Recent studies have reported that rabbit hemorrhagic disease virus (RHDV) infection reduced Nrf2-related antioxidant function. However, the regulatory mechanisms underlying this process remain unclear. The current study showed that the NF-κB p50 subunit partners with Keap1 to form the Keap1-NF-κB complex, which plays a key role in the inhibition of Nrf2 transcriptional activity. More importantly, upregulated Nrf2 activity delayed the death of RHDV-infected rabbits, strongly indicating the importance of oxidative damage during RHDV infection. These findings may provide novel insights into the pathogenesis of RHDV.
KEYWORDS: Keap1, NF-κB, Nrf2, RHDV
ABSTRACT
The rabbit hemorrhagic disease virus (RHDV), which belongs to the family Caliciviridae and the genus Lagovirus, causes lethal fulminant hepatitis in rabbits. RHDV decreases the activity of antioxidant enzymes regulated by Nrf2 in the liver. Antioxidants are important for the maintenance of cellular integrity and cytoprotection. However, the mechanism underlying the regulation of the Nrf2-antioxidant response element (ARE) signaling pathway by RHDV remains unclear. Using isobaric tags for relative and absolute quantification (iTRAQ) technology, the current study demonstrated that RHDV inhibits the induction of ARE-regulated genes and increases the expression of the p50 subunit of the NF-κB transcription factor. We showed that RHDV replication causes a remarkable increase in reactive oxygen species (ROS), which is simultaneously accompanied by a significant decrease in Nrf2. It was found that nuclear translocation of Keap1 plays a key role in the nuclear export of Nrf2, leading to the inhibition of Nrf2 transcriptional activity. The p50 protein partners with Keap1 to form the Keap1-p50/p65 complex, which is involved in the nuclear translocation of Keap1. Moreover, upregulation of Nrf2 protein levels in liver cell nuclei by tert-butylhydroquinone (tBHQ) delayed rabbit deaths due to RHDV infection. Considered together, our findings suggest that RHDV inhibits the Nrf2-dependent antioxidant response via nuclear translocation of Keap1-NF-κB complex and nuclear export of Nrf2 and provide new insight into the importance of oxidative stress during RHDV infection.
IMPORTANCE Recent studies have reported that rabbit hemorrhagic disease virus (RHDV) infection reduced Nrf2-related antioxidant function. However, the regulatory mechanisms underlying this process remain unclear. The current study showed that the NF-κB p50 subunit partners with Keap1 to form the Keap1-NF-κB complex, which plays a key role in the inhibition of Nrf2 transcriptional activity. More importantly, upregulated Nrf2 activity delayed the death of RHDV-infected rabbits, strongly indicating the importance of oxidative damage during RHDV infection. These findings may provide novel insights into the pathogenesis of RHDV.
INTRODUCTION
Rabbit hemorrhagic disease (RHD) is caused by the rabbit hemorrhagic disease virus (RHDV), a nonenveloped single-stranded RNA virus. RHD is a highly contagious and acutely infectious disease which is fatal to rabbits. This disease, first reported in China, has spread worldwide (1–3), resulting in the deaths of millions of domestic and wild adult rabbits.
A suitable cell culture capable of supporting authentic RHDV has not yet been established. This has greatly impeded the investigation of mechanisms underlying the pathogenesis of RHDV. RHDV infections, characterized by disseminated intravascular coagulation (DIC), hepatocellular apoptosis, and fulminant hepatic failure (FHF) (4–7), cause high morbidity and mortality in rabbits (1, 8, 9). Over 90% of RHDV-infected adult rabbits die due to FHF within 3 days of infection (10). More recently, due to the similarity of most representative biochemical and histologic parameters as well as clinical signs to those of human FHF, rabbit FHF, caused by experimental infection of rabbits with RHDV, has become useful as the new animal FHF model (11).
Previous studies have reported that melatonin attenuates oxidative stress, inflammation, and apoptosis in RHDV-infected rabbit livers. The mechanism underlying such attenuation was found to be associated with the activation of antioxidant enzymes and inhibition of endoplasmic reticulum (ER) stress (11, 12). Nuclear factor (erythroid-derived 2)-like 2 (Nrf2) is a redox-sensitive transcription factor and a master regulator of cytoprotective genes, including superoxide dismutase (SOD), glutathione S-transferase (GST), and glutathione peroxidase (GPx) (13–17). Nrf2 has been shown to protect against drug- and hepatitis virus-induced liver injury, indicating that activation of Nrf2 confers protection against oxidative stress in diseases associated with reactive oxygen species (ROS) and inflammation (16). Although it has been reported that Nrf2 is essential for reducing oxidative stress and preventing a reduction of antioxidant enzyme activity in RHDV-induced liver injury, the role of oxidative stress in the occurrence and development of RHD requires clarification. The mechanism by which RHDV infection suppresses Nrf2-mediated antioxidant response in the liver remains unknown.
Keap1 is a key induction suppressor of Nrf2 in the Nrf2-antioxidant response element (ARE) signaling pathway. Keap1 acts as an adaptor protein between Nrf2 and Cul3, which sequesters Nrf2 in the cytoplasm and promotes continuous degradation of Nrf2 by the proteasome under normal conditions (18–21). Covalent modification of Keap1 results in reduced ubiquitination of Nrf2, leading to its accumulation (22). Modification of Keap1 cysteine residues via alkylation also activates the transcription factor, Nrf2 (23). Moreover, upon recovery of cellular redox homeostasis, Keap1 translocates to the nucleus to dissociate Nrf2 from the ARE (24). In addition, the nuclear export sequence (NES) in Keap1 is required for termination of Nrf2-ARE signaling via nuclear export of Nrf2, resulting in the degradation of Nrf2 (24). These reports suggested that Keap1 plays a key role in the nuclear export of Nrf2 as well as termination of the Nrf2 signaling pathway.
The current study demonstrated that the oxidative stress response, induced by ROS during RHDV infection, played a critical role in RHDV pathogenicity. Furthermore, our data indicated that the p50/p65 and Keap1 interaction was involved in the nuclear translocation of Keap1, which is important for the nuclear export and degradation of Nrf2. These findings may provide new mechanistic insights into the pathogenesis of RHDV in rabbits.
RESULTS
Proteomic analysis indicated that oxidative stress in the liver was induced by RHDV at 18 hpi.
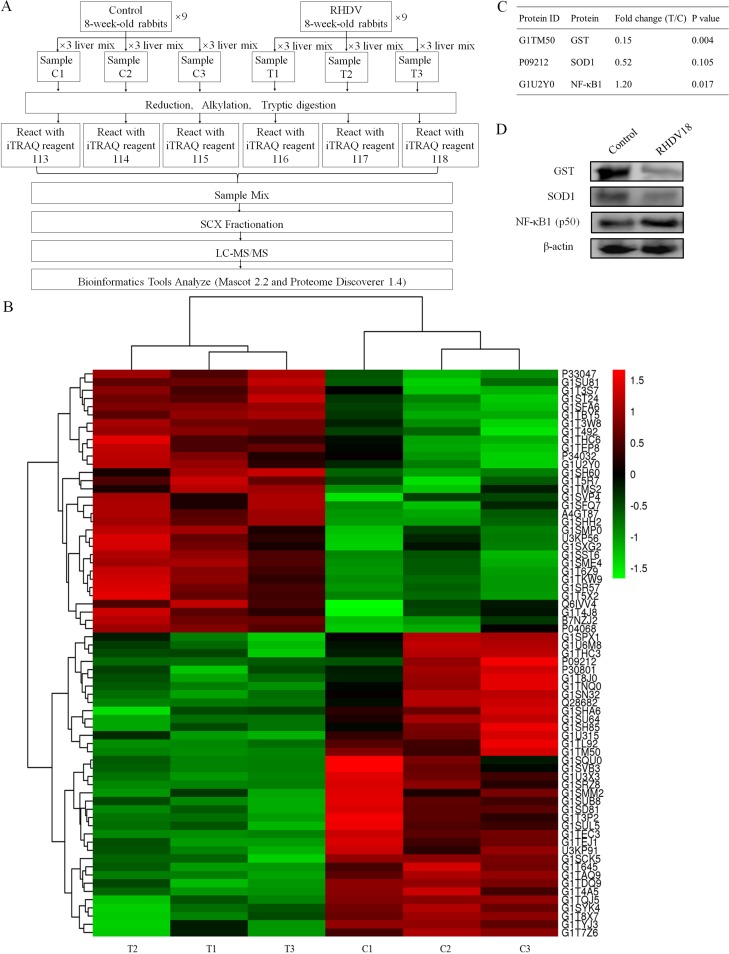
In this trial, 18 rabbits (9 rabbits per experimental group) underwent high-accuracy liquid chromatography-tandem mass spectrometry (LC-MS/MS) isobaric tags for relative and absolute quantification (iTRAQ) analysis combined with strong cation exchange (SCX). Using untargeted proteomic analysis, a total of 27,965 peptides matching 4,169 proteins (1 or more unique peptides with a false-discovery rate [FDR] of less than 0.01) were identified. Fold changes of proteins between normal and RHDV-infected livers were determined via iTRAQ reporter ion ratios. At a threshold of 1.2-fold change and a P value of ≤0.05, significantly different expression levels between RHDV-infected livers and age-matched controls were detected in 68 proteins (32 upregulated and 36 downregulated) at 18 hours postinfection (hpi) (Fig. 1B; see Table S1 in the supplemental material).
