In the title carbohydrazide derivative, the carbohydrazide moiety is almost coplanar with the phenyl ring. The furan ring makes an angle with the phenyl ring of 34.47 (6)°. Hydrogen bonds link the molecules into a two-dimensional network, which develops parallel to bc plane.
Keywords: crystal structure, hydrazide, hydrazone, furoic acid
Abstract
The condensation of 2-furoic hydrazide and 4-dimethyl aminobenzaldehyde in ethanol yielded a yellow solid formulated as the title compound, C14H15N3O2·H2O. The crystal packing is stabilized by intermolecular O(water)—H⋯O,N(carbohydrazide) and N—H⋯O(water) hydrogen bonds, which form a two-dimensional network along the bc plane. Additional C—H⋯O interactions link the molecules into a three-dimensional network. The dihedral angle between the mean planes of the benzene and the furan ring is 34.47 (6)°. The carbohydrazide moiety, i.e., the C=N—N—C=O fragment and the benzene ring are almost coplanar, with an angle of 6.75 (9)° between their mean planes.
Chemical context
Furan is a colorless toxic chemical produced in various food items during heat processing and in some industrial processes (Delatour et al., 2020 ▸; Rehman et al., 2019 ▸; Morehouse et al., 2018 ▸; Sirot et al., 2019 ▸). It has been reported that furan can induce oxidative stress, endocrine disruption and toxic effects on the reproductive system of male rats (Rehman et al., 2019 ▸). However, other studies have shown its ability to inhibit tyrosinase, which is an enzyme responsible for many skin disorders and diseases (Barros et al., 2019 ▸). Furan derivatives, such as hydrazides, are precursors for a large variety of compounds. For example, receptors for carboxylates were prepared from furoic acid hydrazide (de la Torre et al., 1997 ▸). The biological activities of various furoic acid hydrazones have been evaluated against Mycobacterium tuberculosis (Sriram et al., 2010 ▸), myelogenous leukemia cells (Silva et al., 2014 ▸) and for tyrosinase inhibition (Dige et al., 2019 ▸). Hydrazones of this type have also been used in the study of interactions of DNA with small organic or metal–organic molecules to help the development of new drugs. Indeed, the elucidation of the mechanisms involved in the interaction of DNA with these small molecules makes it possible to develop models (Sathyadevi et al., 2012 ▸; Sennappan et al., 2019 ▸). In this paper, we report the synthesis and the characterization of the title compound, obtained from the condensation reaction between furoic acid hydrazide and 4-aminobenzaldehyde.
Structural commentary
The molecular structure of the title compound (I) with the atomic-labeling scheme is shown in Fig. 1 ▸. The asymmetric unit of I contains one molecule of the Schiff base ligand and one water molecule. The molecule adopts an E configuration with respect to the C9=N2 bond. The carbohydrazide moiety, C9=N2—N3—C10=O, is almost coplanar with the benzene ring, with an angle of 6.75 (9)° between their mean planes. The C10=O1 bond length [1.2392 (16) Å], which has double-bond character, shows that the compound did not undergo enolization as observed in some furoic hydrazide derivatives (Rodríguez-Argüelles et al., 2009 ▸). It exists only in the keto form. This form of the Schiff base is further confirmed by the N3—C10 [1.3383 (17) Å] and N2—N3 [1.3846 (14) Å] bond distances, which indicate that these are single bonds and by N2=C9 [1.2832 (17) Å], which is a double bond.
Figure 1.
An ORTEP view of the title compound, showing the atom-numbering scheme and intramolecular contacts. Displacement ellipsoids are plotted at the 50% probability level.
The O1 and N2 atoms are in a syn conformation with respect to the C10—N3 link [O1—C10—N3—N2 = −1.2 (2)°]. The dihedral angle between the benzene and the furan rings is 34.47 (6)°. The presence of the lattice water molecule differentiates the title compound I from that reported by Li & Meng (2010 ▸). In our compound, the oxygen atom of the furan ring and the oxygen atom of the carbonyl group are in a syn orientation with respect to the C10—C11 bond [O1—C10—C11—O2 = −26.44 (19)°], similar to what was observed for the compound (E)-N ’-(2-hydroxybenzylidene)furan-2-carbohydrazide by Bikas et al. (2010 ▸). This is in contrast with most hydrazones from furan-2-carbohydrazide, including the anhydrous form of the title compound, which assume an anti conformation with respect to the link between the carbonyl atom and the Cipso atom of the furan ring (Jiang, 2010 ▸; Li & Jian, 2010a ▸,b ▸,c ▸; Li & Meng, 2010 ▸).
