Abstract
Understanding information processing in the brain requires us to monitor neural activity at high spatiotemporal resolution. Using an ultrafast two-photon fluorescence microscope (2PFM) empowered by all-optical laser scanning, we imaged neural activity in vivo at up to 3,000 frames per second and submicron spatial resolution. This ultrafast imaging method enabled monitoring of both supra- and sub-threshold electrical activity down to 345 μm below the brain surface in head-fixed awake mice.
Monitoring neural signaling with genetically encoded fluorescence reporters [1] at synaptic and cellular resolution in vivo holds the key to dissecting the complex mechanisms of neural activity in behaving animals. Whereas imaging calcium and glutamate sensors with 2PFM enables activity measurements deep in opaque brains with subcellular resolution [2–6], imaging membrane voltage, the most direct measure of neural activity, is more challenging due to its millisecond dynamics. Too fast for conventional 2PFM, voltage imaging was mostly achieved by widefield microscopy with poor spatial resolution and limited to superficial depths [7–9]. To image membrane voltage at high spatial resolution in opaque brains, we need to increase 2PFM frame rate to kHz.
The rate of conventional raster-scanning 2PFM is limited by galvanometric mirrors to tens of frames per second (fps) [5–6]. Random-access 2PFM using acoustic-optical deflectors allows kHz frame rates and was used for voltage imaging [10–11], but can only track pre-selected locations (currently up to ~20). A line-projection 2PFM system detected glutamate release in vivo at kHz [12], but is limited to sparsely labeled samples. Notably, in these implementations, the pixel dwell times (0.1–40 microseconds) are much longer than the nanosecond-scale fluorophore excited-state lifetime, which is the minimum dwell time required for assigning signals to scanned locations unambiguously [13]. Here, by leveraging an all-optical passive laser scanner based on free-space angular-chirp-enhanced delay (FACED) [14], we demonstrated raster-scanning 2PFM at 1,000 fps and 3,000 fps, with the pixel dwell time flexibly configured to reach the fluorescence lifetime limit. We applied it to ultrafast monitoring of calcium activity, glutamate release, and membrane potential, and demonstrated 2PFM imaging of spontaneous and sensory-evoked supra- and sub-threshold electrical activity in awake mouse brains in vivo.
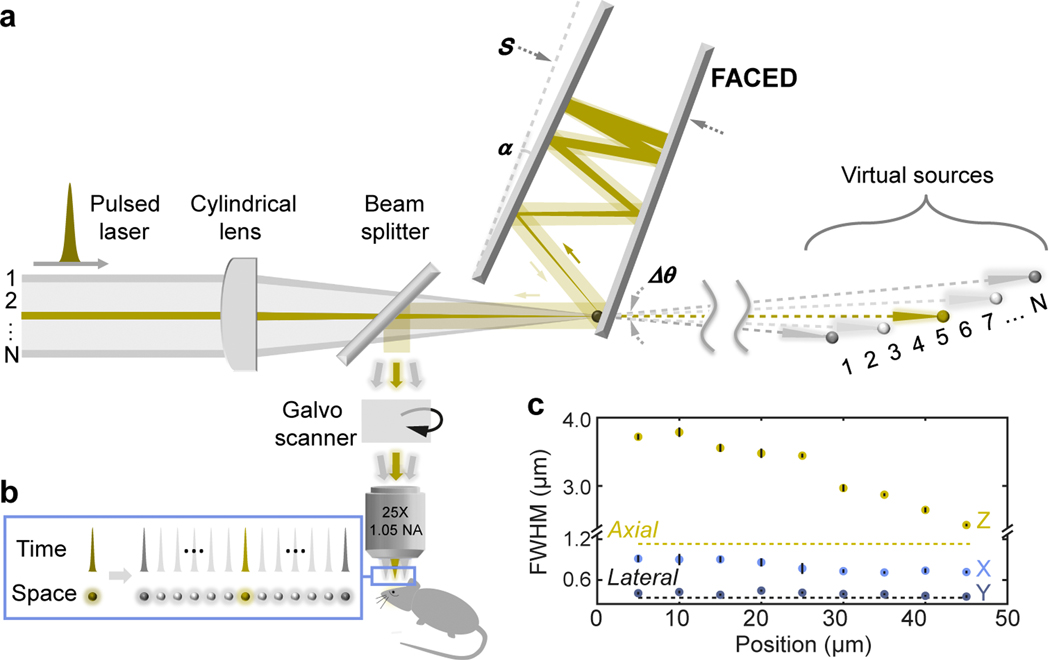
The principle of FACED was detailed previously (Fig. 1a) [14]. Briefly, a pulsed laser beam was focused in 1D by a cylindrical lens and obtained a converging angle Δθ. It was then launched into a pair of almost parallel high-reflectivity mirrors with separation S and misalignment angle α. After multiple reflections by the mirrors, the laser beam/pulse was split into multiple beamlets/subpulses (N=Δθ/α) of distinct propagation directions and retro-reflected with an inter-pulse temporal delay of 2S/c (c: the speed of light). After being relayed to a microscope objective, they formed an array of spatially separated and temporally delayed foci (Fig. 1b). Effectively, this passive FACED module allows line scanning at the laser repetition rate, typically MHz.
