Ducks are frequently infected with avian influenza viruses belonging to multiple subtypes. They represent an important reservoir species of avian influenza viruses, which can occasionally be transmitted to other bird species or mammals, including humans. Ducks thus have a central role in the epidemiology of influenza virus infection. Importantly, ducks usually show little or no clinical signs even following infection with a highly pathogenic avian influenza virus. We provide evidence that the microbiota contributes to the control of influenza virus replication in ducks by modulating the antiviral immune response. Ducks are able to control influenza virus replication more efficiently when they have an intact intestinal microbiota. Therefore, maintaining a healthy microbiota by limiting perturbations to its composition should contribute to the prevention of avian influenza virus spread from the duck reservoir.
KEYWORDS: avian viruses, duck, enteric viruses, immunoglobulins, influenza, innate immunity, interferons, microbiota, microflora, normal flora
ABSTRACT
Ducks usually show little or no clinical signs following highly pathogenic avian influenza virus infection. In order to analyze whether the microbiota could contribute to the control of influenza virus replication in ducks, we used a broad-spectrum oral antibiotic treatment to deplete the microbiota before infection with a highly pathogenic H5N9 avian influenza virus. Antibiotic-treated ducks and nontreated control ducks did not show any clinical signs following H5N9 virus infection. We did not detect any significant difference in virus titers neither in the respiratory tract nor in the brain nor spleen. However, we found that antibiotic-treated H5N9 virus-infected ducks had significantly increased intestinal virus excretion at days 3 and 5 postinfection. This was associated with a significantly decreased antiviral immune response in the intestine of antibiotic-treated ducks. Our findings highlight the importance of an intact microbiota for an efficient control of avian influenza virus replication in ducks.
IMPORTANCE Ducks are frequently infected with avian influenza viruses belonging to multiple subtypes. They represent an important reservoir species of avian influenza viruses, which can occasionally be transmitted to other bird species or mammals, including humans. Ducks thus have a central role in the epidemiology of influenza virus infection. Importantly, ducks usually show little or no clinical signs even following infection with a highly pathogenic avian influenza virus. We provide evidence that the microbiota contributes to the control of influenza virus replication in ducks by modulating the antiviral immune response. Ducks are able to control influenza virus replication more efficiently when they have an intact intestinal microbiota. Therefore, maintaining a healthy microbiota by limiting perturbations to its composition should contribute to the prevention of avian influenza virus spread from the duck reservoir.
INTRODUCTION
Ducks have a central role in the epidemiology of influenza virus (1, 2). They have been shown to shed viruses belonging to multiple subtypes and represent the main reservoir of influenza viruses that can occasionally be transmitted to other animal species, including gallinaceous poultry, and occasionally mammals. Low pathogenic avian influenza viruses (LPAIVs) of the H5 and H7 subtype have the capacity to evolve to highly pathogenic avian influenza viruses (HPAIVs). This evolution is due to the acquisition of a polybasic cleavage site at the hemagglutinin (HA) cleavage site, allowing the HA to be cleaved by intracellular ubiquitous proteases and the virus to spread systematically (3). In contrast, the HA of LPAIV is proteolytically matured by trypsin-like proteases expressed in the respiratory and digestive tract, thus restricting replication of LPAIV to these tissues in birds.
In chickens, HPAIV infection can reach 100% mortality in a few days, whereas ducks usually only exhibit mild, if any, clinical signs following HPAIV infection (4). The mechanisms contributing to the host species-dependent differences in pathogenicity are not fully understood. Ducks express the viral RNA sensor retinoic acid-induced gene I (RIG-I), which is absent in chickens (5). Melanoma differentiation-associated protein 5 (MDA-5) could compensate for the lack of RIG-I expression in chicken cells and mediate type I interferon (IFN) responses to influenza A virus infection (6). However, comparative studies have provided evidence that HPAIV infections are associated with a more rapid type I IFN immune response and reduced proinflammatory cytokines expression in ducks compared to chickens. This may lead to reduced immunopathology and therefore fewer clinical signs in ducks (7–11). Extensive tissue damage due to higher levels of viral replication and inflammation in multiple organs is probably the cause of the higher HPAIV-associated mortality in chickens compared to ducks.
The microbiota is now recognized as a regulator of many physiological functions in the host, reaching far beyond its contribution to digestion (12). Several studies have highlighted the role of the gut microbiota in shaping the antiviral immune response against influenza virus, in mammals, in which the virus replicates in the respiratory tract (13–17), as well as following LPAIV infection in chickens, in which the virus replicates in the respiratory and digestive tract (18, 19).
Altogether, these observations prompted us to investigate to what extent the microbiota could contribute to the control of HPAIV infection in ducks. By treating ducks with a broad-spectrum antibiotic (ABX) cocktail, we achieved a significant depletion of the intestinal and respiratory microbiota. Groups of nontreated and ABX-treated ducks were infected with a H5N9 HPAIV. We revealed that ABX-treated ducks had significantly higher viral excretion in the intestine, which correlated with an impaired intestinal antiviral immune response. This study thus demonstrates that the microbiota contributes to the control of avian virus replication in ducks.
RESULTS
Antibiotic treatment leads to a significant depletion of the microbiota.
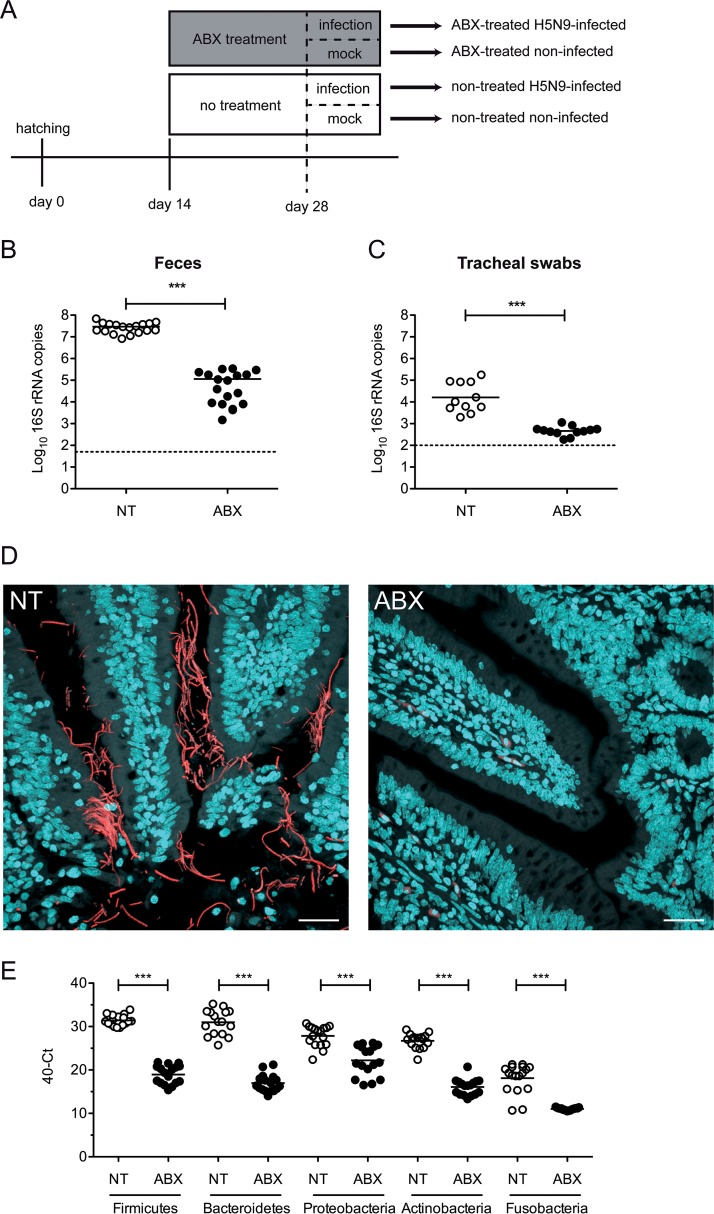
From 2 weeks of age, ABX-treated ducks were continuously treated with a broad-spectrum ABX cocktail via their drinking water (Fig. 1A). To assess the impact of the ABX-treatment on the microbiota, we quantified the number of 16S rRNA gene copies from fresh feces and tracheal swabs from nontreated control ducks and ABX-treated ducks. After 2 weeks of ABX treatment, we detected a >102-fold depletion of the number of bacterial 16S rRNA gene copies detected in feces (Fig. 1B) and a >10-fold depletion in tracheal swabs (Fig. 1C). Using 16S rRNA fluorescence in situ hybridization (FISH), we detected bacteria aggregating as filaments in the vicinity of intestinal epithelial cells on ileal tissue section from nontreated control ducks (Fig. 1D). In contrast, no bacteria were detected by FISH in ileal samples originating from ABX-treated ducks. No bacteria were detected by FISH in respiratory tissues, regardless of the ABX treatment (data not shown), suggesting that the FISH technique is not sensitive enough to detect the respiratory microbiota, which is scarce in comparison to the intestinal microbiota. Altogether, these results demonstrate that the ABX treatment caused a significant reduction in the ducks’ microbiota. Bacterial phylum-specific qPCR revealed that the ABX treatment caused a significant reduction in all major phyla composing the duck microbiota (Fig. 1E) (20). The ABX treatment did not induce any clinical signs or change in the ducks’ behavior, nor did it diminish the quantity of water consumed. There was no significant difference in the average weights of ABX-treated and nontreated animals (data not shown). In addition, there was no difference in fecal consistency between ABX-treated and nontreated animals.
FIG 1.
Antibiotic treatment for microbiota depletion. (A) Treatment timelines for nontreated and antibiotic (ABX)-treated ducks. Ducks received the ABX cocktail in drinking water. Fecal bacterial loads were determined at day 28 after 2 weeks of ABX treatment and prior to H5N9 virus inoculation. (B) 16S rRNA gene copies per g of feces. DNA was extracted from feces harvested on day 28 and analyzed by qPCR. (C) 16S rRNA gene copies per tracheal swab. DNA was extracted from tracheal swabs performed on day 28, vortexed in 500 μl of sterile PBS, and analyzed by qPCR. (D) Ileum sections from NT or ABX-treated ducks were subjected to FISH with eubacterial 16S rRNA-specific Alexa 594-labeled probe and stained with DAPI. Scale bar, 25 μm. (E) Quantification of specific bacterial phylum 16S rRNA gene DNA was performed from DNA extracted from feces harvested on day 28 and analyzed by qPCR. ***, P < 0.001.
