The reaction of 2-(4′,4′-dimethyl-2′-oxazolinyl)aniline (H2-L1) with Na[N(SiMe3)2] afforded colourless crystals of tetrameric Na4(H-L1)4 (2). Reaction of either Na4(H-L1)4 (2) with YbCl3 or reaction of H2-L1 with Yb[N(SiMe3)2]3 afforded yellow crystals of the highly distorted octahedral complex Yb(H-L1)3 (3).
Keywords: crystal structure, synthesis, ytterbium, oxazoline, amide
Abstract
Reaction of 2-(4,4-dimethyl-2-oxazolin-2-yl)aniline (H2-L1) with one equivalent of Na[N(SiMe3)2] in toluene afforded pale-yellow crystals of tetrameric poly[bis[μ3-2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido][μ2-2-(4,4-dimethyl-2-oxazolin-2-yl)aniline]tetrasodium(I)], [Na4(C11H13N2O)4]n or [Na4(H-L1)4]n (2), in excellent yield. Subsequent reaction of [Na4(H-L1)4]n (2) with 1.33 equivalents of anhydrous YbCl3 in a 50:50 mixture of toluene–THF afforded yellow crystals of tris[2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido]ytterbium(III), [Yb(C11H13N2O)3] or Yb(H-L1)3 (3) in moderate yield. Direct reaction of three equivalents of 2-(4′,4′-dimethyl-2′-oxazolinyl)aniline (H2-L1) with Yb[N(SiMe3)2]3 in toluene resulted in elimination of hexamethyldisilazane, HN(SiMe3)2, and produced Yb(H-L1)3 (3) in excellent yield. The structure of 2 consists of tetrameric Na4(H-L1)4 subunits in which each Na+ cation is bound to two H-L1 bridging bidentate ligands and these subunits are connected into a polymeric chain by two of the four oxazoline O atoms bridging to Na+ cations in the adjacent tetramer. This results in two 4-coordinate and two 5-coordinate Na+ cations within each tetrameric unit. The structure of 3 consists of a distorted octahedron where the bite angle of ligand L1 ranges between 74.72 (11) and 77.79 (11) degrees. The oxazoline (and anilide) N atoms occupy meridional sites such that for one ligand an anilide nitrogen is trans to an oxazoline nitrogen while for the other two oxazoline N atoms are trans to each other. This results in a significantly longer Yb—N(oxazoline) distance [2.468 (3) Å] for the bond trans to the anilide compared to those for the oxazoline N atoms trans to one another [2.376 (3), 2.390 (3) Å].
Chemical context
The parent ligand 2-(4′,4′-dimethyl-2′-oxazolinyl)aniline (H2-L1), easily prepared in high yield using established procedures (Gossage, 2009 ▸), has been used as a precursor to biologically active quinilones [see, for example, Hong et al. (2018 ▸)] and to make catalytically active transition-metal complexes (Saiyed et al., 2011 ▸; Resanović et al., 2011 ▸; Decken et al., 2005 ▸). There are many examples of transition metals containing N-substituted variants of L1, either as neutral ligands (HR-L1) or as deprotonated anilido anions (R-L1
−). However, the only example of a transition-metal structure containing the deprotonated and unsubstituted anilido parent ligand (H-L1
−) is an Ru carbonyl hydride dimer (Cabeza et al., 2006 ▸). No lanthanide complexes of this ligand have been reported, although there are several related lanthanide and yttrium complexes bearing oxazoline groups ortho to an anilido-like anionic centre. These complexes fall into two main ligand frameworks: diphenylamido ligands bearing ortho-oxazoline functionality (Fig. 1 ▸
a: Bennett et al., 2013 ▸, 2014 ▸; Liu et al., 2013 ▸) or carbazolide-bis(oxazolines) (Fig. 1 ▸
b: Zou et al., 2011 ▸, 2013 ▸). The crystal structure of the tetrameric sodium salt of this ligand, [Na4(H-L1)4]n (2), and its 6-coordinate, monomeric ytterbium complex, Yb(H-L1)3 (3) are reported in this communication. The ytterbium complex 3 can be prepared by either the salt metathesis reaction between 2 and YbCl3 or by the acid–base (protonolysis) reaction of Yb[N(SiMe3)2]3 with three equivalents of H2-L1. The yields and purity of 3 are better for the protonolysis reaction (Fig. 2 ▸).
Figure 1.
Related ligand types: (a) oxazoline-diphenylamides and (b) carbazolide-bis(oxazolines).
Figure 2.
Synthetic routes to [Na4(H-L1)4]n (2) and Yb(H-L1)3 (3) used in this work.
Structural commentary
The structure of 2 consists of tetrameric Na4(H-L1)4 subunits in which each Na+ cation is bound to two H-L1 bridging bidentate ligands (Fig. 3 ▸ a and 3b). The tetrameric subunits are connected into polymeric chains by two of the four oxazoline oxygens (O9 and O37) bridging to Na+ cations (Na1 and Na3, respectively) in the adjacent tetramer (Fig. 4 ▸). This results in two 4-coordinate (Na2, Na4) and two 5-coordinate (Na1, Na3) Na+ cations within each tetrameric unit. There are only four examples of an oxazoline ligand bonding through the oxygen atom and in all cases this involves an electropositive metal ion [Li: Pawilkowski et al. (2009 ▸) and Mukherjee et al. (2010 ▸); Na: Zou et al. (2013 ▸); Nd: Kanbur et al. (2018 ▸)]. Significant bond lengths and angles for 2 are collected in Table 1 ▸. The bridging Na—O(oxazoline) distances of 2.4003 (15) and 2.4099 (14) Å compare well the Na—O(oxazoline) distance of 2.432 (2) Å in NaCzx [Czx = 1,8-bis(4′,4′-dimethyloxazolin-2′-yl)-3,6-di-tert-butylcarbazole anion; Zou et al., 2013 ▸]. The Na—N distances from the anilide N center to the 4- and 5-coordinate Na ions are essentially the same [2.3411 (18)–2.3701 (18) versus 2.3360 (18)–2.3611 (17) Å, respectively]. In sharp contrast, the distance between the oxazoline N and the Na ions is much shorter for the 4-coordinate Na centres [non-O-bridging oxazoline: 2.3626 (17), 2.3396 (17); O-bridging oxazoline: 2.4407 (17), 2.4035 (17) Å] than for the 5-coordinate Na [non-O-bridging oxazoline: 2.5515 (16), 2.7348 (17); O-bridging oxazoline: 2.9532 (18), 3.0327 (18) Å]. It is clear that the oxazoline nitrogen is much more weakly coordinating to the 5-coordinate Na cation. In fact, for the O-bridging oxazoline, this distance is so long that it is debatable whether there is a significant bonding interaction. However, this result is consistent with localization of electron density on the bridging oxygen at the expense of the nitrogen atom in the same oxazoline ring.
Figure 3.

Molecular structure of polymeric [Na4(H-L1)4 n (2): (a) asymmetric unit, top view and (b) asymmetric unit, side view. Probability ellipsoids are at 50% and hydrogen atoms are omitted for clarity (except the aniline NH).
Figure 4.

Molecular structure of polymeric [Na4(H-L1)4]n (2) showing the polymeric chain structure for three asymmetric units (top view). Probability ellipsoids are at 50% and hydrogen atoms are omitted for clarity (except the aniline NH).
Table 1. Selected geometric parameters (Å, °) for Na4(H-L1)4 .
| Na1—O9i | 2.4003 (15) | Na3—O37ii | 2.4099 (14) |
| Na1—N1 | 2.3465 (18) | Na3—N15 | 2.3360 (18) |
| Na1—N12 | 2.9532 (18) | Na3—N26 | 2.5515 (16) |
| Na1—N43 | 2.3432 (17) | Na3—N29 | 2.3611 (17) |
| Na1—N54 | 2.7348 (17) | Na3—N40 | 3.0327 (18) |
| Na2—N1 | 2.3619 (18) | Na4—N29 | 2.3701 (18) |
| Na2—N12 | 2.3626 (17) | Na4—N40 | 2.3396 (17) |
| Na2—N15 | 2.3411 (18) | Na4—N43 | 2.3519 (18) |
| Na2—N26 | 2.4407 (17) | Na4—N54 | 2.4035 (17) |
| O9i—Na1—N12 | 107.61 (5) | O37ii—Na3—N26 | 107.75 (5) |
| O9i—Na1—N54 | 103.04 (5) | O37ii—Na3—N40 | 104.03 (5) |
| N1—Na1—O9i | 105.11 (6) | N15—Na3—O37ii | 141.85 (6) |
| N1—Na1—N12 | 65.92 (5) | N15—Na3—N26 | 72.56 (5) |
| N1—Na1—N54 | 92.92 (6) | N15—Na3—N29 | 108.71 (6) |
| N43—Na1—O9i | 142.94 (6) | N15—Na3—N40 | 85.90 (5) |
| N43—Na1—N1 | 111.31 (6) | N26—Na3—N40 | 147.75 (5) |
| N43—Na1—N12 | 93.51 (6) | N29—Na3—O37ii | 108.68 (6) |
| N43—Na1—N54 | 69.08 (5) | N29—Na3—N26 | 100.25 (6) |
| N54—Na1—N12 | 146.27 (5) | N29—Na3—N40 | 63.74 (5) |
| N1—Na2—N12 | 76.60 (6) | N29—Na4—N54 | 119.12 (6) |
| N1—Na2—N26 | 125.43 (6) | N40—Na4—N29 | 76.15 (6) |
| N12—Na2—N26 | 135.48 (6) | N40—Na4—N43 | 113.77 (6) |
| N15—Na2—N1 | 138.81 (6) | N40—Na4—N54 | 121.58 (6) |
| N15—Na2—N12 | 116.21 (7) | N43—Na4—N29 | 156.58 (6) |
| N15—Na2—N26 | 74.56 (6) | N43—Na4—N54 | 75.06 (6) |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
The structure of Yb(H-L1)3 (3) is a distorted octahedron where all three H-L1 ligands are distinct (Fig. 5 ▸). Significant geometric parameters for this compound are given in Table 2 ▸. The bite angles of the H-L1 − ligand range from 74.72 (11)–77.79 (11)°, which sits between that of the diphenylamido-oxazoline [see Fig. 1 ▸ a: range 69.97 (11)–80.1 (5)°, median 74.3°; Bennett et al. (2013 ▸, 2014 ▸); Liu et al. (2013 ▸)] and carbazolide-bis(oxazoline) [see Fig. 1 ▸ b: range 77.99 (4)–81.18 (10)°, median 80.3°; Zou et al. (2011 ▸, 2013 ▸)] ligands. The oxazoline (and anilide) nitrogens occupy meridional sites such that for one ligand an anilide nitrogen is trans to an oxazoline nitrogen while the other two oxazoline nitrogens are trans to each other. This results in a significantly longer Yb—N(oxazoline) distance [2.468 (3) Å] for the bond trans to the anilide compared to those for the oxazolines trans to one another [2.376 (3), 2.390 (3) Å]. The Yb—N(anilide) distances [2.234 (3)–2.260 (3) Å] show less variation although the Yb—N(anilide) distance trans to the oxazoline N atom is slightly shorter than those trans to each other. Overall, this is consistent with a stronger trans influence for the anionic anilide nitrogens as might be expected. The torsion angles representing twisting from coplanarity of the oxazoline and benzene units all fall between −9.3 (6) and +8.4 (6)° so the distortions from planarity of the H-L1 ligands in 3 are relatively small.
Figure 5.
Molecular structure of Yb(H-L1)3 (3). Probability ellipsoids are at 50% and hydrogen atoms are omitted for clarity (except the aniline NH).
Table 2. Selected geometric parameters (Å, °) for Yb(H-L1)3 .
| N1—Yb1 | 2.252 (3) | N26—Yb1 | 2.376 (3) |
| N12—Yb1 | 2.468 (3) | N29—Yb1 | 2.234 (3) |
| N15—Yb1 | 2.260 (3) | N40—Yb1 | 2.390 (3) |
| N1—Yb1—N12 | 74.72 (11) | N26—Yb1—N40 | 153.87 (10) |
| N1—Yb1—N15 | 167.59 (12) | N29—Yb1—N1 | 89.28 (12) |
| N1—Yb1—N26 | 109.02 (11) | N29—Yb1—N12 | 159.71 (11) |
| N1—Yb1—N40 | 86.61 (11) | N29—Yb1—N15 | 102.04 (12) |
| N15—Yb1—N12 | 95.23 (11) | N29—Yb1—N26 | 82.94 (11) |
| N15—Yb1—N26 | 77.79 (11) | N29—Yb1—N40 | 76.28 (11) |
| N15—Yb1—N40 | 91.10 (11) | N40—Yb1—N12 | 114.28 (10) |
| N26—Yb1—N12 | 90.47 (10) | ||
| C2—C7—C8—N12 | 8.6 (7) | C30—C35—C36—N40 | −9.3 (6) |
| C16—C21—C22—N26 | 8.4 (6) |
Supramolecular features
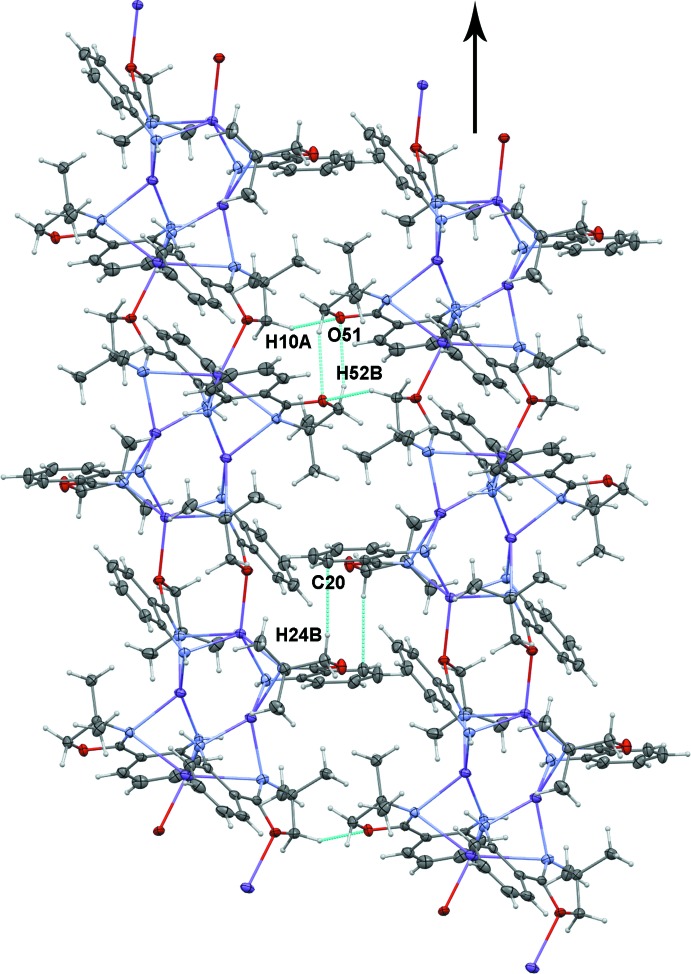
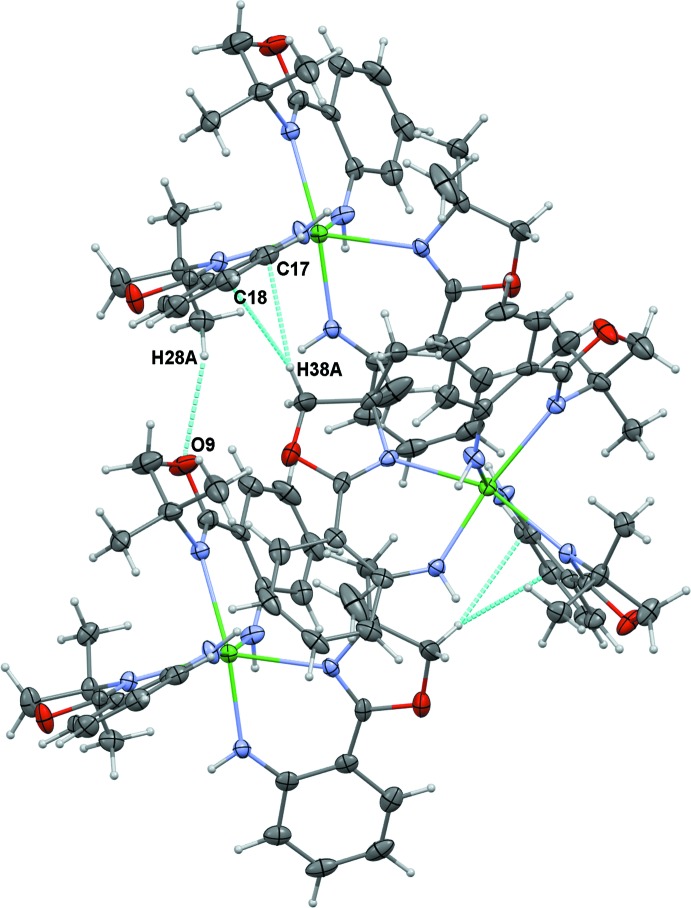
The structure of 2 consists of polymeric chains of Na4(H-L1)4 subunits connected through bridging oxazoline oxygen atoms (Fig. 4 ▸). There are two different types of close contacts between adjacent polymer chains through the non-O-bridging oxazoline rings (Fig. 6 ▸, Table 3 ▸). One type involves the close approach of one H atom of two different oxazoline CH2 groups to a non-bridging O atom of an adjacent chain (H10A⋯O51iii, 2.64 Å; H52B⋯O51iv, 2.58 Å; see Table 3 ▸ for symmetry operators). A C—H⋯π type contact is also observed between one H of the other non-O-bridged oxazoline ring and a carbon of an aromatic ring on a parallel chain (H24B⋯C20v, 2.82 Å; see Table 3 ▸ for symmetry operator). Similarly, the structure of 3 shows two types of close contacts between molecules. One type is between a methyl hydrogen on an oxazoline ring and an oxazoline O atom of an adjacent molecule (H28A⋯O9ii, 2.55 Å; see Table 4 ▸ for symmetry operator). Structure 3 also shows a close C—H⋯π contacts between the H atom of a CH2 group in one oxazoline ring with the aromatic ring of an adjacent molecule (H38A⋯C17i, 3.01 Å; H38A⋯C18i, 2.58 Å; see Table 4 ▸ for symmetry operator), resulting in a zigzag chain of Yb(H-L1)3 units in the solid state (Fig. 7 ▸).
