Picornaviruses, as a large family of human and animal pathogens, cause a bewildering array of disease syndromes. Many host factors are implicated in the pathogenesis of these viruses, and some proteins interact with the viral IRES elements to affect function. Here, we report for the first time that cellular hnRNP L specifically interacts with the IRES of the picornavirus FMDV and negatively regulates FMDV replication through inhibiting viral RNA synthesis. Further, our results showed that hnRNP L coimmunoprecipitates with FMDV 3Dpol in a viral RNA-dependent manner, suggesting that it may remain in the replication complex to function. The data presented here would facilitate further understanding of virus-host interactions and the pathogenesis of picornavirus infections.
KEYWORDS: foot-and-mouth disease virus, internal ribosome entry site, heterogeneous nuclear ribonucleoprotein L, viral RNA synthesis, 3Dpol, virus-host interaction
ABSTRACT
Upon infection, the highly structured 5′ untranslated region (5′ UTR) of picornavirus is involved in viral protein translation and RNA synthesis. As a critical element in the 5′ UTR, the internal ribosome entry site (IRES) binds to various cellular proteins to function in the processes of picornavirus replication. Foot-and-mouth disease virus (FMDV) is an important member in the family Picornaviridae, and its 5′ UTR contains a functional IRES element. In this study, the cellular heterogeneous nuclear ribonucleoprotein L (hnRNP L) was identified as an IRES-binding protein for FMDV by biotinylated RNA pulldown assays, mass spectrometry (MS) analysis, and determination of hnRNP L-IRES interaction regions. Further, we found that hnRNP L inhibited the growth of FMDV through binding to the viral IRES and that the inhibitory effect of hnRNP L on FMDV growth was not due to FMDV IRES-mediated translation, but to influence on viral RNA synthesis. Finally, hnRNP L was demonstrated to coimmunoprecipitate with RNA-dependent RNA polymerase (3Dpol) in an FMDV RNA-dependent manner in the infected cells. Thus, our results suggest that hnRNP L, as a critical IRES-binding protein, negatively regulates FMDV replication by inhibiting viral RNA synthesis, possibly by remaining in the replication complex.
IMPORTANCE Picornaviruses, as a large family of human and animal pathogens, cause a bewildering array of disease syndromes. Many host factors are implicated in the pathogenesis of these viruses, and some proteins interact with the viral IRES elements to affect function. Here, we report for the first time that cellular hnRNP L specifically interacts with the IRES of the picornavirus FMDV and negatively regulates FMDV replication through inhibiting viral RNA synthesis. Further, our results showed that hnRNP L coimmunoprecipitates with FMDV 3Dpol in a viral RNA-dependent manner, suggesting that it may remain in the replication complex to function. The data presented here would facilitate further understanding of virus-host interactions and the pathogenesis of picornavirus infections.
INTRODUCTION
The impact of picornavirus infections on human and animal health has been one of the most threatening problems (1, 2). In the single-stranded positive-sense RNA genome of picornavirus, the 5′ untranslated region (5′ UTR) functions as a platform to recruit host factors for the regulation of viral protein translation and viral RNA synthesis (3, 4). During these processes, the internal ribosomal entry site (IRES) located in the 5′ UTR of the picornavirus genome plays important roles. Foot-and-mouth disease virus (FMDV), the most important veterinary pathogen affecting domestic and wild cloven-hoofed animals, contains a type II IRES in its genomic 5′ UTR (5, 6).
The IRES elements, initially reported in picornaviruses, are characterized by complicated secondary or tertiary structures assembled with stem-loops and pseudoknots, which can serve as ribosome landing pads through multiple RNA-RNA and RNA-protein interactions (7–9). Various cellular proteins binding to viral IRES have been reported to be functionally important for the virus life cycle, and most of them function by affecting viral translation and/or RNA replication. During picornavirus infections, src-associated protein in mitosis (Sam68) interacts with the IRES of enterovirus 71 (EV71) to positively regulate viral protein translation (10), while ribosomal protein L13 (RPL13), and gem nuclear organelle associated protein 5 (Gemin5) associate with the FMDV IRES for participating in IRES-driven translation (11–13). For hepatitis C virus (HCV), heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) and AU-rich element binding factor 1 (AUF1, hnRNP D) positively affect viral translation (14, 15). Poly(rC)-binding protein 2 (PCBP2, hnRNP E2) not only regulates viral IRES-mediated translation but also modulates viral RNA replication in HCV (16–18) and poliovirus (PV) (19, 20).
In addition, studies on the molecular mechanism of virulence and attenuation in picornaviruses have shown that modulating the interaction between cellular factors and IRESs provides a good strategy for developing live attenuated vaccines. For example, mutations in the IRES of PV Sabin vaccine strains reduce its binding affinity for polypyrimidine tract binding protein (PTB)/neural PTB, which may explain the neurovirulence attenuation of Sabin vaccine strains (21, 22). Our previous results determined that a nucleotide mutation in the IRES of FMDV impairs its binding to PTB and thus endows FMDV with a significant temperature-sensitive and attenuated phenotype (23). Thus, knowledge of the cellular proteins that associate with the FMDV IRES will facilitate understanding of virus-host interactions and viral pathogenesis, and these IRES-associated cellular proteins could be crucial molecular targets for antiviral development.
To assess the effects on viral replication of cellular proteins interacting with IRESs, we isolated eight cellular proteins associated with FMDV IRESs using the biotinylated RNA pulldown approach followed by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) analysis. Among these proteins, heterogeneous nuclear ribonucleoprotein L (hnRNP L) has been reported to interact with HCV IRES and promote viral growth (24, 25), but its effect on picornavirus replication remains unknown. In this study, the hnRNP L was identified as an IRES-binding protein interacting with domains 4 to 5 of FMDV IRES through its RNA-binding region RRM3-4. Importantly, we found that hnRNP L inhibits viral growth not by affecting FMDV IRES-mediated translation but by influencing viral RNA synthesis. This heterogeneous nuclear ribonucleoprotein hnRNP L, redistributed from the nucleus to the cytoplasm to associate with FMDV RNA in the infected cells, coimmunoprecipitates with 3Dpol in a viral RNA-dependent manner, suggesting that it possibly functions by staying in the replication complex. Thus, our results indicate that the hnRNP L is an important IRES-binding protein and negatively regulates FMDV replication by inhibiting viral RNA synthesis.
RESULTS
hnRNP L specifically binds to the IRES of FMDV RNA.
The interactions between viral IRESs and cellular proteins in picornavirus-infected cells are crucial for the process of viral replication and translation. To further understand this process in FMDV infection, the cellular proteins that associate with FMDV IRES were isolated using a biotinylated RNA pulldown assay (Fig. 1A), followed by LC-MS/MS analysis (Table 1). As shown in Table 1, the isolated cellular proteins associated with FMDV IRES were named with their accession numbers obtained from the protein database, including polypyrimidine tract binding protein 1 (PTB) (26, 27) in band 5 of Fig. 1A and Ras GTPase-activating protein-binding protein 1 (G3BP1) (28) in band 6 of Fig. 1A, two known cellular proteins that interacts with picornavirus IRES, proving that our RNA-protein pulldown assay was effective.
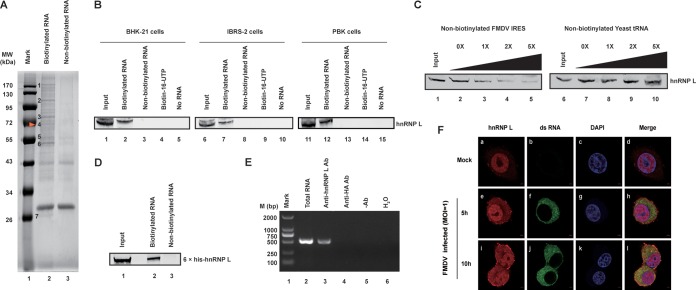
FIG 1.
Interaction of hnRNP L with FMDV IRES. (A) Isolation of cellular proteins associated with FMDV IRES. Biotinylated RNA pulldown assay was performed as described in the Materials and Methods section. (B) FMDV IRES associates with hnRNP L in various cell lines. The inputs of different cell lysates are shown in lanes 1, 6, and 11. The cell lysates of BHK-21, IBRS-2, and PBK were incubated in the absence of RNA (lanes 5, 10, and 15) or the presence of biotin-16-UTP (lanes 4, 9, and 14), nonbiotinylated full-length FMDV IRES (lanes 3, 8, and 13) or biotinylated full-length FMDV IRES (lanes 2, 7, and 12). After the beads were washed, bound proteins were resolved using 12% SDS–PAGE. hnRNP L protein was visualized by immunoblot analysis with anti-hnRNP L protein antibody. (C) The specific association of hnRNP L with FMDV IRES was confirmed by competition assay. Various amounts of unlabeled RNA were added to compete with biotin-labeled FMDV IRES for interacting with hnRNP L. Lanes 1 and 6 contained cell lysate (200 μg) only. Unlabeled FMDV IRES was used in the competition assay (lanes 2 to 5), and unlabeled yeast tRNA was utilized (lanes 7 to 10) as a negative control. (D) FMDV IRES RNA was pulled down with purified hnRNP L. The 6×His-fused hnRNP L protein was purified with Ni-affinity chromatography and then used to perform pulldown assays. (E) FMDV RNA was pulled down with hnRNP L from FMDV-infected cell extracts. BHK-21 cell extracts collected at 8 hpi with FMDV (MOI = 1) were subjected to immunoprecipitation with an antibody against hnRNP L. Two negative controls, a control antibody against HA-tag and a normal mouse IgG, were included in this experiment. The RNA pulled down by the immunocomplexes was extracted and amplified by RT-PCR by using primers specific to the FMDV IRES. (F) hnRNP L localized to the cytoplasm and was associated with viral RNA in FMDV-infected cells. Normal BHK-21 cells (mock-infected) (panels a to d) or BHK-21 cells infected with FMDV (MOI = 1) at 5 hpi (panels e to h) and 10 hpi (panels i to l) were fixed and stained with antibodies against hnRNP L and FMDV viral RNA. Panels a, e, and i were treated with anti-hnRNP L antibody and examined with an Alexa Fluor 633 filter; panels b, f, and j were treated with anti-dsRNA antibody and examined with an FITC filter; panels c, g, and k were treated with Hoechst 33258 and examined with a 40,60-diamidino-2-phenylindole (DAPI) filter. Panels d, h, and l show merged Alexa Fluor, FITC, and Hoechst images.
