The title compound was obtained via a two-step synthesis involving the enole-mediated click Dimroth reaction of 4-azidoanisole with methyl 3-cyclopropyl-3-oxopropanoate leading to the 5-cyclopropyl-1-(4-methoxyphenyl)-1H-1,2,3-triazole-4-carboxylic acid and subsequent acid amidation with 4-chloroaniline by 1,1′-carbonyldiimidazole (CDI). In the extended structure, two molecules arranged in a near coplanar position relative to the triazole ring planes are interconnected by N—H⋯N and C—H⋯N hydrogen bonds into a homodimer. The dimers are linked by C—H⋯O interactions into ribbons.
Keywords: crystal structure; 1,2,3-triazole; DFT calculation; Hirshfeld surface analysis
Abstract
The title compound, C19H17ClN4O2, was obtained via a two-step synthesis involving the enol-mediated click Dimroth reaction of 4-azidoanisole with methyl 3-cyclopropyl-3-oxopropanoate leading to the 5-cyclopropyl-1-(4-methoxyphenyl)-1H-1,2,3-triazole-4-carboxylic acid and subsequent acid amidation with 4-chloroaniline by 1,1′-carbonyldiimidazole (CDI). It crystallizes in space group P21/n, with one molecule in the asymmetric unit. In the extended structure, two molecules arranged in a near coplanar fashion relative to the triazole ring planes are interconnected by N—H⋯N and C—H⋯N hydrogen bonds into a homodimer. The formation of dimers is a consequence of the above interaction and the edge-to-face stacking of aromatic rings, which are turned by 58.0 (3)° relative to each other. The dimers are linked by C—H⋯O interactions into ribbons. DFT calculations demonstrate that the frontier molecular orbitals are well separated in energy and the HOMO is largely localized on the 4-chlorophenyl amide motif while the LUMO is associated with aryltriazole grouping. A Hirshfeld surface analysis was performed to further analyse the intermolecular interactions.
Chemical context
The number of compounds containing a 1,2,3-triazolyl-4-carboxamide motif that are known to exhibit biological activity is increasing rapidly. At present, there are two approved drugs and a number of compounds are undergoing preclinical studies. For instance, rufinamide is a well-known drug among those currently marketed, which is used to treat Lennox–Gastaut syndrome (childhood-onset epilepsy) (Wheless & Vazquez, 2010 ▸). Carboxyamidotriazole is a calcium channel blocker (Figg et al., 1995 ▸) and is currently being actively investigated as an anticancer drug in vitro (Bonnefond et al., 2018 ▸). As an example of preclinical anticancer studies, the cytotoxic activity at nanomolar levels of asymmetric 1-R-N-[(1-R-1H-1,2,3-triazol-4-yl)methyl]-1H-1,2,3-triazole-4-carboxamides in B16 melanoma cells have been estimated (Elamari et al., 2013 ▸).
In our previous studies on the anticancer screening of various 1,2,3-triazoles, compounds based on 1,2,3-triazolyl-4-carboxamide scaffolds possessed the highest antiproliferative activity (Shyyka et al., 2019 ▸; Pokhodylo et al., 2013 ▸, 2014 ▸). Furthermore, a series of 6,7-disubstituted-4-(2-fluorophenoxy)quinoline derivatives possessing the 1,2,3-triazole-4-carboxamide moiety have been evaluated against c-Met kinase and five typical cancer cell lines (A549, H460, HT-29, MKN-45 and U87MG) and exhibited moderate to excellent antiproliferative activity (Zhou et al., 2014 ▸). A library of 1-benzyl-N-(2-(phenylamino)pyridin-3-yl)-1H-1,2,3-triazole-4-carboxamides was screened for their antiproliferative activity and showed promising cytotoxicity against lung cancer cell line A549 (Prasad et al., 2019 ▸). In addition to the antitumor studies, 1H-1,2,3-triazole-4-carboxamides exhibit other biological activities such as fungicidal (Wang et al., 2014 ▸), antiviral (Krajczyk et al., 2014 ▸) and antimicrobial (Jadhav et al., 2017 ▸) activities and were found to be inhibitors of the Wnt/β-catenin signalling pathway (Obianom et al., 2019 ▸). It should be noted that the diversity of such compounds can be obtained by amidation of 1H-1,2,3-triazole-4-carboxylic acids prepared by convenient Dimroth synthesis and further modifications (Pokhodylo et al., 2009 ▸, 2017 ▸, 2018 ▸; Pokhodylo, Matiychuk et al., 2010 ▸; Pokhodylo, Savka et al., 2010 ▸; Pokhodylo & Obushak, 2019 ▸). Given the considerable interest in such scaffolds for drug discovery, a detailed study of their structural features is relevant and the crystal structure of the title compound, C19H17ClN4O2, is described herein.