Impact of microbiota depletion on influenza virus replication.
We inoculated ducks via the ocular, nasal and tracheal route with 3.6 × 106 egg infectious dose 50 (EID50) of the A/Guinea Fowl/France/129/2015(H5N9) HPAIV virus. ABX-treated ducks remained under the same ABX treatment for the whole duration of the experiment. Ducks did not show any clinical signs upon H5N9 infection in the nontreated and ABX-treated groups.
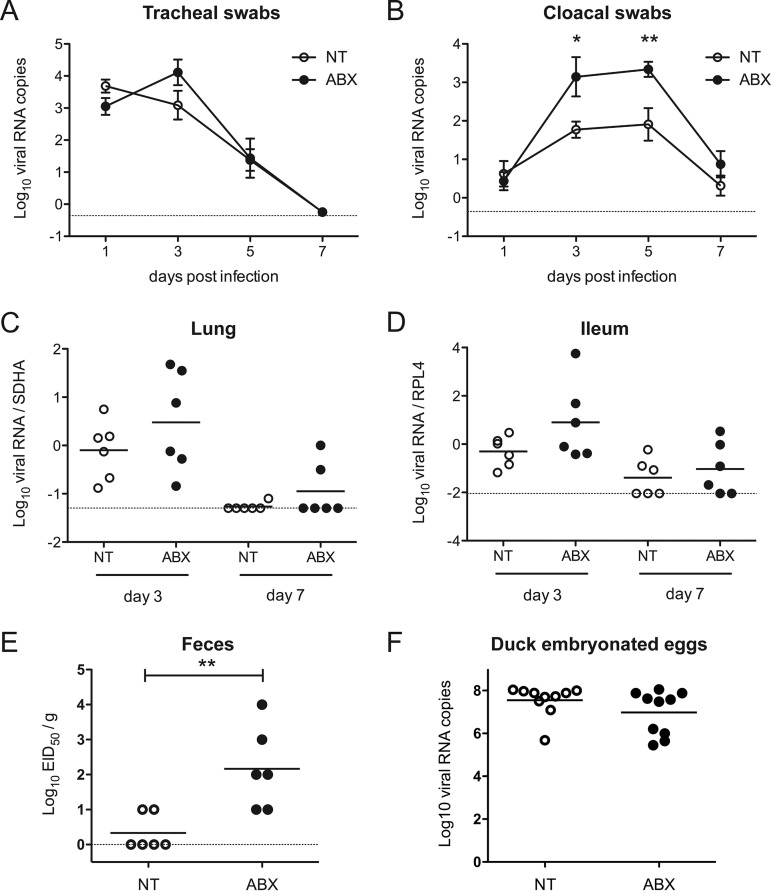
We evaluated the level of virus excretion by quantifying viral nucleic acids by reverse transcription-quantitative PCR (RT-qPCR) from tracheal and cloacal swabs. Tracheal shedding was modestly increased in ABX-treated ducks at 3 days postinfection (dpi) (Fig. 2A). In contrast, cloacal shedding was significantly higher in ABX-treated animals at 3 and 5 dpi (Fig. 2B). We then analyzed viral load in organs harvested from animals autopsied at 3 and 7 dpi. Viral nucleic acid was detected in the brain and in the spleen of infected animals, as expected with a HPAIV, with no difference observed between ABX-treated-ducks and nontreated ducks (data not shown). In the lungs and ileum, the viral nucleic acid load was increased in ABX-treated-ducks compared to nontreated ducks at 3 dpi (Fig. 2C and D); however, the differences did not reach statistical significance. Finally, we measured infectious virus particles from fresh feces collected from individual ducks at 3 dpi. We detected a significantly higher number of infectious virus particles in feces from ABX-treated ducks compared to nontreated ducks (Fig. 2E), thus confirming that ABX-induced depletion of the microbiota is associated with an increase of influenza virus fecal shedding in ducks.
FIG 2.
Consequences of microbiota depletion on H5N9 virus replication. NT or ABX-treated ducks were inoculated via the ocular, nasal, and tracheal routes with 3.6 × 106 EID50 of the H5N9 HPAIV virus. Virus replication was analyzed by quantifying viral RNA by RT-qPCR from RNA extracted from tracheal swabs (A) or cloacal swabs (B). The curves represent the mean viral RNA load, and error bars correspond to the standard errors of the mean (SEM). Viral RNA load was analyzed from total RNA extracted from the lungs (C) and the ileum (D). Viral RNA was normalized with mRNA of the SDHA housekeeping gene in the lungs or the RPL4 housekeeping gene in the ileum. (E) Viral load in the feces of animals autopsied at 3 dpi was titrated on chicken embryonated eggs, and viral titers are expressed as EID50/g. (F) Consequences of ABX treatment on H5N9 virus replication in embryonated duck eggs. Eleven-day-old embryonated duck eggs were injected with antibiotics (ABX) or left untreated (NT). Viral growth was analyzed by quantifying viral RNA by RT-qPCR from RNA extracted from allantoic fluids were collected at 72 h postinfection. Each dot represents an individual value, and the horizontal bar corresponds to the mean. The dotted line represents the limit of detection for each experiment. *, P < 0.05; **, P < 0.01.
To determine whether the ABX-treatment could have a direct microbiota-independent effect on virus replication, embryonated duck eggs were treated with either PBS or the ABX cocktail. One day later, eggs were then inoculated with 104 EID50 H5N9 together with ABX in the ABX-treated eggs or with PBS in the nontreated eggs. We evaluated the level of virus replication by quantifying viral nucleic acids by RT-qPCR from allantoic fluids collected at 3 dpi. We detected equal amounts of viral RNA in nontreated and ABX-treated eggs (Fig. 2F). Since embryonated duck eggs are essentially devoid of bacterial flora (21), this result demonstrates that the ABX treatment-induced increase in H5N9 replication observed in ducks in vivo is not due to a direct effect of ABX on H5N9 replication but rather point to a microbiota-mediated effect.
Impact of microbiota depletion on the respiratory and intestinal epithelia.
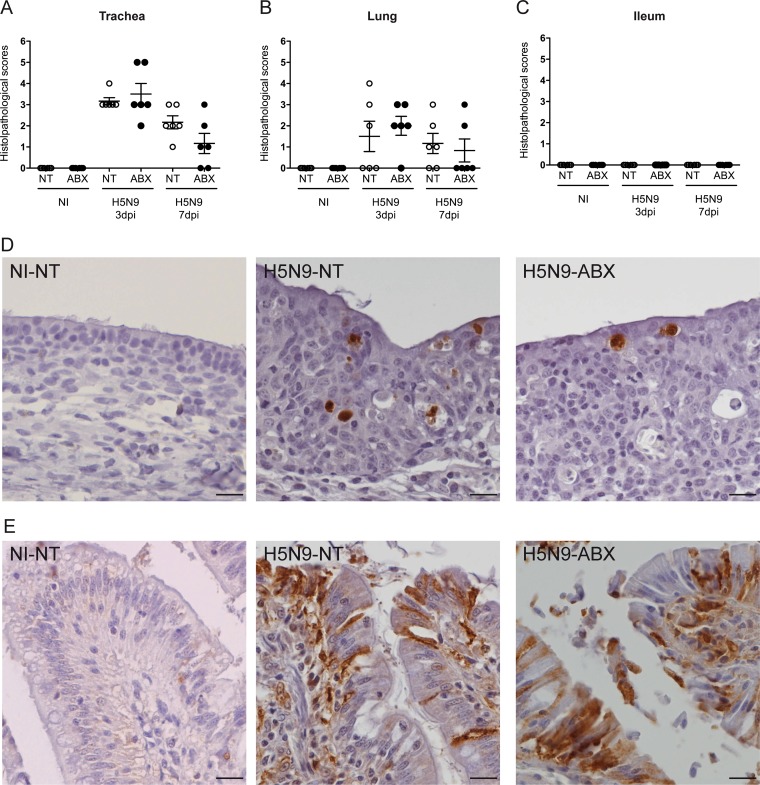
Histopathological analysis performed at 3 and 7 dpi revealed a diffuse to multifocal subacute tracheitis in ducks infected with H5N9 virus, which was equivalent in ABX-treated and nontreated animals. Inflammatory cellular infiltrates in the lamina propria were variably composed of mononuclear cells (lymphocytes, plasmocytes, and macrophages) and a few heterophils. We observed mild focal necrosis and exfoliation of the superficial mucosal epithelium, associated with loss of ciliature, regenerative epithelial hyperplasia and squamous epithelial metaplasia. Histological scores were low for infected animals and yet different from noninfected animals (Fig. 3A). Immunohistochemically, viral antigen was detected in epithelial cells of the trachea, similarly in ABX-treated and nontreated ducks infected with H5N9 virus (Fig. 3D). In the lungs, multifocal to focal subacute bronchitis was observed at 3 and 7 dpi in H5N9-infected ducks (Fig. 3B), characterized by regenerative epithelial hyperplasia and inflammatory cellular infiltrates in the lamina propria composed of mononuclear cells (lymphocytes, plasmocytes, and macrophages) and a few heterophils.
FIG 3.
Histopathological analysis of H5N9 infection in NT and ABX-treated ducks. Histopathological analysis was performed on tracheal, lung, and ileum samples from noninfected (NI) or H5N9-infected ducks that were either nontreated (NT) or treated with ABX. Histological scoring of hematoxylin and eosin stain-colored tracheal (A), lung (B), or ileum (C) sections. Each dot represents an individual value, the horizontal bar corresponds to the mean, and the error bars correspond to the SEM. Immunohistochemical anti-NP staining of hematoxylin-counterstained tracheal (D) or ileum sections (E). Scale bar, 10 μm.