Figure 1: Principles and resolution of a 2PFM with a FACED module.
(a) Schematic of FACED 2PFM. A 1-MHz collimated femtosecond laser was focused into a nearly parallel mirror pair with a converging angle Δθ by a cylindrical lens. After multiple reflections, the misalignment angle α caused the beamlets to retroreflect (e.g., the yellow ray). N = Δθ/α beamlets at different incidence angles (e.g., yellow versus gray rays) emerged with distinct propagation directions and temporal delays. Equivalently, the sequence of beamlets at the output of the FACED module can be treated as light emanating from an array of virtual sources. These beamlets were then coupled into a 2PFM and formed (b) an array of spatially separated and temporally delayed foci at the focal plane of a microscope objective. (c) The focal spot sizes along the X/FACED, Y, and Z axes, measured from 200-nm-diameter fluorescent beads. Error bars show s.d. from 10 beads; dashed lines indicate the expected axial and lateral resolutions at 1.05 NA.
We designed a FACED module and incorporated it into a standard 2PFM with a high-speed data acquisition system (625 MS/s) (Methods, Supplementary Fig. 1). Using a 920-nm laser with 1-MHz repetition rate, our FACED module produced 80 pulsed foci with 2-ns inter-pulse interval spanning 50 μm every microsecond. These foci were images of virtual sources formed after distinct numbers of mirror reflections and located at increasing distances (Fig. 1a), with the more distant virtual sources leading to larger beam sizes at the objective thus smaller foci (Fig. 1c). The beams filled the objective more along the Y than X/FACED axis, resulting in ~0.82-μm (X) and ~0.35-μm (Y) subcellular lateral resolution. By scanning the Y galvanometer at 500 Hz and collecting data bi-directionally, we acquired 80 × 900 pixels per frame at 1,000 Hz.
We first used FACED-2PFM to image calcium dynamics using genetically encoded calcium indicator GCaMP6 [2]. At 1 kHz, neurites were clearly resolved in raw images (Supplementary Fig. 2). We reliably detected calcium transients evoked by extracellular electric stimulation in GCaMP6-expressing cultured neurons and acute mouse brain slices (Supplementary Figs. 3,4, Videos 1,2). We also recorded spontaneous calcium releases in neurites that propagated at 25 μm/s (Supplementary Fig. 5 and Video 3).
Next, we imaged neurons labeled with A184V (medium-affinity) and S72A (low-affinity) variants of the genetically encoded glutamate sensor SF-iGluSnFR [15]. FACED-2PFM reliably reported glutamate release evoked by field stimulation in cultured neurons and spontaneous glutamate release in L2/3 neurons in the primary visual cortex (V1) of head-fixed awake mice (Supplementary Fig. 6, Videos 4-9). We observed faster dynamics from S72A than A184V, consistent with previous characterizations [15]. We also imaged fluorescent particles, likely macrophages in vasculature, moving rapidly at ~1 mm/s (Supplementary Fig. 7, Video 10).
Finally, we imaged neurons expressing the genetically encoded voltage indicator ASAP3 in V1 of head-fixed awake mice with FACED-2PFM. Among GEVIs, ASAP-family indicators are currently the only ones to have reported single spikes in vivo with 2PFM, albeit in a limited number of neurons with random-access scanning [10,11,16]. ASAP3 reports membrane depolarizations and action potentials as downward deflections in fluorescence [10]. In both sparsely (Fig. 2a) and densely labeled preparations (Fig. 2b), we observed fluorescence concentrated to somata of neurons expressing soma-targeted ASAP3-Kv. Whereas imaging at 1,000 fps continuously for six seconds led to substantial photobleaching (Supplementary Fig. 8), 1-kHz imaging carried out in 1-s bouts interleaved with 2.5-s dark periods enabled fluorescence to recover almost completely between bouts (Supplementary Fig. 9), allowing voltage imaging of the same neurons repeatedly.
Figure 2: 1 kHz imaging of supra- and sub-threshold voltage responses with genetically encoded voltage sensor ASAP3 in V1 of awake mice.
