Abstract
The mechanisms whereby Mycobacterium tuberculosis (Mtb) rewires the host metabolism in vivo are surprisingly unexplored. Here, we used three high-resolution mass spectrometry platforms to track altered lung metabolic changes associated with Mtb infection of mice. The multiplatform data sets were merged using consensus orthogonal partial least squares-discriminant analysis (cOPLS-DA), an algorithm that allows for the joint interpretation of the results from a single multivariate analysis. We show that Mtb infection triggers a temporal and progressive catabolic state to satisfy the continuously changing energy demand to control infection. This causes dysregulation of metabolic and oxido-reductive pathways culminating in Mtb-associated wasting. Notably, high abundances of trimethylamine-N-oxide (TMAO), produced by the host from the bacterial metabolite trimethylamine upon infection, suggest that Mtb could exploit TMAO as an electron acceptor under anaerobic conditions. Overall, these new pathway alterations advance our understanding of the link between Mtb pathogenesis and metabolic dysregulation and could serve as a foundation for new therapeutic intervention strategies. Mass spectrometry data has been deposited in the Metabolomics Workbench repository (data-set identifier: ST001328).
Keywords: tuberculosis, metabolomics, pulmonary tuberculosis, tuberculosis progression, functional metabolomics, multiplatform metabolomics, data fusion
Introduction
Tuberculosis (TB) is caused by the obligate pathogen Mycobacterium tuberculosis (Mtb). It is estimated that one-quarter of the world’s population is latently infected with the bacilli, from which 5–10% develop active tuberculosis.1,2 The increased prevalence of multidrug-resistant TB (MDR-TB) and extensively drug-resistant TB (XDR-TB) cases represents a potential threat to global health as the therapeutic arsenal for drug-resistant TB treatment is limited.3 Thus, there is an urgent need for new diagnostic and therapeutic strategies to control this epidemic, including new biomarkers and host-directed therapies, the development of which would be assisted by a comprehensive mechanistic knowledge of host–pathogen interactions.
Metabolomics has been employed for the identification of TB diagnostic biomarkers, the evaluation of potential therapeutics, and the study of the biological mechanisms underlying TB disease onset and progression in both in vitro and in vivo animal models, as well as in human patients.4−6 Characterizing how the host metabolome is altered during Mtb infection is critically important as it may lead to the discovery of new pathways essential for protection against the bacillus and the identification of host-directed therapies.
Previous metabolomic studies in the TB field have contributed to our knowledge of the in vivo carbon sources available to Mtb,7,8 the ability of Mtb to withstand the oxido-reductive stress present in the TB lung,9 and the role of immunometabolism in driving effector functions of the immune cells in tuberculosis.10−14 However, there is a gap in our understanding of how Mtb infection modulates the host metabolome over the course of infection; several reasons are responsible for this breach in our knowledge. First, a limiting factor is that few studies have examined the dynamic metabolic alterations that occur during the course of Mtb infection.15 For example, only three metabolomic studies using nuclear magnetic resonance (NMR) examined the lungs of Mtb-infected animal models,16−18 of which only one guinea pig pulmonary TB model was investigated at multiple time points.17,18 While NMR is robust and well established, it suffers from relatively low sensitivity. Second, another limiting factor is that a single platform metabolomic approach was utilized in all lung metabolomic studies.16−18 The simultaneous use of distinct separation techniques coupled with high-resolution mass spectrometry (HRMS) analyzers enables the separation of specific metabolite subsets according to specific physicochemical properties while permitting accurate mass measurements of the metabolites. Such a multiplatform metabolomic approach can reach higher levels of metabolite coverage, sensitivity, and specificity.19 Unfortunately, to the best of our knowledge, untargeted multiplatform-based metabolomic analyses to examine how Mtb infection affects the lung metabolome have not yet been reported. Likely reasons include the different methodological limitations described for lung metabolomics.4
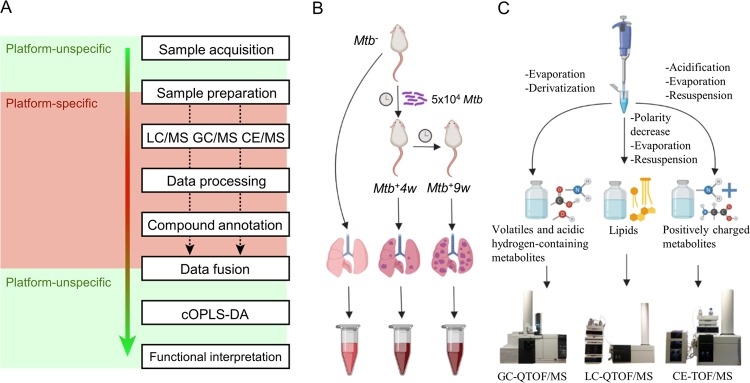
Here, we examined how Mtb affects the host metabolome during infection by exploiting capillary electrophoresis-time-of-flight (CE-TOF/MS), gas chromatography-quadrupole-time-of-flight (GC-QTOF/MS), and liquid chromatography-quadrupole-time-of-flight (LC-QTOF/MS) as analytical platforms. A downstream bioinformatics pipeline employing data fusion algorithms, multivariate statistics, and functional metabolomics was then used to characterize the global metabolomic changes in the lungs of Mtb-infected mice at different time points during infection (Figure 1A–C). This enabled us to identify new and unexpected host disease-associated metabolic pathways including, but not limited to, amino acid, carbohydrate, and fatty acid metabolism and consumption, central carbon metabolism, oxido-reductive stress, and polyamine metabolism of TB modulated by Mtb. Overall, our findings have implications that may contribute toward a better understanding of the mechanisms of the disease and new strategies for the pharmacological control of TB. To the best of our knowledge, this is the first untargeted, MS-based lung metabolomic study characterizing the progression of pulmonary TB in the mouse model for TB.
Figure 1.
Applied metabolomic workflow summary. Simplified metabolomic workflow (A), including consensus orthogonal partial least squares discriminant analysis (cOPLS-DA) data fusion approach. Experimental design, and lung homogenate collection (B). Platform-specific sample treatment and HRMS analysis (C). “Platform unspecific” refers to tasks that are universal regardless of the MS platform being used. “Platform-specific” tasks will vary depending on the MS platform. Note that each metabolomic experiment allows the detection of specific subsets of metabolites.
Experimental Section
Mice and Mtb Infection
Both male and female age-matched C57BL/6 mice (8–10 weeks old) were infected with Mtb H37Rv in an animal BSL-3 laboratory and monitored with food and water ad libitum. Mice were sacrificed by anesthesia with isoflurane followed by gentle cervical dislocation as approved by the institutional Animal Protocol Number (APN): 08591. Mice experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Alabama at Birmingham. For mice studies, we adhered to the national/international regulation of “Public Health Service Policy on Humane Care and Use of Laboratory Animals” (NIH) and “Animal Welfare Act and Animal Welfare Regulations” (USDA). Mouse genotype was confirmed by PCR and Western blotting. Mtb H37Rv was grown at 37 °C with shaking in BD Difco Middlebrook 7H9 media supplemented with 0.2% glycerol and ADS (albumin, dextrose, NaCl) with 0.02% tyloxapol. Mice were infected with 5 × 104Mtb H37Rv via the intratracheal route. Lungs were collected from uninfected (male, n = 2; female, n = 2) and Mtb-infected mice at 4 weeks (male, n = 2; female, n = 2) and 9 weeks (male, n = 2; female, n = 3) postinfection and stored immediately at −80 °C for further processing and metabolite extraction.
Metabolite Extraction
Samples for metabolite analysis were prepared as described previously.11,20 Briefly, 1 mL of 50% methanol was added to 100 mg of Mtb-infected or uninfected lung tissue and homogenized in a dounce homogenizer to prepare a uniform suspension. For CE-TOF/MS, 200 μL of homogenate was mixed with 200 μL of 0.2 M formic acid and vortexed for 2 min. The samples were cleared by centrifugation at 16 000g for 10 min at 4 °C, and the supernatant was filter-sterilized using 0.22 μm spin-X columns (Sigma). For GC-QTOF/MS and LC-QTOF/MS, 200 μL of each sample homogenate was mixed with 800 μL of 80:20 methanol/methyl tert-butyl ether (MTBE) and vortexed for 2 min. Metabolites were then extracted for 1 h with shaking at room temperature and then centrifuged at 4000g at 20 °C for 20 min. Supernatants were sterile-filtered using 0.22 μm spin-X columns. All samples were passed through a Millipore filter (30 kDa cutoff) to remove large proteins. Samples were dried under high vacuum and stored at −80 °C until further platform-specific processing and analysis.
CE-TOF/MS Analysis
The dried samples were resuspended in Milli-Q water containing 0.1 mM formic acid and 0.2 mM methionine sulfone (internal standard) (Sigma-Aldrich, Germany) by vortexing for 1 min. After subsequent centrifugation (12 600g, 15 min), the resulting clear solution was analyzed by CE-TOF/MS using a CE system (Agilent 7100) coupled to a TOF/MS system (Agilent 6224). The separation occurred in a fused-silica capillary (Agilent Technologies) (total length, 96 cm; i.d., 50 μm) under normal polarity with a background electrolyte containing 1.0 M formic acid in 10% (v/v) methanol at 20 °C. Sheath liquid (6 μL·min–1) was methanol/water (1:1, v/v) containing 1.0 mM formic acid with two reference masses to allow correction and high mass resolution in the MS. Samples were hydrodynamically injected at 50 mbar for 35 s and stacked by injecting a background electrolyte at 100 mbar for 10 s. The optimized MS parameters were as follows: fragmentor, 125 V; skimmer, 65 V; octopole, 750 V; nebulizer pressure, 10 psi; drying gas temperature, 200 °C; and flow rate, 10.0 L·min–1. The capillary voltage was 3500 V. Data were acquired in the positive electrospray ionization (ESI) mode with a full scan from m/z 50 to 1000 at a rate of 1 spectra·s–1. The resulting CE-TOF/MS data files were cleaned of background noise and unrelated ions by the Batch Recursive Feature Extraction tool with Agilent MassHunter Profinder version B.06.00 software. Data were extracted using a data-mining algorithm based on the software. To perform an initial selection on disease-associated metabolites, every case group and wild-type comparisons were evaluated by Kruskal−Wallis (KW) analysis of variance (ANOVA) on ranks. This was performed using the software package MATLAB version 9 (The MathWorks, Inc., Natick, MA). Metabolites, whose Benjamini–Hochberg p-values < 0.05, were putatively annotated by comparison of their migration time and spectra with an in-house library of pure standards and the METLIN Metabolomics Database.21
GC-QTOF/MS Analysis
The above-described dried samples were resuspended in 450 μL of MeOH/H2O/MTBE (74:10:16), and after centrifugation at 12 600g, 15 min at 4 °C, the supernatant was transferred to a vial with an insert and evaporated to dryness under high vacuum. The obtained dried extracts were derivatized by an MPS autosampler for GC/MS analysis as previously described by Fiehn.22 Briefly, aldehyde and keto groups were first converted to O-methyloximes by reaction with 10 μL of pyridine containing 15 mg·mL–1O-methoxyamine (Sigma-Aldrich, Germany) for 60 min at 70 °C. In a second step, acid hydrogen-containing metabolites were trimethylsilylated by reaction with 10 μL of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) (Sigma-Aldrich, Germany) to enhance the GC/MS metabolite coverage.
The analysis was performed on an Agilent Technologies 7890B GC system equipped with a Gerstel MPS autosampler and an Agilent Technologies 7200 accurate mass Q/TOF analyzer equipped with an electron ionization (EI) source. Then, 1 μL of the sample was injected into a multimode inlet at 230 °C with the split ratio set at 1:12 with 9.354 mL·min–1 connected to a capillary column (30 m × 0.25 mm × 0.25 μm; Agilent, Germany). Helium was used as the carrier gas, at a flow rate of 0.78 mL·min–1. Column temperature was 60 °C for 1 min and then programmed to increase at a rate of 10 °C·min–1 until 325 °C, which was maintained for 10 min. The total runtime was 37.5 min. The MS scan mode was chosen as the acquisition mode, with the mass range of 50–650 m/z and an acquisition rate of 10 spectra·s–1.
The individual analytical fingerprints obtained were deconvoluted using MassHunter Unknown Analysis version B.07.00. This software also allows for the annotation of metabolites comparing the mass spectrum obtained with those of a target compound library, FiehnLib, and as this FiehnLib library includes retention indices, the retention time was also used as an additional criterion.23
After applying the MassHunter Unknowns Analysis, a.cef file including the compound name, mass, CAS number, formula, and retention times was generated to create a method for the MassHunter Quantitative Analysis version B.07.00 to export a data matrix containing integrated areas for each compound. Signals derived from the column bleed were eliminated; afterward, the abundances were normalized using the mean fold-change method of normalization.
LC-QTOF/MS Analysis
The above-described dried samples were resuspended in 200 μL of methanol/water/MTBE (7.4:1:1.6), vortexed for 1.5 h, and centrifuged (4000g, 10 min, 4 °C). Clear solutions were analyzed by LC-QTOF/MS. An HPLC system (1200 series, Agilent Technologies, Waldbronn, Germany), equipped with a degasser, two binary pumps, and a thermostated autosampler coupled to an Agilent 6520 QTOF/MS system (Agilent Technologies, Waldbronn, Germany), was used in both positive and negative ESI polarity modes to increase the metabolome coverage.