FIG 1.
Proteomic analysis reveals that oxidative stress in the liver is induced by RHDV at 18 h postinfection. (A) Schematic of the experimental design. Nine livers per group were collected; protein was extracted and pooled before iTRAQ labeling and subjected to mass spectrometric analysis. iTRAQ tags 113, 114, and 115 were used for C1, C2, and C3, and iTRAQ tags 116, 117, and 118 were used for T1, T2, and T3, respectively. Samples were fractionated using strong cation exchange (SCX), and MS experiments were executed. (B) 68 differential expression proteins were detected via bioinformatics analysis (≥1.2-fold change and P value ≤0.05). (C) The 2 representative antioxidant proteins and 1 large fold change protein (NF-κB1 or p50) listed were (D) further confirmed by Western blot analysis.
The iTRAQ data showed that antioxidant protein, GST, and NF-κB subunit 1 (p50) were differentially expressed (Fig. 1C). In addition, another important antioxidant protein, SOD1, exhibited a large fold change between the control and infection groups, but the difference was not significant (Fig. 1C). The reason may have been the large differences seen within the infection group itself (Table S1). In order to confirm these results, Western blot analysis with β-actin as the housekeeping protein was used to quantify the expression levels of GST, SOD1, and NF-κB1. Western blot analysis results were consistent with those of the iTRAQ trial (Fig. 1D).
Liver injury was associated with oxidative stress in RHDV-infected livers.
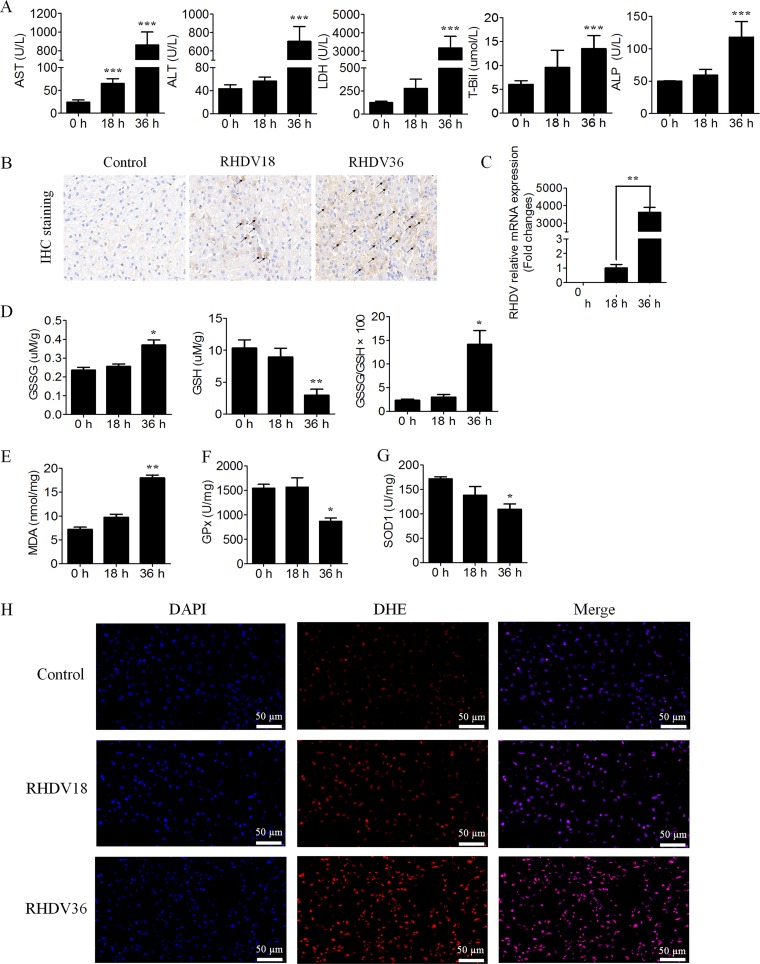
Blood chemistry analyses indicated that alanine aminotransferase (ALT), aspartate transaminase (AST), lactate dehydrogenase (LDH), total bilirubin (T-Bil), and alkaline phosphatase (ALP) activities were all increased in RHDV-infected animals compared with those in the control group. AST showed a significant difference at 18 hpi compared with the control (0 hpi), while the others did so at 36 hpi (Fig. 2A). RHDV antigens were stained brown during the immunohistochemical detection assay, indicating that the virus replicated in a time-dependent manner (Fig. 2B). The mRNA level of RHDV was consistent with that obtained via immunohistochemical detection, which demonstrated again that RHDV replicated in a time-dependent manner (Fig. 2C). These results indicated that liver injury had occurred starting from 18 hpi with RHDV.
FIG 2.
Liver injury is associated with oxidative stress in RHDV-infected livers. (A) Effects of RHDV on blood chemistry (AST, ALT, LDH, T-Bil, and ALP are shown) at indicated time points postinfection were detected. The values are expressed in comparison to the control (0 h). (B) Liver tissues were harvested from RHDV-infected rabbits for antigen detection. Immunohistochemical staining with specific MAb 1B8 against RHDV was used for antigen detection. The antigens are shown by arrows. (C) RHDV relative mRNA expression levels were detected. (D and E) The biochemical markers (GSH, GSSG, GSSG/GSH, and MDA are shown) of liver oxidative stress were detected at different time points. (F and G) Activities of the antioxidant enzymes, GPx and SOD1, were measured at different times postinfection. (H) Intracellular ROS generation was detected at indicated times using DHE, a nonfluorescent cell-permeative compound, as a probe.
Glutathione (GSH) and malondialdehyde (MDA) concentrations in different experimental groups were analyzed as indicators of oxidative stress. GSH concentration was significantly decreased at 36 hpi, while a marked increase was observed in the oxidized glutathione (GSSG)/GSH ratio (Fig. 2D). The liver concentration of MDA also significantly increased in RHDV-infected rabbits at 36 hpi (Fig. 2E). Infected animals showed a progressive decrease in the activity of antioxidant enzymes, GPx and SOD1, at 36 hpi (Fig. 2F and G). Changes in active oxygen content and intracellular ROS generation were further analyzed using ROS fluorescent probe, dihydroethidium (DHE). The results indicated that intracellular ROS generation had significantly increased with RHDV infection (Fig. 2H).
Keap1 plays a key role in the nuclear export and degradation of Nrf2 in RHDV-infected livers.