TABLE 1.
LC-MS/MS analysis of the cellular proteins associated with FMDV IRES
| No. | NCBI description | Accession no. | Percent sequence coverage | Mass (kDa) |
|---|---|---|---|---|
| 1 | Ribosome-binding protein 1 (RRBP1) | Q99PL5 | 4.92 | 173 |
| 2 | Probable ATP-dependent RNA helicase DDX46 (DDX46) | Q569Z5 | 2.71 | 117 |
| 3 | Heterogeneous nuclear ribonucleoprotein M (hnRNP M) | Q9D0E1 | 19.70 | 80 |
| 4 | Heterogeneous nuclear ribonucleoprotein L (hnRNP L) | Q8R081 | 17.06 | 64 |
| 5 | Polypyrimidine tract binding protein 1 (PTB) | Q8K144 | 11.35 | 58 |
| 6-1 | Heterogeneous nuclear ribonucleoprotein K (hnRNP K) | P61979 | 33.05 | 51 |
| 6-2 | Ras GTPase-activating protein-binding protein 1 (G3BP1) | P97855 | 17.20 | 52 |
| 7 | High mobility group protein B1 (HMGB1) | P63158 | 44.19 | 25 |
Among the eight proteins associated with FMDV IRES (Table 1), derived from the seven protein bands (Fig. 1A), some belong to the heterogeneous nuclear ribonucleoproteins (hnRNPs) family, including PTB (hnRNP I), hnRNP L, hnRNP M, and hnRNP K. Although hnRNP L has been reported to interact with HCV IRES and promote viral growth (24, 25), its impact on picornavirus replication remains unclear. Thus, hnRNP L derived from protein band 4 was selected to elucidate its function in FMDV replication. First, the interaction between hnRNP L and FMDV IRES RNA was verified by immunoblotting with an anti-hnRNP L antibody. The 68-kDa hnRNP L protein from several species, including that from hamster-derived BHK-21 cells, porcine-derived IBRS-2 cells, and bovine-derived PBK cells, was separately pulled down by biotin-IRES (Fig. 1B). This result revealed that the association of cellular hnRNP L with FMDV IRES is a common phenomenon among different FMDV-susceptible cells. To confirm the specificity of the interaction between FMDV IRES and hnRNP L, a competition test was performed by the pulldown assay with different amounts of nonbiotinylated FMDV IRES or yeast tRNA. As revealed in Fig. 1C, the interaction between hnRNP L and FMDV IRES was out-competed by nonbiotinylated FMDV IRES (lanes 2, 3, 4, and 5) but not by yeast tRNA (lanes 7, 8, 9, and 10), demonstrating that the interaction is FMDV IRES-specific. In addition, the 6×His-hnRNP L expressed in E. coli clearly bound the FMDV IRES transcripts (Fig. 1D), supporting the notion that the interaction between hnRNP L and FMDV IRES RNA is direct.
To demonstrate this interaction between hnRNP L and FMDV IRES in the FMDV-infected cells, BHK-21 cell lysates collected at 8 h postinfection (hpi) with FMDV (MOI = 1) were subjected to immunoprecipitation with an anti-hnRNP L antibody and an antibody against the HA-tag, while a normal mouse IgG was used as a negative control. The immunocomplexes were extracted and amplified by reverse transcriptase PCR (RT-PCR) using primers specific to the FMDV IRES. A cDNA band with the expected size (500 bp) was noted in immunoprecipitates brought down by anti-hnRNP L antibody but not by anti-HA antibody or normal mouse IgG (Fig. 1E). These data demonstrated that hnRNP L specifically binds to the viral RNA in FMDV-infected cells. As a member of the hnRNP family that shuttles from the nucleus to the cytoplasm, the localization of hnRNP L in FMDV-infected cells was examined to determine its association with the viral RNA in the cytoplasm. As shown in Fig. 1F, hnRNP L is mainly localized in the cell nucleus in mock-infected cells (panel a), but it redistributed to the cytoplasm (panels e and i) after the cells were infected with FMDV (panels f and j). Moreover, the cytoplasmic hnRNP L signals colocalized with those of viral RNA at 5 hpi and 10 hpi (Fig. 1F, panels h and l), supporting the interaction between hnRNP L and FMDV RNA during viral infection.
Taken together, the results in Fig. 1 suggest that hnRNP L interacts with FMDV IRES.
Interaction regions between FMDV IRES element and cellular hnRNP L protein.
It is known that the 450-nt FMDV IRES folds into multiple stem-loops that are organized into four domains (29) (Fig. 2A). To find the IRES domain(s) responsible for binding hnRNP L, full-length IRES (Fig. 2A, row a) and its three truncated forms, domain 2-3 (Fig. 2A, row b), domain 3-4 (Fig. 2A, row c), and domain 4-5 (Fig. 2A, row d), were synthesized by in vitro transcription to evaluate their ability to bind hnRNP L. As presented in Fig. 2B, except for full-length IRES, hnRNP L copurified only with transcripts of IRES domain 4-5, indicating that the domain 4-5 regions of FMDV IRES are responsible for the binding of hnRNP L.
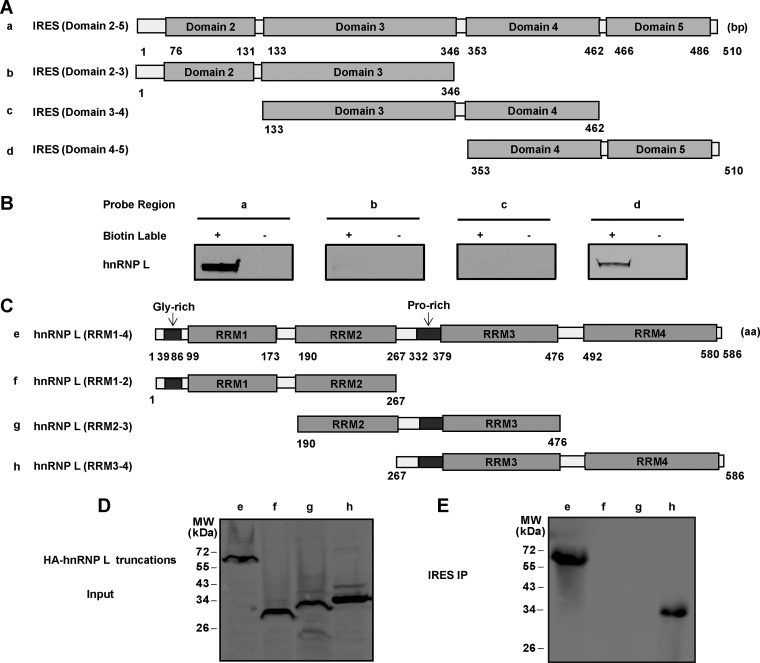
FIG 2.
Identification of interaction regions between FMDV IRES and hnRNP L. (A) Schematic diagram of FMDV IRES and its truncated forms. Four truncated forms of IRES were generated: domain 2-5 (a), domain 2-3 (b), domain 3-4 (c), and domain 4-5 (d). (B) Mapping interaction regions in FMDV IRES for hnRNP L. The truncated forms of IRES RNA were transcribed in vitro and biotinylated. BHK-21 cell lysates were incubated with these biotinylated RNA (lanes a, b, c, and d). Nonbiotinylated RNA was used in this assay as a control. The RNA and protein complex-associated beads were pulled down and resolved by SDS-PAGE (12%). An anti-hnRNP L antibody was used to detect hnRNP L in the pulldown complex. (C) Schematic diagram of hnRNP L and its truncated mutant forms. Four truncated forms of hnRNP L, RRM1-4 (e), RRM1-2 (f), RRM2-3 (g), and RRM3-4 (h) were generated and fused with HA-tags at their N termini. (D) Expression of truncated forms of hnRNP L in HEK-293T cells. Western blotting using an anti-HA antibody was employed to examine the protein expression. (E) Mapping interaction regions between hnRNP L protein and FMDV IRES. Cell extracts of transfected HEK-293T cells were collected at 48 h posttransfection and then incubated with biotinylated FMDV IRES. Streptavidin beads were used in the pulldown assay and an anti-HA antibody was used to detect hnRNP L in the pulldown complex.
The 58×-aa RNA-binding protein hnRNP L contains four RNA recognition motifs (RRMs) (Fig. 2C) that bind RNA regions with CA repeats or CA-rich elements (30). Here, we constructed hnRNP L truncations fused with the HA-tag to identify the RRM(s) involved in the interaction with FMDV IRES by RNA pulldown assay. The complete hnRNP L (Fig. 2C, row e) and its truncated forms RRM1-2 (Fig. 2C, row f), RRM2-3 (Fig. 2C, row g), and RRM3-4 (Fig. 2C, row h) were visualized by Western blotting using an anti-HA antibody (Fig. 2D) in HEK-293T cells. Through binding the biotinylated FMDV IRES, the streptavidin beads captured IRES-associated full-length hnRNP L and its truncated form RRM3-4, but not its truncated forms RRM1-2 or RRM2-3 (Fig. 2E). These results indicated that hnRNP L interacts with FMDV IRES through the RNA-binding region RRM3-4.
hnRNP L inhibits FMDV replication via binding to viral IRES.
To address the role of hnRNP L in FMDV infection via interaction with the viral IRES, BHK-21 cells were transfected with pCAGGS-HA-hnRNP L, pCAGGS-HA-eGFP, or pCAGGS empty vector, followed by infection with FMDV (MOI = 1). As shown in Fig. 3A, compared to overexpression of eGFP and empty vector, the overexpression of hnRNP L resulted in significant decreases in the production of infectious FMDV progeny throughout the experimental period, especially at 8 hpi and 12 hpi. Upon overexpression of hnRNP L, the expression of FMDV VP2 protein and viral RNA level were also reduced compared to that of eGFP and empty vector controls (Fig. 3B and C). This result suggests that hnRNP L negatively regulates FMDV replication in infected cells.
FIG 3.
Overexpression of hnRNP L suppresses the replication of FMDV. BHK-21 cells were transfected with pCAGGS-HA-hnRNP L, pCAGGS-HA-eGFP, or pCAGGS empty vector. The transfected cells were then infected with FMDV at an MOI of 1 and the resulting viruses were harvested at 4 h, 8 h, and 12 h, and the supernatant was analyzed for virus production by TCID50 assay (A), for viral protein expression by Western blotting (B), and for viral RNA synthesis by rRT-PCR (C).
