Abstract
Noncovalent hybrids of single-stranded DNA and single-walled carbon nanotubes (SWCNTs) have demonstrated applications in biomedical imaging and sensing due to their enhanced biocompatibility and photostable, environmentally responsive near-infrared (NIR) fluorescence. The fundamental properties of such DNA-SWCNTs have been studied to determine the correlative relationships between oligonucleotide sequence and length, SWCNT species, and the physical attributes of the resultant hybrids. However, intracellular environments introduce harsh conditions that can change the physical identities of the hybrid nanomaterials, thus altering their intrinsic optical properties. Here, through visible and NIR fluorescence imaging in addition to confocal Raman microscopy, we show that the oligonucleotide length controls the relative uptake, intracellular optical stability, and retention of DNA-SWCNTs in mammalian cells. Although the absolute NIR fluorescence intensity of DNA-SWCNTs in murine macrophages increases with increasing oligonucleotide length (from 12 to 60 nucleotides), we found that shorter oligonucleotide DNA-SWCNTs undergo a greater magnitude of spectral shift and are more rapidly internalized and expelled from the cell after 24 h. Furthermore, by labeling the DNA with a fluorophore that dequenches upon removal from the SWCNT surface, we found that shorter oligonucleotide strands are displaced from the SWCNT within the cell, altering the physical identity and changing the fate of the internalized nanomaterial. Finally, through a pharmacological inhibition study, we identified the mechanism of SWCNT expulsion from the cells as lysosomal exocytosis. These findings provide a fundamental understanding of the interactions between SWCNTs and live cells as well as evidence suggesting the ability to control the biological fate of the nanomaterials merely by varying the type of DNA wrapping.
Keywords: Near-infrared fluorescence, confocal Raman microscopy, live-cell imaging, engineered nanomaterials, nanotoxicity, nanoparticle exocytosis, nanoparticle stability
Graphical Abstract

Single-walled carbon nanotubes (SWCNTs) have attracted substantial attention in the nanotechnology field due to their unique set of electrical,1 physical,2 and optical properties.3 Their electronic band gap energies are dependent on their chiral identity, denoted by integers (n,m), and vary based on diameter and rollup angle,4 resulting in semiconducting species which exhibit band gap photoluminescence.3 Although highly hydrophobic in their raw as-produced form, noncovalent functionalization of SWCNTs using surfactants5,6 or amphiphilic biomolecules7–9 has been shown to effectively disperse SWCNTs into aqueous solutions while preserving their intrinsic optical properties. Single-stranded DNA can noncovalently functionalize SWCNTs via π-stacking of hydrophobic bases onto the SWCNT sidewall, while the hydrophilic phosphate backbone allows for significantly enhanced aqueous solubility.10 These DNA-SWCNT hybrids have shown promise as biological imaging11 and sensing probes12 due to their near-infrared (NIR) photoluminescence which is tunable, photostable, and sensitive to their local environment.13–16
Hybrids of DNA and SWCNTs are preferred over other noncovalent approaches due to their enhanced biocompatibility,17 ability to sort single (n,m)-chiralities from parent mixtures,18,19 and the potential for sensing imparted by the inherent diversity of oligonucleotide sequence.20 Specific sequence formulations of DNA-SWCNTs have been recently used to detect miRNA in vivo21 in addition to reporting lipid concentrations in live cells22 and animals,23 while other approaches have used similar oligonucleotide surface modifications for DNA or siRNA delivery both in vivo24,25 and in plants for controllable gene regulation.26,27 Although these advances are promising displays of the utility of nanoscale technology, fundamental questions relating the identity of these sensors after prolonged exposure within the biological environment remain largely unexplored. The potential instability of such DNA-SWCNT sensors has direct implications on their ability to perform a designated task, yet the indirect consequence is a nanomaterial with altered properties from its original state. The changed identity of such nanomaterials can cause concerns about toxicity and the unknown effects imparted on the immediate biological environment. While many types of DNA-SWCNTs have been studied extensively in situ both computationally28–30 and experimentally,31–36 their direct translation to more complex biological systems cannot be assumed.
Nanomaterials can be designed to enter the body via ingestion, injection, inhalation, etc., yet macrophages are typically the first cells to detect and internalize foreign molecules regardless of entry method.37 Macrophages are the immune system’s first line of defense, whether as a primary response to a wound or to engulf foreign substances such as nanoparticles that enter the bloodstream.38 Various studies have shown that macrophages internalize DNA-SWCNTs via endocytosis and phagocytosis through the endolysosomal pathway, eventually leading to localization within the lysosomes22,39,40 and accumulation in the liver macrophages of mice in vivo.23,41 Once entrapped within the lysosomes, SWCNTs can remain for days where they experience biologically low pH and exposure to more than 60 hydrolases meant for catabolic degradation.42 In these conditions, surface modifications can play a large role on a nanoparticle’s ultimate fate, whether degradation, exocytosis, or lysosomal escape.37 Given their extremely high surface area to volume ratio, small changes in surface functionalization of SWCNTs can make a major impact on their functionality and stability in such environments.
Although oligonucleotide length determines the intrinsic stability of the resultant hybrid with a SWCNT in water,33 little is known how this length of DNA can affect the stability of SWCNTs in complex intracellular environments. Herein, we present an investigation of the physical and optical stabilities of (GT)n-SWCNTs, where n is the number of sequence repeats upon internalization into murine macrophages. Near-infrared hyperspectral microscopy in live cells revealed strong correlations between oligonucleotide length, NIR fluorescence intensity, and spectral stability of the examined SWCNTs. All DNA-SWCNT combinations displayed emission shifts to lower energies (i.e., red shifts) upon interacting with the cells; however, several chiralities of (GT)6-SWCNTs exhibited significant blue shifts over the course of 24 h, indicating molecular adsorption and/or DNA displacement. We quantified SWCNT concentrations in cells using confocal Raman microscopy, which can detect all SWCNTs including nonfluorescent species and revealed significant differences in both internalization and lysosome-mediated expulsion of (GT)6- and (GT)30-SWCNTs over 24 h. Finally, we used fluorophore labeled DNA to probe the condition of the SWCNT hybrids as they were processed through the endolysosomal pathway.
Results and Discussion.
