ABSTRACT
Recent molecular studies have revealed complex bacterial, fungal, archaeal, and viral communities in the gastrointestinal tract of dogs and cats. More than 10 bacterial phyla have been identified, with Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria constituting more than 99% of all gut microbiota. Microbes act as a defending barrier against invading pathogens, aid in digestion, provide nutritional support for enterocytes, and play a crucial role in the development of the immune system. Of significance for gastrointestinal health is their ability to ferment dietary substrates into short-chain fatty acids, predominantly to acetate, propionate, and butyrate. However, microbes can have also a detrimental effect on host health. Specific pathogens (e.g., Salmonella, Campylobacter jejuni, and enterotoxigenic Clostridium perfringens) have been implicated in acute and chronic gastrointestinal disease. Compositional changes in the small intestinal microbiota, potentially leading to changes in intestinal permeability and digestive function, have been suggested in canine small intestinal dysbiosis or antibiotic-responsive diarrhea. There is mounting evidence that microbes play an important role in the pathogenesis of canine and feline inflammatory bowel disease (IBD). Current theories for the development of IBD favor a combination of environmental factors, the intestinal microbiota, and a genetic susceptibility of the host. Recent studies have revealed a genetic susceptibility for defective bacterial clearance in Boxer dogs with granulomatous colitis. Differential expression of pathogen recognition receptors (i.e., Toll-like receptors) were identified in dogs with chronic enteropathies. Similarly to humans, a microbial dysbiosis has been identified in feline and canine IBD. Commonly observed microbial changes are increased Proteobacteria (i.e., Escherichia coli) with concurrent decreases in Firmicutes, especially a reduced diversity in Clostridium clusters XIVa and IV (i.e., Lachnospiraceae, Ruminococcaceae, Faecalibacterium spp.). This would indicate that these bacterial groups, important short-chain fatty acid producers, may play an important role in promoting intestinal health.
Keywords: canine, feline, gastrointestinal tract, inflammatory bowel disease, microbiota, 16S ribosomal RNA gene
INTRODUCTION
The gastrointestinal (GI) microbiota is a complex collection of microorganisms (i.e., bacteria, archaea, fungi, protozoa, and viruses). Recent molecular phylogenetic studies, typically based on comparative 16S rRNA gene analysis, have revealed that the GI tract of mammals harbors several hundred to thousand bacterial phylotypes (Frank et al., 2007; Suchodolski et al., 2009; Handl et al., 2011). It is estimated that the intestine of mammals contains approximately 1010 to 1014 microorganisms, about 10 times more than the number of cells composing the host body. This mutually interacting system composed of the host cells and the resident microbes is called the intestinal microbiome. Gut microbes play a crucial role in host health. They act as a defending barrier against invading pathogens, aid in digestion and energy harvest from the diet, provide nutritional support for enterocytes, and stimulate the development of the immune system. Molecular approaches have improved our understanding about the composition, the dynamics, and the functionality of the intestinal ecosystem in dogs and cats (Ritchie et al., 2008; Desai et al., 2009; Swanson et al., 2010). The composition of the intestinal microbiota can be influenced to some extent by exogenous factors, such as diet (Simpson et al., 2002; Barry et al., 2010; Middelbos et al., 2010). However, the microbiota is resilient to most environmental influences, returning rapidly to its pretreatment state. Antibiotic administration leads to more profound alterations in the intestinal microbiome, with some bacterial groups remaining depressed for several weeks to months (Dethlefsen et al., 2008; Suchodolski et al., 2009; Grønvold et al., 2010). During the last few years, convincing evidence has emerged implicating alterations in the composition of the intestinal microbiota to chronic enteropathies not only in humans, but also in dogs and cats (German et al., 2003; Inness et al., 2007; Janeczko et al., 2008; Craven et al., 2010a; Suchodolski et al., 2010). In addition, extraintestinal disorders (e.g., atopy in infants) have been, due to the interactions of intestinal microbiota with the host immune system, associated with GI dysbiosis (Penders et al., 2007). These findings emphasize the importance of maintaining a balanced intestinal ecosystem. This article will summarize recent work characterizing the intestinal microbiome of healthy cats and dogs, and alterations in the composition of the intestinal microbiota and innate immune system that have been identified in dogs and cats with chronic enteropathies.
CHARACTERIZATION OF THE INTESTINAL MICROBIOTA
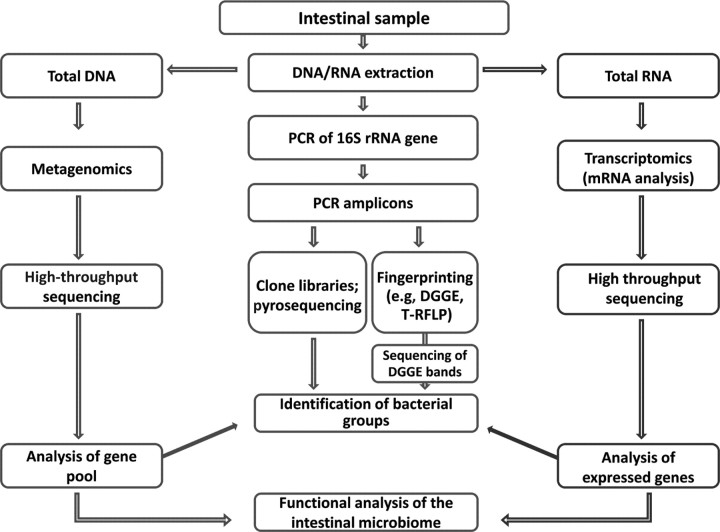
Traditional cultivation techniques have commonly been used to characterize the composition of the canine and feline intestinal microbiota (Benno et al., 1992; Johnston et al., 1993; Mentula et al., 2005). Bacterial culture is useful for the detection of specific intestinal pathogens (e.g., Salmonella spp.), allowing for antibiotic susceptibility testing and genotyping of cultured isolates. However, it is now well established that culture-dependent methods are not well suited for in-depth characterization of complex environments, such as the mammalian GI tract. Limitations of cultivation methods include our inability to culture and correctly classify the vast majority of intestinal microbes (Greetham et al., 2002). Molecular tools have now become the standard approach in microbial ecology (Figure 1) and are now increasingly used for characterization of the intestinal microbiome of dogs and cats (Suchodolski et al., 2005; Desai et al., 2009; Lubbs et al., 2009; Vester et al., 2009). These molecular methods have been reviewed in detail elsewhere (Tannock, 2005).
Figure 1.
Molecular methods for characterization of the intestinal microbiome. Amplification of the 16S rRNA gene by PCR allows either direct identification of bacterial phylotypes or the creation of a molecular fingerprint representing the bacterial diversity in a sample. New metagenomic and transcriptomics approaches, based on high-throughput sequencing of DNA or mRNA without prior amplification of a specific gene, yield an overview of the gene content of the sample and therefore the functional properties of the intestinal microbiome (DGGE = denaturing gradient gel electrophoresis; T-RFLP = terminal RFLP).
THE INTESTINAL MICROBIOME OF DOGS AND CATS
Because of anatomical and physiological differences, each intestinal compartment harbors a unique microbial ecosystem (Suchodolski et al., 2005). Microorganisms reside in specialized niches and provide specialized functions by utilizing host nutrients and, in return, provide metabolites for host uptake (Table 1). Each dog and cat harbors a very unique and individual microbial profile (Suchodolski et al., 2004; Ritchie et al., 2010). The major individual differences occur on a bacterial species and strain level, with typically only minor overlap of bacterial species between individual animals. A recent study has shown that 84% of cats evaluated harbored Bifidobacterium spp. (Ritchie et al., 2010). However, only a minor percentage of cats harbored the same species of Bifidobacteria. Differences in bacterial composition between dogs and cats have been suggested (Johnston et al., 1993; Johnston, 1999). New molecular studies indicate that such differences manifest themselves on a bacterial species and strain level, as most mammals share similar bacterial phyla and genera (Ley et al., 2008).