The notion that hnRNP L is an inhibitor of FMDV infection was further verified in BHK-21 cells by hnRNP L knockout using the CRISPR/Cas9 system (31, 32). A specific guide RNA designed to target the golden hamster-derived hnRNP L locus (Fig. 4A) was subcloned into the pSpCas9(BB)-2A-GFP (PX458) vector and transfected into BHK-21 cells, and a number of clones were screened by limiting dilution. Finally, one cell clone (L3) that did not express any hnRNP L protein (Fig. 4C) was isolated, and a small deletion was confirmed by sequencing the hnRNP L gene and amplifying a 250-bp DNA fragment from the targeted locus in the L3 cell clone (Fig. 4B and D). Importantly, the knockout of hnRNP L did not affect the proliferation of the L3 cell clone (Fig. 4E). These results demonstrated that the hnRNP L gene was successfully knocked out from the L3 clone. The L3 clone was then used to detect the effect of hnRNP L knockout on FMDV replication. As shown in Fig. 4F, enhanced FMDV production was observed in the L3 clone compared to normal BHK-21 cells at 8 hpi and 12 hpi. In addition, the expression of viral VP2 protein was also significantly increased in the FMDV-infected L3 clone (Fig. 4G). These results indicated that hnRNP L negatively regulates FMDV replication, which was consistent with the consequence of hnRNP L overexpression.
FIG 4.
Effect of hnRNP L knockout on picornavirus replication. (A) Schematic illustrating Cas9 inactivation of the hamster hnRNP L locus. The 20-bp guide RNA target sequence is shown in green, and the protospacer-adjacent motif (PAM) is shown in red. The sense primer hnRNP L-KO-S and antisense primer hnRNP L-KO-A sequences used to amplify this gene locus are boxed in purple. (B) Sequence alignment of hnRNP L in BHK-21 cells and L3 clone cells. Numbers indicate the nucleotide positions in the hnRNP L open reading frame. (C) Analysis of hnRNP protein expression in three clones (L2, L3, and L4) isolated from BHK-21 cells transfected with Cas9 and hnRNP L guide RNA expression vectors by Western blotting. (D) A 250-bp DNA fragment was PCR-amplified from the hnRNP L locus of the L3 clone and normal BHK-21 cells using primers hnRNP L-KO-S and hnRNP L-KO-A and analyzed by 10% TBE-polyacrylamide gel. (E) Proliferation test of BHK-21 cells and L3 clones. (F to K) Effects of hnRNP L knockout on viral replication. BHK-21 cells or L3 clones were infected with FMDV, BEV, or EMCV. The resulting viruses were harvested at 4 h, 8 h, and 12 h, and the supernatant was analyzed for virus titer by TCID50 assay (F, H, and J) and for viral protein expression by Western blotting (G, I, and K).
We further explored the effects of hnRNP L knockout on the replication of other picornaviruses. Bovine enterovirus (BEV) infection was similarly enhanced in the L3 clone as measured by viral progeny production in 50% tissue culture infective dose (TCID50) assays (Fig. 4H) and by viral protein expression assessed by Western blotting (Fig. 4I), indicating that hnRNP L also represses the replication of BEV. However, the knockout of hnRNP L did not affect encephalomyocarditis virus (EMCV) replication (Fig. 4J and K), suggesting that the effect of hnRNP L is different for different picornaviruses.
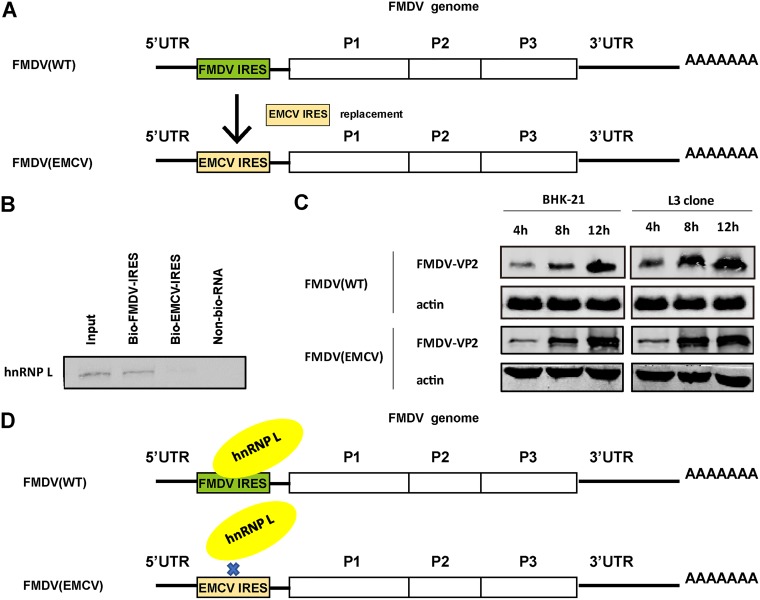
To further determine whether the inhibitory effect of hnRNP L on FMDV replication is due to its binding to the viral IRES, the role of hnRNP L in the viral life cycle was investigated by blocking the binding of hnRNP L to the IRES. As knockout of hnRNP L significantly promoted replication of FMDV (Fig. 4F and G) but not EMCV (Fig. 4J and K), we speculated that the different effects of hnRNP L on FMDV and EMCV replication may be due to the different binding ability of their IRESs to hnRNP L. To test this hypothesis, we first validated the specificity of the interaction between the FMDV IRES and cellular hnRNP L using an RNA-protein pulldown assay. As shown in Fig. 5B, unlike FMDV IRES specifically binding to hnRNP L, the interaction of EMCV IRES with hnRNP L was not observed. Furthermore, we constructed a chimeric virus named FMDV(EMCV) (Fig. 5A), in which the IRES of FMDV was completely replaced with that of EMCV in an infectious clone of serotype O FMDV, and its replication kinetics were analyzed compared to its parental virus FMDV(WT). As shown in Fig. 5C, the VP2 expression of FMDV(WT) at 4, 8, and 12 hpi was significantly enhanced in L3 clone cells compared to normal BHK-21 cells, but the VP2 expression of FMDV(EMCV) had no significant changes in either BHK-21 cells or L3 cells at all time points. These results could explain why the knockout of hnRNP L did not affect replication of FMDV(EMCV) in the L3 cells when the IRES of FMDV was replaced with the IRES of EMCV (Fig. 5A), which did not interact with hnRNP L (Fig. 5D), indicating that negative regulation of FMDV replication by hnRNP L is due to its binding to viral IRES.
FIG 5.
Regulation of FMDV replication by hnRNP L is due to its binding to viral IRES. (A) Schematic representation of the IRES-replaced mutant FMDV(EMCV). FMDV genome showing the encoded polyproteins (P1, P2, and P3) flanked by the 5′ UTR and 3′ UTR. In the mutant FMDV(EMCV) genome, the complete IRES of FMDV was replaced by that of EMCV. The cognate IRES of FMDV(WT) (green) was exchanged with its counterpart from EMCV (yellow). (B) RNA-protein pulldown assay between EMCV IRES and hnRNP L. (C) BHK-21 cells or L3 clones were infected with FMDV(WT) and FMDV(EMCV). The resulting viruses were harvested at 4 h, 8 h, and 12 h, and the supernatant was analyzed for viral protein expression by Western blotting. (D) Schematic diagram of hnRNP L binding to FMDV IRES.
Inhibition of FMDV replication by hnRNP L is not achieved by affecting IRES-dependent translation.
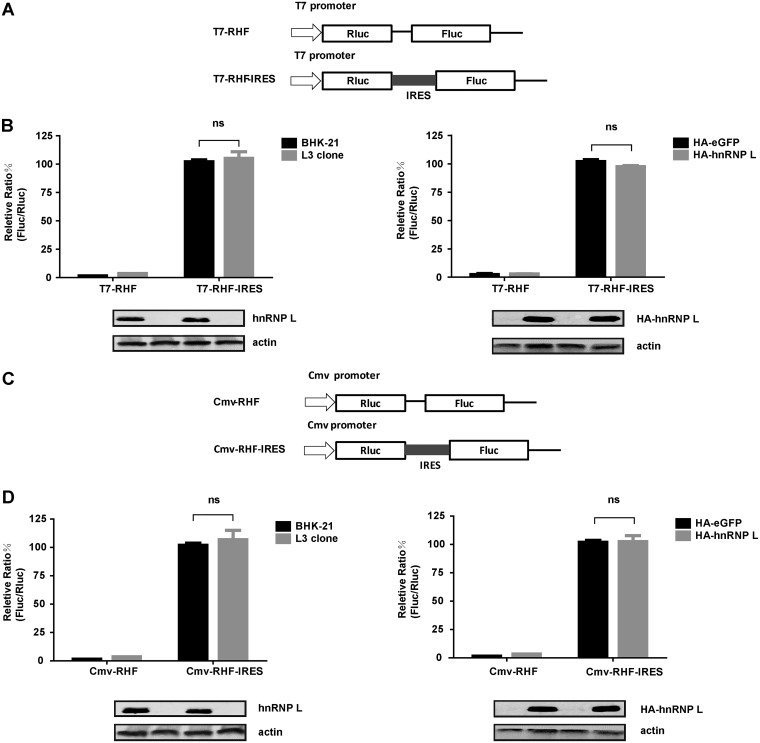
Since the regulation of FMDV replication by hnRNP L is related to its binding to viral IRES, we assumed that hnRNP L inhibits FMDV replication by affecting IRES-dependent translation. To test this hypothesis, we checked the effect of knockout or overexpression of hnRNP L on FMDV IRES activity. Two dicistronic reporter plasmids were used to evaluate FMDV IRES activity using two different promoters (Fig. 6A and C). For these plasmids, translation of the first cistron (Renilla luciferase, RLuc) was cap-dependent, whereas translation of the second cistron (firefly luciferase, FLuc) was engineered to be FMDV IRES-dependent, such that the yield ratio of FLuc expression to RLuc expression allowed a measurement of the relative IRES activity. In the BHK-21 cell lines transfected with the T7 promoter-bicistronic plasmid, knockout (Fig. 6B, left) or overexpression (Fig. 6B, right) of hnRNP L resulted in the same IRES activity. Similarly, in the BHK-21 cell lines transfected with the CMV promoter-dicistronic plasmid, knockout (Fig. 6D, left) or overexpression (Fig. 6D, right) of hnRNP L did not significantly impact FMDV IRES activity. This finding was consistent with the results of the T7 promoter-dicistronic reporter in BHK-21 cells, demonstrating that hnRNP L represses FMDV replication in a manner distinct from viral IRES-dependent translation.