Briefly, 5 μL of extracted lung samples was injected into a thermostated (60 °C) Agilent Poroshell 120 EC-C8 column (150 mm × 2.1 mm, 2.7 μm; Agilent Technologies, CA) with a guard column Ascentis Express C8 (5 mm × 2.1 mm, 2.7 μm; Supelco, Bellefonte, PA). The flow rate was 0.4 mL·min–1 with solvent A (10 mM ammonium formate in Milli-Q water) and solvent B (10 mM ammonium formate in methanol and 15% isopropanol) for analysis in the positive ionization mode and solvent A (Milli-Q water with 0.1% formic acid) and solvent B (methanol with 0.1% formic acid and 15% isopropanol) for analysis in the negative ionization mode. Initial conditions at time 0 were 82% B, increasing to 96% B in 30 min. This was then held until 38 min. The gradient then increased to 100% B by 38.5 min and held until 40.5 min. The conditions were then returned to the starting conditions by 42 min, followed by an 8 min re-equilibration time. The total runtime of the method was 50 min. Capillary voltage was set to 4.5 kV; the drying gas flow rate was 10 L·min–1 at 350 °C and gas nebulizer at 40 psi; fragmentor voltage, skimmer voltage, and octopole radio frequency voltage were set to 175, 65, and 750 V, respectively. Data were collected at a scan rate of 1.05 spectra·s–1. Mass spectrometry detection was performed in both positive and negative ESI modes in a full scan from 100 to 1000 m/z. Samples were analyzed in separate runs (positive and negative ionization modes), in a randomized order. The resulting LC-QTOF/MS data files were cleaned of background noise and unrelated ions by the Batch Recursive Feature Extraction tool with Agilent MassHunter Profinder software version B.06.00. Data were extracted using data-mining algorithms of the software. Putative annotation of metabolites found in positive and negative ionization modes was performed by the CEU Mass Mediator24 for a subset of metabolites as described above for CE-TOF/MS. Univariate statistical significance was determined by the Kruskal–Wallis (KW) test as described above for CE-TOF/MS.
Van Krevelen Diagrams
Oxygen-to-carbon and hydrogen-to-carbon ratios were calculated for subsets of metabolites, whose putative annotations led to one possible chemical formula. The resulting data was plotted in a previously described metabolic map.25
Statistics
To productively mine large data sets from multiplatform HRMS-based metabolomic approaches, a robust and reproducible statistical data pipeline is necessary (Figure 1A).26 To obtain a global view of results arising from different analytical platforms (Supporting Table S1), results were combined using the consensus orthogonal partial least squares-discriminant analysis (cOPLS-DA) data fusion algorithm.27 OPLS-DA-related algorithms calculate mathematical projections, which explain the maximum variability between previously assigned sample groups for a specific metabolite data matrix. In this context, cOPLS-DA is a multivariate statistical test, which allows a joint interpretation of the results from multiple analytical platforms in a single analysis, performed on a merged multiplatform data set. Contrarily to the traditional OPLS-DA, which penalizes the importance of metabolite alterations from smaller metabolic data sets, cOPLS-DA harmonizes the data structure by performing a weighted normalization of each MS platform-specific data matrix, contextualizing and scoring the contribution of individual metabolites of the entire multiplatform data set to an optimally discriminant, group-specific metabolic fingerprint within the data model. Pairwise cOPLS-DA comparisons between uninfected mice and mice 4 and 9 weeks postinfection were generated. Additionally, a cOPLS-DA model including all sample groups was also calculated. Note that cOPLS-DA, as well as OPLS-DA, does not determine a specific cutoff value for determining statistical significance. Then, a quantitative value assessing the variable importance in the projection is assigned to each metabolite in the context of a metabolite data matrix. Therefore, although metabolites with variable importance in the projection (VIP) values > 1 not including 0 in the error confidence interval are generally accepted as statistically relevant, it cannot be assumed that metabolites with lower VIP values do not contribute to the multivariate separation and differences observed between the sample groups. Additionally, a shared and unique structure (SUS) plot was generated to evaluate the differential trends in metabolites across the disease time points.
Bioinformatics
As a first approach to highlight altered biological pathways, MetaboAnalyst (version 4.0)28 was employed to map the metabolites with VIP values higher than 1, using metabolite enrichment analysis (overrepresentation analysis and pathway analysis). To reduce the possible bias induced by signals with more than one tentative annotation, a curated input subset of unique metabolites with their respective KEGG code identifiers (Supporting Table S1) was generated. However, given the inherent bias of enrichment algorithms since metabolomic analyses do not entirely cover the enrichment sets and pathways constitute a classical “dissection” of the metabolome, we also exploited clustering and metabolic network modularity analyses, which can be exploited to explore the influence of metabolites on each other based on their mathematical relationships and the network topology, respectively.
Overrepresentation Analysis
Representation of metabolites with VIP > 1 values determined in cOPLS-DA models, including all pairwise comparisons and a three-group comparison, was obtained by the performance of hypergeometric tests in pathway-associated metabolite sets, using the default reference metabolome. Significance of metabolite sets was assessed by a p-value cutoff of 0.05.
Pathway Analysis
Metabolite representation and pathway impact were assessed by the performance of hypergeometric tests and evaluation of the relative-betweenness centrality of the metabolites in the Mus musculus pathway library for each subset of metabolites with VIP > 1 values determined in all generated cOPLS-DA models. All of the compounds present in the selected pathways were considered. Significance of metabolite sets was assessed by a p-value cutoff of 0.05.
Metabolite Clustering Analysis and Heatmap Generation
The entire data matrix resulting from data processing and annotation, as well as specific subsets of metabolites, was loaded onto the MetaboAnalyst 4.0 server.28 A hierarchical clustering of both samples and metabolites was performed using the whole matrix input and using MetaboAnalyst default parameters (Supporting Figure S1). The different heatmaps highlighted in the Results and Discussion section were generated in parameter consistency with the whole-data matrix heatmap, although no clustering was performed.
Metabolic Subnetwork Generation and Network Clustering Analysis
Metabolite-specific and generic compound KEGG codes29 were assigned to all metabolites with unique annotations. A metabolic subnetwork based on KEGG RPAIR data was generated using MetaboNetworks (version 2.1)30 and converted to .sif format (Supporting Text File S1). After deletion of duplicated edges, the subnetwork modularity was evaluated using the ModuLand (version 2.0)31 Plug-in for Cytoscape (version 3.6.1).32 ModuLand 2.0 employs a community landscape approach, which uses the LinkLand algorithm for calculating influence functions of each node in the whole node data set and the ProportionalHill method to determine the different discrete or overlapping modules present in the subnetwork, besides highlighting central node representatives of the different clusters and nodes bridging between clusters.33 Once the modules are determined, ModuLand 2.0 merges them as metanodes and iteratively runs the algorithm, providing different hierarchical levels of the network. Both discrete and overlapping modularity algorithms were run. After merging of modules with correlation values higher than 0.9, the discrete modularity algorithm was used for careful data interpretation.
Results and Discussion
cOPLS-DA of Multiplatform HRMS-Based Metabolomics Reveals Significant, Time-Dependent Changes in the Lung Metabolome of Mtb-Infected Mice
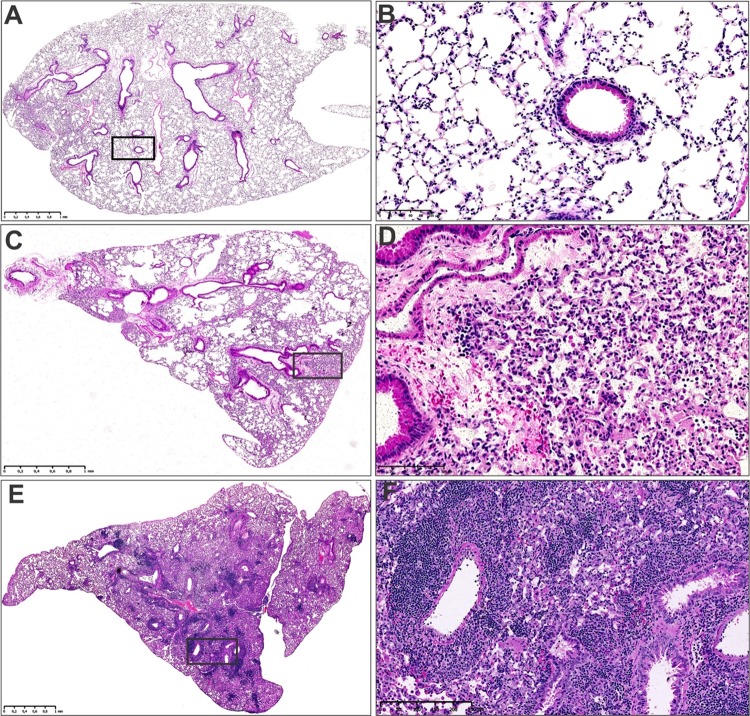
C57BL/6 mice were infected with Mtb H37Rv and euthanized at 4 and 9 weeks postinfection. Metabolome analyses were limited to the 4-week time point, which reflects induction of adaptive immunity, and the 9-week time point, which reflects established, chronic disease. Since multiplatform HRMS requires the processing and analysis of large numbers of samples, for purposes of practicality, only these two time points were chosen. Lungs were removed from uninfected (Mtb–), 4 weeks (Mtb+4w), and 9 weeks (Mtb+9w) postinfection mice. Lesions were clearly visible in the lungs of infected mice, albeit more severe pathology was noted in Mtb+9w compared to Mtb+4w (Figure 2).
Figure 2.
Representative images of the histomorphology of uninfected and Mtb-infected mouse lungs. Low-power (A) and high-power (B) Hematoxylin and Eosin staining (H&E) of uninfected control mouse lungs. Low-power (C) and high-power (D) H&E of Mtb-infected mouse lungs at 4 weeks postinfection. Low-power (E) and high-power (F) H&E of Mtb-infected mouse lungs at 8 weeks postinfection. Note the progressive increase in alveolar consolidation of the infected lung tissue (C, D, E, F), which is absent in the uninfected lung tissue (A, B).
Metabolites were extracted from the lungs and analyzed separately by three HRMS-based platforms (LC-QTOF/MS, GC-QTOF/MS, and CE/TOF-MS). Overall, the different metabolomic platform analyses revealed a notable joined matrix data set of 1215 potential compounds after data processing and curation, where 456 metabolites were putatively annotated (Supporting Table S1). In the cOPLS-DA models, 554, 638, and 546 potential compounds from the entire metabolite data set scored VIP values > 1 for Mtb+4w/Mtb, Mtb+9w/Mtb–, and Mtb+9w/Mtb+4w pairwise comparisons, respectively (Table 1). Additionally, a three-group comparison determined 631 potential compounds with VIP > 1 values. These results suggest that profound changes in the metabolome occur during Mtb infection.
Table 1. Detected, Annotated, and Statistically Significant Potential Compounds.
| variable
importance in the projection (VIP) > 1 |
|||||
|---|---|---|---|---|---|
| analytical platform | annotated/total | Mtb+9w/Mtb+4w/Mtb– | Mtb+4w/Mtb– | Mtb+9w/Mtb– | Mtb+9w/Mtb+4w |
| GC-QTOF/MS | 100/107 | 54 | 46 | 51 | 48 |
| CE-TOF/MS | 127/268 | 153 | 119 | 152 | 119 |
| LC-QTOF/MS+ | 182/643 | 319 | 294 | 337 | 280 |
| LC-QTOF/MS– | 48/197 | 105 | 95 | 98 | 99 |
| sum | 457/1215 | 631 | 554 | 638 | 546 |
Prominent group clustering and separation were observed in the cOPLS-DA score plots (Table 2 and Figure 3A). Given the high model fit (R2Y) and prediction accuracy (Q2Y) values found in cOPLS-DA models, the most striking results that emerged from the data are that strong, time-dependent quantitative metabolic abnormalities occur in the lung of all disease groups. Interestingly, each HRMS-based analysis contributed to explaining additional group separation between the different comparisons. In addition, distinct values were determined for the specific contribution of each HRMS-based metabolomic platform to the strength of the model components. These observations demonstrate the importance of multiplatform analyses in obtaining metabolomic data capable of achieving enhanced metabolome coverage and producing increased phenotype-associated group separation in multivariate analyses (Table 2).
Table 2. Model Fit, Prediction Accuracy, and Technique-Dependent Contribution Values to the Separation Observed in the Different cOPLS-DA Modelsa.
| model | R2Y | Q2Y | A | GC-QTOF/MS | CE-TOF/MS | LC-QTOF/MS+ | LC-QTOF/MS– |
|---|---|---|---|---|---|---|---|
| Mtb+9w/Mtb+4w/Mtb– | 0.976 | 0.886 | tp1 | 0.23 | 0.30 | 0.22 | 0.26 |
| tp2 | 0.25 | 0.19 | 0.27 | 0.29 | |||
| to | 0.26 | 0.20 | 0.34 | 0.20 | |||
| Mtb+4w/Mtb– | 0.999 | 0.919 | tp1 | 0.25 | 0.23 | 0.25 | 0.27 |
| to | 0.27 | 0.27 | 0.26 | 0.20 | |||
| Mtb+9w/Mtb– | 0.994 | 0.936 | tp1 | 0.22 | 0.28 | 0.24 | 0.26 |
| to | 0.19 | 0.15 | 0.43 | 0.23 | |||
| Mtb+9w/Mtb+4w | 0.995 | 0.885 | tp1 | 0.23 | 0.26 | 0.23 | 0.28 |
| to | 0.23 | 0.23 | 0.30 | 0.24 |
R2Y, model fit; Q2Y, predictive accuracy; tp, predictive principal component; to, orthogonal principal component.
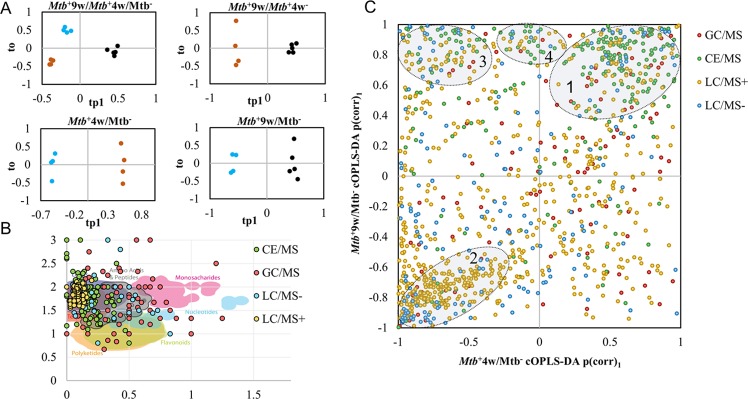
Figure 3.
cOPLS-DA, Van Krevelen, and SUS plots describing the metabolite data set. (A) cOPLS-DA score plots of Mtb+9w/Mtb+4w/Mtb– and pairwise comparisons. (B) SUS plot of potential compounds for Mtb+9w/Mtb– and Mtb+4w/Mtb– comparisons. (C) Van Krevelen diagram of putatively annotated metabolites showing the metabolite distribution according to their molecular formula. In cOPLS-DA score plots, Mtb– represents blue, Mtb+4w represents brown, and Mtb+9w represents black. LC/MS+ and LC/MS– represent positive and negative ESI polarity modes, respectively. In the SUS plot, clusters 1 and 2 represent direct shared structures of metabolites consistently increased and decreased at both time points, respectively. Cluster 3 represents a group of metabolites with an inverse shared structure, while cluster 4 encompasses metabolites increased only in Mtb+9w.