Table 1.
Functions of the intestinal microbiota in the normal gastrointestinal tract
| Microbial activity | Products | Representatives |
|---|---|---|
| Decarboxylation, deamination of AA | Ammonia | Clostridium spp., Peptostreptococcus spp., Peptococcus spp. |
| Deconjugation/dehydroxylation of bile acids | Secondary bile acids (cholate/deoxycholate) | Clostridium hiranonis, Lactobacillus spp. |
| Vitamin synthesis | Vitamin K2, B12, biotin, folate | Enterococcus spp., Pseudomonas spp., Sphingomonas spp., Lactobacillus spp. |
| Carbohydrate fermentation | Lactate, propionate, acetate, butyrate | Clostridium cluster XIVa, Prevotella spp., Faecalibacterium spp., Bifidobacterium spp. |
| AA fermentation | Hydrogen, methane, amines, phenols, NH3, organic acids, hydrogen sulfite | Sulfate-reducing bacteria (SRB), Desulfovibrio spp., Clostridium spp., Peptostreptococcus spp. |
| Degradation of oxalate | Formate and CO2 | Oxalobacter formigenes |
| Inulin and starch degradation | Lactate | Bifidobacterium spp. |
| Metabolism of H2, alcohols, and acetic acid | Methane and CO2 | Methanobacteria |
Differences exist also in composition and in total bacterial numbers in the different compartments of the GI tract (Table 2). Total bacterial counts and species richness increase along the GI tract and vary also between the intestinal lumen and the mucosa (Mentula et al., 2005). The stomach harbors between 101 and 106 cfu/g of bacteria. Bacterial counts in the duodenum and jejunum are typically small (105 cfu/mL of content), but can reach up to 109 cfu/mL in some dogs and cats (Johnston, 1999). This is considerably greater than found in the duodenum of humans, where total bacterial counts >105 cfu/g have been associated with the clinical syndrome small intestinal bacterial overgrowth. Cats appear to have greater counts of anaerobic bacteria in their small intestine compared with dogs (Johnston et al., 1993). The distal small intestine (i.e., ileum) contains a more diverse microbiota and greater bacterial numbers (107 cfu/mL). Bacterial counts in the colon range between 109 and 1011 cfu/g of content (Benno et al., 1992; Mentula et al., 2005). Bacteroides, Clostridium, Lactobacillus, Bifidobacterium spp., and Enterobacteriaceae are the predominant bacterial groups that have been cultured from the canine and feline intestine (Table 2).
Table 2.
Predominant bacterial groups in the canine and feline gastrointestinal tract1
| Location | Culture results1 | 16S rRNA gene results2 | FISH3 | ||||
|---|---|---|---|---|---|---|---|
| Bacterial group | Counts, log cfu/g | Bacterial group | % of total sequences | Bacterial group | Counts, log10 cells/g of feces | ||
| Small intestine | |||||||
| Spiral-shaped rods | 3.0 to 6.8 | Clostridiales | 30 to 50 | N/A4 | |||
| Bacteroides | 0 to 5.5 | Enterobacteriales | 20 to 60 | N/A | |||
| Lactobacillus sp. | 1.0 to 5.4 | Lactobacillales | 5 to 30 | N/A | |||
| Streptococcus spp. | 3.0 to 5.2 | Bacteroidales | 0 to 5 | N/A | |||
| Escherichia coli | 2.3 to 5.0 | Campylobacterales | 0 to 2 | N/A | |||
| Clostridium perfringens | 1.0 to 2.5 | Actinomycetales | 0 to 3 | N/A | |||
| Fusobacteriales | 0 to 10 | N/A | |||||
| Pasteurellales | 2 to 5 | N/A | |||||
| Spirochaetes | 0 to 12 | N/A | |||||
| Large intestine | |||||||
| Bacteroides | 7.3 to 10.2 | ||||||
| Bifidobacterium spp. | 8.0 to 10.0 | Aeromonadales | 0.2 to 0.5 | Bacteroides spp. | 9.1 | ||
| C. perfringens | 5.5 to 8.0 | Bacteroidales | 0.5 to 35 | Bifidobacterium spp. | 8.3 to 9.3 | ||
| Clostridium spp. | 7.3 to 9.5 | Bifidobacterium sp. | N/A | C. cluster IX | 8.3 | ||
| E. coli | 6.4 to 8.6 | Coriobacteriales | 1 to 2.5 | C. cluster XI | 8.03 | ||
| Lactobacillus spp. | 5.5 to 9.0 | Clostridiales | 10 to 78 | C. histolyticum group | 7.9 to 8.0 | ||
| Prevotella | 7.0 to 8.5 | Enterobacteriales | 0.1 to 2 | Desulfovibrio | 7.3 | ||
| Ruminococcus | 7.0 to 8.0 | Erysipelotrichales | 0 to 8 | Escherichia coli | 6.9 | ||
| Staphylococcus spp. | 5.2 to 5.3 | Fusobacteriales | 0.3 to 25 | Eubacterium | 9.2 | ||
| Streptococcus spp. | 8.8 to 9.1 | Lactobacillales | 1 to 5 | Lactobacillus | 8.6 to 9.4 | ||
3 Inness et al., 2007; Jia et al., 2010. FISH = fluorescence in situ hybridization.
4N/A = not applicable.
Molecular tools have greatly expanded our knowledge about the phylogenetic diversity within the canine and feline gut. Recent studies have revealed several hundred bacterial phylotypes in the canine intestinal tract (Suchodolski et al., 2009; Swanson et al., 2010). The phyla Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria constitute more than 99% of all gut microbiota in dogs and cats. The remaining bacterial groups are represented by the phyla Spirochaetes, Tenericutes, Verrucomicrobia, Cyanobacteria, Chloroflexi, and a few unclassified bacterial lineages. Generally, aerobic bacteria or facultative anaerobic bacteria occur in greater abundance in the small intestine, whereas anaerobes predominate in the large intestine. In the stomach, mucosa-adherent Helicobacter spp. predominate, followed by Lactobacillus and Streptococcus spp., and Clostridia spp. Whereas the vast majority of cats and dogs harbor various Helicobacter spp. in their stomach, Helicobacter pylori, the clinically most important Helicobacter species in humans, has not been isolated from pet dogs or cats (Rossi et al., 2008). The proximal small intestine harbors approximately 10 different bacterial phyla, with Clostridia, Lactobacillales, and Proteobacteria being most abundant (Suchodolski et al., 2009). Proteobacteria and Spirochaetes are more abundant in the proximal GI tract and typically represent <1% of obtained sequences in the large intestine of healthy animals. Firmicutes, Bacteroides, and Fusobacteria appear to be the predominant bacterial groups in the large intestine. However, the reported abundance of these bacterial groups differs between studies. For example, percentages of Firmicutes range between 25 and 95% of obtained sequencing tags (Middelbos et al., 2010; Ritchie et al., 2010; Swanson et al., 2010; Handl et al., 2011). It is likely that these discrepancies may be due to differences in DNA extraction methods and PCR protocols between studies. Actinobacteria were identified as highly abundant in fecal samples using comparative chaperonin 60 gene sequence analysis and fluorescence in situ hybridization (Inness et al., 2007; Desai et al., 2009; Jia et al., 2010). This is not surprising, because it has been shown that 16S rRNA gene approaches with universal bacterial primers typically underestimate Actinobacteria (Ritchie et al., 2010). Bifidobacteria, members of the Actinobacteria appear to be less abundant in cats and dogs compared with humans (Inness et al., 2007; Jia et al., 2010).
