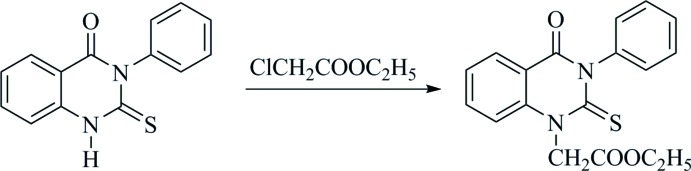In the title compound, C18H16N2O3S, the dihedral angle between the mean planes of the quinazoline and phenyl rings is 86.83 (5)°. In the crystal, C—H⋯O interactions link the molecules into infinite columns along the b-axis direction. Parallel columns interact by additional C—H⋯O hydrogen bonds.
Keywords: crystal structure, quinazolin-4-one, hydrogen bonding, Hirshfeld analysis
Abstract
The title compound, C18H16N2O3S, was synthesized by reaction of 2-mercapto-3-phenylquinazolin-4(3H)-one with ethyl chloroacetate. The quinazoline ring forms a dihedral angle of 86.83 (5)° with the phenyl ring. The terminal methyl group is disordered by a rotation of about 60° in a 0.531 (13): 0.469 (13) ratio. In the crystal, C—H⋯O hydrogen-bonding interactions result in the formation of columns running in the [010] direction. Two parallel columns further interact by C—H⋯O hydrogen bonds. The most important contributions to the surface contacts are from H⋯H (48.4%), C⋯H/H⋯C (21.5%) and O⋯H/H⋯O (18.7%) interactions, as concluded from a Hirshfeld analysis.
Chemical context
Hybrid derivatives, where quinazolin-4-one is incorporated with different heterocycles, possess a variety of biological effects including anticancer (Khalil et al., 2003 ▸; Gursoy & Karal, 2003 ▸; Gawad et al., 2010 ▸; Elfekki et al., 2014 ▸; Alanazi et al., 2016 ▸; El-Sayed et al., 2017 ▸; Nguyen et al., 2019 ▸), anticonvulsant (El-Azab et al., 2013 ▸) and antimicrobial (Pandey et al., 2009 ▸; Al-Khuzaie & Al-Majidi, 2014 ▸; Al-Majidi & Al-Khuzaie, 2015 ▸; Lv et al., 2018 ▸; Godhani et al., 2016 ▸) activities. Some derivatives of 2-mercapto-3-(4-methoxyphenyl)quinazolin-4(3H)-one containing the thiazolidine-4-one moiety have been found to have good antituberculosis activity (Godhani et al., 2016 ▸). In addition, many amide and N-substituted hydrazide compounds derived from 2-mercapto-3-phenylquinazolin-4-one have been demonstrated to have valuable biological activities such as antitumor (Al-Suwaidan et al., 2016 ▸, 2017 ▸; Mohamed et al., 2016 ▸), anticonvulsant (El-Helby & Wahab, 2003 ▸) and antibacterial (Lfta et al., 2016 ▸) activity. The capacity to increase the HDL cholesterol activity of some N-substituted compounds containing a quinazolin-4-one moiety has also been investigated (Deshmukh & Dhongade, 2004 ▸).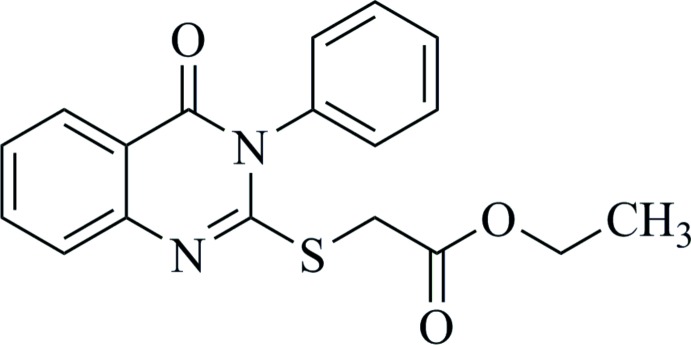
Ethyl 2-[(4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)sulfanyl]acetate is an intermediate compound in the synthesis process of both N-substituted and heterocyclic compounds containing a quinazolin-4-one moiety. The synthesis and properties of ethyl 2-[(4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)thio]acetate have therefore attracted much attention.
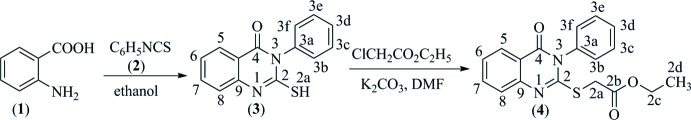
As shown in Fig. 1 ▸, 2-mercapto-3-phenylquinazolin-4(3H)-one (3) was obtained by the reaction of anthranilic acid (1) and phenyl isothiocyanate (2) (Nguyen et al., 2019 ▸). The IR spectrum of (3) shows the stretching vibrations of N—H (3217 and 3134 cm−1) and C=O (1659 cm−1) bonds, indicating that (3) exists in the thione form (Al-Majidi & Al-Khuzaie, 2015 ▸). In the 1H NMR spectrum, besides signals of nine protons in the aromatic area, there is a singlet signal with the intensity of 1H at δ 13.05 ppm attributed to the proton of the thiol group. In an alkaline medium, (3) exists in the thiolate form and reacts easily with ethyl chloroacetate to yield (4). In the IR spectrum of (4), the disappearance of the NH stretching and the presence of a strong C=O absorption at 1732 cm−1 indicate the existence of an ester compound. In the 1H NMR spectrum of (4), the signal at δ 13.05 ppm disappears and three new signals in the aliphatic area [singlet signal at δ 3.99 (2H), quartet signal at δ 4.15 (2H) and triplet signal at δ 1.23 ppm (3H)] are consistent with the presence of the –CH2COOCH2CH3 moiety in (4).
Figure 1.
Reaction scheme for the synthesis of the title compound (4).
As no X-ray crystallographic information is available for this ester, we have determined the crystal structure by single-crystal X-ray diffraction and a Hirshfeld surface analysis has been performed to gain further insight into the intermolecular interactions.