To study the effects of single-stranded DNA length on the intracellular optical properties of DNA-SWCNTs, we first noncovalently functionalized HiPco SWCNTs with one of five different (GT)n oligonucleotides, where n = 6, 9, 12, 15, or 30 repeats (Figure S1, Table S1). Murine macrophages (RAW 264.7 cell line) were incubated with 1 mg/L of each (GT)n-SWCNT sample for 30 min under standard cell culture conditions and replenished with fresh media (hereby referred to as a “pulse” of DNA-SWCNTs). The majority of cells exhibited substantial NIR broadband fluorescence (about 900–1600 nm) when excited by a 730 nm laser (Figure 1a). In agreement with previous studies,15,22,43 NIR fluorescence movies confirmed the internalization of the SWCNTs into endosomal vesicles, which were actively translocated around the cell and could be easily distinguished from background cellular autofluorescence (Movies S1 and S2). The NIR fluorescence images were acquired 0, 6, or 24 h after an initial pulse to assess the DNA length and temporal dependencies on intracellular fluorescence intensity (Figure 1b). In general, the observed NIR fluorescence intensities visibly increased with increasing oligonucleotide length and decreased in time after initial loading into the cells. Histograms constructed from pixel intensity values of the 0 and 24 h images confirmed that the temporal decreases in intensities were similar among all sequences (Figure 1c). Interestingly, the initial intensity distributions were much broader in longer oligonucleotide sequences, suggesting more heterogeneity in the optical response to internalization of these SWCNTs. To quantify the images, the average fluorescence intensities were extracted using a global thresholding analysis to examine the NIR fluorescence from only SWCNTs contained within the cells. We observed significant increases in NIR fluorescence intensities as a function of DNA length (Figure 1d) at each time point. Pearson correlation coefficients (rp) were determined to be 0.846, 0.885, and 0.850 at 0, 6, and 24 h respectively, confirming that the correlation was linear and statistically significant (p < 0.001 for all) between sequence length and fluorescence intensity at any given time point. To mitigate variations in fluorescence quantum yield (Figure S2), the images from Figure 1b were normalized to each (GT)n-SWCNT’s average 0 h intensity (Figure S3a) and the percent change in initial intensity was quantified (Figure S3b). While most DNA-SWCNTs demonstrated significant fluorescence quenching in time, (GT)6-SWCNTs first increased nearly 25% at 6 h before decreasing to more than 25% below the initial intensity after 24 h. Among other factors, it is known that the fluorescence intensity of a SWCNT is inversely correlated to the density of water in the immediate vicinity.44 Thus, a removal of water from the surface of (GT)6-SWCNTs through the adsorption of other amphiphilic molecules can explain the increase in intensity observed at 6 h. Altogether, we propose that these discrepancies in intracellular fluorescence are affected by (1) variations in DNA-SWCNT interaction with and internalization into cells as a function of DNA length, (2) variations in the optical stability of the (GT)n-SWCNT hybrids after interacting with and/or internalizing into the cells, or (3) variable rates of cellular expulsion. Throughout the Letter, we will carefully examine these hypotheses.
Figure 1.
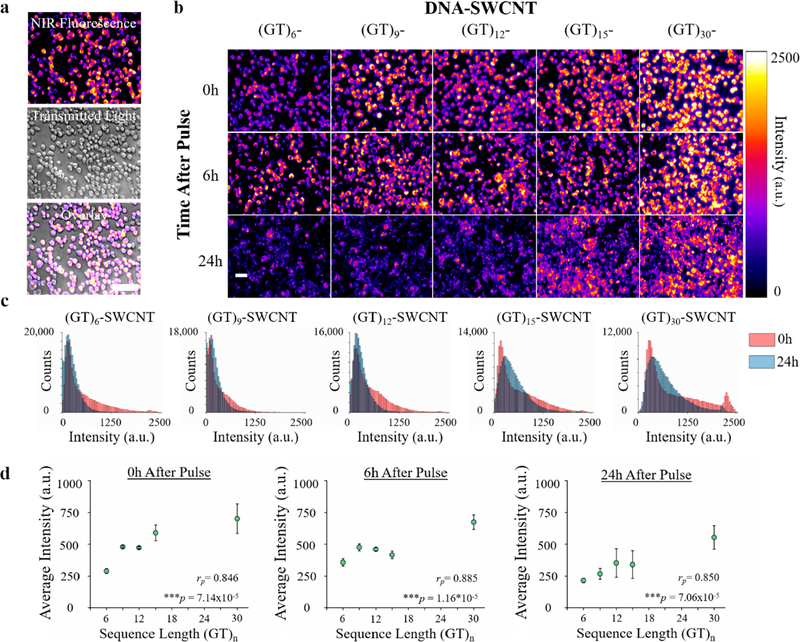
Length-dependent intracellular fluorescence of DNA-SWCNTs. (a) NIR fluorescence image of live macrophages pulsed with (GT)15-SWCNTs, along with respective transmitted light image and merged NIR/transmitted light image. Scale bar = 40 μm. (b) NIR fluorescence images of macrophages after 30 min pulse of (GT)n-SWCNTs, imaged over the course of 24 h. Scale bar = 20 μm. (c) Histograms corresponding to the 0 and 24 h (GT)n-SWCNT images in (b), and (d) average intracellular fluorescence intensities for all examined DNA sequences 0, 6, or 24 h after (GT)n-SWCNT pulse. Experiments were performed in triplicate and are represented as mean ± s.d. (*p < 0.05, **p < 0.01, according to two-tailed two-sample t-test).
The relationship between stability and fluorescence of DNA-SWCNTs is highly dependent on SWCNT chirality as well as oligonucleotide length.33 Therefore, we employed NIR hyperspectral fluorescence microscopy to assess the chirality-resolved intracellular stability of the (GT)n-SWCNTs.15 Using a 730 nm excitation laser, we were able to resolve four distinct bands in the NIR region corresponding to the emission spectra of the four brightest SWCNT chiralities, (10,2), (9,4), (8,6), and (8,7) (Figure S4a).15 Hyperspectral images were acquired immediately following a 30 min pulse of each (GT)n-SWCNT and after an additional 24 h of incubation in SWCNT-free cell media (Figure S4b–f). Upon internalization, we observed two common characteristics of all fluorescence spectra: (1) an initial red shift (i.e., increase in wavelength) of every chirality compared to the spectra acquired in cell culture media and (2) increased intensities of longer wavelength chiralities relative to shorter. To explain the first finding, a red shift in SWCNT emission spectra can be caused by charged species that interact with the phosphate backbone of DNA and induce a conformational change, ultimately modulating the dielectric environment of the SWCNT and thus shifting SWCNT emission to longer wavelengths.45 Surface proteins present on cell membranes with high charge densities have been shown to promote this red shift upon first contact with DNA-SWCNTs before endocytosis.43 Additionally, the exposure of DNA-SWCNTs to serum-containing cell culture media can produce aggregation that causes spectral modulation by protein–DNA electrostatic interactions.46 We attribute the initial red shift observed to a combination of these factors directly following a pulse of (GT)n-SWCNTs in which the macrophages contained both membrane-bound particles that had not yet been internalized as well as newly formed endosomal vesicles that essentially forced DNA-SWCNTs to form small aggregate complexes with other phagocytosed proteins and cargo. Regarding the second finding, changes in the ratiometric intensities between shorter and longer emission wavelength SWCNTs have been described by internanotube exciton energy transfer (INEET),47 a phenomenon that behaves similarly to Förster resonance energy transfer and could be the result of closely packed DNA-SWCNTs contained within lysosomes.