(a,b) Representative images of neurons in V1 (a) sparsely or (b) densely labeled with soma-targeted ASAP3-Kv. (c) Spontaneous voltage traces (SNR, ΔF/√F) from neurons 1 and 2 in (a) and (b), respectively; Green ticks: spikes. (d) Neurons at different depths of a cortical column and their spontaneous voltage traces. (e) Average of 101 spikes from neuron 2 (orange trace) in (d); (f) Rise time, fall time, and ΔF/F of spikes (box-and-whisker plots, n = 45 cells from 3 mice). (g) 1kHz imaging over a 50 × 250 μm2 field of view with four neurons exhibiting distinct spontaneous activity patterns. 7 experiments were repeated independently with similar results. (h) Voltage traces from a V1 neurons showing orientation selectivity, with more sub- and supra-threshold activity for preferred orientations (black traces) than for non-preferred orientations (gray traces). (i) Raster plot of spikes for all trials (10 trials for each of 8 grating stimuli) and (j) polar plot showing the orientation tuning of mean subthreshold ΔF/F response (red) and spiking rate (black) of the neuron in (h). Representative results from 8 neurons. (k) Two neurons at 345 μm depth and (l) their subthreshold ΔF/F (upper traces, average of 52 trials) and spiking (lower histograms, 50-ms bins) responses relative to the onset of grating stimuli (red dashed line). Representative results from 20 neurons. (m) Histogram of the time to reach peak subthreshold voltage responses post stimulus onset from 20 cells in 3 mice. (n) Spiking response relative to stimulus onset. From top to bottom: raster plot, histogram (50-ms bins), and averaged firing rate (50-ms sliding rectangular windows) of 2747 detected spikes from 617 trials of 20 neurons in 3 mice. Shaded areas: s.d.; Scale bars: (a,b) 50 μm; (d,g) 10 μm; (h, k) 20 μm.
We extracted fluorescence from membrane pixels to obtain time-dependent traces for each neuron. Downward signals corresponding to putative individual action potentials (“optical spikes”) were easily detected from single trials. After correcting for photobleaching, we calculated the relative fluorescence change in terms of signal-to-noise ratio (SNR, ΔF/√F) and ΔF/F (Methods, Supplementary Fig. 10, Fig. 2). Detecting both suprathreshold and subthreshold membrane potential variations (Fig. 2c), FACED-2PFM provided high-resolution voltage imaging of neurons located at four depths in the same brain (Fig. 2d). In one neuron (orange trace, ROI2, Fig. 2d), we detected 101 isolated spikes in 48 s of recording and obtained an averaged spiking profile (Fig. 2e). From this and other neurons, we determined the rise time, decay time, and ΔF/F for single action potentials to be 1.93 ± 0.51 ms, 3.73 ± 1.88 ms, and 0.10 ± 0.02 (mean ± s.d., n = 45 cells from three mice), respectively (Fig. 2f), consistent with previous characterizations by random-access 2PFM [10].
We measured both spontaneous spiking and subthreshold activity from four neurons within a 50 × 250 μm2 FOV, and observed burst firing in one neuron (purple trace, ROI1 of Fig. 2g). Because two-photon excitation is restricted to a femtoliter focal volume and fluorescence photons generated at different locations are detected at different times, there was no cross-pixel contamination even in densely labeled brains (Supplementary Fig. 11). Frequently, we observed rhythmic subthreshold oscillations at 4–12 Hz (Supplementary Fig. 12), likely originating from cortical theta oscillations [17,18].
FACED-2PFM also detected sensory-evoked spiking and subthreshold responses of V1 neurons in awake mice presented with drifting grating stimuli. For an orientation-selective neuron preferring gratings along specific orientations, we observed minimal voltage response to non-preferred orientations but strong subthreshold and spiking response to preferred orientations (Fig. 2h). From ten 1-s trials for each of the eight gratings (Fig. 2i), we calculated its tuning curves using its spiking (black curve, Fig. 2j) and subthreshold (red curve, Fig. 2j) activity, and found sharper tuning for spiking activity, a consistent trend observed in other neurons (Supplementary Fig. 13) and previous whole-cell electrophysiological recordings [19,20].
We investigated the latency for visual information to reach V1. For two neurons at 345 μm below dura (Fig. 2k), their subthreshold responses reached their peaks 60 ms and 57 ms after stimulus onset (Fig. 2l). Populationally, the peak subthreshold response had a latency of 57.3 ± 2.3 ms (mean ± s.d., n = 20 cells from 3 mice, Fig. 2m), whereas the spike rate reached its peak 60 ms after stimulus onset (Fig. 2n), consistent with electrophysiological results [21,22].