Three major clusters of metabolites could be observed among statistically significant metabolites in the shared and unique structure (SUS) plot (Figure 3B). The first and second clusters of metabolites encompassed the majority of metabolic alterations and corresponding metabolites whose levels follow a similar trend at both disease stages. A third cluster represented metabolites that decreased at Mtb+4w and increased at Mtb+9w. A smaller, fourth cluster represented metabolites that were exclusively increased at Mtb+9w, therefore representing a specific, late alteration during advanced TB. These results showed that although most of the alterations were consistently increased or decreased at both infection time points, the abundances of a considerable subset of metabolites were altered between the two infection time points. Therefore, a clear phenotypic difference could be detected between Mtb+4w and Mtb+9w mice. This emphasizes the importance of sampling metabolites at multiple time points during infection to describe the phenotypic characteristics of the progression of pulmonary TB. Clustering analysis of samples and potential compounds indicated that Mtb+4w separates from Mtb+9w and Mtb– samples (Supporting Figure S1). These results indicate that the metabolic phenotype of Mtb+4w is notably altered, suggesting an early response to TB infection, which is partially reversed in Mtb+9w. The different clusters of metabolites were in excellent agreement with the data obtained from the SUS plot.
The annotated metabolome was notably enriched in different metabolite classes including carbohydrates, small organic acids, amino acids, peptides, and lipidome-related compounds, thereby representing a broad overview of the metabolome (Figure 3C). In the context of TB, alterations in the levels of these metabolite pools should be carefully interpreted. For example, mass exchange occurs within several animal compartments and the environment, as well as cross-talking between different compartments (i.e., migration and proliferation of macrophages and T lymphocytes in the lung34 and stratified macrophage polarization in Mtb granulomas35,36), to modulate the spatiotemporal distribution of metabolites. In addition, lysis and extraction of a tissue sample prior to analysis trigger a loss of compartmentalization and spatial information.37 As a consequence, metabolite levels in the tissue are not only the result of a superimposition of the different metabolite concentrations in all of the tissue and cellular compartments from both Mtb and mouse metabolomes but also the result of the interaction of both genomes, the so-called cometabolome (i.e., Mtb-secreted proteins that metabolize macrophage metabolites).38 Next, we investigated changes in these metabolite classes in more detail in the context of disease progression.
Systematic Overrepresentation (ORA) and Pathway Analysis (PA) Identified Disease-Specific Pathways Altered in Mtb-Infected Mice
To obtain functional information from differentially regulated metabolic pathways, two enrichment analyses were performed. First, we performed an overrepresentation analysis (ORA), in which significantly different metabolic alterations were identified (Supporting Table S3). These mainly encompassed the metabolism of amino acids and phospholipids. Overall, the ORA results point toward an alteration of phospholipid metabolism in Mtb+4w, which unexpectedly returned to nonsignificant values at Mtb+9w. To further support the data obtained by ORA, pathway analysis was performed as a second enrichment algorithm. In this context, PA mostly detected metabolite sets consistently altered in ORA (Supporting Table S3), including the metabolism of phospholipids, amino acids, and nitrogen. Additional alterations in metabolic pathways not identified by ORA were reported as significant in Mtb+4w/Mtb– (propanoate metabolism) and Mtb+9w/Mtb– (β-alanine metabolism) or both (sphingolipid, glutathione metabolism), demonstrating the need for different algorithms to provide a more holistic data analysis to identify relevant biological processes during the progression of pulmonary TB.
Macronutrient Consumption is Consistent with Mtb-Associated Wasting
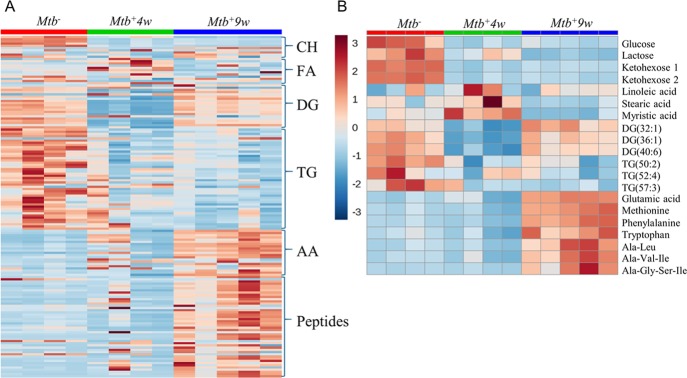
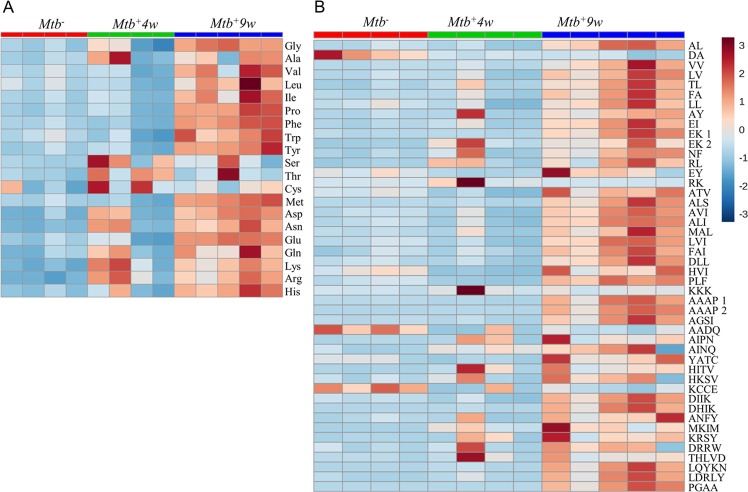
Interestingly, three distinct responses in metabolite levels indicate macronutrient consumption (Figure 4A,B). In the first response, distinct carbohydrates showed a notable decrease in Mtb+4w, which was maintained in Mtb+9w. In the second response, triacylglycerols decreased in Mtb+4w and Mtb+9w, and diacylglycerol metabolites showed an initial decline at Mtb+4w followed by a noticeable increase in Mtb+9w, whereas the levels of several fatty acids increased in Mtb+4w and decreased at Mtb+9w. The third response was represented by amino acids and oligopeptides, which progressively increase throughout the course of infection. Interestingly, the abundance of methylhistidine, which has been proposed as a biomarker of skeletal muscle breakdown and injury,39,40 was notably increased in Mtb+9w (Supporting Table S4). Hence, high levels of methylhistidine suggest that muscle wasting occurs during advanced TB disease and are consistent with previous studies documenting a link between Mtb infection and malnutrition/wasting.41,42 Overall, we posit that these data correspond with the induction of a temporal and progressive catabolic state in Mtb-infected mice, which is elicited to satisfy the continuously changing energy demand to control infection.
Figure 4.
Heatmaps depicting the metabolic changes associated with major nutrient groups during disease progression. (A) Overall metabolic-associated changes. Note the prominent changes in carbohydrates and energy-storage-related lipids, whereas increasing abundances of amino acids and oligopeptides occur with disease progression. (B) Representative selected metabolic changes. CH, carbohydrates; FA, fatty acids; DG, diacylglycerols; TG, triacylglycerols; AA, proteinogenic amino acids annotated in the study at 0 (Mtb–), 4 (Mtb+4w), and 9 (Mtb+9w) weeks postinfection.
TB Disease Progression Correlates with an Increase in Proteolysis-Related Metabolites
Destruction of the extracellular matrix/pulmonary parenchyma is a well-documented phenomenon that occurs during Mtb infection.43,44 Upon Mtb-induced macrophage activation, protein degradation is primarily performed through the activity of secreted matrix metalloproteinases (MMPs, mainly MMP-1 and MMP-943). Surprisingly, a consistent increase in specific and nonspecific protein breakdown products was observed in Mtb-infected mice (Figure 5A,B and Supporting Table S4). Nonspecific metabolites included short oligopeptides, amino acids, and N-glycolylneuraminate. Specific protein degradation-associated metabolites included trans-4-hydroxyproline and galactosylhydroxylysine. These metabolites are related to collagen-like post-translational modifications, suggesting that substantial alterations occur in collagen and surfactant proteins SP-A and SP-D (which are essential components of the lung45,46) during disease. Lastly, increased abundances of a metabolite annotated as desmosine, a breakdown product of elastin, allude to increased elastin degradation in the TB lung. Elastin is a major component of the extracellular matrix of the lung. Upon lung injury, which involves the catabolism of the extracellular matrix and elastin, desmosine is released. Not surprisingly, desmosine has been identified as a potential biomarker for structural lung injury in pulmonary TB and chronic obstructive pulmonary disease.47,48 Overall, we propose that the metabolites here constitute a data subset indicative of alveolar destruction (Figure 2), progressive proteolysis, and establishment of lung fibrosis49,50 occurring during TB progression.
Figure 5.
Detailed heatmap representations depicting Mtb-associated amino acid and peptide abundance changes during disease progression. (A) Proteinogenic amino acid abundances. (B) Oligopeptide abundances annotated in the study. Note that oligopeptide annotations are indicative, since CE-TOF/MS cannot determine the amino acid sequence. A gradual increase in the levels of the majority of metabolites grouped in this section occurs with the disease progression.
Increased Itaconic Acid (ITA) Production Subsequent to Alterations in the Central Carbon Metabolism Occurs in the Lungs of Mtb-Infected Mice
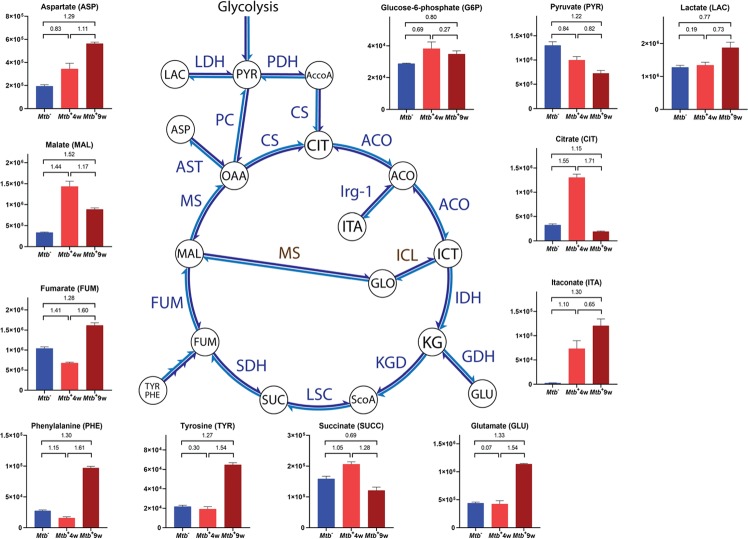
It is widely accepted that the central carbon metabolism constitutes a relevant link between energy production and immunity. In the context of carbohydrate metabolism, we detected carbohydrate depletion occurring upon Mtb infection (Figure 4) and moderate increases in glucose-6-phosphate (Figure 6), which suggested increased substrate availability for the pentose phosphate pathway (PPP), a major biosynthetic pathway for nucleotides and NADPH, which is required as a cofactor for glutathione reductase during oxidative stress and for inducible nitric oxide synthase (iNOS).51 Intriguingly, lactate increased marginally in Mtb+4w and moderately in Mtb+9w (Figure 6), suggesting that most of the pyruvate was being utilized elsewhere at 4 weeks but converted into lactate at 9 weeks postinfection. In contrast, pyruvate progressively decreased with TB disease progression (Figure 6). Previous transcriptomics and immunofluorescence studies of Mtb-infected mouse lungs demonstrated increased expression of glycolytic enzymes, lactate dehydrogenase, and glucose transporters at 4 weeks postinfection, and it was concluded that the Warburg effect is induced in Mtb-infected mouse lungs.52 However, the marginal increase we observed in lactate in Mtb+4w does not fully support these findings and also differs from a previous NMR mouse study,16 where lactate levels were examined at a single, early time point (28 days postinfection). In the guinea pig model for TB, NMR studies have shown a decrease in serum lactate levels but an increase in lung lactate levels.18 Overall, these findings point to the spatiotemporal regulation of lactate, which differs in different animal model systems. The large increase in citrate in Mtb+4w (Figure 6) suggests that most of the pyruvate is being converted into citrate in Mtb+4w for fatty acid synthesis and initial production of itaconate during the early induction of adaptive immunity.53,54 However, the substantial decrease in citrate at 9 weeks postinfection is likely due to the formation of the remarkably high levels of ITA present in Mtb+9w (Figure 6). This observation is consistent with previous NMR studies reporting increased itaconate levels during Mtb infection.16
Figure 6.
Topology of the TCA and related pathways and Mtb-associated semiquantitative changes. Graphs represent arbitrary normalized abundance units and VIP values between pairwise comparisons. Y-axes on graphs represent arbitrary normalized abundance units. VIP values between pairwise comparisons are indicated. Metabolites not appearing on graphs are abbreviated as follows: AccoA, acetyl-coenzyme A; ACO, aconitate; ICT, isocitrate; GLO, glyoxylate; KG, α-ketoglutarate; ScoA, succinyl coenzyme A; OAA, oxaloacetate; enzymes are colored in blue except for Mtb-specific enzymes, which are colored in brown; CS, citrate synthase; LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; PC, pyruvate carboxylase; ACO, aconitase; Irg-1, immune-responsive gene 1 protein; ICL, isocitrate lyase; MS, malate synthase; IDH, isocitrate dehydrogenase; GDH, glutamate dehydrogenase; KGD, α-ketoglutarate dehydrogenase; LSC, succinyl-coA ligase; SDH, succinate dehydrogenase; FUM, fumarase; MS, malate synthase; AST, aspartate aminotransferase.
Itaconate is biosynthesized from cis-aconitic acid by decarboxylation performed by the immune-responsive gene 1 protein (IRG-1), whose gene (Irg155) is upregulated in macrophages upon stimulation with LPS, TNF-α, and IFN-γ.56,57 Not surprisingly, itaconate plays several roles in immunometabolism. ITA has been shown to play a major role in metabolic reprogramming through inhibition of the TCA enzyme succinate dehydrogenase (SDH)58 and to exert direct antibacterial activity through inhibition of bacterial isocitrate lyase.59,60 Given the relevance of itaconate in TB,61 we speculate that at 4 weeks postinfection, pyruvate is being redirected to increase citrate levels necessary for fatty acid synthesis and ITA production. However, when chronic infection sets in at 9 weeks, we surmise that the citrate levels are substantially reduced to generate higher levels of ITA necessary to subdue the inflammatory response.