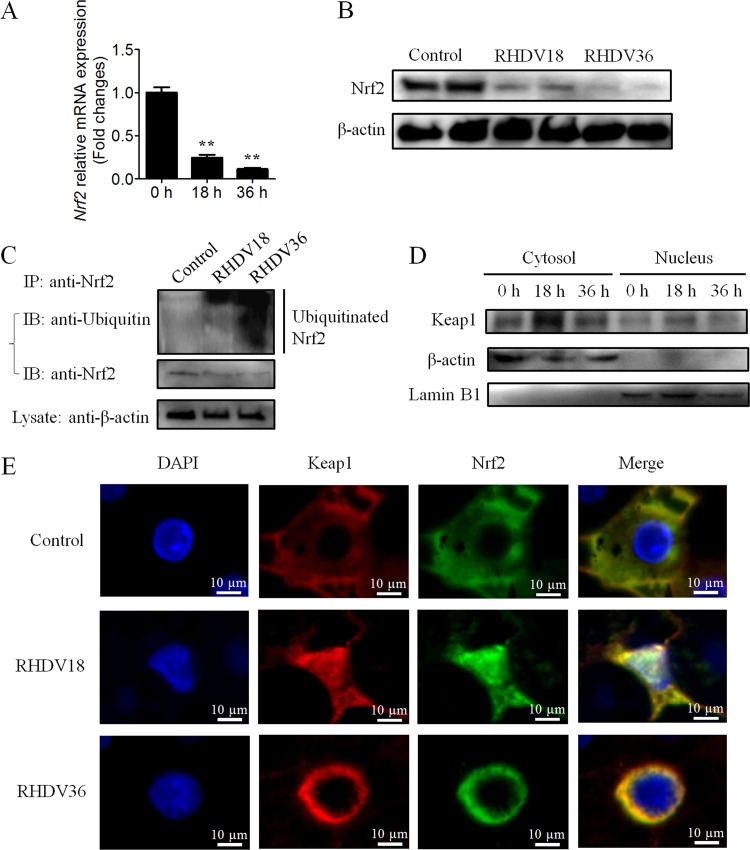
Nrf2 expression in RHDV-infected livers was determined at different time points. The results showed that both mRNA and protein levels of the transcription factor, Nrf2, had undergone a significant reduction following RHDV infection (Fig. 3A and B). Next, we analyzed changes in Nrf2 ubiquitination in the liver. Our results suggested that Nrf2-ARE signaling was regulated via Nrf2 ubiquitination and degradation (Fig. 3C).
FIG 3.
Keap1 is involved in nuclear export and degradation of Nrf2 in RHDV-infected livers. (A) Real-time PCR was performed to detect the mRNA level of Nrf2. (B) Detection of Nrf2 proteins at 0, 18, and 36 h time points postinfection using Western blot analysis. β-actin was employed as an internal control. (C) An IP assay was performed using liver lysates at different time points. Nrf2 was isolated via immunoprecipitation with anti-Nrf2 MAb and analyzed using Western blots with antiubiquitin antibodies. (D) Detection of Keap1 proteins at different time points postinfection in the cytosol and nuclei using Western blots. β-actin and lamin B1 were employed as cytosol and nucleus internal controls, respectively. (E) Immunofluorescence localization analysis of Nrf2 (green) and Keap1 (red). Slides of RHDV-infected liver incubated with Nrf2-specific MAb and Keap1-specific PAb at 0, 18, and 36 h.
Previous studies have reported that Keap1 acts as a key postinduction repressor of Nrf2 (24). In order to determine whether Keap1 is involved in nuclear export and degradation of Nrf2 during RHDV infection, we investigated the localization of Keap1 and Nrf2 in liver cells. At 18 hpi, a significant increase in Keap1 protein levels was observed in the cytosol and nuclei, indicating the importance of Keap1 protein in Nrf2 repression (Fig. 3D). Furthermore, Nrf2 and Keap1 were found to be colocalized in the nucleus at 18 hpi, following which, nuclear export in the form of Nrf2-Keap1 complex was observed at 36 hpi (Fig. 3E).
Nuclear translocation of Keap1 depends on interaction with NF-κB.
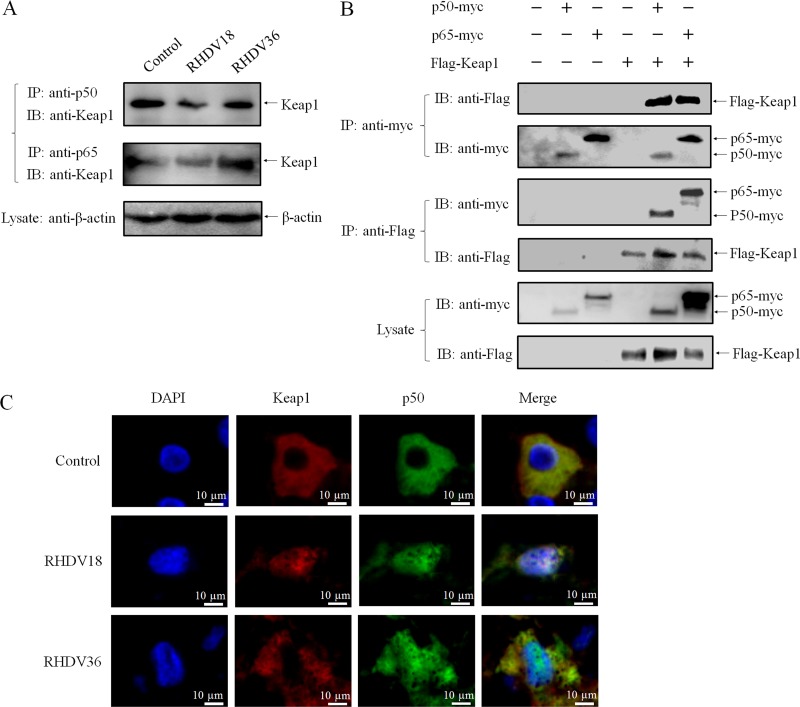
The NF-κB family carries homologous N-terminal Rel homology domain (RHD) for sequence-specific DNA-binding, dimerization, and inhibitory protein binding functions (25–27). The NF-κB subunit, p65, interacts with Keap1 to repress the Nrf2-ARE pathway, and the N-terminal RHD is necessary for both interaction with Keap1 as well as suppression of transcriptional activity (28). In our proteomic analysis, the NF-κB subunit, p50, changed significantly at 18 hpi (Fig. 1B and C). In order to determine whether p50 plays a role in the regulation of Nrf2 by Keap1, we assessed the interaction between p50 and Keap1 in the presence or absence of RHDV infection. An immunoprecipitation (IP) assay using anti-p50 specific polyclonal antibody showed that p50 and Keap1 were associated in infected as well as uninfected liver cells (Fig. 4A). Next, pCI-Flag-Keap1 and pCI-p50-myc eukaryotic expression plasmids were constructed and cotransfected into HEK293T cells, following which, co-IP assays using anti-Myc monoclonal antibody (MAb) and anti-Flag MAb, respectively, were conducted to determine whether p50 directly interacts with Keap1. Western blot analysis using anti-Flag MAb and anti-Myc polyclonal antibodies (PAb) displayed bands that corresponded with each other, indicating direct interaction between p50 and Keap1 (Fig. 4B). In addition, Keap1 and p50 in liver cells were colocalized in the nucleus at 18 hpi, following which, nuclear export of Keap1 was observed at 36 hpi (Fig. 4C). These results showed that p50 specifically and directly interacts with Keap1 and that p50 protein was involved in the nuclear translocation of Keap1.
FIG 4.
Nuclear translocation of Keap1 depends on interaction with NF-κB. (A) p50 and p65 bind to Keap1 in RHDV livers. An IP assay was performed on cell lysates using Keap1-specific PAb that were infected or uninfected with RHDV and then immunoblotted with MAb or PAb against Nrf2 and Keap1. β-actin was employed as an internal control. (B) Validation of the interaction between Keap1 and p50/p65 by co-IP assay. 293T cells were cotransfected with plasmids expressing Flag-Keap1 and p50-myc or p65-myc. Cell lysates were prepared at 48 h posttransfection. The proteins were immunoprecipitated using anti-Flag or anti-Myc MAbs and analyzed via Western blots with MAbs against Myc or Flag. (C) Immunofluorescence localization analysis of Keap1 (red) and p50 (green). Slides of RHDV-infected livers incubated with PAbs against Keap1 and p50 at 0, 18, and 36 h.