To further compare the NIR fluorescence stabilities across SWCNT chiralities, we converted the emission center wavelengths to energies (meV) and computed the change in emission energy relative to solution controls acquired in cell culture media. Of all the examined oligonucleotide lengths, only the (GT)6-SWCNTs exhibited significant increases in emission energies in multiple chiralities over 24 h of intracellular processing (Figure 2a, Table S2), while the other DNA-SWCNTs commonly displayed a moderate loss of energy over the same period of time despite presumably identical intracellular conditions. Previous studies have demonstrated that this increase in emission energy could arise from endosomal lipids binding to the exposed SWCNT surface,22,23 however the maximum shift in emission energy was not observed at the same time for each chirality, indicating the shifts could be a convolution of multiple physical mechanisms. The influence of a lowered pH to mimic the lysosomal environment was also considered as a potential modulator of SWCNT emission. However, most (GT)n-SWCNTs exposed to cell media with pH of 4.5 experienced random, small (<2 meV) shifts compared to physiological pH (Figure S5). We believe these results indicate that longer oligonucleotides protect the SWCNT surface from competitive molecular adsorption.
Figure 2.
DNA length- and SWCNT chirality-dependent intracellular NIR fluorescence stabilities. (a) Heat maps representing average intracellular change in SWCNT emission energy compared to controls in cell culture media, delineated by chirality, as a function of DNA-sequence and time. All experiments were performed in triplicate. Overlay of transmitted light and hyperspectral images of RAW 264.7 pulsed for 30 min with (b) (GT)6- or (c) (GT)30-SWCNTs. Color scale maps to the fitted center emission energy of (9,4)-SWCNTs and histograms represent center emission energies of all SWCNT-containing pixels in each respective image. Bin size = 4 meV. Gaussian functions were fitted to binned data and overlaid with respective R2, fwhm, and center emission energies (Ec). Scale bar = 20 μm.
To assess spectral shifts in SWCNT emission at the single-cell level, we created hyperspectral maps of the shortest and longest oligonucleotide DNA-SWCNTs (i.e., (GT)6 and (GT)30, respectively). By fitting each SWCNT-containing pixel of a hyperspectral image to a Lorentzian curve,15,22 we were able to overlay transmitted light images with center emission energy maps for the (9,4)-SWCNT, i.e., the most abundant and brightest SWCNT under 730 nm excitation in HiPco, and construct histograms for each image to depict the intracellular change in SWCNT emission energy change through time (Figure 2b,c). Immediately following a 30 min pulse (0 h), the average emission energies of (GT)6-SWCNTs and (GT)30-SWCNTs were statistically identical. Additionally, by fitting the pixel histograms to a Gaussian distribution, the heterogeneity in the populations could be assessed by examining the full width at half-maximum (fwhm). In doing so, we uncovered that the fwhm of (GT)6-SWCNTs immediately after internalization was more than double that of (GT)30-SWCNTs. While the emission energy of (GT)30SWCNTs showed little change in time, (GT)6-SWCNTs displayed an 8 meV increase in emission energy and ~50% decrease in fwhm after 6 h. We believe these DNA-length dependent NIR fluorescence modulations are the result of variations in the relative abundance of oligonucleotide strand ends surrounding each SWCNT. For a given weight of DNA in a DNA-SWCNT hybrid, (GT)6-SWCNTs have 5 times the number of oligonucleotide strand ends than (GT)30-SWCNTs assuming a similar degree of surface coverage. We propose that these strand ends can act as initiation sites for amphiphilic biomolecules to interact with and adsorb onto the exposed nanotube surfaces, leading to higher overall surface coverages (reduced water densities) and thus greater emission energies.22,44,48 Consequently, the wide fwhm initially displayed by (GT)6-SWCNTs was likely the result of individual SWCNTs responding to the varying local environments through progressing stages of the endosomal pathway, whereas the reduced fwhm and blue shift after 6 h can be attributed to molecular adsorption by lysosomal molecules and rearrangement or displacement of the oligonucleotide wrapping on the majority of SWCNTs. Interestingly, after 24 h the emission energy slightly decreased closer to its initial value whereas the fwhm increased toward its initial value, revealing that the hybridized (GT)6-SWCNTs observed at 6 h were ultimately unstable.
Because of the large variations in spectral stability of the internalized DNA-SWNCTs, we devised an assay to probe their integrity of the hybrids based on the ability of SWCNTs to quench conventional organic fluorophores.49 We first constructed (GT)6- or (GT)30-SWCNTs with a Cy3 dye attached to the 5′ end of the DNA strand. The initially quenched fluorophore could be restored to a brightly fluorescent state via displacement from the SWCNT surface by a competing molecule (Figure 3a). Note, even partial displacement of the DNA strand can accomplish this process, thus Cy3 dequenching kinetics of the two prepared hybrids are similar despite unequal displacement kinetics by sodium deoxycholate (SDC) determined from NIR fluorescence (Figure 3b).33 When introduced to macrophage cells in the same 30 min pulse method, we observed substantially different dequenching behavior between the two sequences (Figure 3c). The Cy3-(GT)6-SWCNTs significantly dequenched inside of the cells, reaching a maximum intensity 4 h after the pulse (Figure 3d) and decreased to its initial intensity after 24 h. In contrast, dequenching was not observed in the Cy3-(GT)30-SWCNTs at any point, resulting in statistically significant differences in the dequenching behavior of the DNA-SWCNTs, that is, either partial or full displacement of the DNA from the SWCNTs, within the first 6 h.
Figure 3.
Intracellular stability of DNA-SWCNT hybrids. (a) Schematic of experimental design. Cy3-DNA is quenched when wrapping is intact on SWCNTs but highly fluorescent once displaced. (b) Normalized intensity increase as a function of time after Cy3-DNA is displaced with SDC. (c) Overlaid Cy3-DNA and white light images of RAW 264.7 pulsed with Cy3-(GT)6 or Cy3-(GT)30-SWCNTs for 30 min. Scale bar = 10 μm. (d) Average fluorescence intensities (n ≥ 14 cells) normalized to 0 h intensity. Error bars represent mean ± s.d. Five-pointed stars represent significance between Cy3-(GT)6 and Cy3-(GT)30, and six pointed stars represent significance versus initial intensities. (*p < 0.05, **p < 0.01, ***p < 0.001 according to two-tailed two-sample t-test).
While the observed NIR fluorescence of SWCNTs can be modulated by both concentration and local-environment,16,48,50 certain Raman signatures of pristine SWCNTs depend only on concentration,51–54 allowing for all SWCNT chiralities in a sample to be represented regardless of fluorescence ability. Therefore, we assessed the localized intracellular concentrations of SWCNTs using confocal Raman microscopy. Small regions were scanned in 0.5 μm intervals to obtain Raman maps of macrophages pulsed with 1 mg/L (GT)6- or (GT)30-SWCNTs for 30 min (Figure 4a). The intensity of the G-band spectral feature, indicative of sp2 carbon,52,53 was correlated to known SWCNT concentrations in the construction of a calibration curve in order to obtain a mass of SWCNTs per analyzed cell (Figure S6). Although the local concentrations varied greatly within a single cell, on average the cells pulsed with (GT)6-SWCNTs had more than twice the initial intracellular SWCNT weight than those incubated with (GT)30-SWCNTs (Figure 4b). After 24 h of additional incubation in SWCNT-free cell media, the internal SWCNT concentration of cells dosed with (GT)6-SWCNTs decreased by more than 75%, while those dosed with (GT)30-SWCNTs displayed statistically similar initial and final concentrations. Although cellular uptake of nanoparticles can be influenced by a multitude of factors, we attribute the higher uptake of (GT)6-SWCNTs to their higher overall density of DNA per SWCNT as compared to (GT)30-SWCNTs,31 increasing the probability of interactions between DNA and cellular membrane proteins and thus leading to more nanotubes per engulfing phagosome. Conversely, we surmise that changes in the physical identity of internalized (GT)6-SWCNTs are inducing the macrophages to exocytose this sample more rapidly than the stable (GT)30-SWCNTs.