Supramolecular features
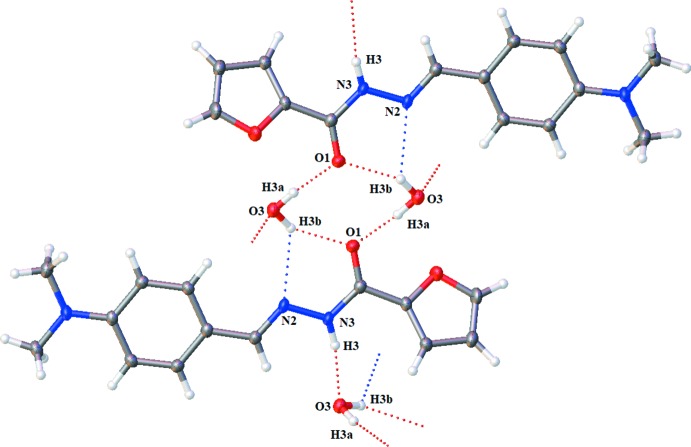
In the crystal, each independent water molecule donates hydrogen bonds to the carbonyl oxygen atom of two ligand molecules, forming a tetramer with  (8) rings (Fig. 2 ▸, Table 1 ▸). One of the hydrogen bonds donated by water is bifurcated between two acceptors, O1 and N2. The structure is built up further around the water molecules by N—H⋯Owater hydrogen bonds, thus producing layers parallel to the bc plane. Additional C—H⋯O interactions interconnect the layers and consolidate the structure into a three-dimensional network (Fig. 3 ▸).
(8) rings (Fig. 2 ▸, Table 1 ▸). One of the hydrogen bonds donated by water is bifurcated between two acceptors, O1 and N2. The structure is built up further around the water molecules by N—H⋯Owater hydrogen bonds, thus producing layers parallel to the bc plane. Additional C—H⋯O interactions interconnect the layers and consolidate the structure into a three-dimensional network (Fig. 3 ▸).
Figure 2.
Rings and connections formed by O—H⋯O, O—H⋯N and N—H⋯O hydrogen bonds (dashed lines).
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3A⋯O1i | 0.87 | 1.92 | 2.7844 (15) | 170 |
| O3—H3B⋯O1 | 0.87 | 2.12 | 2.9033 (12) | 150 |
| O3—H3B⋯N2 | 0.87 | 2.48 | 3.1681 (14) | 137 |
| N3—H3⋯O3ii | 0.88 | 1.95 | 2.7996 (14) | 162 |
| C9—H9⋯O3ii | 0.95 | 2.59 | 3.3724 (15) | 140 |
| C12—H12⋯O1iii | 0.95 | 2.43 | 3.3687 (16) | 170 |
| C7—H7⋯O3 | 0.95 | 2.71 | 3.6295 (16) | 164 |
| C1—H1A⋯O3iv | 0.98 | 2.55 | 3.4057 (17) | 146 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  .
.
Figure 3.
Crystal packing of the title compound, viewed along the b axis.
