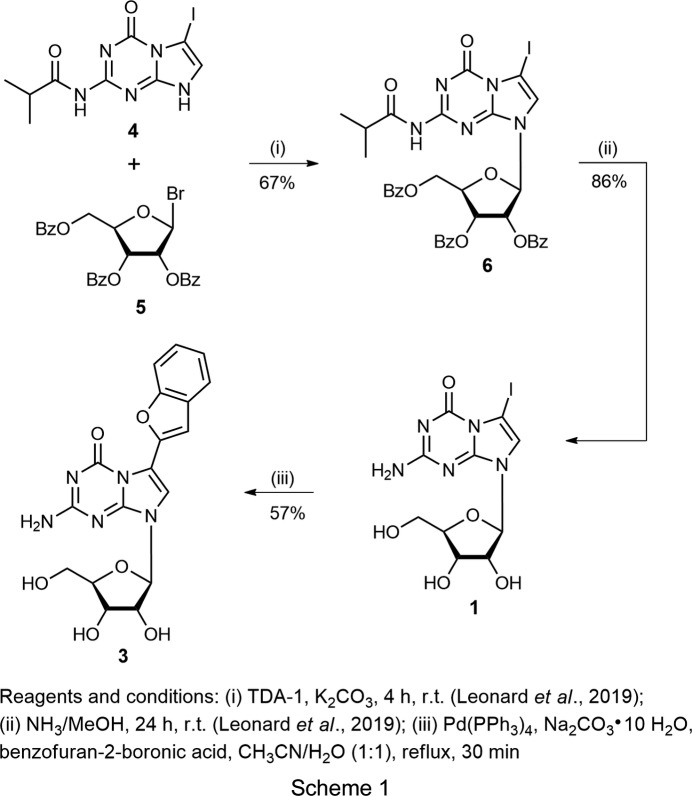
Figure 2.
5-Aza-7-deazaguanine nucleosides.
Contrary to the work performed on ‘all-purine’ DNA, very little is known in the realm of ‘all-purine’ RNA. Iodinated 5-aza-7-deazaguanine ribonucleoside 1 represents a convertible key compound within a series of 5-aza-7-deazaguanine nucleosides such as 2a–2c (Fig. 2 ▸). It has the capability to be functionalized at position-7 of the nucleobase with various functional groups and can be cross-linked at this position. Nucleosides 1 and 2a represent building blocks for the construction of ‘all-purine’ RNA. This work reports the single-crystal X-ray structure of the 7-iodo ribonucleoside 1 (Fig. 2 ▸) and its physical properties. The crystal packing has been studied and a Hirshfeld surface analysis performed to visualize packing interactions. Base-pair stabilities are predicted on the basis of differences of the pKa values (Krishnamurthy, 2012 ▸). Functionalization of 1 with a fluorescent group by cross-coupling has been performed and fluorescence data are collected.
Experimental
Synthesis and crystallization of nucleoside 1
7-Iodo-5-aza-7-deazaguanine (1) was synthesized as reported previously (Leonard et al., 2019 ▸). For crystallization, compound 1 was dissolved in a methanol/water (1:1 v/v) mixture (10 mg in 1 ml) and was obtained as colourless thin needles (m.p. 180–181 °C, decomposition) by slow evaporation of the solvent (room temperature, 24 h). A colourless needle-like specimen of 1 was used for the X-ray crystallographic analysis.
X-ray diffraction and refinement
Crystal data, data collection and structure refinement details are summarized in Table 1 ▸. The H atoms on N2, O2′, O3′ and O5′ were refined freely, but with N—H and O—H distance restraints and U iso(H) values fixed. For atom C1′, the anisotropic displacement parameters were refined with damping in order to hold this atom positive definite.
Table 1. Experimental details.