Figure 4.
SWCNT concentration maps determined by confocal Raman microscopy. (a) Representative confocal Raman microscopy images showing G-band intensity and white light images of RAW 264.7 cells pulsed with (GT)6- or (GT)30-SWCNTs for 30 min. Color map represents local SWCNT pixel concentration derived from G-band intensity calibration (60× objective). Scale bar = 10 μm. (b) Average SWCNT concentration (n ≥ 4 cells) calculated from total pixel concentration within cellular ROIs. (c) Percent of SWCNT dose internalized compared to the initial incubated concentration, determined from cell media supernatant concentration after a 30 min DNA-SWCNT pulse (n = 10). (d) Concentration of exocytosed SWCNTs in cell supernatant 24 h after pulse (n = 4). (e) Representative confocal Raman G-band intensity maps overlaid on white light images of macrophages 24 h after a 30 min pulse of (GT)6- or (GT)30-SWCNTs. Indicated cells were treated with exocytosis inhibitors Bafilomycin A1 (200 nM), Nocodazole (2 μM), Brefeldin A (500 nM), or Exo1 (50 μM) 2 h following DNA-SWCNT pulse. Color map represents local SWCNT pixel concentration derived from G-band intensity calibration (10× objective). Scale bar = 75 μm. (f) Average concentration of SWCNT-containing pixels from all confocal Raman area scans described in (e), (n ≥ 3 area scans). Error bars are represented as mean ± s.d. for all. (*p < 0.05, **p < 0.01, ***p < 0.001, according to two-tailed two-sample t-test).
To corroborate the unexpected results from confocal Raman microscopy, we performed solution-based Raman spectroscopy to determine the SWCNT concentrations in the supernatants at time 0 and 24 h, representing the delivered dose and the amount of exocytosed SWCNTs, respectively. The relative delivered dose, calculated as the percent decrease in supernatant G-band intensity after a 30 min pulse incubation with the cells, confirmed that cells internalized a significantly higher amount of (GT)6-SWCNTs than (GT)30-SWCNTs (Figure 4c). Furthermore, additional agreement with Raman microscopy data was observed at 24 h when significantly more (GT)6-SWCNTs were found in the supernatant as compared to (GT)30-SWCNTs (Figure 4d). We believe these results verified that the decreased intracellular concentration of (GT)6-SWCNTs after 24 h was the result of exocytosis and not cell-mediated degradation of the SWCNT material.
Finally, we sought to better understand the mechanisms dictating retention versus exocytosis by identifying the main pathway in which (GT)6-SWCNTs were being expelled from the cells. Typically, the fate of phagocytosed nanomaterials contained within lysosomes is either regulated secretion in which the contents are further processed and excreted from the Golgi apparatus, or lysosomal exocytosis via direct fusion with the cell membrane.55–57 Therefore, we devised an assay to compare the intracellular DNA-SWCNT concentration of macrophages after treatment with specific pathway inhibiting compounds. Bafilomycin A1 and Nocodazole, both of which inhibit lysosomal exocytosis,58,59 induced retention of (GT)6-SWCNTs within cells at significantly higher average concentrations than the control after 24 h (Figure 4e,f), while little effect was seen on the average concentrations of (GT)30-SWCNTs. Conversely, Brefeldin A and Exo1, inhibitors of Golgi-mediated exocytosis,60,61 did not cause a significant increase in average concentration for either DNA-SWCNT hybrid, suggesting that the main clearance mechanism for DNA-SWCNTs is through lysosomal exocytosis.
One of the main functions of lysosomal exocytosis is to secrete various biomolecules such as proteins, enzymes, or antigens for intercellular communication and illicit an immune response from nearby cells if necessary.42,62 Studies have shown that while SWCNTs with various types of surface functionalization can reduce or prevent cytotoxicity,63 pristine SWCNTs are recognized as pathogenic substances upon interaction with Toll-like receptors present on the cell membrane of macrophages,64 leading to the secretion of inflammatory cytokines as a mechanism of defense.65 Therefore, we believe that the ability of the lysosomal environment to remove the DNA from (GT)6-SWCNTs causes the cell to identify the altered nanomaterial as it would a nonfunctionalized SWCNT (Figure 5a). Once the cell has recognized this material as a foreign body, excretion from the cell via lysosomal exocytosis is initiated in order to illicit an immune response from nearby cells, resulting in a diminished intracellular SWCNT concentration. Conversely, the increased stability provided by a longer DNA wrapping prevents major alterations from occurring in the lysosomal environment and avoids triggering exocytosis, resulting in a high degree of cellular retention (Figure 5b).
Figure 5.
Schematic depicting the DNA length-dependent intracellular processing of DNA-SWCNTs. (a) (GT)6-SWCNTs are internalized into macrophages in large amounts and localize to the lysosomes. There, biomolecules displace DNA from the SWCNT surface and induce lysosomal exocytosis from the cells. (b) (GT)30-SWCNTs are also located to the lysosomes after internalization into the cells but do not experience DNA displacement. These DNA-SWCNTs with enhanced integrity are retained within the cells.
In conclusion, we propose that the intracellular processing and ultimate fate of (GT)n-SWCNTs are controlled by the differential stabilities of the hybrid nanomaterials in the lysosomal environment, which correlate strongly to the length of a given DNA strand. The observed intracellular fluorescence intensities were shown to increase with increasing oligonucleotide length, while only the shortest DNA-SWCNTs (i.e., (GT)6) displayed instabilities in NIR fluorescence spectra in time. We have shown that (GT)30-SWCNTs are mostly retained within the cells over 24 h with minimal exocytosis, while (GT)6-SWCNTs expelled more than 75% of the internalized cargo over the same time period despite nearly a 2-fold higher amount of initial uptake. The correlation between an increase in emission energy and the dequenching of Cy3-(GT)6 strongly suggests that competitive molecular adsorption to the SWCNT sidewall results in a destabilized structure within the lysosome, increasing the probability of complete DNA displacement or degradation from the SWCNT. Without the biocompatibility afforded by the DNA wrapping, the cell is able to recognize a SWCNT as a foreign pathogenic substance and subsequently secrete its lysosomal contents. These findings accentuate the necessity of biocompatible stability when designing any carbon nanotube-based biosensors while highlighting their sensitivity to small changes in surface chemistry.
Materials and Methods.
DNA-SWCNT Sample Preparation.