Structural commentary
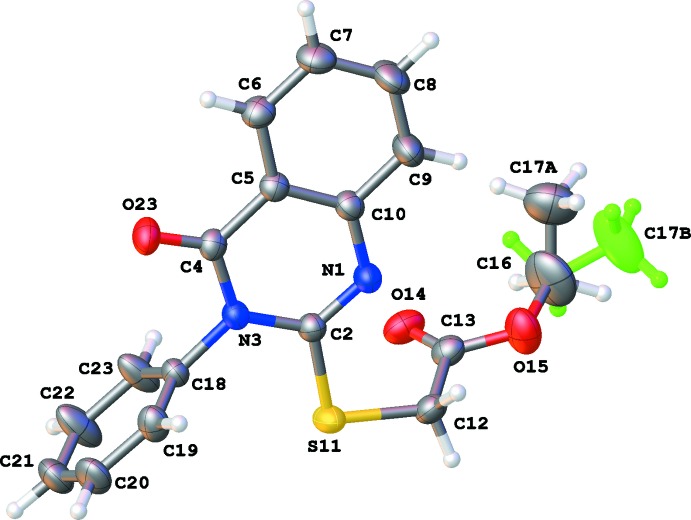
The title compound crystallizes in the space group P21/n with four molecules in the unit cell. The asymmetric unit of the title compound is illustrated in Fig. 2 ▸. The C17 methyl group is disordered over two orientations by a rotation of about 60° about the O15—C16 bond in a 0.531 (13): 0.469 (13) ratio. The quinazoline ring system is almost planar (r.m.s. deviation = 0.0207 Å). The angle between the two fused six-membered rings is 2.05 (9)°. The substituents S11, C18 and O23 deviating by −0.0951 (17), −0.140 (2) and 0.108 (2) Å, respectively, from the best plane through the quinazoline ring system. This plane makes an angle of 86.83 (5)° with the plane of the C18–C23 phenyl ring (r.m.s. deviation = 0.0052 Å). The dihedral angle between the best planes through the acetate atoms (C12, C13, O14 and O15) and the quinazoline ring system is 75.21 (5)°. A short intramolecular C16—H16B⋯O14 contact is observed [C16—H16B = 0.97 Å, H16B⋯O14 = 2.28 Å, C16⋯O14 = 2.655 (4) Å, C16—H16B⋯O14 = 102°].
Figure 2.
The molecular structure of the title compound, showing the atom-labelling scheme and displacement ellipsoids at the 50% probability level. Methyl group C17B [occupancy 0.469 (13)] is shown in green.
Theoretically, compound (3) may exist in the thione form, namely 3-phenyl-2-thioxo-2,3-dihydroquinazolin-4(1H)-one. Therefore, it could react with ethyl chloroacetate to give ethyl 2-(4-oxo-3-phenyl-2-thioxo-3,4-dihydroquinazolin-1(2H)-yl)acetate as illustrated in Fig. 3 ▸. However, our current structure determination indicates that the final product is ethyl 2-[(4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl)sulfanyl]acetate (4), which proves that in the alkaline environment, (3) converts into the thiolate form and then reacts with ethyl chloroacetate to yield the title compound (4).
Figure 3.
Reaction scheme for the thione tautomer of (3) with ethyl chloroacetate resulting in ethyl 2-(4-oxo-3-phenyl-2-thioxo-3,4-dihydroquinazolin-1(2H)-yl)acetate as reaction product.
Supramolecular features and Hirshfeld surface analysis
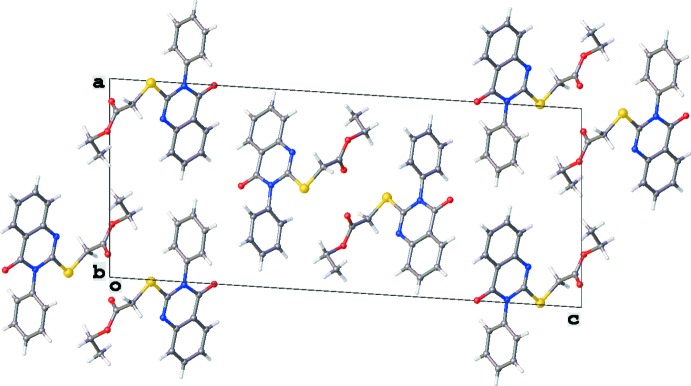
The crystal packing is mainly characterized by C—H⋯O hydrogen-bonding interactions (Table 1 ▸, Figs. 4 ▸ and 5 ▸). Columns running in the [010] direction are formed by C12—H12B⋯O14ii and C19—H19⋯O23ii interactions, which results also in a short S11⋯H23ii contact of 3.020 Å [symmetry code: (ii) x, y + 1, z]. Two parallel columns interact via C7—H7⋯O23i hydrogen-bonding interactions [symmetry code: (i) −x +  , y −
, y − 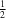 , −z +
, −z + 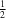 ]. No voids, C—H⋯π interactions or π–π stackings are observed in the crystal packing.
]. No voids, C—H⋯π interactions or π–π stackings are observed in the crystal packing.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C7—H7⋯O23i | 0.93 | 2.59 | 3.452 (3) | 155 |
| C12—H12B⋯O14ii | 0.97 | 2.42 | 3.311 (3) | 153 |
| C19—H19⋯O23ii | 0.93 | 2.41 | 3.236 (2) | 148 |
Symmetry codes: (i)  ; (ii)
; (ii) 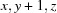 .
.
Figure 4.
View of the crystal packing of the title compound along the [010] direction. Only the major component of the disordered C17 methyl group is shown.
Figure 5.
Partial crystal packing of the title compound showing two parallel columns running in the [010] direction. Intermolecular C—H⋯O interactions are shown as red dashed lines (see Table 1 ▸ for details), C—H⋯S interactions as yellow dashed lines. Only the major component of the disordered C17 methyl group is shown.