| Crystal data | |
| Chemical formula | C10H12IN5O5 |
| M r | 409.15 |
| Crystal system, space group | Monoclinic, P21 |
| Temperature (K) | 100 |
| a, b, c (Å) | 9.1086 (2), 6.3149 (2), 11.0428 (3) |
| β (°) | 95.585 (1) |
| V (Å3) | 632.17 (3) |
| Z | 2 |
| Radiation type | Cu Kα |
| μ (mm−1) | 20.25 |
| Crystal size (mm) | 0.35 × 0.06 × 0.02 |
| Data collection | |
| Diffractometer | Bruker KappaCCD APEXII |
| Absorption correction | Multi-scan (SADABS; Bruker, 2014 ▸) |
| T min, T max | 0.39, 0.69 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 15087, 2216, 2144 |
| R int | 0.072 |
| (sin θ/λ)max (Å−1) | 0.597 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.036, 0.097, 1.07 |
| No. of reflections | 2216 |
| No. of parameters | 205 |
| No. of restraints | 12 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 2.79, −1.19 |
| Absolute structure | Flack x determined using 919 quotients [(I +) − (I −)]/[(I +) + (I −)] (Parsons et al., 2013 ▸) |
| Absolute structure parameter | 0.040 (7) |
Synthesis of benzofuran conjugate 3
For the preparation of 2-amino-6-(benzofuran-2-yl)-8-(β-d-ribofuranosyl)-8H-imidazo[1,2-a]-s-triazin-4-one (3), compound 1 (Leonard et al., 2019 ▸; 100 mg, 0.24 mmol), Pd(PPh3)4 (28 mg, 0.024 mmol), Na2CO3·10H2O (352 mg, 1.23 mmol) and benzofuran-2-boronic acid (158 mg, 0.97 mmol) in CH3CN/H2O (1:1 v/v, 20 ml) were refluxed for 30 min. After consumption of starting material 1 [monitored by thin-layer chromatography (TLC)], the solvent of the reaction mixture was evaporated under reduced pressure, and the residue was applied to flash chromatography (CH2Cl2/MeOH, 90:10→75:25 v/v). Compound 3 was obtained from the main zone as a white solid (yield 55 mg, 57%). TLC (silica gel, CH2Cl2/MeOH, 85:15 v/v) R F 0.4. λ max (MeOH)/nm 312 (∊/dm3 mol−1 cm−1, 22600), 327 (18800). 1H NMR (600 MHz, DMSO-d 6): δ 8.00 (s, 1H, H-8), 7.71–7.65 (m, 2H, benzofuran Ar-H), 7.54 (dq, J = 8.4, 1.0 Hz, 1H, benzofuran Ar-H), 7.33 (ddd, J = 8.4, 7.2, 1.3 Hz, 1H, benzofuran Ar-H), 7.26 (td, J = 7.4, 1.0 Hz, 1H, benzofuran Ar-H), 7.12 (s, 2H, NH2), 5.87 (d, J = 5.8 Hz, 1H, H-1′), 5.51 (d, J = 5.5 Hz, 1H, OH-2′), 5.20 (d, J = 4.6 Hz, 1H, OH-3′), 5.16 (t, J = 5.3 Hz, 1H, OH-5′), 4.37 (q, J = 5.4 Hz, 1H, H-2′), 4.11 (td, J = 4.7, 3.4 Hz, 1H, H-3′), 3.92 (q, J = 3.6 Hz, 1H, H-4′), 3.66 (ddd, J = 11.9, 5.2, 3.8 Hz, 1H, H-5′), 3.58 (ddd, J = 11.9, 5.3, 3.6 Hz, 1H, H-5′). 13C NMR (151 MHz, DMSO-d 6): δ 164.8, 153.9, 151.9, 150.1, 144.7, 128.4, 125.1, 123.2, 121.6, 115.7, 114.7, 110.7, 108.2, 86.5, 85.5, 85.4, 73.7, 70.2, 61.1, 61.0. HRMS (ESI–TOF) m/z: [M + Na+] calculated for C18H17N5NaO6, 422.1071; found, 422.1075.
Results and discussion
Molecular geometry and conformation of 7-iodo-5-aza-7-deazaguanosine (1)
The three-dimensional (3D) structure of 1 is shown in Fig. 3 ▸(a) and selected crystallographic data and structure refinement details are summarized in Table 1 ▸. According to the Flack parameter and the synthetic pathway, the anomeric centre at C1′ is in an R configuration confirming the β-d anomeric structure of 1. The structure of the related 2′-deoxyribonucleoside has been reported previously (for selected geometric parameters of the single-crystal X-ray analysis of 2c, see Table S1 in the supporting information) (Leonard et al., 2019 ▸).
Figure 3.

(a) Perspective view of 1, showing the atom-numbering scheme. Displacement ellipsoids are drawn at the 50% probability level and H atoms are shown as small spheres of arbitrary size. (b) Overlay of ribonucleoside 1 and the related 2′-deoxyribonucleoside 2c.
To evaluate the impact of the 2′-hydroxy group of ribonucleoside 1 on the crystal structure, it was of interest to compare the geometric parameters of compound 1 with those of the 2′-deoxyribonucleoside 2c (Leonard et al., 2019 ▸). An overlay of both molecules visualizes the conformational differences mainly affecting the sugar moiety (Fig. 3 ▸ b).
The nucleobase may adopt two principal orientations with respect to the sugar moiety (syn/anti) and it is defined by the torsion angle χ (O4′—C1′—N9—C4) (IUPAC–IUB Joint Commission on Biochemical Nomenclature, 1983 ▸). Purine nucleosides are found in a wide range of anti conformations (Blackburn et al., 2006 ▸). The 7-iodo-5-aza-7-deazaguanine moiety of both nucleosides also adopts an anti conformation, with χ = −120.6 (9)° for ribonucleoside 1 (Table 2 ▸) and χ = −139.9 (6)° for 2c (Leonard et al., 2019 ▸). The glycosylic bond lengths of 1 (Table 2 ▸) and 2c (Lin et al., 2004 ▸) are within the range of purine nucleosides (Saenger, 1984 ▸).