Raw single-walled carbon nanotubes produced by the HiPco process (Nanointegris) were used throughout this study. For each dispersion, 1 mg of raw nanotubes was added to 2 mg of (GT)n (where n = 6, 9, 12, 15, or 30) oligonucleotide (Integrated DNA Technologies), suspended in 1 mL of 0.1 M NaCl (Sigma-Aldrich), and ultrasonicated using a 1/8 in. tapered microtip for 30 min at 40% amplitude (Sonics Vibracell VCX-130; Sonics and Materials). The resultant suspensions were ultracentrifuged (Sorvall Discovery M120 SE) for 30 min at 250 000×g and the supernatant was collected. Concentrations were determined using a UV/vis/NIR spectrophotometer (Jasco, Tokyo, Japan) and the extinction coefficient of A910 = 0.02554 L mg−1 cm−1.15
Cell Culture.
RAW 264.7 TIB-71 cells (ATCC, Manassas, VA, U.S.A.) were cultured under standard incubation conditions at 37 °C and 5% CO2 in cell culture medium containing sterile filtered high-glucose DMEM with 10% heat-inactivated FBS, 2.5% HEPES, 1% L-glutamine, 1% penicillin/streptomycin, and 0.2% amphotericin B (all acquired from Gibco). For all cell-related studies, cells were allowed to grow until 90% confluency and used up to the 20th passage.
Near-Infrared Fluorescence Microscopy of Live Cells.
A near-infrared hyperspectral fluorescence microscope, similar to a previously described system,15 was used to obtain fluorescence images and hyperspectral data within live cells. In short, a continuous 730 nm diode laser with 1.5 W output power was injected into a multimode fiber to produce an excitation source, which was reflected on the sample stage of an Olympus IX-73 inverted microscope equipped with a 20X LCPlan N, 20x/0.45 IR objective (Olympus, U.S.A.) and a stage incubator (Okolab) to maintain 37 °C and 5% CO2 during imaging. Emission was passed through a volume Bragg Grating and collected with a 2D InGaAs array detector (Photon Etc.) to generate spectral image stacks. For live cell experiments, cells were seeded into tissue culture treated 96-well plates (Fisher Scientific) at a final concentration of 50 00 cells/well and allowed to culture overnight in an incubator. The media was removed from each well, replaced with 1 mg/L each (GT)n-SWCNT diluted in media, and incubated for 30 min (pulsed) to allow for internalization into the cells. After this pulse, the SWCNT-containing media was removed, the cells were rinsed 3X with sterile PBS (Gibco) and fresh media was replenished. Well plates were mounted on the hyperspectral microscope to obtain broadband images, transmitted light images, and fluorescence hyperspectral images at each given time point. Hyperspectral data were processed and extracted using custom codes written with Matlab software. All Gaussian curve fits were generated using OriginPro 2018.
Solution-Based Fluorescence Dequenching Assay.
Cy3-(GT)6- or Cy3-(GT)30 oligonucleotides were purchased from Integrated DNA Technologies and used in the creation of DNA-SWCNTs (see above). After ultrasonication and ultracentrifugation, the Cy3-DNA-SWCNTs were filtered three times using 100 kDa Amicon centrifuge filters (Millipore) to remove free Cy3-DNA from solution, diluted to 2.5 mg/L, and 1 mL was placed in a plastic cuvette under magnetic stirring. The fluorescence intensity of each sample was obtained in 1 s intervals for 3 min using a PerkinElmer LS 55 fluorescence spectrometer set to 532 nm excitation and 569 nm emission with 3 nm bandwidth. A 10 μL aliquot of a 10% sodium deoxycholate solution (Sigma-Aldrich) was spiked into the Cy3-DNA-SWCNTs after a baseline intensity was established for a final concentration of 0.1% SDC in order to temporally displace the Cy3-DNA from the SWCNTs as previously described.33
Visible Fluorescence Microscopy in Live Cells.
Cy3-(GT)6-SWCNTs and Cy3-(GT)30-SWCNTs were first filtered three times using 100 kDa Amicon centrifuge filters (Millipore) to remove free Cy3-DNA from solution. The cells were seeded onto 35 mm glass-bottom Petri dishes (MatTek) to a final concentration of 500 000 cells/dish and allowed to culture overnight in an incubator. The media was removed from each well, replaced with 1 mg/L of filtered Cy3-(GT)6-SWCNTs or Cy3-(GT)30-SWCNTs diluted in media and incubated for 30 min to allow internalization into the cells. The SWCNT-containing media was removed, the cells were rinsed three times with sterile PBS (Gibco), and fresh media was replenished for each sample. The Petri dishes were mounted in a stage incubator (Okolab) on an Olympus IX-73 inverted microscope with a UApo N 100×/1.49 oil immersion objective for epifluorescence imaging with a U-HGLGPS excitation source (Olympus) filtered through a Cy3 filter cube. The fluorescence images were analyzed by extracting average fluorescence intensity values of individual cell ROIs using ImageJ.
Confocal Raman Microscopy.
Cells were seeded into 35 mm glass bottom microwell dishes (MatTek) to a final concentration of 500 000 cells/dish and were allowed to culture overnight in an incubator. The media was removed from each well, replaced with 1 mg/L (GT)6-SWCNT or (GT)30-SWCNT diluted in media, and pulsed for 30 min to allow internalization into the cells. The SWCNT-containing media was removed, the cells were rinsed three times with sterile PBS (Gibco), and fresh media was replenished. The 0 h samples were immediately fixed using 4% paraformaldehyde in PBS for 10 min, rinsed three times with PBS, and covered with PBS to retain an aqueous environment during imaging. The 24 h samples were later fixed using the same procedure. The cells were imaged using an inverted WiTec Alpha300 R confocal Raman microscope (WiTec, Germany) equipped with a Nikon CFI-Achro 60×/0.8 air objective, a 785 nm laser source set to 35 mW sample power, and collected with a CCD detector through a 600 lines/mm grating. The Raman spectra were obtained in 0.5 × 0.5 μm intervals with 1 s integration time to construct hyperspectral Raman area scans of cellular regions. A calibration curve was obtained by recording spectra of known SWCNT concentrations serially diluted in a single pixel volume with identical acquisition settings. Each spectrum was averaged over 20 scans. A global background subtraction and cosmic-ray removal was performed using Witec Control 5.0 software on all acquired confocal Raman data and G-band maximum intensities were extracted and correlated with known concentrations to produce a linear curve fit using OriginPro 2018 analysis software. The cellular SWCNT concentration data were produced by relating the G-band linear equations to each SWCNT-containing pixel and intracellular concentrations were obtained in individual cell ROIs with the correlation
Solution-Based Raman Spectroscopy.