Structural commentary
The title compound crystallizes in the monoclinic centrosymmetric space group P21/n, with one molecule in the asymmetric unit. As shown in Fig. 1 ▸, the 4-methoxyphenyl and 1,2,3-triazole rings are turned relative to each other by 87.77 (7)° because of a significant steric hindrance of the cyclopropyl ring relative to the 4-methoxyphenyl substituent [the N1—C9—C11—C13 and N1—C9—C11—C12 torsion angles are 41.2 (4) and −31.6 (4)°, respectively]. The above angle between the planes is comparable with that for the bulky 5-(2-phenylhydrazineylidene)methyl analogue [73.3 (2)°; Pokhodylo et al., 2018 ▸] but is considerably larger than in the structure of 5-cyclopropyl-1-(3-methoxyphenyl)-1H-1,2,3-triazole-4-carboxylic acid [39.1 (2)°] in which the cyclopropyl ring is oriented to the triazole ring (Pokhodylo et al., 2017 ▸) or in 5-methyl-1-(4-nitrophenyl)-1H-1,2,3-triazol-4-ylphosphonate [45.36 (6)°; Pokhodylo et al., 2020 ▸]. In selected 5-free triazoles, 1-(3-bromo- or 4-fluorophenyl)-1H-1,2,3-triazol-4-yl)methyl methylphosphonates, this angle is 22.9 (3) and 15.7 (2)°, respectively (Pokhodylo, Shyyka et al., 2019 ▸). Within the cyclopropyl ring in the title compound, the three C—C bond lengths differ by an insignificant amount [C11—C12 = 1.491 (3), C11—C13 = 1.475 (3), C12—C13 = 1.457 (3) Å]. The amide group is turned slightly by 7.5 (3)° relative to the triazole ring while the proton of the amide group is involved in an intramolecular hydrogen bond with the heterocyclic N3 atom (Table 1 ▸). The angle between the 4-chlorophenyl and 1,2,3-triazole planes is 29.8 (1)°.
Figure 1.
The molecular structure of the title compound with displacement ellipsoids drawn at the 50% probability level.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N4—H4⋯N3 | 0.86 | 2.24 | 2.680 (3) | 112 |
| N4—H4⋯N2i | 0.86 | 2.68 | 3.491 (2) | 157 |
| C15—H15⋯O1 | 0.93 | 2.39 | 2.936 (2) | 117 |
| C19—H19⋯N2i | 0.93 | 2.68 | 3.475 (3) | 144 |
| C2—H2⋯O1ii | 0.93 | 2.53 | 3.439 (3) | 167 |
| C11—H11⋯O1 | 0.98 | 2.47 | 3.124 (2) | 124 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Supramolecular features
As shown in Fig. 2 ▸ and Table 2 ▸, the extended structure of the title compound is consolidated by a number of intermolecular interactions. Two molecules arranged in a near coplanar manner relative to the triazole ring planes are interconnected by N4—H4⋯N2i and C19—H19⋯N2i hydrogen bonds into a homodimer. Within the dimer, the edge-to-face stacked aromatic rings are tilted by 58.0 (3)°. Atom O1 of the amide group accepts both an intramolecular C—H⋯O link (with the 4-chlorophenyl and cyclopropyl H atoms) and an intermolecular C2—H2⋯O1 interaction with the 4-methoxyphenyl H atom. The last of these links neighbouring dimers into hydrogen-bonded ribbons parallel to the [010] direction (Fig. 3 ▸).
Figure 2.
The hydrogen bonding of molecules in the title compound. Hydrogen bonds are shown as dashed lines. The symmetry codes are as in Table 1 ▸.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C19H17ClN4O2 |
| M r | 368.82 |
| Crystal system, space group | Monoclinic, P21/n |
| Temperature (K) | 293 |
| a, b, c (Å) | 10.5673 (4), 8.0182 (3), 21.2318 (10) |
| β (°) | 95.282 (4) |
| V (Å3) | 1791.35 (13) |
| Z | 4 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.24 |
| Crystal size (mm) | 0.5 × 0.08 × 0.07 |
| Data collection | |
| Diffractometer | Oxford Diffraction Xcalibur3 CCD |
| Absorption correction | Multi-scan (CrysAlis RED; Oxford Diffraction, 2005 ▸) |
| T min, T max | 0.890, 0.982 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 10913, 3475, 1534 |
| R int | 0.046 |
| (sin θ/λ)max (Å−1) | 0.617 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.040, 0.053, 1.05 |
| No. of reflections | 3475 |
| No. of parameters | 236 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.14, −0.19 |
Figure 3.
A view along the a axis of the crystal packing of the title compound.
Hirshfeld surface analysis and computational study
Hirshfeld surface analysis was used to analyse the various intermolecular interactions in the title compound, through mapping the normalized contact distance (d norm) using CrystalExplorer (Turner et al., 2017 ▸; Spackman & Jayatilaka, 2009 ▸). Hirshfeld surfaces enable the visualization of intermolecular interactions by using different colours and colour intensity to represent short or long contacts and indicate the relative strength of the interactions. The most prominent interactions (the ortho-proton of the aryltriazole moiety and the carbonyl group as well as bifurcated interactions among protons of the amide group and the ortho-proton of the aryl group with the triazole ring nitrogen (N2) atoms of neighbouring molecules) can be seen in the Hirshfeld surface plot as red areas (Fig. 4 ▸). Fingerprint plots were produced to show the intermolecular surface bond distances with the regions highlighted for (C)H⋯O and (C, N)H⋯N interactions (Fig. 4 ▸). The contribution to the surface area for such contacts are 11.6% and 10.8%, respectively.
Figure 4.
(a) Hirshfeld surface for the title molecule mapped with d norm over the range −0.171 to 1.473 a.u. showing N—H⋯N, C—H⋯N and C—H⋯O hydrogen-bonded contacts. Fingerprint plots resolved into (b) N⋯H/H⋯N and (c) O⋯H/H⋯O contacts. Neighbouring molecules associated with close contacts are also shown.
The frontier molecular orbitals HOMO and LUMO were analysed to better understand the electronic charge transfer within the molecule and its electron donating and accepting ability. The molecular orbital energies were calculated using the B3LYP functional level with the 6-31+G* basis set in a vacuum with GAMESS software (Schmidt et al., 1993 ▸). The HOMO and LUMO orbitals were found to be well separated in energy and largely localized on the 4-chlorophenyl amide or aryltriazole motifs, respectively (Fig. 5 ▸). Their respective energy values were estimated to be −5.9 eV and −0.8 eV.