To characterize the interaction between Keap1 and p50 protein alone, or Keap1 and the p50/p65 complex, the plasmid pCI-p65-myc was constructed, and p65 protein was also analyzed via assays. Both p50 and p65 interacted directly with Keap1 in RHDV-infected livers and 293T cells (Fig. 4A and B). These results suggested that the 2 subunits of NF-κB (p50 and p65) directly and specifically interacted with Keap1 to form the Keap1-p50/p65 complex during RHDV infections.
Activation of Nrf2 delayed the death of RHDV-infected rabbits.
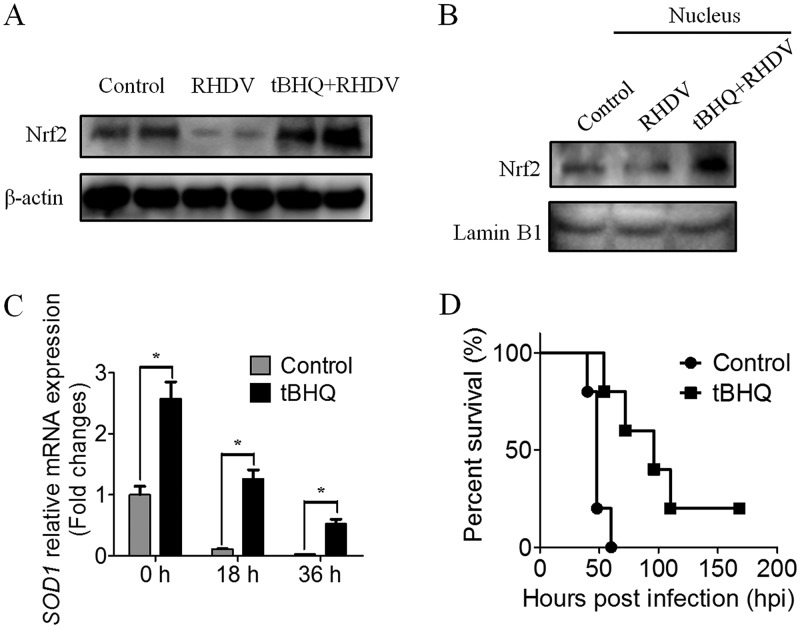
Tert-butylhydroquinone (tBHQ) treatment stabilized and increased Nrf2 protein levels and enhanced Nrf2-mediated transcriptional activation and subsequent antioxidant protection (29). In the challenge study, treating rabbits with tBHQ for 5 days before infection increased the Nrf2 content in the liver compared to that in the untreated group at 36 hpi (Fig. 5A). Activation of Nrf2 was indicated by an increase in Nrf2 protein levels in the nuclei (Fig. 5B). An assessment of SOD1 mRNA expression in the control and tBHQ groups also indicated that the transcriptional activity of Nrf2 was enhanced before infection, although SOD1 mRNA levels decreased with virus infection (Fig. 5C). Evaluation of an observation period of 7 days indicated that the overall survival time of the tBHQ treatment group was significantly prolonged compared to that of the RHDV infection group (Fig. 5D). These data demonstrated that Nrf2 activation provided partial protection and delayed the death of RHDV-infected rabbits, suggesting that the oxidative stress response induced by ROS during RHDV infection played an important role in RHDV pathogenicity.
FIG 5.
tBHQ delaying the death of rabbits against RHDV infection via upregulated Nrf2 expression. (A) Detection of Nrf2 protein in tBHQ-treated and untreated rabbits with RHDV-infected livers by Western blot analysis. β-actin was employed as an internal control. (B) Detection of Nrf2 protein level changes in the nuclei of tBHQ-treated and untreated rabbits. Lamin B1 was employed as an internal control. (C) Detection of mRNA expression levels of the Nrf2-target gene SOD1 in liver tissues. (D) Survival of rabbits infected with RHDV WF/China/2007. The tBHQ group received an intraperitoneal injection of tBHQ once per day before RHDV infection for 5 days. Time points included 0, 18, and 36 h after RHDV infection. The injection solution was prepared fresh every day. All rabbits were challenged subcutaneously with 2 × 104 hemagglutination units of RHDV and subsequently clinically examined daily for 7 days.
DISCUSSION
Previous studies have indicated that inflammation and apoptosis of the liver are associated with RHDV pathogenicity. However, the mechanism underlying liver injury during early infection remains unclear. Here, we provide direct evidence showing that oxidative stress plays a key role in liver injury, especially in the early period of RHDV infection, and that nuclear translocation of Keap1-NF-κB is critical for suppressing the Nrf2-ARE pathway in hepatocytes.
Reportedly, RHDV RNA was found to be present in the liver of RHDV-infected rabbits as early as 18 hpi, although the liver tested positive for viral antigens only at and after 36 hpi (30, 31). Histopathological analyses of the liver revealed that its first significant lesions occurred at 30 hpi (31) and that most rabbits died between 36 and 54 hpi (9). These results indicated that changes that occur in the liver between 18 and 36 h following RHDV infection were critical for the pathogenicity of RHDV. A series of differential proteins between RHDV-infected and uninfected livers were identified at 18 hpi via iTRAQ. It was found that 2 enzymes of the cellular defense system, GST, a phase II detoxification enzyme, and SOD1, an antioxidant enzyme (32–34), both of which are directly involved in ROS removal (35), were downregulated. Further studies demonstrated that ROS in the liver increased substantially with RHDV replication in a time-dependent manner. AST detection indicated liver injury occurring at 18 hpi, although MDA, GSSG/GSH, and antioxidant enzymes were significantly increased only at 36 hpi. These results indicated that oxidative stress injury to the liver was induced in RHDV-infected rabbits at and after 18 hpi.
The expression of a variety of cytoprotective genes is regulated by AREs, wherein Nrf2 is the central regulator of ARE-mediated gene expression. Nrf2 protects against hepatitis virus-induced liver injury by regulating the expression of cytoprotective genes, indicating that activation of Nrf2 confers protection against oxidative stress in virus-induced liver diseases (36). Nrf2 degradation switched Nrf2-activated gene expression off (37). Keap1, which is mainly localized in the cytoplasm, is a known Nrf2 repressor protein that functions as an adaptor protein between Nrf2 and the E3 ligase complex (Cul3) to facilitate Nrf2 degradation (18–21, 38). Keap1 regulates the Nrf2-dependent antioxidant response during induction by modulating Nrf2 levels. Keap1 is also critical for postinduction repression (24). The Keap1 protein is able to translocate to the nucleus and dissociate Nrf2 from the ARE, causing the Nrf2-ARE pathway to turn off (24).
In the current study, significant increases in MDA and the GSSG/GSH ratio demonstrated the occurrence of oxidative stress in the liver. The decrease in GPx and SOD1 indicated suppression of Nrf2 activity. It was observed that Nrf2 protein levels were reduced at 18 and 36 hpi. Nrf2 protein activity was inhibited along with virus replication and liver injury. Moreover, an increase in ubiquitinated Nrf2 levels was observed in RHDV-infected livers. In contrast, Keap1 protein levels in the cytoplasm and nuclei were specifically increased and were also involved in the nuclear export of Nrf2 after RHDV infection. Interestingly, Keap1 expression levels were decreased at 36 hpi compared to those at 18 hpi in livers. Previous work reported that p62 serves as an autophagic cargo receptor for Keap1, sequestering Keap1 protein in the developing phagophore for degradation (39). During infection, RHDV-induced liver autophagy increases at 18 hpi, reaching a maximum at 24 hpi and declining at 30 and 36 hpi (4). These results indicate that autophagy at 36 hpi might be higher than that at 18 hpi, which could lead to a decrease in Keap1 protein levels at 36 hpi compared to those at 18 hpi. This demonstrates that RHDV represses the antioxidant response by regulating Nrf2 protein levels via Keap1 to inhibit the Nrf2-ARE pathway.