The Firmicutes are a heterogeneous bacterial phylum, comprising several distinct phylogenetically Clostridium clusters. These clusters differ in abundance along the intestinal tract. Clusters XIVa and IV encompass many important short-chain fatty acids (SCFA) producing bacteria (e.g., Ruminococcus spp., Faecalibacterium spp., Dorea spp., and Turicibacter spp.) and predominate in the ileum and colon. Cluster XI and Cluster I (Clostridium perfringens-group) are the second most abundant group in the small and large intestine of dogs and cats (Ritchie et al., 2008; Suchodolski et al., 2008a).
Several studies have described the metabolic products generated by the canine and feline intestinal microbiota, including VFA, lactate, ammonia, and other end products (Sunvold et al., 1995; Sparkes et al., 1998; Thompson et al., 1998). Of significance for GI health is the ability of intestinal microbes to ferment dietary products into SCFA (Topping and Clifton, 2001). Of those, acetate, propionate, and butyrate are most abundant, constituting approximately 60, 25, and 10%, respectively, of VFA in the canine and feline fecal samples (Sunvold et al., 1995; Barry et al., 2010). Minor components of the VFA are branched-chain fatty acids, including isobutyric acid, butyric acid, and isovaleric acid, which are formed during protein degradation (Macfarlane and Macfarlane, 2003; Barry et al., 2010).
Although marked differences in the phylogenetic composition of the intestinal microbiota have been observed between individual animals of the same animal species, the metabolic end products are typically quite similar between individuals. Recent metagenomics studies have revealed that despite phylogenetic differences, individuals share a wide array of microbial genes, indicating that the human intestine harbors a core microbiome (Turnbaugh et al., 2009). Furthermore, it is suggested that a functional redundancy exists in the GI tract. Several members of the community are able to perform similar functions, and if one microbial group is displaced because of perturbations (e.g., antibiotic therapy), other members of the community are capable of maintaining a stable ecosystem function (Dethlefsen et al., 2008; Suchodolski et al., 2009). The core microbiome has not been well defined in dogs and cats.
FUNGI, ARCHAEA, AND VIRUSES
The mammalian GI tract harbors complex communities of fungi, archaea, protozoa, and viruses. Recent molecular studies have provided more in-depth analysis about the diversity of these microorganisms in healthy animals, but their interactions, their influences on the host, and their role in GI disease remain unclear. Cultivation studies have reported yeasts and molds in the intestine of approximately 25% healthy Beagle dogs (Davis et al., 1977; Mentula et al., 2005). Molecular tools have revealed fungi in the small intestine of 76% of the dogs evaluated (Suchodolski et al., 2008b). A recent metagenomic approach estimated the abundance of fungi in canine fecal samples as <0.1% of total microbiota (Swanson et al., 2010).
Archaea are single-celled microorganisms and are evolutionary distinct from bacteria and eukaryotes. Archaea are commensals in the intestine of ruminants and have recently been described in the human and canine intestine, with Methanobacteriales most commonly reported, representing approximately 1% of total microbioal sequencing tags (Swanson et al., 2010).
Reported viruses in the GI tract of dogs and cats are limited to a few families, such as rotavirus, coronavirus, and parvovirus (Kempf et al., 2010). New metagenomic studies revealed a highly diverse viral community in the canine intestine, composed of several hundred double-stranded DNA viruses, with the vast majority classified as bacteriophages (Swanson et al., 2010). Future studies will need to evaluate the prevalence and abundance of single-stranded DNA and RNA viruses in the canine and feline GI tract.
CONTRIBUTION OF THE INTESTINAL MICROBIOTA TO GASTROINTESTINAL HEALTH AND DISEASE
A balanced intestinal ecosystem primes and stimulates the immune system, aids in the defense against invading intestinal pathogens, and provides nutritional benefits to the host. Although animals can be raised under germ-free conditions, several morphological and immunological differences between germ-free and conventionally raised animals indicate that commensal microbes contribute significantly to the development and maintenance of gut structure. For example, germ-free rodents have an altered epithelial architecture (e.g., thinner lamina propria, reduced number of Peyer's patches, and lymphoid follicles) and have a reduced turnover time of epithelial cells (Hooper et al., 2001). Dogs raised germ-free show similar growth rates as conventionally raised dogs, but they demonstrate an underdeveloped lymphoid system and have decreased immunoglobulin concentrations (Cohn and Heneghan, 1991). However, germ-free dogs are capable of mounting an adequate immune response to antigenic stimulation. Wound healing and blood-clotting mechanisms in germ-free-raised dogs are similar as in conventionally raised animals (Cohn and Heneghan, 1991). The intestine of germ-free dogs is characterized by thinner villi and a reduction in both the lamina propria and mucosal surface area (Thompson and Trexler, 1971). Interestingly, surgically induced strangulation obstruction of the small intestine and surgically induced bile peritonitis did not lead to death in germ-free-raised dogs, whereas conventionally raised dogs showed 100% mortality, emphasizing the role of intestinal bacteria for the severity of the disease (Cohn and Heneghan, 1991).
The resident microbiota is an integral part of the intestinal barrier, which protects the host from invading pathogens, a mechanism termed colonization resistance. The proposed mechanisms include the competition for oxygen, nutrients, mucosal adhesion sites, and the creation of a physiologically restrictive environment for nonresident bacterial species (e.g., secretion of antimicrobials, alterations in gut pH, production of hydrogen sulfide; Kanauchi et al., 2005). Therefore, young dogs and cats with colonization resistance not yet fully established are typically more susceptible to invading pathogens (e.g., Campylobacterjejuni). The sterile GI tract of newborn puppies and kittens is colonized within hours after birth by bacteria present in the birth canal and surrounding environment. After 24 h, aerobic and anaerobic counts exceed 108 cfu/g of intestinal content (Buddington, 2003). Aerobic and anaerobic bacteria are at similar numbers in the first weeks after birth, but the proportions of anaerobes are greater during adulthood (Buddington, 2003). The presence of intestinal bacteria in early life is necessary to establish oral tolerance to commensal bacteria and food antigens, to prevent onset of an inappropriate immune response, which may lead to chronic GI inflammation (Bauer et al., 2006). Microbes interact and stimulate the immune host system, as has been shown by administration of specific bacterial strains as probiotics in dogs and cats. For example, administration of Lactobacillus acidophilus DSM13241 increased granulocyte phagocytic activity (Marshall-Jones et al., 2006). Growing puppies that received Enterococcus faecium SF68 for 20 wk showed increased plasma IgA concentrations compared with the control group (Benyacoub et al., 2003).
The majority of colonic bacteria are anaerobes and their main functions are to produce energy from food and to help in the competitive exclusion of potentially pathogenic bacteria. The slower flow of ingesta and the increased time and availability of nutrients favors the microbial diversity in the colon. Bacteria within the ecosystem have developed cooperative strategies to transform the complexity of nutrients for their own benefit as well as that of the host. Colonic bacteria provide digestive enzymes that allow the utilization of complex carbohydrates. Microbes metabolize sloughed epithelial cells, endogenous mucus, and nondigested substrates that have passed through the small intestine. The latter are predominantly complex carbohydrates, including starch and dietary fiber, such as cellulose, pectin, and fructans (Topping and Clifton, 2001). The fermentation of these substrates results in the production of SCFA (e.g., acetate, propionate, and butyrate) that provide the energy for bacterial metabolism but also for epithelial cell growth (Sunvold et al., 1995). Up to 7% of the metabolic energy of dogs, and to a lesser extent in cats, is produced by microbial fermentation in the colon (Herschel et al., 1981; Brosey et al., 2000). The fecal concentrations of microbial metabolic products can be modified through dietary modulation, such as varying protein or fiber content (e.g., through prebiotic supplementation) in the diet (Huurinainen, 2009; Barry et al., 2010).