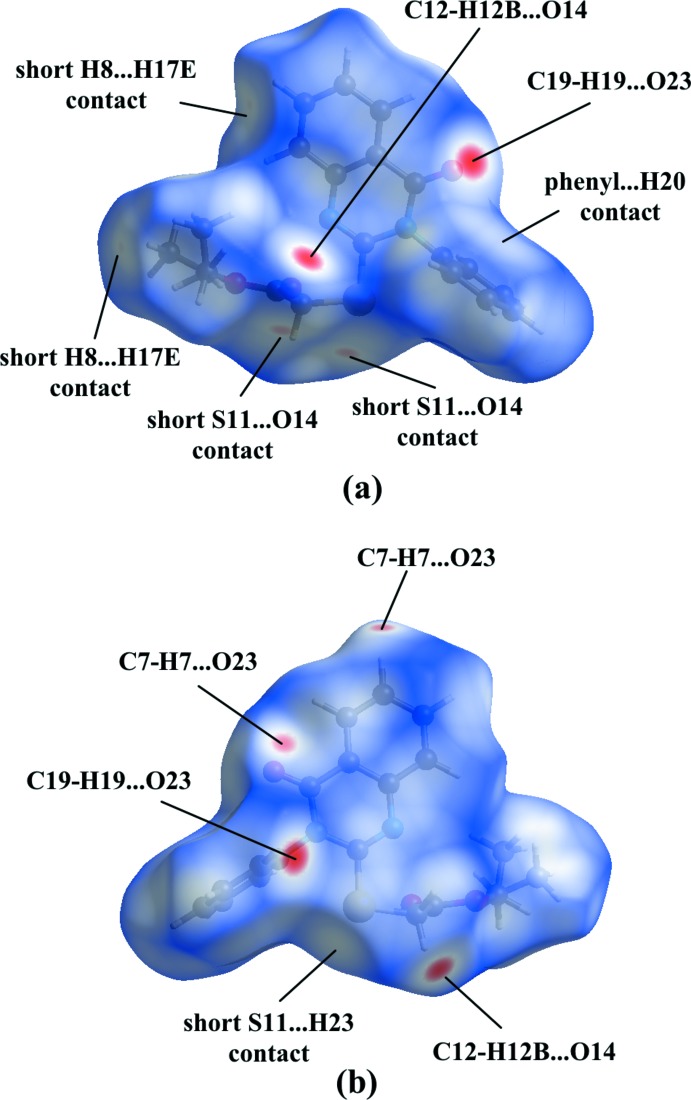
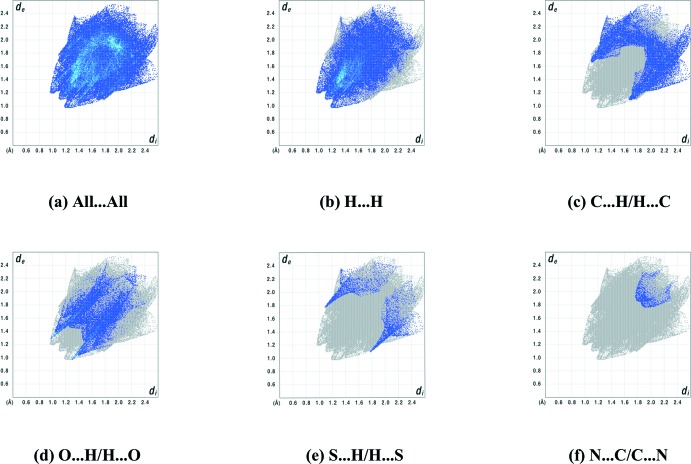
In order to gain further insight into the intermolecular interactions, a Hirshfeld surface and two-dimensional fingerprint plots were calculated using CrystalExplorer (Turner et al., 2017 ▸). The Hirshfeld surface mapped over d norm (Fig. 6 ▸) shows the expected bright-red spots near atoms O14, O23, H7, H12B and H19 involved in the C—H⋯O hydrogen-bonding interactions described above. In addition, the faint-red spots near atoms S11 and O14 indicate a short S⋯O contact [3.2128 (16) Å]. Small faint-red spots appear near atoms H8 and H17E are due to a short H8⋯H17E contact (2.352 Å). The S11⋯H23 contact mentioned is only visible as a white spot, while a white region above the C18–C23 phenyl ring is present because of the proximity of atom H20. The distance between H20 and the centroid of this phenyl ring of 3.204 Å, however, is too long for a C—H⋯π interaction. The fingerprint plots (Fig. 7 ▸) illustrate that the largest contributions to the Hirshfeld surface come from H⋯H contacts (48.4%), followed by significant contributions by reciprocal C⋯H/H⋯C (21.5%) and O⋯H/H⋯O (18.7%) contacts. Smaller contributions are from S⋯H/H⋯S (4.0%), N⋯C/C⋯N (1.6%), C⋯C (1.6%), C⋯S/S⋯C (1.4%), N⋯H/H⋯N (1.3%), S⋯O/O⋯S (1.0%), N⋯S/S⋯N (1.0%) and O⋯O contacts (0.1%).
Figure 6.
The Hirshfeld surface of (4) mapped over d norm for the title compound in the range −0.2419 to 1.2857 a.u.
Figure 7.
Full two-dimensional fingerprint plots for the title compound, showing (a) all interactions, and delineated into (b) H⋯H, (c) C⋯H/H⋯C, (d) O⋯H/H⋯O, (e) S⋯H/H⋯S and (f) N⋯C/C⋯N interactions. The d i and d e values are the closest internal and external distances (in Å) from a given point on the Hirshfeld surface.