The frame rate of FACED-2PFM can be further increased by faster scanning of the Y galvanometer. Using the 4-MHz 1,035-nm output of our laser and scanning the Y galvanometer at 1,500 Hz, we imaged calcium activity in vivo at 3,000 fps with 80 × 1200 pixels per frame (Supplementary Fig. 14, Video 11). New developments in laser systems (e.g., 4-MHz output at 920 nm, now commercially available) or voltage sensors (e.g., yellow or red variants that can be excited at 1,035 nm) should allow us to image voltage at 3,000 fps as well. With 2-ns inter-pulse interval, FACED-2PFM reaches the limit on pixel rate imposed by the excited-state lifetime of high-quantum-yield fluorophores. Further reduction of dwell time would cause substantial cross-talk between neighboring pixels and decrease spatial resolution along the FACED axis. However, for activity imaging of spatially extended ROIs where signals from multiple pixels are averaged, minor crosstalk can be tolerated.
In summary, using an all-optical passive laser scanner based on FACED, we achieved kilohertz-rate full-frame 2PFM imaging of neural activity with subcellular resolution in the mouse brain in vivo. We measured both spontaneous and sensory-evoked supra- and sub-threshold electrical activity of V1 neurons in awake mice, using post-objective average laser power of 10–85 mW (Supplementary Table 1), a value similar to those employed by conventional 2PFM. We did not observe signs of photodamages (e.g., blabbing of dendrites), suggesting higher-order nonlinear photodamage processes to be minimal. Furthermore, subsequent excitation pulses arrived at the same sample positions after 1-ms delays, providing ample time for the fluorophores to return from their damage-prone dark states back to their ground state, a process that was previously shown to reduce photobleaching substantially [23].
Transforming a standard galvanometer-based 2PFM into a kHz imaging system by adding a FACED module, our approach is readily compatible with existing systems. Following raster-scanning strategy, FACED-2PFM requires minimal computational processing, works in both sparsely and densely labeled samples, and is immune to crosstalk effects or source localization ambiguities observed in computation-based approaches [12,24]. Sampling an entire image plane allows post hoc motion correction, therefore making FACED-2PFM more resistant to sample motion than random-access 2PFMs. With existing sensors, FACED-2PFM offers sufficient speed and sensitivity to detect calcium and glutamate transients from neuronal processes, as well as both spiking and subthreshold voltage events from cell bodies. Future improvement in brightness and sensitivity of voltage indicators would enhance detection of voltage signals in subcellular compartments and allow FACED imaging to fulfill its full potential in interrogating electrical activity at synaptic resolution.
ONLINE METHODS
Animals
All animal experiments were conducted according to the National Institutes of Health guidelines for animal research. Procedures and protocols on mice were approved by the Institutional Animal Care and Use Committee at Janelia Research Campus, Howard Hughes Medical Institute. Additional information can be found in Life Sciences Reporting Summary.
FACED two-photon fluorescence microscope (2PFM)
The simplified schematic of FACED-2PFM is shown in Supplementary Fig. 1a. The two-photon excitation laser source at 920 nm (1 MHz repetition rate, 2 W maximal average output power, < 100 fs pulse width) was generated by an optical parametric amplifier (Opera-F, Coherent Inc.) that was pumped by a fiber laser at 1035 nm (Monaco 1035–40-40, 1 MHz or 4 MHz repetition rate, < 315 fs pulse width, 35 W, Coherent Inc.). After dispersion compensation [25], the laser beam was expanded with a 2× beam expander (BE02M-B, Thorlabs) to 8 mm in diameter. The beam then passed through a 4 to 5-mm wide slit and was one-dimensionally focused (Δθ = 1°) by a cylindrical lens (LJ1267RM-B, Thorlabs, effective input NA 0.01) into a nearly parallel mirror pair (α = 0.0125°, reflectivity > 99.9% and GDD<10 fs2 per reflection at 920 nm, fused silica substrate, 250 mm long and 20 mm wide, Layertec GmbH) with a separation of 300 mm. Because of the misalignment angle α, all the light rays eventually reflected back, following a set of zig-zig paths determined by their angles of incidence. After the FACED module, the rays (e.g. red rays in Fig. 1a) subjected to the same number of reflections by the mirror pair formed a single beamlet, and can be considered as emanating from a virtual light source located far away from the mirror pair. In this work, the retroreflected light rays formed 80 beamlets, and had their propagation distances within the FACED module (or their distance from their respective virtual sources) monotonically increase from ~10 m to ~60 m. The power throughput of the FACED module was ~40%.