Normal intestinal motility is a major defense mechanism against attachment of pathogenic bacteria in the small intestine, and subnormal intestinal motility has been associated with small intestinal dysbiosis. Physiological concentrations of SCFA stimulated intestinal motility in the canine ileum, emphasizing the importance of microbial fermentation products on host health (Kamath et al., 1987). In vitro studies in dogs and cats revealed that physiological concentrations of SCFA stimulate contraction of longitudinal, but not circular smooth muscles in the canine and feline colon, indicating that SCFA may also contribute to in vivo colonic motility (McManus et al., 2002; Rondeau et al., 2003). However, no effects of SCFA on canine colonic motility were observed in in vivo studies (Flourie et al., 1989).
It is obvious that the close relationship between the intestinal microbiota and host cells will have a significant impact on GI health. Gastrointestinal disease may develop due to colonization with transient pathogens, due to an overgrowth by opportunistic resident bacterial groups, or due to an altered cross-talk between the intestinal innate immune system and the commensal microbiota. Invasion with specific pathogens may profoundly disturb the intestinal epithelium by altering the structure of the GI mucosa. Enteric pathogens can penetrate into the submucosa and Peyer's patches, or produce exo- or enterotoxins that alter enterocyte function. Enterotoxins produced by pathogenic bacteria (e.g., enterotoxigenic Clostridium perfringens, and toxigenic Clostridium difficile) can stimulate mucosal fluid secretions, whereas villus effacement and loss of surface area will diminish mucosal absorptive capacity, resulting in diarrhea (Marks and Kather, 2003). A dysfunction of the mucosal barrier can lead to an increase in intestinal permeability and clinically significant bacterial translocation. Intestinal pathogens that have been associated with acute or chronic diarrhea in dogs and cats include enterotoxigenic C. perfringens, toxigenic C. difficile, Salmonella, Escherichia coli (i.e., enteropathogenic, enterotoxigenic, enterohemorrhagic, and enteroinvasive), and specific Campylobacter spp. (i.e., Campylobacter jejuni; Olson and Sandstedt, 1987; Marks et al., 2002; Sancak et al., 2004).
Several GI diseases of dogs and cats are associated with nonspecific alterations in the intestinal microbiota. Small intestinal dysbiosis is a common disorder that is suspected to be caused by an intestinal dysbiosis. This disorder commonly has been referred to as small intestinal bacterial overgrowth or antibiotic-responsive diarrhea, as it has been previously suggested that affected dogs harbor increased bacterial counts (i.e., >105 cfu/ml) in their duodenum (Batt et al., 1983). Patients respond favorably to antibiotics (e.g., tylosin), but diarrhea usually returns shortly after cessation of therapy (Westermarck et al., 2005). Recent studies found no correlation between increased duodenal bacterial counts and disease status of dogs (German et al., 2003). Failure of host control mechanisms that regulate bacterial counts in the small intestine may lead to more general changes in bacterial populations, causing a dysbiosis in the small intestine. These regulatory mechanisms include intestinal motility and antimicrobial substances in pancreatic and biliary secretions. Therefore, spontaneous changes in GI motility or procedures that alter the architecture of the intestine (e.g., surgical creation of intestinal loops and resection of the ileocolic valve) can predispose animals to intestinal dysbiosis (Thompson et al., 1998). In exocrine pancreatic insufficiency, the defective pancreas is not capable of secreting antimicrobial peptides, and exocrine pancreatic insufficiency is often associated with an increase or compositional changes in small intestinal microbiota (Williams et al., 1987; Simpson et al., 1990). These changes may lead to various mechanisms that negatively affect the function of the GI tract. Examples are the dehydroxylation of fatty acids leading to impaired fat absorption, alterations in the intestinal barrier with increased intestinal permeability, the destruction of brush border enzymes and epithelial carrier proteins, and competition for substrates leading to nutrient and vitamin malabsorption (e.g., vitamin B12; Rutgers et al., 1993, 1996; Melgarejo et al., 2000).
MICROBIOTA AND INFLAMMATORY BOWEL DISEASE
There is emerging evidence implicating commensal intestinal microbiota in the pathogenesis of inflammatory bowel disease (IBD) in humans, dogs, and cats (Janeczko et al., 2008; Suchodolski et al., 2010). The currently proposed pathogenic mechanism behind IBD involves an abnormal interaction between commensal intestinal microbiota and the intestinal immune system in genetically predisposed individuals (Packey and Sartor, 2009). For example, genome-wide association studies in humans with Crohn's disease have revealed at least 33 susceptibility genes, and many of these genes are associated with a defective bacterial killing of the innate immune system (e.g., nucleotide-binding oligomerization domain containing protein 2, also known as caspase activation and recruitment domain 15; Packey and Sartor, 2009). The microbiota is implicated in human IBD because inflammation is present in gut compartments with the greatest bacterial counts. Studies in engineered animal models with susceptibility for inflammation indicate that IBD develops only if bacteria are present (Packey and Sartor, 2009). The cause-effect relationship between microbial alterations and inflammation is not well determined. It is suspected that intestinal inflammation causes a dysbiosis toward gram-negative bacteria (i.e., Proteobacteria), and a depletion of commensal bacterial groups may lead to a reduced capability of the intestinal microbiome to downregulate an aberrant intestinal immune response, leading to a perturbation of intestinal inflammation (Sokol et al., 2008). New hypotheses indicate that Campylobacter jejuni and Salmonella gastroenteritis trigger changes in mucosal architecture and in the innate immune system, which diminish the colonization resistance of resident microbes (Stecher and Hardt, 2008). In human IBD, decreases in the phyla Firmicutes and Bacteroidetes, and increases in Proteobacteria are commonly observed. Furthermore, reductions in the diversity of Clostridium clusters XIVa and IV (i.e., Ruminococcaceae, Faecalibacterium prausnitzii, and C. coccoides subgroups) in IBD patients indicate that these bacterial groups, important producers of SCFA, play an important role in maintenance of GI health (Sokol et al., 2008; Packey and Sartor, 2009).
Recent molecular studies performed in dogs and cats have also revealed differences in the intestinal microbiome between healthy animals and IBD patients (Table 3). Dogs and cats with idiopathic small intestinal IBD were significantly enriched in Enterobacteriaceae compared with controls (Janeczko et al., 2008; Xenoulis et al., 2008). Two other studies revealed an increase in Proteobacteria (i.e., Pseudomonas spp.) in the duodenum of IBD dogs (Jergens et al., 2010; Suchodolski et al., 2010). Similar to humans, IBD dogs showed a reduction in the proportions of Bacteroidales and Clostridiales (e.g., genus Faecalibacterium, Ruminococcus, and Dorea within the Clostridium clusters IV and XIVa; Jergens et al., 2010). Reduced bacterial species richness was identified in the small intestine of IBD dogs (Xenoulis et al., 2008; Craven et al., 2009). Compositional changes have also been observed in the large intestine of dogs and cats with chronic enteropathies. Fluorescence in situ hybridization analysis revealed greater microscopic counts of total bacteria, Bifidobacterium spp. and Bacteroides spp., in healthy cats, whereas cats with IBD had greater microscopic counts of Desulfovibrio spp., potential producers of toxic sulfides (Inness et al., 2007).