Database survey
A search of the Cambridge Structural Database (CSD, Version 5.41, update of November 2019; Groom et al., 2016 ▸) for 4-oxo-3,4-dihydroquinazoline gave 645 hits, of which 141 have a phenyl group at position N3 and 27 have a sulfur atom at position C2. A combination of both substitutions (without a link between the two) results in a set of 10 hits, which was used for further analysis. The dihedral angle between the least-squares planes through the quinazoline and phenyl rings varies between 71.99° (CSD refcode MUDGID; Saeed et al., 2014 ▸) and 86.46° (CSD refcode GUWDIM; Rimaz et al., 2009 ▸) with an average of 81.63°. The dihedral angle does not depend on eventual ortho subsitution of the phenyl ring, as illustrated by the structures MUDGID (71.99°) and MUDNAC (85.90°; Saeed et al., 2014 ▸), which both have an o-toluidine substituent at position N3. The almost perpendicular mutual orientation of both rings is also observed for the title compound.
Synthesis and crystallization
Anthranilic acid, phenyl isothiocyanate and ethyl chloroacetate were purchased from Acros and used without purification. Melting points were measured in open capillary tubes on a Gallenkamp melting point apparatus. IR spectra (ν, cm−1) were recorded on FTIR-8400S-SHIMADZU spectrometer using KBr pellets. The NMR spectra were recorded on a Bruker Avance III spectrometer (500 MHz for 1H NMR) using residual solvent DMSO-d 6 signals as internal reference. The spin–spin coupling constants (J) are given in Hz. Peak multiplicity is reported as s (singlet), d (doublet), dd (doublet-doublet), t (triplet), q (quartet), m (multiplet). The synthetic protocol for title compound (4) is shown in Fig. 1 ▸ (Nguyen et al., 2019 ▸).
Synthesis of 2-mercapto-3-phenylquinazolin-4-one (3):
Phenyl isothiocyanate (2) (0.1 mol) was added to the solution of anthranilic acid (1) (0.1 mol) and triethylamine (3.0 mL) in absolute ethanol (200 mL). The reaction mixture then was refluxed for 4 h. After cooling to room temperature, the reaction mixture was poured into cold water. The resulting solid was filtered and recrystallized from a mixture of DMF and water, then washed with cold ethanol to give the product (3). M.p. 569 K; yield 80%. IR (KBr, cm−1): 3217, 3134 (N—H), 3028 (C—H aromatic), 1659 (C=O), 1618, 1524, 1485 (C=N, C=C aromatic). 1H NMR [Bruker XL-500, 500 MHz, d 6-DMSO, δ (ppm), J (Hz)]: 13.05 (s, 1H, H2a), 7.96 (d, 1H, 3 J = 8.0 Hz, H5), 7.80 (dd, 1H, 3 J 1 = 3 J 2 = 8.0 Hz, H7), 7.50–7.40 (m, 3H, H8,3c,3e), 7.42 (dd, 1H, 3 J 1 = 3 J 2 = 7.5 Hz, H6), 7.36 (dd, 1H, 3 J 1 = 3 J 2 = 7.5 Hz, H3d), 7.29 (d, 2H, 3 J = 7.5 Hz, H3b,3f).
Synthesis of ethyl 2-[(4-oxo-3-phenyl-3,4-dihydroquinazolin-2-yl) sulfanyl]acetate (4):
A mixture of (3) (20 mmol) and anhydrous potassium carbonate (20 mmol) in dry DMF (30 mL) was stirred for 30 min, ethyl chloroacetate (20 mmol) was then added. After refluxing for 5 h, the reaction mixture was cooled to room temperature and poured into ice-cold water. The white precipitate was filtered off and recrystallized from ethanol to afford crystals of (4). Colourless crystals, m.p. 485 K, yield 65%. IR (KBr, cm−1): 3059 (C—H aromatic), 2976, 2906 (C—H aliphatic), 1732 (C=O ester), 1680 (C=O ketone), 1607, 1598, 1468 (C=N, C=C aromatic). 1H NMR [Bruker XL-500, 500 MHz, d 6-DMSO, δ (ppm), J (Hz)]: 8.09 (d, 1H, 3 J = 8.0 Hz, H5), 7.84 (d, 1H, 3 J = 7.5 Hz, H8), 7.61-7.48 (m, 7H, H6,7,3b,3c,3d,3e,3f), 4.15 (q, 2H, 3 J = 7.0 Hz, H2c), 3.99 (s, 2H, H2a), 1.23 (t, 3H, 3 J = 7.0 Hz, H2d).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. The methyl group C17 is disordered over two positions with population parameters 0.531 (13) and 0.469 (13)]. The H atoms were placed in idealized positions and included as riding contributions with U iso(H) values of 1.2U eq or 1.5U eq of the parent atoms, with C—H distances of 0.93 (aromatic), 0.97 (CH2) and 0.96 Å (CH3). In the final cycles of refinement, two outliers were omitted.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C18H16N2O3S |