Figure 5.
Frontier molecular orbital energies.
Database survey
The closest related compounds containing a similar 1-aryl-1H-1,2,3-triazole-4-carboxamide skeleton to the title compound but with different substituents on the amide are: (S)-1-(4-chlorophenyl)-N-(−1-hydroxy-3-phenylpropan-2-yl)-5-methyl-1H-1,2,3-triazole-4-carboxamide (I) (CCDC refcode: ZIPSEY; Shen et al., 2013 ▸), 1-(4-chlorophenyl)-5-methyl-N-[(3-phenyl-1,2-oxazol-5-yl)methyl]-1H-1,2,3-triazole-4-carboxamide (II) (LELHOB; Niu et al., 2013 ▸), (5-methyl-1-(8-[trifluoromethyl)quinolin-4-yl]-1H-1,2,3-triazol-4-yl)morpholino)methanone (III) (LOHWIP; Anuradha et al., 2008 ▸) and 1-(3-amino-5-(3-hydroxy-3-methylbut-1-yn-1-yl)phenyl)-N-butyl-1H-1,2,3-triazole-4-carboxamide (IV) (BEBJEZ; Li et al., 2012 ▸).
Compounds (I) and (II) crystallize in the monoclinic crystal system [non-centrosymmetric space group P21 in (I) and centrosymmetric P21/c in (II)], while compounds (III) and (IV) crystallize in the triclinic space group P
 . Structure (I) contains two crystallographically independent molecules, the hydroxyl groups of which participate in intermolecular O—H⋯O hydrogen bonds. In contrast to the structure of title compound, the dihedral angles between the phenyl rings and triazole rings in (I) are −45.2 (6)° (C5—C6—N1—N2) and 39.9 (6)° (C1′—C6′—N1′—N2′). The analogous angle in (II) is 19.2 (2)°. In structure (II), the carboxamide groups connect neighbouring molecules into infinite hydrogen-bonded chains by means of N—H⋯O hydrogen bonds: these are linked by N—H⋯O (oxazole) contacts into a three-dimensional framework. Similarly to (I) and (II), structure (III) contains a 5-methyl substituent at the triazole ring and, because of significant steric hindrance of the 8-(trifluoromethyl)quinoline group, the dihedral angle between the rings is 54.7°. The phenyl and triazole rings in (IV) are close to coplanar (7.5°), while the hydroxyl, carboxamide and amino groups participate in O—H⋯O and N—H⋯O hydrogen bonds. Finally, two copper(I) π-complexes with compositions [Cu(C12H13N5O)(NO3)]·0.5H2O and [Cu(C12H13N5O)(CF3COO)] (C12H13N5O is N-allyl-5-amino-1-phenyl-1H-1,2,3-triazole-4-carboxamide) were obtained by electrochemical synthesis (ZEQTOG and ZEQTUM; Slyvka et al., 2012 ▸). Crystals of both compounds are monoclinic, space group C2/c. In both structures, the N-allyl-1H-1,2,3-triazole-4-carboxamide moiety acts as a bridging chelating ligand and forms, with the copper(I) atoms, infinite chains containing [CuC4NO] seven-membered rings.
. Structure (I) contains two crystallographically independent molecules, the hydroxyl groups of which participate in intermolecular O—H⋯O hydrogen bonds. In contrast to the structure of title compound, the dihedral angles between the phenyl rings and triazole rings in (I) are −45.2 (6)° (C5—C6—N1—N2) and 39.9 (6)° (C1′—C6′—N1′—N2′). The analogous angle in (II) is 19.2 (2)°. In structure (II), the carboxamide groups connect neighbouring molecules into infinite hydrogen-bonded chains by means of N—H⋯O hydrogen bonds: these are linked by N—H⋯O (oxazole) contacts into a three-dimensional framework. Similarly to (I) and (II), structure (III) contains a 5-methyl substituent at the triazole ring and, because of significant steric hindrance of the 8-(trifluoromethyl)quinoline group, the dihedral angle between the rings is 54.7°. The phenyl and triazole rings in (IV) are close to coplanar (7.5°), while the hydroxyl, carboxamide and amino groups participate in O—H⋯O and N—H⋯O hydrogen bonds. Finally, two copper(I) π-complexes with compositions [Cu(C12H13N5O)(NO3)]·0.5H2O and [Cu(C12H13N5O)(CF3COO)] (C12H13N5O is N-allyl-5-amino-1-phenyl-1H-1,2,3-triazole-4-carboxamide) were obtained by electrochemical synthesis (ZEQTOG and ZEQTUM; Slyvka et al., 2012 ▸). Crystals of both compounds are monoclinic, space group C2/c. In both structures, the N-allyl-1H-1,2,3-triazole-4-carboxamide moiety acts as a bridging chelating ligand and forms, with the copper(I) atoms, infinite chains containing [CuC4NO] seven-membered rings.