Figure 6.
Close contacts between polymeric chains of Na4(H-L1)4 (2): interchain contacts consisting of C—H⋯O and C—H⋯π interactions are shown in teal; the chain direction is indicated by the arrow.
Table 3. Significant intermolecular interactions (Å) in (2) and (3).
| Compound | D—H⋯A | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| Na4(H-L1)4 (2) | C10—H10A⋯O51iii | 2.64 | 3.413 (2) | 135.3 |
| C52—H52B⋯O51iv | 2.58 | 3.439 (2) | 145.2 | |
| C24—H24B⋯C20v | 2.82 | 3.720 (3) | 151.4 | |
| Yb(H-L1)3 (3) | C28—H28A⋯O9ii | 2.55 | 3.382 (5) | 142.8 |
| C38—H38A⋯C17i | 3.01 | 3.501 (6) | 149.0 | |
| C38—H38A⋯C18i | 2.58 | 3.548 (6) | 165.8 |
Symmetry codes: (i) −x +  , y +
, y +  , −z +
, −z +  ; (ii) x, y − 1, z; (iii) x, y + 1, z; (iv) −x + 2, −y, −z + 1; (v) −x + 2, −y + 1, −z.
; (ii) x, y − 1, z; (iii) x, y + 1, z; (iv) −x + 2, −y, −z + 1; (v) −x + 2, −y + 1, −z.
Table 4. Experimental details.
| Na4(H-L1)4 | Yb(H-L1)3 | |
|---|---|---|
| Crystal data | ||
| Chemical formula | [Na4(C11H13N2O)4] | [Yb(C11H13N2O)3] |
| M r | 848.89 | 740.74 |
| Crystal system, space group | Triclinic, P

|
Monoclinic, P21/n |
| Temperature (K) | 87 | 86 |
| a, b, c (Å) | 10.9545 (5), 11.8785 (5), 18.8415 (8) | 10.9428 (5), 9.8253 (5), 28.6089 (14) |
| α, β, γ (°) | 105.266 (1), 97.446 (1), 106.120 (1) | 90, 94.722 (1), 90 |
| V (Å3) | 2217.20 (17) | 3065.5 (3) |
| Z | 2 | 4 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.12 | 3.10 |
| Crystal size (mm) | 0.26 × 0.18 × 0.15 | 0.22 × 0.18 × 0.05 |
| Data collection | ||
| Diffractometer | SMART APEX CCD area detector | SMART APEX CCD area detector |
| Absorption correction | Multi-scan (SADABS; Bruker, 2016 ▸) | Multi-scan (SADABS; Bruker, 2016 ▸) |
| T min, T max | 0.776, 0.983 | 0.219, 0.262 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 33551, 12487, 8858 | 39651, 7061, 5882 |
| R int | 0.054 | 0.059 |
| (sin θ/λ)max (Å−1) | 0.704 | 0.651 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.066, 0.158, 1.05 | 0.032, 0.073, 1.07 |
| No. of reflections | 12487 | 7061 |
| No. of parameters | 565 | 394 |
| No. of restraints | 4 | 0 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.49, −0.31 | 0.83, −1.19 |
Figure 7.
Close contacts between molecules of Yb(H-L1)3 (3) in the solid state: C—H⋯O and C—H⋯π interactions between adjacent molecules are shown in teal.
Database survey
There are 68 structure in the CSD (version 5.39, update of November, 2018; Groom et al., 2016 ▸) containing a substituted anilido-oxazoline ligand (R-L1 −) coordinated to a transition or main group metal. [COZFIH (Coeffard et al., 2009 ▸); DEJHIK (Cabeza et al., 2006 ▸); EDEBOG (Niwa & Nakada, 2012 ▸); EFICON (Bian et al., 2014 ▸); FONYAI (Mikami et al., 1999 ▸); GIWYES (Chen et al., 2014 ▸); GUTTOF (Inagaki et al., 2010 ▸); ISEWAG (Abbina et al., 2016 ▸); LUNGOS (Bauer et al., 2015a ▸); LUNHAF (Bauer et al., 2015a ▸); MALVUS, MALWAZ and MALWED (Kieltsch et al., 2010 ▸); MICTID, MICTOJ, MICTUP, MICVAX and MICVEB (Lu et al., 2013 ▸); MUQNAN and MUQNER (Wan et al., 2002 ▸); NANFEP, NANFIT, NANFOZ, NANFUF, NANGAM, NANGEQ, NANGIU and NANGOA (Peng & Chen, 2011 ▸); OCIHOX, OCIHUD and OCIJAL (Cabaleiro et al., 2001 ▸); PUDKUV, PUDLAC, PUDLEG and PUDLIK (Chen et al., 2009a ▸); QIFFES and QIFFIW (Abbina & Du, 2012 ▸); RAKTAA (McKeon et al., 2011 ▸); RAMFIW, RAMFOC, RAMFUI, RAMGAP and RAMGET (Chen & Chen, 2011 ▸); RAMKEY (Huang et al., 2017 ▸); ROGWAM (Nakada & Inoue, 2007 ▸); SELVIQ (Nixon & Ward, 2012 ▸); SUYQOS, SUYQUY, SUYRAF and SUYREJ (Castro et al., 2001 ▸); TIMLIL and TIMLOR (Chen et al., 2007 ▸); VOZZOB, VOZZUH and VUBBAX (Bauer et al., 2015b ▸); VUQZAK (O’Reilly et al., 2015 ▸); WUGQOF, WUGQUL, WUGRAS and WUGREW (Chen et al., 2009b ▸); XIGYEU (Wu et al., 2018 ▸); XOQVEG and XOQVIK (He et al., 2014 ▸); XOYVOW, XOYVUC, XOYWAJ, XOYWEN and XOYWIR (Castro et al., 2002 ▸)]. In contrast, there is only one structure of an unsubstituted anilido-oxazoline ligand (H-L1 −) coordinated to a transition metal (Cabeza et al., 2006 ▸) and there are no structures of this type with a lanthanide metal. There are 10 lanthanide complexes that have been structurally characterized with the related ligands shown in Fig. 1 ▸ a and 1b as discussed in the Chemical context.
Synthesis and crystallization
General. All solvents were purchased from Sigma–Aldrich Chemicals and dried by distillation from sodium under nitrogen. 2-(4′,4′-Dimethyl-2′-oxazolinyl)aniline was prepared according to Gossage (2009 ▸) and purified by recrystallization from hot toluene. Yb[N(SiMe)3)2]3 was prepared by analogy to the procedure of Bradley et al. (1973 ▸) using NaN(SiMe3)2 and YbCl3 and was recrystallized from a hot mixture of hexane and toluene. NMR spectra were recorded on a Bruker AV III 300 MHz Spectrometer in sealable Teflon-valved tubes and were referenced to residual solvent resonances. The line widths at half maximum (ν1/2 in Hz) were measured for all paramagnetic resonances in 3 and are reported below. Elemental analyses were performed by Canadian Microanalytical Ltd.
Synthesis of [Na4(L1)4]n. One equivalent of Na[N(SiMe3)2] (0.183 g, 1.00 mmol) was dissolved in toluene (10 mL) and to this was added 1 equivalent of 2-(4′,4′-dimethyl-2′-oxazolinyl)aniline (H2-L1, 0.190 g, 1.00 mmol) in 20 mL toluene under vigorous stirring. The colourless reaction mixture was stirred overnight, filtered through Celite on a sintered glass frit and the solvent removed under reduced pressure to leave a tacky white solid. Recrystallization of the product from a hot mixture of toluene and hexane afforded clear pale-yellow crystals of [Na4(L1)4]n (2). Yield: 0.178 g (84%). 1H NMR (THF-d 8, 300 MHz, 296 K): δ 7.568 (1H, d, 3-arylH), 7.067 (1H, t, 5-arylH), 6.635 (1H, d, 6-arylH), 6.62 (1H, br s, NH, overlaps previous resonance), 6.459 (1H, t, 4-arylH), 3.954 (2H, s, OCH2), 1.315 (6H, s, C(CH3)2); 13C{1H} (THF-d 8, 75 MHz, 296 K): δ 163.12 (C=N), 150.79 (arylCNH), 132.31 (5-arylCH), 130.04 (3-arylCH), 115.90 (6-arylCH), 115.15 (4-arylCH), 109.27 (2-arylC—C=N), 77.82 (OCH2), 68.58 [NC(CH3)2], 28.99 [NC(CH3)2].
Synthesis of Yb(H-L1)3 (3) Method A: A solution of [Na4(L1)4]n (2) (0.250 g, 0.295 mmol) in THF (10 mL) was added to a suspension of YbCl3 (0.062 g, 0.22 mmol) in THF (5 mL) under vigorous stirring. The suspension was stirred overnight at room temperature, filtered through Celite on a sintered glass frit and the filtrate was evaporated to dryness under reduced pressure. The yellow solid was recrystallized from a mixture of toluene and hexane at 243 K overnight. Yield: 0.102 g (63%). Method B: A solution of 2-(4′,4′-dimethyl-2′-oxazolinyl)aniline (0.250 g, 1.31 mmol) in 25 mL toluene was prepared in the glovebox and added by Pasteur pipette to a vigorously stirred solution of Yb[N(SiMe)3)2]3 (0.287 g, 0.438 mmol) in 15 mL of toluene. The pale-yellow solution darkened to golden yellow on stirring overnight. The solution was filtered through Celite on a sintered glass frit and the filtrate was evaporated to dryness under reduced pressure. The orange–yellow solid was recrystallized from a mixture of toluene and hexane at 243 K yielding yellow crystals. Yield: 0.301 g (93%). 1H NMR (C6D6, 300 MHz, 296 K): δ 88.4 (6H, ν1/2 = 700 Hz), 49.4 (3H, overlaps next resonance), 47.9 (6H, ν1/2 = 350 Hz, overlaps previous resonance), 12.86 (2H, ν1/2 = 9 Hz), 11.70 (4H, ν1/2 = 12 Hz), 10.93 (4H, ν1/2 = 12 Hz), 10.00 (4H, ν1/2 = 25 Hz), 9.30 (4H, ν1/2 = 70 Hz), 1.26 (2H, t), 0.96 (2H, t), −2.77 (3H, ν1/2 = 100 Hz), −3.89 (2H, ν1/2 = 14 Hz), −5.38 (2H, ν1/2 = 20 Hz), −11.2 (6H, ν1/2 ∼150 Hz, overlaps next resonance), −11.4 (6H, ν1/2 ∼300 Hz, overlaps previous resonance), −16.0 (3H, ν1/2 = 140 Hz), −24.4 (3H, ν1/2 = 800 Hz), −77.2 (3H, ν1/2 = 600 Hz). Analysis calculated for C33H39N6O3Yb (%): C, 53.49; H, 5.31; N, 11.35. Found: C, 53.39; H, 5.22; N, 11.11.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. In Na4(H-L1)4 (2), the H atoms on N1, N15, N29 and N43 were located in a difference map and refined with distance restraints of 0.88 (1) Å. U iso(H) were freely refined. In Yb(H-L1)3 (3), the H atoms on N1, N15 were added geometrically and refined with distance restraints of 0.88 (1) Å, with U iso(H) = 1.2U eq(N). H29 was located in the difference map for geometrical considerations and refined with coordinates riding on N29 with U iso(H) = 1.2U eq(N). All the H atoms bonded to carbon were refined in geometrically calculated positions, with C—H= 0.95 (methine), 0.99 (methylene), and 0.98 Å (methyl), and with U iso(H) = 1.2U eq(C) (methine and methylene) or 1.5U eq(C) (methyl).