FIG 6.
Effects of downregulation and upregulation of hnRNP L on FMDV IRES activity. (A and C) Schematic diagrams of dicistronic reporter plasmids T7-RHF, T7-RHF-IRES, CMV-RHF, and CMV-RHF-IRES. Plasmids express dicistronic mRNA, consisting of the T7 promoter or CMV promoter, Renilla luciferase (RLuc) gene, the FMDV IRES, and firefly luciferase (FLuc) gene. (B and D) Effects of hnRNP L on FMDV IRES activity using dicistronic reporter plasmids. The cells whose hnRNP L was knocked out or overexpressed as described above were transfected with either the T7-RHF, T7-RHF-IRES, CMV-RHF, or CMV-RHF-IRES plasmid. At 24 h posttransfection, the activities of the RLuc and FLuc reporter genes were measured. The bars in the histogram represent percentages of FLuc/RLuc activity. Experiments were performed in triplicate and represented in the bar graph. The expression levels of hnRNP L, HA-hnRNP L, and actin were analyzed by Western blotting.
Inhibition of FMDV replication by hnRNP L is achieved by inhibiting viral RNA synthesis.
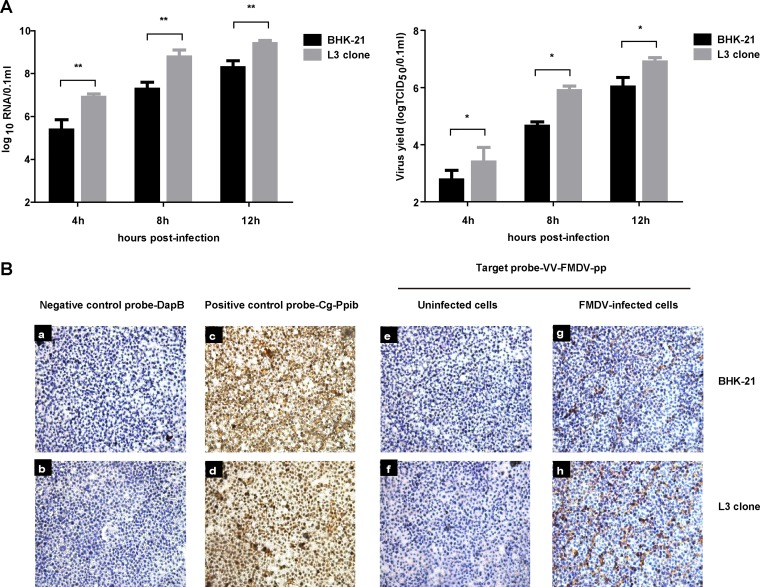
As the repression of FMDV replication by hnRNP L was not via IRES-dependent translation, we assessed the effect of hnRNP L on viral RNA synthesis in FMDV-infected cells. Normal BHK-21 cells or hnRNP L gene-knockout L3 clones were inoculated with FMDV (MOI = 1) in parallel, and the cell cultures were harvested at different time points for real-time reverse transcription-PCR (rRT-PCR) amplification and TCID50 assay. As shown in Fig. 7A, the RNA synthesis level in the FMDV-infected L3 clone was significantly higher than that of FMDV-infected BHK-21 cells at different time points, and the viral titer in the L3 clone was also obviously increased compared to that of normal BHK-21 cells. Taken together, these results suggested that hnRNP L negatively regulates FMDV replication by inhibiting viral RNA synthesis.
FIG 7.
Knockout of hnRNP L in cells promoted FMDV viral RNA synthesis. (A) BHK-21 cells or L3 clones were infected with FMDV at an MOI of 1. The resulting viruses were harvested at 4 h, 8 h, and 12 h and the supernatant was analyzed for viral RNA and virus titer. (B) Seed cells of BHK-21 cells or L3 clone cells in growth medium on chamber slides were infected with FMDV at an MOI of 1. At 8 h postinfection, the cells were used for the RNAscope assay. Target probes were hybridized for 2 h at 40°C, followed by a series of signal amplification and washing steps. Hybridization signals were detected by chromogenic reactions using DAB chromogen followed by 1:1 (vol/vol)-diluted hematoxylin counterstaining. Only in vitro samples with an average of at least 1 positive (brown) dot per cell were included for analysis. Slides were examined and captured for each section using Leica Application Suite (LAS) v3.8 (Leica Microsystems).
To confirm these results, an RNAscope assay was used to detect viral RNA levels in FMDV-infected BHK-21 cells. This assay is a novel RNA in situ hybridization method in which single-molecule visualization in individual cells is achieved through the use of a novel probe design strategy and a hybridization-based signal amplification system to simultaneously amplify signals and suppress background. Hybridization signals were detected by chromogenic reactions using DAB chromogen followed by 1:1 (vol/vol)-diluted hematoxylin counterstaining, which enabled target RNA molecules to be visualized as brown chromogenic dots and cell nuclei to be visualized as blue chromogenic dots. As shown in Fig. 7B, normal BHK-21 cells and hnRNP L knockout L3 cells showed no background staining with the negative-control probe targeting the bacterial gene dapB (Fig. 7B, panels a and b), while amplification was clearly visible as brown staining with the positive-control probe targeting the common housekeeping gene Cg-Ppib (Fig. 7B, panels c and d). Nonspecific staining was not observed with the FMDV-target probe on FMDV-uninfected BHK-21 cells or the L3 clone (Fig. 7B, panels e and f). Specific brown staining was detected only in FMDV-infected BHK-21 cells or the L3 clone with the FMDV-target probe (Fig. 7B, panels g and h). These results indicated that this RNAscope assay is highly specific for detecting viral RNA in FMDV-infected BHK-21 cells. Furthermore, specific brown staining was visibly increased with the FMDV-target probe in the FMDV-infected L3 clone (Fig. 7B, panels g and h) compared to the FMDV-infected BHK-21 cells, demonstrating that hnRNP L affects FMDV RNA synthesis during infection. Since hnRNP L did not influence IRES-dependent translation (Fig. 6), we concluded that hnRNP L negatively regulates FMDV replication through inhibition of viral RNA synthesis.
hnRNP L may be present in the viral RNA replication complex.
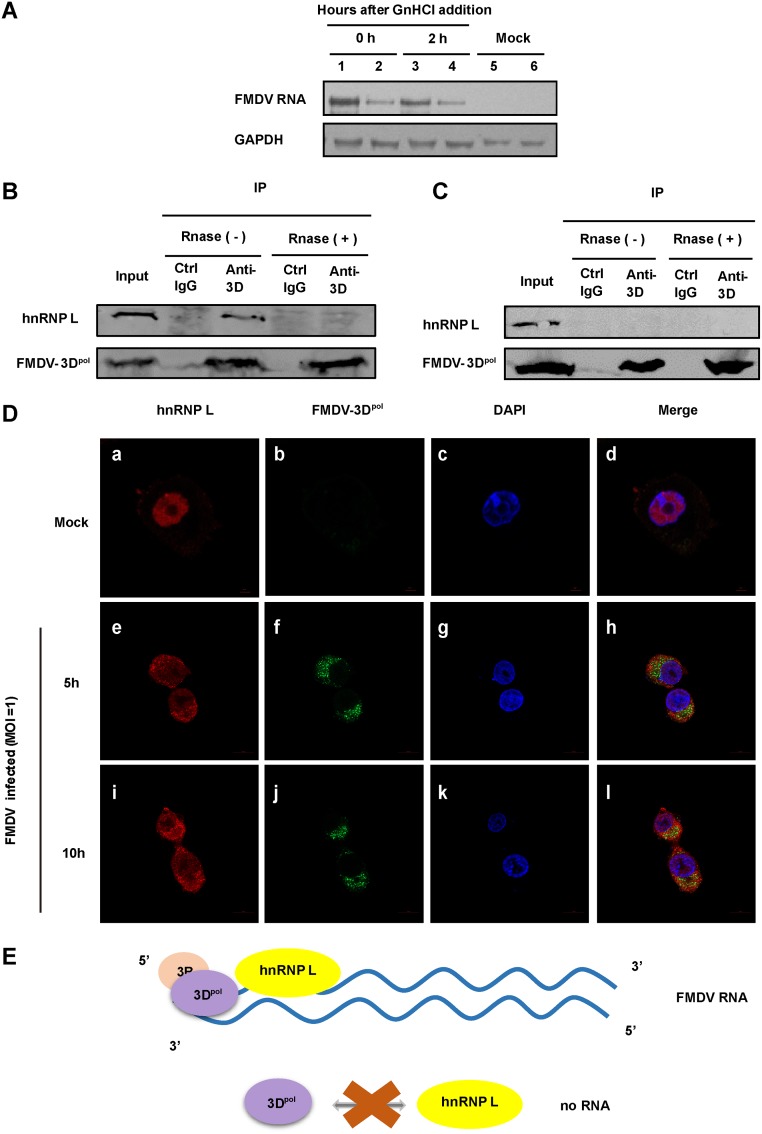
Our results demonstrated that hnRNP L inhibits FMDV replication by affecting viral RNA synthesis rather than IRES-dependent translation, although inhibition of FMDV replication by hnRNP L is related to its binding to viral IRES. Since hnRNP L is involved in the alternative splicing of genes (33), and may regulate the stability of inducible nitric oxide synthase mRNA (34), we further explored the influence of hnRNP L on viral RNA stability. Northern blot analysis was used to monitor the fate of viral RNA in FMDV-infected normal BHK-21 cells versus the hnRNP L knockout L3 clone after treating the cells at 8 hpi with 2 mM guanidine hydrochloride (GnHCl), an inhibitor of FMDV RNA synthesis (35). As shown in Fig. 8A, no significant differences were observed between the stabilities of viral RNA in these two cells following the addition of GnHCl, revealing that hnRNP L is not required for maintaining FMDV RNA stability during infection. To further confirm that the negative regulation of hnRNP L on FMDV replication is through inhibiting viral RNA synthesis, we examined the association of hnRNP L with viral RNA replication complexes. The RNA-dependent RNA polymerase 3Dpol, an essential component of the viral RNA replication complex, was used as an indicator of viral RNA replication complexes in this study. As shown in Fig. 8B, hnRNP L was found to coprecipitate with 3Dpol in FMDV-infected cells by immunoblot assay. The association between hnRNP L and 3Dpol was RNA-dependent, since RNase treatment completely abolished this coimmunoprecipitation (co-IP) (Fig. 8B). To determine whether the RNA-mediated 3Dpol-hnRNP L association was FMDV specific, we repeated this assay using BHK-21 cells transfected with p3×Flag-FMDV-3Dpol and found that no association was evident between FMDV 3Dpol and hnRNP L under this condition (Fig. 8C). Thus, the co-IP assay showed that hnRNP L does not interact directly with FMDV 3Dpol, and it can coimmunoprecipitate with 3Dpol in an FMDV RNA-dependent manner (Fig. 8E), suggesting that hnRNP L may be present in the replication complex to inhibit viral RNA synthesis. Further, the results from confocal microscopy showed that hnRNP L and 3Dpol are not colocalized in the FMDV-infected cells (Fig. 8D, panels h and l), supporting that hnRNP L does not interact directly with FMDV 3Dpol. These results suggested that hnRNP L associates with 3Dpol in the presence of FMDV RNA and possibly functions by staying in the replication complex to inhibit synthesis of viral RNA in one way or another to repress FMDV replication.