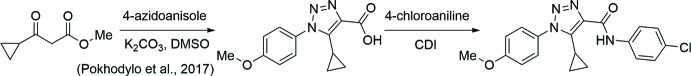
Synthesis and crystallization
The title compound was synthesized from 5-cyclopropyl-1-(4-methoxyphenyl)-1H-1,2,3-triazole-4-carboxylic acid (Pokhodylo et al., 2017 ▸) by the following procedure (Fig. 6 ▸). 5-Cyclopropyl-1-(4-methoxyphenyl)-1H-1,2,3-triazole-4-carboxylic acid 1 (1.3 g, 5.0 mmol) was added to a solution of 1,1′-carbonyldiimidazole (0.81 g, 5.0 mmol) in dry acetonitrile (25 ml) and the mixture was kept for 30 min at 323 K. Then 4-chloroaniline 2 (0.64 g, 5.0 mmol) was added, and the mixture was heated at 343 K for 1 h. After cooling to room temperature, water (30 ml) was added. The precipitate was filtered off, washed with water on a filter, recrystallized from ethanol solution, and dried in air to give the title compound as colourless prismatic crystals, m.p. 422–423 K; 1H NMR (500 MHz, DMSO-d 6) δ 10.56 (s, 1H, NH), 7.89 (d, J = 8.6 Hz, 2H, HAr), 7.58 (d, J = 8.6 Hz, 2H, HAr), 7.39 (d, J = 8.6 Hz, 2H, HAr), 7.16 (d, J = 8.6 Hz, 2H, HAr), 3.86 (s, 3H, MeO), 2.10–1.99 (m, 1H, cPrCH), 0.95–0.80 (m, 4H, cPrCH2); 13C NMR (126 MHz, DMSO-d 6) δ 160.62 (C=O or CAr—O), 159.60 (C=O or CAr—O), 142.26 (CTriazole-4), 138.87 (CTriazole-5), 138.21 (CClAr-1), 129.08 (CAr-1), 128.91 (2 × CClAr-3,5), 127.77 (2 × CAr-2,6), 127.70 (CClAr-4), 122.25 (2 × CClAr-2,6), 115.00 (2 × CAr-3,5), 56.06 (MeO), 8.09 (2 × CH2 cPr), 5.75 (CHcPr); MS m/z = 369 (M ++1); Analysis calculated for C19H17ClN4O2 (M r = 368.82), (%): C 61.88, H 4.65, N 15.19; found (%): C 61.91, H 4.74, N 15.21.
Figure 6.
Synthesis of N-(4-chlorophenyl)-5-cyclopropyl-1-(4-methoxyphenyl)-1H-1,2,3-triazole-4-carboxamide.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All H atoms were positioned geometrically with N—H = 0.86 Å and C—H = 0.93–0.98 Å and refined as riding atoms. The constraint U iso(H) = 1.2U eq(carrier) or 1.5U eq(C-methyl carrier) was applied in all cases.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989020005848/hb7901sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020005848/hb7901Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989020005848/hb7901Isup3.mol
Supporting information file. DOI: 10.1107/S2056989020005848/hb7901Isup4.cml
CCDC reference: 1999643
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
Crystal data
| C19H17ClN4O2 | F(000) = 768 |
| Mr = 368.82 | Dx = 1.368 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.5673 (4) Å | Cell parameters from 1540 reflections |
| b = 8.0182 (3) Å | θ = 0.9–1.0° |
| c = 21.2318 (10) Å | µ = 0.24 mm−1 |
| β = 95.282 (4)° | T = 293 K |
| V = 1791.35 (13) Å3 | Prism, colourless |
| Z = 4 | 0.5 × 0.08 × 0.07 mm |
Data collection
| Oxford Diffraction Xcalibur3 CCD diffractometer | 1534 reflections with I > 2σ(I) |
| ω scans | Rint = 0.046 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2005) | θmax = 26.0°, θmin = 2.7° |
| Tmin = 0.890, Tmax = 0.982 | h = −12→12 |
| 10913 measured reflections | k = −5→9 |
| 3475 independent reflections | l = −25→26 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: inferred from neighbouring sites |
| R[F2 > 2σ(F2)] = 0.040 | H-atom parameters constrained |
| wR(F2) = 0.053 | w = 1/[σ2(Fo2) + (0.0071P)2 + 0.050P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.05 | (Δ/σ)max = 0.001 |
| 3475 reflections | Δρmax = 0.14 e Å−3 |
| 236 parameters | Δρmin = −0.19 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 1.19945 (6) | 0.04278 (8) | 0.35044 (3) | 0.0860 (2) | |
| O1 | 0.80102 (13) | 0.41861 (18) | 0.54511 (7) | 0.0627 (5) | |