The TCA cycle in inflammatory macrophages has breakpoints at SDH and isocitrate dehydrogenase.54,62 In addition, it has been described that ATP generation in effector T-cells relies on glycolysis, rather than OXPHOS.63 Alterations in several TCA-related metabolites (Figure 6 and Supporting Table S4) propose similar observations at Mtb+4w, including (i) increased citrate accumulation in Mtb+4w; (ii) possible SDH inhibition supported by increased succinate and itaconate abundances in Mtb+4w; (iii) possible malate dehydrogenase (MDH) inhibition, supported by reduced fumarate levels and increased malate and citrate in Mtb+4w (which act as MDH inhibitors);64 and (iv) NAD+ modulation (Supporting Table S2). During the early stages of infection (Mtb+4w), notable alterations in the levels of NAD+ occur in the lung of infected mice, which has been also reported to occur in C57BL/6 mice using NMR.16 More interestingly, the levels of nicotinamide, determined as the breakdown product of two Mtb NAD+ glycohydrolases (MbcT and TNT,65,66 the latter identified as relevant in Mtb pathogenesis),67 were notably increased in Mtb+4w (Supporting Table S4). However, these changes in NAD+ and nicotinamide could also be a consequence of the host-related metabolic activity.68 At Mtb+9w, succinate levels and malate levels were decreased with a contrasting increase in fumarate levels. The high glutamate levels are possibly due to the accumulation of succinate from the inhibition of succinate dehydrogenase and the proteolysis discussed above (Figure 5A,B and Supporting Table S4). The high aspartate level, which is a proxy for oxaloacetate, is probably due to transamination reactions with high levels of glutamate.
Taken together, these results suggest that the observed TCA metabolic alterations are the outcome of a complex host-pathogen regulatory network (Figure 6) wherein the lung in Mtb+4w resembles an inflammatory immunometabolic response, which are not fully maintained in Mtb+9w as citrate levels are significantly decreased. In this context, it is important to point out that itaconate production is a marker for anti-inflammatory cellular responses induced following proinflammatory stimulation.69 Thus, the metabolic alterations observed in the TCA cycle between the two time points suggest transitions between inflammatory and anti-inflammatory responses described in several studies.63,65 Given the notable alterations in NAD-related metabolites occurring in Mtb+4w, we suggest that the potential modulatory role of Mtb in NAD+ metabolism occurring in the TB lung should be addressed further in future studies.
Mtb Disease Causes Alterations in Oxido-Reductive Stress-Related Metabolites
Mtb-induced inflammatory activation leads to the generation of reactive oxygen and reactive nitrogen species (ROS and RNS, respectively).70 Consistently, the increase in arginine and citrulline levels in Mtb+4w and Mtb+9w is likely a consequence of the increased expression of inducible nitric oxide synthase (iNOS) in the lung (Supporting Table S4).71 However, the presence of increased nitric oxide in infected cells is controversial, since it is also known that Mtb induces the expression of host arginase in infected macrophages.72 Additionally, altered levels of xanthine and hypoxanthine (Supporting Table S4) point to substantial modulation in xanthine oxidase (XO) activity, which constitutes a major regulator of the superoxide ion (O2•–). Interestingly, the abundances of xanthine and hypoxanthine levels were decreased in Mtb+4w, whereas they were notably increased in Mtb+9w, suggesting a differential behavior of XO with disease progression. During Mtb infection, NADPH oxidase (NADPHox) is another key enzyme that produces O2•– during the oxidative burst.73 Although metabolites involved in NADPH production could not be detected, it is reasonable to infer that enhanced glucose-6-phosphate levels generate more substrates for NADPH production through the PPP.74 However, it has been reported that MbcT hydrolyzes NAD+,67 and TNT is able to degrade both NAD+ and NADP+,66 as mentioned above. Given that NADPHox and XO/iNOS require NADPH and NAD+ as cofactors, respectively, we suggest that Mtb-mediated NAD(P)+ glycohydrolase activity in modulating ROS/RNS production should be assessed in further studies. Overall, we identified a distinct subset of metabolites involved in ROS/RNS biosynthetic pathways, which provides compelling evidence that Mtb triggers time-dependent alterations in oxidative stress-related metabolites during infection.
Glutathione- and glutathione-related compounds play a key role in maintaining redox cellular homeostasis. Here, altered levels of reduced glutathione (GSH) and glutathione-derived products (oxidized glutathione (GSSG), S-lactoylglutathione, S-hydroxymethylglutathione) as well as glutathione precursors in Mtb+4w and Mtb+9w were detected (Supporting Table S4 and Figure 5A). Altered levels of these metabolites suggest modifications of the redox environment in the Mtb-infected lung. These results intimate an increase in de novo GSH synthesis, which could be an ROS-protective mechanism of the host while providing GSH-mediated direct antimycobacterial activity.75,76 Generally, GSSG abundances were similar in Mtb+4w and Mtb+9w. The GSH/GSSG ratio returned to Mtb– levels in Mtb+9w, suggesting the depletion of reduced glutathione with disease progression. This is supported by the alterations in metabolites related to the generation of ROS/RNS and the increase in glutathione-derived compounds observed in Mtb+9w. Intriguingly, the abundances of S-lactoylglutathione were notably increased in Mtb+4w and to a lesser degree in Mtb+9w. This compound is primarily biosynthesized in the glyoxalase system, a metabolic pathway that is able to detoxify the cells from methylglyoxal, a glycolysis-derived compound. S-Lactoylglutathione is related to a plethora of immunity-related functions including phagocyte activation, anti-IgE-induced secretion of histamine in basophils, microtubule assembly, neutrophil granule secretion, and chemotaxis.77 Thus, the levels found in our results suggest that dysregulation occurs in the glyoxalase system upon Mtb infection. Glyoxalase inhibition causes an accumulation of methylglyoxal, which has been described as cytotoxic for certain microorganisms such as Staphylococcus aureus(78) and Plasmodium falciparum.77 Given this ex vivo observation, the pharmacological inhibition of the glyoxalase system-associated enzymes (GLO-I and GLO-II) may have modulatory activity in TB, as was previously proposed.78
Ergothioneine (EGT) is an antioxidant taken up from the environment by mammalian cells. However, Mtb also produces EGT, and it has been shown to be essential for survival in macrophages79 and in mice.80 Importantly, we found increased EGT levels at the late stage of infection (Mtb+9w) (Supporting Table S4), which highlights the therapeutic potential of this antioxidant pathway.
Mtb Infection Drives Alterations in the Urea Cycle and Polyamine Metabolism
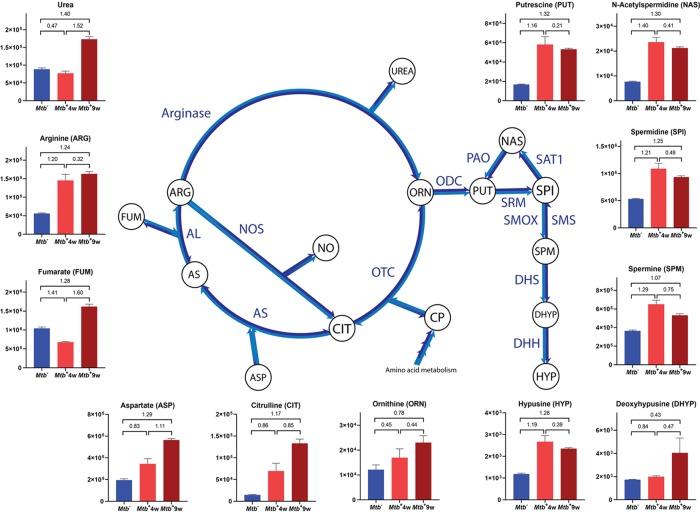
Important alterations in the urea cycle were found in both Mtb+4w and Mtb+9w (Figure 7 and Supporting Table S4). As explained above, augmented concentrations of arginine and citrulline may be regulated by altered iNOS activity, which generates citrulline and NO from arginine, thereby “bypassing” the urea cycle. However, the progressive increase in the expression of arginase mentioned previously72 would favor the catabolism of arginine by arginase, thereby reducing RNS generation.81 Therefore, increases in the levels of urea and ornithine observed in Mtb+9w suggest a progressive anti-inflammatory metabolism. Alternatively, Mtb-associated wasting and the subsequent enhancement of protein catabolism for energy generation might explain a substantial part of the alterations in the urea cycle.
Figure 7.
Topology of the urea cycle and polyamine biosynthesis pathways and Mtb-associated semiquantitative changes. Y-axes on graphs represent arbitrary normalized abundance units. VIP values between pairwise comparisons are indicated. Metabolites not appearing on graphs are abbreviated as follows: CP, carbamoyl phosphate; NO, nitric oxide; AS, argininosuccinate. Enzyme abbreviations are colored in blue: OTC, ornithine transcarbamoylase; AS, argininosuccinate synthase; AL, argininosuccinate lyase; NOS, nitric oxide synthase; ODC, ornithine decarboxylase; SRM, spermidine synthase; SMS, spermine synthase; SMOX, spermine oxidase; SAT1, spermidine/spermine-N1-acetyltransferase; PAO, polyamine oxidase; DHS, deoxyhypusine synthase; DHH, deoxyhypusine hydrolase.
Consistent with these findings, increased levels of polyamines (i.e., spermidine, spermine, and putrescine) were found in both Mtb+4w and Mtb+9w (Figure 7 and Supporting Table S4). Polyamines are positively charged alkyl amines that interact with DNA and are essential for cell proliferation and adequate macrophage function,82,83 although an immunosuppressive role has also been suggested.84 Polyamines are also capable of binding iron, and interestingly, a link between ferritin H and polyamines during Mtb infection was recently established.11 Endogenous polyamines have also been described as beneficial for Mtb, increasing the activity of Mtb RNA polymerase85 and conferring fluoroquinolone resistance to the bacteria.86 The increase in polyamine levels suggests that anti-inflammatory responses are present at both disease time points. Anti-inflammatory responses are associated with arginine catabolism through arginase activity, reducing NO production by iNOS.11 Further studies on the modulation of enzymes involved in these pathways are expected to provide new insights into the polyamine-related immunometabolism of TB.
Polyamines also play an important role in protein translation since spermidine is a precursor for the synthesis of hypusine, which is increased in Mtb+4w and Mtb+9w. Hypusine is an amino acid exclusively found in the eukaryotic translation initiation factor 5A-1 (eIF-5A), which plays an important role in protein translation, particularly in the elongation step. Cell proliferation, ROS tolerance, mitochondrial function, and endoplasmic reticulum stress are a few examples of the variety of biological processes in which eIF-5A is involved.87−89 Thus, increased levels of polyamines leading to increased levels of hypusine suggest altered activity of eIF-5A in Mtb-infected mice.
Mtb Infection Regulates Lipid Metabolism to Modulate Signaling and Immunity
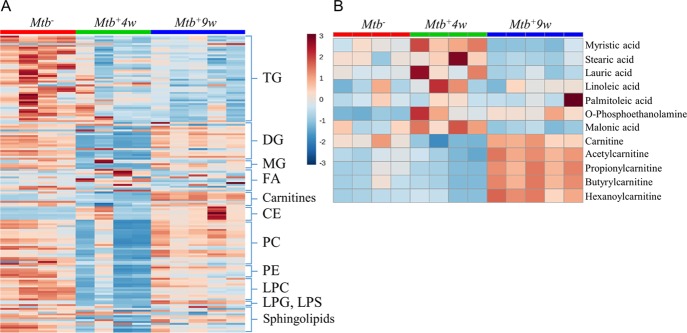
We observed substantial variation in the lipidome over the course of infection (Figure 8A). Mtb+4w mice show an overall decrease in the levels of triglycerides (TGs), diglycerides (DGs), monoglycerides (MGs), phosphatidylcholines (PCs), phosphatidylethanolamines (PEs), and lysophosphatidylcholines (LPCs), together with an increase in the levels of several free fatty acids (i.e., myristic, stearic, lauric, linoleic, palmitoleic) and O-phosphoethanolamine (Figure 8A,B and Supporting Table S4). These results suggest an increase in phospholipase and different lipase activities, which may contribute to the degradation of the main pulmonary surfactant lipid constituents.90 These observations are of particular significance, as it could be argued that some of these lipids function as an in vivo carbon source for Mtb.91 Hence, the depletion of triacylglycerols could be due to its hydrolysis by Mtb.92 Our findings are consistent with previous studies demonstrating an increase in phospholipase A2 (PLA2) activity upon Mtb infection.93 Furthermore, an increase in the abundance of malonic acid, a compound proposed as an indirect biomarker of fatty acid synthesis,94 was detected in Mtb+4w but decreased in Mtb+9w (Figure 8B). Enhanced fatty acid synthesis in Mtb+4w mice is also supported by the reduced levels of L-carnitine and acylcarnitines found in Mtb+4w mice (Figure 8A,B). This correlates with the increase in the fatty acid synthase (FASN) activity that has been described in inflammatory macrophages and is regulated by the sterol regulatory element-binding transcription factor (SREBP1c).95 Both PLA2 and FASN activities have been demonstrated to exert proinflammatory effects.93−96 Furthermore, fatty acid anabolism constitutes alternative pathways for NADPH generation and subsequent ROS formation, which is upregulated in inflammatory macrophages.97,98 However, except for most TGs, the changes in the above-mentioned metabolites reflecting fatty acid synthesis are considerably reversed in Mtb+9w, suggesting a possible attenuation of PLA2 activity. Considering the carbohydrate depletion occurring in Mtb+4w (Figure 4A,B), it is likely that Mtb causes a shift in metabolism toward fatty acid oxidation during disease. This is further supported by highly increased levels of carnitines (Figure 8A,B) found in Mtb+9w. Such metabolic shifts correlate with the described capacity of Mtb to shift the macrophage from an inflammatory to an anti-inflammatory-like phenotype.98,99
Figure 8.