FIG 8.
hnRNP L appears to be associated with the viral RNA replication complex. (A) hnRNP L is not required for maintaining FMDV RNA stability during infection. Seed cells of BHK-21 (lanes 2, 4, and 6) and L3 clone cells (lanes 1, 3, and 5) were infected with FMDV(WT) at an MOI of 1. After 8 hpi, the cells were treated with 2 mM guanidine hydrochloride (GnHCl) and harvested at 0 h and 2 h, and total RNA was extracted with TRIzol reagent. Northern blotting was performed by running approximately 30 mg of total RNA on 12% acrylamide denaturing (urea) gels and then transferring to a Hybond-N+ nylon membrane by electrophoresis using a semidry transfer cell. Hybridization was performed according to a standard protocol. Digoxin-labeled oligonucleotide probes complementary to FMDV were used in the hybridization. (B) Viral RNA-dependent association of FMDV 3Dpol with hnRNP L. FMDV 3Dpol protein was immunoprecipitated with FMDV-infected BHK-21 lysate with or without RNase treatment. 3Dpol and hnRNP L proteins in the precipitates were detected by immunoblotting using specific antibody or control IgG. The coimmunoprecipitation of hnRNP L with 3Dpol occurred only in the non-RNase-treated samples. (C) The 3Dpol immunoprecipitation was carried out as in panel B for BHK-21 cells transfected with p3×Flag-FMDV-3Dpol. No coprecipitation of hnRNP L with 3Dpol was observed. (D) hnRNP L does not interact directly with FMDV 3Dpol in virus-infected cells. Normal BHK-21 cells (mock) (panels a to d) or FMDV-infected BHK-21 cells (MOI = 1) at 5 hpi (panels e to h) and 10 hpi (panels i to l) were fixed and stained with antibodies against hnRNP L and FMDV 3Dpol. Panels a, e, and i were treated with anti-hnRNP L antibody and examined with an Alexa Fluor 633 filter; panels b, f, and j were treated with anti-3Dpol antibody and examined with an FITC filter; panels c, g, and k were treated with Hoechst 33258 and examined with a 40,60-diamidino-2-phenylindole (DAPI) filter. Panels d, h, and l show merged Alexa Fluor, FITC, and Hoechst images. (E) Schematic diagram of the association between hnRNP L and FMDV 3Dpol. hnRNP L does not interact directly with 3Dpol. The coprecipitation of hnRNP L with 3Dpol in infected cells is mediated by FMDV RNA.
DISCUSSION
The highly structured 5′ UTR of picornaviruses functions as a platform to recruit host factors, which directs viral protein translation and regulates viral RNA synthesis (4). As a critical element in the 5′ UTR, the IRES binds to various cellular proteins to participate in the viral life cycle (6). Thus, knowledge of the cellular proteins that associate with the IRES of picornaviruses would facilitate insight into virus-host interactions, and these proteins are crucial molecular targets for antiviral development. Here, we used the biotinylated RNA pulldown approach followed by LC-MS/MS analysis to identify eight cellular proteins that are associated with FMDV IRES (Table 1). Among them, hnRNP L, as we report for the first time here, specifically interacts with FMDV IRES (Fig. 1) to regulate FMDV replication negatively through inhibition of viral RNA synthesis.
As an important member of the hnRNP family, hnRNP L is involved in the formation, packaging, and processing of mammalian mRNA (33, 36). Both hnRNP L and PTB cooperatively regulate the translation of Cat-1 mRNA during amino acid starvation (37). Moreover, hnRNP L has been reported to interact with HCV IRES and promote viral growth (24, 25). However, whether hnRNP L regulates picornavirus replication and how it regulates viral replication has remained unclear. Here, we determined the effect of hnRNP L on FMDV infection to be interaction with the viral IRES. Overexpression of hnRNP L decreased viral protein expression and viral production within FMDV-infected cells (Fig. 3), suggesting that hnRNP L is a negative regulator of FMDV replication, which was further verified by hnRNP L knockout (Fig. 4F and Fig. 4G). Interestingly, BEV infection was also enhanced in the hnRNP L knockout L3 clone (Fig. 4H and Fig. 4I), but knockout of hnRNP L did not affect the replication of EMCV (Fig. 4J and K). These results suggested that the effect of hnRNP L on different picornaviruses is not the same. In further exploration, we found that the different effects of hnRNP L on FMDV and EMCV replication were due to differing abilities to bind to the two IRESs (Fig. 5), although more elaborate mechanisms need to be elucidated. In contrast to the situation for picornavirus infection, hnRNP L interacts with the HCV IRES to promote efficient viral replication (25). Based on the characteristics of different types of viral IRESs (38), we speculated that the distinct IRES structures between picornaviruses and HCV may contribute to the converse function of hnRNP L during infection by the two families of viruses.
Surprisingly, the inhibitory effect of hnRNP L on FMDV replication proved to be due to its binding to viral IRES but not via IRES-dependent translation (Fig. 6). Similarly, hnRNP L interacts specifically with HCV IRES, and depletion of hnRNP L in HCV-infected cells impairs viral replication but does not affect HCV IRES translation (25). In another similar study, a novel HCV RNA-binding factor, HMGB1, associates with the HCV IRES to promote HCV RNA replication without regulating viral protein translation (39). These studies have demonstrated that some IRES-binding proteins regulate the replication of IRES-associated viruses by methods other than IRES-mediated translation, but the mechanisms remain unclear. In our study, similarly, it is puzzling why hnRNP L binds to the FMDV IRES but does not regulate IRES-dependent translation. Most likely, the IRES of FMDV provides the binding site for hnRNP L, which facilitates interaction with other host or viral factors to restrain FMDV RNA replication without affecting the translation functions of viral IRES. Thus, we explored the step of FMDV RNA replication in the viral life cycle that might be impacted by hnRNP L and tried to identify other host and viral proteins involved. Our results by rRT-PCR and RNAscope (Fig. 7) indicated that hnRNP L affects viral RNA synthesis during FMDV infection and thereby inhibits FMDV replication.
Since hnRNP L is involved in the alternative splicing of genes via different mechanisms (33), and may regulate the stability of inducible nitric oxide synthase mRNA (34), we further explored the influence of hnRNP L on viral RNA stability. Northern blotting showed that hnRNP L did not play a role in maintaining FMDV RNA stability in infected BHK-21 cells treated with GnHCl (Fig. 8A), thereby arguing against hnRNP L inhibiting FMDV RNA synthesis by a mechanism related to viral RNA stability.
In addition, we examined the association of hnRNP L with viral RNA replication complexes for further support of the conclusion that the hnRNP L negatively regulates FMDV replication by inhibiting viral RNA synthesis. The RNA-dependent RNA polymerase 3Dpol, an essential component of the viral RNA replication complex, was used as an indicator of viral RNA replication complexes in this study. In the co-IP experiments, we found that hnRNP L can coimmunoprecipitate with FMDV 3Dpol in an FMDV RNA-dependent manner (Fig. 8B and C), suggesting that hnRNP L may be present in the replication complex to function. In general, the replication complex of RNA viruses is made up of viral proteins, viral RNA, and cellular factors (40). Here, we have confirmed that the association of cellular hnRNP L protein with FMDV 3Dpol is mediated by viral RNA during FMDV infection, but how hnRNP L functions in the FMDV replication complex to impact viral RNA synthesis remains to be determined.
Our results showed that hnRNP L interacts with the FMDV IRES through its RNA-binding region RRM3-4 (Fig. 2), and the RRM3-4 region has been reported to bind two appropriately separate binding sites within the same RNA by inducing RNA looping (41). RNA looping could be a widespread mechanism for RNA-binding proteins to change RNA secondary structure for special functions (41). Based on these data, we speculate that the FMDV IRES bound by hnRNP L may change its own conformation, thereby forming a new platform to recruit a series of crucial proteins or long noncoding RNAs to impact viral RNA synthesis in the replication complex. It is known that hnRNP L can interact with other members of the hnRNP group, such as PTB, PCBP2, and hnRNP K (42); thus, another possibility is that the protein-protein interaction between hnRNPs may assist two separate parts of an RNA molecule to come near to one another and contribute to the reorganization of RNA molecules that impacts viral RNA synthesis.
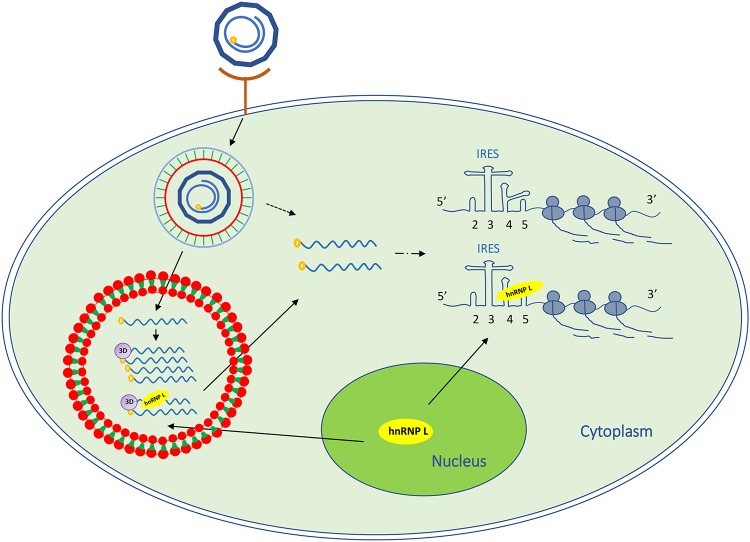
In summary, our findings demonstrated that the cellular protein hnRNP L specifically binds to the FMDV IRES and negatively regulates viral replication (Fig. 9). During infection, the inhibitory effect of hnRNP L on FMDV replication is due to its binding to viral IRES to inhibit viral RNA synthesis without affecting IRES-dependent translation. Importantly, hnRNP L associates with FMDV 3Dpol in an FMDV RNA-dependent manner, suggesting that the protein may function by remaining in the replication complex. These results show that hnRNP L negatively regulates FMDV replication by interacting with IRES to inhibit viral RNA synthesis.
