Abstract
The novel human coronavirus disease COVID-19 has become the fifth documented pandemic since the 1918 flu pandemic. COVID-19 was first reported in Wuhan, China, and subsequently spread worldwide. The coronavirus was officially named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the International Committee on Taxonomy of Viruses based on phylogenetic analysis. SARS-CoV-2 is believed to be a spillover of an animal coronavirus and later adapted the ability of human-to-human transmission. Because the virus is highly contagious, it rapidly spreads and continuously evolves in the human population. In this review article, we discuss the basic properties, potential origin, and evolution of the novel human coronavirus. These factors may be critical for studies of pathogenicity, antiviral designs, and vaccine development against the virus.
Keywords: COVID-19, SARS-COV-2, Coronavirus, Pandemic
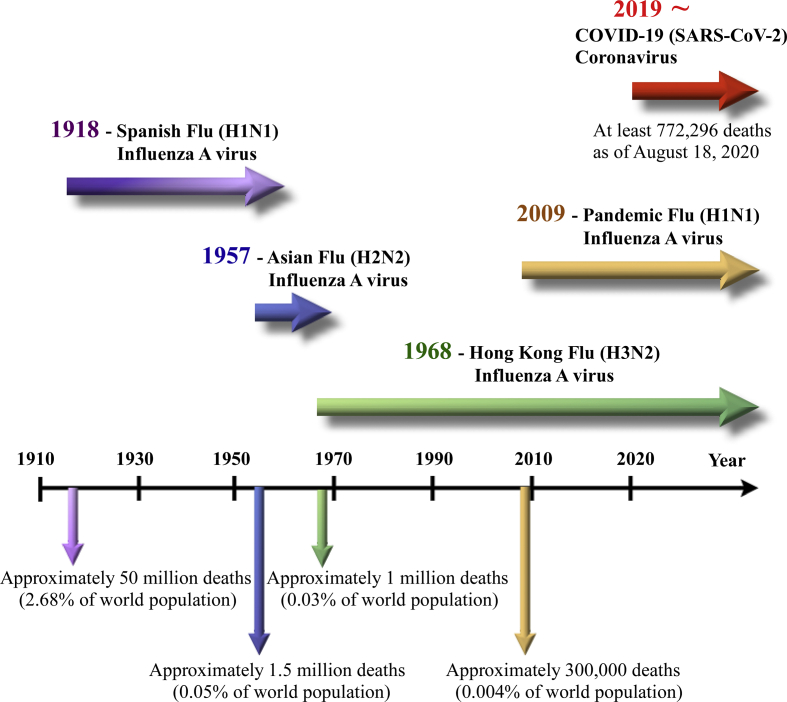
Currently, people all over the world have been affected by coronavirus disease 2019 (COVID-19), which is the fifth pandemic after the 1918 flu pandemic. As of now, we can trace the first report and subsequent outbreak from a cluster of novel human pneumonia cases in Wuhan City, China, since late December 2019. The earliest date of symptom onset was 1 December 2019. The symptomatology of these patients, including fever, malaise, dry cough, and dyspnea, was diagnosed as viral pneumonia [1,2]. Initially, the disease was called Wuhan pneumonia by the press because of the area and pneumonia symptoms. Whole-genome sequencing results showed that the causative agent is a novel coronavirus. Therefore, this virus is the seventh member of the coronavirus family to infect humans [3]. The World Health Organization (WHO) temporarily termed the new virus 2019 novel coronavirus (2019-nCoV) on 12 January 2020 and then officially named this infectious disease coronavirus disease 2019 (COVID-19) on 12 February 2020. Later, the International Committee on Taxonomy of Viruses (ICTV) officially designated the virus as SARS-CoV-2 based on phylogeny, taxonomy and established practice [4]. Subsequently, human-to-human transmission of COVID-19 occurring within Hong Kong has been shown in clinical data [5]. Since COVID-19 initially emerged in China, the virus has evolved for four months and rapidly spread to other countries worldwide as a global threat. On 11 March 2020, the WHO finally made the assessment that COVID-19 can be characterized as a pandemic, following 1918 Spanish flu (H1N1), 1957 Asian flu (H2N2), 1968 Hong Kong flu (H3N2), and 2009 Pandemic flu (H1N1), which caused an estimated 50 million, 1.5 million, 1 million, and 300,000 human deaths, respectively [[6], [7], [8], [9]] [Fig. 1].