| O2 | 0.01792 (15) | 0.44313 (19) | 0.73741 (7) | 0.0686 (5) | |
| N4 | 0.75389 (15) | 0.1763 (2) | 0.49150 (8) | 0.0524 (5) | |
| H4 | 0.695819 | 0.101374 | 0.485272 | 0.063* | |
| N1 | 0.43143 (16) | 0.3069 (2) | 0.60195 (9) | 0.0556 (5) | |
| C14 | 0.8626 (2) | 0.1540 (2) | 0.45845 (11) | 0.0443 (6) | |
| C1 | 0.3243 (2) | 0.3477 (2) | 0.63661 (12) | 0.0492 (6) | |
| C9 | 0.5403 (2) | 0.3903 (2) | 0.59331 (10) | 0.0479 (6) | |
| C4 | 0.1154 (2) | 0.4142 (3) | 0.70082 (12) | 0.0512 (6) | |
| C10 | 0.7294 (2) | 0.3016 (3) | 0.53206 (11) | 0.0505 (6) | |
| N3 | 0.53626 (18) | 0.1428 (2) | 0.54590 (10) | 0.0763 (7) | |
| C15 | 0.9796 (2) | 0.2218 (2) | 0.47738 (10) | 0.0520 (6) | |
| H15 | 0.989197 | 0.290445 | 0.512804 | 0.062* | |
| C8 | 0.6050 (2) | 0.2842 (3) | 0.55768 (11) | 0.0486 (6) | |
| C3 | 0.11310 (19) | 0.4532 (2) | 0.63777 (11) | 0.0546 (6) | |
| H3 | 0.041147 | 0.501519 | 0.616770 | 0.066* | |
| N2 | 0.43009 (18) | 0.1551 (2) | 0.57229 (11) | 0.0831 (7) | |
| C19 | 0.84942 (19) | 0.0551 (3) | 0.40452 (10) | 0.0539 (6) | |
| H19 | 0.770296 | 0.011077 | 0.390662 | 0.065* | |
| C2 | 0.2188 (2) | 0.4199 (3) | 0.60555 (10) | 0.0546 (6) | |
| H2 | 0.218048 | 0.446667 | 0.562890 | 0.066* | |
| C16 | 1.0831 (2) | 0.1881 (3) | 0.44386 (11) | 0.0573 (7) | |
| H16 | 1.162198 | 0.233351 | 0.456901 | 0.069* | |
| C11 | 0.57821 (19) | 0.5584 (3) | 0.61661 (11) | 0.0596 (6) | |
| H11 | 0.658079 | 0.596910 | 0.601492 | 0.072* | |
| C18 | 0.9525 (2) | 0.0218 (2) | 0.37150 (10) | 0.0585 (7) | |
| H18 | 0.943421 | −0.045418 | 0.335686 | 0.070* | |
| C17 | 1.0685 (2) | 0.0881 (3) | 0.39158 (11) | 0.0533 (6) | |
| C5 | 0.2213 (2) | 0.3398 (3) | 0.73129 (11) | 0.0635 (7) | |
| H5 | 0.222148 | 0.312257 | 0.773873 | 0.076* | |
| C6 | 0.3263 (2) | 0.3056 (3) | 0.69943 (12) | 0.0623 (7) | |
| H6 | 0.397430 | 0.254642 | 0.720160 | 0.075* | |
| C13 | 0.5580 (2) | 0.6216 (3) | 0.68014 (12) | 0.0782 (8) | |
| H13A | 0.511849 | 0.550992 | 0.707204 | 0.094* | |
| H13B | 0.625552 | 0.687084 | 0.702118 | 0.094* | |
| C12 | 0.4869 (2) | 0.6965 (3) | 0.62524 (13) | 0.0809 (8) | |
| H12A | 0.510672 | 0.808059 | 0.613084 | 0.097* | |
| H12B | 0.396882 | 0.671863 | 0.618174 | 0.097* | |
| C7 | −0.0993 (2) | 0.4984 (3) | 0.70643 (12) | 0.0970 (9) | |
| H7A | −0.128382 | 0.419658 | 0.674241 | 0.146* | |
| H7B | −0.087613 | 0.605304 | 0.687433 | 0.146* | |
| H7C | −0.161144 | 0.507674 | 0.736641 | 0.146* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0723 (5) | 0.0988 (5) | 0.0925 (5) | 0.0004 (4) | 0.0379 (4) | −0.0087 (4) |
| O1 | 0.0573 (10) | 0.0568 (10) | 0.0765 (12) | −0.0187 (8) | 0.0189 (9) | −0.0171 (9) |
| O2 | 0.0585 (11) | 0.0879 (12) | 0.0627 (12) | 0.0026 (9) | 0.0231 (10) | 0.0082 (9) |
| N4 | 0.0475 (12) | 0.0478 (12) | 0.0639 (14) | −0.0110 (9) | 0.0167 (11) | −0.0108 (10) |
| N1 | 0.0512 (13) | 0.0499 (12) | 0.0682 (15) | −0.0072 (11) | 0.0186 (12) | −0.0094 (11) |
| C14 | 0.0454 (15) | 0.0413 (14) | 0.0468 (15) | −0.0044 (11) | 0.0087 (14) | −0.0008 (12) |
| C1 | 0.0468 (16) | 0.0458 (14) | 0.0565 (18) | −0.0048 (12) | 0.0130 (15) | −0.0036 (13) |
| C9 | 0.0480 (15) | 0.0428 (14) | 0.0534 (16) | −0.0059 (12) | 0.0067 (14) | −0.0038 (12) |
| C4 | 0.0516 (17) | 0.0527 (15) | 0.0507 (17) | −0.0039 (12) | 0.0127 (15) | 0.0017 (13) |
| C10 | 0.0562 (17) | 0.0463 (15) | 0.0501 (16) | 0.0002 (13) | 0.0103 (15) | −0.0024 (13) |
| N3 | 0.0591 (14) | 0.0633 (15) | 0.1115 (19) | −0.0171 (11) | 0.0347 (14) | −0.0363 (12) |
| C15 | 0.0512 (15) | 0.0488 (15) | 0.0553 (18) | −0.0033 (12) | 0.0013 (15) | −0.0112 (12) |