Table 3.
Microbial changes in cats and dogs with gastrointestinal disease1
| Species | Sampling location | Tissue type | Disease | Method | Microbial changes in diseased animals | Reference |
|---|---|---|---|---|---|---|
| Cats | Small intestine | Biopsies | IBD | FISH | Increase in Enterobacteriaceae | Janeczko et al., 2008 |
| Dogs | Duodenum | Biopsies | IBD | 16S rRNA gene clone libraries | Increase in Proteobacteria; decrease in Clostridia | Suchodolski et al., 2010 |
| Dogs | Duodenum | Mucosal/luminal brushings | IBD | 16S rRNA gene clone libraries | Increase in Enterobacteriaceae (Escherichia coli); reduction in biodiversity | Xenoulis et al., 2008 |
| Dogs | Duodenum | Mucosal/luminal brushings | Chronic enteropathies (FRD, ARD) | 16S rRNA gene clone libraries | Increase in Lactobacillales (Streptococcus and Abiotrophia) | Allenspach et al., 2010 |
| Dogs | Duodenum | Biopsies | IBD | 454-pyrosequencing of the 16S rRNA gene | Increase in Proteobacteria; decrease in Faecalibacterium, Ruminococcus, and Dorea spp. within the Clostridium clusters IV and XIVa | Jergens et al., 2010 |
| Dogs | Duodenum | Biopsies | Chronic enteropathies (SRD, FRD, ARD) | 454-pyrosequencing of the 16S rRNA gene | Reduced biodiversity | Craven et al., 2009 |
| Cats | Feces | Fecal samples | Small and large bowel IBD | FISH | Decreased total bacteria, Bifidobacterium spp. and Bacteroides spp.; increase in Desulfovibrio spp. | Inness et al., 2007 |
| Dogs | Feces | Fecal samples | Chronic diarrhea | FISH | Increase in Bacteroides | Jia et al., 2010 |
| Dogs | Feces | Fecal samples | Diarrhea | T-RFLP | Increases in Clostridium perfringens, Enterococcus faecalis, and Enterococcus faecium | Bell et al., 2008 |
| Dogs | Colon | Biopsies | Granulomatous colitis of Boxer dogs | FISH | Intracellular translocation of adherent and invasive E. coli | Simpson et al., 2006 |
1IBD = inflammatory bowel disease; FISH = fluorescence in situ hybridization; ARD = antibiotic responsive diarrhea; FRD = food-responsive diarrhea; SRD = steroid-responsive diarrhea; T-RFLP = terminal RFLP.
Similar to humans, studies have indicated that feline and canine IBD are likely associated with an immune dysregulation, as differential cytokine expressions have been identified in dogs and cats with chronic enteropathies (Nguyen Van et al., 2006; Janeczko et al., 2008; Luckschander et al., 2010). Several studies have evaluated expression of Toll-like receptors (TLR) in dogs with chronic enteropathies. Toll-like receptors are crucial members of the innate immune system. They are located on cell surfaces, recognize microbe-associated molecular patterns, and activate immune responses. Mucosal expression of TLR-2, TLR-4, and TLR-9 was increased in various dog breeds with IBD (Burgener et al., 2008; McMahon et al., 2010). German Shepherd dogs are predisposed to chronic enteropathies, and a recent study has revealed an increased TLR-2 and a decreased TLR-5 expression when compared with healthy Greyhound dogs (Allenspach et al., 2010). Furthermore, the microbiota of affected German Shepherd dogs differed from the control dogs, and was enriched in Streptococcus and Abiotrophia spp., indicating a potential interplay between resident microbiota and the innate immune system (Allenspach et al., 2010).
Granulomatous colitis of Boxer dogs, sometimes also referred to as histiocytic ulcerative colitis of Boxer dogs, has recently been associated with the presence of adherent and invasive Escherichia coli (AIEC; Simpson et al., 2006). These AIEC isolates share similarities to AIEC isolates obtained from ileal tissues of humans with Crohn's disease. The inflammation responds well to antimicrobial treatment, and improvement of clinical signs correlates with the intracellular clearance of bacteria (Craven et al., 2010b). Because this disease occurs almost exclusively in Boxer dogs, a genetic susceptibility has been hypothesized. A recent genome-wide analysis of affected and unaffected Boxer dogs has identified SNP in the gene encoding neutrophil cytosolic factor 2, a subunit of the NADPH complex in phagocytes (Craven et al., 2010b). The defect in the NADPH complex may result in an inability to eliminate intracellular pathogens, predisposing the host to chronic infections.
SUMMARY AND CONCLUSIONS
The use of molecular tools has vastly improved our understanding of the complexity and the diversity of the intestinal microbiome of dogs and cats. These methods have allowed us to monitor and to understand microbial dynamics in response to various environmental influences (e.g., dietary changes, antibiotic treatment) and to identify bacterial groups that appear to play a key role in maintaining homeostasis and balance of the intestinal ecosystem (e.g., members of the Clostridium clusters XIVa and IV). These bacterial groups are often depleted in intestinal inflammation. Furthermore, recent studies have revealed differential immune responses in dogs and cats with chronic enteropathies and have also identified underlying genetic defects in the host innate immunity. Together, these findings clearly implicate an interaction of the innate immune system and commensal intestinal microbiota in the pathogenesis of canine and feline enteropathies.
Footnotes
Based on a presentation at the Companion Animals Symposium titled “Microbes and Health,” at the Joint Annual Meeting, July 11 to 15, 2010, Denver, Colorado. The symposium was sponsored, in part, by Hill's Pet Nutrition Inc. (Topeka, KS) and The Procter & Gamble Company (Cincinnati, OH), with publication sponsored by the Journal of Animal Science and the American Society of Animal Science.