Supplementary Material
Crystal structure: contains datablock(s) Na4H-L142, YbH-L133, global. DOI: 10.1107/S2056989020005034/zl2777sup1.cif
Structure factors: contains datablock(s) Na4H-L142. DOI: 10.1107/S2056989020005034/zl2777Na4H-L142sup2.hkl
Supporting information file. DOI: 10.1107/S2056989020005034/zl2777Na4H-L142sup5.mol
Structure factors: contains datablock(s) YbH-L133. DOI: 10.1107/S2056989020005034/zl2777YbH-L133sup3.hkl
Supporting information file. DOI: 10.1107/S2056989020005034/zl2777YbH-L133sup6.mol
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors wish to acknowledge the assistance of Mrs Chris Greenwood in obtaining the NMR spectra and Karen Button for preliminary investigations of this ligand with lanthanide metals.
supplementary crystallographic information
Poly[bis[µ3-2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido][µ2-2-(4,4-dimethyl-2-oxazolin-2-yl)aniline]tetrasodium(I)] (Na4H-L142) . Crystal data
| [Na4(C11H13N2O)4] | Z = 2 |
| Mr = 848.89 | F(000) = 896 |
| Triclinic, P1 | Dx = 1.272 Mg m−3 |
| a = 10.9545 (5) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 11.8785 (5) Å | Cell parameters from 9632 reflections |
| c = 18.8415 (8) Å | θ = 2.2–29.9° |
| α = 105.266 (1)° | µ = 0.12 mm−1 |
| β = 97.446 (1)° | T = 87 K |
| γ = 106.120 (1)° | Needle, pale yellow |
| V = 2217.20 (17) Å3 | 0.26 × 0.18 × 0.15 mm |
Poly[bis[µ3-2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido][µ2-2-(4,4-dimethyl-2-oxazolin-2-yl)aniline]tetrasodium(I)] (Na4H-L142) . Data collection
| SMART APEX CCD area detector diffractometer | 12487 independent reflections |
| Radiation source: sealed X-ray tube | 8858 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.054 |
| Detector resolution: 8.3 pixels mm-1 | θmax = 30.1°, θmin = 1.9° |
| φ and ω scans | h = −15→14 |
| Absorption correction: multi-scan (SADABS; Bruker, 2016) | k = −16→16 |
| Tmin = 0.776, Tmax = 0.983 | l = −26→26 |
| 33551 measured reflections |
Poly[bis[µ3-2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido][µ2-2-(4,4-dimethyl-2-oxazolin-2-yl)aniline]tetrasodium(I)] (Na4H-L142) . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.066 | Hydrogen site location: mixed |
| wR(F2) = 0.158 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0734P)2 + 0.1127P] where P = (Fo2 + 2Fc2)/3 |
| 12487 reflections | (Δ/σ)max < 0.001 |
| 565 parameters | Δρmax = 0.49 e Å−3 |
| 4 restraints | Δρmin = −0.31 e Å−3 |
Poly[bis[µ3-2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido][µ2-2-(4,4-dimethyl-2-oxazolin-2-yl)aniline]tetrasodium(I)] (Na4H-L142) . Special details
| Experimental. The data collection nominally covered a full sphere of reciprocal space by a combination of 5 sets of ω scans each set at different φ and/or 2θ angles and each scan (10 s exposure) covering -0.300° degrees in ω. The crystal to detector distance was 5.0 cm. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Donor N-H hydrogen atoms located on the difference map and refined with restraints (DFIX). |
Poly[bis[µ3-2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido][µ2-2-(4,4-dimethyl-2-oxazolin-2-yl)aniline]tetrasodium(I)] (Na4H-L142) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Na1 | 0.95553 (8) | 0.27949 (7) | 0.39918 (4) | 0.02273 (18) | |
| Na2 | 1.00695 (7) | 0.40238 (7) | 0.25656 (4) | 0.01977 (17) | |
| Na3 | 1.01274 (8) | 0.22441 (7) | 0.09593 (4) | 0.01954 (17) | |
| Na4 | 0.96731 (7) | 0.11951 (7) | 0.24031 (4) | 0.01974 (17) | |
| O9 | 0.97838 (14) | 0.66218 (12) | 0.46616 (7) | 0.0223 (3) | |
| O23 | 1.05206 (13) | 0.61390 (12) | 0.11145 (8) | 0.0240 (3) | |
| O37 | 0.92170 (12) | −0.17535 (11) | 0.02263 (7) | 0.0172 (3) | |
| O51 | 0.93031 (13) | −0.09390 (12) | 0.40245 (7) | 0.0190 (3) | |
| N1 | 1.13690 (16) | 0.38393 (14) | 0.36039 (9) | 0.0194 (3) | |
| H1 | 1.1910 (17) | 0.3417 (18) | 0.3515 (12) | 0.023 (6)* | |
| N12 | 0.93307 (16) | 0.49954 (15) | 0.36081 (9) | 0.0210 (3) | |
| N15 | 0.84608 (16) | 0.27468 (14) | 0.14907 (9) | 0.0188 (3) | |
| H15 | 0.7835 (15) | 0.2151 (14) | 0.1541 (11) | 0.016 (5)* | |
| N26 | 1.08503 (15) | 0.45903 (13) | 0.15111 (8) | 0.0154 (3) | |
| N29 | 1.14745 (16) | 0.17884 (14) | 0.18512 (9) | 0.0187 (3) | |
| H29 | 1.2125 (16) | 0.2441 (15) | 0.2135 (11) | 0.032 (6)* | |
| N40 | 0.90467 (15) | −0.02419 (14) | 0.11936 (9) | 0.0188 (3) | |
| N43 | 0.79137 (16) | 0.14194 (14) | 0.29563 (9) | 0.0177 (3) | |
| H43 | 0.7403 (18) | 0.1721 (19) | 0.2724 (11) | 0.025 (6)* | |
| N54 | 0.99785 (15) | 0.05812 (14) | 0.35133 (8) | 0.0157 (3) | |
| C2 | 1.21029 (19) | 0.49537 (17) | 0.40943 (10) | 0.0185 (4) | |
| C3 | 1.3480 (2) | 0.52674 (19) | 0.43478 (11) | 0.0240 (4) | |
| H3 | 1.387659 | 0.465981 | 0.417427 | 0.029* | |
| C4 | 1.4255 (2) | 0.6402 (2) | 0.48287 (12) | 0.0294 (5) | |
| H4 | 1.516605 | 0.656142 | 0.497231 | 0.035* | |
| C5 | 1.3727 (2) | 0.7324 (2) | 0.51099 (11) | 0.0307 (5) | |
| H5 | 1.426790 | 0.811193 | 0.543908 | 0.037* | |
| C6 | 1.2407 (2) | 0.70662 (18) | 0.48998 (11) | 0.0244 (4) | |
| H6 | 1.204232 | 0.768938 | 0.509515 | 0.029* | |
| C7 | 1.1568 (2) | 0.59127 (17) | 0.44049 (10) | 0.0192 (4) | |
| C8 | 1.01968 (19) | 0.57690 (16) | 0.41890 (10) | 0.0179 (4) | |
| C10 | 0.8393 (2) | 0.62951 (19) | 0.43632 (11) | 0.0235 (4) | |
| H10A | 0.816298 | 0.703301 | 0.433271 | 0.028* | |
| H10B | 0.787910 | 0.591249 | 0.468432 | 0.028* | |
| C11 | 0.8141 (2) | 0.53719 (18) | 0.35752 (11) | 0.0210 (4) | |
| C13 | 0.6923 (2) | 0.4263 (2) | 0.34069 (13) | 0.0300 (5) | |
| H13A | 0.682024 | 0.369426 | 0.290390 | 0.045* | |
| H13B | 0.616121 | 0.454057 | 0.342445 | 0.045* | |
| H13C | 0.700305 | 0.384032 | 0.378366 | 0.045* | |
| C14 | 0.8081 (2) | 0.59894 (19) | 0.29623 (11) | 0.0285 (5) | |
| H14A | 0.886600 | 0.670811 | 0.308121 | 0.043* | |
| H14B | 0.731041 | 0.625564 | 0.293703 | 0.043* | |
| H14C | 0.802784 | 0.540086 | 0.247512 | 0.043* | |
| C16 | 0.78884 (18) | 0.34087 (16) | 0.11669 (10) | 0.0159 (4) | |
| C17 | 0.64991 (19) | 0.30802 (17) | 0.09628 (11) | 0.0214 (4) | |
| H17 | 0.598324 | 0.236507 | 0.105205 | 0.026* | |
| C18 | 0.5880 (2) | 0.37338 (19) | 0.06493 (12) | 0.0256 (4) | |
| H18 | 0.495521 | 0.347610 | 0.053331 | 0.031* | |
| C19 | 0.6594 (2) | 0.4787 (2) | 0.04951 (13) | 0.0289 (5) | |
| H19 | 0.616576 | 0.524673 | 0.027426 | 0.035* | |
| C20 | 0.7927 (2) | 0.51362 (19) | 0.06716 (12) | 0.0251 (4) | |
| H20 | 0.841448 | 0.584382 | 0.056354 | 0.030* | |
| C21 | 0.86026 (18) | 0.44926 (17) | 0.10054 (10) | 0.0169 (4) | |
| C22 | 1.00186 (19) | 0.50168 (16) | 0.12171 (10) | 0.0165 (4) | |
| C24 | 1.19172 (19) | 0.64358 (18) | 0.12865 (12) | 0.0226 (4) | |
| H24A | 1.234644 | 0.730969 | 0.159843 | 0.027* | |
| H24B | 1.225242 | 0.628660 | 0.081893 | 0.027* | |
| C25 | 1.21513 (18) | 0.55680 (17) | 0.17229 (11) | 0.0191 (4) | |
| C27 | 1.2462 (2) | 0.6203 (2) | 0.25726 (12) | 0.0300 (5) | |
| H27A | 1.176504 | 0.652862 | 0.270847 | 0.045* | |
| H27B | 1.328909 | 0.688172 | 0.271962 | 0.045* | |
| H27C | 1.253007 | 0.560475 | 0.283555 | 0.045* | |
| C28 | 1.3196 (2) | 0.5025 (2) | 0.14974 (13) | 0.0284 (5) | |
| H28A | 1.325909 | 0.442906 | 0.176312 | 0.043* | |
| H28B | 1.403489 | 0.568677 | 0.163127 | 0.043* | |
| H28C | 1.297027 | 0.460680 | 0.095216 | 0.043* | |
| C30 | 1.20008 (18) | 0.09568 (17) | 0.15045 (10) | 0.0157 (4) | |
| C31 | 1.33777 (19) | 0.12026 (18) | 0.16195 (11) | 0.0215 (4) | |
| H31 | 1.393351 | 0.198458 | 0.194834 | 0.026* | |
| C32 | 1.3932 (2) | 0.03672 (19) | 0.12792 (12) | 0.0253 (4) | |
| H32 | 1.485371 | 0.058376 | 0.137784 | 0.030* | |
| C33 | 1.3172 (2) | −0.08002 (19) | 0.07886 (12) | 0.0254 (4) | |
| H33 | 1.356161 | −0.138164 | 0.055791 | 0.031* | |
| C34 | 1.18429 (19) | −0.10787 (18) | 0.06512 (11) | 0.0200 (4) | |
| H34 | 1.131534 | −0.186644 | 0.031580 | 0.024* | |
| C35 | 1.12306 (18) | −0.02447 (16) | 0.09871 (10) | 0.0153 (4) | |
| C36 | 0.98178 (18) | −0.06778 (16) | 0.08327 (10) | 0.0148 (4) | |
| C38 | 0.78300 (18) | −0.19763 (17) | 0.01536 (10) | 0.0176 (4) | |
| H38A | 0.736096 | −0.285692 | 0.007473 | 0.021* | |
| H38B | 0.747054 | −0.173973 | −0.027352 | 0.021* | |
| C39 | 0.77194 (18) | −0.11577 (17) | 0.09051 (11) | 0.0192 (4) | |
| C41 | 0.7463 (2) | −0.1871 (2) | 0.14685 (12) | 0.0301 (5) | |
| H41A | 0.810457 | −0.229695 | 0.150809 | 0.045* | |
| H41B | 0.658504 | −0.247779 | 0.129470 | 0.045* | |
| H41C | 0.753363 | −0.129417 | 0.196360 | 0.045* | |
| C42 | 0.6701 (2) | −0.0533 (2) | 0.08068 (13) | 0.0298 (5) | |
| H42A | 0.667093 | −0.001814 | 0.130065 | 0.045* | |
| H42B | 0.584663 | −0.116216 | 0.057219 | 0.045* | |
| H42C | 0.692872 | −0.001556 | 0.048415 | 0.045* | |
| C44 | 0.71576 (18) | 0.05709 (16) | 0.32088 (10) | 0.0165 (4) | |
| C45 | 0.57763 (19) | 0.03168 (18) | 0.31010 (11) | 0.0233 (4) | |
| H45 | 0.538590 | 0.074141 | 0.282748 | 0.028* | |
| C46 | 0.4998 (2) | −0.05047 (19) | 0.33726 (13) | 0.0285 (5) | |
| H46 | 0.408827 | −0.063596 | 0.328368 | 0.034* | |
| C47 | 0.5513 (2) | −0.11577 (19) | 0.37792 (12) | 0.0274 (5) | |
| H47 | 0.496904 | −0.171305 | 0.397908 | 0.033* | |
| C48 | 0.68180 (19) | −0.09766 (17) | 0.38817 (11) | 0.0210 (4) | |
| H48 | 0.717283 | −0.142442 | 0.415335 | 0.025* | |
| C49 | 0.76637 (18) | −0.01501 (16) | 0.36004 (10) | 0.0157 (4) | |
| C50 | 0.90186 (18) | −0.01006 (16) | 0.37005 (9) | 0.0149 (4) | |
| C52 | 1.07034 (19) | −0.06290 (19) | 0.41603 (11) | 0.0210 (4) | |
| H52A | 1.095885 | −0.138279 | 0.404131 | 0.025* | |
| H52B | 1.111240 | −0.012808 | 0.469199 | 0.025* | |
| C53 | 1.10936 (18) | 0.01097 (17) | 0.36303 (10) | 0.0172 (4) | |
| C55 | 1.1135 (2) | −0.07327 (18) | 0.28694 (11) | 0.0220 (4) | |
| H55A | 1.030049 | −0.140126 | 0.266136 | 0.033* | |
| H55B | 1.183594 | −0.108335 | 0.293956 | 0.033* | |
| H55C | 1.129591 | −0.025254 | 0.252043 | 0.033* | |
| C56 | 1.23701 (19) | 0.11658 (18) | 0.39697 (12) | 0.0262 (5) | |
| H56A | 1.260152 | 0.157776 | 0.359531 | 0.039* | |
| H56B | 1.305850 | 0.084140 | 0.411950 | 0.039* | |
| H56C | 1.227343 | 0.175941 | 0.441303 | 0.039* |
Poly[bis[µ3-2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido][µ2-2-(4,4-dimethyl-2-oxazolin-2-yl)aniline]tetrasodium(I)] (Na4H-L142) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Na1 | 0.0299 (4) | 0.0196 (4) | 0.0135 (4) | 0.0026 (3) | 0.0046 (3) | 0.0024 (3) |