Database survey
Reflecting the interest in compounds similar to I, no fewer than 43 associated structures are included in the Cambridge Structural Database (CSD version 5.40, last update November 2018; Groom et al., 2016 ▸). Of these, KABNOS (Li & Meng, 2010 ▸) has the most similar structure to the title compound, the only differences being the presence of the water molecule and the rotation of the furan ring around the link between the carbonyl C atom and the Cipso atom of the furan ring in the title compound (see Structural commentary). Several hydrazones hits are found with the fragment furan-2-carbohydrazide. The difference between them is the substitution of the aromatic ring by a variety of groups, such as NO2 for AZILOM (Wang & Tai, 2016 ▸), hydroxyl for CEDZIX (Mohanraj et al., 2016 ▸) and DUSZEX (Bikas et al., 2010 ▸), a CH3 group for DUTJOS (Li & Jian, 2010b ▸), a methoxy group for EMOMUP (Cui et al., 2010 ▸) or a halogen atom for GAQKEQ (Bikas et al., 2012 ▸). These kinds of Schiff bases were used for preparing complexes with transition-metal or lanthanide ions. The ligand acts in a bidentate or tridentate fashion, as reported in the literature [ABUKIU (Singh et al., 2017 ▸), DAZMEX (Haba et al., 2005 ▸), FIGMEO (Maurya et al., 2005 ▸), and VIVGOY (Alagesan et al., 2014 ▸)]. One organometallic palladium complex was found containing a metal–carbon bond in a six-membered ring (TAPXEQ; Qian et al., 2017 ▸). One hit corresponds to a calcium complex, in which only the carbonyl oxygen atom is coordinated to the calcium ion (YEDCIW; Tai & Wang, 2017 ▸).
Synthesis and crystallization
All purchased chemicals and solvents were of reagent grade and were used without further purification. The melting point was determined with a Büchi 570 melting-point apparatus and is uncorrected. To a mixture of 0.5 g (3.96 mmol) of 2-furoic hydrazide and 25 ml of ethanol were added a few drops of glacial acetic acid. A solution of 0.59 g (3.96 mmol) of 4-dimethyl aminobenzaldehyde in 25 ml of ethanol was added dropwise. The resulting mixture was stirred at 323 K for 24 h. On cooling in an ice bath, a yellow solid appeared after a few minutes. The compound was filtered off, washed with water and diethyl ether, and dried at room temperature; 0.42 g of solid was obtained (yield: 37.96%). A small quantity was purified by recrystallization from a dimethylformamide solution and yellow single crystals suitable for XRD grew within a few weeks.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All H atoms of the ligand were located by HFIX, positioned geometrically and allowed to ride on their respective parent atoms, with C—H = 0.95 Å (CarH), 0.98 Å (CH3) or 0.88 Å (NH). Both H atoms of the water molecule were located in a difference-Fourier map, positioned geometrically and refined as a free rotating group with idealized geometry.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C14H15N3O2·H2O |
| M r | 275.30 |
| Crystal system, space group | Monoclinic, P21/c |
| Temperature (K) | 100 |
| a, b, c (Å) | 12.9328 (5), 11.2551 (4), 9.8092 (3) |
| β (°) | 106.245 (4) |
| V (Å3) | 1370.82 (9) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.10 |
| Crystal size (mm) | 0.20 × 0.06 × 0.06 |
| Data collection | |
| Diffractometer | XtaLAB AFC12 (RCD3) |
| Absorption correction | Gaussian (CrysAlis PRO; Rigaku OD, 2019 ▸) |
| T min, T max | 0.536, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 14982, 3102, 2649 |
| R int | 0.059 |
| (sin θ/λ)max (Å−1) | 0.649 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.048, 0.136, 1.06 |
| No. of reflections | 3102 |
| No. of parameters | 186 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.37, −0.35 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S205698902000465X/fy2143sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698902000465X/fy2143Isup2.hkl
Supporting information file. DOI: 10.1107/S205698902000465X/fy2143Isup3.cml
CCDC reference: 1994610
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are grateful to the Sonatel Foundation for financial support.