| M r | 340.39 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 293 |
| a, b, c (Å) | 11.8865 (6), 5.1298 (3), 28.2942 (14) |
| β (°) | 93.667 (4) |
| V (Å3) | 1721.72 (16) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.21 |
| Crystal size (mm) | 0.5 × 0.15 × 0.15 |
| Data collection | |
| Diffractometer | Rigaku Oxford Diffraction SuperNova, single source at offset/far, Eos |
| Absorption correction | Multi-scan (CrysAlis PRO; Rigaku OD, 2018 ▸) |
| T min, T max | 0.715, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 18522, 3533, 2875 |
| R int | 0.024 |
| (sin θ/λ)max (Å−1) | 0.625 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.043, 0.111, 1.08 |
| No. of reflections | 3533 |
| No. of parameters | 229 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.15, −0.22 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989020005071/dj2002sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020005071/dj2002Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989020005071/dj2002Isup3.cml
CCDC reference: 1996127
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C18H16N2O3S | F(000) = 712 |
| Mr = 340.39 | Dx = 1.313 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.8865 (6) Å | Cell parameters from 7343 reflections |
| b = 5.1298 (3) Å | θ = 2.9–26.9° |
| c = 28.2942 (14) Å | µ = 0.21 mm−1 |
| β = 93.667 (4)° | T = 293 K |
| V = 1721.72 (16) Å3 | Needle, colourless |
| Z = 4 | 0.5 × 0.15 × 0.15 mm |
Data collection
| Rigaku Oxford Diffraction SuperNova, Single source at offset/far, Eos diffractometer | 3533 independent reflections |
| Radiation source: micro-focus sealed X-ray tube, SuperNova (Mo) X-ray Source | 2875 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.024 |
| Detector resolution: 15.9631 pixels mm-1 | θmax = 26.4°, θmin = 2.7° |
| ω scans | h = −14→14 |
| Absorption correction: multi-scan (CrysAlisPro; Rigaku OD, 2018) | k = −6→6 |
| Tmin = 0.715, Tmax = 1.000 | l = −35→35 |
| 18522 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.043 | H-atom parameters constrained |
| wR(F2) = 0.111 | w = 1/[σ2(Fo2) + (0.0409P)2 + 0.5306P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.08 | (Δ/σ)max < 0.001 |
| 3533 reflections | Δρmax = 0.14 e Å−3 |
| 229 parameters | Δρmin = −0.22 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| N1 | 0.68674 (11) | 0.7908 (3) | 0.38689 (5) | 0.0521 (4) | |
| C2 | 0.58088 (13) | 0.8340 (3) | 0.37790 (6) | 0.0453 (4) | |
| N3 | 0.51456 (10) | 0.7160 (3) | 0.34201 (5) | 0.0438 (3) | |
| C4 | 0.55985 (14) | 0.5351 (3) | 0.31132 (6) | 0.0456 (4) | |
| C5 | 0.67846 (13) | 0.4761 (3) | 0.32214 (6) | 0.0450 (4) | |
| C6 | 0.73246 (16) | 0.2867 (4) | 0.29607 (7) | 0.0577 (5) | |
| H6 | 0.692325 | 0.197199 | 0.271885 | 0.069* | |
| C7 | 0.84377 (17) | 0.2334 (4) | 0.30609 (8) | 0.0702 (6) | |
| H7 | 0.879740 | 0.107617 | 0.288799 | 0.084* | |
| C8 | 0.90293 (17) | 0.3672 (5) | 0.34210 (9) | 0.0794 (7) | |
| H8 | 0.979020 | 0.331142 | 0.348609 | 0.095* | |
| C9 | 0.85182 (16) | 0.5518 (5) | 0.36837 (8) | 0.0709 (6) | |
| H9 | 0.893080 | 0.639601 | 0.392451 | 0.085* | |
| C10 | 0.73754 (14) | 0.6078 (4) | 0.35893 (6) | 0.0489 (4) | |
| S11 | 0.50921 (4) | 1.05531 (10) | 0.41245 (2) | 0.06194 (18) | |
| C12 | 0.62397 (16) | 1.1534 (4) | 0.45253 (7) | 0.0564 (5) | |
| H12A | 0.686170 | 1.209133 | 0.434376 | 0.068* | |
| H12B | 0.600486 | 1.301892 | 0.470701 | 0.068* | |
| C13 | 0.66465 (17) | 0.9421 (4) | 0.48617 (7) | 0.0563 (5) | |
| O14 | 0.61472 (13) | 0.7513 (3) | 0.49620 (5) | 0.0728 (4) | |
| O15 | 0.76681 (14) | 1.0046 (3) | 0.50483 (6) | 0.0907 (5) | |
| C16 | 0.8156 (3) | 0.8266 (9) | 0.54064 (14) | 0.1386 (14) | |
| H16A | 0.856216 | 0.924744 | 0.565576 | 0.166* | 0.531 (13) |
| H16B | 0.756086 | 0.729319 | 0.554614 | 0.166* | 0.531 (13) |
| H16C | 0.780435 | 0.857679 | 0.570144 | 0.166* | 0.469 (13) |
| H16D | 0.798109 | 0.649159 | 0.530875 | 0.166* | 0.469 (13) |
| C17A | 0.8872 (9) | 0.6589 (19) | 0.5203 (4) | 0.129 (4) | 0.531 (13) |
| H17A | 0.844520 | 0.538073 | 0.500469 | 0.194* | 0.531 (13) |
| H17B | 0.930264 | 0.565109 | 0.544649 | 0.194* | 0.531 (13) |