FIG 9.
Schematic representation of the hnRNP L involved in FMDV replication. FMDV binds to a cellular receptor and its genome is released into the cytoplasm. hnRNP L redistributes from the cellular nucleus to the cytoplasm to bind to the IRES in the 5′ UTR of viral genomic RNA, but does not regulate IRES-mediated translation. hnRNP L associated with FMDV 3Dpol in an FMDV RNA-dependent manner may be present in the viral replication complex to inhibit viral RNA synthesis.
MATERIALS AND METHODS
Ethics statement.
The animal work for preparation of primary fetal bovine kidney cells was carried out in strict accordance with the Chinese Regulations of Laboratory Animals—The Guidelines for the Care of Laboratory Animals (Ministry of Science and Technology of People’s Republic of China) and Laboratory Animal-Requirements of Environment and Housing Facilities (GB 14925-2010, National Laboratory Animal Standardization Technical Committee). Protocols for the animal studies were approved by the Committee on the Ethics of Animal Experiments of the Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences (Protocol number 100515-01).
Cells, viruses, and antibodies.
BHK-21 (baby hamster kidney cells), IBRS-2 (porcine kidney cells), PBK (primary fetal bovine kidney cells), and HEK293T cells (human embryonic kidney cells) were cultured in Dulbecco’s minimal essential medium (DMEM) (Gibco) with 10% fetal bovine serum (FBS) (HyClone).
The FMDV O/YS/CHA/05 strain (HM008917), the bovine enterovirus (BEV) BHM26 strain (HQ917060), and the encephalomyocarditis virus (EMCV) HB10 strain (JQ864080.1) were propagated in BHK-21 cells as described previously (43–45). Viral titers were determined by 50% tissue culture infective dose (TCID50) assays by the Reed-Muench method (46).
Anti-FMDV VP2 monoclonal antibody (MAb) 4B2 (47), anti-FMDV 3Dpol polyclonal antibody, and anti-BEV-B serum (45) were prepared in our laboratory. Anti-EMCV VP1 MAb was a gift from Changjiang Weng (Harbin Veterinary Research Institute, Harbin, China). Anti-dsRNA J2 MAb was purchased from English & Scientific Consulting (Szirák, Hungary), while anti-HA, anti-GST, and anti-β-actin MAb were obtained from GenScript (Nanjing, China). Rabbit anti-hnRNP L polyclonal antibody was purchased from Proteintech (Wuhan, China). Mouse anti-hnRNP L MAb was obtained from Novus Biological. IRDye 800CW goat anti-rabbit antibody and IRDye 800CW goat anti-mouse antibody were purchased from LI-COR. Alexa Fluor 633-labeled goat anti-rabbit IgG and Alexa Fluor 488-labeled goat anti-mouse IgG were obtained from Thermo Fisher Scientific. Alexa Fluor 633-conjugated anti-mouse IgG (Invitrogen) and Alexa Fluor 488-labeled goat anti-rabbit IgG (Beyotime) were also used in this study.
Plasmid construction and transfections.
The gene encoding hnRNP L was PCR-amplified from BHK-21 cells and cloned into plasmid pet-28a (Novagen). Competent cells of E. coli Rosetta (DE3) pLys-S (Invitrogen) were transformed with a prokaryotic expression plasmid (pET-28a-hnRNP L) and then induced by 0.25 mmol isopropyl β-D-1-thiogalactopyranoside (IPTG) for 8 h at 37°C. The fusion protein 6×His-hnRNP L was expressed and purified with Ni-affinity chromatography.
To construct plasmids expressing HA-tagged full-length hnRNP L and different truncated forms of hnRNP L, the corresponding cDNAs were amplified by RT-PCR using total RNA extracted from BHK-21 cells as a template and subcloned into the pCAGGS vector (Clontech) (using the primers 1 to 6 in Table 2). For DNA transfections, HEK293T cells were transfected with 1 to 2 μg of plasmid using Lipofectamine 3000 transfection reagent (Invitrogen). The protein expression was validated by Western blotting. To construct the plasmid expressing p3×Flag-FMDV-3Dpol, the cDNA of FMDV 3Dpol was amplified by RT-PCR from FMDV O/YS/CHA/05 strain and inserted into the p3×Flag-CMV10 vector (Sigma). Then, this plasmid was transfected into BHK-21 cells using Lipofectamine 3000 transfection reagent.
TABLE 2.
Primers used in this study
| No. | Primer name | Sequence (5′–3′) |
|---|---|---|
| 1 | HA-L1-4-F | CCGGAATTCATGTCGCGGAGGCTGCTGCCCCGGGCGGAGAAGCGGCGT |
| 2 | HA-L1-4-R | CTAGCTAGCTTAGGAGGCGTGCTGTGCAGTGGAGAAGCACAGCTTCAGAG |
| 3 | HA-L1-2-R | CTAGCTAGCTTAAGGCTTCGCATACTCGATCTTCAGAGTGCAACAGCCAG |
| 4 | HA-L3-4-F | CGGAATTCACCCGTTTAAATGTGTTCAAGAATGACCAAGATACT |
| 5 | HA-L2-3-F | CGGAATTCAGTGTGCTTCTGTTCACCATCCTGAACCCC |
| 6 | HA-L2-3-R | CTAGCTAGCTTACTCGCTGAAGTCTTTGTAACTGCAGGACCCATCTTCT |
| 7 | IRES-D2-5-F | CGGAATTCTAATACGACTCACTATAGGGCACGAAACGCGCCGTCGCTTGAGGAGGACT |
| 8 | IRES-D2-3-R | AAAGATATCGTGGTTGAGTACCAGTATCAGTGTCACCTTAAAGTGGTTTTCACAAT |
| 9 | IRES-D3-4-F | CGGAATTCTAATACGACTCACTATAGGGTGCTTGACTCCACGCTCGGTCCACT |
| 10 | IRES-D3-4-R | AAGATATCCAGGCGTAGAAGCTTTTTAAACCAGGCGCTTTT |
| 11 | IRES-D4-5-F | CGGAATTCTAATACGACTCACTATAGGCAGGCTAAGGATGCCCTTCAGGT |
| 12 | IRES-D4-5-R | AAAGATATCTTAAAGACAGTTGTTCGAAGGAAAGGTGCCGGCCTC |
| 13 | RIP-F | GATCTGCACGAAACGCGCCGTCGCT |
| 14 | RIP-R | CATGGTTAAAGACAGTTGTTCGAAG |
| 15 | rRT-PCR-F | AAT GCA CTC AAA CAA CGG AC |
| 16 | rRT-PCR-R | GCA GTG GTT AGC ATCAAA GG |
| 17 | NB-FMDV-F | GAC CGC ATC CTC ACT ACC C |
| 18 | NB-FMDV-R | ACG CCT CAG CCA CAT CAA |
| 19 | NB-GAPDH-F | CGT ATT GGA CGC CTG GTT |
| 20 | NB-GAPDH-R | GTC TTC TGG GTG GCA GTG AT |
To prepare the T7-FMDV-IRES and its various deletion constructs, cDNA fragments of the corresponding IRES sequences were amplified by RT-PCR using an FMDV sense primer containing the T7 promoter sequence (TAATACGACTCACTATAG) and cloned into the pVAX1 vector (Invitrogen) (using the primers 7 to 12 in Table 2). As these cDNA sequences are located downstream of the T7 polymerase promoter, the corresponding RNAs can be produced and labeled with biotinylated UTP, Biotin-16-UTP (Roche), using the RiboMAX Large Scale RNA Production Systems-T7 kit (Promega). A biotinylated FMDV IRES and its various deletion constructs were synthesized in a 20-μl transcription reaction by adding 1.25 μl of 20 mM Biotin-16-UTP. The synthesized RNA was purified using the MEGA Clear kit (Ambion).
T7-RHF was constructed as follows: the Renilla luciferase gene (RLuc) was inserted in the BglII site of the pGL-3 vector (Addgene) that contains the T7 promoter; the firefly luciferase gene (FLuc) was inserted in the NcoI site of pGL-3. The dicistronic reporter plasmid T7-RHF-IRES that contains FMDV IRES between Renilla and firefly luciferase was constructed by inserting a BglII-FMDV-IRES–NcoI fragment into T7-RHF. CMV-RHF-IRES was constructed as follows: the BGH pA-CMV promoter of the pVAX1 vector (Addgene) was amplified with EcoRI and BamHI sites and ligated to RLuc-IRES-FLuc from the T7-RHF-IRES plasmid with the same sites. The construction of the CMV-RHF plasmid also follows this approach. All constructs used in this study were validated by DNA sequencing.
Biotinylated RNA pulldown assay.
The biotinylated RNA pulldown assay was performed based on previously published methods (48, 49). Total cell lysates were centrifuged at 16,000 × g for 10 min at 4°C, and to the supernatants were added egg white avidin (EMD chemicals) and yeast RNA (Sigma) to block endogenous biotinylated proteins and nonspecific RNPs. After blocking, the lysates were again centrifuged at 16,000 × g for 10 min at 4°C, and the supernatants were collected in new tubes with 200 U/ml RNasin (Promega). The biotinylated FMDV IRES RNA was heated to 90°C for 2 min in RNA folding buffer (10 mM Tris [pH 7], 0.1 M KCl, and 10 mM MgCl2) and the mixture was shifted to room temperature for 20 min to allow proper secondary structure formation. For the biotinylated RNA-binding assay, a reaction mixture containing 200 μg of cell extract and 5 μg of biotinylated RNA was prepared. The mixture, at a final volume of 100 μl, was incubated in RNA mobility-shift buffer (5 mM HEPES [pH 7.1], 40 mM KCl, 2 mM MgCl2, 1 U RNasin, and 0.25 mg/ml heparin) for 60 min at 30°C and then added to 100 μl of Dynabeads M-280 Streptavidin (Invitrogen) for 10 min at room temperature. The RNA-protein complexes were washed five times with RNA mobility-shift buffer without heparin. After the last wash, 30 μl of 1× SDS-PAGE sample buffer was added to the beads, and the captured proteins were analyzed by SDS-PAGE, and further visualized by Coomassie blue staining or Western blot analysis. Protein bands were excised and identified by in-gel trypsin digestion and analyzed by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS).