NF-κB is activated in response to a wide range of stimuli, such as proinflammatory cytokines and viral infections. NF-κB is a transcription factor that regulates various cellular processes, including immune stress response and proliferation (40, 41). Several studies have demonstrated that NF-κB activation antagonizes the Nrf2 pathway (28, 42). The NF-κB subunit, p65, deprives CBP of Nrf2 and facilitates the recruitment of histone deacetylase 3 (HDAC3) to ARE (42) or connects with Keap1 to promote the nuclear translocation of Keap1 to functionally inactivate Nrf2 (28). Follow-up studies are warranted in order to elucidate the role of oxidative stress and NF-κB in RHDV pathogenicity. Using iTRAQ, the current study found that the NF-κB p50 protein was upregulated at 18 hpi with RHDV, indicating a change in NF-κB activity in the liver during the early period of RHDV infection. Further studies showed that p50 and Keap1 were colocated in the nucleus, indicating an interaction between them. Furthermore, co-IP experiments demonstrated that p50 directly and specifically interacts with Keap1, indicating that p50 is a new partner of Keap1 and that not only p50, but also p65, directly binds Keap1. These results indicated that NF-κB family proteins were important regulatory factors of special biological functions associated with the Nrf2-ARE pathway. Considered together, these results suggest that NF-κB (p50/p65) binds to Keap1, forming the Keap1-p50/p65 complex, which plays a critical role in Nrf2-ARE pathway regulation.
Antioxidants cause stabilization and nuclear translocation of Nrf2, which binds to AREs and upregulates defensive genes that protect cells against oxidative stress. Several studies have described the cytoprotective action of tBHQ under pathological conditions in vitro as well as in vivo. Previous work reported that 50 to 100 mg/kg tBHQ treatment or pretreatment can induce Nrf2 activation, preventing hepatic ischemia/reperfusion injury and scrotal heat-induced damage (43–45). Nrf2 activation induced by tBHQ also protects hepatocytes from lipotoxicity (46) and activates the augmenter of liver regeneration (47). Cytoprotective actions of tBHQ were also confirmed by our study via animal experiments. We detected antioxidant activities of tBHQ by inducing the activation of Nrf2 and upregulating SOD1 expression. Following tBHQ injection, rabbits were partially protected from RHDV assault, and their death was delayed. Thus, we speculate that tBHQ may reduce oxidative stress injury in the liver, thereby protecting rabbits during RHDV infection.
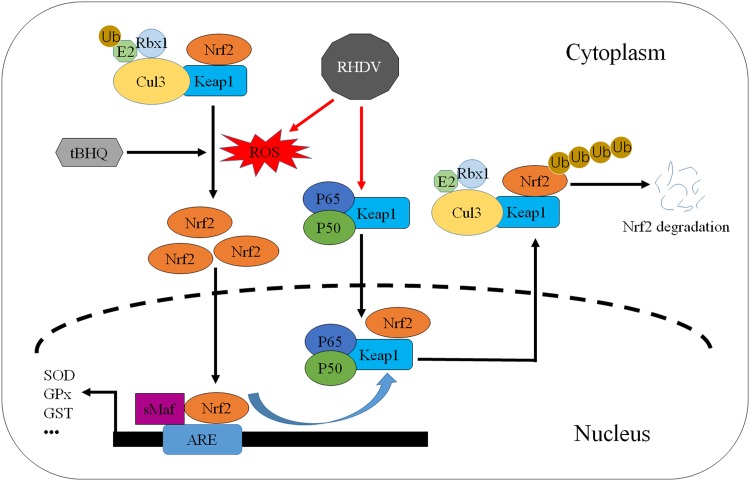
In conclusion, the findings of this study indicated that nuclear export of Nrf2 mediated by Keap1 plays a key role in the pathogenic mechanism underlying RHD. Keap1 binds with p50/p65 and translocates to the nucleus, where Keap1 induces the nuclear export of Nrf2, leading to the inhibition of Nrf2 activity as well as antioxidant response (Fig. 6). These results may enrich existing knowledge regarding the pathogenic mechanisms of RHDV and provide important clues for the development of antioxidant-based therapies of RHDV infection.
FIG 6.
The model of Nrf2-mediated repression of antioxidant responses in RHDV-infected livers. First, Keap1 interacts with NF-κB (p50/p65) to form a complex in liver cells. RHDV infection induces oxidative stress in livers, and the Keap1-p50/p65 complex translocates into the nucleus to facilitate the dissociation of Nrf2 from the ARE. Then, the Keap1-Nrf2 complex separates from p50/p65 and is exported from the nucleus. Finally, ubiquitination and degradation of Nrf2, mediated by Keap1, occur. In addition, activation of Nrf2 by the antioxidant tBHQ upregulates defensive genes that protect cells from oxidative stress.
MATERIALS AND METHODS
Viruses and antibodies.
WF/China/2007 (GenBank accession no. FJ794180), a virulent strain of RHDV (48), was used to induce infection in rabbits. The VP60-specific MAb, 1B8, was prepared in our laboratory as previously described (49). Nrf2 MAb and Keap1 PAb were purchased from Cell Signaling Technology (Danvers, MA, USA). SOD1, GST, and p50 PAbs were purchased from Nanjing Sciben Biotechnology Co. Ltd. (Nanjing, China). P65 and ubiquitin PAbs were purchased from Shanghai Beyotime Biotechnology Co. Ltd. (Shanghai, China).
Cell culture and plasmid constructs.
HEK293T cells were purchased from the American Tissue Culture Collection (ATCC; Manassas, VA, USA). They were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% (vol/vol) fetal bovine serum (FBS; Gibco). The Keap1 (GenBank accession no. XM_008251548), p50 (GenBank accession no. XM_017347386), and p65 (GenBank accession no. NM_021975) sequences were amplified using reverse transcription PCR (RT-PCR) from a rabbit liver cDNA library. Total RNA was extracted from liver tissues using a total RNA kit I (Omega Bio-Tek, Inc., Norcross, GA), and 1 μg of RNA was used for cDNA synthesis using a PrimeScript RT reagent kit (TaKaRa, Dalian, China) according to the manufacturer’s instructions. The Keap1 gene was cloned into the pCMV-3Tag-1a vector (GenScript, Nanjing, China). The p50 and p65 genes were cloned into the pCMV-3Tag-4a vector (GenScript). The resultant plasmids pCMV-Flag-Keap1, pCMV-p50-myc, and pCMV-p65-myc were used to transfect the HEK293T cells with Lipofectamine 3000 reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The primers used in this study are listed in Table 1.
TABLE 1.
Primers for the amplification of p50, p65, and Keap1 genesa
| Primer | Sequence (5′-3′) |
|---|---|
| P50-F1 | TATACAGCTCCAGAATGGC |
| P50-R1 | CACGTCTTCCTGCTTAGTG |
| P50-F2 | TTTTGGATCCATGGCAGACGACGACC |
| P50-R2 | TTTCTCGAGATGGTTCATCCCAGC |
| P65-F1 | ATTTCCGCCTCTGGCGAATG |
| P65-R1 | CAAACGCTGGTGTTAGGC |
| P65-F2 | TTTGGATCCATGGACGAACTGTTCC |
| P65-R2 | TTTTCTCGAGGGAGCTGATCTGACTC |
| Keap1-F1 | GCGTGATCCCTTGTCACTT |
| Keap1-R1 | CACGTCAACAGGTGCAGTT |
| Keap1-F2 | TTTTGGATCCATGCAGCCGGAAGCC |
| Keap1-R2 | TTTCTCGAGTCAACAGGTGCAGTTC |
Restriction site sequences are underlined. GGATCC, BamH I; CTCGAG, Xho I.
RHDV infection in rabbits.