| H17C | 0.937310 | 0.755551 | 0.501607 | 0.194* | 0.531 (13) |
| C17B | 0.9293 (6) | 0.850 (3) | 0.5485 (4) | 0.152 (7) | 0.469 (13) |
| H17D | 0.964924 | 0.820026 | 0.519528 | 0.228* | 0.469 (13) |
| H17E | 0.955908 | 0.724381 | 0.571772 | 0.228* | 0.469 (13) |
| H17F | 0.947221 | 1.022473 | 0.559888 | 0.228* | 0.469 (13) |
| C18 | 0.39382 (13) | 0.7640 (3) | 0.33627 (6) | 0.0454 (4) | |
| C19 | 0.35129 (17) | 0.9418 (4) | 0.30424 (7) | 0.0620 (5) | |
| H19 | 0.399266 | 1.040013 | 0.286589 | 0.074* | |
| C20 | 0.23496 (18) | 0.9749 (5) | 0.29822 (9) | 0.0763 (6) | |
| H20 | 0.205352 | 1.096917 | 0.276528 | 0.092* | |
| C21 | 0.16481 (17) | 0.8324 (5) | 0.32340 (9) | 0.0773 (6) | |
| H21 | 0.087168 | 0.852870 | 0.318649 | 0.093* | |
| C22 | 0.20844 (17) | 0.6590 (6) | 0.35575 (11) | 0.0981 (9) | |
| H22 | 0.160210 | 0.563733 | 0.373781 | 0.118* | |
| O23 | 0.50149 (10) | 0.4422 (3) | 0.27855 (5) | 0.0628 (4) | |
| C23 | 0.32353 (16) | 0.6215 (5) | 0.36235 (9) | 0.0804 (7) | |
| H23 | 0.352638 | 0.500489 | 0.384337 | 0.096* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| N1 | 0.0442 (8) | 0.0581 (9) | 0.0531 (9) | 0.0025 (7) | −0.0027 (6) | −0.0094 (7) |
| C2 | 0.0450 (9) | 0.0470 (9) | 0.0436 (9) | 0.0019 (7) | 0.0000 (7) | −0.0009 (7) |
| N3 | 0.0390 (7) | 0.0495 (8) | 0.0429 (7) | −0.0008 (6) | 0.0014 (6) | −0.0007 (6) |
| C4 | 0.0448 (9) | 0.0500 (10) | 0.0426 (9) | −0.0079 (7) | 0.0059 (7) | −0.0003 (7) |
| C5 | 0.0436 (8) | 0.0490 (9) | 0.0432 (9) | −0.0020 (7) | 0.0087 (7) | 0.0011 (7) |
| C6 | 0.0592 (11) | 0.0621 (12) | 0.0530 (10) | 0.0015 (9) | 0.0133 (9) | −0.0057 (9) |
| C7 | 0.0640 (12) | 0.0763 (14) | 0.0722 (14) | 0.0156 (11) | 0.0198 (10) | −0.0067 (11) |
| C8 | 0.0448 (10) | 0.1025 (18) | 0.0911 (16) | 0.0192 (11) | 0.0054 (10) | −0.0125 (14) |
| C9 | 0.0454 (10) | 0.0877 (15) | 0.0784 (14) | 0.0082 (10) | −0.0050 (9) | −0.0172 (12) |
| C10 | 0.0415 (9) | 0.0551 (10) | 0.0503 (10) | 0.0012 (8) | 0.0051 (7) | −0.0006 (8) |
| S11 | 0.0562 (3) | 0.0648 (3) | 0.0638 (3) | 0.0166 (2) | −0.0040 (2) | −0.0173 (2) |
| C12 | 0.0662 (11) | 0.0442 (10) | 0.0583 (11) | 0.0019 (9) | −0.0007 (9) | −0.0083 (8) |
| C13 | 0.0682 (12) | 0.0521 (11) | 0.0486 (10) | 0.0027 (9) | 0.0039 (9) | −0.0064 (9) |
| O14 | 0.0954 (11) | 0.0510 (8) | 0.0736 (10) | −0.0013 (8) | 0.0189 (8) | −0.0009 (7) |
| O15 | 0.0849 (11) | 0.0986 (12) | 0.0845 (11) | −0.0084 (9) | −0.0259 (9) | 0.0233 (10) |
| C16 | 0.130 (3) | 0.164 (3) | 0.115 (3) | 0.017 (3) | −0.037 (2) | 0.058 (3) |
| C17A | 0.131 (7) | 0.112 (6) | 0.143 (7) | 0.033 (5) | 0.006 (5) | 0.020 (5) |
| C17B | 0.093 (5) | 0.221 (15) | 0.137 (9) | 0.000 (6) | −0.030 (5) | 0.083 (10) |
| C18 | 0.0390 (8) | 0.0472 (9) | 0.0495 (9) | 0.0018 (7) | −0.0019 (7) | 0.0005 (7) |
| C19 | 0.0591 (11) | 0.0629 (12) | 0.0625 (12) | −0.0047 (9) | −0.0084 (9) | 0.0135 (10) |
| C20 | 0.0663 (13) | 0.0759 (14) | 0.0830 (15) | 0.0141 (11) | −0.0229 (12) | 0.0171 (12) |
| C21 | 0.0445 (10) | 0.0862 (16) | 0.0997 (17) | 0.0125 (11) | −0.0074 (11) | 0.0044 (14) |
| C22 | 0.0431 (11) | 0.113 (2) | 0.139 (2) | 0.0074 (12) | 0.0148 (13) | 0.0576 (19) |
| O23 | 0.0525 (7) | 0.0796 (9) | 0.0558 (8) | −0.0115 (7) | 0.0002 (6) | −0.0174 (7) |
| C23 | 0.0441 (10) | 0.0888 (16) | 0.1085 (18) | 0.0097 (10) | 0.0063 (11) | 0.0494 (14) |
Geometric parameters (Å, º)
| N1—C2 | 1.287 (2) | S11—C12 | 1.7896 (19) |
| N1—C10 | 1.390 (2) | C12—H12A | 0.9700 |
| C17Aa—H17A | 0.9600 | C12—H12B | 0.9700 |
| C17Aa—H17B | 0.9600 | C12—C13 | 1.502 (3) |
| C17Aa—H17C | 0.9600 | C13—O14 | 1.188 (2) |
| C17Bb—H17D | 0.9600 | C13—O15 | 1.332 (2) |
| C17Bb—H17E | 0.9600 | O15—C16 | 1.456 (3) |
| C17Bb—H17F | 0.9600 | C16—H16A | 0.9700 |
| C2—N3 | 1.384 (2) | C16—H16B | 0.9700 |
| C2—S11 | 1.7541 (17) | C16—H16C | 0.9700 |
| N3—C4 | 1.402 (2) | C16—H16D | 0.9700 |
| N3—C18 | 1.4550 (19) | C16—C17A | 1.363 (8) |
| C4—C5 | 1.455 (2) | C16—C17B | 1.361 (9) |
| C4—O23 | 1.219 (2) | C18—C19 | 1.360 (2) |
| C5—C6 | 1.400 (2) | C18—C23 | 1.362 (3) |
| C5—C10 | 1.393 (2) | C19—H19 | 0.9300 |