A polarization beam splitter (CCM1-PBS253, Thorlabs) in combination with a half-wave plate (AHWP05M-980, Thorlabs) and quarter-wave plate (AQWP05M-980, Thorlabs) were used to direct the spatially and temporally separated pulse trains into a 2PFM. A pair of achromatic doublets (AC508–500-B and AC508–250-B, Thorlabs) was used to conjugate the focal plan of the cylindrical lens and the midpoint of a pair of closely and orthogonally arranged X and Y galvo mirrors (6215H, Cambridge Technology).A scan lens / tube lens pair (SL50–2P2 and TTL200MP, Thorlabs) were used to conjugate the galvos to the back focal plane of a 25×/1.05 NA water-dipping objective lens (XLPLN25XWMP2, Olympus) that was mounted on a piezo stage (P-725K094, Physik Instrumente). The two-photon excited fluorescence signal was collected by the same microscope objective, reflected by a dichroic mirror (FF665-Di02–25×36, Semrock), focused by two lenses (AC508–080-A and LA1951-A Thorlabs), and after passing through an emission filter (FF01–680/SP, Semrock), detected by a photomultiplier tube without a current protection circuit (PMT, H7422P-40 MOD, Hamamatsu). The PMT signal was sampled at 625 MS/s with a high-speed digitizer (2G onboard memory, PXIe-5160, National Instruments) embedded in a chassis (PXIe-1071, National Instruments). From the chassis, the data was transferred to and saved by a desktop computer through a PCIe 16× interface.
By tuning the misalignment angle between the two mirrors (α = 0.0125°, see above), we generated a sequence of 80 focal spots extending 50 μm along the X axis. With the separation between the two mirrors set at 300 mm, the time delay between adjacent pulses was 2 ns. With 1 MHz repetition rate of the 920 nm output, the FACED module gave rise to a line scan rate of 1 MHz. Using the Y galvo to scan the foci along the direction orthogonal to the X/FACED axis at 500 Hz, and collecting the data bidirectionally, we achieved a frame rate of 1,000 fps with an effective image size of 80 × 900 pixels. 80 was given by the number of foci in the FACED axis and 900 was the product of the effective frame time (1 ms frame time minus a 100 μs dead time during mirror turns) and line scan rate. To increase the number of pixels in the FACED/X axis, X galvo was stepped to tile FACED images and increase the field of view. With 4 MHz 1035 nm excitation, we scanned the Y galvo at 1,500 Hz and achieved a frame rate of 3,000 Hz with an effective image size of 80 × 1200 pixels. For morphological imaging, we scanned the Y galvo at 50 Hz, resulting in a FACED imaging frame rate of 100 fps. (Supplementary Table 1 listed all the major imaging parameters used in this work).
The raw data from the digitizer was saved as 1D waveforms. The Gen-1 PCIe 16× slot on the data acquisition computer had a maximal streaming rate of 250 Mb/s, which caused the data to overflow the on-chip memory of the digitizer after 6 s of data acquisition and thus limited the data collection to up to 6-s bouts. Upgrading the computer with Gen-3 PCIe 16× interface should allow us to stream data continuously. At 1 kHz frame rate, each image frame had 625 × 900 sampling points, with 100 × 900 data points sampling actual fluorescence excitation by the FACED foci and used to reconstruct a single image. If desired, multi-line scans were averaged to generate a single X line in the final image: for a Y-axis range of 150 μm, 3 line scans were averaged to form a single row; for a Y-axis range of 50 μm, 9 line scans are averaged. The final images were motion-registered with an iterative cross-correlation-based registration algorithm [26]. For morphological imaging, each FACED image frame had 625 × 9000 sampling points (10× exposure time), and a 10× increase in line averaging to form the final image.
Analysis of activity data
We manually selected regions of interests (ROIs) from the averaged images of the registered image sequence. The mean fluorescent intensity within the ROIs was used to calculate the ΔF/F time traces, with F being the baseline fluorescence and ΔF being the fluorescence change due to neural activity. In the calcium and glutamate indicator datasets, to calculate the ΔF/F, we calculated the baseline fluorescence F by fitting the data points away from the transients (e.g. the first and last 1000 data points in Supplementary Fig. 6a) with a single exponential function.
In the voltage indicator datasets, the ROI was selected to cover the cell membrane. Adjoining membrane segments between neighboring neurons were excluded from the ROI. A rolling percentile filter (50%, 500 ms window) was applied to the mean-intensity trace of the ROI to get the fluorescence baseline F (Supplementary Fig. 10) and rare periods with uncorrectable motion artifacts were discarded. In addition to ΔF/F traces, we calculated SNR traces as the ratio between ΔF (functional change) and √F (Poisson noise) [27]. For spike detection, the SNR traces were further subjected to a 250-Hz 12th order low-pass Butterworth filter. The threshold for spikes was initially set at SNR = 7.5 and was manually adjusted if needed. Spikes were identified as local maxima that were at least 3 ms apart. To calculate the subthreshold activity, raw ΔF/F traces were low-pass filtered with a 50-Hz 12th order low-pass Butterworth filter and the mean ΔF/F value during the entire duration of sensory stimulation (1-s of drifting grating) was used as the strength of subthreshold activity. To quantify the temporal dynamics of the optical voltage response, we aligned the optical spikes from the same neuron at peak response, and measured the rise (10% to 90%), decay (90% to 10%) and FWHM time from their averaged traces.