LITERATURE CITED
- Allenspach K., House A., Smith K., McNeill F. M., Hendricks A., Elson-Riggins J., Riddle A., Steiner J. M., Werling D., Garden O. A., Catchpole B., Suchodolski J. S. 2010. Evaluation of mucosal bacteria and histopathology, clinical disease activity and expression of toll-like receptors in German shepherd dogs with chronic enteropathies. Vet. Microbiol. 10.1016/j.vetmic.2010.05.025. [DOI] [PubMed] [Google Scholar]
- Barry K. A., Wojcicki B. J., Middelbos I. S., Vester B. M., Swanson K. S., Fahey G. C. 2010. Dietary cellulose, fructooligosaccharides, and pectin modify fecal protein catabolites and microbial populations in adult cats. J. Anim. Sci. 88:2978–2987. 10.2527/jas.2009-2464 [DOI] [PubMed] [Google Scholar]
- Batt R. M., Needham J. R., Carter M. W. 1983. Bacterial overgrowth associated with a naturally occurring enteropathy in the German Shepherd dog. Res. Vet. Sci. 35:42–46. [PubMed] [Google Scholar]
- Bauer E., Williams B. A., Smidt H., Verstegen M. W., Mosenthin R. 2006. Influence of the gastrointestinal microbiota on development of the immune system in young animals. Curr. Issues Intest. Microbiol. 7:35–51. [PubMed] [Google Scholar]
- Bell J. A., Kopper J. J., Turnbull J. A., Barbu N. I., Murphy A. J., Mansfield C. S. 2008. Ecological characterization of the colonic microbiota of normal and diarrheic dogs. Interdiscip. Perspect. Infect. Dis. 2008:149694. 10.1155/2008/149694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benno Y., Nakao H., Uchida K., Mitsuoka T. 1992. Impact of the advances in age on the gastrointestinal microflora of beagle dogs. J. Vet. Med. Sci. 54:703–706. 10.1292/jvms.54.703 [DOI] [PubMed] [Google Scholar]
- Benyacoub J., Czarnecki-Maulden G. L., Cavadini C., Sauthier T., Anderson R. E., Schiffrin E. J., von der Weid T. 2003. Supplementation of food with Enterococcus faecium (SF68) stimulates immune functions in young dogs. J. Nutr. 133:1158–1162. 10.1093/jn/133.4.1158 [DOI] [PubMed] [Google Scholar]
- Brosey B. P., Hill R. C., Scott K. C. 2000. Gastrointestinal volatile fatty acid concentrations and pH in cats. Am. J. Vet. Res. 61:359–361. 10.2460/ajvr.2000.61.359 [DOI] [PubMed] [Google Scholar]
- Buddington R. K. 2003. Postnatal changes in bacterial populations in the gastrointestinal tract of dogs. Am. J. Vet. Res. 64:646–651. 10.2460/ajvr.2003.64.646 [DOI] [PubMed] [Google Scholar]
- Burgener I. A., König A., Allenspach K., Sauter S. N., Boisclair J., Dorherr M. G., Jungi T. W. 2008. Upregulation of toll-like receptors in chronic enteropathies in dogs. J. Vet. Intern. Med. 22:553–560. 10.1111/j.1939-1676.2008.0093.x [DOI] [PubMed] [Google Scholar]
- Cohn I., Jr., Heneghan J. B. 1991. Germfree animals and technics in surgical research. Am. J. Surg. 161:279–283. 10.1016/0002-9610(91)91145-9 [DOI] [PubMed] [Google Scholar]
- Craven M., Acland G. M., Mezey J. G., Boyko A. R., Wang W., Meurs K., McDonough S. P., Simpson K. W. 2010b. Genome-wide analysis of granulomatous colitis in the boxer dog. J. Vet. Intern. Med. 24:725 (Abstr.) [DOI] [PubMed] [Google Scholar]
- Craven M., Dogan B., Schukken A., Volkman M., Chandler A., McDonough P. L., Simpson K. W. 2010a. Antimicrobial resistance impacts clinical outcome of granulomatous colitis in Boxer dogs. J. Vet. Intern. Med. 24:819–824. 10.1111/j.1939-1676.2010.0527.x [DOI] [PubMed] [Google Scholar]
- Craven M., Dowd S. E., McDonough S. P., Simpson K. W. 2009. High throughput pyrosequencing reveals reduced bacterial diversity in the duodenal mucosa of dogs with IBD. J. Vet. Intern. Med. 23:731 (Abstr.) [Google Scholar]
- Davis C. P., Cleven D., Balish E., Yale C. E. 1977. Bacterial association in the gastrointestinal tract of Beagle dogs. Appl. Environ. Microbiol. 34:194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A. R., Musil K. M., Carr A. P., Hill J. E. 2009. Characterization and quantification of feline fecal microbiota using cpn60 sequence-based methods and investigation of animal to animal variation in microbial population structure. Vet. Microbiol. 137:120–128. 10.1016/j.vetmic.2008.12.019 [DOI] [PubMed] [Google Scholar]
- Dethlefsen L., Huse S., Sogin M. L., Relman D. A. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280. 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flourie B., Phillips S., Richter H., Azpiroz F. 1989. Cyclic motility in the canine colon: Responses to feeding and perfusion. Dig. Dis. Sci. 34:1185–1192. 10.1007/BF01537266 [DOI] [PubMed] [Google Scholar]
- Frank D. N., Amand A. L. S., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. 2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 104:13780–13785. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- German A. J., Day M. J., Ruaux C. G., Steiner J. M., Williams D. A., Hall E. J. 2003. Comparison of direct and indirect tests for small intestinal bacterial overgrowth and antibiotic-responsive diarrhea in dogs. J. Vet. Intern. Med. 17:33–43. 10.1111/j.1939-1676.2003.tb01321.x [DOI] [PubMed] [Google Scholar]
- Greetham H. L., Giffard C., Hutson R. A., Collins M. D., Gibson G. R. 2002. Bacteriology of the Labrador dog gut: A cultural and genotypic approach. J. Appl. Microbiol. 93:640–646. 10.1046/j.1365-2672.2002.01724.x [DOI] [PubMed] [Google Scholar]
- Grønvold A. M. R., L'Abee-Lund T. M., Sorum H., Skancke E., Yannarell A. C., Mackie R. I. 2010. Changes in fecal microbiota of healthy dogs administered amoxicillin. FEMS Microbiol. Ecol. 71:313–326. 10.1111/j.1574-6941.2009.00808.x [DOI] [PubMed] [Google Scholar]
- Handl S., Dowd S. E., Garcia-Mazcorro J. F., Steiner J. M., Suchodolski J. S. 2011. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol. Ecol. (In press) [DOI] [PubMed] [Google Scholar]
- Herschel D. A., Argenzio R. A., Southworth M., Stevens C. E. 1981. Absorption of volatile fatty acid, Na, and H2O by the colon of the dog. Am. J. Vet. Res. 42:1118–1124. [PubMed] [Google Scholar]
- Hooper L. V., Wong M. H., Thelin A., Hansson L., Falk P. G., Gordon J. I. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881–884. 10.1126/science.291.5505.881 [DOI] [PubMed] [Google Scholar]
- Huurinainen O. 2009. Dietary protein and carbohydrate modify volatile fatty acid (VFA) profile in canine faeces. Licentiate Thesis. Faculty of Veterinary Medicine, University of Helsinki, Finland. [Google Scholar]
- Inness V. L., McCartney A. L., Khoo C., Gross K. L., Gibson G. R. 2007. Molecular characterisation of the gut microflora of healthy and inflammatory bowel disease cats using fluorescence in situ hybridisation with special reference to Desulfovibrio spp. J. Anim. Physiol. Anim. Nutr. (Berl.) 91:48–53. 10.1111/j.1439-0396.2006.00640.x [DOI] [PubMed] [Google Scholar]
- Janeczko S., Atwater D., Bogel E., Greiter-Wilke A., Gerold A., Baumgart M., Bender H., McDonough P. L., McDonough S. P., Goldstein R. E., Simpson K. W. 2008. The relationship of mucosal bacteria to duodenal histopathology, cytokine mRNA, and clinical disease activity in cats with inflammatory bowel disease. Vet. Microbiol. 128:178–193. 10.1016/j.vetmic.2007.10.014 [DOI] [PubMed] [Google Scholar]