| C6—H6 | 0.9300 | C19—C20 | 1.393 (3) |
| C6—C7 | 1.363 (3) | C20—H20 | 0.9300 |
| C7—H7 | 0.9300 | C20—C21 | 1.346 (3) |
| C7—C8 | 1.383 (3) | C21—H21 | 0.9300 |
| C8—H8 | 0.9300 | C21—C22 | 1.356 (3) |
| C8—C9 | 1.370 (3) | C22—H22 | 0.9300 |
| C9—H9 | 0.9300 | C22—C23 | 1.382 (3) |
| C9—C10 | 1.397 (2) | C23—H23 | 0.9300 |
| C2—N1—C10 | 117.28 (15) | H16Aa—C16—H16B | 108.2 |
| N1—C2—N3 | 124.99 (15) | C17Bb—C16—H16C | 108.8 |
| N1—C2—S11 | 120.31 (13) | C17Bb—C16—H16D | 108.8 |
| H17Aa—C17Aa—H17B | 109.5 | H16Cb—C16—H16D | 107.7 |
| H17Aa—C17Aa—H17C | 109.5 | H12A—C12—H12B | 107.7 |
| N3—C2—S11 | 114.70 (11) | C13—C12—S11 | 113.59 (13) |
| C2—N3—C4 | 121.38 (13) | C13—C12—H12A | 108.8 |
| C2—N3—C18 | 121.27 (13) | C13—C12—H12B | 108.8 |
| C4—N3—C18 | 117.26 (13) | O14—C13—C12 | 126.89 (19) |
| N3—C4—C5 | 114.37 (14) | O14—C13—O15 | 124.07 (19) |
| O23—C4—N3 | 120.51 (15) | O15—C13—C12 | 109.01 (17) |
| H17Ba—C17Aa—H17C | 109.5 | C13—O15—C16 | 115.9 (2) |
| H17Db—C17Bb—H17E | 109.5 | O15—C16—H16A | 109.8 |
| H17Db—C17Bb—H17F | 109.5 | O15—C16—H16B | 109.8 |
| H17Eb—C17Bb—H17F | 109.5 | O15—C16—H16C | 108.8 |
| O23—C4—C5 | 125.12 (16) | O15—C16—H16D | 108.8 |
| C6—C5—C4 | 120.27 (16) | C16—C17Aa—H17A | 109.5 |
| C10—C5—C4 | 119.46 (15) | C19—C18—N3 | 120.65 (16) |
| C10—C5—C6 | 120.27 (16) | C16—C17Aa—H17B | 109.5 |
| C5—C6—H6 | 120.0 | C19—C18—C23 | 120.39 (17) |
| C7—C6—C5 | 120.09 (19) | C23—C18—N3 | 118.92 (15) |
| C7—C6—H6 | 120.0 | C16—C17Aa—H17C | 109.5 |
| C6—C7—H7 | 120.2 | C16—C17Bb—H17D | 109.5 |
| C6—C7—C8 | 119.64 (19) | C18—C19—H19 | 120.4 |
| C8—C7—H7 | 120.2 | C18—C19—C20 | 119.14 (19) |
| C7—C8—H8 | 119.3 | C16—C17Bb—H17E | 109.5 |
| C9—C8—C7 | 121.39 (19) | C16—C17Bb—H17F | 109.5 |
| C9—C8—H8 | 119.3 | C20—C19—H19 | 120.4 |
| C8—C9—H9 | 120.1 | C19—C20—H20 | 119.6 |
| C8—C9—C10 | 119.9 (2) | C21—C20—C19 | 120.9 (2) |
| C10—C9—H9 | 120.1 | C21—C20—H20 | 119.6 |
| N1—C10—C5 | 122.43 (15) | C20—C21—H21 | 120.3 |
| N1—C10—C9 | 118.83 (17) | C20—C21—C22 | 119.37 (19) |
| C17Aa—C16—O15 | 109.5 (5) | C22—C21—H21 | 120.3 |
| C5—C10—C9 | 118.73 (17) | C21—C22—H22 | 119.5 |
| C2—S11—C12 | 99.06 (8) | C21—C22—C23 | 121.0 (2) |
| C17Bb—C16—O15 | 113.9 (5) | C23—C22—H22 | 119.5 |
| S11—C12—H12A | 108.8 | C18—C23—C22 | 119.24 (19) |
| C17Aa—C16—H16A | 109.8 | C18—C23—H23 | 120.4 |
| S11—C12—H12B | 108.8 | C22—C23—H23 | 120.4 |
| C17Aa—C16—H16B | 109.8 | ||
| N1—C2—N3—C4 | −0.6 (3) | C7—C8—C9—C10 | 0.0 (4) |
| N1—C2—N3—C18 | 176.02 (16) | C8—C9—C10—N1 | 178.3 (2) |
| N1—C2—S11—C12 | 0.18 (17) | C8—C9—C10—C5 | −1.0 (3) |
| C2—N1—C10—C5 | 0.7 (3) | C10—N1—C2—N3 | −1.3 (3) |
| C2—N1—C10—C9 | −178.59 (18) | C10—N1—C2—S11 | 178.30 (13) |
| C2—N3—C4—C5 | 2.9 (2) | C10—C5—C6—C7 | −1.1 (3) |
| C2—N3—C4—O23 | −176.56 (16) | S11—C2—N3—C4 | 179.77 (12) |
| C2—N3—C18—C19 | 97.9 (2) | S11—C2—N3—C18 | −3.6 (2) |
| C2—N3—C18—C23 | −84.4 (2) | S11—C12—C13—O14 | −19.1 (3) |
| C2—S11—C12—C13 | −68.94 (15) | S11—C12—C13—O15 | 162.70 (14) |
| N3—C2—S11—C12 | 179.82 (13) | C12—C13—O15—C16 | 176.3 (3) |
| N3—C4—C5—C6 | 176.06 (15) | O14—C13—O15—C16 | −1.9 (4) |
| N3—C4—C5—C10 | −3.3 (2) | C13—O15—C16—C17Bb | 161.5 (9) |
| N3—C18—C19—C20 | 177.27 (18) | C13—O15—C16—C17Aa | 97.7 (7) |
| N3—C18—C23—C22 | −177.5 (2) | C18—N3—C4—C5 | −173.88 (14) |
| C4—N3—C18—C19 | −85.4 (2) | C18—N3—C4—O23 | 6.7 (2) |
| C4—N3—C18—C23 | 92.4 (2) | C18—C19—C20—C21 | −0.4 (4) |
| C4—C5—C6—C7 | 179.52 (18) | C19—C18—C23—C22 | 0.3 (4) |
| C4—C5—C10—N1 | 1.7 (3) | C19—C20—C21—C22 | 1.5 (4) |
| C4—C5—C10—C9 | −179.01 (18) | C20—C21—C22—C23 | −1.7 (5) |
| C5—C6—C7—C8 | 0.0 (3) | C21—C22—C23—C18 | 0.8 (5) |
| C6—C5—C10—N1 | −177.70 (16) | O23—C4—C5—C6 | −4.5 (3) |
| C6—C5—C10—C9 | 1.6 (3) | O23—C4—C5—C10 | 176.07 (17) |
| C6—C7—C8—C9 | 0.6 (4) | C23—C18—C19—C20 | −0.5 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C7—H7···O23i | 0.93 | 2.59 | 3.452 (3) | 155 |
| C12—H12B···O14ii | 0.97 | 2.42 | 3.311 (3) | 153 |
| C19—H19···O23ii | 0.93 | 2.41 | 3.236 (2) | 148 |
Symmetry codes: (i) −x+3/2, y−1/2, −z+1/2; (ii) x, y+1, z.