| Na2 | 0.0246 (4) | 0.0207 (4) | 0.0128 (4) | 0.0069 (3) | 0.0036 (3) | 0.0040 (3) |
| Na3 | 0.0284 (4) | 0.0190 (4) | 0.0132 (4) | 0.0111 (3) | 0.0049 (3) | 0.0044 (3) |
| Na4 | 0.0248 (4) | 0.0210 (4) | 0.0134 (4) | 0.0075 (3) | 0.0063 (3) | 0.0044 (3) |
| O9 | 0.0364 (8) | 0.0185 (7) | 0.0140 (6) | 0.0137 (6) | 0.0065 (6) | 0.0028 (5) |
| O23 | 0.0216 (7) | 0.0206 (7) | 0.0333 (8) | 0.0055 (6) | 0.0045 (6) | 0.0163 (6) |
| O37 | 0.0198 (7) | 0.0163 (6) | 0.0137 (6) | 0.0057 (5) | 0.0036 (5) | 0.0019 (5) |
| O51 | 0.0212 (7) | 0.0197 (6) | 0.0201 (7) | 0.0083 (6) | 0.0038 (5) | 0.0111 (5) |
| N1 | 0.0244 (9) | 0.0151 (7) | 0.0181 (8) | 0.0072 (7) | 0.0044 (7) | 0.0037 (6) |
| N12 | 0.0238 (9) | 0.0190 (8) | 0.0191 (8) | 0.0081 (7) | 0.0062 (7) | 0.0025 (6) |
| N15 | 0.0197 (8) | 0.0137 (7) | 0.0219 (8) | 0.0022 (6) | 0.0036 (7) | 0.0076 (6) |
| N26 | 0.0159 (8) | 0.0129 (7) | 0.0157 (7) | 0.0030 (6) | 0.0023 (6) | 0.0039 (6) |
| N29 | 0.0207 (8) | 0.0160 (8) | 0.0162 (8) | 0.0050 (7) | 0.0011 (6) | 0.0022 (6) |
| N40 | 0.0178 (8) | 0.0199 (8) | 0.0160 (8) | 0.0048 (7) | 0.0033 (6) | 0.0025 (6) |
| N43 | 0.0234 (9) | 0.0169 (7) | 0.0152 (8) | 0.0108 (7) | 0.0015 (6) | 0.0057 (6) |
| N54 | 0.0153 (8) | 0.0162 (7) | 0.0158 (7) | 0.0058 (6) | 0.0023 (6) | 0.0052 (6) |
| C2 | 0.0253 (10) | 0.0165 (9) | 0.0153 (9) | 0.0040 (8) | 0.0053 (8) | 0.0100 (7) |
| C3 | 0.0244 (11) | 0.0277 (10) | 0.0219 (10) | 0.0082 (9) | 0.0048 (8) | 0.0114 (8) |
| C4 | 0.0234 (11) | 0.0357 (12) | 0.0221 (10) | −0.0005 (9) | −0.0001 (9) | 0.0108 (9) |
| C5 | 0.0346 (12) | 0.0255 (11) | 0.0178 (10) | −0.0072 (9) | 0.0033 (9) | 0.0029 (8) |
| C6 | 0.0349 (12) | 0.0184 (9) | 0.0143 (9) | 0.0016 (9) | 0.0069 (8) | 0.0029 (7) |
| C7 | 0.0281 (11) | 0.0155 (8) | 0.0115 (8) | 0.0024 (8) | 0.0045 (7) | 0.0051 (7) |
| C8 | 0.0311 (11) | 0.0117 (8) | 0.0135 (8) | 0.0076 (8) | 0.0084 (8) | 0.0057 (7) |
| C10 | 0.0350 (12) | 0.0239 (10) | 0.0184 (9) | 0.0161 (9) | 0.0109 (9) | 0.0085 (8) |
| C11 | 0.0279 (11) | 0.0195 (9) | 0.0200 (9) | 0.0124 (8) | 0.0084 (8) | 0.0071 (8) |
| C13 | 0.0275 (12) | 0.0257 (11) | 0.0405 (13) | 0.0120 (9) | 0.0083 (10) | 0.0125 (10) |
| C14 | 0.0452 (14) | 0.0234 (10) | 0.0173 (10) | 0.0123 (10) | 0.0067 (9) | 0.0057 (8) |
| C16 | 0.0197 (9) | 0.0127 (8) | 0.0124 (8) | 0.0050 (7) | 0.0026 (7) | 0.0000 (7) |
| C17 | 0.0194 (10) | 0.0155 (9) | 0.0254 (10) | 0.0028 (8) | 0.0047 (8) | 0.0032 (8) |
| C18 | 0.0170 (10) | 0.0257 (10) | 0.0317 (11) | 0.0089 (8) | 0.0022 (8) | 0.0046 (9) |
| C19 | 0.0252 (11) | 0.0305 (11) | 0.0373 (12) | 0.0143 (9) | 0.0038 (9) | 0.0168 (10) |
| C20 | 0.0267 (11) | 0.0251 (10) | 0.0291 (11) | 0.0108 (9) | 0.0064 (9) | 0.0150 (9) |
| C21 | 0.0203 (10) | 0.0164 (8) | 0.0135 (8) | 0.0063 (7) | 0.0034 (7) | 0.0037 (7) |
| C22 | 0.0227 (10) | 0.0123 (8) | 0.0142 (8) | 0.0036 (7) | 0.0062 (7) | 0.0048 (7) |
| C24 | 0.0217 (10) | 0.0189 (9) | 0.0267 (10) | 0.0031 (8) | 0.0046 (8) | 0.0104 (8) |
| C25 | 0.0184 (9) | 0.0153 (9) | 0.0198 (9) | 0.0011 (7) | 0.0004 (7) | 0.0059 (7) |
| C27 | 0.0312 (12) | 0.0237 (10) | 0.0235 (11) | −0.0032 (9) | −0.0030 (9) | 0.0058 (9) |
| C28 | 0.0193 (10) | 0.0258 (10) | 0.0429 (13) | 0.0077 (9) | 0.0075 (9) | 0.0147 (10) |
| C30 | 0.0188 (9) | 0.0173 (8) | 0.0132 (8) | 0.0051 (7) | 0.0029 (7) | 0.0095 (7) |
| C31 | 0.0181 (10) | 0.0207 (9) | 0.0227 (10) | 0.0023 (8) | 0.0009 (8) | 0.0080 (8) |
| C32 | 0.0165 (10) | 0.0299 (11) | 0.0329 (11) | 0.0081 (8) | 0.0057 (8) | 0.0145 (9) |
| C33 | 0.0244 (11) | 0.0261 (10) | 0.0318 (11) | 0.0137 (9) | 0.0102 (9) | 0.0111 (9) |
| C34 | 0.0228 (10) | 0.0194 (9) | 0.0183 (9) | 0.0077 (8) | 0.0044 (8) | 0.0059 (7) |
| C35 | 0.0172 (9) | 0.0178 (9) | 0.0131 (8) | 0.0065 (7) | 0.0036 (7) | 0.0074 (7) |
| C36 | 0.0214 (9) | 0.0135 (8) | 0.0105 (8) | 0.0063 (7) | 0.0023 (7) | 0.0054 (7) |
| C38 | 0.0168 (9) | 0.0178 (9) | 0.0156 (9) | 0.0035 (7) | 0.0009 (7) | 0.0045 (7) |
| C39 | 0.0177 (9) | 0.0172 (9) | 0.0185 (9) | 0.0032 (7) | 0.0046 (7) | 0.0015 (7) |
| C41 | 0.0381 (13) | 0.0269 (11) | 0.0210 (10) | 0.0044 (10) | 0.0120 (9) | 0.0045 (9) |
| C42 | 0.0164 (10) | 0.0243 (10) | 0.0412 (13) | 0.0054 (8) | 0.0034 (9) | 0.0007 (9) |
| C44 | 0.0197 (9) | 0.0156 (8) | 0.0106 (8) | 0.0066 (7) | 0.0021 (7) | −0.0018 (7) |
| C45 | 0.0201 (10) | 0.0223 (10) | 0.0234 (10) | 0.0098 (8) | −0.0018 (8) | 0.0007 (8) |
| C46 | 0.0149 (10) | 0.0254 (10) | 0.0363 (12) | 0.0032 (8) | 0.0040 (9) | −0.0007 (9) |
| C47 | 0.0217 (11) | 0.0253 (10) | 0.0290 (11) | −0.0001 (9) | 0.0083 (9) | 0.0055 (9) |
| C48 | 0.0233 (10) | 0.0183 (9) | 0.0179 (9) | 0.0031 (8) | 0.0033 (8) | 0.0045 (7) |
| C49 | 0.0180 (9) | 0.0152 (8) | 0.0107 (8) | 0.0040 (7) | 0.0021 (7) | 0.0009 (7) |
| C50 | 0.0200 (9) | 0.0132 (8) | 0.0100 (8) | 0.0055 (7) | 0.0005 (7) | 0.0025 (6) |
| C52 | 0.0203 (10) | 0.0259 (10) | 0.0191 (9) | 0.0110 (8) | 0.0017 (8) | 0.0083 (8) |
| C53 | 0.0179 (9) | 0.0161 (8) | 0.0178 (9) | 0.0077 (7) | 0.0019 (7) | 0.0041 (7) |
| C55 | 0.0232 (10) | 0.0239 (10) | 0.0214 (10) | 0.0116 (8) | 0.0062 (8) | 0.0066 (8) |
| C56 | 0.0169 (10) | 0.0215 (10) | 0.0355 (12) | 0.0059 (8) | 0.0026 (9) | 0.0031 (9) |
Poly[bis[µ3-2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido][µ2-2-(4,4-dimethyl-2-oxazolin-2-yl)aniline]tetrasodium(I)] (Na4H-L142) . Geometric parameters (Å, º)
| Na1—O9i | 2.4003 (15) | C16—C17 | 1.434 (3) |
| Na1—N1 | 2.3465 (18) | C16—C21 | 1.439 (3) |
| Na1—N12 | 2.9532 (18) | C17—H17 | 0.9500 |
| Na1—N43 | 2.3432 (17) | C17—C18 | 1.357 (3) |
| Na1—N54 | 2.7348 (17) | C18—H18 | 0.9500 |
| Na2—N1 | 2.3619 (18) | C18—C19 | 1.403 (3) |
| Na2—N12 | 2.3626 (17) | C19—H19 | 0.9500 |
| Na2—N15 | 2.3411 (18) | C19—C20 | 1.370 (3) |
| Na2—N26 | 2.4407 (17) | C20—H20 | 0.9500 |
| Na3—O37ii | 2.4099 (14) | C20—C21 | 1.405 (3) |
| Na3—N15 | 2.3360 (18) | C21—C22 | 1.458 (3) |
| Na3—N26 | 2.5515 (16) | C24—H24A | 0.9900 |
| Na3—N29 | 2.3611 (17) | C24—H24B | 0.9900 |
| Na3—N40 | 3.0327 (18) | C24—C25 | 1.531 (3) |
| Na4—N29 | 2.3701 (18) | C25—C27 | 1.526 (3) |
| Na4—N40 | 2.3396 (17) | C25—C28 | 1.516 (3) |
| Na4—N43 | 2.3519 (18) | C27—H27A | 0.9800 |
| Na4—N54 | 2.4035 (17) | C27—H27B | 0.9800 |
| O9—C8 | 1.381 (2) | C27—H27C | 0.9800 |
| O9—C10 | 1.457 (3) | C28—H28A | 0.9800 |
| O23—C22 | 1.369 (2) | C28—H28B | 0.9800 |
| O23—C24 | 1.443 (2) | C28—H28C | 0.9800 |
| O37—C36 | 1.393 (2) | C30—C31 | 1.430 (3) |
| O37—C38 | 1.450 (2) | C30—C35 | 1.444 (2) |
| O51—C50 | 1.380 (2) | C31—H31 | 0.9500 |
| O51—C52 | 1.445 (2) | C31—C32 | 1.366 (3) |
| N1—H1 | 0.882 (9) | C32—H32 | 0.9500 |
| N1—C2 | 1.351 (2) | C32—C33 | 1.398 (3) |
| N12—C8 | 1.288 (2) | C33—H33 | 0.9500 |
| N12—C11 | 1.490 (3) | C33—C34 | 1.373 (3) |
| N15—H15 | 0.873 (9) | C34—H34 | 0.9500 |
| N15—C16 | 1.346 (2) | C34—C35 | 1.407 (3) |
| N26—C22 | 1.291 (2) | C35—C36 | 1.452 (3) |
| N26—C25 | 1.493 (2) | C38—H38A | 0.9900 |
| N29—H29 | 0.878 (10) | C38—H38B | 0.9900 |
| N29—C30 | 1.345 (2) | C38—C39 | 1.533 (3) |
| N40—C36 | 1.285 (2) | C39—C41 | 1.530 (3) |
| N40—C39 | 1.483 (2) | C39—C42 | 1.519 (3) |
| N43—H43 | 0.873 (9) | C41—H41A | 0.9800 |
| N43—C44 | 1.349 (2) | C41—H41C | 0.9800 |
| N54—C50 | 1.288 (2) | C42—H42A | 0.9800 |
| N54—C53 | 1.493 (2) | C42—H42B | 0.9800 |
| C2—C3 | 1.432 (3) | C42—H42C | 0.9800 |
| C2—C7 | 1.446 (3) | C44—C45 | 1.434 (3) |
| C3—H3 | 0.9500 | C44—C49 | 1.441 (3) |
| C3—C4 | 1.371 (3) | C45—H45 | 0.9500 |
| C4—H4 | 0.9500 | C45—C46 | 1.362 (3) |
| C4—C5 | 1.393 (3) | C46—H46 | 0.9500 |
| C5—H5 | 0.9500 | C46—C47 | 1.397 (3) |
| C5—C6 | 1.372 (3) | C47—H47 | 0.9500 |
| C6—H6 | 0.9500 | C47—C48 | 1.365 (3) |
| C6—C7 | 1.413 (3) | C48—H48 | 0.9500 |
| C7—C8 | 1.454 (3) | C48—C49 | 1.414 (3) |
| C10—H10A | 0.9900 | C49—C50 | 1.454 (3) |
| C10—H10B | 0.9900 | C52—H52A | 0.9900 |
| C10—C11 | 1.533 (3) | C52—H52B | 0.9900 |
| C11—C13 | 1.519 (3) | C52—C53 | 1.521 (3) |
| C11—C14 | 1.526 (3) | C53—C55 | 1.532 (3) |
| C13—H13A | 0.9800 | C53—C56 | 1.520 (3) |
| C13—H13B | 0.9800 | C55—H55B | 0.9800 |
| C13—H13C | 0.9800 | C55—H55C | 0.9800 |
| C14—H14A | 0.9800 | C56—H56A | 0.9800 |
| C14—H14B | 0.9800 | C56—H56B | 0.9800 |
| C14—H14C | 0.9800 | ||
| O9i—Na1—N12 | 107.61 (5) | N15—C16—C21 | 123.45 (17) |
| O9i—Na1—N54 | 103.04 (5) | C17—C16—C21 | 114.92 (17) |
| N1—Na1—O9i | 105.11 (6) | C16—C17—H17 | 118.2 |
| N1—Na1—N12 | 65.92 (5) | C18—C17—C16 | 123.67 (18) |
| N1—Na1—N54 | 92.92 (6) | C18—C17—H17 | 118.2 |
| N43—Na1—O9i | 142.94 (6) | C17—C18—H18 | 119.7 |
| N43—Na1—N1 | 111.31 (6) | C17—C18—C19 | 120.53 (19) |
| N43—Na1—N12 | 93.51 (6) | C19—C18—H18 | 119.7 |
| N43—Na1—N54 | 69.08 (5) | C18—C19—H19 | 120.9 |
| N54—Na1—N12 | 146.27 (5) | C20—C19—C18 | 118.25 (19) |
| N1—Na2—N12 | 76.60 (6) | C20—C19—H19 | 120.9 |
| N1—Na2—N26 | 125.43 (6) | C19—C20—H20 | 118.6 |
| N12—Na2—N26 | 135.48 (6) | C19—C20—C21 | 122.87 (19) |
| N15—Na2—N1 | 138.81 (6) | C21—C20—H20 | 118.6 |
| N15—Na2—N12 | 116.21 (7) | C16—C21—C22 | 122.91 (16) |
| N15—Na2—N26 | 74.56 (6) | C20—C21—C16 | 119.76 (18) |
| O37ii—Na3—N26 | 107.75 (5) | C20—C21—C22 | 117.22 (17) |
| O37ii—Na3—N40 | 104.03 (5) | O23—C22—C21 | 114.87 (16) |
| N15—Na3—O37ii | 141.85 (6) | N26—C22—O23 | 116.13 (16) |
| N15—Na3—N26 | 72.56 (5) | N26—C22—C21 | 128.95 (16) |
| N15—Na3—N29 | 108.71 (6) | O23—C24—H24A | 110.9 |
| N15—Na3—N40 | 85.90 (5) | O23—C24—H24B | 110.9 |
| N26—Na3—N40 | 147.75 (5) | O23—C24—C25 | 104.31 (15) |
| N29—Na3—O37ii | 108.68 (6) | H24A—C24—H24B | 108.9 |
| N29—Na3—N26 | 100.25 (6) | C25—C24—H24A | 110.9 |
| N29—Na3—N40 | 63.74 (5) | C25—C24—H24B | 110.9 |
| N29—Na4—N54 | 119.12 (6) | N26—C25—C24 | 102.30 (14) |
| N40—Na4—N29 | 76.15 (6) | N26—C25—C27 | 108.12 (16) |
| N40—Na4—N43 | 113.77 (6) | N26—C25—C28 | 111.12 (15) |
| N40—Na4—N54 | 121.58 (6) | C27—C25—C24 | 111.40 (16) |
| N43—Na4—N29 | 156.58 (6) | C28—C25—C24 | 112.81 (16) |
| N43—Na4—N54 | 75.06 (6) | C28—C25—C27 | 110.72 (17) |
| C8—O9—Na1i | 121.72 (11) | C25—C27—H27A | 109.5 |
| C8—O9—C10 | 106.14 (14) | C25—C27—H27B | 109.5 |
| C10—O9—Na1i | 115.61 (11) | C25—C27—H27C | 109.5 |
| C22—O23—C24 | 106.11 (14) | H27A—C27—H27B | 109.5 |
| C36—O37—Na3ii | 125.75 (11) | H27A—C27—H27C | 109.5 |
| C36—O37—C38 | 105.65 (13) | H27B—C27—H27C | 109.5 |
| C38—O37—Na3ii | 114.12 (10) | C25—C28—H28A | 109.5 |