Table 2. Selected geometric parameters (Å, °).
| I7—C7 | 2.081 (10) | N9—C1′ | 1.439 (12) |
| C4—N9—C1′ | 123.7 (8) | O6—C6—N1 | 125.1 (8) |
| C1′—O4′—C4′ | 103.7 (7) | N5—C7—I7 | 126.9 (7) |
| N2—C2—N1 | 117.1 (9) | O5′—C5′—C4′ | 110.3 (8) |
| C4′—O4′—C1′—C2′ | −41.3 (9) | C1′—O4′—C4′—C3′ | 47.8 (9) |
| C4—N9—C1′—O4′ | −120.6 (9) | C2′—C3′—C4′—O4′ | −34.8 (9) |
| O4′—C1′—C2′—C3′ | 18.1 (9) | C3′—C4′—C5′—O5′ | −172.9 (8) |
| C1′—C2′—C3′—C4′ | 9.8 (10) |
The ribose ring is twisted out of plane to minimize steric interactions of the substituents, referred to as sugar puckering (Altona & Sundaralingam, 1972 ▸). The more electronegative substituents of C2′ and C3′ prefer an axial orientation. Nucleosides can adopt two principal sugar puckering modes, namely C3′-endo (N) and C2′-endo (S) corresponding to the major displacement of C3′ or C2′ from the median C1′/O4′/C4′ plane. The C2′-endo conformation is the preferred puckering mode of 2′-deoxyribonucleosides (Saenger, 1984 ▸), whereas ribonucleosides show a preference for the C3′-endo sugar pucker. Also, 7-iodo ribonucleoside 1 shows an N conformation, with a pseudorotational phase angle P = 77.87° and a maximum amplitude τm = 47.14°. However, the typical C3′-endo conformation is less pronounced and the sugar pucker is shifted towards O4′-endo. This is a sugar conformation where the nucleobase and the exocyclic 5′-OH group are arranged equatorially. The sugar moiety of 2′-deoxyribonucleoside 2c also adopts an N-type sugar conformation (C3′-endo–C4′-exo, 3 T 4, P = 28.55° and τm = 34.28°) (Leonard et al., 2019 ▸).
In many cases, the solid-state conformation of nucleosides differs from that in solution and is also different from that of nucleosides as constituents of a single- or double-stranded nucleic acid. Sugar puckering of 1 was determined in solution from the coupling constants of high-resolution 1H NMR spectra (600 MHz) (for details, see Table S2 in the supporting information). The population of S versus N conformers was calculated on the basis of coupling constants using the program PSEUROT (Version 6.3; Van Wijk et al., 1999 ▸). Notably, the conformation found for ribonucleoside 1 in solution is S-type (69%) and is different from that in the solid state (N-type). The closely related 2′-deoxyribonucleoside 2c also prefers an S conformation (62%) in solution (Lin et al., 2004 ▸).
The orientation of the exocyclic 5′-hydroxy group relative to the sugar ring is described by the torsion angle γ (O5′—C5′—C4′—C3′). Surprisingly, γ is identical in both nucleosides despite the fact of their having different sugar moieties (ribose versus 2′-deoxyribose). The 5′-hydroxy group adopts, in both cases, an antiperiplanar (trans) conformation, with γ = −172.9 (8)° for 1 and γ = −172.7 (4)° for 2c (Leonard et al., 2019 ▸).
Hydrogen bonding and molecular packing
In nucleoside solid-state structures, hydrogen bonds are formed between N and/or O atoms functioning as proton donors or acceptors. They can be provided by the nucleobases and/or the sugar moieties. Fig. 4 ▸(a) displays the hydrogen-bonding pattern of 7-iodo-5-aza-7-deazaguanosine (1) within one particular layer.
Figure 4.
(a) Hydrogen bonding of 1, viewed along the b axis. (b) Multilayered molecular packing of 1, shown along the slightly inclined b axis. (c) Detailed view of part (a), showing the nucleobase–sugar interaction forming a central core.
Nucleobase-to-nucleobase interactions, such as self-pairing in a Watson–Crick-like fashion, are not observed for ribonucleoside 1. This is different from the solid-state structure of the 5-aza-7-deazaguanine nucleobase reported recently (Laos et al., 2019 ▸). The free nucleobase is able to adopt tautomeric forms with the H atom located either at N9 or at N3. The N9—H tautomer can base pair with the N3—H tautomer, forming a tridentate base pair (Laos et al., 2019 ▸). However, in the context of DNA or RNA structure, this is an artificial situation, as nucleosides glycosylated at N9 lose the proton at the heterocycle and thus the ability to form these tautomers.