Funding Statement
This work was funded by The Ministry of Education and Training of Vietnam grant B2019-SPS-02. Hercules Foundation grant AKUL/09/0035.
References
- Alanazi, A. M., Abdel-Aziz, A. A.-M., Shawer, T. Z., Ayyad, R. R., Al-Obaid, A. M., Al-Agamy, M. H. M., Maarouf, A. R. & El-Azab, A. S. (2016). J. Enzyme Inhib. Med. Chem. 31, 721–735. [DOI] [PubMed]
- Al-Khuzaie, M. G. A. & Al-Majidi, S. M. H. (2014). Iraqi J. Sci. 55, 582–593.
- Al-Majidi, S. M. H. & Al-Khuzaie, M. G. A. (2015). Asian J. Chem. 27, 756–762.
- Al-Suwaidan, I. A., Abdel-Aziz, A. A. M., Shawer, T. Z., Ayyad, R. R., Alanazi, A. M., El-Morsy, A. M., Mohamed, M. A., Abdel-Aziz, A. I., El-Sayed, M. A. A. & El-Azab, A. S. (2016). J. Enzyme Inhib. Med. Chem. 31, 78–89. [DOI] [PubMed]
- Al-Suwaidan, I. A., Abdel-Aziz, A. A. M., Shawer, T. Z., Ayyad, R. R., Alanazi, A. M., El-Morsy, A. M., Mohamed, M. A., Abdel-Aziz, A. I., El-Sayed, M. A. A. & El-Azab, A. S. (2017). J. Enzyme Inhib. Med. Chem. 32, 1229–1239.
- Deshmukh, M. B. & Dhongade, S. (2004). E-J. Chem. 1, 17–31.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- El-Azab, A. S., Abdel-Hamide, S. G., Sayed-Ahmed, M. M., Hassan, G. S., El-Hadiyah, T. M., Al-Shabanah, O. A., Al-Deeb, O. A. & El-Subbagh, H. I. (2013). Med. Chem. Res. 22, 2815–2827.
- Elfekki, I. M., Hassan, W. F. M., Elshihawy, H. E. A. E., Ali, I. A. I. & Eltamany, E. H. M. (2014). Chem. Pharm. Bull. 62, 675–694. [DOI] [PubMed]
- El-Helby, A. G. A. & Wahab, M. H. A. (2003). Acta Pharm. 53, 127–138. [PubMed]
- El-Sayed, S., Metwally, K., El-Shanawani, A. A., Abdel-Aziz, L. M., Pratsinis, H. & Kletsas, D. (2017). Chem. Cent. J. 11, 102–111. [DOI] [PMC free article] [PubMed]
- Gawad, N. M. A., Georgey, H. H., Youssef, R. M. & El-Sayed, N. A. (2010). Eur. J. Med. Chem. 45, 6058–6067. [DOI] [PubMed]
- Godhani, D. R., Jogel, A. A., Sanghani, A. M. & Mehta, J. P. (2016). Indian J. Chem. 55B, 734–746.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Gursoy, A. & Karal, N. (2003). Eur. J. Med. Chem. 38, 633–643. [DOI] [PubMed]
- Khalil, A. A., Hamide, S. G. A., Al-Obaid, A. M. & El-Subbagh, H. I. (2003). Arch. Pharm. Med. Chem. 2, 95–103. [DOI] [PubMed]
- Lfta, S. J., Ayram, N. B. & Baqer, S. M. (2016). Al-Nahrain J. Sci. 19, 1–12.
- Lv, X., Yang, L., Fan, Z. & Bao, X. (2018). J. Saudi Chem. Soc. 22, 101–109.
- Mohamed, M. A., Ayyad, R. R., Shawer, T. Z., Abdel-Aziz, A. A. M. & El-Azab, A. S. (2016). Eur. J. Med. Chem. 112, 106–113. [DOI] [PubMed]
- Nguyen, C. T., Nguyen, Q. T., Dao, P. H., Nguyen, T. L., Nguyen, P. T. & Nguyen, H. H. (2019). J. Chem., Article ID 1492316, 8 pp (https://doi. org/10.1155/2019/1492316)
- Pandey, S. K., Singh, A. & Nizamuddin, A. S. (2009). Eur. J. Med. Chem. 44, 1188–1197. [DOI] [PubMed]
- Rigaku OD (2018). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, UK.
- Rimaz, M., Khalafy, J., Tavana, K., Slepokura, K., Lis, T., Souldozi, A., Mahyari, A. T., Shajari, N. & Ramazani, A. (2009). Z. Naturforsch. Teil B, 64, 1065–1069.
- Saeed, A., Mahmood, S. & Florke, U. (2014). Turk. J. Chem. 38, 275–287.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. University of Western Australia. http://hirshfeldsurface.net
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989020005071/dj2002sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020005071/dj2002Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989020005071/dj2002Isup3.cml
CCDC reference: 1996127
Additional supporting information: crystallographic information; 3D view; checkCIF report