- Jergens A. E., Nettleton D., Suchodolski J. S., Wymore M., Wilke V., Dowd S. E., Steiner J. M., Wang C., Wannemuehler M. J. 2010. Relationship of mucosal gene expression to microbiota composition in dogs with inflammatory bowel disease. J. Vet. Intern. Med. 24:725 (Abstr.) [Google Scholar]
- Jia J., Frantz N., Khoo C., Gibson G. R., Rastall R. A., McCartney A. L. 2010. Investigation of the faecal microbiota associated with canine chronic diarrhoea. FEMS Microbiol. Ecol. 71:304–312. 10.1111/j.1574-6941.2009.00812.x [DOI] [PubMed] [Google Scholar]
- Johnston K. L. 1999. Small intestinal bacterial overgrowth. Vet. Clin. North Am. Small Anim. Pract. 29:523–550. [PubMed] [Google Scholar]
- Johnston K. L., Lamport A., Batt R. M. 1993. An unexpected bacterial flora in the proximal small intestine of normal cats. Vet. Rec. 132:362–363. 10.1136/vr.132.14.362 [DOI] [PubMed] [Google Scholar]
- Kamath P. S., Hoepfner M. T., Phillips S. F. 1987. Short-chain fatty acids stimulate motility of the canine ileum. Am. J. Physiol. 253:G427–G433. 10.1152/ajpgi.1987.253.4.G427 [DOI] [PubMed] [Google Scholar]
- Kanauchi O., Matsumoto Y., Matsumura M., Fukuoka M., Bamba T. 2005. The beneficial effects of microflora, especially obligate anaerobes, and their products on the colonic environment in inflammatory bowel disease. Curr. Pharm. Des. 11:1047–1053. 10.2174/1381612053381675 [DOI] [PubMed] [Google Scholar]
- Kempf C., Schulz B., Strauch C., Sauter-Louis C., Tuyen U., Hartmann K. 2010. Virusnachweis in Kotproben und klinische sowie labordiagnostische Befunde von Hunden mit akutem haemorrhagischem Durchfall. Tierarztl. Prax. 38:79–86. [PubMed] [Google Scholar]
- Ley R. E., Hamady M., Lozupone C., Turnbaugh P. J., Ramey R. R., Bircher J. S., Schlegel M. L., Tucker T. A., Schrenzel M. D., Knight R., Gordon J. I. 2008. Evolution of mammals and their gut microbes. Science 320:1647–1651. 10.1126/science.1155725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubbs D. C., Vester B. M., Fastinger N. D., Swanson K. S. 2009. Dietary protein concentration affects intestinal microbiota of adult cats: A study using DGGE and qPCR to evaluate differences in microbial populations in the feline gastrointestinal tract. J. Anim. Physiol. Anim. Nutr. (Berl.) 93:113–121. 10.1111/j.1439-0396.2007.00788.x [DOI] [PubMed] [Google Scholar]
- Luckschander N., Hall J. A., Gaschen F., Forster U., Wenzlow N., Hermann P., Allenspach K., Dobbelaere D., Burgener I. A., Welle M. 2010. Activation of nuclear factor-kappaß in dogs with chronic enteropathies. Vet. Immunol. Immunopathol. 133:228–236. 10.1016/j.vetimm.2009.08.014 [DOI] [PubMed] [Google Scholar]
- Macfarlane S., Macfarlane G. T. 2003. Regulation of short-chain fatty acid production. Proc. Nutr. Soc. 62:67–72. 10.1079/PNS2002207 [DOI] [PubMed] [Google Scholar]
- Marks S. L., Kather E. J. 2003. Bacterial-associated diarrhea in the dog: A critical appraisal. Vet. Clin. North Am. Small Anim. Pract. 33:1029–1060. 10.1016/s0195-5616(03)00091-3 [DOI] [PubMed] [Google Scholar]
- Marks S. L., Kather E. J., Kass P. H., Melli A. C. 2002. Genotypic and phenotypic characterization of Clostridium perfringens and Clostridium difficile in diarrheic and healthy dogs. J. Vet. Intern. Med. 16:533–540. 10.1111/j.1939-1676.2002.tb02383.x [DOI] [PubMed] [Google Scholar]
- Marshall-Jones Z. V., Baillon M. L., Croft J. M., Butterwick R. F. 2006. Effects of Lactobacillus acidophilus DSM13241 as a probiotic in healthy adult cats. Am. J. Vet. Res. 67:1005–1012. 10.2460/ajvr.67.6.1005 [DOI] [PubMed] [Google Scholar]
- McMahon L. A., House A. K., Catchpole B., Elson-Riggins J., Riddle A., Smith K., Werling D., Burgener I. A., Allenspach K. 2010. Expression of toll-like receptor 2 in duodenal biopsies from dogs with inflammatory bowel disease is associated with severity of disease. Vet. Immunol. Immunopathol. 135:158–163. 10.1016/j.vetimm.2009.11.012 [DOI] [PubMed] [Google Scholar]
- McManus C. M., Michel K. E., Simon D. M., Washabau R. J. 2002. Effect of short-chain fatty acids on contraction of smooth muscle in the canine colon. Am. J. Vet. Res. 63:295–300. 10.2460/ajvr.2002.63.295 [DOI] [PubMed] [Google Scholar]
- Melgarejo T., Williams D. A., O'Connell N. C., Setchell K. D. 2000. Serum unconjugated bile acids as a test for intestinal bacterial overgrowth in dogs. Dig. Dis. Sci. 45:407–414. 10.1023/a:1005493416946 [DOI] [PubMed] [Google Scholar]
- Mentula S., Harmoinen J., Heikkila M., Westermarck E., Rautio M., Huovinen P., Kononen E. 2005. Comparison between cultured small-intestinal and fecal microbiotas in Beagle dogs. Appl. Environ. Microbiol. 71:4169–4175. 10.1128/AEM.71.8.4169-4175.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelbos I. S., Boler B. M. V., Qu A., White B. A., Swanson K. S., Fahey G. C. 2010. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS ONE 5:e9768. 10.1371/journal.pone.0009768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Van N. N., Taglinger K., Helps C. R., Tasker S., Gruffydd-Jones T. J., Day M. J. 2006. Measurement of cytokine mRNA expression in intestinal biopsies of cats with inflammatory enteropathy using quantitative real-time RT-PCR. Vet. Immunol. Immunopathol. 113:404–414. 10.1016/j.vetimm.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Olson P., Sandstedt K. 1987. Campylobacter in the dog—A clinical and experimental study. Vet. Rec. 121:99–101. 10.1136/vr.121.5.99 [DOI] [PubMed] [Google Scholar]
- Packey C. D., Sartor R. B. 2009. Commensal bacteria, traditional and opportunistic pathogens, dybiosis and bacterial killing in inflammatory bowel diseases. Curr. Opin. Infect. Dis. 22:292–301. 10.1097/QCO.0b013e32832a8a5d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penders J., Thijs C., van den Brandt P. A., Kummeling I., Snijders B., Stelma F., Adams H., van Ree R., Stobberingh E. E. 2007. Gut microbiota composition and development of atopic manifestations in infancy: The koala birth cohort study. Gut 56:661–667. 10.1136/gut.2006.100164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie L. E., Burke K. F., Garcia-Mazcorro J. F., Steiner J. M., Suchodolski J. S. 2010. Characterization of fecal microbiota in cats using universal 16S rRNA gene and group-specific primers for Lactobacillus and Bifidobacterium spp. Vet. Microbiol. 144:140–146. 10.1016/j.vetmic.2009.12.045 [DOI] [PubMed] [Google Scholar]
- Ritchie L. E., Steiner J. M., Suchodolski J. S. 2008. Assessment of microbial diversity along the feline intestinal tract using 16S rRNA gene analysis. FEMS Microbiol. Ecol. 66:590–598. 10.1111/j.1574-6941.2008.00609.x [DOI] [PubMed] [Google Scholar]
- Rondeau M. P., Meltzer K., Michel K. E., McManus C. M., Washabau R. J. 2003. Short chain fatty acids stimulate feline colonic smooth muscle contraction. J. Feline Med. Surg. 5:167–173. 10.1016/S1098-612X(03)00002-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M., Hanninen M. L., Revez J., Hannula M., Zanoni R. G. 2008. Occurrence and species level diagnostics of Campylobacter spp., enteric Helicobacter spp. and Anaerobiospirillum spp. in healthy and diarrheic dogs and cats. Vet. Microbiol. 129:304–314. 10.1016/j.vetmic.2007.11.014 [DOI] [PubMed] [Google Scholar]