Compound 1 forms in the crystal a core of four molecules displaying a ‘parallelogram’-like arrangement, with the O2′—H2′ and C2′—H2′1 groups as proton donors and atom O6 of the nucleobase as a bi-acceptor (Fig. 4 ▸ a and Table 3 ▸). This structure is surrounded by ‘cyclohexane’-like motifs formed by two molecules connected via N2—H2A⋯O4′i and C1′—H1′⋯N2v hydrogen bonds. Further stabilization of the solid-state structure is achieved by hydrogen bonds formed by the exocyclic 5′-OH group (N2—H2B⋯O5′ii and O5′—H5′⋯O3′iv).
Table 3. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N2—H2A⋯O4′i | 0.90 (3) | 2.25 (10) | 2.941 (11) | 133 (11) |
| N2—H2B⋯O5′ii | 0.90 (3) | 2.04 (5) | 2.905 (12) | 160 (12) |
| O2′—H2′⋯O6iii | 0.87 (3) | 1.93 (8) | 2.689 (9) | 144 (12) |
| O5′—H5′⋯O3′iv | 0.87 (3) | 1.87 (7) | 2.673 (10) | 151 (13) |
| C1′—H1′⋯N2v | 1.0 | 2.63 | 3.345 (15) | 128 |
| C2′—H2′1⋯O6vi | 1.0 | 2.57 | 3.541 (12) | 163 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  ; (vi)
; (vi)  .
.
It is well documented that the iodo substituents of halogenated nucleosides can participate in molecular interactions, sometimes even playing a predominant role. Strong iodo–iodo interactions have been reported for 5-iodo-α-2′-deoxycytidine (Müller et al., 2019 ▸), while the iodo substituent of 2′-deoxyribonucleoside 2c shows a strong contact to atom O3′ [I7⋯O3′ = 2.794 (4) Å and C7—I7⋯O3′ = 169.2 (2)°] (Leonard et al., 2019 ▸). In the crystal structure of 7-iodo-5-aza-7-deazaguanosine (1), the iodo substituent I7 shows a contact to atom O2′ [2.936 (7) Å and C7—I7⋯O2′vi = 170.4 (3)°; for symmetry code, see Table 3 ▸]. However, the I⋯O contact of ribonucleoside 1 is less strong than that in compound 2c.
The individual nucleoside layers are stacked upon each other in a very regular fashion, as demonstrated in Fig. 4 ▸(b). Fig. 5 ▸ shows that compound 1 forms chains which are arranged in an alternating reverse fashion (A B A B…). Two chains (A and B) are connected to each other by hydrogen bonds (C1′—H1′⋯N2v, N2—H2B⋯O5′ii and O5′—H5′⋯O3′iv; Table 3 ▸). However, as was discussed above, the ribonucleoside molecules are not arranged in a Watson–Crick-like fashion. Instead, the nucleobase of one chain is placed opposite the sugar residue of the next chain. As a result, the iodo substituent points towoards the outside of each double chain (composed of A and B chains). The double chains are connected to each other by the C2′—H2′1⋯O6vi hydrogen bond. This arrangement can also be visualized by the space-filling model shown in Fig. 5 ▸.
Figure 5.
(Top) The crystal packing of 1, showing the hydrogen bonds along the a axis. (Bottom) Space-filling model of 1, viewed along the a axis.
Hirshfeld surface analysis of 1
Hirshfeld surface analysis is a valuable tool to obtain additional insight into the role of crystal packing forces and to visualize the intermolecular interactions of crystalline compounds. The CrystalExplorer program (Version 17; Spackman & Jayatilaka, 2009 ▸; Turner et al., 2017 ▸) was used to conduct a Hirshfeld surface analysis mapped over d norm (−0.5 to 1.5 Å), shape index (−1.0 to 1.0 Å) and curvedness (−4.0 to 0.4 Å) (Fig. S2 in the supporting information), as well as a two-dimensional (2D) fingerprint plot analysis. The Hirshfeld surface of compound 1 is shown in Figs. 6 ▸(a) and 6(b). Several red spots are visible on the Hirshfeld surface corresponding to the close O—H⋯O and N—H⋯O contacts of the nucleobase and sugar moieties. The shape index indicates π–π stacking interactions by the presence of adjacent red and blue triangles. Their absence clearly suggests that there are no π–π interactions in compound 1, as demonstrated by Figs. 6 ▸(c) and 6(d).
Figure 6.
Hirshfeld surface of compound 1, mapped over d norm (−0.5 to 1.5 Å), shown in (a) a front and (b) a back view. The shape index, shown in (c) a front and (d) a back view.
2D fingerprint plots can be resolved to highlight particular atom pair interactions. This enables the separation of contributions from different contact types that overlap in the full fingerprint plot. The overall 2D fingerprint plot and those resolved into H⋯C/C⋯H, H⋯H, I⋯O/O⋯I, N⋯H/H⋯N and O⋯H/H⋯O contacts are illustrated in Fig. 7 ▸, together with their relative contributions to the Hirshfeld surface. The proportions of O⋯H/H⋯O, H⋯H and N⋯H/H⋯N interactions comprise 28.7, 19.1 and 15.9%, respectively, of the total Hirshfeld surfaces. Lower percentages are found for the H⋯C/C⋯H contacts (9.2%). Moreover, the interaction between iodo substituent I7 and atom O2′ is confirmed and appears as two wings in the fingerprint plot, with a proportion of 4.1%.