supplementary crystallographic information
Crystal data
| C14H15N3O2·H2O | F(000) = 584 |
| Mr = 275.30 | Dx = 1.334 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71075 Å |
| a = 12.9328 (5) Å | Cell parameters from 7776 reflections |
| b = 11.2551 (4) Å | θ = 2.4–31.3° |
| c = 9.8092 (3) Å | µ = 0.10 mm−1 |
| β = 106.245 (4)° | T = 100 K |
| V = 1370.82 (9) Å3 | Block, yellow |
| Z = 4 | 0.20 × 0.06 × 0.06 mm |
Data collection
| XtaLAB AFC12 (RCD3) diffractometer | 3102 independent reflections |
| Radiation source: Rotating-anode X-ray tube, Rigaku (Mo) X-ray Source | 2649 reflections with I > 2σ(I) |
| Mirror monochromator | Rint = 0.059 |
| Detector resolution: 10.0000 pixels mm-1 | θmax = 27.5°, θmin = 2.4° |
| ω scans | h = −16→15 |
| Absorption correction: gaussian (CrysAlis Pro; Rigaku OD, 2019) | k = −12→14 |
| Tmin = 0.536, Tmax = 1.000 | l = −12→12 |
| 14982 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: dual |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.048 | H-atom parameters constrained |
| wR(F2) = 0.136 | w = 1/[σ2(Fo2) + (0.0786P)2 + 0.3872P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.06 | (Δ/σ)max = 0.001 |
| 3102 reflections | Δρmax = 0.37 e Å−3 |
| 186 parameters | Δρmin = −0.35 e Å−3 |
| 0 restraints |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.94791 (8) | 0.57248 (8) | 0.63770 (9) | 0.0190 (2) | |
| O3 | 0.86017 (8) | 0.55049 (8) | 0.33216 (9) | 0.0176 (2) | |
| H3A | 0.915716 | 0.504655 | 0.340015 | 0.026* | |
| H3B | 0.867014 | 0.575836 | 0.417953 | 0.026* | |
| O2 | 1.06257 (9) | 0.57949 (9) | 0.92208 (10) | 0.0229 (3) | |
| N2 | 0.81295 (9) | 0.75421 (10) | 0.52944 (11) | 0.0157 (3) | |
| N3 | 0.88775 (9) | 0.75945 (10) | 0.66155 (11) | 0.0159 (3) | |
| H3 | 0.893005 | 0.823175 | 0.715043 | 0.019* | |
| N1 | 0.39936 (10) | 0.87858 (10) | 0.00561 (12) | 0.0199 (3) | |
| C9 | 0.74829 (11) | 0.84311 (11) | 0.50238 (13) | 0.0159 (3) | |
| H9 | 0.758000 | 0.905931 | 0.569470 | 0.019* | |
| C11 | 1.02928 (11) | 0.68189 (11) | 0.84694 (13) | 0.0154 (3) | |
| C12 | 1.07767 (11) | 0.77766 (12) | 0.92134 (14) | 0.0178 (3) | |
| H12 | 1.068840 | 0.858404 | 0.892168 | 0.021* | |
| C6 | 0.66077 (11) | 0.85061 (11) | 0.37235 (13) | 0.0151 (3) | |
| C3 | 0.48293 (11) | 0.86842 (11) | 0.12594 (13) | 0.0148 (3) | |
| C7 | 0.63960 (11) | 0.76157 (11) | 0.26809 (13) | 0.0153 (3) | |
| H7 | 0.685817 | 0.694446 | 0.280113 | 0.018* | |
| C10 | 0.95190 (11) | 0.66568 (11) | 0.70625 (13) | 0.0152 (3) | |
| C8 | 0.55339 (11) | 0.76949 (11) | 0.14894 (13) | 0.0153 (3) | |