Fig. 1.
A timeline of five pandemics since 1918 and the globally circulating viruses afterward.
Virology – morphology, gene structure and replication
SARS-CoV-2 is an enveloped and spherical particle approximately 120 nm in diameter containing a positive-sense single-stranded RNA genome. It belongs to the subfamily Coronavirinae, family Coronavirdiae, and order Nidovirales. The RNA genome of SARS-CoV-2 contains a 5′ methyl-guanosine cap, poly (A)-tail, and 29,903 nucleotides according to WH-Human 1 coronavirus (WHCV) [3,10]. It is classified as a beta-coronavirus (βCoV) [lineage B] and is the seventh coronavirus to infect humans, following 2 αCoV (HCoV-229E and HKU-NL63) and 4 βCoV (HCoV-OC43 [lineage A], HCoV-HKU1 [lineage A], severe acute respiratory syndrome SARS-CoV [lineage B] and Middle East respiratory syndrome MERS-CoV [lineage C]) [[11], [12], [13], [14]]. It has been shown that bats and rodents are the gene sources of most αCoVs and βCoVs, however, avian species are the gene sources of most δCoVs and γCoVs by evolutionary analyses. The human coronavirus (HCoV) strains HCoV-NL63, HCoV-229E, HCoV-HKU1, and HCoV-OC43 usually cause mild, self-limiting upper respiratory tract infections, such as the common cold [15,16]. However, SARS-CoV, MERS-CoV, and SARS-CoV-2 can cause severe acute respiratory syndrome and result in life-threatening disease [[17], [18], [19]] [Table 1].
Table 1.
Classification of human coronaviruses.
| Strain | Discovery | Genera-Lineage | Cellular Receptor | Natural Host | Intermediate Host | Respiratory Symptom |
|---|---|---|---|---|---|---|
| HcoV-229E | 1966 | α | Aminopeptidase N (CD13) | Bats | Camelids | Mild |
| HcoV-OC43 | 1967 | β-A | 9-O-Acetylated sialic acid (SA) | Rodents | Cattle | Mild |
| SARS-CoV | 2003 | β-B | ACE2 | Bats | Masked palm civets | Severe acute |
| HcoV-NL63 | 2004 | α | ACE2 | Bats | Unknown | Mild |
| HcoV-HKU1 | 2005 | β-A | 9-O-Acetylated sialic acid (SA) | Rodents | Unknown | Mild |
| MERS-CoV | 2012 | β-C | DPP4 | Bats | Dromedary camels | Severe acute |
| SARS-CoV-2 | 2019 | β-B | ACE2 | Bats | Pangolin? | Severe acute |
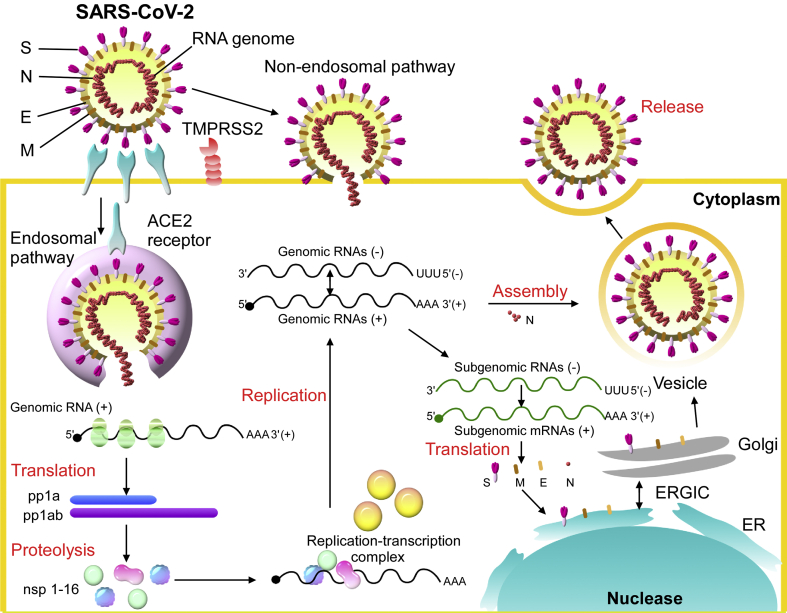
SARS-CoV-2 transcribes nine subgenomic RNAs, and its genome comprises a 5′ untranslated region including a 5′ leader sequence; an open reading frame (ORF) 1a/ab encoding nonstructural proteins (nsp) for replication; four structural proteins including spike (S), envelope (E), membrane (M) and nucleocapsid (N); several accessory proteins such as ORF 3a, 6, 7a/b, and 8; and a 3′ untranslated region. The replicase polyprotein pp1a/ab encodes and is proteolytically cleaved into 16 putative nsps, including nsp3 (papain-like protease), nsp5 (3C-like protease), nsp12 (RNA-dependent RNA polymerase [RdRp]), nsp13 (helicase), and other nsps [10,13,20]. The spike glycoprotein of SARS-CoV-2 binds to