In the ileum, neither ABX treatment nor H5N9 infection was associated with any change in cell morphology and histological organization, both in the epithelium and in the chorion (Fig. 3C). Viral antigen was detected in differentiated epithelial cells of the ileum, as well as in desquamated cells in the intestinal lumen (Fig. 3E).
To gain further insight into the potential consequences of increased intestinal replication of H5N9 associated with depletion of the bacterial flora, we analyzed the expression of tight junction genes on ileal samples. The expression of zona occludens 1 (Fig. 4A) and Claudin-2 (Fig. 4B) mRNA was similar in all groups. In addition, we did not detect any change in the expression of lysozyme mRNA (Fig. 4C), a marker of Paneth cell function. We also analyzed the expression of the gene encoding mucin 2, a secreted high-molecular-weight glycoprotein, which is a component of the intestinal mucus. Mucin 2 expression was reduced specifically in H5N9-infected ducks treated with ABX (Fig. 4D). These results indicate that depletion of the bacterial flora may compromise intestinal integrity in response to HPAIV infection. Finally, we analyzed the expression of the mitochondrial function markers Mitochondrial transcription factor A (TFAM) and mitochondrial DNA-directed RNA polymerase (POLRMT), whose expression is commonly altered in response to intestinal injury (22, 23). We did not detect any change in the expression of TFAM (Fig. 4E) or POLRMT (Fig. 4F), suggesting that H5N9 infection and ABX treatment did not alter mitochondrial function.
FIG 4.
Analysis of intestinal integrity following H5N9 infection in NT and ABX-treated ducks. Analysis of mRNA gene expression from total RNA extracted from the ilea of noninfected (NI) or H5N9-infected ducks that were either nontreated (NT) or treated with ABX. RT-qPCR analysis of Zona occludens 1 (A), Claudin-2 (B), lysozyme (C), mucin 2 (D), mitochondrial transcription factor A (E), and mitochondrial DNA-directed RNA polymerase (F) mRNA levels in the ilea of ducks. mRNA levels were normalized to RPL4 levels. The results are expressed as means ± the SEM. **, P < 0.01.
To determine whether depletion of the intestinal bacterial flora could affect influenza virus receptor expression, we generated fluorescent trimeric H5 HA to detect H5N9 virus receptor expression levels and distribution on tissue sections (24). The intensity and distribution of H5N9 receptors were similar in the tracheas (Fig. 5A) and intestines (Fig. 5B) of ABX-treated and nontreated ducks. This result suggests that the increased replication of H5N9 in the intestinal tracts of ABX-treated ducks is not due to a difference in virus receptor expression.
FIG 5.
Analysis of H5N9 virus receptor expression. (A and B) Staining of tracheas (A) and ilea (B) from nontreated (NT) or ABX-treated ducks with trimeric HA from the H5N9 virus, precomplexed with α-strep-tag-488 conjugated mouse antibody and goat-α-mouse-488 conjugated antibody. Tissue sections were stained with DAPI (blue signal). Scale bar, 25 μm. Quantification of H5 HA fluorescent tissue staining is shown on the right. Each dot represents the mean H5 HA fluorescence intensity for an animal. The horizontal bar corresponds to the mean of all animals within a group, and error bars correspond to the SEM.
Impact of microbiota depletion on the antiviral immune response.
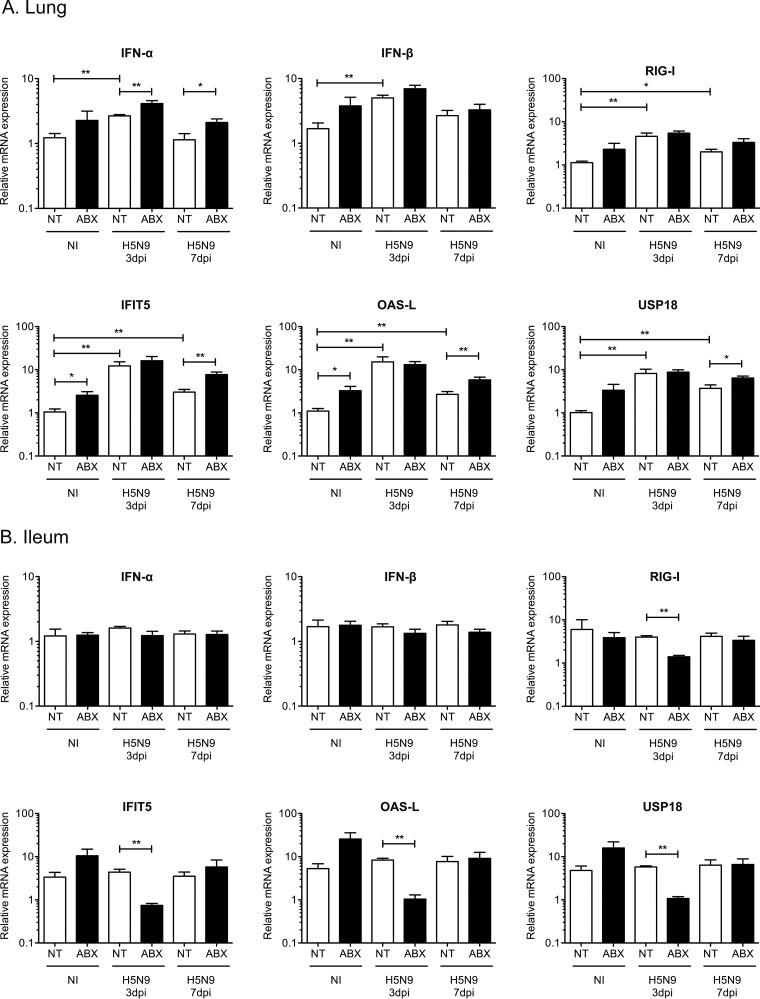
We then evaluated the impact of ABX-mediated microbiota depletion on the innate antiviral immune response. In the lungs, we observed an upregulation of IFN-α and IFN-β mRNA expression in H5N9-infected ducks at 3 dpi, which correlated with increased levels of the cytosolic RNA sensor retinoic acid inducible gene 1 (RIG-I) (Fig. 6A). This was associated with an upregulation of interferon-induced protein with tetratricopeptide repeat 5 (IFIT5), oligoadenylate synthetase-like (OAS-L), and ubiquitin specific peptidase 18 (USP18) mRNA expression (Fig. 6A). IFIT5, OAS-L, and USP18 are type I IFN-induced genes whose expression is a good indicator of the amount of type I IFN active locally. Interestingly, ABX treatment was associated with a modest upregulation of IFN-α/β mRNA and type I IFN-induced genes expression regardless of the infection status.
FIG 6.
Analysis of the type I IFN immune response following H5N9 infection in NT and ABX-treated ducks. Analysis of mRNA gene expression from total RNA extracted from the lung (A) and ileum (B) of noninfected (NI) or H5N9-infected ducks that were either nontreated (NT) or treated with ABX. The mRNA expression levels of IFN-α, IFN-β, RIG-I, and of three type I IFN-induced genes—IFIT5, OAS-L, and USP18—were analyzed by RT-qPCR. The mRNA levels were normalized to the geometric means of RPL4/SDHA mRNA levels in the lung and RPL4/RPL30 mRNA levels in the ileum. The results are expressed as means ± the SEM. *, P < 0.05; **, P < 0.01.
In the ileum, we did not detect significant upregulation of alpha interferon (IFN-α) or IFN-β mRNA expression in ducks infected with HPAIV, regardless of the status of the microbiota (Fig. 6B). However, we observed an upregulation of type I IFN-induced genes in noninfected ABX-treated ducks (Fig. 6B) but the differences did not reach statistical significance. Interestingly, RIG-I mRNA transcripts were significantly reduced at 3 dpi in H5N9-infected ABX-treated ducks compared to H5N9-infected nontreated ducks (Fig. 6B), and this downregulation correlated with a significant reduction in type I IFN-induced gene expression at 3 dpi in H5N9-infected ABX-treated ducks compared to H5N9-infected nontreated ducks (Fig. 6B). Since the microbiota is required for IgA production in the intestine (25, 26), we measured IgA heavy-chain constant region mRNA expression in the intestine. We observed strongly reduced IgA heavy-chain constant region mRNA expression in the intestines of ABX-treated ducks, regardless of the H5N9 infection status (Fig. 7). Altogether, these results indicate that increased H5N9 replication in the intestine observed upon microbiota depletion is associated with an impaired intestinal immune response.
FIG 7.
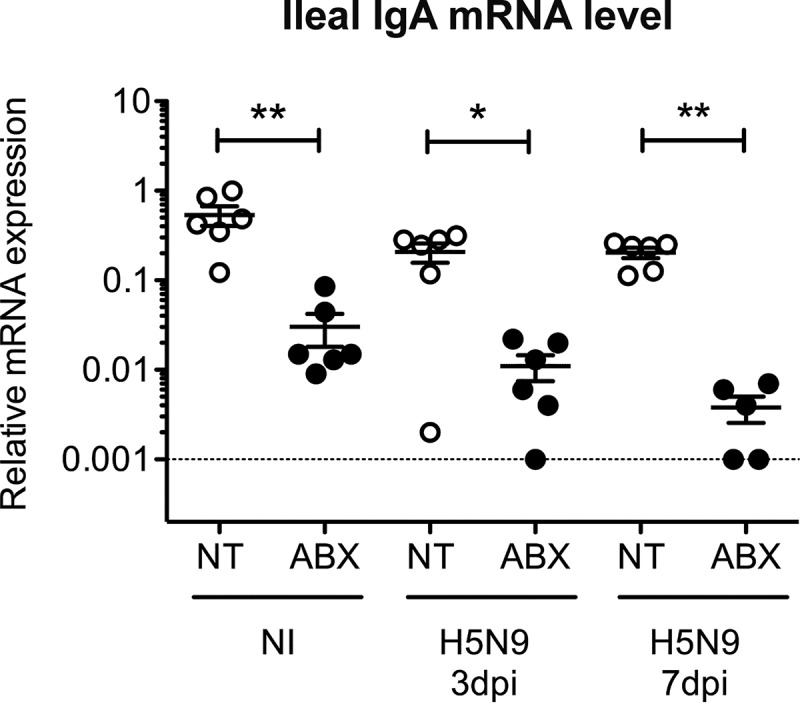
Analysis of the intestinal IgA heavy-chain mRNA expression after H5N9 infection in NT and ABX-treated ducks. IgA heavy-chain constant region mRNA expression levels were analyzed by RT-qPCR from total RNA extracted from the ileum of noninfected (NI) or H5N9-infected ducks that were either not treated (NT) or treated with ABX. IgA expression levels were normalized to RPL4 mRNA levels. Each dot represents an individual value, the horizontal bar corresponds to the mean, and error bars correspond to the SEM. *, P < 0.05; **, P < 0.01.