| C50—O51—C52 | 105.58 (14) | C25—C28—H28B | 109.5 |
| Na1—N1—Na2 | 93.03 (7) | C25—C28—H28C | 109.5 |
| Na1—N1—H1 | 113.0 (14) | H28A—C28—H28B | 109.5 |
| Na2—N1—H1 | 118.3 (14) | H28A—C28—H28C | 109.5 |
| C2—N1—Na1 | 115.17 (12) | H28B—C28—H28C | 109.5 |
| C2—N1—Na2 | 111.80 (12) | N29—C30—C31 | 122.19 (17) |
| C2—N1—H1 | 105.6 (14) | N29—C30—C35 | 123.07 (17) |
| Na2—N12—Na1 | 79.12 (5) | C31—C30—C35 | 114.74 (17) |
| C8—N12—Na1 | 94.47 (11) | C30—C31—H31 | 118.4 |
| C8—N12—Na2 | 117.61 (13) | C32—C31—C30 | 123.14 (18) |
| C8—N12—C11 | 108.22 (15) | C32—C31—H31 | 118.4 |
| C11—N12—Na1 | 124.85 (12) | C31—C32—H32 | 119.3 |
| C11—N12—Na2 | 126.23 (12) | C31—C32—C33 | 121.42 (19) |
| Na2—N15—H15 | 117.9 (13) | C33—C32—H32 | 119.3 |
| Na3—N15—Na2 | 87.12 (6) | C32—C33—H33 | 121.1 |
| Na3—N15—H15 | 118.1 (13) | C34—C33—C32 | 117.83 (19) |
| C16—N15—Na2 | 111.22 (11) | C34—C33—H33 | 121.1 |
| C16—N15—Na3 | 115.06 (12) | C33—C34—H34 | 118.7 |
| C16—N15—H15 | 106.7 (14) | C33—C34—C35 | 122.67 (18) |
| Na2—N26—Na3 | 80.39 (5) | C35—C34—H34 | 118.7 |
| C22—N26—Na2 | 106.64 (12) | C30—C35—C36 | 122.34 (16) |
| C22—N26—Na3 | 110.53 (11) | C34—C35—C30 | 120.20 (17) |
| C22—N26—C25 | 107.27 (15) | C34—C35—C36 | 117.32 (16) |
| C25—N26—Na2 | 114.75 (11) | O37—C36—C35 | 115.12 (15) |
| C25—N26—Na3 | 132.60 (12) | N40—C36—O37 | 115.49 (16) |
| Na3—N29—Na4 | 88.98 (6) | N40—C36—C35 | 129.31 (16) |
| Na3—N29—H29 | 112.6 (15) | O37—C38—H38A | 110.9 |
| Na4—N29—H29 | 115.7 (15) | O37—C38—H38B | 110.9 |
| C30—N29—Na3 | 110.70 (12) | O37—C38—C39 | 104.18 (14) |
| C30—N29—Na4 | 121.06 (12) | H38A—C38—H38B | 108.9 |
| C30—N29—H29 | 106.8 (16) | C39—C38—H38A | 110.9 |
| Na4—N40—Na3 | 74.97 (5) | C39—C38—H38B | 110.9 |
| C36—N40—Na3 | 91.91 (11) | N40—C39—C38 | 102.51 (14) |
| C36—N40—Na4 | 125.10 (13) | N40—C39—C41 | 107.69 (16) |
| C36—N40—C39 | 107.94 (15) | N40—C39—C42 | 111.29 (16) |
| C39—N40—Na3 | 129.29 (11) | C41—C39—C38 | 111.86 (16) |
| C39—N40—Na4 | 121.69 (11) | C42—C39—C38 | 112.55 (17) |
| Na1—N43—Na4 | 83.79 (6) | C42—C39—C41 | 110.59 (17) |
| Na1—N43—H43 | 117.7 (15) | C39—C41—H41A | 109.5 |
| Na4—N43—H43 | 114.4 (15) | C39—C41—H41B | 109.5 |
| C44—N43—Na1 | 106.88 (11) | C39—C41—H41C | 109.5 |
| C44—N43—Na4 | 125.00 (12) | H41A—C41—H41B | 109.5 |
| C44—N43—H43 | 107.5 (15) | H41A—C41—H41C | 109.5 |
| Na4—N54—Na1 | 74.89 (5) | H41B—C41—H41C | 109.5 |
| C50—N54—Na1 | 99.29 (11) | C39—C42—H42A | 109.5 |
| C50—N54—Na4 | 121.60 (12) | C39—C42—H42B | 109.5 |
| C50—N54—C53 | 107.20 (15) | C39—C42—H42C | 109.5 |
| C53—N54—Na1 | 136.32 (11) | H42A—C42—H42B | 109.5 |
| C53—N54—Na4 | 116.25 (11) | H42A—C42—H42C | 109.5 |
| N1—C2—C3 | 121.90 (18) | H42B—C42—H42C | 109.5 |
| N1—C2—C7 | 123.10 (18) | N43—C44—C45 | 121.95 (17) |
| C3—C2—C7 | 115.00 (17) | N43—C44—C49 | 123.11 (17) |
| C2—C3—H3 | 118.4 | C45—C44—C49 | 114.95 (17) |
| C4—C3—C2 | 123.2 (2) | C44—C45—H45 | 118.5 |
| C4—C3—H3 | 118.4 | C46—C45—C44 | 123.07 (19) |
| C3—C4—H4 | 119.5 | C46—C45—H45 | 118.5 |
| C3—C4—C5 | 121.0 (2) | C45—C46—H46 | 119.4 |
| C5—C4—H4 | 119.5 | C45—C46—C47 | 121.23 (19) |
| C4—C5—H5 | 120.8 | C47—C46—H46 | 119.4 |
| C6—C5—C4 | 118.36 (19) | C46—C47—H47 | 120.9 |
| C6—C5—H5 | 120.8 | C48—C47—C46 | 118.22 (19) |
| C5—C6—H6 | 118.6 | C48—C47—H47 | 120.9 |
| C5—C6—C7 | 122.8 (2) | C47—C48—H48 | 118.6 |
| C7—C6—H6 | 118.6 | C47—C48—C49 | 122.72 (19) |
| C2—C7—C8 | 122.39 (16) | C49—C48—H48 | 118.6 |
| C6—C7—C2 | 119.62 (19) | C44—C49—C50 | 122.67 (16) |
| C6—C7—C8 | 117.86 (18) | C48—C49—C44 | 119.72 (17) |
| O9—C8—C7 | 114.85 (16) | C48—C49—C50 | 117.57 (17) |
| N12—C8—O9 | 115.72 (17) | O51—C50—C49 | 114.72 (15) |
| N12—C8—C7 | 129.38 (17) | N54—C50—O51 | 115.49 (16) |
| O9—C10—H10A | 110.8 | N54—C50—C49 | 129.76 (17) |
| O9—C10—H10B | 110.8 | O51—C52—H52A | 111.0 |
| O9—C10—C11 | 104.51 (15) | O51—C52—H52B | 111.0 |
| H10A—C10—H10B | 108.9 | O51—C52—C53 | 103.90 (14) |
| C11—C10—H10A | 110.9 | H52A—C52—H52B | 109.0 |
| C11—C10—H10B | 110.8 | C53—C52—H52A | 111.0 |
| N12—C11—C10 | 102.57 (16) | C53—C52—H52B | 111.0 |
| N12—C11—C13 | 110.96 (16) | N54—C53—C52 | 102.01 (14) |
| N12—C11—C14 | 108.21 (16) | N54—C53—C55 | 108.32 (15) |
| C13—C11—C10 | 112.78 (17) | N54—C53—C56 | 111.15 (15) |
| C13—C11—C14 | 110.34 (18) | C52—C53—C55 | 110.98 (16) |
| C14—C11—C10 | 111.67 (16) | C56—C53—C52 | 112.86 (16) |
| C11—C13—H13A | 109.5 | C56—C53—C55 | 111.09 (16) |
| C11—C13—H13B | 109.5 | C53—C55—H55A | 109.5 |
| C11—C13—H13C | 109.5 | C53—C55—H55B | 109.5 |
| H13A—C13—H13B | 109.5 | C53—C55—H55C | 109.5 |
| H13A—C13—H13C | 109.5 | H55A—C55—H55B | 109.5 |
| H13B—C13—H13C | 109.5 | H55A—C55—H55C | 109.5 |
| C11—C14—H14A | 109.5 | H55B—C55—H55C | 109.5 |
| C11—C14—H14B | 109.5 | C53—C56—H56A | 109.5 |
| C11—C14—H14C | 109.5 | C53—C56—H56B | 109.5 |
| H14A—C14—H14B | 109.5 | C53—C56—H56C | 109.5 |
| H14A—C14—H14C | 109.5 | H56A—C56—H56B | 109.5 |
| H14B—C14—H14C | 109.5 | H56A—C56—H56C | 109.5 |
| N15—C16—C17 | 121.62 (17) | H56B—C56—H56C | 109.5 |
| Na1i—O9—C8—N12 | 139.42 (14) | N43—C44—C45—C46 | −177.71 (18) |
| Na1i—O9—C8—C7 | −43.15 (19) | N43—C44—C49—C48 | 176.76 (16) |
| Na1i—O9—C10—C11 | −151.40 (11) | N43—C44—C49—C50 | −5.4 (3) |
| Na1—N1—C2—C3 | 127.34 (16) | C2—C3—C4—C5 | 0.9 (3) |
| Na1—N1—C2—C7 | −52.4 (2) | C2—C7—C8—O9 | 164.80 (16) |
| Na1—N12—C8—O9 | −121.99 (13) | C2—C7—C8—N12 | −18.2 (3) |
| Na1—N12—C8—C7 | 61.0 (2) | C3—C2—C7—C6 | 2.0 (2) |
| Na1—N12—C11—C10 | 94.69 (15) | C3—C2—C7—C8 | 177.76 (16) |
| Na1—N12—C11—C13 | −26.0 (2) | C3—C4—C5—C6 | 0.6 (3) |
| Na1—N12—C11—C14 | −147.17 (13) | C4—C5—C6—C7 | −0.7 (3) |
| Na1—N43—C44—C45 | 125.50 (15) | C5—C6—C7—C2 | −0.6 (3) |
| Na1—N43—C44—C49 | −54.82 (19) | C5—C6—C7—C8 | −176.61 (18) |
| Na1—N54—C50—O51 | −137.84 (12) | C6—C7—C8—O9 | −19.3 (2) |
| Na1—N54—C50—C49 | 44.4 (2) | C6—C7—C8—N12 | 157.67 (19) |
| Na1—N54—C53—C52 | 105.62 (16) | C7—C2—C3—C4 | −2.1 (3) |
| Na1—N54—C53—C55 | −137.24 (14) | C8—O9—C10—C11 | −13.18 (18) |
| Na1—N54—C53—C56 | −14.9 (2) | C8—N12—C11—C10 | −14.5 (2) |
| Na2—N1—C2—C3 | −128.14 (16) | C8—N12—C11—C13 | −135.21 (18) |
| Na2—N1—C2—C7 | 52.2 (2) | C8—N12—C11—C14 | 103.60 (18) |
| Na2—N12—C8—O9 | 157.96 (12) | C10—O9—C8—N12 | 4.4 (2) |
| Na2—N12—C8—C7 | −19.0 (3) | C10—O9—C8—C7 | −178.22 (15) |
| Na2—N12—C11—C10 | −162.31 (12) | C11—N12—C8—O9 | 7.0 (2) |
| Na2—N12—C11—C13 | 77.02 (19) | C11—N12—C8—C7 | −169.98 (18) |
| Na2—N12—C11—C14 | −44.2 (2) | C16—C17—C18—C19 | 0.9 (3) |
| Na2—N15—C16—C17 | −130.62 (15) | C16—C21—C22—O23 | −171.28 (16) |
| Na2—N15—C16—C21 | 48.8 (2) | C16—C21—C22—N26 | 5.9 (3) |
| Na2—N26—C22—O23 | 128.93 (14) | C17—C16—C21—C20 | 0.0 (2) |
| Na2—N26—C22—C21 | −48.2 (2) | C17—C16—C21—C22 | 175.84 (16) |
| Na2—N26—C25—C24 | −133.65 (12) | C17—C18—C19—C20 | −0.2 (3) |
| Na2—N26—C25—C27 | −15.98 (19) | C18—C19—C20—C21 | −0.6 (3) |
| Na2—N26—C25—C28 | 105.72 (15) | C19—C20—C21—C16 | 0.8 (3) |
| Na3ii—O37—C36—N40 | 142.02 (13) | C19—C20—C21—C22 | −175.37 (19) |
| Na3ii—O37—C36—C35 | −40.80 (19) | C20—C21—C22—O23 | 4.7 (2) |
| Na3ii—O37—C38—C39 | −158.32 (11) | C20—C21—C22—N26 | −178.11 (19) |
| Na3—N15—C16—C17 | 132.37 (15) | C21—C16—C17—C18 | −0.8 (3) |
| Na3—N15—C16—C21 | −48.3 (2) | C22—O23—C24—C25 | −16.83 (19) |
| Na3—N26—C22—O23 | −145.32 (13) | C22—N26—C25—C24 | −15.39 (19) |
| Na3—N26—C22—C21 | 37.5 (2) | C22—N26—C25—C27 | 102.28 (18) |
| Na3—N26—C25—C24 | 126.32 (14) | C22—N26—C25—C28 | −136.02 (17) |
| Na3—N26—C25—C27 | −116.02 (16) | C24—O23—C22—N26 | 7.7 (2) |
| Na3—N26—C25—C28 | 5.7 (2) | C24—O23—C22—C21 | −174.71 (15) |
| Na3—N29—C30—C31 | 122.47 (16) | C25—N26—C22—O23 | 5.5 (2) |
| Na3—N29—C30—C35 | −57.2 (2) | C25—N26—C22—C21 | −171.62 (17) |
| Na3—N40—C36—O37 | −124.35 (13) | C30—C31—C32—C33 | 0.0 (3) |
| Na3—N40—C36—C35 | 58.96 (19) | C30—C35—C36—O37 | 169.03 (15) |
| Na3—N40—C39—C38 | 90.53 (16) | C30—C35—C36—N40 | −14.3 (3) |
| Na3—N40—C39—C41 | −151.36 (13) | C31—C30—C35—C34 | 1.2 (2) |
| Na3—N40—C39—C42 | −30.0 (2) | C31—C30—C35—C36 | 176.71 (16) |
| Na4—N29—C30—C31 | −135.69 (15) | C31—C32—C33—C34 | 0.8 (3) |
| Na4—N29—C30—C35 | 44.7 (2) | C32—C33—C34—C35 | −0.5 (3) |
| Na4—N40—C36—O37 | 162.73 (11) | C33—C34—C35—C30 | −0.5 (3) |
| Na4—N40—C36—C35 | −14.0 (3) | C33—C34—C35—C36 | −176.24 (18) |
| Na4—N40—C39—C38 | −173.06 (11) | C34—C35—C36—O37 | −15.3 (2) |
| Na4—N40—C39—C41 | −54.96 (19) | C34—C35—C36—N40 | 161.36 (19) |
| Na4—N40—C39—C42 | 66.40 (19) | C35—C30—C31—C32 | −1.0 (3) |
| Na4—N43—C44—C45 | −140.23 (15) | C36—O37—C38—C39 | −16.11 (17) |
| Na4—N43—C44—C49 | 39.4 (2) | C36—N40—C39—C38 | −17.54 (19) |
| Na4—N54—C50—O51 | 144.26 (12) | C36—N40—C39—C41 | 100.57 (18) |
| Na4—N54—C50—C49 | −33.5 (2) | C36—N40—C39—C42 | −138.07 (17) |
| Na4—N54—C53—C52 | −158.89 (11) | C38—O37—C36—N40 | 5.6 (2) |
| Na4—N54—C53—C55 | −41.75 (17) | C38—O37—C36—C35 | −177.25 (14) |
| Na4—N54—C53—C56 | 80.56 (17) | C39—N40—C36—O37 | 8.2 (2) |
| O9—C10—C11—N12 | 16.47 (18) | C39—N40—C36—C35 | −168.45 (17) |
| O9—C10—C11—C13 | 135.88 (17) | C44—C45—C46—C47 | 0.0 (3) |
| O9—C10—C11—C14 | −99.20 (19) | C44—C49—C50—O51 | −174.02 (15) |
| O23—C24—C25—N26 | 19.30 (18) | C44—C49—C50—N54 | 3.8 (3) |
| O23—C24—C25—C27 | −96.01 (18) | C45—C44—C49—C48 | −3.5 (2) |
| O23—C24—C25—C28 | 138.75 (16) | C45—C44—C49—C50 | 174.26 (16) |
| O37—C38—C39—N40 | 20.13 (18) | C45—C46—C47—C48 | −1.7 (3) |
| O37—C38—C39—C41 | −94.98 (18) | C46—C47—C48—C49 | 0.6 (3) |
| O37—C38—C39—C42 | 139.79 (16) | C47—C48—C49—C44 | 2.1 (3) |
| O51—C52—C53—N54 | 23.61 (17) | C47—C48—C49—C50 | −175.82 (18) |
| O51—C52—C53—C55 | −91.59 (18) | C48—C49—C50—O51 | 3.8 (2) |
| O51—C52—C53—C56 | 142.96 (16) | C48—C49—C50—N54 | −178.38 (18) |
| N1—C2—C3—C4 | 178.15 (18) | C49—C44—C45—C46 | 2.6 (3) |
| N1—C2—C7—C6 | −178.33 (17) | C50—O51—C52—C53 | −20.39 (18) |
| N1—C2—C7—C8 | −2.5 (3) | C50—N54—C53—C52 | −19.05 (18) |
| N15—C16—C17—C18 | 178.63 (19) | C50—N54—C53—C55 | 98.09 (17) |
| N15—C16—C21—C20 | −179.45 (17) | C50—N54—C53—C56 | −139.59 (17) |
| N15—C16—C21—C22 | −3.6 (3) | C52—O51—C50—N54 | 9.1 (2) |
| N29—C30—C31—C32 | 179.32 (18) | C52—O51—C50—C49 | −172.80 (15) |
| N29—C30—C35—C34 | −179.10 (17) | C53—N54—C50—O51 | 7.0 (2) |
| N29—C30—C35—C36 | −3.6 (3) | C53—N54—C50—C49 | −170.76 (17) |
Symmetry codes: (i) −x+2, −y+1, −z+1; (ii) −x+2, −y, −z.