Heatmap representations of Mtb-associated lipidomic changes during disease progression. (A) Overall changes in different lipids, phospholipids, and carnitines. (B) Detailed changes in the abundances of selected fatty acids, carnitines, O-phosphorylethanolamine, and malonate. TG, triacylglycerols; DG, diacylglycerols; MG, monoacylglycerols; FA, fatty acids; CE, cholesteryl esters; PC, phosphatidylcholines; PE, phosphatidylethanolamines; LPC, lysophosphatidylcholines; LPG, lysophosphatidylglycerols; LPS, lysophosphatidylserines. Note that for most PC, LPC, DG, MG, and sphingolipids, the alterations in metabolite levels in Mtb+4w are notably reversed in Mtb+9w.
Another interesting feature of Mtb-infected macrophages is the generation of cytosolic lipid droplets, which has been recently proposed to be a defense mechanism against infection, sequestering lipids to prevent carbon fueling of Mtb.100 The composition of certain lipid droplets is rich in cholesteryl esters,101 which have been found to be notably increased with disease progression. The notable increase found in cholesteryl esters suggests that foam cells accumulating cholesteryl esters in lipid droplets progressively increase in our disease model.102,103 Consequently, it is highly likely that the Acyl-CoA cholesterol acyl transferase activity (ACAT) is increased in the lung of Mtb-infected mice.11
Sphingolipids, which are related to different immunomodulatory properties,104 follow different patterns of expression in the diseased mice (Figure 8A). Regarding sphingolipid alterations occurring during TB disease, a decrease in the activity of sphingosine kinase (SphK) has been documented to occur in the lung of Mtb-infected mice, thereby blocking the Ca2+ influx toward the macrophage cytoplasm and thereby inhibiting the maturation of the phagosome.105 Given the alterations found in our model and the reported evidence of sphingolipid metabolism modulation as a therapeutic strategy,106 more lipidomic studies are needed to assess the importance of sphingolipids in Mtb-induced immune response.
Mtb Infection Dysregulates Purine and Pyrimidine-Related Metabolism
Overall, the levels of pyrimidine-related compounds increased, while purine-related compounds showed a complex profile (Supporting Table S4). Purine and pyrimidine-derived compounds participate in different biological processes including (i) DNA replication and RNA synthesis, and their demand is increased when cellular proliferation occurs; (ii) enzyme cofactors in reactions that require chemical energy (especially ATP) or a methyl donor (especially S-adenosylmethionine (SAM)); and (iii) signaling molecules through purinergic receptors. Our results suggest that these processes are strongly altered during Mtb infection. Interestingly, adenosine levels were decreased upon infection in both Mtb+4w and Mtb+9w. Adenosine binds with high affinity to A1, A2B, and A3 receptors, which are expressed by different cellular types of the immune system.107 Activation of such receptors induces a variety of cellular functions, including secretion of pro- and anti-inflammatory cytokines, IgE production, mucous production, suppression of TNFα release, and bronchoconstriction.107 Increased levels of adenosine might be the consequence of an increase in adenosine deaminase activity (ADA), which has been reported to occur in the bronchoalveolar lavage and serum of TB-infected patients.108 In our model, depletion of adenosine implies diminishing ADO-mediated signaling through purinergic receptors while generating substrates for XO activity and subsequent ROS generation. AMP levels were increased in Mtb+4w and notably decreased in Mtb+9w. Therefore, AMP could also be implicated in differential signaling through purinergic receptors with disease progression.109
Mtb Infection Dysregulates Smooth Muscle Physiology
In addition to the bronchodilator effect of NO release subsequent to increased iNOS activity,110,111 significant alterations in metabolites that modulate lung smooth muscle tone were found (Supporting Table S4). With respect to bronchoconstrictors, the levels of acetylcholine, the main lung physiological bronchoconstrictor, were reduced in Mtb+4w and increased in Mtb+9w, while histamine showed the opposite trend. Interestingly, serotonin and adenosine, which trigger bronchoconstriction in mice,107,112 were decreased at both infection time points. Altogether, these results suggest an interplay between metabolite bronchoconstrictor and bronchodilator signals in which complex regulatory mechanisms are involved.
Trimethylamine-N-oxide (TMAO) Establishes a Metabolic Link between Mtb Infection and Cardiovascular Risk
The metabolic-associated changes mentioned in the above sections consider Mtb and the host as the sole metabolomes present in our study. However, this simplistic assumption does not consider the different bacterial communities that are continuously present in mice, including the gut and lung microbiomes.113,114 This potentially expands the repertoire of bacterial-specific biosynthetic pathways during TB. For example, we detected high levels of TMAO in the lungs of Mtb-infected mice, especially in Mtb+4w. Increases in TMAO were consistent with alterations in topologically related metabolites (Supporting Table S4). TMAO is a host flavin monooxygenase 3 (FMO3) degradation product of trimethylamine, a bacterial metabolite, which can be biosynthesized from different substrates, including choline, betaine, carnitine, and EGT.115 Interestingly, RNA-seq experiments have pointed FMO3 to be related to the regulation of iron homeostasis during TB.11 Although TMAO has been classically associated with the gut microbiome,116 our data suggest that Mtb could be the biosynthetic source of trimethylamine. Hence, it seems reasonable that TMAO may serve as an electron acceptor for alternative Mtb respiration under anaerobic conditions, which was previously described for other bacteria (i.e., Salmonella spp., Alteromonas spp., Vibrio spp.117). Furthermore, increased TMAO levels have also been associated with induction of adhesion molecules and inflammation via NF-kB activation,118 increased expression of murine macrophage scavenger receptors A and CD36, and inhibition of reverse cholesterol transport.119 Lastly, our results suggest that TMAO is linked to lipid droplet and foam cell development during Mtb granuloma formation, which is consistent with recent studies assessing the role of TMAO in cardiovascular disease120 and in line with the increased levels of cholesteryl esters as described above (Figure 8A). Overall, our findings identified a multifunctional metabolite, TMAO, which, on the one hand, could be exploited by Mtb to modulate its own respiration and, on the other hand, induces deleterious effects on the host, including increased cardiovascular risk and renal insufficiency.121,122
Mtb Infection Dysregulates Host Immunometabolism
Innate immunity plays a key role in the pathophysiological course of pulmonary TB. Canonically, the innate response includes recognition of pathogen-associated molecular patterns (PAMPs), which induces diverse events, including inflammation, cell differentiation, and proliferation. Several findings in this study suggest that host immunometabolism is dysregulated following Mtb infection. For example, the following metabolic shifts (Supporting Table S4) could contribute to inflammation: (i) alterations in iNOS-related metabolites;123 (ii) alterations in XO-related metabolites, which may contribute to IL-1β secretion through regulation of the NRLP3 inflammasome;124 and (iii) increases in succinate levels, which has been shown to induce the release of proinflammatory cytokines through stabilization of HIF-1α.62
We found altered abundances of amino acids, which have been found to play a key role in immune-related processes (Supporting Table S4). For example, increased levels of several amino acids (i.e., arginine, glutamine, tryptophan) can potentially be sensed by the mechanistic target of rapamycin (mTOR) in mice, which is involved in a plethora of functions, including T-cell and monocyte differentiation and lipid synthesis.125 Increased arginine and citrulline levels in our disease model have been related to T-cell proliferation and survival against infection.126,127 Tryptophan and its degradation metabolites through the LPS-induced kynurenine pathway are key players in immunomodulation and immune proliferation.128 The highly increased levels of kynurenine found in Mtb+4w and Mtb+9w suggest an alteration of the kynurenine pathway, which may exert deleterious effects on the immune system given that inhibition of indoleamine 2,3-dioxygenase (IDO) activity promotes control of TB infection.129
Subnetwork Modularity Reveals Connections between Distinct Mtb-Associated Pathobiological Processes
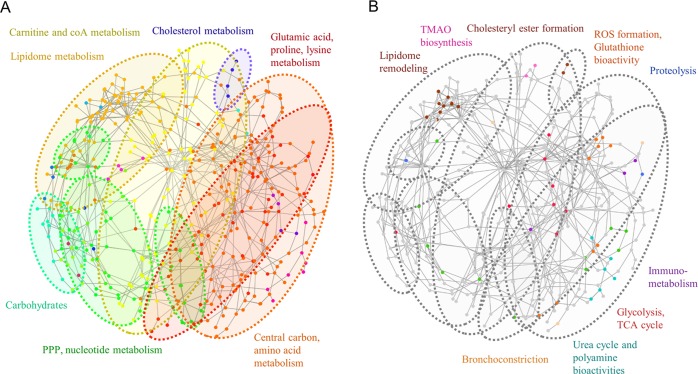
A subnetwork matrix containing the metabolites found in the study as well as bridging nodes was parsed and curated from MetaboNetworks. The resulting metabolic subnetwork was composed of 435 nodes and 739 edges. Using discrete ModuLand algorithms, we identified 21 modules. High ModuLand betweenness centrality values were assigned to metabolites including pyruvate, AMP, aspartate, serine, glutamate, glycine, glutathione, and cysteine, which were previously identified in our study as notably altered upon disease onset and progression. These results suggest that alterations in the abundances of such metabolites may have an impact on several biological processes arising from different metabolic pathways gathered in the network structure. In addition, different clusters could be observed (Figure 9A). Small clusters matched to specific pathways such as nicotinic acid metabolism and cholesterol biosynthesis. Overall, several network modules encompassed chemically similar metabolites involved in functionally described pathways. These included modules grouping carbohydrates, lipidome-related compounds, nucleotides and PPP compounds, carnitine and coA-related metabolites, glutamate and lysine-related compounds, and central carbon and amino acid metabolism. These clustering results provide evidence that clusters present in the network share a common biological role and suggest that alterations in one module are more prone to causing alterations in the modules with a higher number of intermodular edges, thereby providing a holistic view of cluster-specific metabolic alterations during TB. Overlapping of the different Mtb-associated biological alterations reflected in the metabolites from the previous sections (Figure 9B) reveals that, with the exception of cholesteryl esters and TMAO-related metabolites, the majority of lipidome remodeling-associated compounds conform to a tight cluster notably distant from the rest of the perturbations. In addition, metabolites related to disease-associated processes were mostly represented in the glutamate, proline, and lysine cluster or in the central carbon metabolism cluster, both being highly interconnected. These results suggest that proteolysis, and immunometabolism, alterations in the urea cycle and the polyamine metabolism are functional processes strongly related to the central carbon metabolism, which can have a joint impact via the metabolome.
Figure 9.
Metabolic subnetwork topology and network clustering analysis. (A) Color-coded map indicating the discrete clustering analysis results and major metabolite families found in clusters. (B) Superimposition of the different TB disease-associated alterations described in previous sections with the distinct network clusters.
Conclusions
Overall, our data emphasize that critical metabolic changes occur in the mouse lung metabolome of Mtb-infected mice, which are substantially altered or even reversed with disease progression, manifesting the dynamic nature of metabolic alterations. The non-targeted metabolomic approach in this study allowed the discovery of metabolic signatures of distinct pathobiological processes, several of which confirm previous observations during Mtb infection. Furthermore, network analysis has permitted the evaluation of comprehensive interconnections between previously and newly described TB-related metabolic processes. Also, specific metabolic alterations canonically described as inflammatory and anti-inflammatory macrophage polarization markers were found in both Mtb+4w and Mtb+9w, suggesting that different macrophage polarization subpopulations may coexist in the Mtb-infected lung. Clustering analysis of samples revealed closer clustering of Mtb+9w and Mtb– mice, suggesting the return of several metabolite abundances to control levels, after their alteration during the acute immune response occurring in Mtb+4w mice. Nonetheless, the levels of metabolites indicate highly non-compensated metabolic processes, such as proteolysis and macronutrient consumption, which are likely to reflect disease progression and worsening in Mtb+9w. Importantly, to the best of our knowledge, this is the first report indicating that high abundances of TMAO found in the TB lung may be linked to a negative impact on the host and a possible positive impact on Mtb.
To contextualize the vast amount of general data generated in this study, consensus OPLS-DA has allowed data fusion and significance assessment in a technique-independent manner, providing a high-throughput-oriented statistical test for determining alterations in the metabolome. Both cOPLS-DA and SUS plots highlighted the importance of performing multiplatform studies in our model, given the specific metabolic coverage and group separation achieved by each MS-based metabolomic platform. Hence, cOPLS-DA models could be used to determine altered metabolic pathways in both ORA and PA and highlight possible altered metabolic pathways. Given the notable consistency between cOPLS-DA VIP values and percentages of change, both parameters could be used to interpret manually the alterations in specific pathways not detected by metabolite representation algorithms. Collectively, these advances allowed us to accurately contextualize and interpret metabolic changes triggered by Mtb, which was not previously possible.
Finally, in this study, new and previously described metabolic alterations arising from pathological processes that were modulated by TB disease were integrated into a network model that demonstrated the collective interdependent metabolic modulations induced with TB disease progression. This will contribute to our understanding of the progression of Mtb pathogenesis and potentially serve as a foundation for new host-directed therapeutic strategies.
Acknowledgments
The co-authors would like to acknowledge funding from the Spanish Ministry of Economy and Competitiveness (RTI2018-095166-B-I00). M.F-G. acknowledges Fundación Universitaria San Pablo CEU for his Ph.D. fellowship. This work was supported by NIH grants R01Al111940, R61AI138280, R01AI137043 and the Bill and Melinda Gates Foundation award OPP1130017 (AJCS). The research was also co-funded by the South African Medical Research Council (AJCS). We acknowledge Joanna Godzien, Estanislao Nistal Villán and Francisco Javier Rupérez Pascualena (San Pablo CEU University) for useful discussions and their valuable contributions. Figure 1B and the TOC graphic were created with BioRender.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jproteome.9b00868.
