We have recently shown that the amounts of most gene products are precisely regulated during the development of hematopoietic cells in the bone marrow (1, 2). Comparing the intensities of antigens (gene products) on cells of the same maturational stages, the variability within an individual exceeds that of the variability of the means between individuals. These data show that not only is the timing of appearance of gene product expression during hematopoiesis precise, but the absolute amounts of expression of most gene products are invariant from individual to individual, independent of age and marrow stress. The single antigen studied that differed from all others was CD33, which demonstrated a broad variability in quantitative expression from individual to individual even though the within patient variability was low.
The intensity of CD33 expression has been identified to significantly affect the response of AML patients to treatment with Gemtuzumab ozogamicin (GO, Mylotarg)(3). Pediatric patients with the lowest amounts of CD33 expression did not show a benefit when treated with this antibody-toxin conjugate. We have recently demonstrated that a pair of SNPs, in linkage disequilibrium (LD), are also related to the cell surface intensity expression of CD33 on the diagnostic leukemia cells as well as response to therapy (4). The CD33 SNP rs12459419 (C<T) is present in exon 2 and results in an Alanine to Valine change at codon 14. This SNP is found within 4bp of the intron-1/exon-2 junction, and resides within exon splicing enhancer binding site for splicing factor -SRSF2. Presence of the variant T results in altered splicing resulting in skipping of exon2, and thus a shorter CD33-isoform (D2-CD33) that lacks the IgV-domain.(5–7) Interestingly, exon2-encoding IgV-domain contains the epitope for the p67.6-CD33 antibody, which is used both for diagnostic immunophenotyping and also for the humanized antibody that is conjugated to calicheamicin in GO. This splicing SNP also occurs in LD with a second SNP, rs3865444, which is in the non-coding, promotor region of the CD33 gene. In this study, we examined the relationship between the quantitative expression of CD33 on both the diagnostic leukemia cells and the normal regenerating monocytes following initial chemotherapy on those same patients with respect to the underlying genetic presence of these specific SNPs.
Patients in this study participated in Children’s Oncology Group (COG) study AAML0531, a randomized evaluation of GO used as an initial chemotherapy (8). Supplementary Table 1 provides characteristics of the patient population included in the study. For this study bone marrow aspirates were obtained at diagnosis and end of initial induction. Specimens were processed utilizing routine clinical guidelines as described before (9). The quantitative expression of CD33 on the diagnostic acute leukemia cells was compared to the amounts identified on the CD14 positive monocytes in the normal recovering bone marrows following the initial induction. Patients without signed consent forms, qualitatively poor specimens and specimens identified as having residual disease were excluded from the study. A total of 414 patients were included in this analysis. Bone marrow aspirates were collected and processed as previously described (9). The technique to determine mean fluorescence intensity (MFI) of CD33 on the leukemia cells (identified using CD45 and log side scatter gating) and on the mature monocytes on the corresponding regenerating marrow following induction chemotherapy (defined using support vector machines) have been described (3, 10). Quantification of the expression of CD33 on mature monocytes was performed as described previously (1,2). Correlation of MFI to number of molecules was performed by defining the MFI of CD4 conjugated 1:1 to phycoerythrin to be 46,000 molecules (11,12). SNP analysis for splicing CD33 SNP rs12459419 and the linked promoter SNP rs3865444 was performed on genomic DNA obtained from samples obtained at remission using the Sequenome platform at the Biomedical Genomics Center, University of Minnesota. Both SNPs had a call rate of more than 0.98 and were in Hardy–Weinberg equilibrium.