Figure 7.
2D fingerprint plots showing the percentages of the contribution of various interactions to the total Hirshfeld surface area of compound 1. Full interactions (upper row, left) and resolved contacts (upper row: middle, C⋯H/H⋯C; right, H⋯H; lower row: left, I⋯O/O⋯I; middle, N⋯H/H⋯N; right, O⋯H/H⋯O).
The stability of 5-aza-7-deazaguanine base pairs controlled by pK value differences of complementary bases
The Watson–Crick base-pair recognition of canonical DNA and RNA constituents depends on the hydrogen-bonding donor–acceptor pattern of purine and pyrimidine nucleosides, as well as on environmental influences caused by base-to-base stacking interactions, helix formation and various other factors. Among these, the pK values of nucleobases play an important role as they control the stability and lifetime of base pairs. Recently, the role of the pK values of nucleobases has been discussed with regard to the origin of chemical evolution. Strong base pairs are formed when the pK value difference (ΔpK) between the acceptor and donor sites of nucleobases is at least five units or more. A ΔpK < 5 results in weaker base pairs (Krishnamurthy, 2012 ▸).
Early studies on the physical properties of 5-aza-7-deaza-2′-deoxyguanosine (2b) showed that the nucleobase is protonated on the s-triazine moiety having a pKa value of 3.7 (Rosemeyer & Seela, 1987 ▸). Nitrogen-1 was established as the protonation site using 13C NMR spectroscopy (Seela & Melenewski, 1999 ▸). The pKa value and the protonation site is different from the closely related 2′-deoxyguanosine (pKa value of protonation = 1.6 and deprotonation = 9.2) (Seela et al., 2003 ▸), as well as to 7-deaza-2′-deoxyguanosine (1.1 and 10.3) (Ramzaeva & Seela, 1996 ▸), and is a direct consequence of the transposition of the nitrogen-7 atom to bridgehead position-5.
The positional change of the N atom leads to the absence of a proton at N1 which now becomes a proton-acceptor site and not a donor site as in 2′-deoxyguanosine or 7-deaza-2′-deoxyguanosine. Furthermore, position-7 cannot interact with protons or metal ions anymore, and similar to 7-deaza-2′-deoxyguanosine, Hoogsteen base pairs cannot be formed. Consequently, 5-aza-7-deaza-2′-deoxyguanosine exhibits a Watson–Crick recognition face similar to that of isocytidine. Thus, the complementary base has to provide a proton, as observed for base pairing with guanine or isoguanine, or with protonated cytidines or cytidine derivatives which carry a proton at nitrogen-1 (Fig. 8 ▸ a) (Hoshika et al., 2018 ▸). Silver ions can function as proton substitutes forming metal-mediated base pairs (Fig. 8 ▸ b) (Guo et al., 2018 ▸).
Figure 8.
(a) Protonated base pair of 5-aza-7-deaza-2′-deoxyguanosine (2b) and 2′-deoxycytidine formed in acidic medium. (b) Silver-mediated base pair of 2b and dC.
As ribonucleoside 1 is a promising candidate for the synthesis of ‘all-purine’ RNA, it is of particular importance to identify the pKa value of 1 and to compare this value to related purine and 7-deazapurine nucleosides. The pKa value of compound 1 was determined by UV spectroscopic titration from absorbance changes at 280 nm at different pH values (Fig. S3 in the supporting information). According to Fig. 9 ▸, the pKa value of 1 is 3.6. This pKa value is almost identical to that of the corresponding 2′-deoxyribonucleoside 2c (pKa = 3.7) (Lin et al., 2004 ▸) and the purine nucleoside adenosine (pKa = 3.6). Contrary to the related purine and 7-deazapurine nucleosides, the molecule does not show a second pK value (for deprotonation), as the heterocyclic skeleton does not carry a proton which can be released in alkaline medium. Atom N1 is the protonation site, as was determined for 5-aza-7-deazaguanine 2′-deoxyribonucleoside 2b (Seela & Melenewski, 1999 ▸).
Figure 9.
pKa value of 1 determined by UV titration (1st derivative, red line). Spectroscopic changes determined at 280 nm at different pH values (black line) were measured in 0.1 M sodium phosphate buffer.
Table S3 (see supporting information) summarizes the pK values and protonation positions for 5-aza-7-deazaguanine, 7-deazaguanine, guanine and isoguanine nucleosides. It is obvious that the iodo substituent has no significant influence on the pK value of the nucleosides. However, compared to guanosine and 7-deazaguanosine, the pK for protonation of the unsubstituted and iodinated 5-aza-7-deazaguanosine base is strongly shifted and the molecule is much easier to protonate. Also, the position of protonation (N1) is different from that in guanosine (N7) and 7-deazaguanosine (N3).