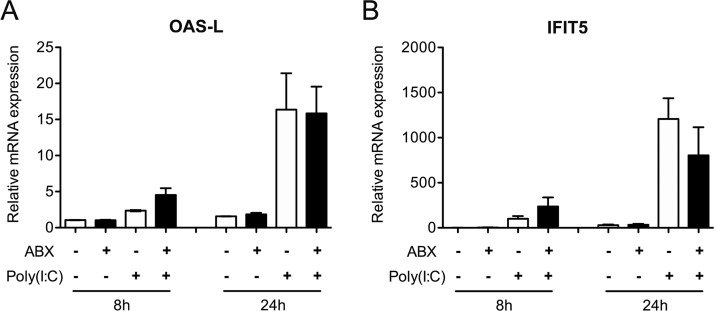
To determine whether the ABX-treatment could have a direct microbiota-independent effect on the innate antiviral immune response, we treated duck embryonic fibroblasts with the ABX cocktail and stimulated them with intracellularly delivered poly(I·C). We observed no change in IFIT5 and OAS-L mRNA expression following ABX treatment alone and no effect of ABX treatment on the level of upregulation of IFIT5 and OAS-L mRNA transcripts levels following poly(I·C) stimulation (Fig. 8). This result indicates that the ABX-treatment induced decrease in the intestinal antiviral innate immune response is not due to a direct effect of ABX treatment but rather points to a microbiota-mediated effect.
FIG 8.
Analysis of the type I IFN immune response after poly(I·C) treatment in NT and ABX-treated duck embryonic fibroblasts. Duck embryonic fibroblasts (CCL-141 cell line) were untreated (NT) or treated with antibiotics (ABX) and subsequently stimulated with intracellularly delivered poly(I·C) for 8 or 24 h. The mRNA expression levels of the type I IFN-induced genes OAS-L and IFIT5 were analyzed by RT-qPCR. mRNA levels were normalized to GAPDH mRNA levels. The results are expressed as means ± the SEM of two independent experiments.
DISCUSSION
The A/Guinea Fowl/France/129/2015(H5N9) HPAIV virus studied here is a field isolate collected from a duck farm in France during the 2015-2019 H5Nx epizootics. The first HPAIV detected during this epizootic was of the H5N1 subtype. Genetic analyses revealed that the HA was independent from the Gs/Gd/1/96-like H5 lineage and thus represented a new HPAIV emergence event (27). The virus has undergone multiple reassortment events during its circulation in France, leading to the cocirculation of H5N1, H5N2, and H5N9 HPAIV subtypes (28). Field observations provided evidence that clinical signs depended on the HPAIV subtype and the host species. Gallinaceous poultry exhibited a moderate increase in mortality, while waterfowl mostly showed no clinical signs of infection (28). Here, we performed the first experimental H5N9 HPAIV infection in ducks (Anas platyrhynchos), which confirmed that H5N9 HPAIV infection in ducks was not associated with clinical signs and only produced mild lesions detected microscopically in the respiratory tract.
The contribution of the microbiota to the control of influenza virus replication has been studied in mice, as well as in chickens infected with a LPAIV (13, 14, 16–19, 29). How the microbiota modulates HPAIV infection, or more generally influenza virus infection in ducks has to our knowledge never been investigated. Here, we provide evidence that ABX-mediated depletion of the microbiota caused an increase in H5N9 HPAIV replication in the digestive tract of ducks and was associated with a reduction of the antiviral immune response in the intestine. Similar results were observed in the digestive tract of chickens infected with a LPAIV (18, 19). Depletion of the microbiota in mice and chickens was associated with an increase in influenza virus replication in the respiratory tract (13, 14, 18, 19). In contrast, we did not detect a significant increase in influenza virus replication in the respiratory tracts of ducks, suggesting that this difference could be due to the viral strain or host factors.
We did not detect any effect of ABX cocktail treatment on H5N9 growth in duck embryonic eggs. Since duck embryonic eggs are essentially devoid of microbiota, this result suggests that the increased H5N9 virus replication observed in 4-week-old ducks upon ABX treatment is not due to a direct toxic effect of the ABX cocktail on cell viability but rather to consequences of the ABX cocktail treatment on the microbiota. Several mechanisms could account for the increased virus replication observed in the digestive tract of ducks upon depletion of the microbiota with ABX (30). Bacteria (31) or bacterium-derived products, such as lipopolysaccharide (32), have been shown to trap or destabilize influenza virus and thus could directly impair influenza virus infection in the bacteria dense intestine. Upon depletion of the microbiota with ABX, we observed a reduction of RIG-I mRNA expression, which correlated with a reduction of the type I IFN-induced gene expression in the intestine following influenza virus infection. Thus, by impairing the innate antiviral immune response, depletion of the microbiota could indirectly promote influenza virus replication. The gut microbiota has been shown to stimulate the immune response to pathogens most likely through the constant low-level exposure of epithelial cells and immune cells present in the intestinal mucosa to microbiota-derived pathogen-associated molecular patterns, as well as to microbial metabolites (29, 33–35). In addition, an RNA-binding protein secreted by Listeria monocytogenes has recently been shown to stimulate RIG-I signaling, suggesting that proteins with similar function, produced by resident bacteria, could promote antiviral innate immune signaling in the intestine (36). This constant low-level stimulation is thought to maintain the immune system in an optimal state of reactivity allowing it to respond to pathogens via a timely innate immune response (17, 37, 38). The microbiota is also required for IgA production in the intestine (25, 26). In line with this observation, we detected a strong reduction of IgA heavy-chain constant region mRNA expression in the intestine of ABX-treated ducks. Decreased IgA levels could impair the clearance of influenza virus during the later stages of infection or upon reinfection.
While we detected a significant upregulation of type I IFN-induced genes expression in the respiratory tract of ducks infected with the H5N9 HPAIV, no upregulation could be detected in the digestive tract following H5N9 HPAIV infection. Since a significant reduction of type I IFN-induced gene expression was observed at 3 dpi in H5N9-infected ABX-treated ducks, the expression of type I IFN-induced genes in the digestive tract is not at its minimal level in noninfected ducks. These observations thus suggest that the basal expression level of type I IFN-induced genes in the digestive tracts of ducks may be constitutively high and could contribute to the efficient control of influenza virus replication in ducks through early inhibition of the viral life cycle.
Interestingly, type I IFN-induced gene expression was increased in the digestive and respiratory tracts of noninfected animals treated with ABX compared to nontreated noninfected animals. The ABX cocktail used in our experiments contained gentamicin, a member of the aminoglycoside class of ABX, which have been shown recently to induce type I IFN-induced gene expression through a Toll-like receptor 3-dependent pathway, independently of the microbiota (39). However, aminoglycosides stimulated type I IFN-induced gene expression locally and did not lead to increased type I IFN-induced gene expression in the respiratory tract when administered orally (39). In addition, we did not detect any effect of ABX cocktail treatment alone or in combination with poly(I·C) treatment on the expression of type I IFN-induced gene expression in duck embryonic fibroblasts. Therefore, in our experiments, how the ABX-cocktail led to an increase of type I IFN-induced gene expression remains unknown.
In association with the respiratory tract and internal organs, the digestive tract represents a major site of HPAIV replication and excretion in birds (7, 8, 11). Moreover, the digestive tract is the main site of LPAIV replication in ducks (9, 40–42). Thus, avian influenza viruses can be considered as adapted to the digestive tract in ducks. A number of studies have shown that the microbiota promotes infection by enteric viruses such as poliovirus, norovirus, or rotavirus through a variety of processes, including promotion of virus stability, attachment and entry, or evasion of the immune response (43–47). Our results thus highlight another type of interaction between an enteric virus and the microbiota since we observed impairment of avian influenza virus replication in the intestine of ducks harboring an intact microbiota compared to ABX-treated ducks.
Increased influenza virus replication and excretion in chickens (18, 19) and ducks treated with ABX raises questions about the risks associated with the use of ABX treatments in farmed animals. However, the ABX treatment used to deplete the gut microbiota in both our and the vast majority of experimental studies consists of a broad-spectrum ABX cocktail. This ABX cocktail is composed of six antimicrobial molecules, including molecules, which are not permitted to be used in veterinary medicine since their use is restricted to human medicine in hospital settings or to research. ABX treatments given to farmed animals mostly consist of one or two antimicrobial molecules and therefore should not have such a profound effect on the microbiota composition. Considering that ABX consumption for food animal production is forecasted to increase worldwide (48), further studies are needed to investigate whether narrow-spectrum ABX treatments given to farmed animals could also have consequences on virus replication and spread. In that respect, it is crucial to keep in mind that ABX treatments administered to farmed animals are critical to the well being of animals and the economic profitability of the vast majority of farms worldwide (49–51). ABX treatments are not the only cause of microbiota dysbiosis in food animals. Nutrition and environmental parameters at birth and during rearing have also been shown to be associated with microbiota dysbiosis (52, 53). In addition, microbiota dysbiosis can also indirectly promote the spread and burden of infectious diseases by impairing the response to vaccines (54–57). In conclusion, further studies are needed to evaluate the consequences of microbiota dysbiosis and possible interventions with probiotics on the burden of infectious diseases (55, 58).
MATERIALS AND METHODS
Ethics statement.