| C8 | 0.0436 (15) | 0.0443 (15) | 0.0589 (17) | −0.0099 (12) | 0.0110 (14) | −0.0123 (12) |
| C3 | 0.0493 (15) | 0.0617 (15) | 0.0540 (17) | 0.0043 (12) | 0.0109 (14) | 0.0055 (13) |
| N2 | 0.0665 (16) | 0.0618 (14) | 0.127 (2) | −0.0215 (11) | 0.0420 (15) | −0.0392 (13) |
| C19 | 0.0503 (15) | 0.0561 (14) | 0.0564 (16) | −0.0130 (12) | 0.0109 (14) | −0.0093 (13) |
| C2 | 0.0619 (17) | 0.0589 (15) | 0.0434 (15) | −0.0041 (14) | 0.0065 (15) | 0.0055 (12) |
| C16 | 0.0456 (16) | 0.0629 (16) | 0.0644 (19) | −0.0073 (13) | 0.0105 (15) | −0.0063 (14) |
| C11 | 0.0580 (16) | 0.0533 (15) | 0.0704 (18) | −0.0040 (13) | 0.0210 (14) | −0.0185 (14) |
| C18 | 0.0657 (17) | 0.0595 (16) | 0.0520 (16) | −0.0095 (14) | 0.0143 (15) | −0.0114 (12) |
| C17 | 0.0523 (16) | 0.0561 (15) | 0.0542 (17) | 0.0004 (13) | 0.0198 (14) | 0.0034 (13) |
| C5 | 0.0602 (18) | 0.0838 (18) | 0.0474 (17) | −0.0001 (14) | 0.0095 (16) | 0.0168 (14) |
| C6 | 0.0478 (17) | 0.0717 (17) | 0.067 (2) | 0.0024 (13) | 0.0025 (16) | 0.0143 (15) |
| C13 | 0.083 (2) | 0.0669 (18) | 0.085 (2) | −0.0152 (15) | 0.0110 (19) | −0.0197 (16) |
| C12 | 0.073 (2) | 0.0489 (16) | 0.119 (2) | 0.0045 (14) | −0.0003 (19) | −0.0099 (17) |
| C7 | 0.0580 (18) | 0.136 (3) | 0.102 (2) | 0.0258 (17) | 0.0314 (17) | 0.0219 (19) |
Geometric parameters (Å, º)
| Cl1—C17 | 1.742 (2) | C3—H3 | 0.9300 |
| O1—C10 | 1.221 (2) | C3—C2 | 1.389 (3) |
| O2—C4 | 1.366 (2) | C19—H19 | 0.9300 |
| O2—C7 | 1.419 (2) | C19—C18 | 1.375 (3) |
| N4—H4 | 0.8600 | C2—H2 | 0.9300 |
| N4—C14 | 1.412 (2) | C16—H16 | 0.9300 |
| N4—C10 | 1.364 (2) | C16—C17 | 1.367 (3) |
| N1—C1 | 1.443 (2) | C11—H11 | 0.9800 |
| N1—C9 | 1.358 (2) | C11—C13 | 1.475 (3) |
| N1—N2 | 1.370 (2) | C11—C12 | 1.491 (3) |
| C14—C15 | 1.376 (3) | C18—H18 | 0.9300 |
| C14—C19 | 1.389 (2) | C18—C17 | 1.367 (3) |
| C1—C2 | 1.370 (3) | C5—H5 | 0.9300 |
| C1—C6 | 1.374 (3) | C5—C6 | 1.379 (3) |
| C9—C8 | 1.363 (2) | C6—H6 | 0.9300 |
| C9—C11 | 1.478 (3) | C13—H13A | 0.9700 |
| C4—C3 | 1.373 (3) | C13—H13B | 0.9700 |
| C4—C5 | 1.376 (3) | C13—C12 | 1.457 (3) |
| C10—C8 | 1.476 (3) | C12—H12A | 0.9700 |
| N3—C8 | 1.357 (2) | C12—H12B | 0.9700 |
| N3—N2 | 1.303 (2) | C7—H7A | 0.9600 |
| C15—H15 | 0.9300 | C7—H7B | 0.9600 |
| C15—C16 | 1.385 (3) | C7—H7C | 0.9600 |
| C4—O2—C7 | 117.47 (18) | C15—C16—H16 | 120.1 |
| C14—N4—H4 | 115.9 | C17—C16—C15 | 119.8 (2) |
| C10—N4—H4 | 115.9 | C17—C16—H16 | 120.1 |
| C10—N4—C14 | 128.10 (18) | C9—C11—H11 | 113.1 |
| C9—N1—C1 | 132.18 (19) | C9—C11—C12 | 124.1 (2) |
| C9—N1—N2 | 110.40 (17) | C13—C11—C9 | 124.2 (2) |
| N2—N1—C1 | 117.40 (17) | C13—C11—H11 | 113.1 |
| C15—C14—N4 | 123.7 (2) | C13—C11—C12 | 58.84 (14) |
| C15—C14—C19 | 119.1 (2) | C12—C11—H11 | 113.1 |
| C19—C14—N4 | 117.2 (2) | C19—C18—H18 | 120.2 |
| C2—C1—N1 | 119.5 (2) | C17—C18—C19 | 119.7 (2) |
| C2—C1—C6 | 120.7 (2) | C17—C18—H18 | 120.2 |
| C6—C1—N1 | 119.8 (2) | C16—C17—Cl1 | 119.63 (19) |
| N1—C9—C8 | 103.98 (17) | C16—C17—C18 | 120.8 (2) |
| N1—C9—C11 | 127.6 (2) | C18—C17—Cl1 | 119.56 (18) |
| C8—C9—C11 | 128.4 (2) | C4—C5—H5 | 119.6 |
| O2—C4—C3 | 124.7 (2) | C4—C5—C6 | 120.8 (2) |
| O2—C4—C5 | 115.5 (2) | C6—C5—H5 | 119.6 |
| C3—C4—C5 | 119.9 (2) | C1—C6—C5 | 119.1 (2) |
| O1—C10—N4 | 124.08 (19) | C1—C6—H6 | 120.5 |
| O1—C10—C8 | 122.9 (2) | C5—C6—H6 | 120.5 |
| N4—C10—C8 | 113.0 (2) | C11—C13—H13A | 117.7 |
| N2—N3—C8 | 108.94 (17) | C11—C13—H13B | 117.7 |
| C14—C15—H15 | 119.9 | H13A—C13—H13B | 114.8 |