RNA-protein coimmunoprecipitation and RT-PCR.
For RNA-protein coimmunoprecipitation analysis, BHK-21 cells were infected with FMDV at a multiplicity of infection (MOI) of 1.0 for 8 h and the cell lysates were preincubated with protein A/G-agarose (GE Healthcare) on ice for 1 h to bind nonspecific protein. The nonspecific protein complexes were pelleted by centrifugation at 1,000 × g at 4°C for 10 min. The supernatant was mixed with mouse anti-hnRNP L, mouse anti-HA, or control mouse IgG antibody and incubated at 4°C for 4 h. Subsequently, prewashed protein A/G-agarose beads were added to each sample and incubated overnight at 4°C. Immune complexes were then washed three times with RNA mobility-shift buffer and the RNA was extracted from the immunoprecipitated complexes with a Simply P total RNA extraction kit (BioFlux) according to the manufacturer’s instructions. cDNA was synthesized using PrimeScript reverse transcriptase (TaKaRa), and PCR analysis was performed by using specific primers for the FMDV IRES (RIP-F and RIP-R in Table 2).
Overexpression of hnRNP L in BHK-21 cells.
The HA-tagged hnRNP L gene was ligated into the pCAGGS vector (pCAGGS-HA-hnRNP L), and the HA-tagged eGFP gene was ligated into the same vector (pCAGGS-HA-eGFP) as a control. Next, BHK-21 cells were transfected with 2 μg of pCAGGS-HA-hnRNP L, pCAGGS-HA-eGFP, or empty pCAGGS vector using Lipofectamine 3000 transfection reagent. The protein expression of HA-hnRNP L or HA-eGFP was validated by Western blotting.
Establishment of hnRNP L knockout BHK-21 cells using CRISPR/Cas9 technology.
hnRNP L guide RNA was subcloned into the pSpCas9(BB)-2A-GFP (PX458) vector (Addgene plasmid number 48138). BHK-21 cells grown in a 6-well plate were transfected with 500 ng of pSpCas9(BB)-2A-GFP (PX458)-hnRNP L using Lipofectamine 3000 transfection reagent. Cells were trypsinized 2 days after transfection and resuspended in DMEM. Single cells were isolated into 96-well plates using a BD Influx sorter gated on GFP fluorescence. Clonal cell populations were expanded, and whole-cell extracts were analyzed by Western blotting. In addition, genomic DNA samples were extracted from 1 × 105 cells by lysing and digesting cells in 25 μl of buffer (10 mM Tris-HCl [pH 8.0], 50 mM NaCl, 5 mM EDTA, 0.1% SDS, 60 μg/ml proteinase K) and incubation at 55°C for 1 h. Following another incubation at 98°C for 10 min, samples were diluted at a 1:10 ratio with water, and 2 μl was used to amplify the 250-bp fragment encompassing the gene inactivation locus in 20 μl of PCR buffer using hnRNP L-KO-S and hnRNP L-KO-A as a primer pair. The PCR-amplified fragments were analyzed in a 10% polyacrylamide gel using 1× Tris-borate-EDTA (TBE) as a buffer after ethidium bromide staining.
Virus infection.
Normal BHK-21 cells and cells with hnRNP L knocked out or overexpressed as described above were infected with FMDV strain O/YS/CHA/05, EMCV strain HB10, or BEV strain BHM26 at an MOI of 1. After 1 h, the viral inoculum was removed and the infected cells were washed twice with 1× phosphate-buffered saline (PBS; pH 7.4) and reincubated with DMEM containing 2% FBS. At various time points postinfection, cell-free culture supernatants and cell lysates were harvested to detect the infectious virus titer using the TCID50 assay and analyze viral protein expression by Western blotting.
Western blotting.
Western blotting was performed as described previously (50). Protein samples were separated by 12% SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked with 5% skim milk in PBS and then incubated with primary antibodies followed by IRDye 800CW secondary antibodies, and signal detection was performed using a near-infrared fluorescence scanning imaging system (Licor Odyssey).
Confocal microscopy.
BHK-21 cells grown on glass coverslips were infected with FMDV at an MOI of 1. After 5 h or 10 h postinfection, the FMDV-infected or mock-treated cells were washed three times with PBS. The cells on the coverslip were fixed with 3.7% (wt/vol) formaldehyde for 20 min. After washing three times with PBS, the cells on the coverslip were permeabilized in 0.1% Triton X-100 for 15 min and washed again three times with PBS. Then, the samples were blocked in solution (PBS, containing 5% bovine serum albumin [BSA]) for 60 min. Subsequently, appropriate primary antibodies were incubated with the samples for 60 min followed by secondary antibodies for 40 min. The nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI) for 15 min and examined using a Leica SP2 confocal system (Leica Microsystems). The colocalization signal was analyzed using a confocal laser-scanning microscope (Zeiss LSM800).
Real-time RT-PCR.
To detect FMDV RNA synthesis levels during virus replication, FMDV genome copy number was quantified by using a previously described real-time reverse transcription-PCR (rRT-PCR) assay (51). Briefly, total RNA was extracted from FMDV-infected cells by using TRIzol (Invitrogen), and rRT-PCR amplification was performed on Platinum SYBR Green qPCR Super Mix-UDG with ROX (TOYOBO). The mean values and standard deviations were derived from triplicate measurements. The primers used to perform rRT-PCRs are listed in Table 2.
Luciferase reporter assay.
Cells with hnRNP L knocked out or overexpressed as described above were transfected with CMV-RHF-IRES, CMV-RHF, T7-RHF-IRES, or T7-RHF using Lipofectamine 3000 transfection reagent. At 24 h posttransfection, the luciferase activity of the RLuc and FLuc reporter genes was measured using a dual-luciferase reporter assay kit (Promega). As expression of RLuc is driven by the CMV or T7 promoter while expression of FLuc is dependent on FMDV IRES activity, the ratio of the FLuc expression level to the RLuc expression level in a sample represents its relative translation efficiency driven by the FMDV IRES.
In situ hybridization.
In situ hybridization was performed using the RNAscope 2.5 HD Brown Chromogenic reagent kit according to the manufacturer’s instructions (Advanced Cell Diagnostics, Hayward, CA). GenBank accession numbers, target regions, and catalog order numbers for the proprietary target probes are FMDV (GenBank HM008917.1; nucleotides 3770 to 4640; Advanced Cell Diagnostics number 402481). RNAScope bacterial dihydrodipicolinate reductase (dapB; Advanced Cell Diagnostics number 310043) was used as a negative control for the target probe, and RNAscope housekeeping gene peptidylprolyl isomerase B (Cg-Ppib; Advanced Cell Diagnostics number 450461) was used as a positive control for the target probe. Briefly, seed cells of BHK-21 or L3 hnRNP L knockout cells in growth medium on chamber slides were infected with FMDV(WT) at an MOI of 1. At 8 h postinfection, cells were treated according to Sample Preparation Technical Note for Cultured Adherent Cells Using RNAscope 2.5 Chromogenic Assay (single plex and duplex). Target probes were hybridized for 2 h at 40°C, followed by a series of signal amplification and washing steps. Hybridization signals were detected by chromogenic reactions using DAB chromogen followed by 1:1 (vol/vol)-diluted hematoxylin (Thermo Fisher Scientific, Pittsburg, PA) counterstaining. Only in vitro samples with an average of at least 1 positive (brown) dot per cell were included for analysis. Slides were examined and captured for each section using Leica Application Suite (LAS) v3.8 (Leica Microsystems).
Immunoprecipitation.
The lysates of FMDV-infected BHK-21 cells were incubated with anti-hnRNP L, rabbit anti-3D, or isotype control IgG at 4°C for 2 h, followed by the addition of 30 μl of Protein A/G MagBeads (GenScript) for 1 h. The lysate was treated with or without RNase A. The Protein A/G MagBeads were washed three times in lysis buffer. Proteins were eluted with SDS-PAGE sample buffer and subjected to SDS-PAGE, followed by immunoblotting.
Northern blotting.
Cells of BHK-21 and hnRNP L knockout cells were infected with FMDV(WT) at an MOI of 1. After 8 hpi, the cells were treated with 2 mM guanidine hydrochloride (GnHCl) and harvested at 0 h and 2 h, and total RNA was extracted with TRIzol. Approximately 30 mg of total RNA was run on 12% acrylamide denaturing (urea) gels and then transferred to a Hybond-N + nylon membrane (Amersham Biosciences) by electrophoresis using a semidry transfer cell (Bio-Rad). Hybridization was performed according to a standard protocol. Digoxin-labeled oligonucleotide probes complementary to FMDV VP2 were used in the hybridization. The probes (NB-FMDV-F, NB-FMDV-R, NB-GAPDH-F, and NB-GAPDH-R) are listed in Table 2.
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (grant no. 31770173 and 31400138) and the National Key Research and Development Program of China (grant no. 2016YFD0501505).