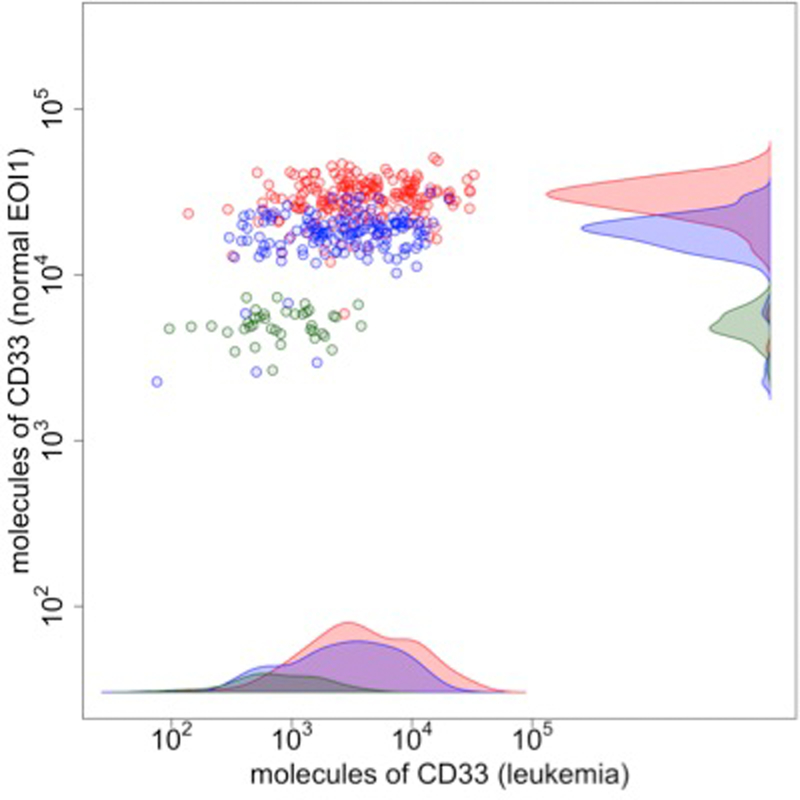
Although a difference in expression of CD33 on the leukemia cells is related to the specific SNP, Figure #1, this quantitative difference is more clearly observed on the corresponding normal monocytes from those same patients after induction chemotherapy. It is evident that these SNPs are highly correlated to differential expression amounts of CD33 on both the normal monocytes and the leukemia cells. The mean number of molecules of CD33 for CC, CT and TT on leukemia cells was 3.8 X103, 2.6 X 103 and 8.0 X102 respectively, with rather broad histograms. In comparison the amount of CD33 on the normal monocytes was higher 2.8 X104, 1.7 X 104 and 5.0 X 103 for CC, CT and TT respectively (p<10−10) with much smaller variation between individuals with the same genotype (Figure 1). Six outliers for CC (n=1) and CT (n=5)(red and blue dots, respectively) were identified within the grouping of low CD33 intensities (green dots, which primarily represent TT genotype). Further testing for other low expressing CD33 genotypes described in a previous study (13) demonstrated that all of these six were heterozygous carriers for at least one another low expressing CD33 allele. Occurrence of other CD33 SNPs distinguish these patients from the rest of the patients suggesting additional changes in the genotype can also affect the amounts of CD33 expressed on the cell surfaces. These data span 3.8 years, demonstrating the stability of the assay to quantify small differences in cell surface gene product expression.
Figure 1:
Correlation between intensity of CD33 expression on AML cells (X –axis) and the normal monocytes (Y axis) in the recovering bone marrow specimens post induction chemotherapy, and the SNP for rs12459419. The intensity level of CD33 on the monocytes is directly related to the rs12459419 SNP [CC, red, vs CT, blue, vs TT, green, p<10−10). Essentially identical data is obtained for rs3865444 in the LD non-coding promoter region (data not shown).
In separate experiments using HIM3–4 (a monoclonal antibody recognizing CD33 that does not cross block p67.6) also showed similar variation in intensities between CC, CT and TT when tested on normal bone marrow monocytes. At maximum titration concentrations, the number of CD33 molecules identified by p67.6 were 3.8 X 104, 1.9 X 104, and 4.2 X 103 for individuals genotyped for CC, CT, and TT, respectively. Quantification using HIM3–4 produced 1.7 X 104, 7.4 X 103, and 1.7 X 103 molecules for the same corresponding genotypes. These data suggest that the SNP genotype affects the quantitative display of both epitopes identified by both p67.6 and HIM3–4 although the absolute amounts of CD33 defined by the two antibodies were approximately 2 fold different.