Curated data matrix representing qualitative and quantitative parameters of the entire metabolomics data set, along with information on metabolic feature characteristics, annotations, normalized abundances per sample, cOPLS-DA variable importance in the projection and SUS plot correlation loading values (detailed legend provided in an additional spreadsheet found in the file) (XLSX)
Qualitative and quantitative NAD+ properties in CE-TOF/MS analysis of uninfected and Mtb-infected mice (XLSX)
Overrepresentation analysis and pathway analysis results showing the associated p-value for the altered metabolic pathways in each specific group comparison (XLSX)
Data matrix containing percentage of change and VIP values for each specific metabolite present in the experimental data and mentioned in the manuscript, sorted by the Results and Discussion subsection (XLSX)
Metabolic clustering analysis of the entire metabolomics data set showing the differences in normalized abundances of metabolites from uninfected (Mtb–), and 4 (Mtb+4w) and 9 (Mtb+9w) weeks postinfection mice (PDF)
Pulmonary TB metabolic subnetwork following the .sif format indicating the connectivity between different enzymatic reactions, including the different metabolites annotated in the resource (TXT)
Author Contributions
M.F.-G. and F.R.-S. contributed equally to this work. J. B. and S. R. contributed with statistical aspects related to the multiblock data analysis. V.P.R. contributed with the animal model. M.F.-G., F.R.-S, A.G., and C.B. (CEMBIO) contributed with the multiplatform metabolomics formal analysis and data processing. All the authors contributed to the discussion of results and writing. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Notes
The mass spectrometry data have been deposited to the Metabolomics Workbench Data Repository (https://www.metabolomicsworkbench.org/) via the NIH Data Repository with the data set identifier ST001328.
Supplementary Material
References
- Centers for Disease Control and Prevention. The Difference Between Latent TB Infection and TB Disease. https://www.cdc.gov/tb/publications/factsheets/general/ltbiandactivetb.html (accessed March 21, 2018).
- World Health Organization. Global Tuberculosis Report; 2017. https://www.who.int/tb/publications/global_report/gtbr2017_main_text.pdf.
- Dheda K.; Gumbo T.; Maartens G.; Dooley K. E.; McNerney R.; Murray M.; Furin J.; Nardell E. A.; London L.; Lessem E.; Theron G.; van Helden P.; Niemann S.; Merker M.; Dowdy D.; Van Rie A.; Siu G. K.; Pasipanodya J. G.; Rodrigues C.; Clark T. G.; Sirgel F. A.; Esmail A.; Lin H. H.; Atre S. R.; Schaaf H. S.; Chang K. C.; Lange C.; Nahid P.; Udwadia Z. F.; Horsburgh C. R.; Churchyard G. J.; Menzies D.; Hesseling A. C.; Nuermberger E.; McIlleron H.; Fennelly K. P.; Goemaere E.; Jaramillo E.; Low M.; Jara C. M.; Padayatchi N.; Warren R. M. The epidemiology, pathogenesis, transmission, diagnosis, and management of multidrug-resistant, extensively drug-resistant, and incurable tuberculosis. Lancet Respir. Med. 2017, 5, 291–360. 10.1016/S2213-2600(17)30079-6. [DOI] [PubMed] [Google Scholar]
- du Preez I.; Luies L.; Loots D. T. The application of metabolomics toward pulmonary tuberculosis research. Tuberculosis 2019, 115, 126–139. 10.1016/j.tube.2019.03.003. [DOI] [PubMed] [Google Scholar]
- Fernández-García M.; Rojo D.; Rey-Stolle F.; García A.; Barbas C.. Metabolomic-Based Methods in Diagnosis and Monitoring Infection Progression. In Metabolic Interaction in Infection, 1st ed.;Silvestre R.; Torrado E., Eds.; Springer: 2018; pp 283–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirsaeidi M.; Banoei M. M.; Winston B. W.; Schraufnagel D. E. Metabolomics: Applications and Promise in Mycobacterial Disease. Ann. Am. Thorac. Soc. 2015, 12, 1278–1287. 10.1513/AnnalsATS.201505-279PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee K. Y.; de Carvalho L. P.; Bryk R.; Ehrt S.; Marrero J.; Park S. W.; Schnappinger D.; Venugopal A.; Nathan C. Central carbon metabolism in Mycobacterium tuberculosis: an unexpected frontier. Trends Microbiol. 2011, 19, 307–314. 10.1016/j.tim.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beste D. J.; Nöh K.; Niedenführ S.; Mendum T. A.; Hawkins N. D.; Ward J. L.; Beale M. H.; Wiechert W.; McFadden J. 13C-flux spectral analysis of host-pathogen metabolism reveals a mixed diet for intracellular Mycobacterium tuberculosis. Chem. Biol. 2013, 20, 1012–1021. 10.1016/j.chembiol.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.; Crossman D. K.; Mai D.; Guidry L.; Voskuil M. I.; Renfrow M. B.; Steyn A. J. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 2009, 5, e1000545 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cumming B. M.; Addicott K. W.; Adamson J. H.; Steyn A. J. Mycobacterium tuberculosis induces decelerated bioenergetic metabolism in human macrophages. eLife 2018, 7, e39169 10.7554/eLife.39169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. P.; Chinta K. C.; Saini V.; Glasgow J. N.; Hull T. D.; Traylor A.; Rey-Stolle F.; Soares M. P.; Madansein R.; Rahman M. A.; Barbas C.; Nargan K.; Naidoo T.; Ramdial P. K.; George J. F.; Agarwal A.; Steyn A. J. C. Ferritin H Deficiency in Myeloid Compartments Dysregulates Host Energy Metabolism and Increases Susceptibility to Mycobacterium tuberculosis infection. Front. Immunol. 2018, 9, 860 10.3389/fimmu.2018.00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson L. E.; Sheedy F. J.; Palsson-McDermott E. M.; Triglia D.; O’Leary S. M.; O’Sullivan M. P.; O’Neill L. A.; Keane J. Cutting Edge: Mycobacterium tuberculosis Induces Aerobic Glycolysis in Human Alveolar Macrophages That Is Required for Control of Intracellular Bacillary Replication. J. Immunol. 2016, 196, 2444–2449. 10.4049/jimmunol.1501612. [DOI] [PubMed] [Google Scholar]
- Braverman J.; Stanley S. A. Nitric Oxide Modulates Macrophage Responses to Mycobacterium tuberculosis Infection through Activation of HIF-1 α and repression of NF-κB. J. Immunol. 2017, 199, 1805–1816. 10.4049/jimmunol.1700515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualls J. E.; Murray P. J. Immunometabolism within the tuberculosis granuloma: amino acids, hypoxia, and cellular respiration. Semin. Immunopathol. 2016, 38, 139–152. 10.1007/s00281-015-0534-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J.; Maertzdorf J.; Sutherland J. S.; Duffy F. J.; Thompson E.; Suliman S.; McEwen G.; Thiel B.; Parida S. K.; Zyla J.; Hanekom W. A.; Mohney R. P.; Boom W. H.; Mayanja-Kizza H.; Howe R.; Dockrell H. M.; Ottenhoff T. H. M.; Scriba T. J.; Zak D. E.; Walzl G.; Kaufmann S. H. E. The GC6-74 consortium. Metabolite changes in blood predict the onset of tuberculosis. Nat. Commun. 2018, 9, 5208 10.1038/s41467-018-07635-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. H.; Yang J. Y.; Jeon B. Y.; Yoon Y. J.; Cho S. N.; Kang Y. H.; Ryu D. H.; Hwang G. S. 1H NMR-based metabolomic profiling in mice infected with Mycobacterium tuberculosis. J. Proteome Res. 2011, 10, 2238–2247. 10.1021/pr101054m. [DOI] [PubMed] [Google Scholar]
- Somashekar B. S.; Amin A. G.; Rithner C. D.; Troudt J.; Basaraba R.; Izzo A.; Crick D. C.; Chatterjee D. Metabolic profiling of lung granuloma in Mycobacterium tuberculosis infected guinea pigs: ex vivo 1H magic angle spinning NMR studies. J. Proteome Res. 2011, 10, 4186–4195. 10.1021/pr2003352. [DOI] [PubMed] [Google Scholar]
- Somashekar B. S.; Amin A. G.; Tripathi P.; MacKinnon N.; Rithner C. D.; Shanley C. A.; Basaraba R.; Henao-Tamayo M.; Kato-Maeda M.; Ramamoorthy A.; Orme I. M.; Ordway D. J.; Chatterjee D. Metabolomic signatures in guinea pigs infected with epidemic-associated W-Beijing strains of Mycobacterium tuberculosis. J. Proteome Res. 2012, 11, 4873–4884. 10.1021/pr300345x. [DOI] [PubMed] [Google Scholar]
- Emwas A.-H. M. The strengths and weaknesses of NMR spectroscopy and mass spectrometry with particular focus on metabolomics research. Methods Mol. Biol. 2015, 1277, 161–193. [DOI] [PubMed] [Google Scholar]
- Naz S.; García A.; Barbas C. Multiplatform analytical methodology for metabolic fingerprinting of lung tissue. Anal. Chem. 2013, 85, 10941–10948. 10.1021/ac402411n. [DOI] [PubMed] [Google Scholar]
- Guijas C.; Montenegro-Burke J. R.; Domingo-Almenara X.; Palermo A.; Warth B.; Hermann G.; Koellensperger G.; Huan T.; Uritboonthai W.; Aisporna A. E.; Wolan D. W.; Spilker M. E.; Benton H. P.; Siuzdak G. METLIN: A Technology Platform for Identifying Knowns and Unknowns. Anal. Chem. 2018, 90, 3156–3164. 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O.Metabolite profiling in Arabidopsis. In Arabidopsis Protocols, 2nd ed.; Salinas J.; Sanchez-Serrano J. J., Eds.; Springer: Berlin, 2006; pp 439–447. [DOI] [PubMed] [Google Scholar]
- Kind T.; Wohlgemuth G.; Lee D. Y.; Lu Y.; Palazoglu M.; Shahbaz S.; Fiehn O. FiehnLib: mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. 10.1021/ac9019522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil de la Fuente A.; Godzien J.; Fernández López M.; Rupérez F. J.; Barbas C.; Otero A. Knowledge-based metabolite annotation tool: CEU Mass Mediator. J. Pharm. Biomed. Anal. 2018, 154, 138–149. 10.1016/j.jpba.2018.02.046. [DOI] [PubMed] [Google Scholar]
- Brockman S. A.; Roden E. V.; Hegeman A. D. Van Krevelen diagram visualization of high resolution-mass spectrometry metabolomics data with OpenVanKrevelen. Metabolomics 2018, 14, 48 10.1007/s11306-018-1343-y. [DOI] [PubMed] [Google Scholar]
- Godzien J.; Alonso-Herranz V.; Barbas C.; Grace Armitage E. Controlling the quality of metabolomics data: new strategies to get the best out of the QC sample. Metabolomics 2015, 11, 518–528. 10.1007/s11306-014-0712-4. [DOI] [Google Scholar]
- Boccard J.; Rutledge D. N. A consensus orthogonal partial least squares discriminant analysis (OPLS-DA) strategy for multiblock Omics data fusion. Anal. Chim. Acta 2013, 769, 30–39. 10.1016/j.aca.2013.01.022. [DOI] [PubMed] [Google Scholar]
- Chong J.; Soufan O.; Li C.; Caraus I.; Li S.; Bourque G.; Wishart D. S.; Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. 10.1093/nar/gky310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M.; Furumichi M.; Tanabe M.; Sato Y.; Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posma J. M.; Robinette S. L.; Holmes E.; Nicholson J. K. MetaboNetworks, an interactive Matlab-based toolbox for creating, customizing and exploring sub-networks from KEGG. Bioinformatics 2014, 30, 893–895. 10.1093/bioinformatics/btt612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szalay-Beko M.; Palotai R.; Szappanos B.; Kovács I. A.; Papp B.; Csermely P. ModuLand plug-in for Cytoscape: determination of hierarchical layers of overlapping network modules and community centrality. Bioinformatics 2012, 28, 2202–2204. 10.1093/bioinformatics/bts352. [DOI] [PubMed] [Google Scholar]
- Shannon P.; Markiel A.; Ozier O.; Baliga N. S.; Wang J. T.; Ramage D.; Amin N.; Schwikowski B.; Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács I. A.; Palotai R.; Szalay M. S.; Csermely P. Community landscapes: an integrative approach to determine overlapping network module hierarchy, identify key nodes and predict network dynamics. PLoS One 2010, 5, e12528 10.1371/journal.pone.0012528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stek C.; Allwood B.; Walker N. F.; Wilkinson R. J.; Lynen L.; Meintjes G. The Immune Mechanisms of Lung Parenchymal Damage in Tuberculosis and the Role of Host-Directed Therapy. Front. Microbiol. 2018, 9, 2603 10.3389/fmicb.2018.02603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo-Villarino G.; Hudrisier D.; Benard A.; Neyrolles O. Emerging trends in the formation and function of tuberculosis granulomas. Front. Immunol. 2012, 3, 405 10.3389/fimmu.2012.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marakalala M. J.; Raju R. M.; Sharma K.; Zhang Y. J.; Eugenin E. A.; Prideaux B.; Daudelin I. B.; Chen P. Y.; Booty M. G.; Kim J. H.; Eum S. Y.; Via L. E.; Behar S. M.; Barry C. E.; Mann M.; Dartois V.; Rubin E. J. Inflammatory signaling in human tuberculosis granulomas is spatially organized. Nat. Med. 2016, 22, 531–538. 10.1038/nm.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Locasale J. W. Metabolomics: A Primer. Trends Biochem. Sci. 2017, 42, 274–284. 10.1016/j.tibs.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.; Wilson I. D.; Nicholson J. K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717. 10.1016/j.cell.2008.08.026. [DOI] [PubMed] [Google Scholar]
- Aranibar N.; Vassallo J. D.; Rathmacher J.; Stryker S.; Zhang Y.; Dai J.; Janovitz E. B.; Robertson D.; Reily M.; Lowe-Krentz L.; Lehman-McKeeman L. Identification of 1- and 3-methylhistidine as biomarkers of skeletal muscle toxicity by nuclear magnetic resonance-based metabolic profiling. Anal. Biochem. 2011, 410, 84–91. 10.1016/j.ab.2010.11.023. [DOI] [PubMed] [Google Scholar]