Tris[2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido]ytterbium(III) (YbH-L133). Crystal data
| [Yb(C11H13N2O)3] | F(000) = 1492 |
| Mr = 740.74 | Dx = 1.605 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.9428 (5) Å | Cell parameters from 7270 reflections |
| b = 9.8253 (5) Å | θ = 2.2–24.6° |
| c = 28.6089 (14) Å | µ = 3.10 mm−1 |
| β = 94.722 (1)° | T = 86 K |
| V = 3065.5 (3) Å3 | Plate, yellow |
| Z = 4 | 0.22 × 0.18 × 0.05 mm |
Tris[2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido]ytterbium(III) (YbH-L133). Data collection
| SMART APEX CCD area detector diffractometer | 7061 independent reflections |
| Radiation source: sealed X-ray tube | 5882 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.059 |
| Detector resolution: 8.3 pixels mm-1 | θmax = 27.6°, θmin = 1.9° |
| φ and ω scans | h = −14→14 |
| Absorption correction: multi-scan (SADABS; Bruker, 2016) | k = −12→12 |
| Tmin = 0.219, Tmax = 0.262 | l = −37→37 |
| 39651 measured reflections |
Tris[2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido]ytterbium(III) (YbH-L133). Refinement
| Refinement on F2 | Primary atom site location: heavy-atom method |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.032 | H-atom parameters constrained |
| wR(F2) = 0.073 | w = 1/[σ2(Fo2) + (0.0248P)2 + 4.5847P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.07 | (Δ/σ)max = 0.002 |
| 7061 reflections | Δρmax = 0.83 e Å−3 |
| 394 parameters | Δρmin = −1.19 e Å−3 |
| 0 restraints |
Tris[2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido]ytterbium(III) (YbH-L133). Special details
| Experimental. The data collection nominally covered a full sphere of reciprocal space by a combination of 5 sets of ω scans each set at different φ and/or 2θ angles and each scan (20 s exposure) covering -0.300° degrees in ω. The crystal to detector distance was 5.0 cm. |
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Tris[2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido]ytterbium(III) (YbH-L133). Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C2 | 1.0818 (3) | 0.2384 (4) | 0.10704 (13) | 0.0200 (8) | |
| C3 | 1.2109 (4) | 0.2483 (4) | 0.11734 (15) | 0.0262 (9) | |
| H3 | 1.253065 | 0.173718 | 0.132394 | 0.031* | |
| C4 | 1.2766 (4) | 0.3596 (5) | 0.10659 (16) | 0.0316 (10) | |
| H4 | 1.362244 | 0.362906 | 0.115191 | 0.038* | |
| C5 | 1.2190 (4) | 0.4686 (5) | 0.08310 (17) | 0.0340 (11) | |
| H5 | 1.265345 | 0.544940 | 0.074364 | 0.041* | |
| C6 | 1.0945 (4) | 0.4649 (4) | 0.07264 (16) | 0.0283 (10) | |
| H6 | 1.055257 | 0.539088 | 0.056247 | 0.034* | |
| C7 | 1.0229 (4) | 0.3534 (4) | 0.08564 (13) | 0.0199 (8) | |
| C8 | 0.8911 (4) | 0.3647 (4) | 0.07860 (14) | 0.0240 (9) | |
| C10 | 0.7215 (4) | 0.4633 (5) | 0.0446 (2) | 0.0415 (13) | |
| H10A | 0.700654 | 0.427434 | 0.012561 | 0.050* | |
| H10B | 0.679548 | 0.551716 | 0.047774 | 0.050* | |
| C11 | 0.6856 (4) | 0.3624 (4) | 0.08159 (16) | 0.0261 (9) | |
| C13 | 0.6499 (4) | 0.4350 (5) | 0.12536 (18) | 0.0402 (12) | |
| H13A | 0.717992 | 0.492735 | 0.137852 | 0.060* | |
| H13B | 0.631465 | 0.367551 | 0.149013 | 0.060* | |
| H13C | 0.577374 | 0.491481 | 0.117436 | 0.060* | |
| C14 | 0.5842 (4) | 0.2703 (4) | 0.06114 (15) | 0.0267 (9) | |
| H14A | 0.512628 | 0.325250 | 0.050405 | 0.040* | |
| H14B | 0.561617 | 0.206075 | 0.085192 | 0.040* | |
| H14C | 0.612722 | 0.219744 | 0.034575 | 0.040* | |
| C16 | 0.5330 (3) | −0.0138 (4) | 0.14097 (13) | 0.0178 (8) | |
| C17 | 0.4277 (3) | −0.0117 (4) | 0.16733 (14) | 0.0215 (8) | |
| H17 | 0.428448 | 0.045669 | 0.194088 | 0.026* | |
| C18 | 0.3261 (4) | −0.0887 (4) | 0.15562 (15) | 0.0244 (9) | |
| H18 | 0.258888 | −0.085095 | 0.174572 | 0.029* | |
| C19 | 0.3196 (4) | −0.1728 (4) | 0.11627 (15) | 0.0270 (9) | |
| H19 | 0.248910 | −0.226754 | 0.108288 | 0.032* | |
| C20 | 0.4175 (4) | −0.1757 (4) | 0.08933 (14) | 0.0238 (9) | |
| H20 | 0.413171 | −0.232525 | 0.062388 | 0.029* | |
| C21 | 0.5241 (3) | −0.0979 (4) | 0.09994 (13) | 0.0178 (8) | |
| C22 | 0.6183 (4) | −0.0995 (4) | 0.06692 (13) | 0.0195 (8) | |
| C24 | 0.6760 (4) | −0.1389 (5) | −0.00564 (14) | 0.0298 (10) | |
| H24A | 0.643468 | −0.071623 | −0.029244 | 0.036* | |
| H24B | 0.700649 | −0.221940 | −0.022058 | 0.036* | |
| C25 | 0.7847 (3) | −0.0797 (4) | 0.02494 (13) | 0.0211 (8) | |
| C27 | 0.8359 (4) | 0.0464 (4) | 0.00316 (14) | 0.0243 (9) | |
| H27A | 0.870264 | 0.021891 | −0.026275 | 0.036* | |
| H27B | 0.900484 | 0.085664 | 0.024844 | 0.036* | |
| H27C | 0.770182 | 0.113260 | −0.003178 | 0.036* | |
| C28 | 0.8841 (4) | −0.1848 (4) | 0.03572 (15) | 0.0270 (9) | |
| H28A | 0.849832 | −0.263196 | 0.051309 | 0.041* | |
| H28B | 0.950454 | −0.144768 | 0.056380 | 0.041* | |
| H28C | 0.916454 | −0.214532 | 0.006452 | 0.041* | |
| C30 | 0.9880 (3) | −0.1928 (4) | 0.18242 (13) | 0.0203 (8) | |
| C31 | 1.0439 (4) | −0.3196 (4) | 0.17370 (15) | 0.0272 (9) | |
| H31 | 1.013838 | −0.370665 | 0.146990 | 0.033* | |
| C32 | 1.1392 (4) | −0.3713 (5) | 0.20210 (16) | 0.0311 (10) | |
| H32 | 1.175725 | −0.454914 | 0.194121 | 0.037* | |
| C33 | 1.1835 (4) | −0.3025 (5) | 0.24272 (16) | 0.0309 (10) | |
| H33 | 1.249963 | −0.338361 | 0.262385 | 0.037* | |
| C34 | 1.1293 (4) | −0.1827 (5) | 0.25362 (15) | 0.0267 (9) | |
| H34 | 1.157426 | −0.137272 | 0.281780 | 0.032* | |
| C35 | 1.0322 (3) | −0.1236 (4) | 0.22427 (14) | 0.0207 (8) | |
| C36 | 0.9814 (3) | 0.0058 (4) | 0.23845 (13) | 0.0191 (8) | |
| C38 | 0.9638 (5) | 0.1729 (5) | 0.29017 (16) | 0.0433 (13) | |
| H38A | 1.024830 | 0.244297 | 0.299462 | 0.052* | |
| H38B | 0.908083 | 0.162360 | 0.315504 | 0.052* | |
| C39 | 0.8918 (4) | 0.2105 (4) | 0.24440 (13) | 0.0230 (8) | |
| C41 | 0.9493 (5) | 0.3302 (4) | 0.22070 (15) | 0.0340 (11) | |
| H41A | 0.906794 | 0.344554 | 0.189583 | 0.051* | |
| H41B | 1.036097 | 0.311041 | 0.217469 | 0.051* | |
| H41C | 0.942135 | 0.412284 | 0.239752 | 0.051* | |
| C42 | 0.7588 (4) | 0.2413 (6) | 0.2513 (2) | 0.0558 (17) | |
| H42A | 0.719510 | 0.159755 | 0.262947 | 0.084* | |
| H42B | 0.716179 | 0.269040 | 0.221383 | 0.084* | |
| H42C | 0.754580 | 0.315123 | 0.274210 | 0.084* | |
| N1 | 1.0186 (3) | 0.1251 (3) | 0.11733 (12) | 0.0216 (7) | |
| H1 | 1.065691 | 0.052590 | 0.117092 | 0.026* | |
| N12 | 0.8051 (3) | 0.2885 (3) | 0.09365 (11) | 0.0186 (7) | |
| N15 | 0.6330 (3) | 0.0598 (3) | 0.15501 (11) | 0.0215 (7) | |
| H15 | 0.619157 | 0.118443 | 0.177229 | 0.026* | |
| N26 | 0.7256 (3) | −0.0444 (3) | 0.06940 (11) | 0.0177 (7) | |
| N29 | 0.8955 (3) | −0.1416 (3) | 0.15281 (11) | 0.0190 (7) | |
| H29 | 0.891440 | −0.201964 | 0.130248 | 0.023* | |
| N40 | 0.9045 (3) | 0.0857 (3) | 0.21535 (11) | 0.0176 (7) | |
| O9 | 0.8521 (3) | 0.4775 (3) | 0.05403 (12) | 0.0371 (8) | |
| O23 | 0.5856 (3) | −0.1705 (3) | 0.02681 (9) | 0.0294 (7) | |
| O37 | 1.0238 (3) | 0.0466 (3) | 0.28185 (10) | 0.0272 (7) | |
| Yb1 | 0.82829 (2) | 0.06815 (2) | 0.13487 (2) | 0.01617 (6) |
Tris[2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido]ytterbium(III) (YbH-L133). Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C2 | 0.0199 (19) | 0.026 (2) | 0.0154 (19) | −0.0011 (16) | 0.0074 (15) | −0.0038 (15) |
| C3 | 0.019 (2) | 0.034 (2) | 0.027 (2) | 0.0026 (17) | 0.0050 (16) | −0.0058 (18) |
| C4 | 0.022 (2) | 0.038 (3) | 0.036 (3) | −0.0077 (19) | 0.0092 (19) | −0.013 (2) |
| C5 | 0.031 (2) | 0.028 (2) | 0.046 (3) | −0.0154 (19) | 0.019 (2) | −0.012 (2) |
| C6 | 0.030 (2) | 0.022 (2) | 0.034 (2) | −0.0019 (17) | 0.0083 (19) | −0.0054 (18) |
| C7 | 0.027 (2) | 0.0137 (18) | 0.020 (2) | −0.0045 (16) | 0.0076 (16) | −0.0064 (15) |
| C8 | 0.027 (2) | 0.024 (2) | 0.021 (2) | 0.0000 (17) | 0.0040 (17) | 0.0031 (17) |
| C10 | 0.028 (2) | 0.036 (3) | 0.060 (3) | −0.002 (2) | −0.004 (2) | 0.020 (2) |
| C11 | 0.021 (2) | 0.020 (2) | 0.037 (2) | 0.0027 (17) | 0.0009 (18) | 0.0040 (18) |
| C13 | 0.027 (2) | 0.041 (3) | 0.051 (3) | 0.010 (2) | −0.007 (2) | −0.019 (2) |
| C14 | 0.021 (2) | 0.027 (2) | 0.031 (2) | 0.0041 (17) | −0.0044 (17) | −0.0010 (18) |
| C16 | 0.0172 (18) | 0.0188 (19) | 0.0175 (19) | 0.0030 (15) | 0.0036 (15) | 0.0049 (15) |
| C17 | 0.0190 (19) | 0.023 (2) | 0.023 (2) | 0.0040 (16) | 0.0076 (16) | 0.0048 (16) |
| C18 | 0.0184 (19) | 0.029 (2) | 0.027 (2) | 0.0004 (17) | 0.0082 (16) | 0.0060 (18) |
| C19 | 0.019 (2) | 0.029 (2) | 0.033 (2) | −0.0079 (17) | 0.0036 (17) | 0.0047 (18) |
| C20 | 0.024 (2) | 0.026 (2) | 0.022 (2) | −0.0082 (17) | 0.0027 (16) | −0.0005 (17) |
| C21 | 0.0153 (17) | 0.0195 (19) | 0.0188 (19) | −0.0020 (15) | 0.0025 (14) | 0.0048 (15) |
| C22 | 0.021 (2) | 0.022 (2) | 0.0148 (18) | −0.0023 (15) | −0.0011 (15) | −0.0010 (15) |
| C24 | 0.026 (2) | 0.045 (3) | 0.020 (2) | −0.011 (2) | 0.0099 (17) | −0.0054 (19) |