Preparation and electric stimulation of primary neuronal culture
Primary neuronal cultures from neonatal rat pups were prepared as described previously [28]. AAV2/1.syn.GCaMP6s (1.8 × 1013 GC/ml), AAV.DJ.syn.iGluSnFR.A184V (3 × 1012 GC/ml), and AAV.DJ.syn.iGluSnFR.S72A (8.0 × 1012 GC/ml) were used to label cultured neurons by adding 1 μl of viral solution to each well in 24 well plates with 300 μl medium inside, respectively. After incubation overnight, 1 ml culture medium was added to each well. Neurons were imaged between 10 – 21 days post-transfection at room temperature in imaging buffer (145 mM NaCl, 2.5 mM KCl, 10 mM glucose, 10 mM HEPES, 2 mM CaCl2, 1 mM MgCl2, pH7.4).
For electric stimulation, cultured neurons in imaging buffer were positioned between two parallel electrodes separated at ~10 mm. A stimulus isolator (NPIISO-01D100, ALA Scientific Instruments Inc.) and functional generator (AFG1022, Tektronix Inc.) was used to generate the electric field. For each stimulation, a train of 10 pulses (pulse duration: 1 ms; period: 12 ms; voltage: 50 V) was used to drive the neurons.
Preparation and electric stimulation of acute brain slices
25-week-old male transgenic mice expressing GCaMP6f (scnn1a-TG3-cre x Ai93 x ACTB-tTA) [29, 30] were decapitated under deep isoflurane anesthesia, and the brain was transferred to an ice-cold dissection solution containing (in mM): 204.5 sucrose, 2.5 KCl, 1.25 NaH2PO4, 28 NaHCO3, 7 dextrose, 3 Na-pyruvate, 1 Na-ascorbate, 0.5 CaCl2, 7 MgCl2 (pH 7.4, oxygenated with 95% CO2 and 5% O2). 350-μm-thick coronal slices of the primary visual cortex (V1) were sectioned using a vibrating tissue slicer (Leica VT 1200S, Leica Microsystems, Wetzlar, Germany). The slices were then transferred to a suspended mesh within an incubation chamber filled with artificial cerebrospinal fluid (ACSF) containing (in mM): 125 NaCl, 2.5 KCl, 1.25 NaH2PO4, 25 NaHCO3, 25 dextrose, 1.3 CaCl2, 1 MgCl2 (pH 7.4, oxygenated with 95% CO2 and 5% O2). After 30 – 60 minutes of recovery at 35°C, the chamber was maintained at room temperature.
During imaging, slices were submerged in a recording chamber constantly perfused with oxygenated ACSF. A micropipette filled with ACSF were used for monopolar stimulation via a stimulus isolator (NPIISO-01D100, ALA Scientific Instruments Inc.) and a function generator (AFG1022, Tektronix Inc.). To provide extracellular stimulation, the stimulating electrode was placed in the proximity of the recorded cell, and a train of 10 pulses (pulse duration: 1 ms; period: 12 ms; current: 300 μA) was applied.
Mouse preparation for in vivo imaging
Mice (females or males, >2-months-old) were housed in cages (in groups of 1 – 5 before surgeries and in pairs or single housed after) under reverse light cycle. Wild-type (Jackson Laboratories, Black 6, stock #:000664) as well as Gad2-IRES-cre (Jackson Laboratories, Gad2tm2 (cre) Zjh/J, stock #: 010802) mice were used.
Virus injection and cranial window implantation procedures have been described previously [26]. Briefly, mice were anaesthetized with isoflurane (1 – 2% by volume in O2) and given the analgesic buprenorphine (SC, 0.3 mg per kg of body weight). Animals were head fixed in a stereotaxic apparatus (Model 1900, David Kopf Instruments). A 3.5-mm diameter craniotomy was made over the left V1 with dura left intact. A glass pipette (Drummond Scientific Company) beveled at 45° with a 15 – 20 μm opening was back-filled with mineral oil. A fitted plunger controlled by a hydraulic manipulator (Narishige, MO10) was inserted into the pipette and used to load and slowly inject 30 nl viral solution into the brain (~200 – 400 μm below pia). 3 – 6 injection sites were chosen in the left V1 with 0.3 to 0.5 mm space between injection sites. The following viral vectors were used to label neurons with different sensors. Labeling with calcium sensor: AAV2/1.syn.GCaMP6s (1.8 × 1013 GC/ml); Dense labeling with glutamate sensors: AAV.DJ.syn.iGluSnFR.A184V (3 × 1012 GC/ml); AAV.DJ.syn.iGluSnFR.S72A (8.0 × 1012 GC/ml); Sparse labeling with glutamate sensors: AAV.DJ.syn.FLEX.iGluSnFR.A184V (2.8 × 1013 GC/ml) 1:1 mixed with AAV2/1.syn.Cre (500 times diluted from 1.5 × 1013 GC/ml); AAV.DJ.syn.FLEX.iGluSnFR.S72A (5.2 × 1012 GC/ml) 1:1 mixed with AAV2/1.syn.Cre (500 times diluted from 1.5 × 1013 GC/ml); labeling with the voltage sensor: AAV2/9.syn.ASAP3-Kv (1.55 × 1012 GC/ml). At the completion of viral injections, a glass window made of a single coverslip (Fisher Scientific No. 1.5) was embedded in the craniotomy and sealed in place with dental acrylic. A titanium head-post was then attached to the skull with cyanoacrylate glue and dental acrylic. In vivo imaging was carried out after at least two weeks of recovery with single or paired housing and habituation for head fixation. All imaging experiments were carried out on head-fixed awake mice.