It has been shown that based on pK value differences of nucleosides participating in base-pair formation, predictions regarding base-pair stability can be made (Krishnamurthy, 2012 ▸). To this end, the ΔpK of base pairs of 7-iodo-5-aza-7-deazaguanosine with different nucleosides acting as donor or acceptor were calculated and compared (Fig. 10 ▸). Antiparallel (aps) and parallel (ps) strand orientation of oligonucleotides within a duplex were considered.
Figure 10.
Base-pairing motifs of 5-aza-7-deazaguanine nucleosides, pKa values of the base-pairing nucleosides and their pK value difference (ΔpK), as well as strand orientation of the respective duplex. Notes: R = β-d-ribofuranosyl, C = cytidine, iG = isoguanosine, iC = isocytidine, G = guanosine, c7G = 7-deazaguanosine, aps = antiparallel strands and ps = parallel strands.
The ΔpK of the 7-iodo-5-aza-7-deazaguanosine–isoguanosine base pair is 6.2 (motif I, Fig. 10 ▸) referring to an aps orientation of oligonucleotides. As a result, the stability of this purine–purine base pair is expected to be higher than for the Watson–Crick purine–pyrimidine G–C base pair (ΔpK = 4.8; motif VII, Fig. 10 ▸). A parallel arrangement of 1 with guanosine (ΔpK = 5.7; motif II) or 7-deazaguanosine (ΔpK = 6.6; motif III) also leads to stable purine–purine pairs. However, the formation of a purine–pyrimidine base pair with 1 results in a mismatch situation in an aps alignment with cytidine (ΔpK = 0.9; motif IV) or isocytidine (ΔpK = 0.7; motif VI). Consequently, the RNA constituent 1 is a promising candidate for the construction of stable purine–purine base pairs as building blocks for ‘all-purine’ RNA with a conventional antiparallel orientation of the strands. Moreover, even a parallel orientation of oligonucleotides might be applicable. The 7-iodo substituent allows the introduction of reporter groups to monitor structural characteristics of aps or ps ‘all-purine’ RNA.
However, self-pairing of 1, as demonstrated by motif V (ps alignment; Fig. 10 ▸) or even in a shifted arrangement, leads to a mismatch situation. Moreover, none of the reported crystal structures of 5-aza-7-deazaguanine nucleosides show self-pairing of the nucleobases (Kojić-Prodić et al., 1982 ▸; Seela et al., 2002 ▸; Leonard et al., 2019 ▸). Only for the isolated 5-aza-7-deazaguanine nucleobase was self-pairing observed when one of the nucleobases forms a N3—H tautomer (Laos et al., 2019 ▸). In nucleosides, as well as in nucleic acids, this arrangement is not possible as the ribose moiety is attached to N9 and the N3 atom lacks a proton.
Functionalization of nucleoside 1 and fluorescence properties of the conjugate 3
The ability of 7-iodoribonucleoside 1 to act as a target for functionalization with functional reporter groups is demonstrated by use of the Suzuki–Miyaura cross-coupling reaction (Agrofoglio et al., 2003 ▸). As the benzofuran residue is sensitive to molecular crowding and the viscosity of solvents (Greco & Tor, 2007 ▸; Leonard et al., 2019 ▸), this residue was chosen for functionalization of ribonucleoside 1.
For this, compound 1 was used as the starting material (see Scheme 1). The synthesis of ribonucleoside 1 by nucleobase anion glycosylation was reported recently, employing isobutyrylated iodo base 4 and bromo sugar 5 (Leonard et al., 2019 ▸). In order to introduce the benzofuran side chain, Suzuki–Miyaura cross-coupling of ribonucleoside 1 and benzofuran-2-boronic acid was performed. The reaction was carried out under reflux for 30 min in a CH3CN/H2O mixture (1:1 v/v) in the presence of Pd(PPh3)4 and Na2CO3·H2O (see Scheme 1), yielding benzofuran derivative 3 in 57%.
The new compound 3 was characterized by 1H and 13C NMR spectroscopy, as well as by ESI–TOF mass spectroscopy. 1H–13C correlated (HMBC and HSQC) NMR spectra were used to assign the 13C NMR signals (for spectra, see the supporting information, and for details, see the Experimental section).
The UV and fluorescence spectra of 3 were recorded in solvents of different polarity, and the corresponding Stokes shift and quantum yields were determined (Table S4 in the supporting information). The UV spectra of compound 3 show two maxima at around 312 and 327 nm in MeOH, MeCN and water (Fig. S4 in the supporting information). The maxima are bathochromally shifted (around 5 nm) in the less polar solvents dimethyl sulfoxide (DMSO) and dimethylformamide (DMF).
The fluorescence emission spectra for benzofuran conjugate 3 in various solvents are displayed in Fig. 11 ▸(a). The highest quantum yields are obtained in DMF and DMSO (Table S4 in the supporting information), with emission maxima which are red-shifted compared to the other solvents. The fluorescence of 3 is very low in water compared to the other polar solvents.