- Rutgers H. C., Batt R. M., Proud F. J., Sorensen S. H., Elwood C. M., Petrie G., Matthewman L. A., Forster-van Hijfte M. A., Boswood A., Entwistle M., Fensome R. H. 1996. Intestinal permeability and function in dogs with small intestinal bacterial overgrowth. J. Small Anim. Pract. 37:428–434. 10.1111/j.1748-5827.1996.tb02443.x [DOI] [PubMed] [Google Scholar]
- Rutgers H. C., Lamport A., Simpson K. W., Elwood C. E., Batt R. M. 1993. Bacterial overgrowth in dogs with chronic intestinal disease. J. Vet. Intern. Med. 7:133–135. [PubMed] [Google Scholar]
- Sancak A. A., Rutgers H. C., Hart C. A., Batt R. M. 2004. Prevalence of enteropathic Escherichia coli in dogs with acute and chronic diarrhoea. Vet. Rec. 154:101–106. 10.1136/vr.154.4.101 [DOI] [PubMed] [Google Scholar]
- Simpson J. M., Martineau B., Jones W. E., Ballam J. M., Mackie R. I. 2002. Characterization of fecal bacterial populations in canines: Effects of age, breed and dietary fiber. Microb. Ecol. 44:186–197. 10.1007/s00248-002-0001-z [DOI] [PubMed] [Google Scholar]
- Simpson K. W., Batt R. M., Jones D., Morton D. B. 1990. Effects of exocrine pancreatic insufficiency and replacement therapy on the bacterial flora of the duodenum in dogs. Am. J. Vet. Res. 51:203–206. [PubMed] [Google Scholar]
- Simpson K. W., Dogan B., Rishniw M., Goldstein R. E., Klaessig S., McDonough P. L., German A. J., Yates R. M., Russell D. G., Johnson S. E., Berg D. E., Harel J., Bruant G., McDonough S. P., Schukken Y. H. 2006. Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect. Immun. 74:4778–4792. 10.1128/IAI.00067-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermudez-Humaran L. G., Gratadoux J. J., Blugeon S., Bridonneau C., Furet J. P., Corthier G., Grangette C., Vasquez N., Pochart P., Trugnan G., Thomas G., Blottiere H. M., Dore J., Marteau P., Seksik P., Langella P. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 105:16731–16736. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes A. H., Papasouliotis K., Sunvold G., Werrett G., Clarke C., Jones M., Gruffydd-Jones T. J., Reinhart G. 1998. Bacterial flora in the duodenum of healthy cats, and effect of dietary supplementation with fructo-oligosaccharides. Am. J. Vet. Res. 59:431–435. [PubMed] [Google Scholar]
- Stecher B., Hardt W. D. 2008. The role of microbiota in infectious disease. Trends Microbiol. 16:107–114. 10.1016/j.tim.2007.12.008 [DOI] [PubMed] [Google Scholar]
- Suchodolski J. S., Camacho J., Steiner J. M. 2008a. Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16s rRNA gene analysis. FEMS Microbiol. Ecol. 66:567–578. 10.1111/j.1574-6941.2008.00521.x [DOI] [PubMed] [Google Scholar]
- Suchodolski J. S., Dowd S. E., Westermarck E., Steiner J. M., Wolcott R. D., Spillman T., Harmoinen J. A. 2009. The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16s rRNA gene sequencing. BMC Microbiol. 9:210. 10.1186/1471-2180-9-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski J. S., Morris E. K., Allenspach K., Jergens A. E., Harmoinen J. A., Westermarck E., Steiner J. M. 2008b. Prevalence and identification of fungal DNA in the small intestine of healthy dogs and dogs with chronic enteropathies. Vet. Microbiol. 132:379–388. 10.1016/j.vetmic.2008.05.017 [DOI] [PubMed] [Google Scholar]
- Suchodolski J. S., Ruaux C. G., Steiner J. M., Fetz K., Williams D. A. 2004. Application of molecular fingerprinting for qualitative assessment of small-intestinal bacterial diversity in dogs. J. Clin. Microbiol. 42:4702–4708. 10.1128/JCM.42.10.4702-4708.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchodolski J. S., Ruaux C. G., Steiner J. M., Fetz K., Williams D. A. 2005. Assessment of the qualitative variation in bacterial microflora among compartments of the intestinal tract of dogs by use of a molecular fingerprinting technique. Am. J. Vet. Res. 66:1556–1562. 10.2460/ajvr.2005.66.1556 [DOI] [PubMed] [Google Scholar]
- Suchodolski J. S., Xenoulis P. G., Paddock C. G., Steiner J. M., Jergens A. E. 2010. Molecular analysis of the bacterial microbiota in duodenal biopsies from dogs with idiopathic inflammatory bowel disease. Vet. Microbiol. 142:394–400. 10.1016/j.vetmic.2009.11.002 [DOI] [PubMed] [Google Scholar]
- Sunvold G. D., Hussein H. S., Fahey G. C., Jr., Merchen N. R., Reinhart G. A. 1995. In vitro fermentation of cellulose, beet pulp, citrus pulp, and citrus pectin using fecal inoculum from cats, dogs, horses, humans, and pigs and ruminal fluid from cattle. J. Anim. Sci. 73:3639–3648. 10.2527/1995.73123639x [DOI] [PubMed] [Google Scholar]
- Swanson K. S., Dowd S. E., Suchodolski J. S., Middelbos I. S., Vester B. M., Barry K. A., Nelson K. E., Cann I. K., White B. A., Fahey G. C. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2010 doi: 10.1038/ismej.2010.162. doi: 10.1038/ismej.2010.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock G. W. 2005. New perceptions of the gut microbiota: Implications for future research. Gastroenterol. Clin. North Am. 34:361–382. 10.1016/j.gtc.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Thompson G. R., Trexler P. C. 1971. Gastrointestinal structure and function in germ-free or gnotobiotic animals. Gut 12:230–235. 10.1136/gut.12.3.230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. S., Quigley E. M., Adrian T. E. 1998. Qualitative changes in enteric flora and short-chain fatty acids after intestinal resection. Dig. Dis. Sci. 43:624–631. 10.1023/a:1018831728734 [DOI] [PubMed] [Google Scholar]
- Topping D. L., Clifton P. M. 2001. Short-chain fatty acids and human colonic function: Roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031–1064. 10.1152/physrev.2001.81.3.1031 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., Sogin M. L., Jones W. J., Roe B. A., Affourtit J. P., Egholm M., Henrissat B., Heath A. C., Knight R., Gordon J. I. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester B. M., Dalsing B. L., Middelbos I. S., Apanavicius C. J., Lubbs D. C., Swanson K. S. 2009. Faecal microbial populations of growing kittens fed high or moderate protein diets. Arch. Anim. Nutr. 63:254–265. [Google Scholar]
- Westermarck E., Frias R., Skrzypczak T. 2005. Effect of diet and tylosin on chronic diarrhea in Beagles. J. Vet. Intern. Med. 19:822–827. 10.1111/j.1939-1676.2005.tb02771.x [DOI] [PubMed] [Google Scholar]
- Williams D. A., Batt R. M., McLean L. 1987. Bacterial overgrowth in the duodenum of dogs with exocrine pancreatic insufficiency. J. Am. Vet. Med. Assoc. 191:201–206. [PubMed] [Google Scholar]
- Xenoulis P. G., Palculict B., Allenspach K., Steiner J. M., VanHouse A. M., Suchodolski J. S. 2008. Molecular-phylogenetic characterization of microbial communities imbalances in the small intestine of dogs with inflammatory bowel disease. FEMS Microbiol. Ecol. 66:579–589. 10.1111/j.1574-6941.2008.00556.x [DOI] [PubMed] [Google Scholar]