This study was carried out in compliance with European animal welfare regulation. The protocols were approved by the Animal Care and Use Committee (Comité d’Éthique en Science et Santé Animales—115) under protocol 13205-2018012311319709.
Animals and antibiotic treatment.
One-day-old Pekin ducklings (Anas platyrhynchos domesticus, ST5 heavy) were obtained from a commercial hatchery (ORVIA–Couvoir de la Seigneurtière, Vieillevigne, France). The study employed only female birds to minimize between-individual variability arising from sex. Birds were fed ad libitum with a starter diet. Ducks were housed for 2 weeks in a litter-covered floor pen in a biosafety level II facility at the National Veterinary School of Toulouse, France. They were then transferred into a biosafety level III facility, equipped with poultry isolators (I-Box; Noroit, Nantes, France) that were ventilated under negative pressure with HEPA-filtered air. A total of 36 ducks were randomly assigned to four groups: 6 nontreated noninfected animals (NT, NI), 6 ABX-treated noninfected animals (ABX, NI), 12 ABX-treated infected animals (ABX, H5N9), and 12 nontreated infected animals (NT, H5N9). Ducks of the ABX groups were treated through drinking water. ABX was administered at the following daily doses: 80 mg/kg vancomycin, 300 mg/kg neomycin, 200 mg/kg metronidazole, 0.2g/kg ampicillin, and 24 mg/kg colistin. To prevent fungal overgrowth, ABX-treated ducks also received 2 mg/kg amphotericin B daily. The composition and posology of the ABX cocktail are similar to the ones described in other publications (14, 18, 59, 60). Water was changed every 2 days. ABX treatment started 2 weeks prior to infection and continued for the whole duration of the experiment. No difference in water consumption was observed between nontreated and ABX-treated ducks.
Virus and experimental infection.
One day prior to infection, serum was collected from all the birds to ensure that they were serologically negative to influenza virus by using a commercial influenza A NP antibody competition enzyme-linked immunosorbent assay kit (ID Screen; ID-Vet, Montpellier, France) according to the manufacturer’s instructions. The A/Guinea Fowl/France/129/2015(H5N9) (GenBank accession numbers MN400993 to MN401000) HPAIV was propagated in 10-day-old specific-pathogen-free (SPF) embryonated chicken eggs (INRA PFIE, Nouzilly, France). Infectious allantoic fluid was harvested at 72 h postinoculation and titrated in 10-day-old SPF embryonated chicken eggs to determine the 50% egg infective dose (EID50)/ml using the Reed-Muench method. When they were 4 weeks old, the ducks were infected with 3.6 × 106 EID50 of virus via the ocular, nasal, and tracheal routes. Ducks of the noninfected groups received the equivalent volume of allantoic fluid collected from noninfected embryonated chicken eggs. Birds were observed for 7 days, and clinical signs were recorded. Tracheal and cloacal swabs were performed on all animals at days 0, 1, 3, 5, and 7 dpi. Half of the infected animals were euthanized and necropsied at 3 dpi, and the other half were euthanized and necropsied at 7 dpi.
Analysis of bacterial depletion.
Fecal samples were collected 1 day prior to infection by placing ducks in individual clean plastic crates. DNA was extracted from 500 mg of feces and tracheal swabs vortexed in 500 μl of sterile phosphate-buffered saline (PBS; PureLink microbiome DNA purification kit; Invitrogen, Thermo Fisher Scientific Inc., Canada) according to the manufacturer’s instruction.
For 16S rRNA gene DNA quantification, microbial qPCR was performed with 2 μl of extracted DNA according to the manufacturer’s instructions (QuantiFast SYBR green PCR; Qiagen, Canada) using 16S-V5-F (AGCRAACAGGATTAGATAC) and 16S-V5-R (TGTGCGGGCCCCCGTCAAT) primers to amplify the V5 region from 16S rRNA genes. Absolute quantification was performed using a standard curve based on a 10-fold serial dilution of plasmid containing the 16S-V5 region of Escherichia coli. Quantification of the specific phylum 16S rRNA gene DNA was performed in the same way using specific primers of five major bacteria phyla (Table 1), and quantification was performed using the 40-Ct method.
TABLE 1.
Primers used for qPCRa
| Gene | Accession no. | Primer sequence (5′–3′) | Reference |
|---|---|---|---|
| CLDN1 | XM_013108556.1 | F: TCATGGTATGGCAACAGAGTGG | 64 |
| R: TCATGGTATGGCAACAGAGTGG | |||
| IAV-M | F: CTTCTAACCGAGGTCGAAACG | 65 | |
| R: AGGGCATTTTGGACAAAGCGTCTA | |||
| IFIT5 | KF956064 | F: TCCTGCGATATGCTGCTATATTTTAT | 66 |
| R: GGTGTCACTGTTAAGGCTTTTCTCA | |||
| IFN-α | AB128861.1 | F: CAACGACACGCAGCAAGC | 40 |
| R: GGGTGTCGAAGAGGTGTTGG | |||
| IFN-β | KT428159 | F: TCTACAGAGCCTTGCCTGCAT | 67 |
| R: TGTCGGTGTCCAAAAGGATGT | |||
| IgA | U27222.1 | F: TCGCTCAAGGAACCCATCGT | 64 |
| R: GCGGGACCACGAGAACTTCA | |||
| GAPDH | AY436595.1 | F: CCACTTCCGGGGCACTGTCA | 63 |
| R: AGCACCAGCATCTGCCCACT | |||
| LYZ | XM_005008880.2 | F: TAACACGCAGGCTACAAACCG | 64 |
| R: TTCCATCGCTGACAATCCTCTT | |||
| MUC2 | XM_005024513.2 | F: GGGCGCTCAATTCAACATAAGTA | 64 |
| R: TAAACTGATGGCTTCTTATGCGG | |||
| OAS-L | KU569293.1 | F: CCGCCAAGCTGAAGAACCTG | 68 |
| R: CGCCCTGCTCCCAGGTATAG | |||
| Pan-Actinobacteria | F: TACGGCCGCAAGGCTA | 69 | |
| R: TCRTCCCCACCTTCCTCCG | |||
| Pan-Bacteroides | F: CRAACAGGATTAGATACCCT | 69 | |
| R: GGTAAGGTTCCTCGCGTAT | |||
| Pan-Firmicutes | F: TGAAACTYAAAGGAATTGACG | 69 | |
| R: ACCATGCACCACCTGTC | |||
| Pan-Fusobacteria | F: GCCTCACAGNTAGGGACAACAT | 69 | |
| R: GCYYACCTCTCCAGTACTCTAG | |||
| Pan-Gammaproteobacteria | F: TCGTCAGCTCGTGTYGTGA | 69 | |
| R: CGTAAGGGCCATGATG | |||
| POLRMT | XM_027472517.1 | F: CCACCAGAAGATCATGCAGC | 70 |
| R: CGATGGTCAGCAGCTTTCTC | |||
| RIG-I | NM_001310380 | F: GTGTATGGAGGAAAACCCTATTCTTAACT | 71 |
| R: GGAGGGTCATACCTGTTGTTTGAT | |||
| RPL4 | XM_027465643.1 | F: AAGCTGAACCCATACGCCAA | 63 |
| R: CCTGGGCCTTAGCTGTAACC | |||
| RPL30 | XM_027452502.1 | F: GCAAAGCCAAGCTGGTCATC | 63 |
| R: CTCAATGTTGTTGCCGCTGT | |||
| SDHA | XM_027451817.1 | F: GACACAGTGAAAGGCTCCGA | 63 |
| R: CTCCAGCTCTATCACGGCAG | |||
| TFAM | XM_013107580.1 | F: AGCATCACAGAAGCAGGTTTA | 70 |
| R: TGCCAGTCGTTTCCTCCT | |||
| USP18 | XM_005009931 | F: AACCTGACGGCAGAAGAAGA | 72 |
| R: GCACCGTGATCCTTCGTAGT | |||
| ZO-1 | XM_021276271.2 | F: TCAGCGAGATGAACGAGCC | 70 |
| R: TCTGAAGGCTCTGACCTCTGG |
CLDN1, Claudin-1; IAV-M: gene M of influenza A virus; IFIT5, interferon induced protein with tetratricopeptide repeats 5; IFN-α/β, interferons alpha and beta; IgA, immunoglobulin A; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; LYZ: lysozyme; MUC2, mucin 2; OAS-L, oligoadenylate synthetase-like; POLRMT, RNA polymerase mitochondrial; RIG-I, retinoic acid-inducible gene I; RPL4/30, ribosomal protein L4/L30; TFAM, mitochondrial transcription factor A; SDHA, succinate dehydrogenase complex flavoprotein subunit A; USP18, ubiquitin specific peptidase 18; ZO-1, zonula occludens-1. F, forward; R, reverse.
Fluorescence in situ hybridization (FISH) staining of the ileum was perfromed as described previously (61). The tissue sections were incubated with the universal bacterial probe EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′; Eurogentec, Liège, Belgium) conjugated to Alexa Fluor 594. A “nonsense” probe (5′-CGACGGAGGGCATCCTCA-3′) conjugated to Alexa Fluor 594 was used as a negative control. Tissue sections were mounted with Vectashield mounting medium containing DAPI (4′,6′-diamidino-2-phenylindole). Images were acquired on a Zeiss LSM710 confocal microscope (Carl Zeiss MicroImaging GmbH, Jena, Germany) at the cellular imaging facility of the CPTP (Toulouse, France).
Influenza virus quantification.
Cloacal and tracheal swabs were briefly vortexed in 500 μl of sterile PBS, and viral RNA was extracted from 140 μl according to the manufacturer’s instructions (QIAamp viral RNA; Qiagen, Toronto, Canada). Influenza virus nucleic load was determined by RT-qPCR using influenza virus segment M specific primers (IAV-M; Table 1) in 96-well plates according to the manufacturer’s instructions (OneStep RT-PCR; Qiagen). Absolute quantification was performed using a standard curve based on a 10-fold serial dilution of a plasmid containing the A/Guinea Fowl/129/2015(H5N9) M gene. The EID50 was determined by diluting 0.1 g of feces into 100 μl of PBS containing the antibiotics vancomycin (500 mg/liter), neomycin (500 mg/liter), metronidazole (500 mg/liter), ampicillin (1 g/liter), and colistin (80 mg/liter) to avoid bacterial growth. Samples were centrifuged at 1,000 × g at 4°C for 2 min to pellet the debris, and supernatants were diluted in antibiotic-complemented PBS with a 10× dilution factor. For each sample, 100-μl portions were injected into three 10-day-old SPF embryonated chicken eggs, which were incubated for 72 h before the allantoic fluids were harvested. The presence of virus was revealed by a hemagglutination test.