- Sugawara T.; Ito Y.; Nishizawa N.; Suzuki H.; Kobayashi H.; Nagasawa T. Measurement of the rate of myofibrillar protein degradation using the arteriovenous difference in plasma 3-methylhistidine concentration of rats. J. Nutr. Sci. Vitaminol. 2009, 55, 381–384. 10.3177/jnsv.55.381. [DOI] [PubMed] [Google Scholar]
- Gupta K. B.; Gupta R.; Atreja A.; Verma M.; Vishvkarma S. Tuberculosis and nutrition. Lung India 2009, 26, 9–16. 10.4103/0970-2113.45198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macallan D. C.; McNurlan M. A.; Kurpad A. V.; de Souza G.; Shetty P. S.; Calder A. G.; Griffin G. E. Whole body protein metabolism in human pulmonary tuberculosis and undernutrition: evidence for anabolic block in tuberculosis. Clin. Sci. 1998, 94, 321–331. 10.1042/cs0940321. [DOI] [PubMed] [Google Scholar]
- Ong C. W.; Elkington P. T.; Friedland J. S. Tuberculosis, pulmonary cavitation, and matrix metalloproteinases. Am. J. Respir. Crit. Care Med. 2014, 190, 9–18. 10.1164/rccm.201311-2106PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev S. A.; Cieplak P.; Aleshin A. E.; Sun Q.; Zhu W.; Motamedchaboki K.; Sloutsky A.; Strongin A. Y. Matrix metalloproteinase proteolysis of the mycobacterial HSP65 protein as a potential source of immunogenic peptides in human tuberculosis. FEBS J. 2011, 278, 3277–3286. 10.1111/j.1742-4658.2011.08244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindskog C.; Fagerberg L.; Hallström B.; Edlund K.; Hellwig B.; Rahnenführer J.; Kampf C.; Uhlén M.; Pontén F.; Micke P. The lung-specific proteome defined by integration of transcriptomics and antibody-based profiling. FASEB J. 2014, 28, 5184–5196. 10.1096/fj.14-254862. [DOI] [PubMed] [Google Scholar]
- Toshima M.; Ohtani Y.; Ohtani O. Three-dimensional architecture of elastin and collagen fiber networks in the human and rat lung. Arch. Histol. Cytol. 2004, 67, 31–40. 10.1679/aohc.67.31. [DOI] [PubMed] [Google Scholar]
- Kim C.; Ko Y.; Kim S. H.; Yoo H. J.; Lee J. S.; Rhee C. K.; Lee J. H.; Kim T. H.; Lim S. Y.; Yoo K. H.; Seo J. B.; Oh Y. M.; Lee S. D.; Park Y. B.; Lee J.-H. Urinary desmosine is associated with emphysema severity and frequent exacerbation in patients with COPD. Respirology 2018, 23, 176–181. 10.1111/resp.13170. [DOI] [PubMed] [Google Scholar]
- Seddon J.; Kasprowicz V.; Walker N. F.; Yuen H. M.; Sunpath H.; Tezera L.; Meintjes G.; Wilkinson R. J.; Bishai W. R.; Friedland J. S.; Elkington P. T. Procollagen III N-terminal propeptide and desmosine are released by matrix destruction in pulmonary tuberculosis. J. Infect. Dis. 2013, 208, 1571–1579. 10.1093/infdis/jit343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taooka Y.; Maeda A.; Hiyama K.; Ishioka S.; Yamakido M. Effects of neutrophil elastase inhibitor on bleomycin-induced pulmonary fibrosis in mice. Am. J. Respir. Crit. Care Med. 1997, 156, 260–265. 10.1164/ajrccm.156.1.9612077. [DOI] [PubMed] [Google Scholar]
- Toyoki Y.; Sasaki M.; Narumi S.; Yoshihara S.; Morita T.; Konn M. Semiquantitative evaluation of hepatic fibrosis by measuring tissue hydroxyproline. Hepatogastroenterology 1998, 45, 2261–2264. [PubMed] [Google Scholar]
- Hothersall J. S.; Gordge M.; Noronha-Dutra A. A. Inhibition of NADPH supply by 6-aminonicotinamide: effect on glutathione, nitric oxide and superoxide in J774 cells. FEBS Lett. 1998, 434, 97–100. 10.1016/S0014-5793(98)00959-4. [DOI] [PubMed] [Google Scholar]
- Shi L.; Salamon H.; Eugenin E. A.; Pine R.; Cooper A.; Gennaro M. L. Infection with Mycobacterium tuberculosis induces the Warburg effect in mouse lungs. Sci. Rep. 2016, 5, 18176 10.1038/srep18176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N. C.; O’Neill L. A. J. A Role for the Krebs Cycle Intermediate Citrate in Metabolic Reprogramming in Innate Immunity and Inflammation. Front. Immunol. 2018, 9, 141 10.3389/fimmu.2018.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A. K.; Huang S. C.; Sergushichev A.; Lampropoulou V.; Ivanova Y.; Loginicheva E.; Chmielewski K.; Stewart K. M.; Ashall J.; Everts B.; Pearce E. J.; Driggers E. M.; Artyomov M. N. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 2015, 42, 419–430. 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Michelucci A.; Cordes T.; Ghelfi J.; Pailot A.; Reiling N.; Goldmann O.; Binz T.; Wegner A.; Tallam A.; Rausell A.; Buttini M.; Linster C. L.; Medina E.; Balling R.; Hiller K. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 7820–7825. 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelko C. L.; Lu W.; Dufort F. J.; Seyfried T. N.; Chiles T. C.; Rabinowitz J. D.; Roberts M. F. Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 2011, 133, 16386–16389. 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallam A.; Perumal T. M.; Antony P. M.; Jäger C.; Fritz J. V.; Vallar L.; Balling R.; Del Sol A.; Michelucci A. Gene Regulatory Network Inference of Immunoresponsive Gene 1 (IRG1) Identifies Interferon Regulatory Factor 1 (IRF1) as Its Transcriptional Regulator in Mammalian Macrophages. PLoS One 2016, 11, e0149050 10.1371/journal.pone.0149050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampropoulou V.; Sergushichev A.; Bambouskova M.; Nair S.; Vincent E. E.; Loginicheva E.; Cervantes-Barragan L.; Ma X.; Huang S. C.; Griss T.; Weinheimer C. J.; Khader S.; Randolph G. J.; Pearce E. J.; Jones R. G.; Diwan A.; Diamond M. S.; Artyomov M. N. Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab. 2016, 24, 158–166. 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krátký M.; Vinšová J. Advances in mycobacterial isocitrate lyase targeting and inhibitors. Curr. Med. Chem. 2012, 19, 6126–6137. 10.2174/0929867311209066126. [DOI] [PubMed] [Google Scholar]
- Pham T. V.; Murkin A. S.; Moynihan M. M.; Harris L.; Tyler P. C.; Shetty N.; Sacchettini J. C.; Huang H. L.; Meek T. D. Mechanism-based inactivator of isocitrate lyases 1 and 2 from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 7617–7622. 10.1073/pnas.1706134114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S.; Huynh J. P.; Lampropoulou V.; Loginicheva E.; Esaulova E.; Gounder A. P.; Boon A. C. M.; Schwarzkopf E. A.; Bradstreet T. R.; Edelson B. T.; Artyomov M. N.; Stallings C. L.; Diamond M. S. Irg1 expression in myeloid cells prevents immunopathology during M. tuberculosis infection. J. Exp. Med. 2018, 215, 1035–1045. 10.1084/jem.20180118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannahill G. M.; Curtis A. M.; Adamik J.; Palsson-McDermott E. M.; McGettrick A. F.; Goel G.; Frezza C.; Bernard N. J.; Kelly B.; Foley N. H.; Zheng L.; Gardet A.; Tong Z.; Jany S. S.; Corr S. C.; Haneklaus M.; Caffrey B. E.; Pierce K.; Walmsley S.; Beasley F. C.; Cummins E.; Nizet V.; Whyte M.; Taylor C. T.; Lin H.; Masters S. L.; Gottlieb E.; Kelly V. P.; Clish C.; Auron P. E.; Xavier R. J.; O’Neill L. A. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature 2013, 496, 238–242. 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L. A.; Kishton R. J.; Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016, 16, 553–565. 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahien L. A.; Kmiotek E. H.; MacDonald M. J.; Fibich B.; Mandic M. Regulation of malate dehydrogenase activity by glutamate, citrate, alpha-ketoglutarate, and multienzyme interaction. J. Biol. Chem. 1988, 263, 10687–10697. [PubMed] [Google Scholar]
- Freire D. M.; Gutierrez C.; Garza-Garcia A.; Grabowska A. D.; Sala A. J.; Ariyachaokun K.; Panikova T.; Beckham K. S. H.; Colom A.; Pogenberg V.; Cianci M.; Tuukkanen A.; Boudehen Y. M.; Peixoto A.; Botella L.; Svergun D. I.; Schnappinger D.; Schneider T. R.; Genevaux P.; de Carvalho L. P. S.; Wilmanns M.; Parret A. H. A.; Neyrolles O. An NAD+ Phosphorylase Toxin Triggers Mycobacterium tuberculosis Cell Death. Mol. Cell 2019, 73, 1282–1291.e8. 10.1016/j.molcel.2019.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tak U.; Vlach J.; Garza-Garcia A.; William D.; Danilchanka O.; de Carvalho L. P. S.; Saad J. S.; Niederweis M. The tuberculosis necrotizing toxin is an NAD+ and NADP+ glycohydrolase with distinct enzymatic properties. J. Biol. Chem. 2018, 294, 3024–3026. 10.1074/jbc.RA118.005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajuelo D.; Gonzalez-Juarbe N.; Tak U.; Sun J.; Orihuela C. J.; Niederweis M. NAD+ Depletion Triggers Macrophage Necroptosis, a Cell Death Pathway Exploited by Mycobacterium tuberculosis. Cell Rep. 2018, 24, 429–440. 10.1016/j.celrep.2018.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinathan K. P.; Sirsi M.; Vaidyanathan C. S. Nicotinamide-Adenine Dinucleotide-Glycohydrolase Activity in Experimental Tuberculosis. Biochem. J. 1965, 94, 446–451. 10.1042/bj0940446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. H.; Zhang D. W.; Zheng X. L.; Tang C. K. Itaconate: an emerging determinant of inflammation in activated macrophages. Immunol. Cell Biol. 2019, 97, 134–141. 10.1111/imcb.12218. [DOI] [PubMed] [Google Scholar]
- Tan H. Y.; Wang N.; Li S.; Hong M.; Wang X.; Feng Y. The Reactive Oxygen Species in Macrophage Polarization: Reflecting Its Dual Role in Progression and Treatment of Human Diseases. Oxid. Med. Cell. Longevity 2016, 2016, 2795090 10.1155/2016/2795090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamaati H.; Mortaz E.; Pajouhi Z.; Folkerts G.; Movassaghi M.; Moloudizargari M.; Adcock I. M.; Garssen J. Nitric Oxide in the Pathogenesis and Treatment of Tuberculosis. Front. Microbiol. 2017, 8, 2008 10.3389/fmicb.2017.02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque-Correa M. A.; Kühl A. A.; Rodriguez P. C.; Zedler U.; Schommer-Leitner S.; Rao M.; Weiner J.; Hurwitz R.; Qualls J. E.; Kosmiadi G. A.; Murray P. J.; Kaufmann S. H.; Reece S. T. Macrophage arginase-1 controls bacterial growth and pathology in hypoxic tuberculosis granulomas. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, E4024–E4032. 10.1073/pnas.1408839111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.; Peng Y.; Yin Y.; Zhou Z.; Zhou W.; Dai Y. The involvement of NADPH oxidase-mediated ROS in cytokine secretion from macrophages induced by Mycobacterium tuberculosis ESAT-6. Inflammation 2014, 37, 880–892. 10.1007/s10753-013-9808-7. [DOI] [PubMed] [Google Scholar]
- Granger D. N.; Kvietys P. R. Reperfusion injury and reactive oxygen species: The evolution of a concept. Redox Biol. 2015, 6, 524–551. 10.1016/j.redox.2015.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R.; Teskey G.; Islamoglu H.; Abrahem R.; Munjal S.; Gyurjian K.; Zhong L.; Venketaraman V. Characterizing the Effects of Glutathione as an Immunoadjuvant in the Treatment of Tuberculosis. Antimicrob. Agents Chemother. 2018, 62, e01132-18 10.1128/AAC.01132-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venketaraman V.; Dayaram Y. K.; Amin A. G.; Ngo R.; Green R. M.; Talaue M. T.; Mann J.; Connell N. D. Role of glutathione in macrophage control of mycobacteria. Infect. Immun. 2003, 71, 1864–1871. 10.1128/IAI.71.4.1864-1871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley P. J. The glyoxalase system in health and disease. Mol. Aspects Med. 1993, 14, 287–371. 10.1016/0098-2997(93)90002-U. [DOI] [PubMed] [Google Scholar]
- Edagwa B.; Wang Y.; Narayanasamy P. Synthesis of azide derivative and discovery of glyoxalase pathway inhibitor against pathogenic bacteria. Bioorg. Med. Chem. Lett. 2013, 23, 6138–6140. 10.1016/j.bmcl.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Greenblatt M.; Bach H.; Adamson J.; Peña-Diaz S.; Li W.; Steyn A. J.; Av-Gay Y. Regulation of Ergothioneine Biosynthesis and Its Effect on Mycobacterium tuberculosis Growth and Infectivity. J. Biol. Chem. 2015, 290, 23064–23076. 10.1074/jbc.M115.648642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini V.; Cumming B. M.; Guidry L.; Lamprecht D. A.; Adamson J. H.; Reddy V. P.; Chinta K. C.; Mazorodze J. H.; Glasgow J. N.; Richard-Greenblatt M.; Gomez-Velasco A.; Bach H.; Av-Gay Y.; Eoh H.; Rhee K.; Steyn A. J. C. Ergothioneine Maintains Redox and Bioenergetic Homeostasis Essential for Drug Susceptibility and Virulence of Mycobacterium tuberculosis. Cell Rep. 2016, 14, 572–585. 10.1016/j.celrep.2015.12.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardbower D. M.; Asim M.; Luis P. B.; Singh K.; Barry D. P.; Yang C.; Steeves M. A.; Cleveland J. L.; Schneider C.; Piazuelo M. B.; Gobert A. P.; Wilson K. T. Ornithine decarboxylase regulates M1 macrophage activation and mucosal inflammation via histone modifications. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, E751–E760. 10.1073/pnas.1614958114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A. P.; Wilson K. T. Editorial: Orchestration of macrophage polarization by polyamines. J. Leukocyte Biol. 2012, 91, 677–679. 10.1189/jlb.0112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg A. E. Functions of Polyamines in Mammals. J. Biol. Chem. 2016, 291, 14904–14912. 10.1074/jbc.R116.731661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes C. S.; Shicora A. C.; Keough M. P.; Snook A. E.; Burns M. R.; Gilmour S. K. Polyamine-blocking therapy reverses immunosuppression in the tumor microenvironment. Cancer Immunol. Res. 2014, 2, 274–285. 10.1158/2326-6066.CIR-13-0120-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A.; Tyagi A. K. Role of polyamines in the synthesis of RNA in mycobacteria. Mol. Cell. Biochem. 1987, 78, 3–8. 10.1007/BF00224418. [DOI] [PubMed] [Google Scholar]