REFERENCES
- 1.Whitton JL, Cornell CT, Feuer R. 2005. Host and virus determinants of picornavirus pathogenesis and tropism. Nat Rev Microbiol 3:765–776. doi: 10.1038/nrmicro1284. [DOI] [PubMed] [Google Scholar]
- 2.Jiang P, Liu Y, Ma HC, Paul AV, Wimmer E. 2014. Picornavirus morphogenesis. Microbiol Mol Biol Rev 78:418–437. doi: 10.1128/MMBR.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pilipenko EV, Blinov VM, Chernov BK, Dmitrieva TM, Agol VI. 1989. Conservation of the secondary structure elements of the 5′-untranslated region of cardio- and aphthovirus RNAs. Nucleic Acids Res 17:5701–5711. doi: 10.1093/nar/17.14.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agol VI. 1991. The 5′-untranslated region of picornaviral genomes. Adv Virus Res 40:103–180. doi: 10.1016/s0065-3527(08)60278-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grubman MJ, Baxt B. 2004. Foot-and-mouth disease. Clin Microbiol Rev 17:465–493. doi: 10.1128/cmr.17.2.465-493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martinez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, Lozano G, Diaz-Toledano R. 2015. Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res 206:62–73. doi: 10.1016/j.virusres.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Salas E, Lozano G, Fernandez-Chamorro J, Francisco-Velilla R, Galan A, Diaz R. 2013. RNA-binding proteins impacting on internal initiation of translation. Int J Mol Sci 14:21705–21726. doi: 10.3390/ijms141121705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filbin ME, Kieft JS. 2009. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol 19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee KM, Chen CJ, Shih SR. 2017. Regulation mechanisms of viral IRES-driven translation. Trends Microbiol 25:546–561. doi: 10.1016/j.tim.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Song L, Cong H, Tien P. 2015. Nuclear protein Sam68 interacts with the enterovirus 71 internal ribosome entry site and positively regulates viral protein translation. J Virol 89:10031–10043. doi: 10.1128/JVI.01677-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pineiro D, Fernandez N, Ramajo J, Martinez-Salas E. 2013. Gemin5 promotes IRES interaction and translation control through its C-terminal region. Nucleic Acids Res 41:1017–1028. doi: 10.1093/nar/gks1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pacheco A, López de Quinto S, Ramajo J, Fernández N, Martínez-Salas E. 2009. A novel role for Gemin5 in mRNA translation. Nucleic Acids Res 37:582–590. doi: 10.1093/nar/gkn979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han S, Sun S, Li P, Liu Q, Zhang Z, Dong H, Sun M, Wu W, Wang X, Guo H. 2020. Ribosomal protein L13 promotes IRES-driven translation of foot-and-mouth disease virus in a helicase DDX3-dependent manner. J Virol 94. doi: 10.1128/JVI.01679-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim CS, Seol SK, Song OK, Park JH, Jang SK. 2007. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis C virus replication. J Virol 81:3852–3865. doi: 10.1128/JVI.01311-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paek KY, Kim CS, Park SM, Kim JH, Jang SK. 2008. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J Virol 82:12082–12093. doi: 10.1128/JVI.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Jeng KS, Lai MM. 2011. Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5′ untranslated region and directs HCV RNA replication through circularizing the viral genome. J Virol 85:7954–7964. doi: 10.1128/JVI.00339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang KS, Luo G. 2006. The polypyrimidine tract-binding protein (PTB) is required for efficient replication of hepatitis C virus (HCV) RNA. Virus Res 115:1–8. doi: 10.1016/j.virusres.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 18.Domitrovich AM, Diebel KW, Ali N, Sarker S, Siddiqui A. 2005. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology 335:72–86. doi: 10.1016/j.virol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Walter BL, Nguyen JH, Ehrenfeld E, Semler BL. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5:1570–1585. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walter BL, Parsley TB, Ehrenfeld E, Semler BL. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J Virol 76:12008–12022. doi: 10.1128/jvi.76.23.12008-12022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guest S, Pilipenko E, Sharma K, Chumakov K, Roos RP. 2004. Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3. J Virol 78:11097–11107. doi: 10.1128/JVI.78.20.11097-11107.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pilipenko EV, Viktorova EG, Guest ST, Agol VI, Roos RP. 2001. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J 20:6899–6908. doi: 10.1093/emboj/20.23.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu L, Yang D, Wang H, Zhou G. November 2017. Temperature-sensitive attenuated FMDV strains, construction method and application thereof. US patent application PCT/CN2017/111936.
- 24.Hahm B, Kim YK, Kim JH, Kim TY, Jang SK. 1998. Heterogeneous nuclear ribonucleoprotein L interacts with the 3′ border of the internal ribosomal entry site of hepatitis C virus. J Virol 72:8782–8788. doi: 10.1128/JVI.72.11.8782-8788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Masaki T, Shimakami T, Lemon SM. 2014. hnRNP L and NF90 interact with hepatitis C virus 5′-terminal untranslated RNA and promote efficient replication. J Virol 88:7199–7209. doi: 10.1128/JVI.00225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luz N, Beck E. 1991. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J Virol 65:6486–6494. doi: 10.1128/JVI.65.12.6486-6494.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc Natl Acad Sci U S A 90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galan A, Lozano G, Pineiro D, Martinez-Salas E. 2017. G3BP1 interacts directly with the FMDV IRES and negatively regulates translation. FEBS J 284:3202–3217. doi: 10.1111/febs.14184. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Salas E, Fernandez-Miragall O. 2004. Picornavirus IRES: structure function relationship. CPD 10:3757–3767. doi: 10.2174/1381612043382657. [DOI] [PubMed] [Google Scholar]
- 30.Hui J, Reither G, Bindereif A. 2003. Novel functional role of CA repeats and hnRNP L in RNA stability. RNA 9:931–936. doi: 10.1261/rna.5660803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. 2013. RNA-guided human genome engineering via Cas9. Science 339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hung LH, Heiner M, Hui J, Schreiner S, Benes V, Bindereif A. 2008. Diverse roles of hnRNP L in mammalian mRNA processing: a combined microarray and RNAi analysis. RNA 14:284–296. doi: 10.1261/rna.725208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soderberg M, Raffalli-Mathieu F, Lang MA. 2002. Inflammation modulates the interaction of heterogeneous nuclear ribonucleoprotein (hnRNP) I/polypyrimidine tract binding protein and hnRNP L with the 3′untranslated region of the murine inducible nitric-oxide synthase mRNA. Mol Pharmacol 62:423–431. doi: 10.1124/mol.62.2.423. [DOI] [PubMed] [Google Scholar]
- 35.Pringle CR. 1964. Inhibition of multiplication of foot-and-mouth disease virus by guanidine hydrochloride. Nature 204:1012–1013. doi: 10.1038/2041012a0. [DOI] [PubMed] [Google Scholar]
- 36.Han SP, Tang YH, Smith R. 2010. Functional diversity of the hnRNPs: past, present and perspectives. Biochem J 430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 37.Majumder M, Yaman I, Gaccioli F, Zeenko VV, Wang C, Caprara MG, Venema RC, Komar AA, Snider MD, Hatzoglou M. 2009. The hnRNA-binding proteins hnRNP L and PTB are required for efficient translation of the Cat-1 arginine/lysine transporter mRNA during amino acid starvation. Mol Cell Biol 29:2899–2912. doi: 10.1128/MCB.01774-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niepmann M. 2009. Internal translation initiation of picornaviruses and hepatitis C virus. Biochim Biophys Acta 1789:529–541. doi: 10.1016/j.bbagrm.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Yu R, Yang D, Lei S, Wang X, Meng X, Xue B, Zhu H. 2015. HMGB1 promotes hepatitis C virus replication by interaction with stem-loop 4 in the viral 5′ untranslated region. J Virol 90:2332–2344. doi: 10.1128/JVI.02795-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid CR, Airo AM, Hobman TC. 2015. The virus-host interplay: biogenesis of +RNA replication complexes. Viruses 7:4385–4413. doi: 10.3390/v7082825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Zeng F, Liu Y, Zhao Y, Lv H, Niu L, Teng M, Li X. 2013. Crystal structures and RNA-binding properties of the RNA recognition motifs of heterogeneous nuclear ribonucleoprotein L: insights into its roles in alternative splicing regulation. J Biol Chem 288:22636–22649. doi: 10.1074/jbc.M113.463901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim JH, Hahm B, Kim YK, Choi M, Jang SK. 2000. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J Mol Biol 298:395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- 43.Yang DC, Tu YB, Wang HW, Zhou GH, Yu L. 2009. Construction of infectious cDNA clone for PanAsia strain of FMDV serotype O. Chin J Preve Vet Med 31:1–5. [Google Scholar]
- 44.Huang L, Liu Q, Zhang L, Zhang Q, Hu L, Li C, Wang S, Li J, Zhang Y, Yu H, Wang Y, Zhong Z, Xiong T, Xia X, Wang X, Yu L, Deng G, Cai X, Cui S, Weng C. 2015. Encephalomyocarditis virus 3C protease relieves TRAF family member-associated NF-kappaB activator (TANK) inhibitory effect on TRAF6-mediated NF-kappaB signaling through cleavage of TANK. J Biol Chem 290:27618–27632. doi: 10.1074/jbc.M115.660761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang J, Li Y, Yang D, Wang F, Jiang Z, Yu L. 2013. VP1 B-C and D-E loops of bovine enterovirus cluster B can effectively display foot-and-mouth disease virus type O-conserved neutralizing epitope. J Gen Virol 94:2691–2699. doi: 10.1099/vir.0.057745-0. [DOI] [PubMed] [Google Scholar]
- 46.Pizzi M. 1950. Sampling variation of the fifty percent end-point, determined by the Reed-Muench (Behrens) method. Hum Biol 22:151–190. [PubMed] [Google Scholar]
- 47.Yu Y, Wang H, Zhao L, Zhang C, Jiang Z, Yu L. 2011. Fine mapping of a foot-and-mouth disease virus epitope recognized by serotype-independent monoclonal antibody 4B2. J Microbiol 49:94–101. doi: 10.1007/s12275-011-0134-1. [DOI] [PubMed] [Google Scholar]
- 48.Lin JY, Li ML, Shih SR. 2009. Far upstream element binding protein 2 interacts with enterovirus 71 internal ribosomal entry site and negatively regulates viral translation. Nucleic Acids Res 37:47–59. doi: 10.1093/nar/gkn901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iioka H, Loiselle D, Haystead TA, Macara IG. 2011. Efficient detection of RNA-protein interactions using tethered RNAs. Nucleic Acids Res 39:e53. doi: 10.1093/nar/gkq1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun C, Yang D, Gao R, Liang T, Wang H, Zhou G, Yu L. 2016. Modification of the internal ribosome entry site element impairs the growth of foot-and-mouth disease virus in porcine-derived cells. J Gen Virol 97:901–911. doi: 10.1099/jgv.0.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaw AE, Reid SM, Ebert K, Hutchings GH, Ferris NP, King DP. 2007. Implementation of a one-step real-time RT-PCR protocol for diagnosis of foot-and-mouth disease. J Virol Methods 143:81–85. doi: 10.1016/j.jviromet.2007.02.009. [DOI] [PubMed] [Google Scholar]