Figure 11.
Fluorescence emission spectra of compound 3 (1 µM) measured in (a) solvents of different polarity, (b) ethylene glycol and (c) glycerol. Excitation wavelength of nucleoside 3 was 315 nm in ethylene glycol and 345 nm in glycerol.
In the RNA constituent 3, the benzofuran moiety and the 5-aza-7-deazaguanine base are separated by a freely rotatable aryl–aryl bond. The spatial positioning of both aromatic systems with respect to each other (π–π conjugation) has a direct impact on the fluorescence properties of conjugate 3. In order to study this effect for 3, fluorescence measurements were performed in solvents of similar polarity but different viscosity. Fig. 11 ▸(b) shows the fluorescence emission spectra of conjugate 3 determined in ethylene glycol, water and their mixtures. The fluorescence of 3 is highest in ethylene glycol and decreases with reduced viscosity of ethylene glycol/water mixtures. However, the highest fluorescence is observed in a 50% glycerol–water mixture (Fig. 11 ▸ c). Apparently, a rotational barrier exists for the benzofuran residue which is sensitive to solvent viscosity, a phenomenon which has been reported for other nucleoside–benzofuran conjugates (Tanpure & Srivatsan, 2018 ▸; Tokugawa et al., 2016 ▸; Manna et al., 2018 ▸).
Conclusion and outlook
The single-crystal X-ray structure of 7-iodo-5-aza-7-deazaguanosine (1) was determined and the geometric parameters of this RNA constituent were analyzed. This includes an anti conformation of the 5-aza-7-deazaguanine base, an N conformation (O4′-endo) of the ribose moiety and an antiperiplanar orientation of the 5′-hydroxy group.
The crystal packing is controlled by interactions between the nucleobase and sugar moieties, while self-pairing of the nucleobase does not take place. The 7-iodo substituent forms a contact to oxygen-O2′ of the ribofuranosyl moiety. A Hirshfeld surface analysis of 1 highlights the contacts of the nucleobase and sugar moieties (O—H⋯O and N—H⋯O). The concept of pK value differences was applied to predict base pairing of 1. Ribonucleoside 1 has the ability to form extraordinary stable antiparallel-stranded purine–purine pairs with isoguanosine or base pairs with guanosine displaying a parallel-strand arrangement. Related base pairs were described for ‘all-purine’ DNA (Seela et al., 2001 ▸).
Due to the large surface area of purine–purine pairs, base stacking in ‘all-purine’ RNA is expected to be stronger, as in canonical RNA formed by pyrimidine–purine pairs (Figs. 12 ▸ a and 12b). Furthermore, it was shown that the 7-iodo substituent of ribonucleoside 1 could be functionalized. Benzofuran was used in this work to detect restrictions of rotational motion by fluorescence spectroscopy.
Figure 12.
Illustration of RNA duplexes with pyrimidine–purine and purine–purine pairs with (a)/(c) antiparallel-stranded (aps) and (b)/(d) parallel-stranded (ps) orientations. Notes: Cyt = cytosine, Gua = guanine, iCyt = isocytosine, z5Gua = 5-aza-7-deazaguanine and iGua = isoguanine.
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2053229620004684/yf3200sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2053229620004684/yf3200Isup2.hkl
Geometric parameters of 2c, curvedness of 1, pKa values, photophysical data of 3, NMR spectra of compound 1. DOI: 10.1107/S2053229620004684/yf3200sup3.pdf
CCDC reference: 1950946
Acknowledgments
We thank Dr Peter Leonard for a critical reading of the manuscript. We would like to thank Dr Letzel, Organisch-Chemisches Institut, Universität Münster, Germany, for the measurement of the mass spectra and Professor Dr B. Wünsch, Institut für Pharmazeutische und Medizinische Chemie, Universität Münster, for providing us with 600 MHz NMR spectra. Funding by ChemBiotech, Münster, Germany, is gratefully acknowledged.
References
- Agrofoglio, L. A., Gillaizeau, I. & Saito, Y. (2003). Chem. Rev. 103, 1875–1916. [DOI] [PubMed]
- Altona, C. & Sundaralingam, M. (1972). J. Am. Chem. Soc. 94, 8205–8212. [DOI] [PubMed]
- Blackburn, G. M., Gait, M. J., Loakes, D. & Williams, D. M. (2006). Editors. Nucleic Acids in Chemistry and Biology, 3rd ed. Cambridge: RSC Publishing.
- Bruker (1998). XP. Version 5.1. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2014). SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2015). SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2016). APEX3. Bruker AXS Inc., Madison, Wisconsin, USA.