RAW 264.7 cells were seeded into tissue culture treated 96-well plates (Fisher Scientific) at a final concentration of 50 000 cells/well and allowed to culture overnight in an incubator. The media was removed from each well, replaced with 200 μL of 1 mg/L each (GT)n-SWCNT diluted in media, and incubated for 30 min (pulsed) to allow for internalization into the cells. After this pulse, the SWCNT-containing media was collected, the cells were rinsed three times with sterile PBS (Gibco) and 200 μL of fresh media was replenished. Twenty-four hours later, the supernatant was again collected from the cells. All supernatant was placed into new 96-well plates and Raman spectra were obtained using a WiTec Alpha300 R confocal Raman microscope (WiTec, Germany) equipped with a Zeiss Epiplan-Neofluar 10×/0.25 objective, a 785 nm laser source set to 35 mW sample power, and collected with a CCD detector through a 600 lines/mm grating. A calibration curve was obtained by recording spectra of known SWCNT concentrations serially diluted in a single pixel volume with identical acquisition settings. A global background subtraction and cosmic-ray removal was performed using Witec Control 5.0 software on all acquired confocal Raman data and G-band maximum intensities were extracted and correlated with known concentrations to produce a linear curve fit using OriginPro 2018 analysis software. The intensity of the G-band was extracted from each spectrum and related to G-band linear fit equations to determined average supernatant SWCNT concentrations.
Pharmacological Inhibition of Exocytosis Pathways.
RAW 264.7 cells were cultured and dosed with (GT)6-or (GT)30-SWCNTs following the same procedure previously described, however 2 h after SWCNT removal cells were spiked with either 200 nM Bafilomycin A1,58 2 μM Nocodazole,59 500 nM Brefeldin A,61 50 μM Exo1,60 or an equal volume of media. At 24 h after the initial SWCNT dose (22 h post inhibitor treatment), cells were fixed using 4% paraformaldehyde in PBS for 10 min, rinsed three times with PBS, and covered with PBS to retain an aqueous environment during imaging. Large cellular regions were scanned in 10 μm intervals using a WiTec Alpha300 R confocal Raman microscope (WiTec, Germany) equipped with a Zeiss Epiplan-Neofluar 10×/0.25 objective, a 785 nm laser source set to 35 mW sample power, and collected with a CCD detector through a 600 lines/mm grating with a 0.5s integration time. A calibration curve was obtained by recording spectra of known SWCNT concentrations serially diluted in a single pixel volume with identical acquisition settings. A global background subtraction and cosmic-ray removal was performed using Witec Control 5.0 software on all acquired confocal Raman data and G-band maximum intensities were extracted and correlated with known concentrations to produce a linear curve fit using OriginPro 2018 analysis software. The cellular SWCNT concentration data were produced by relating the G-band linear equations to each SWCNT-containing pixel and average cellular SWCNT concentrations were extracted from each area scan using a custom Matlab script.
Statistical Analysis.
All statistical measures for hypothesis testing were carried out using two-sample two-tailed unequal variance t-tests in Microsoft Office Excel 2016. All curve fitting and related statistics were performed in OriginPro 2018.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation CAREER Award #1844536, the RI-INBRE Early Career Development Award Grant P20GM103430 from the National Institute of General Medical Sciences of the National Institutes of Health, the Rhode Island Foundation-Medical Research Fund, and the URI College of Engineering. The confocal Raman data was acquired at the RI Consortium for Nanoscience and Nanotechnology, a URI College of Engineering core facility partially funded by the National Science Foundation EPSCoR, Cooperative Agreement #OIA-1655221. We acknowledge P. V. Jena for the Matlab analysis codes used in Figure 2.
Footnotes
ASSOCIATED CONTENT
Supporting Information
TThe Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.9b02267.
Table of DNA-SWCNT physical properties, solution-based optical characterization, time-dependent intracellular fluorescence, fluorescence spectroscopy of DNA-SWCNTs, confocal Raman concentration-intensity calibration, table of peak emission energy shifts (PDF)
NIR fluorescence movie (AVI)
NIR fluorescence movie (AVI)
The authors declare no competing financial interest.
REFERENCES
- (1).Ebbesen T; Lezec H; Hiura H; Bennett J; Ghaemi H; Thio T. Electrical conductivity of individual carbon nanotubes. Nature 1996, 382 (6586), 54. [Google Scholar]
- (2).Dresselhaus G; Riichiro S. Physical properties of carbon nanotubes; World Scientific, 1998. [Google Scholar]
- (3).Lefebvre J; Homma Y; Finnie P. Bright band gap photoluminescence from unprocessed single-walled carbon nanotubes. Phys. Rev. Lett 2003, 90 (21), 217401. [DOI] [PubMed] [Google Scholar]
- (4).Bachilo SM; Strano MS; Kittrell C; Hauge RH; Smalley RE; Weisman RB Structure-assigned optical spectra of single-walled carbon nanotubes. Science 2002, 298 (5602), 2361–2366. [DOI] [PubMed] [Google Scholar]
- (5).Moore VC; Strano MS; Haroz EH; Hauge RH; Smalley RE; Schmidt J; Talmon Y. Individually suspended single-walled carbon nanotubes in various surfactants. Nano Lett. 2003, 3 (10), 1379–1382. [Google Scholar]
- (6).Strano MS; Moore VC; Miller MK; Allen MJ; Haroz EH; Kittrell C; Hauge RH; Smalley R. The role of surfactant adsorption during ultrasonication in the dispersion of single-walled carbon nanotubes. J. Nanosci. Nanotechnol 2003, 3 (1–2), 81–86. [DOI] [PubMed] [Google Scholar]
- (7).Hashida Y; Tanaka H; Zhou S; Kawakami S; Yamashita F; Murakami T; Umeyama T; Imahori H; Hashida M. Photothermal ablation of tumor cells using a single-walled carbon nanotube–peptide composite. J. Controlled Release 2014, 173, 59–66. [DOI] [PubMed] [Google Scholar]
- (8).Heller DA; Pratt GW; Zhang J; Nair N; Hansborough AJ; Boghossian AA; Reuel NF; Barone PW; Strano MS Peptide secondary structure modulates single-walled carbon nanotube fluorescence as a chaperone sensor for nitroaromatics. Proc. Natl. Acad. Sci. U. S. A 2011, 108 (21), 8544–8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Antonucci A; Kupis-Rozmysłowicz J; Boghossian AA Noncovalent protein and peptide functionalization of single-walled carbon nanotubes for biodelivery and optical sensing applications. ACS Appl. Mater. Interfaces 2017, 9 (13), 11321–11331. [DOI] [PubMed] [Google Scholar]