Visual stimulation in head-fixed awake mice
Visual stimuli were presented by a liquid crystal display (22-inch diagonal and 1920 × 1080 pixels). The screen was positioned at 15 cm from the eye of the mice and orientated at ∼40° to the long axis of the mice. Drifting sinusoidal gratings were presented for 1.5 s at 8 orientations (0° to 315° at 45° steps) in pseudorandom sequences. Between the grating stimulus, 6s dark screen were presented. Gratings had 100% contrast and 0.06 cycle per degree and drifted at 2 Hz. During each 1.5 s stimulation period, a sequence of 1,000 images were recorded from 0 s to 1 s; during each 6 s dark adaptation period, a sequence of 1,000 images were recorded from 2.5 s to 3.5 s. A total of 5 or 10 trials were repeated for each stimulus.
Data processing
Unless stated otherwise, all images and data presented here were unprocessed raw images/data, without smoothing, denoising, or deconvolution. Supplementary Videos 1-10 were collected at 1,000 fps but were binned every 20 or 50 frames (no binning in Supplementary Video 10) and saved at 20 binned fps for video output by Fiji [31], which was not capable of saving videos at 1,000 fps. Supplementary Video 11 were collected at 3,000 fps but were binned every 60 frames and saved at 20 binned fps for video output. “Green hot” lookup table in ImageJ was used for all images.
Statistics
All statistical values are presented as mean ± s.d. unless otherwise noted. Box-and-whisker plots in Fig. 2f and Supplementary Fig. 9b: center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank C. Rodriguez for help with the laser system; E. Carroll for help with electronics; R. Lu for help with visual stimulation experiments; G. Cao at Janelia Research Campus for providing cultured neuron samples; J. Marvin, L. Looger at Janelia for providing glutamate sensors; and the Janelia JET team for designing and assembling the dispersion compensation unit. This work was supported by Howard Hughes Medical Institute (J.W., Y.L., C.-L.H., N.J.), and American Epilepsy Society predoctoral fellowship (M.C.); the China Scholarship Council Joint PhD Training Program (D.S.); Stanford Neuroscience PhD Program training grant 5T32MH020016 and the Post-9/11 GI Bill (S.W.E.); Research Grants Council of the Hong Kong Special Administrative Region of China (17209017, 17259316, 17207715) (J.W., K.K.T.); and NIH BRAIN Initiative grants 1U01NS103464 (M.Z.L.), 1RF1MH114105 (M.Z.L.), and 1UF1NS107696 (J.W., N.J., K.K.T.).
Footnotes
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare the following competing interests: KKT and The University of Hong Kong have filed a U.S. patent application (14/733,454) that relates to the all-optical laser-scanning imaging methods.
DATA AVAILABILITY STATEMENT
Raw images used for data in Fig. 2 are deposited at https://figshare.com/articles/Image_sequences_for_Fig_2/11492712. All other data are available upon reasonable requests to the correspondence authors.