| C14—C15—C16 | 120.2 (2) | C12—C13—C11 | 61.14 (16) |
| C16—C15—H15 | 119.9 | C12—C13—H13A | 117.7 |
| C9—C8—C10 | 130.9 (2) | C12—C13—H13B | 117.7 |
| N3—C8—C9 | 109.70 (18) | C11—C12—H12A | 117.8 |
| N3—C8—C10 | 119.4 (2) | C11—C12—H12B | 117.8 |
| C4—C3—H3 | 120.2 | C13—C12—C11 | 60.02 (15) |
| C4—C3—C2 | 119.6 (2) | C13—C12—H12A | 117.8 |
| C2—C3—H3 | 120.2 | C13—C12—H12B | 117.8 |
| N3—N2—N1 | 106.97 (17) | H12A—C12—H12B | 114.9 |
| C14—C19—H19 | 119.8 | O2—C7—H7A | 109.5 |
| C18—C19—C14 | 120.4 (2) | O2—C7—H7B | 109.5 |
| C18—C19—H19 | 119.8 | O2—C7—H7C | 109.5 |
| C1—C2—C3 | 120.0 (2) | H7A—C7—H7B | 109.5 |
| C1—C2—H2 | 120.0 | H7A—C7—H7C | 109.5 |
| C3—C2—H2 | 120.0 | H7B—C7—H7C | 109.5 |
| O1—C10—C8—C9 | 6.7 (4) | C4—C5—C6—C1 | 0.4 (3) |
| O1—C10—C8—N3 | −173.6 (2) | C10—N4—C14—C15 | 23.3 (4) |
| O2—C4—C3—C2 | 179.10 (19) | C10—N4—C14—C19 | −158.0 (2) |
| O2—C4—C5—C6 | −179.5 (2) | C15—C14—C19—C18 | 1.7 (3) |
| N4—C14—C15—C16 | 177.1 (2) | C15—C16—C17—Cl1 | −178.77 (17) |
| N4—C14—C19—C18 | −177.10 (19) | C15—C16—C17—C18 | 0.6 (3) |
| N4—C10—C8—C9 | −171.8 (2) | C8—C9—C11—C13 | −139.8 (3) |
| N4—C10—C8—N3 | 7.9 (3) | C8—C9—C11—C12 | 147.4 (3) |
| N1—C1—C2—C3 | 177.23 (19) | C8—N3—N2—N1 | −0.5 (3) |
| N1—C1—C6—C5 | −177.6 (2) | C3—C4—C5—C6 | 0.8 (3) |
| N1—C9—C8—C10 | 179.6 (2) | N2—N1—C1—C2 | −86.6 (2) |
| N1—C9—C8—N3 | −0.1 (3) | N2—N1—C1—C6 | 89.8 (3) |
| N1—C9—C11—C13 | 41.2 (4) | N2—N1—C9—C8 | −0.2 (2) |
| N1—C9—C11—C12 | −31.6 (4) | N2—N1—C9—C11 | 179.1 (2) |
| C14—N4—C10—O1 | 1.2 (4) | N2—N3—C8—C9 | 0.4 (3) |
| C14—N4—C10—C8 | 179.7 (2) | N2—N3—C8—C10 | −179.4 (2) |
| C14—C15—C16—C17 | 0.5 (3) | C19—C14—C15—C16 | −1.6 (3) |
| C14—C19—C18—C17 | −0.7 (3) | C19—C18—C17—Cl1 | 178.88 (17) |
| C1—N1—C9—C8 | 178.1 (2) | C19—C18—C17—C16 | −0.5 (3) |
| C1—N1—C9—C11 | −2.7 (4) | C2—C1—C6—C5 | −1.2 (3) |
| C1—N1—N2—N3 | −178.2 (2) | C11—C9—C8—C10 | 0.4 (4) |
| C9—N1—C1—C2 | 95.2 (3) | C11—C9—C8—N3 | −179.3 (2) |
| C9—N1—C1—C6 | −88.4 (3) | C5—C4—C3—C2 | −1.2 (3) |
| C9—N1—N2—N3 | 0.4 (3) | C6—C1—C2—C3 | 0.8 (3) |
| C9—C11—C13—C12 | −112.4 (3) | C7—O2—C4—C3 | 8.1 (3) |
| C9—C11—C12—C13 | 112.6 (3) | C7—O2—C4—C5 | −171.63 (19) |
| C4—C3—C2—C1 | 0.4 (3) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N4—H4···N3 | 0.86 | 2.24 | 2.680 (3) | 112 |
| N4—H4···N2i | 0.86 | 2.68 | 3.491 (2) | 157 |
| C15—H15···O1 | 0.93 | 2.39 | 2.936 (2) | 117 |
| C19—H19···N2i | 0.93 | 2.68 | 3.475 (3) | 144 |
| C2—H2···O1ii | 0.93 | 2.53 | 3.439 (3) | 167 |
| C11—H11···O1 | 0.98 | 2.47 | 3.124 (2) | 124 |
Symmetry codes: (i) −x+1, −y, −z+1; (ii) −x+1, −y+1, −z+1.
Funding Statement
This work was funded by Ministry of Education and Science of Ukraine grant .
References
- Anuradha, N., Thiruvalluvar, A., Mahalinga, M. & Butcher, R. J. (2008). Acta Cryst. E64, o2375. [DOI] [PMC free article] [PubMed]
- Bonnefond, M., Florent, R., Lenoir, S., Lambert, B., Abeilard, E., Giffard, F., Louis, M., Elie, N., Briand, M., Vivien, D., Poulain, L., Gauduchon, P. & N’Diaye, M. (2018). Oncotarget, 9, 33896–33911. [DOI] [PMC free article] [PubMed]
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Elamari, H., Slimi, R., Chabot, G. G., Quentin, L., Scherman, D. & Girard, C. (2013). Eur. J. Med. Chem. 60, 360–364. [DOI] [PubMed]
- Figg, W. D., Cole, K. A., Reed, E., Steinberg, S. M., Piscitelli, S. C., Davis, P. A., Soltis, M. J., Jacob, J., Boudoulas, S. & Goldspiel, B. (1995). Clin. Cancer Res. 1, 797–803. [PubMed]
- Jadhav, R. P., Raundal, H. N., Patil, A. A. & Bobade, V. D. (2017). J. Saudi Chem. Soc. 21, 152–159.