For iTRAQ analysis, eighteen 8-week-old New Zealand white rabbits (specific pathogen free [SPF]) purchased from Qingdao Kangda Rabbit Industry Development Co. Ltd. (Qingdao, China) were randomly divided into 2 groups, the control group and the infection group. Rabbits in the infection group were injected subcutaneously with 2 × 104 hemagglutination units (HAU; approximately 125 50% lethal doses [LD50]) (5, 9, 11, 50) of the RHDV isolate WF/China/2007. At 18 h postinfection (hpi), 9 rabbits in the control and infection groups, respectively, were euthanized, and their livers were immediately collected for iTRAQ analysis. In order to investigate molecular changes taking place in the liver tissues of infected animals, 3 more rabbits from each group were euthanized at 0, 18, and 36 hpi, following which, their liver samples were stored at −80°C or in liquid nitrogen. To study the survival of antioxidant treatment, eleven 8-week-old rabbits were administered an intraperitoneal injection of tBHQ (100 mg/kg body weight [43, 44] dissolved in 10 ml of 5% alcohol saline; Sigma, USA) once a day for 5 days prior to RHDV infection. Next, the tBHQ-treated animals and 11 untreated rabbits (control) were challenged with WF/China/2007. At 0, 18, and 36 hpi, 2 rabbits each of the tBHQ-treated group and untreated group were euthanized, and their liver samples were stored at −80°C. The 5 remaining animals of both groups were left to die spontaneously for purposes of survival calculation.
The animals were handled according to the Standards for Laboratory Animals (GB14925-2001) and Guideline on the Humane Treatment of Laboratory Animals (MOST 2006a) established by the People’s Republic of China.
iTRAQ and LC-MS/MS.
The workflow of the study is presented (Fig. 1A). Protein digestion, iTRAQ labeling, and LC-MS/MS analysis were performed as previously described (51). Briefly, 9 RHDV-positive and 9 negative-control liver tissues were ground with liquid nitrogen and dissolved in HU buffer (20 mM HEPES, 8 M urea, 150 mM tris-HCl, pH 7.8). After centrifuging at 14,000 × g for 10 min at 4°C, the protein content was determined using the Bradford protein assay reagent (Beyotime Institute of Biotechnology, Shanghai, China). Protein samples (30 μg) were separated via electrophoresis on 12% SDS-PAGE and stained to confirm parallelism among samples.
An eight-plex iTRAQ reagent was used to label the resultant peptide mixture according to the manufacturer’s instructions (Applied Biosystems). To reduce individual variation, 9 controls were pooled into 3 biological replicates of 3 samples each (C1, C2, and C3) labeled with iTRAQ tags 113, 114, and 115, respectively, and 9 RHDVs were pooled into 3 biological replicates of 3 samples each (T1, T2, and T3) labeled with iTRAQ tags 116, 117, and 118, respectively. Next, iTRAQ-labeled peptides were fractionated via strong cation exchange (SCX) chromatography using an AKTA purifier 100 (GE Healthcare, Chalfont St. Giles, UK). MS experiments were executed on a Q-Exactive mass spectrometer (Thermo Finnigan) connected to a Thermo Fisher Scientific EASY-nLC autosampler. MS data were acquired, and the instrument was run with the peptide-recognition mode enabled.
Finally, MS/MS spectra were searched using a Mascot engine 2.2 (Matrix Science, London, UK) embedded in Proteome Discoverer 1.4 (Thermo Electron, San Jose, CA, USA) against an Oryctolagus cuniculus protein sequence database (UniProt Oryctolagus cuniculus; FASTA, 22,911 sequences; downloaded on 18 May 2015) from the Universal Protein Resource (http://www.uniprot.org/). For protein identification, the following parameters were used: a peptide mass tolerance of 20 ppm, MS/MS tolerance of 0.1 Da, enzyme specificity set to trypsin with 2 missed cleavages, fixed modification of carbamidomethyl at C and iTRAQ 8-plex at K and the N terminus, variable modification of oxidation at M, and a false-discovery rate (FDR) of peptide identification set to be ≤0.01. Protein identification was based on the identification of at least one unique peptide. Relative quantitative analyses of proteins in the samples were performed using proteome Discoverer 1.4, on the basis of iTRAQ reporter ion ratios from all unique peptides representing each protein. Sample REF was used as reference on the basis of the weighted average of the intensity of reporter ions of each identified peptide. Then, the final protein ratios were normalized by the median average protein ratio for an unequal mix of the different labeled samples.
Blood chemistry.
ALT, AST, LDH, T-Bil, and ALP were detected via standard techniques using an automatic biochemical analyzer (Spotchem EZ SP-4430; Arkray Ltd., Kyoto, Japan).
Activities of MDA and antioxidant enzymes.
Liver tissues were homogenized in 0.01 μM phosphate buffer (pH 7.4). Following centrifugation at 12,000 × g for 20 min at 4°C, the MDA, SOD1, and GPx contents in the supernatant were assessed spectrophotometrically using corresponding kits (Nanjing Jiancheng Biochemistry Co., China). The Bradford method was used to determine protein concentrations. MDA levels were expressed as nmol/g protein, and SOD1 and GPx levels were expressed as U/mg protein.
Measurement of ROS and immunofluorescence assays.
Liver tissues were collected at different postinfection time points, fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned (4 μm thick) for measurement of ROS and viral antigens or cell target protein labeling. Intracellular ROS generation was detected using dihydroethidium (DHE), a nonfluorescent cell-permeative compound, as a probe. Antigen labeling was performed via immunohistochemistry using MAb 1B8 prepared in our laboratory. Nrf2, Keap1, and p50 proteins were labeled using specific MAb or PAb antibodies. The dilutions of Nrf2, Keap1, and p50 antibodies were 1:3,000, 1:4,000 and 1:1,000, respectively. Images were acquired using an ortho-fluorescence microscope (Nikon Eclipse C1; Nikon, Japan) and imaging system (Nikon DS-U3; Nikon, Japan).
Gene expression analysis using RT-qPCR.
Total RNA was extracted from liver tissues using a total RNA kit I (Omega), and 1 μg of RNA was used for cDNA synthesis using a PrimeScript RT reagent kit (TaKaRa). Specific primers for RHDV and rabbit SOD1 gene were used (11, 52) for quantitative real-time PCR (RT-qPCR) analyses using a real-time SYBR master mix (TaKaRa) and a Light Cycler 480 II system (Roche) according to previously reported methods. The expression of antioxidant enzymes was normalized to that of the β-actin (53), and the results were presented as fold induction relative to normal control. The primers used for RT-qPCR are described in Table 2.
TABLE 2.
Sequences of oligonucleotide primers used for RT-qPCR
| Gene | Sequence (5′-3′) | Reference or source |
|---|---|---|
| RHDV VP60 | Forward: GTTCCCACACTGGTCCTTAG | 23 |
| Reverse: TGAGGACTGGGGTCGTGAG | ||
| Nrf2 | Forward: AGTGGATCTGCCAACTACTC | This study |
| Reverse: CTCTGCCAAAAGCTGCAT | ||
| SOD1 | Forward: ACCATCCACTTCGAGCAGAAG | 3 |
| Reverse: GTCCTGTTATGCGTCCCTTGA | ||
| β-actin | Forward: GGACCTGACCGACTACCTCA | 34 |
| Reverse: CAGCTTCTCCTTGATGTCCC |
Western blot analysis.
Western blot analyses were performed on total cell proteins or cytosolic and nuclear extracts. Cytosolic and nuclear extracts were prepared from liver homogenates using a Minute cytoplasmic and nuclear extraction kit (Invent Biotechnologies, Inc., Beijing, China) according to the manufacturer’s instructions. Protein samples were collected and stored in aliquots at −80°C until use. Protein concentrations in cytosolic and nuclear liver fractions were measured using Bradford assays. Equal amounts of protein were separated by 12% SDS-PAGE and blotted onto polyvinylidene difluoride (PVDF) membranes. Target proteins were probed with special MAbs or PAbs at each suitable dilution. After being washed and incubated with anti-mouse or anti-rabbit secondary horseradish peroxidase (HRP)-conjugated antibodies (Jackson ImmunoResearch Laboratories, Inc., USA), the membranes were visualized using an ECL detection kit (Vazyme Biotech, Nanjing, China).
Immunoprecipitation and immunoblotting.