Antibiotics treatment and infection of embryonated duck eggs.
Eleven-day-old embryonated duck eggs were injected with antibiotics (ABX) to obtain the following concentrations in allantoic fluids: vancomycin (500 mg/liter), neomycin (500 mg/liter), metronidazole (500 mg/liter), ampicillin (1g/l) and colistin (80 mg/liter) or with sterile PBS (NT). After 24 h of incubation at 37°C, 104 EID50 of H5N9 virus were injected with 100 μl of the ABX cocktail (ABX) or with PBS (NT). Allantoic fluids were collected at 72 h postinfection, and viral RNA was extracted from 140 μl according to the manufacturer’s instructions (QIAamp viral RNA; Qiagen). Influenza virus nucleic acid load was determined by RT-qPCR using influenza virus segment M-specific primers (IAV-M; Table 1) in 96-well plates according to the manufacturer’s instructions (OneStep RT-PCR; Qiagen). Absolute quantification was performed using a standard curve based on a 10-fold serial dilution of a plasmid containing the A/Guinea Fowl/129/2015(H5N9) M gene.
Antibiotics and Poly(I·C) cell treatment.
Duck embryonic fibroblasts (ATCC, CCL-141) were cultured in cell culture medium supplemented or not with the following antibiotics: vancomycin (500 mg/liter), neomycin (500 mg/liter), metronidazole (500 mg/liter), ampicillin (1g/l) and colistin (80 mg/liter). After 24h, cell culture medium was renewed and cells were treated or not with Poly(I·C) (LMW)/LyoVec (Invivogen) at a final concentration of 1 μg/ml during 8 or 24 h and subsequently harvested for RNA extraction according to the manufacturer’s instructions (NucleoSpin RNA, Macherey-Nagel GmbH&Co, Germany). cDNA was synthesized by reverse transcription of 100 ng of total RNA using both oligo(dT)18 (0.25 μg) and random hexamer (0.1 μg) and the RevertAid First Strand cDNA Synthesis kit (Invitrogen, Thermo Fisher Scientific, Inc., Ontario, Canada) according to manufacturer’s instruction. Quantitative PCR for the analysis of IFIT5 and OAS-L mRNAs expression was performed in 96-well plates in a 20-μl final volume according to the manufacturer’s instructions (QuantiFast SYBR green PCR; Qiagen), along with 2 μl of cDNA and a 1 μM final concentration of each primer (Table 1). qPCR was performed on a LightCycler 96 (Roche, Mannheim, Germany) and relative quantification was carried out using the 2−ΔΔCT method with GAPDH mRNA expression for normalization.
RNA extraction from tissue samples and cDNA synthesis.
For each organ, 30-mg portions of tissue were placed in tubes with beads (Precellys lysis kit; Stretton Scientific, Ltd., Stretton, United Kingdom) filled with 600 μl of TRIzol reagent (Invitrogen, Carlsbad, CA) and mixed for 5 s at 6,000 rpm three times in a bead beater (Precellys 24; Bertin Technologies, Montigny-le-Bretonneux, France). After TRIzol extraction, the aqueous phase was transferred to an RNA extraction column and processed according to the manufacturer’s instructions (NucleoSpin RNA; Macherey-Nagel GmbH & Co, Germany). cDNA was synthesized by reverse transcription of 500 ng of total RNA using both oligo(dT)18 (0.25 μg) and random hexamer (0.1 μg) and a RevertAid first-strand cDNA synthesis kit (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s instructions.
Quantitative PCR from tissue samples.
Quantitative PCR for the analysis of host genes expression was performed in 384-well plates in a final volume of 5 μl using a Bravo automated liquid handling platform (Agilent Technologies, Palo Alto, CA) and a ViiA 7 real-time PCR system (Applied Biosystems, Foster City, CA) at the GeT-TRiX platform (GénoToul, Génopole, Toulouse, France). Mixes were prepared according to the manufacturer’s instructions (QuantiFast SYBR green PCR; Qiagen) with 1 μl of 1:20 diluted cDNA and a final 1 μM concentration of each primer (Table 1). Relative quantification was carried out by using the 2−ΔΔCT method (62), and the geometric means of two couples of housekeeping genes were validated in specific duck tissues, RPL4/RPL30 and RPL4/SDHA, for ileum and lung samples, respectively (63). Quantification of influenza virus nucleic acid load in tissues was performed in 96-well plates with a 20-μl final volume according to the manufacturer’s instructions (QuantiFast SYBR green PCR; Qiagen), along with 2 μl of cDNA and a final 1 μM concentration of each primer. We used influenza virus segment M-specific primers. For normalization, we used RPL30 for ileum samples and SDHA for lung samples, since they provided similar results to the geometric means of either RPL4/RPL30 or RPL4/SDHA. qPCR was performed on a LightCycler 96 (Roche), and relative quantification was carried out using the 2−ΔΔCT method.
Histopathological examination.
All animals were subjected to a complete postmortem examination. Tissue samples of the trachea (one transversal section in the proximal portion and another one in the terminal portion), lungs, ileum, cecum, colon, and brain were taken and stored in 10% neutral formalin. After fixation, tissues were processed in paraffin blocks, sectioned at 4 μm, and stained with hematoxylin and eosin for microscopic examination. A board-certified veterinary pathologist who was blind to the experimental conditions assessed lesions histologically. Lesion intensity was graded as follows: 0, no lesion; 1, minimal; 2, slight; 3, moderate; 4, marked, or 5, severe.
Immunohistochemistry.
Immunohistochemistry was performed on paraffin-embedded sections of trachea with a monoclonal mouse anti-nucleoprotein influenza A virus antibody (Argene 11-030; pronase 0.05% retrieval solution, 10 min at 37°C: antibody dilution 1/50, incubation overnight at 4°C). The immunohistochemical staining was revealed with a biotinylated polyclonal goat anti-mouse immunoglobulin conjugated with horseradish peroxidase (HRP; Dako, LSAB2 system-HRP, K0675) and the diaminobenzidine HRP chromogen (Thermo Scientific, TA-125-HDX). Negative controls comprised sections incubated either without specific primary antibody or with another monoclonal antibody of the same isotype (IgG2).
H5 HA fluorescent tissue staining and quantification.
A pCD5 plasmid was constructed with the HA sequence originating from A/Guinea Fowl/France/150207n/2015(H5N9) (GenBank KU320887.1), which has exactly the same amino acid sequence as the HA of A/Guinea Fowl/France/129/2015(H5N9). The plasmid encodes for a GCN4 leucine zipper trimerization motif, followed by a 7-amino-acid cleavage recognition sequence of tobacco etch virus and a sfGFP fused to Strep-tag II (IBA, Germany) C terminally. The vector was transfected into HEK293S GNT1(–) cells (which are modified HEK293S cells lacking glucosaminyltransferase I activity [ATCC CRL-3022]) with polyethyleneimine I (PEI) in a 1:8 ratio (μg of DNA:μg of PEI). Tissues were stained with purified recombinant multimeric H5N9 as previously described (24). Quantification of H5 HA fluorescent tissue staining was done on coded images so that the investigator was blind to experimental conditions. For each animal, a minimum of 40 regions of interest corresponding to the plasma membrane of individual cells were manually drawn using the 6-pixel-thick curved line tool in ImageJ software. The fluorescence intensity was measured for each region of interest, and the mean fluorescence intensity of the regions of interest for each animal was then calculated and plotted.
Statistical analysis.
Statistical significance was determined by using the Mann-Whitney test for individual time points. Statistical analyses were performed with Prism GraphPad software v5.01 (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
ACKNOWLEDGMENTS
We thank Jean-Luc Guérin and Luc Robertet (Université de Toulouse, ENVT, INRA, UMR 1225, Toulouse, France) for providing clinical samples that enabled the isolation of the A/Guinea Fowl/France/129/2015(H5N9) virus. We also thank Marie Souvestre and Luc Robertet for their assistance in the animal facility. We thank Sophie Allart and Astrid Canivet for technical assistance at the cellular imaging facility of INSERM UMR 1043, Toulouse, France. We thank the GeT-TRiX platform (GénoToul, Génopole, Toulouse, France) for access to the liquid handling platform and real-time PCR system.
This study was funded by the French National Agency for Research (ANR), project ANR-16-CE35-0005-01 Rule of Three to R.V. P.B. is supported by a Ph.D. scholarship funded by the Region Occitanie (France) and by the Chaire de Biosécurité at the École Nationale Vétérinaire de Toulouse (French Ministry of Agriculture). R.P.D.V. is a recipient of an ERC starting grant (802780) and a Beijerinck Premium of the Royal Dutch Academy of Sciences (KNAW).