| H8 | 0.540804 | 0.707402 | 0.080663 | 0.018* | |
| C4 | 0.50366 (11) | 0.95729 (12) | 0.23173 (13) | 0.0173 (3) | |
| H4 | 0.457358 | 1.024256 | 0.220967 | 0.021* | |
| C5 | 0.59054 (11) | 0.94761 (11) | 0.35049 (13) | 0.0174 (3) | |
| H5 | 0.603117 | 1.008900 | 0.419786 | 0.021* | |
| C13 | 1.14452 (12) | 0.73294 (13) | 1.05223 (14) | 0.0213 (3) | |
| H13 | 1.189178 | 0.778064 | 1.127900 | 0.026* | |
| C1 | 0.32241 (11) | 0.97532 (13) | −0.00914 (14) | 0.0213 (3) | |
| H1A | 0.290556 | 0.973484 | 0.070497 | 0.032* | |
| H1B | 0.265496 | 0.966333 | −0.098568 | 0.032* | |
| H1C | 0.359355 | 1.051331 | −0.009268 | 0.032* | |
| C14 | 1.13200 (12) | 0.61440 (13) | 1.04763 (14) | 0.0240 (3) | |
| H14 | 1.166921 | 0.561822 | 1.122047 | 0.029* | |
| C2 | 0.37893 (12) | 0.78798 (13) | −0.10364 (14) | 0.0227 (3) | |
| H2A | 0.446986 | 0.764697 | −0.121504 | 0.034* | |
| H2B | 0.329883 | 0.819423 | −0.191193 | 0.034* | |
| H2C | 0.346096 | 0.718529 | −0.072104 | 0.034* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0221 (6) | 0.0142 (5) | 0.0170 (5) | 0.0019 (4) | −0.0008 (4) | −0.0029 (3) |
| O3 | 0.0184 (5) | 0.0183 (5) | 0.0142 (4) | 0.0004 (4) | 0.0014 (4) | 0.0006 (3) |
| O2 | 0.0303 (6) | 0.0168 (5) | 0.0160 (5) | 0.0017 (4) | −0.0030 (4) | 0.0030 (3) |
| N2 | 0.0145 (6) | 0.0170 (5) | 0.0120 (5) | −0.0006 (4) | −0.0022 (4) | −0.0007 (4) |
| N3 | 0.0162 (6) | 0.0155 (5) | 0.0122 (5) | 0.0013 (4) | −0.0022 (4) | −0.0032 (4) |
| N1 | 0.0179 (6) | 0.0215 (6) | 0.0159 (5) | 0.0059 (4) | −0.0023 (4) | −0.0005 (4) |
| C9 | 0.0165 (7) | 0.0154 (6) | 0.0142 (6) | −0.0014 (5) | 0.0015 (5) | −0.0011 (4) |
| C11 | 0.0145 (7) | 0.0169 (6) | 0.0136 (6) | 0.0032 (5) | 0.0016 (5) | 0.0022 (4) |
| C12 | 0.0169 (7) | 0.0157 (6) | 0.0179 (6) | 0.0029 (5) | 0.0002 (5) | −0.0004 (5) |
| C6 | 0.0151 (7) | 0.0150 (6) | 0.0136 (6) | −0.0006 (5) | 0.0016 (5) | 0.0019 (4) |
| C3 | 0.0134 (7) | 0.0169 (6) | 0.0131 (6) | −0.0002 (5) | 0.0023 (5) | 0.0028 (4) |
| C7 | 0.0161 (7) | 0.0135 (6) | 0.0154 (6) | 0.0017 (5) | 0.0029 (5) | 0.0024 (4) |
| C10 | 0.0153 (7) | 0.0155 (6) | 0.0137 (6) | −0.0014 (5) | 0.0023 (5) | −0.0004 (4) |
| C8 | 0.0169 (7) | 0.0144 (6) | 0.0138 (6) | 0.0004 (5) | 0.0031 (5) | −0.0011 (4) |
| C4 | 0.0176 (7) | 0.0145 (6) | 0.0187 (6) | 0.0037 (5) | 0.0031 (5) | 0.0023 (5) |
| C5 | 0.0196 (7) | 0.0140 (6) | 0.0167 (6) | 0.0004 (5) | 0.0018 (5) | −0.0019 (4) |
| C13 | 0.0187 (7) | 0.0247 (7) | 0.0167 (6) | 0.0029 (5) | −0.0017 (5) | −0.0025 (5) |