Liver tissues or HEK293T cells were washed with cold phosphate-buffered saline (PBS) once and lysed in lysis buffer (reagent of Capturem IP and Co-IP kit; TaKaRa) by adding an appropriate amount of protease inhibitor cocktail. Lysates were incubated on ice for 15 min and centrifuged at 17,000 × g for 10 min at 4°C. Following incubation with recommended amounts of antibodies and clarified lysates for up to 20 min at room temperature, immunoprecipitation (IP) assays were performed using a Capturem IP and Co-IP kit (TaKaRa) according to the manufacturer’s instructions. Collected samples were then analyzed using SDS-PAGE and Western blot analysis, as described above.
Statistical analysis.
All results are expressed as mean ± standard error of the mean. The levels of significance for differences between infection and control groups are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Differences between groups were determined using Student’s t tests with GraphPad Prism software 5.0 (San Diego, CA).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grants no. 31600130 and 31702274) and funds earmarked for the China Agriculture Research System (grant no. CARS-43-C-1).
We thank Shanghai Applied Protein Technology Co. Ltd. and Wuhan Servicebio Technology Co. Ltd. for providing technical support. We thank Editage for English language editing.
B.H. and H.W. designed and performed the experiments and wrote the manuscript. B.H., Y.S., and R.Q. analyzed the data. M.C., Z.F., W.Z., and W.X. assisted in performing some experiments. F.W. contributed essential ideas and discussion. All authors read and approved the final manuscript.
We declare no conflict of interest regarding the publication of the manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Abrantes J, van der Loo W, Le Pendu J, Esteves PJ. 2012. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): a review. Vet Res 43:12. doi: 10.1186/1297-9716-43-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forrester NL, Abubakr MI, Abu Elzein EM, Al-Afaleq AI, Housawi FM, Moss SR, Turner SL, Gould EA. 2006. Phylogenetic analysis of rabbit haemorrhagic disease virus strains from the Arabian Peninsula: did RHDV emerge simultaneously in Europe and Asia? Virology 344:277–282. doi: 10.1016/j.virol.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Rodak L, Smid B, Valicek L, Vesely T, Stepanek J, Hampl J, Jurak E. 1990. Enzyme-linked immunosorbent assay of antibodies to rabbit haemorrhagic disease virus and determination of its major structural proteins. J Gen Virol 71:1075–1080. doi: 10.1099/0022-1317-71-5-1075. [DOI] [PubMed] [Google Scholar]
- 4.Vallejo D, Crespo I, San-Miguel B, Alvarez M, Prieto J, Tuñón MJ, González-Gallego J. 2014. Autophagic response in the rabbit hemorrhagic disease, an animal model of virally-induced fulminant hepatic failure. Vet Res 45:15. doi: 10.1186/1297-9716-45-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.San-Miguel B, Crespo I, Vallejo D, Álvarez M, Prieto J, González-Gallego J, Tuñón MJ. 2014. Melatonin modulates the autophagic response in acute liver failure induced by the rabbit hemorrhagic disease virus. J Pineal Res 56:313–321. doi: 10.1111/jpi.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alonso C, Oviedo JM, Martin-Alonso JM, Diaz E, Boga JA, Parra F. 1998. Programmed cell death in the pathogenesis of rabbit hemorrhagic disease. Arch Virol 143:321–332. doi: 10.1007/s007050050289. [DOI] [PubMed] [Google Scholar]
- 7.Ueda K, Park JH, Ochiai K, Itakura C. 1992. Disseminated intravascular coagulation (DIC) in rabbit haemorrhagic disease. Jpn J Vet Res 40:133–141. [PubMed] [Google Scholar]
- 8.Hukowska-Szematowicz B, Tokarz-Deptuła B, Deptuła W. 2012. Genetic variation and phylogenetic analysis of rabbit haemorrhagic disease virus (RHDV) strains. Acta Biochim Pol 59:459–465. [PubMed] [Google Scholar]
- 9.Tunon MJ, Sanchez-Campos S, Garcia-Ferreras J, Alvarez M, Jorquera F, Gonzalez-Gallego J. 2003. Rabbit hemorrhagic viral disease: characterization of a new animal model of fulminant liver failure. J Lab Clin Med 141:272–278. doi: 10.1067/mlc.2003.30. [DOI] [PubMed] [Google Scholar]
- 10.Park JH, Lee YS, Itakura C. 1995. Pathogenesis of acute necrotic hepatitis in rabbit hemorrhagic disease. Lab Anim Sci 45:445–449. [PubMed] [Google Scholar]
- 11.Crespo I, Miguel BS, Laliena A, Alvarez M, Culebras JM, González-Gallego J, Tuñón MJ. 2010. Melatonin prevents the decreased activity of antioxidant enzymes and activates nuclear erythroid 2-related factor 2 signaling in an animal model of fulminant hepatic failure of viral origin. J Pineal Res 49:193–200. doi: 10.1111/j.1600-079X.2010.00787.x. [DOI] [PubMed] [Google Scholar]
- 12.Tuñón MJ, San-Miguel B, Crespo I, Laliena A, Vallejo D, Álvarez M, Prieto J, González-Gallego J. 2013. Melatonin treatment reduces endoplasmic reticulum stress and modulates the unfolded protein response in rabbits with lethal fulminant hepatitis of viral origin. J Pineal Res 55:221–228. doi: 10.1111/jpi.12063. [DOI] [PubMed] [Google Scholar]
- 13.Guo Y, Yu S, Zhang C, Kong AN. 2015. Epigenetic regulation of Keap1-Nrf2 signaling. Free Radic Biol Med 88:337–349. doi: 10.1016/j.freeradbiomed.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. 2003. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol 23:8786–8794. doi: 10.1128/mcb.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skibinski G, Hwang V, Ando DM, Daub A, Lee AK, Ravisankar A, Modan S, Finucane MM, Shaby BA, Finkbeiner S. 2017. Nrf2 mitigates LRRK2- and alpha-synuclein-induced neurodegeneration by modulating proteostasis. Proc Natl Acad Sci U S A 114:1165–1170. doi: 10.1073/pnas.1522872114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dodson M, de la Vega MR, Cholanians AB, Schmidlin CJ, Chapman E, Zhang DD. 2019. Modulating NRF2 in disease: timing is everything. Annu Rev Pharmacol Toxicol 59:555–575. doi: 10.1146/annurev-pharmtox-010818-021856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. 1997. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun 236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 18.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. 1999. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev 13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xue F, Cooley L. 1993. kelch encodes a component of intercellular bridges in Drosophila egg chambers. Cell 72:681–693. doi: 10.1016/0092-8674(93)90397-9. [DOI] [PubMed] [Google Scholar]