REFERENCES
- 1.Munster VJ, Baas C, Lexmond P, Waldenström J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WEP, Schutten M, Olsen B, Osterhaus A, Fouchier R. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog 3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Osterhaus A, Munster VJ, Fouchier R. 2008. Epidemiology of avian influenza, p 1–10. In Klenk H-D, Matrosovich MN, Stech J (ed), Monographs in virology. Karger, Basel, Switzerland. [Google Scholar]
- 3.Böttcher-Friebertshäuser E, Klenk H-D, Garten W. 2013. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog Dis 69:87–100. doi: 10.1111/2049-632X.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swayne DE, Pantin-Jackwood M. 2009. Pathobiology of avian influenza virus infections in birds and mammals, p 87–122. In Avian influenza. John Wiley & Sons, Ltd, New York, NY. [Google Scholar]
- 5.Barber MRW, Aldridge JR, Webster RG, Magor KE. 2010. Association of RIG-I with innate immunity of ducks to influenza. Proc Natl Acad Sci U S A 107:5913–5918. doi: 10.1073/pnas.1001755107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liniger M, Summerfield A, Zimmer G, McCullough KC, Ruggli N. 2012. Chicken cells sense influenza A virus infection through MDA5 and CARDIF signaling involving LGP2. J Virol 86:705–717. doi: 10.1128/JVI.00742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelissen J, Vervelde L, Post J, Rebel J. 2013. Differences in highly pathogenic avian influenza viral pathogenesis and associated early inflammatory response in chickens and ducks. Avian Pathol 42:347–364. doi: 10.1080/03079457.2013.807325. [DOI] [PubMed] [Google Scholar]
- 8.Kuchipudi SV, Tellabati M, Sebastian S, Londt BZ, Jansen C, Vervelde L, Brookes SM, Brown IH, Dunham SP, Chang K-C. 2014. Highly pathogenic avian influenza virus infection in chickens but not ducks is associated with elevated host immune and proinflammatory responses. Vet Res 45:118. doi: 10.1186/s13567-014-0118-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann TW, Munier S, Larcher T, Soubieux D, Ledevin M, Esnault E, Tourdes A, Croville G, Guérin J-L, Quéré P, Volmer R, Naffakh N, Marc D. 2012. Length variations in the NA stalk of an H7N1 influenza virus have opposite effects on viral excretion in chickens and ducks. J Virol 86:584–588. doi: 10.1128/JVI.05474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soubies SM, Hoffmann TW, Croville G, Larcher T, Ledevin M, Soubieux D, Quéré P, Guérin J-L, Marc D, Volmer R. 2013. Deletion of the C-terminal ESEV domain of NS1 does not affect the replication of a low-pathogenic avian influenza virus H7N1 in ducks and chickens. J Gen Virol 94:50–58. doi: 10.1099/vir.0.045153-0. [DOI] [PubMed] [Google Scholar]
- 11.Burggraaf S, Karpala AJ, Bingham J, Lowther S, Selleck P, Kimpton W, Bean A. 2014. H5N1 infection causes rapid mortality and high cytokine levels in chickens compared to ducks. Virus Res 185:23–31. doi: 10.1016/j.virusres.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sommer F, Bäckhed F. 2013. The gut microbiota: masters of host development and physiology. Nat Rev Microbiol 11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 13.Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, Wherry EJ, Artis D. 2012. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ichinohe T, Pang IK, Kumamoto Y, Peaper DR, Ho JH, Murray TS, Iwasaki A. 2011. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A 108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh JZ, Ravindran R, Chassaing B, Carvalho FA, Maddur MS, Bower M, Hakimpour P, Gill KP, Nakaya HI, Yarovinsky F, Sartor RB, Gewirtz AT, Pulendran B. 2014. TLR5-mediated sensing of gut microbiota is necessary for antibody responses to seasonal influenza vaccination. Immunity 41:478–492. doi: 10.1016/j.immuni.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, Hickman HD, McCulloch JA, Badger JH, Ajami NJ, Trinchieri G, Pardo-Manuel de Villena F, Yewdell JW, Rehermann B. 2017. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell 171:1015–1028.e13. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bradley KC, Finsterbusch K, Schnepf D, Crotta S, Llorian M, Davidson S, Fuchs SY, Staeheli P, Wack A. 2019. Microbiota-driven tonic interferon signals in lung stromal cells protect from influenza virus infection. Cell Rep 28:245–256.e4. doi: 10.1016/j.celrep.2019.05.105. [DOI] [PubMed] [Google Scholar]
- 18.Yitbarek A, Taha-Abdelaziz K, Hodgins DC, Read L, Nagy É, Weese JS, Caswell JL, Parkinson J, Sharif S. 2018. Gut microbiota-mediated protection against influenza virus subtype H9N2 in chickens is associated with modulation of the innate responses. Sci Rep 8:13189. doi: 10.1038/s41598-018-31613-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yitbarek A, Alkie T, Taha-Abdelaziz K, Astill J, Rodriguez-Lecompte JC, Parkinson J, Nagy É, Sharif S. 2018. Gut microbiota modulates type I interferon and antibody-mediated immune responses in chickens infected with influenza virus subtype H9N2. Benef Microbes 9:417–427. doi: 10.3920/BM2017.0088. [DOI] [PubMed] [Google Scholar]
- 20.Vasaï F, Brugirard Ricaud K, Bernadet MD, Cauquil L, Bouchez O, Combes S, Davail S. 2014. Overfeeding and genetics affect the composition of intestinal microbiota in Anas platyrhynchos (Pekin) and Cairina moschata (Muscovy) ducks. FEMS Microbiol Ecol 87:204–216. doi: 10.1111/1574-6941.12217. [DOI] [PubMed] [Google Scholar]
- 21.Grond K, Sandercock BK, Jumpponen A, Zeglin LH. 2018. The avian gut microbiota: community, physiology and function in wild birds. J Avian Biol 49:e01788. doi: 10.1111/jav.01788. [DOI] [Google Scholar]
- 22.Srivillibhuthur M, Warder BN, Toke NH, Shah PP, Feng Q, Gao N, Bonder EM, Verzi MP. 2018. TFAM is required for maturation of the fetal and adult intestinal epithelium. Dev Biol 439:92–101. doi: 10.1016/j.ydbio.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Zhang Y, Li Y, Yan H, Zhang H. 2019. l-Tryptophan enhances intestinal integrity in diquat-challenged piglets associated with improvement of redox status and mitochondrial function. Animals (Basel) 9:E266. doi: 10.3390/ani9050266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nemanichvili N, Tomris I, Turner HL, McBride R, Grant OC, van der Woude R, Aldosari MH, Pieters RJ, Woods RJ, Paulson JC, Boons G-J, Ward AB, Verheije MH, de Vries RP. 2019. Fluorescent trimeric hemagglutinins reveal multivalent receptor binding properties. J Mol Biol 431:842–856. doi: 10.1016/j.jmb.2018.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crabbé PA, Bazin H, Eyssen H, Heremans JF. 1968. The normal microbial flora as a major stimulus for proliferation of plasma cells synthesizing IgA in the gut: the germ-free intestinal tract. Int Arch Allergy Appl Immunol 34:362–375. doi: 10.1159/000230130. [DOI] [PubMed] [Google Scholar]
- 26.Robak OH, Heimesaat MM, Kruglov AA, Prepens S, Ninnemann J, Gutbier B, Reppe K, Hochrein H, Suter M, Kirschning CJ, Marathe V, Buer J, Hornef MW, Schnare M, Schneider P, Witzenrath M, Bereswill S, Steinhoff U, Suttorp N, Sander LE, Chaput C, Opitz B. 2018. Antibiotic treatment-induced secondary IgA deficiency enhances susceptibility to Pseudomonas aeruginosa pneumonia. J Clin Invest 128:3535–3545. doi: 10.1172/JCI97065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briand F-X, Schmitz A, Ogor K, Le Prioux A, Guillou-Cloarec C, Guillemoto C, Allée C, Le Bras M-O, Hirchaud E, Quenault H, Touzain F, Cherbonnel-Pansart M, Lemaitre E, Courtillon C, Gares H, Daniel P, Fediaevsky A, Massin P, Blanchard Y, Eterradossi N, van der Werf S, Jestin V, Niqueux E. 2017. Emerging highly pathogenic H5 avian influenza viruses in France during winter 2015/16: phylogenetic analyses and markers for zoonotic potential. Euro Surveill 22:30473. doi: 10.2807/1560-7917.ES.2017.22.9.30473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briand FX, Niqueux E, Schmitz A, Hirchaud E, Quenault H, Allée C, Le Prioux A, Guillou-Cloarec C, Ogor K, Le Bras MO, Gares H, Daniel P, Fediaevsky A, Martenot C, Massin P, Le Bouquin S, Blanchard Y, Eterradossi N. 2018. Emergence and multiple reassortments of French 2015–2016 highly pathogenic H5 avian influenza viruses. Infect Genet Evol 61:208–214. doi: 10.1016/j.meegid.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 29.Steed AL, Christophi GP, Kaiko GE, Sun L, Goodwin VM, Jain U, Esaulova E, Artyomov MN, Morales DJ, Holtzman MJ, Boon ACM, Lenschow DJ, Stappenbeck TS. 2017. The microbial metabolite desaminotyrosine protects from influenza through type I interferon. Science 357:498–502. doi: 10.1126/science.aam5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li N, Ma W-T, Pang M, Fan Q-L, Hua J-L. 2019. The commensal microbiota and viral infection: a comprehensive review. Front Immunol 10:1551. doi: 10.3389/fimmu.2019.01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Chai W, Burwinkel M, Twardziok S, Wrede P, Palissa C, Esch B, Schmidt M. 2013. Inhibitory influence of Enterococcus faecium on the propagation of swine influenza A virus in vitro. PLoS One 8:e53043. doi: 10.1371/journal.pone.0053043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bandoro C, Runstadler JA. 2017. Bacterial lipopolysaccharide destabilizes influenza viruses. mSphere 2:e00267-17. doi: 10.1128/mSphere.00267-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belkaid Y, Harrison OJ. 2017. Homeostatic immunity and the microbiota. Immunity 46:562–576. doi: 10.1016/j.immuni.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature 535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 35.Trompette A, Gollwitzer ES, Pattaroni C, Lopez-Mejia IC, Riva E, Pernot J, Ubags N, Fajas L, Nicod LP, Marsland BJ. 2018. Dietary fiber confers protection against flu by shaping Ly6c– patrolling monocyte hematopoiesis and CD8+ T cell metabolism. Immunity 48:992–1005.e8. doi: 10.1016/j.immuni.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 36.Pagliuso A, Tham TN, Allemand E, Robertin S, Dupuy B, Bertrand Q, Bécavin C, Koutero M, Najburg V, Nahori M-A, Tangy F, Stavru F, Bessonov S, Dessen A, Muchardt C, Lebreton A, Komarova AV, Cossart P. 2019. An RNA-binding protein secreted by a bacterial pathogen modulates RIG-I signaling. Cell Host Microbe 26:823–835.e11. doi: 10.1016/j.chom.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazel-Sanchez B, Yildiz S, Schmolke M. 2019. Ménage à trois: virus, host, and microbiota in experimental infection models. Trends Microbiol 27:440–452. doi: 10.1016/j.tim.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 38.Libertucci J, Young VB. 2019. The role of the microbiota in infectious diseases. Nat Microbiol 4:35–45. doi: 10.1038/s41564-018-0278-4. [DOI] [PubMed] [Google Scholar]