| C1 | 0.0168 (7) | 0.0230 (7) | 0.0213 (7) | 0.0059 (5) | 0.0009 (5) | 0.0033 (5) |
| C14 | 0.0274 (8) | 0.0249 (7) | 0.0143 (6) | 0.0045 (6) | −0.0028 (5) | 0.0025 (5) |
| C2 | 0.0210 (8) | 0.0241 (7) | 0.0181 (6) | 0.0018 (5) | −0.0026 (5) | −0.0015 (5) |
Geometric parameters (Å, º)
| O1—C10 | 1.2392 (16) | C6—C5 | 1.3979 (18) |
| O3—H3A | 0.8701 | C3—C8 | 1.4163 (18) |
| O3—H3B | 0.8694 | C3—C4 | 1.4120 (18) |
| O2—C11 | 1.3709 (15) | C7—H7 | 0.9500 |
| O2—C14 | 1.3633 (16) | C7—C8 | 1.3740 (17) |
| N2—N3 | 1.3846 (14) | C8—H8 | 0.9500 |
| N2—C9 | 1.2832 (17) | C4—H4 | 0.9500 |
| N3—H3 | 0.8800 | C4—C5 | 1.3780 (18) |
| N3—C10 | 1.3383 (17) | C5—H5 | 0.9500 |
| N1—C3 | 1.3641 (16) | C13—H13 | 0.9500 |
| N1—C1 | 1.4547 (17) | C13—C14 | 1.343 (2) |
| N1—C2 | 1.4491 (17) | C1—H1A | 0.9800 |
| C9—H9 | 0.9500 | C1—H1B | 0.9800 |
| C9—C6 | 1.4517 (17) | C1—H1C | 0.9800 |
| C11—C12 | 1.3530 (18) | C14—H14 | 0.9500 |
| C11—C10 | 1.4720 (17) | C2—H2A | 0.9800 |
| C12—H12 | 0.9500 | C2—H2B | 0.9800 |
| C12—C13 | 1.4242 (18) | C2—H2C | 0.9800 |
| C6—C7 | 1.4030 (18) | ||
| H3A—O3—H3B | 104.5 | N3—C10—C11 | 113.95 (11) |
| C14—O2—C11 | 105.77 (10) | C3—C8—H8 | 119.4 |
| C9—N2—N3 | 113.86 (10) | C7—C8—C3 | 121.29 (11) |
| N2—N3—H3 | 120.7 | C7—C8—H8 | 119.4 |
| C10—N3—N2 | 118.67 (10) | C3—C4—H4 | 119.7 |
| C10—N3—H3 | 120.7 | C5—C4—C3 | 120.50 (12) |
| C3—N1—C1 | 120.22 (11) | C5—C4—H4 | 119.7 |
| C3—N1—C2 | 121.15 (11) | C6—C5—H5 | 118.9 |
| C2—N1—C1 | 118.37 (11) | C4—C5—C6 | 122.18 (12) |
| N2—C9—H9 | 119.0 | C4—C5—H5 | 118.9 |
| N2—C9—C6 | 121.99 (11) | C12—C13—H13 | 126.8 |
| C6—C9—H9 | 119.0 | C14—C13—C12 | 106.48 (12) |
| O2—C11—C10 | 115.39 (11) | C14—C13—H13 | 126.8 |
| C12—C11—O2 | 110.58 (11) | N1—C1—H1A | 109.5 |
| C12—C11—C10 | 134.03 (11) | N1—C1—H1B | 109.5 |
| C11—C12—H12 | 127.0 | N1—C1—H1C | 109.5 |
| C11—C12—C13 | 106.10 (12) | H1A—C1—H1B | 109.5 |
| C13—C12—H12 | 127.0 | H1A—C1—H1C | 109.5 |
| C7—C6—C9 | 122.97 (12) | H1B—C1—H1C | 109.5 |
| C5—C6—C9 | 119.58 (11) | O2—C14—H14 | 124.5 |
| C5—C6—C7 | 117.39 (12) | C13—C14—O2 | 111.06 (12) |
| N1—C3—C8 | 121.51 (11) | C13—C14—H14 | 124.5 |
| N1—C3—C4 | 121.20 (12) | N1—C2—H2A | 109.5 |
| C4—C3—C8 | 117.28 (12) | N1—C2—H2B | 109.5 |
| C6—C7—H7 | 119.3 | N1—C2—H2C | 109.5 |
| C8—C7—C6 | 121.34 (12) | H2A—C2—H2B | 109.5 |
| C8—C7—H7 | 119.3 | H2A—C2—H2C | 109.5 |
| O1—C10—N3 | 124.19 (12) | H2B—C2—H2C | 109.5 |
| O1—C10—C11 | 121.86 (11) | ||
| O2—C11—C12—C13 | −0.78 (16) | C12—C11—C10—N3 | −27.5 (2) |
| O2—C11—C10—O1 | −26.44 (19) | C12—C13—C14—O2 | 0.68 (18) |