- 20.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. 2006. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “tethering” mechanism: a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem 281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi A, Kang MI, Watai Y, Tong KI, Shibata T, Uchida K, Yamamoto M. 2006. Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol 26:221–229. doi: 10.1128/MCB.26.1.221-229.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mills EL, Ryan DG, Prag HA, Dikovskaya D, Menon D, Zaslona Z, Jedrychowski MP, Costa ASH, Higgins M, Hams E, Szpyt J, Runtsch MC, King MS, McGouran JF, Fischer R, Kessler BM, McGettrick AF, Hughes MM, Carroll RG, Booty LM, Knatko EV, Meakin PJ, Ashford MLJ, Modis LK, Brunori G, Sévin DC, Fallon PG, Caldwell ST, Kunji ERS, Chouchani ET, Frezza C, Dinkova-Kostova AT, Hartley RC, Murphy MP, O’Neill LA. 2018. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Z, Zhang S, Chan JY, Zhang DD. 2007. Keap1 controls postinduction repression of the Nrf2-mediated antioxidant response by escorting nuclear export of Nrf2. Mol Cell Biol 27:6334–6349. doi: 10.1128/MCB.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayden MS, Ghosh S. 2012. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev 26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smale ST. 2012. Dimer-specific regulatory mechanisms within the NF-kappaB family of transcription factors. Immunol Rev 246:193–204. doi: 10.1111/j.1600-065X.2011.01091.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q, Lenardo MJ, Baltimore D. 2017. 30 years of NF-kappaB: a blossoming of relevance to human pathobiology. Cell 168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu M, Li H, Liu Q, Liu F, Tang L, Li C, Yuan Y, Zhan Y, Xu W, Li W, Chen H, Ge C, Wang J, Yang X. 2011. Nuclear factor p65 interacts with Keap1 to repress the Nrf2-ARE pathway. Cell Signal 23:883–892. doi: 10.1016/j.cellsig.2011.01.014. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Johnson D, Calkins M, Wright L, Svendsen C, Johnson J. 2005. Stabilization of Nrf2 by tBHQ confers protection against oxidative stress-induced cell death in human neural stem cells. Toxicol Sci 83:313–328. doi: 10.1093/toxsci/kfi027. [DOI] [PubMed] [Google Scholar]
- 30.Shien JH, Shieh HK, Lee LH. 2000. Experimental infections of rabbits with rabbit haemorrhagic disease virus monitored by polymerase chain reaction. Res Vet Sci 68:255–259. doi: 10.1053/rvsc.1999.0372. [DOI] [PubMed] [Google Scholar]
- 31.Guittré C, Ruvoen-Clouet N, Barraud L, Cherel Y, Baginski I, Prave M, Ganiere JP, Trépo C, Cova L. 1996. Early stages of rabbit haemorrhagic disease virus infection monitored by polymerase chain reaction. Zentralbl Veterinarmed B 43:109–118. doi: 10.1111/j.1439-0450.1996.tb00294.x. [DOI] [PubMed] [Google Scholar]
- 32.Pool-Zobel B, Veeriah S, Böhmer F-D. 2005. Modulation of xenobiotic metabolising enzymes by anticarcinogens: focus on glutathione S-transferases and their role as targets of dietary chemoprevention in colorectal carcinogenesis. Mutat Res 591:74–92. doi: 10.1016/j.mrfmmm.2005.04.020. [DOI] [PubMed] [Google Scholar]
- 33.Lee SB, Kim CY, Lee HJ, Yun JH, Nho CW. 2009. Induction of the phase II detoxification enzyme NQO1 in hepatocarcinoma cells by lignans from the fruit of Schisandra chinensis through nuclear accumulation of Nrf2. Planta Med 75:1314–1318. doi: 10.1055/s-0029-1185685. [DOI] [PubMed] [Google Scholar]
- 34.Kumaraguruparan R, Seshagiri PB, Hara Y, Nagini S. 2007. Chemoprevention of rat mammary carcinogenesis by black tea polyphenols: modulation of xenobiotic-metabolizing enzymes, oxidative stress, cell proliferation, apoptosis, and angiogenesis. Mol Carcinog 46:797–806. doi: 10.1002/mc.20309. [DOI] [PubMed] [Google Scholar]
- 35.Jung KA, Kwak MK. 2010. The Nrf2 system as a potential target for the development of indirect antioxidants. Molecules 15:7266–7291. doi: 10.3390/molecules15107266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramezani A, Nahad MP, Faghihloo E. 2018. The role of Nrf2 transcription factor in viral infection. J Cell Biochem 119:6366–6382. doi: 10.1002/jcb.26897. [DOI] [PubMed] [Google Scholar]
- 37.Niture SK, Khatri R, Jaiswal AK. 2014. Regulation of Nrf2: an update. Free Radic Biol Med 66:36–44. doi: 10.1016/j.freeradbiomed.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. 2004. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol 24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simon HU, Friis R, Tait SW, Ryan KM. 2017. Retrograde signaling from autophagy modulates stress responses. Sci Signal 10:eaag2791. doi: 10.1126/scisignal.aag2791. [DOI] [PubMed] [Google Scholar]
- 40.Oeckinghaus A, Ghosh S. 2009. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pfeffer LM, Kim JG, Pfeffer SR, Carrigan DJ, Baker DP, Wei L, Homayouni R. 2004. Role of nuclear factor-kappaB in the antiviral action of interferon and interferon-regulated gene expression. J Biol Chem 279:31304–31311. doi: 10.1074/jbc.M308975200. [DOI] [PubMed] [Google Scholar]
- 42.Liu GH, Qu J, Shen X. 2008. NF-kappaB/p65 antagonizes Nrf2-ARE pathway by depriving CBP from Nrf2 and facilitating recruitment of HDAC3 to MafK. Biochim Biophys Acta 1783:713–727. doi: 10.1016/j.bbamcr.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 43.Cheung KL, Yu S, Pan Z, Ma J, Wu TY, Kong AN. 2011. tBHQ-induced HO-1 expression is mediated by calcium through regulation of Nrf2 binding to enhancer and polymerase II to promoter region of HO-1. Chem Res Toxicol 24:670–676. doi: 10.1021/tx1004369. [DOI] [PubMed] [Google Scholar]
- 44.Li YS, Piao YG, Nagaoka K, Watanabe G, Taya K, Li CM. 2013. Preventive effect of tert-butylhydroquinone on scrotal heat-induced damage in mouse testes. Genet Mol Res 12:5433–5441. doi: 10.4238/2013.November.11.5. [DOI] [PubMed] [Google Scholar]
- 45.Zeng XP, Li XJ, Zhang QY, Liu QW, Li L, Xiong Y, He CX, Wang YF, Ye QF. 2017. Tert-butylhydroquinone protects liver against ischemia/reperfusion injury in rats through Nrf2-activating anti-oxidative activity. Transplant Proc 49:366–372. doi: 10.1016/j.transproceed.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Li S, Li J, Shen C, Zhang X, Sun S, Cho M, Sun C, Song Z. 2014. tert-Butylhydroquinone (tBHQ) protects hepatocytes against lipotoxicity via inducing autophagy independently of Nrf2 activation. Biochim Biophys Acta 1841:22–33. doi: 10.1016/j.bbalip.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dayoub R, Vogel A, Schuett J, Lupke M, Spieker SM, Kettern N, Hildt E, Melter M, Weiss TS. 2013. Nrf2 activates augmenter of liver regeneration (ALR) via antioxidant response element and links oxidative stress to liver regeneration. Mol Med 19:237–244. doi: 10.2119/molmed.2013.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu B, Fan Z, Wang F, Song Y, Wei H, Liu X, Qiu R, Xu W, Yuan W, Xue J. 2016. A new variant of rabbit hemorrhagic disease virus G2-like strain isolated in China. Virus Res 215:20–24. doi: 10.1016/j.virusres.2016.01.018. [DOI] [PubMed] [Google Scholar]
- 49.Song Y, Wang F, Fan Z, Hu B, Liu X, Wei H, Xue J, Xu W, Qiu R. 2016. Identification of novel rabbit hemorrhagic disease virus B-cell epitopes and their interaction with host histo-blood group antigens. J Gen Virol 97:356–365. doi: 10.1099/jgv.0.000355. [DOI] [PubMed] [Google Scholar]
- 50.Farnos O, Boue O, Parra F, Martin-Alonso JM, Valdes O, Joglar M, Navea L, Naranjo P, Lleonart R. 2005. High-level expression and immunogenic properties of the recombinant rabbit hemorrhagic disease virus VP60 capsid protein obtained in Pichia pastoris. J Biotechnol 117:215–224. doi: 10.1016/j.jbiotec.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 51.Liu X, Wang J, Gao L, Liu H, Liu C. 2017. iTRAQ-based proteomic analysis of neonatal kidney from offspring of protein restricted rats reveals abnormalities in intraflagellar transport proteins. Cell Physiol Biochem 44:185–199. doi: 10.1159/000484626. [DOI] [PubMed] [Google Scholar]
- 52.Liu W, Dang R, Wang X. 2015. Development of a SYBR-based real-time PCR to detect rabbit hemorrhagic disease virus (RHDV) and analyze its tissue distribution in experimentally infected rabbits. Virol Sin 30:228–230. doi: 10.1007/s12250-015-3560-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sánchez-Campos S, Alvarez M, Culebras JM, Gonzalez-Gallego J, Tuñón MJ. 2004. Pathogenic molecular mechanisms in an animal model of fulminant hepatic failure: rabbit hemorrhagic viral disease. J Lab Clin Med 144:215–222. doi: 10.1016/j.lab.2004.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.