- Sarathy J. P.; Lee E.; Dartois V. Polyamines inhibit porin-mediated fluoroquinolone uptake in mycobacteria. PLoS One 2013, 8, e65806 10.1371/journal.pone.0065806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal A.; Mandal S.; Park M. H. Global quantitative proteomics reveal up-regulation of endoplasmic reticulum stress response proteins upon depletion of eIF5A in HeLa cells. Sci. Rep. 2016, 6, 25795 10.1038/srep25795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis N.; Rubera I.; Cougnon M.; Giraud S.; Mograbi B.; Belaid A.; Pisani D. F.; Huber S. M.; Lacas-Gervais S.; Fragaki K.; Blondeau N.; Vigne P.; Frelin C.; Hauet T.; Duranton C.; Tauc M. Targeting eIF5A Hypusination Prevents Anoxic Cell Death through Mitochondrial Silencing and Improves Kidney Transplant Outcome. J. Am. Soc. Nephrol. 2017, 28, 811–822. 10.1681/ASN.2016010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K.; Murozumi K.; Shirahata A.; Park M. H.; Kashiwagi K.; Igarashi K. Independent roles of eIF5A and polyamines in cell proliferation. Biochem. J. 2005, 385, 779–785. 10.1042/BJ20041477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fessler M. B.; Summer R. S. Surfactant Lipids at the Host-Environment Interface. Metabolic Sensors, Suppressors, and Effectors of Inflammatory Lung Disease. Am. J. Respir. Cell Mol. Biol. 2016, 54, 624–635. 10.1165/rcmb.2016-0011PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.; VanderVen B. C.; Fahey R. J.; Russell D. G. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J. Biol. Chem. 2013, 288, 6788–6800. 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh G.; Singh G.; Jadeja D.; Kaur J. Lipid hydrolizing enzymes in virulence: Mycobacterium tuberculosis as a model system. Crit. Rev. Microbiol. 2010, 36, 259–269. 10.3109/1040841X.2010.482923. [DOI] [PubMed] [Google Scholar]
- Schneider B. E.; Behrends J.; Hagens K.; Harmel N.; Shayman J. A.; Schaible U. E. Lysosomal phospholipase A2: a novel player in host immunity to Mycobacterium tuberculosis. Eur. J. Immunol. 2014, 44, 2394–2404. 10.1002/eji.201344383. [DOI] [PubMed] [Google Scholar]
- Honda A.; Yamashita K.; Ikegami T.; Hara T.; Miyazaki T.; Hirayama T.; Numazawa M.; Matsuzaki Y. Highly sensitive quantification of serum malonate, a possible marker for de novo lipogenesis, by LC-ESI-MS/MS. J. Lipid Res. 2009, 50, 2124–2130. 10.1194/jlr.D800054-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker J.; Liebisch G.; Englmaier M.; Grandl M.; Robenek H.; Schmitz G. Induction of fatty acid synthesis is a key requirement for phagocytic differentiation of human monocytes. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 7817–7822. 10.1073/pnas.0912059107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon J. S.; Lee S.; Park M. A.; Siempos I. I.; Haslip M.; Lee P. J.; Yun M.; Kim C. K.; Howrylak J.; Ryter S. W.; Nakahira K.; Choi A. M. UCP2-induced fatty acid synthase promotes NLRP3 inflammasome activation during sepsis. J. Clin. Invest. 2015, 125, 665–680. 10.1172/JCI78253. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kol S.; Kehat I.; Adashi E. Y. Ovarian interleukin-1-induced gene expression: privileged genes threshold theory. Med. Hypotheses 2002, 58, 6–8. 10.1054/mehy.2001.1389. [DOI] [PubMed] [Google Scholar]
- Gaber T.; Strehl C.; Buttgereit F. Metabolic regulation of inflammation. Nat. Rev. Rheumatol. 2017, 13, 267–279. 10.1038/nrrheum.2017.37. [DOI] [PubMed] [Google Scholar]
- Huang Z.; Luo Q.; Guo Y.; Chen J.; Xiong G.; Peng Y.; Ye J.; Li J. Mycobacterium tuberculosis-Induced Polarization of Human Macrophage Orchestrates the Formation and Development of Tuberculous Granulomas In Vitro. PLoS One 2015, 10, e0129744 10.1371/journal.pone.0129744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight M.; Braverman J.; Asfaha K.; Gronert K.; Stanley S. Lipid droplet formation in Mycobacterium tuberculosis infected macrophages requires IFN-γ/HIF-1α signaling and supports host defense. PLoS Pathog. 2018, 14, e1006874 10.1371/journal.ppat.1006874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khor V. K.; Ahrends R.; Lin Y.; Shen W. J.; Adams C. M.; Roseman A. N.; Cortez Y.; Teruel M. N.; Azhar S.; Kraemer F. B. The proteome of cholesteryl-ester-enriched versus triacylglycerol-enriched lipid droplets. PLoS One 2014, 9, e105047 10.1371/journal.pone.0105047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cáceres N.; Tapia G.; Ojanguren I.; Altare F.; Gil O.; Pinto S.; Vilaplana C.; Cardona P. J. Evolution of foamy macrophages in the pulmonary granulomas of experimental tuberculosis models. Tuberculosis 2009, 89, 175–182. 10.1016/j.tube.2008.11.001. [DOI] [PubMed] [Google Scholar]
- Ghosh S.; Zhao B.; Bie J.; Song J. Macrophage cholesteryl ester mobilization and atherosclerosis. Vascul. Pharmacol. 2010, 52, 1–10. 10.1016/j.vph.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hla T.; Dannenberg A. J. Sphingolipid signaling in metabolic disorders. Cell Metab. 2012, 16, 420–434. 10.1016/j.cmet.2012.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik Z. A.; Thompson C. R.; Hashimi S.; Porter B.; Iyer S. S.; Kusner D. J. Cutting edge: Mycobacterium tuberculosis blocks Ca2+ signaling and phagosome maturation in human macrophages via specific inhibition of sphingosine kinase. J. Immunol. 2003, 170, 2811–2815. 10.4049/jimmunol.170.6.2811. [DOI] [PubMed] [Google Scholar]
- Sharma L.; Prakash H. Sphingolipids Are Dual Specific Drug Targets for the Management of Pulmonary Infections: Perspective. Front. Immunol. 2017, 8, 378 10.3389/fimmu.2017.00378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vass G.; Horváth I. Adenosine and adenosine receptors in the pathomechanism and treatment of respiratory diseases. Curr. Med. Chem. 2008, 15, 917–922. 10.2174/092986708783955392. [DOI] [PubMed] [Google Scholar]
- Afrasiabian S.; Mohsenpour B.; Bagheri K. H.; Sigari N.; Aftabi K. Diagnostic value of serum adenosine deaminase level in pulmonary tuberculosis. J. Res. Med. Sci. 2013, 18, 252–254. [PMC free article] [PubMed] [Google Scholar]
- Volonté C.; D’Ambrosi N. Membrane compartments and purinergic signalling: the purinome, a complex interplay among ligands, degrading enzymes, receptors and transporters. FEBS J. 2009, 276, 318–329. 10.1111/j.1742-4658.2008.06793.x. [DOI] [PubMed] [Google Scholar]
- Chan J.; Xing Y.; Magliozzo R. S.; Bloom B. R. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J. Exp. Med. 1992, 175, 1111–1122. 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J. D.; North R. J.; LaCourse R.; Mudgett J. S.; Shah S. K.; Nathan C. F. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 1997, 94, 5243–5248. 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer W.; Wiegand S.; Akinci S.; Schinkel A. H.; Wess J.; Koepsell H.; Haberberger R. V.; Lips K. S. Role of acetylcholine and muscarinic receptors in serotonin-induced bronchoconstriction in the mouse. J. Mol. Neurosci. 2006, 30, 67–68. 10.1385/JMN:30:1:67. [DOI] [PubMed] [Google Scholar]
- Barfod K. K.; Vrankx K.; Mirsepasi-Lauridsen H. C.; Hansen J. S.; Hougaard K. S.; Larsen S. T.; Ouwenhand A. C.; Krogfelt K. A. The Murine Lung Microbiome Changes During Lung Inflammation and Intranasal Vancomycin Treatment. Open Microbiol. J. 2015, 9, 167–179. 10.2174/1874285801509010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel T.; Lagkouvardos I.; Blaut M.; Stecher B. The mouse gut microbiome revisited: From complex diversity to model ecosystems. Int. J. Med. Microbiol. 2016, 306, 316–327. 10.1016/j.ijmm.2016.03.002. [DOI] [PubMed] [Google Scholar]
- Fennema D.; Phillips I. R.; Shephard E. A. Trimethylamine and Trimethylamine N-Oxide, a Flavin-Containing Monooxygenase 3 (FMO3)-Mediated Host-Microbiome Metabolic Axis Implicated in Health and Disease. Drug Metab. Dispos. 2016, 44, 1839–1850. 10.1124/dmd.116.070615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Waiz M.; Mikov M.; Mitchell S. C.; Smith R. L. The exogenous origin of trimethylamine in the mouse. Metabolism 1992, 41, 135–136. 10.1016/0026-0495(92)90140-6. [DOI] [PubMed] [Google Scholar]
- Barrett E. L.; Kwan H. S. Bacterial reduction of trimethylamine oxide. Annu. Rev. Microbiol. 1985, 39, 131–149. 10.1146/annurev.mi.39.100185.001023. [DOI] [PubMed] [Google Scholar]
- Seldin M. M.; Meng Y.; Qi H.; Zhu W.; Wang Z.; Hazen S. L.; Lusis A. J.; Shih D. M. Trimethylamine N-Oxide Promotes Vascular Inflammation Through Signaling of Mitogen-Activated Protein Kinase and Nuclear Factor-κB. J. Am. Heart Assoc. 2016, 5, e002767 10.1161/JAHA.115.002767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ierardi E.; Sorrentino C.; Principi M.; Giorgio F.; Losurdo G.; Di Leo A. Intestinal microbial metabolism of phosphatidylcholine: a novel insight in the cardiovascular risk scenario. Hepatobiliary Surg. Nutr. 2015, 4, 289–292. 10.3978/j.issn.2304-3881.2015.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.; Klipfell E.; Bennett B. J.; Koeth R.; Levison B. S.; Dugar B.; Feldstein A. E.; Britt E. B.; Fu X.; Chung Y. M.; Wu Y.; Schauer P.; Smith J. D.; Allayee H.; Tang W. H.; DiDonato J. A.; Lusis A. J.; Hazen S. L. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011, 472, 57–63. 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaysen G. A.; Johansen K. L.; Chertow G. M.; Dalrymple L. S.; Kornak J.; Grimes B.; Dwyer T.; Chassy A. W.; Fiehn O. Associations of Trimethylamine N-Oxide With Nutritional and Inflammatory Biomarkers and Cardiovascular Outcomes in Patients New to Dialysis. J. Ren. Nutr. 2015, 25, 351–356. 10.1053/j.jrn.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. H. W.; Wang Z.; Kennedy D. J.; Wu Y.; Buffa J. A.; Agatisa-Boyle B.; Li X. S.; Levison B. S.; Hazen S. L. Gut microbiota-dependent trimethylamine N-oxide (TMAO) pathway contributes to both development of renal insufficiency and mortality risk in chronic kidney disease. Circ. Res. 2015, 116, 448–455. 10.1161/CIRCRESAHA.116.305360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng X.; Zhang H.; Snead C.; Catravas J. D. Molecular mechanisms of iNOS induction by IL-1 beta and IFN-gamma in rat aortic smooth muscle cells. Am. J. Physiol.: Cell Physiol. 2002, 282, C144–52. 10.1152/ajpcell.2002.282.1.C144. [DOI] [PubMed] [Google Scholar]
- Ives A.; Nomura J.; Martinon F.; Roger T.; LeRoy D.; Miner J. N.; Simon G.; Busso N.; So A. Xanthine oxidoreductase regulates macrophage IL1β secretion upon NLRP3 inflammasome activation. Nat. Commun. 2015, 6, 6555 10.1038/ncomms7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichhart T.; Hengstschläger M.; Linke M. Regulation of innate immune cell function by mTOR. Nat. Rev. Immunol. 2015, 15, 599–614. 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger R.; Rieckmann J. C.; Wolf T.; Basso C.; Feng Y.; Fuhrer T.; Kogadeeva M.; Picotti P.; Meissner F.; Mann M.; Zamboni N.; Sallusto F.; Lanzavecchia A. L-Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity. Cell 2016, 167, 829–842.e13. 10.1016/j.cell.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S. M.; McKell M. C.; Schmidt S. M.; Hossfeld A. P.; Chaturvedi V.; Kinder J. M.; McAlees J. W.; Lewkowich I. P.; Way S. S.; Turner J.; Qualls J. E. L-Citrulline Metabolism in Mice Augments CD4+ T Cell Proliferation and Cytokine Production In Vitro, and Accumulation in the Mycobacteria-infected lung. Front. Immunol. 2017, 8, 1561 10.3389/fimmu.2017.01561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy J. P.; Routy B.; Graziani G. M.; Mehraj V. The Kynurenine Pathway Is a Double-Edged Sword in Immune-Privileged Sites and in Cancer: Implications for Immunotherapy. Int. J. Tryptophan Res. 2016, 9, 67–77. 10.4137/IJTR.S38355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam U. S.; Foreman T. W.; Bucsan A. N.; Veatch A. V.; Alvarez X.; Adekambi T.; Golden N. A.; Gentry K. M.; Doyle-Meyers L. A.; Russell-Lodrigue K. E.; Didier P. J.; Blanchard J. L.; Kousoulas K. G.; Lackner A. A.; Kalman D.; Rengarajan J.; Khader S. A.; Kaushal D.; Mehra S. In vivo inhibition of tryptophan catabolism reorganizes the tuberculoma and augments immune-mediated control of Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 2018, 115, E62–E71. 10.1073/pnas.1711373114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.