| O2—C11—C10—N3 | 153.34 (12) | C6—C7—C8—C3 | 0.6 (2) |
| N2—N3—C10—O1 | −1.2 (2) | C3—C4—C5—C6 | −0.5 (2) |
| N2—N3—C10—C11 | 179.07 (11) | C7—C6—C5—C4 | −0.2 (2) |
| N2—C9—C6—C7 | 0.7 (2) | C10—C11—C12—C13 | 179.99 (15) |
| N2—C9—C6—C5 | 177.86 (13) | C8—C3—C4—C5 | 1.20 (19) |
| N3—N2—C9—C6 | −176.75 (11) | C4—C3—C8—C7 | −1.24 (19) |
| N1—C3—C8—C7 | 177.79 (12) | C5—C6—C7—C8 | 0.16 (19) |
| N1—C3—C4—C5 | −177.83 (12) | C1—N1—C3—C8 | 174.21 (12) |
| C9—N2—N3—C10 | 173.35 (12) | C1—N1—C3—C4 | −6.80 (19) |
| C9—C6—C7—C8 | 177.35 (12) | C14—O2—C11—C12 | 1.18 (16) |
| C9—C6—C5—C4 | −177.48 (12) | C14—O2—C11—C10 | −179.43 (12) |
| C11—O2—C14—C13 | −1.14 (17) | C2—N1—C3—C8 | 0.3 (2) |
| C11—C12—C13—C14 | 0.07 (17) | C2—N1—C3—C4 | 179.28 (13) |
| C12—C11—C10—O1 | 152.75 (15) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3A···O1i | 0.87 | 1.92 | 2.7844 (15) | 170 |
| O3—H3B···O1 | 0.87 | 2.12 | 2.9033 (12) | 150 |
| O3—H3B···N2 | 0.87 | 2.48 | 3.1681 (14) | 137 |
| N3—H3···O3ii | 0.88 | 1.95 | 2.7996 (14) | 162 |
| C9—H9···O3ii | 0.95 | 2.59 | 3.3724 (15) | 140 |
| C12—H12···O1iii | 0.95 | 2.43 | 3.3687 (16) | 170 |
| C7—H7···O3 | 0.95 | 2.71 | 3.6295 (16) | 164 |
| C1—H1A···O3iv | 0.98 | 2.55 | 3.4057 (17) | 146 |
Symmetry codes: (i) −x+2, −y+1, −z+1; (ii) x, −y+3/2, z+1/2; (iii) −x+2, y+1/2, −z+3/2; (iv) −x+1, y+1/2, −z+1/2.
References
- Alagesan, M., Bhuvanesh, N. S. P. & Dharmaraj, N. (2014). Dalton Trans. 43, 6087–6099. [DOI] [PubMed]
- Barros, M. R., Menezes, T. M., da Silva, L. P., Pires, D. S., Princival, J. L., Seabra, G. & Neves, J. L. (2019). Int. J. Biol. Macromol. 136, 1034–1041. [DOI] [PubMed]
- Bikas, R., Anarjan, P. M., Ng, S. W. & Tiekink, E. R. T. (2012). Acta Cryst. E68, o413–o414. [DOI] [PMC free article] [PubMed]
- Bikas, R., Hosseini Monfared, H., Kazak, C., Arslan, N. B. & Bijanzad, K. (2010). Acta Cryst. E66, o2015. [DOI] [PMC free article] [PubMed]
- Cui, Z., Li, Y., Ling, Y., Huang, J., Cui, J., Wang, R. & Yang, X. (2010). Eur. J. Med. Chem. 45, 5576–5584. [DOI] [PubMed]
- Delatour, T., Huertas-Pérez, J. F., Dubois, M., Theurillat, X., Desmarchelier, A., Ernest, M. & Stadler, R. H. (2020). Food Chem. 303, 125406. [DOI] [PubMed]
- Dige, N. C., Mahajan, P. G., Raza, H., Hassan, M., Vanjare, B. D., Hong, H., Hwan Lee, K., Latip, J. & Seo, S.-Y. (2019). Bioorg. Chem. 92, 103201. [DOI] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Haba, P. M., Diouf, O., Gaye, M., Sall, A. S., Barry, A. H., Weller, R. & Chahrazed, B. (2005). Z. Kristallogr. New Cryst. Struct. 220, 421–422.