- (10).Zheng M; Jagota A; Semke ED; Diner BA; McLean RS; Lustig SR; Richardson RE; Tassi NG DNA-assisted dispersion and separation of carbon nanotubes. Nat. Mater 2003, 2 (5), 338. [DOI] [PubMed] [Google Scholar]
- (11).Hong G; Diao S; Antaris AL; Dai H. Carbon nanomaterials for biological imaging and nanomedicinal therapy. Chem. Rev 2015, 115 (19), 10816–10906. [DOI] [PubMed] [Google Scholar]
- (12).Krus S; Hilmer AJ.; Zhang J; Reuel NF; Mu B; Strano MS. Carbon nanotubes as optical biomedical sensors. Adv. Drug Delivery Rev 2013, 65 (15), 1933–1950. [DOI] [PubMed] [Google Scholar]
- (13).O’connell MJ; Bachilo SM; Huffman CB; Moore VC; Strano MS; Haroz EH; Rialon KL; Boul PJ; Noon WH; Kittrell C. Band gap fluorescence from individual single-walled carbon nanotubes. Science 2002, 297 (5581), 593–596. [DOI] [PubMed] [Google Scholar]
- (14).Jena PV; Galassi TV; Roxbury D; Heller DA Progress toward Applications of Carbon Nanotube Photoluminescence. ECS J. Solid State Sci. Technol 2017, 6 (6), M3075–M3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Roxbury D; Jena PV; Williams RM; Enyedi B; Niethammer P; Marcet S; Verhaegen M; Blais-Ouellette S; Heller DA Hyperspectral microscopy of near-infrared fluorescence enables 17-chirality carbon nanotube imaging. Sci. Rep 2015, 5, 14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Choi JH; Strano MS Solvatochromism in single-walled carbon nanotubes. Appl. Phys. Lett 2007, 90 (22), 223114. [Google Scholar]
- (17).Gao Z; Varela JA; Groc L; Lounis B; Cognet L. Toward the suppression of cellular toxicity from single-walled carbon nanotubes. Biomater. Sci 2016, 4 (2), 230–244. [DOI] [PubMed] [Google Scholar]
- (18).Zheng M. Sorting carbon nanotubes. Topics in Current Chemistry 2017, 375 (1), 13. [DOI] [PubMed] [Google Scholar]
- (19).Ao G; Streit JK; Fagan JA; Zheng M. Differentiating leftand right-handed carbon nanotubes by DNA. J. Am. Chem. Soc 2016, 138 (51), 16677–16685. [DOI] [PubMed] [Google Scholar]
- (20).Salem DP; Landry MP; Bisker G; Ahn J; Kruss S; Strano MS Chirality dependent corona phase molecular recognition of DNA-wrapped carbon nanotubes. Carbon 2016, 97, 147–153. [Google Scholar]
- (21).Harvey JD; Jena PV; Baker HA; Zerze GH; Williams RM; Galassi TV; Roxbury D; Mittal J; Heller DA A carbon nanotube reporter of microRNA hybridization events in vivo. Nature biomedical engineering 2017, 1 (4), 0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Jena PV; Roxbury D; Galassi TV; Akkari L; Horoszko CP; Iaea DB; Budhathoki-Uprety J; Pipalia N; Haka AS; Harvey JD; et al. A carbon nanotube optical reporter maps endolysosomal lipid flux. ACS Nano 2017, 11 (11), 10689–10703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Galassi TV; Jena PV; Shah J; Ao G; Molitor E; Bram Y; Frankel A; Park J; Jessurun J; Ory DS; et al. An optical nanoreporter of endolysosomal lipid accumulation reveals enduring effects of diet on hepatic macrophages in vivo. Sci. Transl. Med 2018, 10 (461), No. eaar2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Siu KS; Chen D; Zheng X; Zhang X; Johnston N; Liu Y; Yuan K; Koropatnick J; Gillies ER; Min W-P Non-covalently functionalized single-walled carbon nanotube for topical siRNA delivery into melanoma. Biomaterials 2014, 35 (10), 3435–3442. [DOI] [PubMed] [Google Scholar]
- (25).Bartholomeusz G; Cherukuri P; Kingston J; Cognet L; Lemos R; Leeuw TK; Gumbiner-Russo L; Weisman RB; Powis G. In vivo therapeutic silencing of hypoxia-inducible factor 1 alpha (HIF-1α) using single-walled carbon nanotubes noncovalently coated with siRNA. Nano Res. 2009, 2 (4), 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zhang H; Demirer GS; Zhang H; Ye T; Goh NS; Aditham AJ; Cunningham FJ; Fan C; Landry MP DNA nanostructures coordinate gene silencing in mature plants. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Demirer GS; Zhang H; Matos JL; Goh NS; Cunningham FJ; Sung Y; Chang R; Aditham AJ; Chio L; Cho M-J High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol 2019, 14, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Roxbury D; Mittal J; Jagota A. Molecular-basis of single-walled carbon nanotube recognition by single-stranded DNA. Nano Lett. 2012, 12 (3), 1464–1469. [DOI] [PubMed] [Google Scholar]
- (29).Roxbury D; Jagota A; Mittal J. Sequence-specific selfstitching motif of short single-stranded DNA on a single-walled carbon nanotube. J. Am. Chem. Soc 2011, 133 (34), 13545–13550. [DOI] [PubMed] [Google Scholar]
- (30).Yang Y; Zheng M; Jagota A. Learning to predict single-wall carbon nanotube-recognition DNA sequences. npj Computational Materials 2019, 5 (1), 3. [Google Scholar]
- (31).Safaee MM; Gravely M; Rocchio C; Simmeth M; Roxbury D. DNA Sequence Mediates Apparent Length Distribution in Single-Walled Carbon Nanotubes. ACS Appl. Mater. Interfaces 2019, 11 (2), 2225–2233. [DOI] [PubMed] [Google Scholar]
- (32).Yang Y; Shankar A; Aryaksama T; Zheng M; Jagota A. Quantification of DNA/SWCNT solvation differences by aqueous two-phase separation. Langmuir 2018, 34 (5), 1834–1843. [DOI] [PubMed] [Google Scholar]
- (33).Jena PV; Safaee MM; Heller DA; Roxbury D. DNA–Carbon Nanotube Complexation Affinity and Photoluminescence Modulation Are Independent. ACS Appl. Mater. Interfaces 2017, 9 (25), 21397–21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Gillen AJ; Kupis-Rozmysłowicz J; Gigli C; Schuergers N; Boghossian AA Xeno Nucleic Acid Nanosensors for Enhanced Stability Against Ion-Induced Perturbations. J. Phys. Chem. Lett 2018, 9 (15), 4336–4343. [DOI] [PubMed] [Google Scholar]
- (35).Salem DP; Gong X; Liu AT; Koman VB; Dong J; Strano MS Ionic strength-mediated phase transitions of surface-adsorbed DNA on single-walled carbon nanotubes. J. Am. Chem. Soc 2017, 139 (46), 16791–16802. [DOI] [PubMed] [Google Scholar]
- (36).Roxbury D; Tu X; Zheng M; Jagota A. Recognition ability of DNA for carbon nanotubes correlates with their binding affinity. Langmuir 2011, 27 (13), 8282–8293. [DOI] [PubMed] [Google Scholar]
- (37).Gustafson HH; Holt-Casper D; Grainger DW; Ghandehari H. Nanoparticle uptake: the phagocyte problem. Nano Today 2015, 10 (4), 487–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Smith BR; Ghosn EEB; Rallapalli H; Prescher JA; Larson T; Herzenberg LA; Gambhir SS Selective uptake of single-walled carbon nanotubes by circulating monocytes for enhanced tumour delivery. Nat. Nanotechnol 2014, 9 (6), 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Porter AE; Gass M; Muller K; Skepper JN; Midgley PA; Welland M. Direct imaging of single-walled carbon nanotubes in cells. Nat. Nanotechnol 2007, 2 (11), 713. [DOI] [PubMed] [Google Scholar]