| C25 | 0.0186 (18) | 0.030 (2) | 0.0156 (18) | −0.0018 (17) | 0.0047 (14) | −0.0025 (16) |
| C27 | 0.022 (2) | 0.032 (2) | 0.021 (2) | 0.0011 (17) | 0.0077 (16) | 0.0048 (17) |
| C28 | 0.029 (2) | 0.026 (2) | 0.027 (2) | 0.0031 (18) | 0.0125 (18) | −0.0013 (18) |
| C30 | 0.0196 (19) | 0.021 (2) | 0.0220 (19) | −0.0042 (16) | 0.0103 (15) | 0.0046 (16) |
| C31 | 0.029 (2) | 0.023 (2) | 0.032 (2) | 0.0011 (17) | 0.0144 (18) | 0.0046 (18) |
| C32 | 0.028 (2) | 0.026 (2) | 0.042 (3) | 0.0071 (18) | 0.019 (2) | 0.015 (2) |
| C33 | 0.020 (2) | 0.035 (2) | 0.039 (3) | 0.0008 (18) | 0.0069 (18) | 0.021 (2) |
| C34 | 0.0170 (19) | 0.038 (3) | 0.025 (2) | −0.0059 (18) | 0.0050 (16) | 0.0090 (19) |
| C35 | 0.0177 (19) | 0.022 (2) | 0.024 (2) | −0.0038 (15) | 0.0083 (16) | 0.0059 (16) |
| C36 | 0.0165 (18) | 0.028 (2) | 0.0130 (18) | −0.0071 (16) | 0.0037 (14) | 0.0002 (16) |
| C38 | 0.082 (4) | 0.021 (2) | 0.025 (2) | −0.004 (2) | −0.006 (2) | −0.0028 (19) |
| C39 | 0.023 (2) | 0.026 (2) | 0.020 (2) | −0.0036 (17) | 0.0037 (16) | −0.0083 (17) |
| C41 | 0.054 (3) | 0.022 (2) | 0.026 (2) | −0.004 (2) | −0.001 (2) | −0.0008 (18) |
| C42 | 0.032 (3) | 0.072 (4) | 0.066 (4) | −0.006 (3) | 0.018 (3) | −0.048 (3) |
| N1 | 0.0143 (16) | 0.0217 (17) | 0.0293 (19) | 0.0022 (13) | 0.0052 (14) | 0.0059 (14) |
| N12 | 0.0147 (15) | 0.0190 (16) | 0.0220 (17) | −0.0015 (13) | 0.0012 (12) | 0.0013 (13) |
| N15 | 0.0207 (16) | 0.0261 (18) | 0.0183 (16) | −0.0050 (14) | 0.0057 (13) | −0.0072 (14) |
| N26 | 0.0175 (16) | 0.0219 (18) | 0.0143 (15) | 0.0006 (13) | 0.0040 (12) | −0.0007 (13) |
| N29 | 0.0249 (17) | 0.0176 (16) | 0.0149 (16) | 0.0008 (13) | 0.0035 (13) | −0.0014 (13) |
| N40 | 0.0203 (16) | 0.0175 (16) | 0.0154 (15) | −0.0047 (13) | 0.0038 (12) | −0.0019 (13) |
| O9 | 0.0285 (17) | 0.0294 (17) | 0.053 (2) | −0.0036 (13) | −0.0018 (15) | 0.0204 (15) |
| O23 | 0.0247 (15) | 0.0445 (19) | 0.0197 (15) | −0.0152 (13) | 0.0070 (12) | −0.0109 (13) |
| O37 | 0.0274 (15) | 0.0358 (18) | 0.0178 (14) | −0.0039 (13) | −0.0018 (12) | −0.0018 (12) |
| Yb1 | 0.01452 (8) | 0.01806 (9) | 0.01624 (9) | −0.00202 (7) | 0.00307 (6) | 0.00040 (7) |
Tris[2-(4,4-dimethyl-2-oxazolin-2-yl)anilinido]ytterbium(III) (YbH-L133). Geometric parameters (Å, º)
| C2—C3 | 1.423 (5) | C25—C28 | 1.513 (6) |
| C2—C7 | 1.414 (5) | C25—N26 | 1.514 (5) |
| C2—N1 | 1.356 (5) | C27—H27A | 0.9800 |
| C3—H3 | 0.9500 | C27—H27B | 0.9800 |
| C3—C4 | 1.358 (6) | C27—H27C | 0.9800 |
| C4—H4 | 0.9500 | C28—H28A | 0.9800 |
| C4—C5 | 1.388 (7) | C28—H28B | 0.9800 |
| C5—H5 | 0.9500 | C28—H28C | 0.9800 |
| C5—C6 | 1.372 (6) | C30—C31 | 1.419 (6) |
| C6—H6 | 0.9500 | C30—C35 | 1.427 (6) |
| C6—C7 | 1.414 (5) | C30—N29 | 1.361 (5) |
| C7—C8 | 1.445 (6) | C31—H31 | 0.9500 |
| C8—N12 | 1.303 (5) | C31—C32 | 1.366 (6) |
| C8—O9 | 1.362 (5) | C32—H32 | 0.9500 |
| C10—H10A | 0.9900 | C32—C33 | 1.396 (7) |
| C10—H10B | 0.9900 | C33—H33 | 0.9500 |
| C10—C11 | 1.526 (6) | C33—C34 | 1.365 (6) |
| C10—O9 | 1.439 (5) | C34—H34 | 0.9500 |
| C11—C13 | 1.519 (6) | C34—C35 | 1.423 (6) |
| C11—C14 | 1.512 (6) | C35—C36 | 1.458 (6) |
| C11—N12 | 1.511 (5) | C36—N40 | 1.292 (5) |
| C13—H13A | 0.9800 | C36—O37 | 1.350 (4) |
| C13—H13B | 0.9800 | C38—H38A | 0.9900 |
| C13—H13C | 0.9800 | C38—H38B | 0.9900 |
| C14—H14A | 0.9800 | C38—C39 | 1.517 (6) |
| C14—H14B | 0.9800 | C38—O37 | 1.433 (5) |
| C14—H14C | 0.9800 | C39—C41 | 1.520 (6) |
| C16—C17 | 1.429 (5) | C39—C42 | 1.516 (6) |
| C16—C21 | 1.432 (5) | C39—N40 | 1.495 (5) |
| C16—N15 | 1.345 (5) | C41—H41A | 0.9800 |
| C17—H17 | 0.9500 | C41—H41B | 0.9800 |
| C17—C18 | 1.364 (6) | C41—H41C | 0.9800 |
| C18—H18 | 0.9500 | C42—H42A | 0.9800 |
| C18—C19 | 1.393 (6) | C42—H42B | 0.9800 |
| C19—H19 | 0.9500 | C42—H42C | 0.9800 |
| C19—C20 | 1.371 (5) | N1—H1 | 0.8800 |
| C20—H20 | 0.9500 | N1—Yb1 | 2.252 (3) |
| C20—C21 | 1.406 (5) | N12—Yb1 | 2.468 (3) |
| C21—C22 | 1.455 (5) | N15—H15 | 0.8800 |
| C22—N26 | 1.289 (5) | N15—Yb1 | 2.260 (3) |
| C22—O23 | 1.366 (4) | N26—Yb1 | 2.376 (3) |
| C24—H24A | 0.9900 | N29—H29 | 0.8749 |
| C24—H24B | 0.9900 | N29—Yb1 | 2.234 (3) |
| C24—C25 | 1.532 (5) | N40—Yb1 | 2.390 (3) |
| C24—O23 | 1.445 (5) | Yb1—H29 | 2.7484 |
| C25—C27 | 1.515 (5) | ||
| C7—C2—C3 | 116.4 (4) | H28B—C28—H28C | 109.5 |
| N1—C2—C3 | 121.8 (4) | C31—C30—C35 | 116.4 (4) |
| N1—C2—C7 | 121.8 (3) | N29—C30—C31 | 121.6 (4) |
| C2—C3—H3 | 118.6 | N29—C30—C35 | 122.0 (4) |
| C4—C3—C2 | 122.9 (4) | C30—C31—H31 | 118.6 |
| C4—C3—H3 | 118.6 | C32—C31—C30 | 122.8 (4) |
| C3—C4—H4 | 119.9 | C32—C31—H31 | 118.6 |
| C3—C4—C5 | 120.3 (4) | C31—C32—H32 | 119.6 |
| C5—C4—H4 | 119.9 | C31—C32—C33 | 120.7 (4) |
| C4—C5—H5 | 120.4 | C33—C32—H32 | 119.6 |
| C6—C5—C4 | 119.2 (4) | C32—C33—H33 | 120.7 |
| C6—C5—H5 | 120.4 | C34—C33—C32 | 118.7 (4) |
| C5—C6—H6 | 119.2 | C34—C33—H33 | 120.7 |
| C5—C6—C7 | 121.7 (4) | C33—C34—H34 | 118.8 |
| C7—C6—H6 | 119.2 | C33—C34—C35 | 122.3 (4) |
| C2—C7—C8 | 122.4 (3) | C35—C34—H34 | 118.8 |
| C6—C7—C2 | 119.4 (4) | C30—C35—C36 | 122.6 (4) |
| C6—C7—C8 | 118.2 (4) | C34—C35—C30 | 119.1 (4) |
| N12—C8—C7 | 130.5 (4) | C34—C35—C36 | 118.4 (4) |
| N12—C8—O9 | 115.7 (4) | N40—C36—C35 | 129.4 (4) |
| O9—C8—C7 | 113.7 (3) | N40—C36—O37 | 116.7 (4) |
| H10A—C10—H10B | 109.0 | O37—C36—C35 | 113.9 (3) |
| C11—C10—H10A | 111.0 | H38A—C38—H38B | 108.7 |
| C11—C10—H10B | 111.0 | C39—C38—H38A | 110.5 |
| O9—C10—H10A | 111.0 | C39—C38—H38B | 110.5 |
| O9—C10—H10B | 111.0 | O37—C38—H38A | 110.5 |
| O9—C10—C11 | 103.9 (4) | O37—C38—H38B | 110.5 |
| C13—C11—C10 | 111.4 (4) | O37—C38—C39 | 106.3 (3) |
| C14—C11—C10 | 110.1 (4) | C38—C39—C41 | 111.7 (4) |
| C14—C11—C13 | 111.7 (4) | C42—C39—C38 | 111.8 (4) |
| N12—C11—C10 | 101.8 (3) | C42—C39—C41 | 110.0 (4) |
| N12—C11—C13 | 108.3 (3) | N40—C39—C38 | 102.4 (3) |
| N12—C11—C14 | 113.2 (3) | N40—C39—C41 | 109.0 (3) |
| C11—C13—H13A | 109.5 | N40—C39—C42 | 111.8 (3) |
| C11—C13—H13B | 109.5 | C39—C41—H41A | 109.5 |
| C11—C13—H13C | 109.5 | C39—C41—H41B | 109.5 |
| H13A—C13—H13B | 109.5 | C39—C41—H41C | 109.5 |
| H13A—C13—H13C | 109.5 | H41A—C41—H41B | 109.5 |
| H13B—C13—H13C | 109.5 | H41A—C41—H41C | 109.5 |
| C11—C14—H14A | 109.5 | H41B—C41—H41C | 109.5 |
| C11—C14—H14B | 109.5 | C39—C42—H42A | 109.5 |
| C11—C14—H14C | 109.5 | C39—C42—H42B | 109.5 |
| H14A—C14—H14B | 109.5 | C39—C42—H42C | 109.5 |
| H14A—C14—H14C | 109.5 | H42A—C42—H42B | 109.5 |
| H14B—C14—H14C | 109.5 | H42A—C42—H42C | 109.5 |
| C17—C16—C21 | 115.9 (3) | H42B—C42—H42C | 109.5 |
| N15—C16—C17 | 120.4 (4) | C2—N1—H1 | 110.7 |
| N15—C16—C21 | 123.7 (3) | C2—N1—Yb1 | 138.6 (3) |
| C16—C17—H17 | 118.7 | Yb1—N1—H1 | 110.7 |
| C18—C17—C16 | 122.6 (4) | C8—N12—C11 | 106.4 (3) |
| C18—C17—H17 | 118.7 | C8—N12—Yb1 | 127.8 (3) |
| C17—C18—H18 | 119.5 | C11—N12—Yb1 | 125.7 (2) |
| C17—C18—C19 | 121.0 (4) | C16—N15—H15 | 112.5 |
| C19—C18—H18 | 119.5 | C16—N15—Yb1 | 134.9 (3) |
| C18—C19—H19 | 120.8 | Yb1—N15—H15 | 112.5 |
| C20—C19—C18 | 118.4 (4) | C22—N26—C25 | 107.9 (3) |
| C20—C19—H19 | 120.8 | C22—N26—Yb1 | 127.4 (3) |
| C19—C20—H20 | 118.7 | C25—N26—Yb1 | 124.2 (2) |
| C19—C20—C21 | 122.7 (4) | C30—N29—H29 | 101.5 |
| C21—C20—H20 | 118.7 | C30—N29—Yb1 | 134.3 (3) |
| C16—C21—C22 | 122.2 (3) | Yb1—N29—H29 | 117.4 |
| C20—C21—C16 | 119.3 (3) | C36—N40—C39 | 107.5 (3) |
| C20—C21—C22 | 118.3 (3) | C36—N40—Yb1 | 127.6 (3) |
| N26—C22—C21 | 130.6 (3) | C39—N40—Yb1 | 123.6 (2) |
| N26—C22—O23 | 115.8 (3) | C8—O9—C10 | 106.4 (3) |
| O23—C22—C21 | 113.6 (3) | C22—O23—C24 | 106.5 (3) |
| H24A—C24—H24B | 108.9 | C36—O37—C38 | 106.4 (3) |
| C25—C24—H24A | 110.8 | N1—Yb1—N12 | 74.72 (11) |
| C25—C24—H24B | 110.8 | N1—Yb1—N15 | 167.59 (12) |
| O23—C24—H24A | 110.8 | N1—Yb1—N26 | 109.02 (11) |
| O23—C24—H24B | 110.8 | N1—Yb1—H29 | 89.3 |
| O23—C24—C25 | 104.8 (3) | N1—Yb1—N40 | 86.61 (11) |
| C27—C25—C24 | 111.8 (3) | N12—Yb1—H29 | 147.2 |
| C28—C25—C24 | 111.6 (4) | N15—Yb1—N12 | 95.23 (11) |
| C28—C25—C27 | 111.0 (3) | N15—Yb1—N26 | 77.79 (11) |
| C28—C25—N26 | 109.6 (3) | N15—Yb1—H29 | 103.0 |
| N26—C25—C24 | 101.6 (3) | N15—Yb1—N40 | 91.10 (11) |
| N26—C25—C27 | 110.9 (3) | N26—Yb1—N12 | 90.47 (10) |
| C25—C27—H27A | 109.5 | N26—Yb1—H29 | 67.5 |
| C25—C27—H27B | 109.5 | N26—Yb1—N40 | 153.87 (10) |
| C25—C27—H27C | 109.5 | N29—Yb1—N1 | 89.28 (12) |
| H27A—C27—H27B | 109.5 | N29—Yb1—N12 | 159.71 (11) |
| H27A—C27—H27C | 109.5 | N29—Yb1—N15 | 102.04 (12) |
| H27B—C27—H27C | 109.5 | N29—Yb1—N26 | 82.94 (11) |
| C25—C28—H28A | 109.5 | N29—Yb1—H29 | 16.4 |
| C25—C28—H28B | 109.5 | N29—Yb1—N40 | 76.28 (11) |
| C25—C28—H28C | 109.5 | N40—Yb1—N12 | 114.28 (10) |
| H28A—C28—H28B | 109.5 | N40—Yb1—H29 | 92.7 |
| H28A—C28—H28C | 109.5 | ||
| C2—C3—C4—C5 | −2.3 (7) | C30—C35—C36—O37 | 171.8 (3) |