- Jiang, J.-H. (2010). Acta Cryst. E66, o627. [DOI] [PMC free article] [PubMed]
- Li, Y.-F. & Jian, F.-F. (2010a). Acta Cryst. E66, o2157. [DOI] [PMC free article] [PubMed]
- Li, Y.-F. & Jian, F.-F. (2010b). Acta Cryst. E66, o2061. [DOI] [PMC free article] [PubMed]
- Li, Y.-F. & Jian, F.-F. (2010c). Acta Cryst. E66, o1670. [DOI] [PMC free article] [PubMed]
- Li, Y.-F. & Meng, F.-Y. (2010). Acta Cryst. E66, o2696. [DOI] [PMC free article] [PubMed]
- Maurya, M. R., Agarwal, S., Bader, C., Ebel, M. & Rehder, D. (2005). Dalton Trans. pp. 537–544. [DOI] [PubMed]
- Mohanraj, M., Ayyannan, G., Raja, G. & Jayabalakrishnan, C. (2016). J. Coord. Chem. 69, 3545–3559.
- Morehouse, K. M., Perez, G. & McNeal, T. P. (2018). Radiat. Phys. Chem. 152, 81–88.
- Qian, H., Zhang, T., Song, L., Yu, S., Yuan, Q., Sun, L., Zhang, D., Yin, Z. & Dai, Y. (2017). Eur. J. Org. Chem. 2017, 1337–1342.
- Rehman, H., Jahan, S., Ullah, I. & Winberg, S. (2019). Chemosphere, 230, 327–336. [DOI] [PubMed]
- Rigaku OD (2019). CrysAlis PRO. Rigaku Oxford Diffraction, Yarnton, England.
- Rodríguez-Argüelles, M. C., Cao, R., García-Deibe, A. M., Pelizzi, C., Sanmartín-Matalobos, J. & Zani, F. (2009). Polyhedron, 28, 2187–2195.
- Sathyadevi, P., Krishnamoorthy, P., Jayanthi, E., Butorac, R. R., Cowley, A. H. & Dharmaraj, N. (2012). Inorg. Chim. Acta, 384, 83–96.
- Sennappan, M., Krishna, P. M. & Krishna, R. H. (2019). J. Mol. Struct. 1178, 333–340.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Silva, P. P., Guerra, W., Dos Santos, G. C., Fernandes, N. G., Silveira, J. N., da Costa Ferreira, A. M., Bortolotto, T., Terenzi, H., Bortoluzzi, A. J., Neves, A. & Pereira-Maia, E. C. (2014). J. Inorg. Biochem. 132, 67–76. [DOI] [PubMed]
- Singh, Y. P., Patel, R. N., Singh, Y., Butcher, R. J., Vishakarma, P. K. & Singh, R. K. B. (2017). Polyhedron, 122, 1–15.
- Sirot, V., Rivière, G., Leconte, S., Vin, K., Traore, T., Jean, J., Carne, G., Gorecki, S., Veyrand, B., Marchand, P., Le Bizec, B., Jean-Pierre, C., Feidt, C., Vasseur, P., Lambert, M., Inthavong, C., Guérin, T. & Hulin, M. (2019). Food Chem. Toxicol. 130, 308–316. [DOI] [PubMed]
- Sriram, D., Yogeeswari, P., Vyas, D. R. K., Senthilkumar, P., Bhat, P. & Srividya, M. (2010). Bioorg. Med. Chem. Lett. 20, 4313–4316. [DOI] [PubMed]
- Tai, X.-S. & Wang, X. (2017). Crystallogr. Rep. 62, 242–245.
- Torre, M. F. de la, González, S., Campos, E. G., Mussons, M. L., Morán, J. R. & Caballero, M. C. (1997). Tetrahedron Lett. 38, 8591–8594.
- Wang, L.-H. & Tai, X.-S. (2016). Crystals, 6, 57–63.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S205698902000465X/fy2143sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698902000465X/fy2143Isup2.hkl
Supporting information file. DOI: 10.1107/S205698902000465X/fy2143Isup3.cml
CCDC reference: 1994610
Additional supporting information: crystallographic information; 3D view; checkCIF report