- 39.Gopinath S, Kim MV, Rakib T, Wong PW, van Zandt M, Barry NA, Kaisho T, Goodman AL, Iwasaki A. 2018. Topical application of aminoglycoside antibiotics enhances host resistance to viral infections in a microbiota-independent manner. Nat Microbiol 3:611–621. doi: 10.1038/s41564-018-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volmer C, Soubies SM, Grenier B, Guérin J-L, Volmer R. 2011. Immune response in the duck intestine following infection with low-pathogenic avian influenza viruses or stimulation with a Toll-like receptor 7 agonist administered orally. J Gen Virol 92:534–543. doi: 10.1099/vir.0.026443-0. [DOI] [PubMed] [Google Scholar]
- 41.Kida H, Yanagawa R, Matsuoka Y. 1980. Duck influenza lacking evidence of disease signs and immune response. Infect Immun 30:547–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Webster RG, Yakhno M, Hinshaw VS, Bean WJ, Murti KG. 1978. Intestinal influenza: replication and characterization of influenza viruses in ducks. Virology 84:268–278. doi: 10.1016/0042-6822(78)90247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson CM. 2019. Enteric viruses exploit the microbiota to promote infection. Curr Opin Virol 37:58–62. doi: 10.1016/j.coviro.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, Chervonsky AV, Golovkina TV. 2011. Successful transmission of a retrovirus depends on the commensal microbiota. Science 334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, Grau KR, Gonzalez-Hernandez MB, Iovine NM, Wobus CE, Vinjé J, Tibbetts SA, Wallet SM, Karst SM. 2014. Enteric bacteria promote human and mouse norovirus infection of B cells. Science 346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, Diamond MS, Ivanova Y, Artyomov M, Virgin HW. 2015. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science 347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Boeckel TP, Brower C, Gilbert M, Grenfell BT, Levin SA, Robinson TP, Teillant A, Laxminarayan R. 2015. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A 112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Adam CJM, Fortané N, Coviglio A, Delesalle L, Ducrot C, Paul MC. 2019. Epidemiological assessment of the factors associated with antimicrobial use in French free-range broilers. BMC Vet Res 15:219. doi: 10.1186/s12917-019-1970-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Fels-Klerx HJ, Puister-Jansen LF, van Asselt ED, Burgers S. 2011. Farm factors associated with the use of antibiotics in pig production. J Anim Sci 89:1922–1929. doi: 10.2527/jas.2010-3046. [DOI] [PubMed] [Google Scholar]
- 51.Pardon B, Catry B, Dewulf J, Persoons D, Hostens M, De Bleecker K, Deprez P. 2012. Prospective study on quantitative and qualitative antimicrobial and anti-inflammatory drug use in white veal calves. J Antimicrob Chemother 67:1027–1038. doi: 10.1093/jac/dkr570. [DOI] [PubMed] [Google Scholar]
- 52.Gresse R, Chaucheyras-Durand F, Fleury MA, Van de Wiele T, Forano E, Blanquet-Diot S. 2017. Gut microbiota dysbiosis in postweaning piglets: understanding the keys to health. Trends Microbiol 25:851–873. doi: 10.1016/j.tim.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 53.Yeoman CJ, White BA. 2014. Gastrointestinal tract microbiota and probiotics in production animals. Annu Rev Anim Biosci 2:469–486. doi: 10.1146/annurev-animal-022513-114149. [DOI] [PubMed] [Google Scholar]
- 54.Harris VC, Haak BW, Handley SA, Jiang B, Velasquez DE, Hykes BL, Droit L, Berbers GAM, Kemper EM, van Leeuwen EMM, Boele van Hensbroek M, Wiersinga WJ. 2018. Effect of antibiotic-mediated microbiome modulation on rotavirus vaccine immunogenicity: a human, randomized-control proof-of-concept trial. Cell Host Microbe 24:197–207.e4. doi: 10.1016/j.chom.2018.07.005. [DOI] [PubMed] [Google Scholar]
- 55.Belkacem N, Serafini N, Wheeler R, Derrien M, Boucinha L, Couesnon A, Cerf-Bensussan N, Gomperts Boneca I, Di Santo JP, Taha M-K, Bourdet-Sicard R. 2017. Lactobacillus paracasei feeding improves immune control of influenza infection in mice. PLoS One 12:e0184976. doi: 10.1371/journal.pone.0184976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hagan T, Cortese M, Rouphael N, Boudreau C, Linde C, Maddur MS, Das J, Wang H, Guthmiller J, Zheng N-Y, Huang M, Uphadhyay AA, Gardinassi L, Petitdemange C, McCullough MP, Johnson SJ, Gill K, Cervasi B, Zou J, Bretin A, Hahn M, Gewirtz AT, Bosinger SE, Wilson PC, Li S, Alter G, Khurana S, Golding H, Pulendran B. 2019. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 178:1313–1328.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yitbarek A, Astill J, Hodgins DC, Parkinson J, Nagy É, Sharif S. 2019. Commensal gut microbiota can modulate adaptive immune responses in chickens vaccinated with whole inactivated avian influenza virus subtype H9N2. Vaccine 37:6640–6647. doi: 10.1016/j.vaccine.2019.09.046. [DOI] [PubMed] [Google Scholar]
- 58.Vitetta L, Saltzman ET, Thomsen M, Nikov T, Hall S. 2017. Adjuvant probiotics and the intestinal microbiome: enhancing vaccines and immunotherapy outcomes. Vaccines (Basel) 5. doi: 10.3390/vaccines5040050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Simon K, Verwoolde MB, Zhang J, Smidt H, de Vries Reilingh G, Kemp B, Lammers A. 2016. Long-term effects of early life microbiota disturbance on adaptive immunity in laying hens. Poult Sci 95:1543–1554. doi: 10.3382/ps/pew088. [DOI] [PubMed] [Google Scholar]
- 60.Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, McCoy KD, Macpherson AJ, Meza-Zepeda LA, Johansen F-E. 2011. Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One 6:e17996. doi: 10.1371/journal.pone.0017996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elderman M, Sovran B, Hugenholtz F, Graversen K, Huijskes M, Houtsma E, Belzer C, Boekschoten M, P de V, Dekker J, Wells J, Faas M. 2017. The effect of age on the intestinal mucus thickness, microbiota composition and immunity in relation to sex in mice. PLoS One 12:e0184274. doi: 10.1371/journal.pone.0184274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔC(T) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 63.Chapman JR, Helin AS, Wille M, Atterby C, Järhult JD, Fridlund JS, Waldenström J. 2016. A panel of stably expressed reference genes for real-time qPCR gene expression studies of mallards (Anas platyrhynchos). PLoS One 11:e0149454. doi: 10.1371/journal.pone.0149454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wen M, Zhao H, Liu G, Chen X, Wu B, Tian G, Cai J, Jia G. 2018. Effect of zinc supplementation on growth performance, intestinal development, and intestinal barrier-related gene expression in Pekin ducks. Biol Trace Elem Res 183:351–360. doi: 10.1007/s12011-017-1143-7. [DOI] [PubMed] [Google Scholar]
- 65.Fouchier RA, Bestebroer TM, Herfst S, Van Der Kemp L, Rimmelzwaan GF, Osterhaus AD. 2000. Detection of influenza A viruses from different species by PCR amplification of conserved sequences in the matrix gene. J Clin Microbiol 38:4096–4101. doi: 10.1128/JCM.38.11.4096-4101.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vanderven HA, Petkau K, Ryan-Jean KEE, Aldridge JR, Webster RG, Magor KE. 2012. Avian influenza rapidly induces antiviral genes in duck lung and intestine. Mol Immunol 51:316–324. doi: 10.1016/j.molimm.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H, Zhai Y, Fan Y, Chen H, Zhang A, Jin H, Luo R. 2016. Molecular cloning and functional characterization of duck mitochondrial antiviral-signaling protein (MAVS). Dev Comp Immunol 56:1–6. doi: 10.1016/j.dci.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 68.Saito LB, Diaz-Satizabal L, Evseev D, Fleming-Canepa X, Mao S, Webster RG, Magor KE. 2018. IFN and cytokine responses in ducks to genetically similar H5N1 influenza A viruses of varying pathogenicity. J Gen Virol 99:464–474. doi: 10.1099/jgv.0.001015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bacchetti De Gregoris T, Aldred N, Clare AS, Burgess JG. 2011. Improvement of phylum- and class-specific primers for real-time PCR quantification of bacterial taxa. J Microbiol Methods 86:351–356. doi: 10.1016/j.mimet.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 70.Ruan D, Wang WC, Lin CX, Fouad AM, Chen W, Xia WG, Wang S, Luo X, Zhang WH, Yan SJ, Zheng CT, Yang L. 2019. Effects of curcumin on performance, antioxidation, intestinal barrier and mitochondrial function in ducks fed corn contaminated with ochratoxin A. Animal 13:42–52. doi: 10.1017/S1751731118000678. [DOI] [PubMed] [Google Scholar]
- 71.Cornelissen J, Post J, Peeters B, Vervelde L, Rebel J. 2012. Differential innate responses of chickens and ducks to low-pathogenic avian influenza. Avian Pathol 41:519–529. doi: 10.1080/03079457.2012.732691. [DOI] [PubMed] [Google Scholar]
- 72.Qian W, Wei X, Zhou H, Jin M. 2016. Molecular cloning and functional analysis of duck ubiquitin-specific protease 18 (USP18) gene. Dev Comp Immunol 62:39–47. doi: 10.1016/j.dci.2016.04.008. [DOI] [PubMed] [Google Scholar]