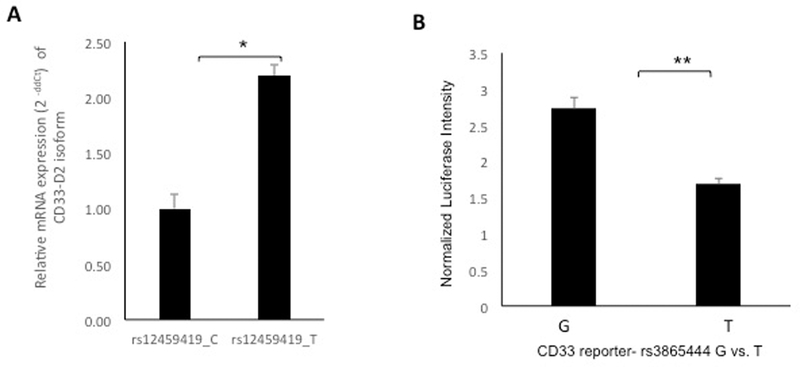
We further performed in vitro functional studies to investigate the mechanism for these differences by performing a mini-gene experiment to confirm the altered splicing due to the C>T change for rs12459419 and luciferase assays to study the impact of the linked promoter polymorphism rs3865444 (Figure 2). Supplementary Note 1 provides details of the methods used. Using transiently transfected HEK293 cells containing rs12459419, the RNA expression level of the altered CD33-D2 isoform increased 2 fold for T as compared to the C genotype (p<0.05) thus confirming C>T change to be associated with increased production of alternatively spliced D2 isoform (Figure 2A). Luciferase reporter assays were performed using CD33 promoter vector with G or T allele for rs3865444 in K562 cells. In agreement with the association of the SNP with low CD33 cell surface intensity on monocytes, T allele was associated with lower transcriptional activation (Figure 2B, p<0.005).
Figure 2.
A) RNA expression level of CD33-D2 isoform in HEK293 cells transiently transfected with CD33 mini-gene containing rs12459419 C or T genotype. The values calculated from quantitative PCR were normalized with rs12459419 C genotype expression. The CD33 rs12459419 T genotype generates approximately twofold transcript of CD33-D2 isoform. B) Luciferase assay for CD33 SNP promoter SNP-rs3865444 G>T, CD33 reporter construct (pGL3basci) with G or T allele was transfected into K562 cells. The values from Firefly luciferase intensity normalized with Renilla luciferase activity (n=3 separate experiments done in triplicate) and p values were two tailed and calculated by student’s t-test. (**p <0.005, *p <0.05)
These results show that the variability of amounts of CD33 expressed on normal hematopoietic cells are the result of a genetic polymorphism predominantly related to two SNPs that are LD. CC patients express approximately 1.6 times as many CD33 molecules in comparison to CT patients and 5.6 times greater compared to TT individuals. The TT splice variant does not result in a complete loss of the expression of CD33, therefore, the epitope identified by p67.6 is still expressed but at a lower level. Interestingly these changes do not affect the within patient variability of expression (1, 2).
The intensity difference of CD33 between CC and CT is minimal on the leukemia cells with broad overlap. Therefore, the differential response to GO observed between the homozygous and heterozygous individuals cannot be completely explained by quantitative epitope expression (4). The differences in response to therapy may be caused by a secondary phenomenon, possibly differential internalization.
The ratio in CD33 expression between the monocytes and leukemic blasts is similar to that observed in normal hematopoiesis between the uncommitted progenitor cells and monocytes(1, 2). CD33 increases approximately 5X during maturation from the uncommitted progenitor cells to mature monocytes. Therefore, the lower expression of CD33 on leukemia cells as compared to that detected on mature monocytes may simply be a result of the choice of which normal cell population is used for intensity comparison. These results illustrate an important concept in neoplastic transformation. We demonstrate a single SNP can cause a 5.6X reduction in the expression of a specific gene product. Therefore, point mutations targeting a single SNP may have a dramatic affect on the cell development by changing the amounts of expressed gene products. Therefore, very small genetic mutations may have large consequences in the ability of cells to develop properly.