| C2—C7—C8—N12 | 8.6 (7) | C31—C30—C35—C34 | 1.3 (5) |
| C2—C7—C8—O9 | −174.9 (4) | C31—C30—C35—C36 | −178.4 (3) |
| C3—C2—C7—C6 | 4.7 (5) | C31—C30—N29—Yb1 | −152.8 (3) |
| C3—C2—C7—C8 | −172.0 (4) | C31—C32—C33—C34 | 0.3 (6) |
| C3—C2—N1—Yb1 | 152.0 (3) | C32—C33—C34—C35 | −2.1 (6) |
| C3—C4—C5—C6 | 2.7 (7) | C33—C34—C35—C30 | 1.2 (6) |
| C4—C5—C6—C7 | 0.7 (7) | C33—C34—C35—C36 | −179.0 (4) |
| C5—C6—C7—C2 | −4.5 (6) | C34—C35—C36—N40 | 171.0 (4) |
| C5—C6—C7—C8 | 172.4 (4) | C34—C35—C36—O37 | −7.9 (5) |
| C6—C7—C8—N12 | −168.2 (4) | C35—C30—C31—C32 | −3.2 (6) |
| C6—C7—C8—O9 | 8.3 (5) | C35—C30—N29—Yb1 | 28.5 (5) |
| C7—C2—C3—C4 | −1.5 (6) | C35—C36—N40—C39 | −175.0 (4) |
| C7—C2—N1—Yb1 | −27.8 (6) | C35—C36—N40—Yb1 | −7.7 (6) |
| C7—C8—N12—C11 | 171.3 (4) | C35—C36—O37—C38 | −179.2 (4) |
| C7—C8—N12—Yb1 | −7.9 (6) | C38—C39—N40—C36 | −7.3 (4) |
| C7—C8—O9—C10 | 172.1 (4) | C38—C39—N40—Yb1 | −175.2 (3) |
| C10—C11—N12—C8 | 17.7 (4) | C39—C38—O37—C36 | −6.4 (5) |
| C10—C11—N12—Yb1 | −163.1 (3) | C41—C39—N40—C36 | 111.1 (4) |
| C11—C10—O9—C8 | 21.3 (5) | C41—C39—N40—Yb1 | −56.9 (4) |
| C13—C11—N12—C8 | −99.8 (4) | C42—C39—N40—C36 | −127.1 (4) |
| C13—C11—N12—Yb1 | 79.4 (4) | C42—C39—N40—Yb1 | 64.9 (4) |
| C14—C11—N12—C8 | 135.8 (4) | N1—C2—C3—C4 | 178.7 (4) |
| C14—C11—N12—Yb1 | −45.0 (5) | N1—C2—C7—C6 | −175.5 (4) |
| C16—C17—C18—C19 | −1.1 (6) | N1—C2—C7—C8 | 7.8 (6) |
| C16—C21—C22—N26 | 8.4 (6) | N12—C8—O9—C10 | −10.8 (5) |
| C16—C21—C22—O23 | −171.1 (3) | N15—C16—C17—C18 | −177.3 (4) |
| C17—C16—C21—C20 | −2.6 (5) | N15—C16—C21—C20 | 177.2 (4) |
| C17—C16—C21—C22 | 173.4 (3) | N15—C16—C21—C22 | −6.8 (6) |
| C17—C16—N15—Yb1 | 168.1 (3) | N26—C22—O23—C24 | −11.2 (5) |
| C17—C18—C19—C20 | −0.4 (6) | N29—C30—C31—C32 | 178.0 (4) |
| C18—C19—C20—C21 | 0.3 (6) | N29—C30—C35—C34 | −179.9 (3) |
| C19—C20—C21—C16 | 1.3 (6) | N29—C30—C35—C36 | 0.4 (6) |
| C19—C20—C21—C22 | −174.8 (4) | N40—C36—O37—C38 | 1.8 (5) |
| C20—C21—C22—N26 | −175.7 (4) | O9—C8—N12—C11 | −5.1 (5) |
| C20—C21—C22—O23 | 4.9 (5) | O9—C8—N12—Yb1 | 175.7 (3) |
| C21—C16—C17—C18 | 2.6 (6) | O9—C10—C11—C13 | 91.9 (4) |
| C21—C16—N15—Yb1 | −11.7 (6) | O9—C10—C11—C14 | −143.6 (4) |
| C21—C22—N26—C25 | 179.8 (4) | O9—C10—C11—N12 | −23.3 (5) |
| C21—C22—N26—Yb1 | 7.3 (6) | O23—C22—N26—C25 | −0.8 (5) |
| C21—C22—O23—C24 | 168.3 (3) | O23—C22—N26—Yb1 | −173.3 (2) |
| C24—C25—N26—C22 | 11.5 (4) | O23—C24—C25—C27 | −135.6 (4) |
| C24—C25—N26—Yb1 | −175.7 (3) | O23—C24—C25—C28 | 99.4 (4) |
| C25—C24—O23—C22 | 17.6 (4) | O23—C24—C25—N26 | −17.3 (4) |
| C27—C25—N26—C22 | 130.5 (3) | O37—C36—N40—C39 | 3.8 (4) |
| C27—C25—N26—Yb1 | −56.8 (4) | O37—C36—N40—Yb1 | 171.2 (2) |
| C28—C25—N26—C22 | −106.7 (4) | O37—C38—C39—C41 | −108.3 (4) |
| C28—C25—N26—Yb1 | 66.1 (4) | O37—C38—C39—C42 | 128.0 (4) |
| C30—C31—C32—C33 | 2.5 (6) | O37—C38—C39—N40 | 8.2 (5) |
| C30—C35—C36—N40 | −9.3 (6) |
Funding Statement
This work was funded by Natural Sciences and Engineering Research Council of Canada grant .
References
- Abbina, S., Chidara, V. K., Bian, S., Ugrinov, A. & Du, G. (2016). Chem. Sel. 1, 3175–3183.
- Abbina, S. & Du, G. (2012). Organometallics, 31, 7394–7403.
- Bauer, G., Scopelliti, R. & Hu, X. (2015a). Private communications (refcodes LUNGOS and LUNHAF). CCDC, Cambridge, England.
- Bauer, G., Wodrich, M. D., Scopelliti, R. & Hu, X. (2015b). Organometallics, 34, 289–298.
- Bennett, S. D., Core, B. A., Blake, M. P., Pope, S. J. A., Mountford, P. & Ward, B. D. (2014). Dalton Trans. 43, 5871–5885. [DOI] [PubMed]
- Bennett, S. D., Pope, S. J. A. & Ward, B. D. (2013). Chem. Commun. 49, 6072–6074. [DOI] [PubMed]
- Bian, S., Abbina, S., Lu, Z., Kolodka, E. & Du, G. (2014). Organometallics, 33, 2489–2495. [DOI] [PMC free article] [PubMed]
- Bradley, D. C., Ghotra, J. S. & Hart, F. A. (1973). J. Chem. Soc. Dalton Trans. pp. 1021–1023.
- Bruker (2003). SAINT and SMART. Bruker AXS Inc., Madison, Wisconsin, USA.
- Bruker (2016). SADABS. Bruker AXS Inc., Madison, Wisconsin, USA.
- Cabaleiro, S., Pérez–Lourido, P., Castro, J., Romero, J., García–Vázquez, J. A. & Sousa, A. (2001). Transition Met. Chem. 26, 709–716.
- Cabeza, J. A., da Silva, I., del Río, I., Gossage, R. A., Miguel, D. & Suárez, M. (2006). Dalton Trans. pp. 2450–2455. [DOI] [PubMed]
- Castro, J., Cabaleiro, S., Pérez-Lourido, P., Romero, J., García-Vázquez, J. A. & Sousa, A. (2001). Polyhedron, 20, 2329–2337.
- Castro, J., Cabaleiro, S., Pérez-Lourido, P., Romero, J., García-Vázquez, J. A. & Sousa, A. (2002). Z. Anorg. Allg. Chem. 628, 1210–1210.
- Chen, C.-T., Chan, C.-Y., Huang, C.-A., Chen, M.-T. & Peng, K.-F. (2007). Dalton Trans. pp. 4073–4078. [DOI] [PubMed]
- Chen, C.-T., Liao, C.-H., Peng, K.-F., Chen, M.-T. & Huang, T.-L. (2014). J. Organomet. Chem. 753, 9–19.
- Chen, C.-T., Weng, H.-J., Chen, M.-T., Huang, C.-A. & Peng, K.-F. (2009a). Eur. J. Inorg. Chem. pp. 2129–2135.
- Chen, M.-T., Chang, P.-J., Huang, C.-A., Peng, K.-F. & Chen, C.-T. (2009b). Dalton Trans. pp. 9068–9074. [DOI] [PubMed]
- Chen, M.-T. & Chen, C.-T. (2011). Dalton Trans. 40, 12886–12894. [DOI] [PubMed]
- Coeffard, V., Müller-Bunz, H. & Guiry, P. J. (2009). Org. Biomol. Chem. 7, 1723–1734. [DOI] [PubMed]
- Decken, A., Gossage, R. A. & Yadav, P. N. (2005). Can. J. Chem. 83, 1185–1189.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Gossage, R. A. (2009). Experiments in Green and Sustainable Chemistry, edited by H. W. Roesky & D. K. Kennepohl, pp. 19–24. Weinheim: Wiley-VCH.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- He, J., Liu, Z., Du, G., Fu, Y., Zhang, S. & Li, X. (2014). Organometallics, 33, 6103–6112.
- Hong, W. D., Leung, S. C., Amporndanai, K., Davies, J., Priestley, R. S., Nixon, G. L., Berry, N. G., Samar Hasnain, S., Antonyuk, S., Ward, S. A., Biagini, G. A. & O’Neill, P. M. (2018). ACS Med. Chem. Lett. 9, 1205–1210. [DOI] [PMC free article] [PubMed]
- Huang, Y., He, J., Liu, Z., Cai, G., Zhang, S. & Li, X. (2017). Polym. Chem. 8, 1217–1222.
- Inagaki, T., Phong, le T., Furuta, A., Ito, J. & Nishiyama, H. (2010). Chem. Eur. J. 16, 3090–3096. [DOI] [PubMed]
- Kanbur, U., Ellern, A. & Sadow, A. D. (2018). Organometallics, 37, 4409–4414.
- Kieltsch, I., Dubinina, G. G., Hamacher, C., Kaiser, A., Torres-Nieto, J., Hutchison, J. M., Klein, A., Budnikova, Y. & Vicic, D. A. (2010). Organometallics, 29, 1451–1456.
- Liu, H., He, J., Liu, Z., Lin, Z., Du, G., Zhang, S. & Li, X. (2013). Macromolecules, 46, 3257–3265.
- Lu, Z., Abbina, S., Sabin, J. R., Nemykin, V. N. & Du, G. (2013). Inorg. Chem. 52, 1454–1465. [DOI] [PubMed]
- McKeon, S. C., Muller-Bunz, H. & Guiry, P. J. (2011). Eur. J. Org. Chem. pp. 7107–7115.
- Mikami, K., Hatano, H. & Terada, M. (1999). Chem. Lett. 28, 55–56.
- Mukherjee, D., Ellern, A. & Sadow, A. D. (2010). J. Am. Chem. Soc. 132, 7582–7583. [DOI] [PubMed]
- Nakada, M. & Inoue, M. (2007). Heterocycles, 72, 133–138.
- Niwa, T. & Nakada, M. (2012). J. Am. Chem. Soc. 134, 13538–13541. [DOI] [PubMed]
- Nixon, T. D. & Ward, B. D. (2012). Chem. Commun. 48, 11790–11792. [DOI] [PubMed]
- O’Reilly, S., Aylward, M., Keogh-Hansen, C., Fitzpatrick, B., McManus, H. A., Müller-Bunz, H. & Guiry, P. J. (2015). J. Org. Chem. 80, 10177–10186. [DOI] [PubMed]
- Pawlikowski, A. V., Gray, T. S., Schoendorff, G., Baird, B., Ellern, A., Windus, T. & Sadow, A. D. (2009). Inorg. Chim. Acta, 362, 4517–4525.
- Peng, K.-F. & Chen, C.-T. (2011). Eur. J. Inorg. Chem. pp. 5182–5195.
- Resanović, S., Wylie, R. S., Quail, J. W., Foucher, D. A. & Gossage, R. A. (2011). Inorg. Chem. 50, 9930–9932. [DOI] [PubMed]
- Saiyed, A. S., Joshi, R. S. & Bedekar, A. V. (2011). J. Chem. Res. (S), 35, 408–411.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Wan, Z.-K., Choi, H.-W., Kang, F.-A., Nakajima, K., Demeke, D. & Kishi, Y. (2002). Org. Lett. 4, 4431–4434. [DOI] [PubMed]
- Wu, X., Yang, Z., Yan, X., Zhang, P., Wang, L., Guo, G., Dong, Y. & Li, X. (2018). Polym. Chem. 9, 4856–4865.
- Zou, J., Berg, D. J., Oliver, A. & Twamley, B. (2013). Organometallics, 32, 6532–6540.
- Zou, J., Berg, D. J., Stuart, D., McDonald, R. & Twamley, B. (2011). Organometallics, 30, 4958–4967.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) Na4H-L142, YbH-L133, global. DOI: 10.1107/S2056989020005034/zl2777sup1.cif
Structure factors: contains datablock(s) Na4H-L142. DOI: 10.1107/S2056989020005034/zl2777Na4H-L142sup2.hkl
Supporting information file. DOI: 10.1107/S2056989020005034/zl2777Na4H-L142sup5.mol
Structure factors: contains datablock(s) YbH-L133. DOI: 10.1107/S2056989020005034/zl2777YbH-L133sup3.hkl
Supporting information file. DOI: 10.1107/S2056989020005034/zl2777YbH-L133sup6.mol
Additional supporting information: crystallographic information; 3D view; checkCIF report