REFERENCES:
- 1.Lin MZ & Schnitzer MJ Genetically encoded indicators of neuronal activity. Nat. Neurosci.19, 1142–1153 (2016). DOI: 10.1038/nn.4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen T, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013). DOI: 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marvin JS, et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nature Methods 10, 162–170 (2013). DOI: 10.1038/nmeth.2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helmchen F & Denk W Deep tissue two-photon microscopy. Nat. Methods 2, 932–940 (2005). DOI: 10.1038/nmeth818 [DOI] [PubMed] [Google Scholar]
- 5.Ji N, Freeman J & Smith SL Technologies for imaging neural activity in large volumes. Nat. Neurosci. 19, 1154–1164 (2016). DOI: 10.1038/nn.4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang W & Yuste R In vivo imaging of neural activity. Nat. Methods 14, 349–359 (2017). DOI: 10.1038/nmeth.4230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao G, et al. Genetically targeted optical electrophysiology in intact neural circuits. Cell 154, 904–913 (2013). DOI: 10.1016/j.cell.2013.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong Y, et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350, 1361–1366 (2015). DOI: 10.1126/science.aab0810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adam Y, et al. Voltage imaging and optogenetics reveal behaviour-dependent changes in hippocampal dynamics. Nature 569, 413–417 (2019). DOI: 10.1038/s41586-019-1166-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villette V, et al. Ultrafast two-photon imaging of a high-gain voltage indicator in awake behaving mice. Cell 179, 1590–1608 (2019). DOI: 10.1016/j.cell.2019.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chamberland S, et al. Fast two-photon imaging of subcellular voltage dynamics in neuronal tissue with genetically encoded indicators. eLife 6, e25690 (2017). DOI: 10.7554/eLife.25690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kazemipour A, et al. Kilohertz frame-rate two-photon tomography. Nat. Methods 16, 778–786 (2019). DOI: 10.1038/s41592-019-0493-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan ACS, et al. Speed-dependent resolution analysis of ultrafast laser-scanning fluorescence microscopy. J. Opt. Soc. Am. B 31, 755–764 (2014). DOI: 10.1364/JOSAB.31.000755 [DOI] [Google Scholar]
- 14.Wu JL, et al. Ultrafast laser-scanning time-stretch imaging at visible wavelengths. Light Sci. Appl. 6, e16196 (2016). DOI: 10.1038/lsa.2016.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marvin JS, et al. Stability, affinity and chromatic variants of the glutamate sensor iGluSnFR. Nat. Methods 15, 936–939 (2018). DOI: 10.1038/s41592-018-0171-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang HH, et al. Subcellular imaging of voltage and calcium signals reveals neural processing in vivo. Cell 166, 245–257 (2016). DOI: 10.1016/j.cell.2016.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buzsáki G Theta oscillations in the hippocampus. Neuron 33, 325–340 (2002). DOI: 10.1016/S0896-6273(02)00586-X [DOI] [PubMed] [Google Scholar]
- 18.Alonso A & Llinás RR. Subthreshold Na+-dependent theta-like rhythmicity in stellate cells of entorhinal cortex layer II. Nature 342, 175–177 (1989). DOI: 10.1038/342175a0 [DOI] [PubMed] [Google Scholar]
- 19.Tan A et al. Orientation Selectivity of Synaptic Input to Neurons in Mouse and Cat Primary Visual Cortex, J. Neurosci. 31, 12339–12350 (2011). DOI: 10.1523/JNEUROSCI.2039-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li YT et al. Synaptic Basis for Differential Orientation Selectivity between Complex and Simple Cells in Mouse Visual Cortex. J. Neurosci. 35, 11081–11093 (2015). DOI: 10.1523/JNEUROSCI.5246-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Resulaj A, Ruediger S, Olsen SR & Scanziani M First spikes in visual cortex enable perceptual discrimination. eLife 7, e34044 (2018). DOI: 10.7554/eLife.34044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma W. p., et al. Visual representations by cortical somatostatin inhibitory neurons—selective but with weak and delayed responses. J. Neurosci. 30, 14371–14379 (2010). DOI: 10.1523/JNEUROSCI.3248-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donnert G, Eggeling C & Hell SW Major signal increase in fluorescence microscopy through dark-state relaxation. Nat. Methods 4, 81–86 (2007). DOI: 10.1038/nmeth986 [DOI] [PubMed] [Google Scholar]
- 24.Tsyboulski D, Orlova N, Ledochowitsch P & Saggau P Two-photon frequency division multiplexing for functional in vivo imaging: a feasibility study. Optics Express 27, 4488–4503 (2019). DOI: 10.1364/OE.27.004488 [DOI] [PubMed] [Google Scholar]
- 25.Chauhan V, Bowlan P, Cohen J & Trebino R Single-diffraction-grating and prism pulse compressors. App. Optics 4, 619–624 (2010). DOI: 10.1364/JOSAB.27.000619 [DOI] [Google Scholar]
- 26.Lu R, et al. Video-rate volumetric functional imaging of the brain at synaptic resolution. Nat. Neurosci. 20, 620–628 (2017). DOI: 10.1038/nn.4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilt BA, Fitzgerald JE & Schnitzer MJ Photon shot noise limits on optical detection of neuronal spikes and estimation of spike timing. Biophys. J. 104, 51–62 (2013). DOI: 10.1016/j.bpj.2012.07.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wardill TJ, et al. A neuron-based screening platform for optimizing genetically-encoded calcium indicators. PLoS One 8, e77728 (2013). DOI: 10.1371/journal.pone.0077728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madisen L, et al. Transgenic mice for intersectional targeting of neural sensors and effectors with high specificity and performance. Neuron 85, 942–958 (2015). DOI: 10.1016/j.neuron.2015.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133–140 (2010). DOI: 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). DOI: 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.