- (40).Zhou F; Xing D; Wu B; Wu S; Ou Z; Chen WR New insights of transmembranal mechanism and subcellular localization of noncovalently modified single-walled carbon nanotubes. Nano Lett. 2010, 10 (5), 1677–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Iverson NM; Barone PW; Shandell M; Trudel LJ; Sen S; Sen F; Ivanov V; Atolia E; Farias E; McNicholas TP; et al. In vivo biosensing via tissue-localizable near-infrared-fluorescent single-walled carbon nanotubes. Nat. Nanotechnol 2013, 8 (11), 873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Xu H; Ren D. Lysosomal physiology. Annu. Rev. Physiol 2015, 77, 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Roxbury D; Jena PV; Shamay Y; Horoszko CP; Heller DA Cell membrane proteins modulate the carbon nanotube optical bandgap via surface charge accumulation. ACS Nano 2016, 10 (1), 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Cambré S; Santos SM; Wenseleers W; Nugraha AR; Saito R; Cognet L; Lounis B. Luminescence properties of individual empty and water-filled single-walled carbon nanotubes. ACS Nano 2012, 6 (3), 2649–2655. [DOI] [PubMed] [Google Scholar]
- (45).Heller DA; Jeng ES; Yeung T-K; Martinez BM; Moll AE; Gastala JB; Strano MS Optical detection of DNA conformational polymorphism on single-walled carbon nanotubes. Science 2006, 311 (5760), 508–511. [DOI] [PubMed] [Google Scholar]
- (46).Gong X; Sharma AK; Strano MS; Mukhopadhyay D. Selective assembly of DNA-conjugated single-walled carbon nanotubes from the vascular secretome. ACS Nano 2014, 8 (9), 9126–9136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Budhathoki-Uprety J; Jena PV; Roxbury D; Heller DA Helical polycarbodiimide cloaking of carbon nanotubes enables internanotube exciton energy transfer modulation. J. Am. Chem. Soc 2014, 136 (44), 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Harvey.; Zerze G.l. H.; Tully KM; Mittal J; Heller DA. Electrostatic screening modulates analyte binding and emission of carbon nanotubes. J. Phys. Chem. C 2018, 122 (19), 10592–10599. [Google Scholar]
- (49).Yang R; Jin J; Chen Y; Shao N; Kang H; Xiao Z; Tang Z; Wu Y; Zhu Z; Tan W. Carbon nanotube-quenched fluorescent oligonucleotides: probes that fluoresce upon hybridization. J. Am. Chem. Soc 2008, 130 (26), 8351–8358. [DOI] [PubMed] [Google Scholar]
- (50).Larsen BA; Deria P; Holt JM; Stanton IN; Heben MJ; Therien MJ; Blackburn JL Effect of solvent polarity and electrophilicity on quantum yields and solvatochromic shifts of single-walled carbon nanotube photoluminescence. J. Am. Chem. Soc 2012, 134 (30), 12485–12491. [DOI] [PubMed] [Google Scholar]
- (51).Jorio A; Pimenta M; Souza Filho A; Saito R; Dresselhaus G; Dresselhaus M. Characterizing carbon nanotube samples with resonance Raman scattering. New J. Phys 2003, 5 (1), 139. [Google Scholar]
- (52).Dresselhaus MS; Jorio A; Hofmann M; Dresselhaus G; Saito R. Perspectives on carbon nanotubes and graphene Raman spectroscopy. Nano Lett. 2010, 10 (3), 751–758. [DOI] [PubMed] [Google Scholar]
- (53).Dresselhaus MS; Dresselhaus G; Saito R; Jorio A. Raman spectroscopy of carbon nanotubes. Phys. Rep 2005, 409 (2), 47–99. [Google Scholar]
- (54).Jin S; Wijesekara P; Boyer PD; Dahl KN; Islam MF Length-dependent intracellular bundling of single-walled carbon nanotubes influences retention. J. Mater. Chem. B 2017, 5 (32), 6657–6665. [DOI] [PubMed] [Google Scholar]
- (55).Fröhlich E. Cellular elimination of nanoparticles. Environ. Toxicol. Pharmacol 2016, 46, 90–94. [DOI] [PubMed] [Google Scholar]
- (56).Sakhtianchi R; Minchin RF; Lee K-B; Alkilany AM; Serpooshan V; Mahmoudi M. Exocytosis of nanoparticles from cells: role in cellular retention and toxicity. Adv. Colloid Interface Sci 2013, 201, 18–29. [DOI] [PubMed] [Google Scholar]
- (57).Park JH; Oh N. Endocytosis and exocytosis of nanoparticles in mammalian cells. Int. J. Nanomed 2014, 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Yanes RE; Tarn D; Hwang AA; Ferris DP; Sherman SP; Thomas CR; Lu J; Pyle AD; Zink JI; Tamanoi F. Involvement of lysosomal exocytosis in the excretion of mesoporous silica nanoparticles and enhancement of the drug delivery effect by exocytosis inhibition. Small 2013, 9 (5), 697–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Strobel C; Oehring H; Herrmann R; Förster M; Reller A; Hilger I Fate of cerium dioxide nanoparticles in endothelial cells: exocytosis. J. Nanopart. Res 2015, 17 (5), 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Feng Y; Yu S; Lasell TK; Jadhav AP; Macia E; Chardin P; Melancon P; Roth M; Mitchison T; Kirchhausen T. Exo1: a new chemical inhibitor of the exocytic pathway. Proc. Natl. Acad. Sci. U. S. A 2003, 100 (11), 6469–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Jackson CL Brefeldin A revealing the fundamental principles governing membrane dynamics and protein transport In Fusion of biological membranes and related problems; Springer, 2002; pp 233–272. [DOI] [PubMed] [Google Scholar]
- (62).Medina DL; Fraldi A; Bouche V; Annunziata F; Mansueto G; Spampanato C; Puri C; Pignata A; Martina JA; Sardiello M; et al. Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell 2011, 21 (3), 421–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Dumortier H; Lacotte S; Pastorin G; Marega R; Wu W; Bonifazi D; Briand J-P; Prato M; Muller S; Bianco A. Functionalized carbon nanotubes are non-cytotoxic and preserve the functionality of primary immune cells. Nano Lett. 2006, 6 (7), 1522–1528. [DOI] [PubMed] [Google Scholar]
- (64).Mukherjee SP; Bondarenko O; Kohonen P; Andón FT; Brzicová T; Gessner I; Mathur S; Bottini M; Calligari P; Stella L; et al. Macrophage sensing of single-walled carbon nanotubes via Toll-like receptors. Sci. Rep 2018, 8 (1), 1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Shvedova AA; Kisin ER; Mercer R; Murray AR; Johnson VJ; Potapovich AI; Tyurina YY; Gorelik O; Arepalli S; Schwegler-Berry D; et al. Unusual inflammatory and fibrogenic pulmonary responses to single-walled carbon nanotubes in mice. American Journal of Physiology-Lung Cellular and Molecular Physiology 2005, 289 (5), L698–L708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