- Greco, N. J. & Tor, Y. (2007). Tetrahedron, 63, 3515–3527. [DOI] [PMC free article] [PubMed]
- Guo, X., Leonard, P., Ingale, S. A., Liu, J., Mei, H., Sieg, M. & Seela, F. (2018). Chem. Eur. J. 24, 8883–8892. [DOI] [PubMed]
- Hoshika, S., Singh, I., Switzer, C., Molt, R. W. Jr, Leal, N. A., Kim, M.-J., Kim, M.-S., Kim, H.-J., Georgiadis, M. M. & Benner, S. A. (2018). J. Am. Chem. Soc. 140, 11655–11660. [DOI] [PubMed]
- IUPAC–IUB Joint Commission on Biochemical Nomenclature (1983). Eur. J. Biochem. 131, 9–15. [DOI] [PubMed]
- Kojić-Prodić, B., Ružić-Toroš, Ž., Golič, L., Brdar, B. & Kobe, J. (1982). Biochim. Biophys. Acta, 698, 105–110. [DOI] [PubMed]
- Krishnamurthy, R. (2012). Acc. Chem. Res. 45, 2035–2044. [DOI] [PMC free article] [PubMed]
- Laos, R., Lampropoulos, C. & Benner, S. A. (2019). Acta Cryst. C75, 22–28. [DOI] [PubMed]
- Leonard, P. (2020). Personal communication.
- Leonard, P., Kondhare, D., Jentgens, X., Daniliuc, C. & Seela, F. (2019). J. Org. Chem. 84, 13313–13328. [DOI] [PubMed]
- Lin, W., Zhang, X. & Seela, F. (2004). Helv. Chim. Acta, 87, 2235–2244.
- Manna, S., Sarkar, D. & Srivatsan, S. G. (2018). J. Am. Chem. Soc. 140, 12622–12633. [DOI] [PMC free article] [PubMed]
- Müller, S. L., Zhou, X., Leonard, P., Korzhenko, O., Daniliuc, C. & Seela, F. (2019). Chem. Eur. J. 25, 3077–3090. [DOI] [PubMed]
- Parsons, S., Flack, H. D. & Wagner, T. (2013). Acta Cryst. B69, 249–259. [DOI] [PMC free article] [PubMed]
- Ramzaeva, N. & Seela, F. (1996). Helv. Chim. Acta, 79, 1549–1558.
- Rosemeyer, H. & Seela, F. (1987). J. Org. Chem. 52, 5136–5143.
- Saenger, W. (1984). In Principles of Nucleic Acid Structure, edited by C. R. Cantor. New York: Springer-Verlag.
- Seela, F., Amberg, S., Melenewski, A. & Rosemeyer, H. (2001). Helv. Chim. Acta, 84, 1996–2014.
- Seela, F. & Melenewski, A. (1999). Eur. J. Org. Chem. 1999, 485–496.
- Seela, F., Melenewski, A. & Wei, C. (1997). Bioorg. Med. Chem. Lett. 7, 2173–2176.
- Seela, F., Ramzaeva, N. & Rosemeyer, H. (2003). Science of Synthesis, Vol. 16, edited by Y. Yamamoto, pp. 945–1108. Stuttgart: Georg Thieme Verlag.
- Seela, F. & Rosemeyer, H. (2002). Recent Advances in Nucleosides: Chemistry and Chemotherapy, edited by C. K. Chu, pp. 505–533. Amsterdam: Elsevier.
- Seela, F., Rosemeyer, H., Melenewski, A., Heithoff, E.-M., Eickmeier, H. & Reuter, H. (2002). Acta Cryst. C58, o142–o144. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Tanpure, A. A. & Srivatsan, S. G. (2018). ChemBioChem, 13, 2392–2399. [DOI] [PubMed]
- Tokugawa, M., Masaki, Y., Canggadibrata, J. C., Kaneko, K., Shiozawa, T., Kanamori, T., Grøtli, M., Wilhelmsson, L. M., Sekine, M. & Seio, K. (2016). Chem. Commun. 52, 3809–3812. [DOI] [PubMed]
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. University of Western Australia. http://hirshfeldsurface.net.
- Van Wijk, L., Haasnoot, C. A. G., de Leeuw, F. A. A. M., Huckriede, B. D., Westra Hoekzema, A. J. A. & Altona, C. (1999). PSEUROT6.3. Leiden Institute of Chemistry, Leiden University, The Netherlands.
- Wächtershäuser, G. (1988). Proc. Natl Acad. Sci. USA, 85, 1134–1135. [DOI] [PMC free article] [PubMed]
- Watson, J. D. & Crick, F. H. (1953). Nature, 171, 737–738. [DOI] [PubMed]
- Winnacker, M. & Kool, E. T. (2013). Angew. Chem. Int. Ed. 52, 12498–12508. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2053229620004684/yf3200sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2053229620004684/yf3200Isup2.hkl
Geometric parameters of 2c, curvedness of 1, pKa values, photophysical data of 3, NMR spectra of compound 1. DOI: 10.1107/S2053229620004684/yf3200sup3.pdf
CCDC reference: 1950946