Supplementary Material
Acknowledgements:
This research was supported by NIH under the award numbers: U10CA180899, U10CA180886, U10CA98413, and U10CA098543 as well as by NCI-R21CA155524 (Lamba and Walter), NCI (R01CA132946) and R01CA133881 (Aplenc). We are thankful to Yi-Cheng Wang and Dr. Todd Alonzo for their help in the manuscript.
Footnotes
Conflict of Interest:
Loken MR, Eidenschink Brodersen L and Voigt AP are employees of Hematologics Inc. Loken MR, holds leadership position and has stocks/ownership of Hematologics Inc. Aplenc R: Honoraria Sigma-Tau; Gamis A, Consultant/Advisory role Pfizer. Other authors have no conflicts to disclose.
References:
- 1.Loken MR, Voigt AP, Eidenschink Brodersen L, Fritschle W, Menssen AJ, Wells DA. Consistent quantitative gene product expression: #3. Invariance with age. Cytometry A. 2016;89(11):997–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loken MR, Voigt AP, Eidenschink Brodersen L, Fritschle W, Menssen AJ, Meshinchi S, et al. Consistent quantitative gene product expression: #2. Antigen intensities on bone marrow cells are invariant between individuals. Cytometry A. 2016;89(11):987–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard JA, Loken M, Gerbing RB, Raimondi SC, Hirsch BA, Aplenc R, et al. CD33 Expression and Its Association With Gemtuzumab Ozogamicin Response: Results From the Randomized Phase III Children’s Oncology Group Trial AAML0531. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016;34(7):747–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamba JK, Chauhan L, Shin M, Loken MR, Pollard JA, Wang Y-C, Ries R et al. CD33 Splicing polymorphism determines gemtuzumab ozogamicin response in de novo AML: report from randomized phase III Children’s Oncology Group Trial AAML0531 Journal of Clinical Oncology. : official journal of the American Society of Clinical Oncology. 2017;35(23):2674–2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malik M, Chiles J 3rd, Xi HS, Medway C, Simpson J, Potluri S, et al. Genetics of CD33 in Alzheimer’s disease and acute myeloid leukemia. Hum Mol Genet. 2015;24(12):3557–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik M, Simpson JF, Parikh I, Wilfred BR, Fardo DW, Nelson PT, et al. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 2013;33(33):13320–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raj T, Ryan KJ, Replogle JM, Chibnik LB, Rosenkrantz L, Tang A, et al. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet. 2014;23(10):2729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gamis AS, Alonzo TA, Meshinchi S, Sung L, Gerbing RB, Raimondi SC, et al. Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: results from the randomized phase III Children’s Oncology Group trial AAML0531. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2014;32(27):3021–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loken MR, Alonzo TA, Pardo L, Gerbing RB, Raimondi SC, Hirsch BA, et al. Residual disease detected by multidimensional flow cytometry signifies high relapse risk in patients with de novo acute myeloid leukemia: a report from Children’s Oncology Group. Blood. 2012;120(8):1581–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voigt AP, Eidenschink Brodersen L, Pardo L, Meshinchi S, Loken MR. Consistent quantitative gene product expression: #1. Automated identification of regenerating bone marrow cell populations using support vector machines. Cytometry A. 2016;89(11):978–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poncelet P, George F, Papa S, Lanza F. Quantitation of hemopoietic cell antigens in flow cytometry. Eur J Histochem. 1996;40 Suppl 1:15–32. [PubMed] [Google Scholar]
- 12.Loken MR Quantized Hematopoiesis, in. Acute Leukemia, Emadi A; Karp JE eds, Demosmedical, New York, 2018, p. 243. [Google Scholar]
- 13.Mortland L, Alonzo TA, Walter RB, Gerbing RB, Mitra AK, Pollard JA, et al. Clinical significance of CD33 nonsynonymous single-nucleotide polymorphisms in pediatric patients with acute myeloid leukemia treated with gemtuzumab-ozogamicin-containing chemotherapy. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19(6):1620–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.