- Krajczyk, A., Kulinska, K., Kulinski, T., Hurst, B. L., Day, C. W., Smee, D. F., Ostrowski, T., Januszczyk, P. & Zeidler, J. (2014). Antivir. Chem. Chemother. 23, 161–171. [DOI] [PubMed]
- Li, Y.-J., Xu, L., Yang, W.-L., Liu, H.-B., Lai, S.-W., Che, C.-M. & Li, Y.-L. (2012). Chem. Eur. J. 18, 4782–4790. [DOI] [PubMed]
- Niu, T.-F., Lv, M.-F., Wang, L., Yi, W.-B. & Cai, C. (2013). Org. Biomol. Chem. 11, 1040–1048. [DOI] [PubMed]
- Obianom, O. N., Ai, Y., Li, Y., Yang, W., Guo, D., Yang, H., Sakamuru, S., Xia, M., Xue, F. & Shu, Y. (2019). J. Med. Chem. 62, 727–741. [DOI] [PMC free article] [PubMed]
- Oxford Diffraction (2005). CrysAlis PRO. Oxford Diffraction, Abingdon, England.
- Pokhodylo, N. T., Matiychuk, V. S. & Obushak, M. D. (2010). Synth. Commun. 40, 1932–1938.
- Pokhodylo, N. T. & Obushak, M. D. (2019). Russ. J. Org. Chem. 55, 1241–1243.
- Pokhodylo, N. T., Savka, R. D., Pidlypnyi, N. I., Matiychuk, V. S. & Obushak, M. D. (2010). Synth. Commun. 40, 391–399.
- Pokhodylo, N. T., Shyyka, O. Ya., Goreshnik, E. A. & Obushak, M. D. (2020). ChemistrySelect, 5, 260–264.
- Pokhodylo, N. T., Shyyka, O. Ya. & Matiychuk, V. S. (2014). Med. Chem. Res. 23, 2426–2438.
- Pokhodylo, N. T., Shyyka, O. Ya., Matiychuk, V. S., Obushak, M. D. & Pavlyuk, V. V. (2017). ChemistrySelect, 2, 5871–5876.
- Pokhodylo, N. T., Shyyka, O. Ya. & Obushak, M. D. (2018). Chem. Heterocycl. Compd, 54, 773–779.
- Pokhodylo, N. T., Shyyka, O. Ya., Skrobala, V. E. & Matiychuk, V. S. (2013). Clinical Pharmacy, Pharmacotherapy & Medical Standardization, Vol. 16–17, pp. 92–97. (In Ukrainian) http://nbuv.gov.ua/UJRN/Kff_2012_3_15
- Pokhodylo, N. T., Shyyka, O. Ya., Tupychak, M. A., Slyvka, Yu. I. & Obushak, M. D. (2019). Chem. Heterocycl. C. 55, 374–378.
- Pokhodylo, N. T., Teslenko, Y. O., Matiychuk, V. S. & Obushak, M. D. (2009). Synthesis, pp. 2741–2748.
- Prasad, B., Lakshma Nayak, V., Srikanth, P. S., Baig, M. F., Subba Reddy, N. V., Babu, K. S. & Kamal, A. (2019). Bioorg. Chem. 83, 535–548. [DOI] [PubMed]
- Schmidt, M. W., Baldridge, K. K., Boatz, J. A., Elbert, S. T., Gordon, M. S., Jensen, J. H., Koseki, S., Matsunaga, N., Nguyen, K. A., Su, S. J., Windus, T. L., Dupuis, M. & Montgomery, J. A. (1993). J. Comput. Chem. 14, 1347–1363.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Shen, G.-L., Chen, Z.-B., Wu, Z.-F. & Dong, H.-S. (2013). J. Heterocycl. Chem. 50, 781–786.
- Shyyka, O. Ya., Pokhodylo, N. T. & Finiuk, N. S. (2019). Biopolym. Cell, 35, 321–330.
- Slyvka, Yu. I., Pavlyuk, A. V., Ardan, B. R., Pokhodilo, N. T., Goreshnik, E. A. & Demchenko, P. Yu. (2012). Russ. J. Inorg. Chem. 57, 815–821.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Turner, M. J., Mckinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17. The University of Western Australia. http://hirshfeldsurface.net
- Wang, Z., Gao, Y., Hou, Y., Zhang, C., Yu, S. J., Bian, Q., Li, Z. M. & Zhao, W. G. (2014). Eur. J. Med. Chem. 86, 87–94. [DOI] [PubMed]
- Wheless, J. W. & Vazquez, B. (2010). Epilepsy Curr. 10, 1–6. [DOI] [PMC free article] [PubMed]
- Zhou, S., Liao, H., Liu, M., Feng, G., Fu, B., Li, R., Cheng, M., Zhao, Y. & Gong, P. (2014). Bioorg. Med. Chem. 22, 6438–6452. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989020005848/hb7901sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989020005848/hb7901Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989020005848/hb7901Isup3.mol
Supporting information file. DOI: 10.1107/S2056989020005848/hb7901Isup4.cml
CCDC reference: 1999643
Additional supporting information: crystallographic information; 3D view; checkCIF report