angiotensin-converting enzyme 2 (ACE2) in human and Chinese horseshoe bats, civet for cell entry, that is also dependent on S protein priming by the serine protease TMPRSS2. A similar panel of mammalian cell lines can be infected with SARS-CoV-2-S and SARS-CoV-S [[21], [22], [23], [24]]. The spike protein could be cleaved by host proteases into the S1 and S2 subunits, which are responsible for receptor recognition and membrane fusion, respectively. S1 also can be divided into an N-terminal domain (NTD) and a C-terminal domain (CTD). The S1 CTD of SARS-CoV-2, but not the NTD, showed strong affinity for human ACE2 (hACE2). The receptor-binding domain (RBD) within SARS-CoV-2 CTD is the key region that interacts with the hACE2 receptor with higher affinity than the RBD of SARS-CoV by 10- to 20-fold with kinetic quantification [23,25]. The putative life cycle of SARS-CoV-2 in host cells begins from spike protein and hACE2 receptor binding. The conformational change in the S protein after receptor binding facilitates viral envelope fusion with the cell membrane through the endosomal pathway. The viral RNA genome is then released into the cytoplasm and translated into viral replicase polyproteins pp1a and 1 ab, which can be cleaved into small products by virus-encoded proteinases. The polymerase transcribes a series of subgenomic mRNAs by discontinuous transcription. The subgenomic mRNAs are finally translated into viral structural proteins. The S, E and M proteins enter the endoplasmic reticulum (ER) and Golgi apparatus, and the N protein is combined with the positive-stranded genomic RNA to form a nucleoprotein complex. The structural proteins and nucleoprotein complex are assembled with the viral envelope at the ER–Golgi intermediate compartment. The newly assembled viral particles are then released from the infected cell [Fig. 2].
Fig. 2.
The putative life cycle of SARS-CoV-2.
Ecology - the potential origin of the virus
All human coronaviruses have animal origins, namely, natural hosts. Bats may be the natural hosts of HCoV-229E, SARS-CoV, HCoV-NL63, and MERS-CoV. Furthermore, HCoV-OC43 and HKU1 probably originated from rodents [[26], [27], [28]]. Bats are undoubtedly important and the major natural reservoirs of alpha-coronaviruses and beta-coronaviruses [29]. Domestic animals can suffer from disease as intermediate hosts that cause virus transmission from natural hosts to humans; for example, SARS-CoV and MERS-CoV crossed the species barriers into masked palm civets and camels, respectively [30,31] [Table 1]. SARS-CoV-2 sequenced at the early stage of the COVID-19 outbreak only shares 79.6% sequence identity with SARS-CoV through early full-length genomic comparisons. However, it is highly identical (96.2%) at the whole-genome level to Bat-CoV RaTG13, which was previously detected in Rhinolophus affinis from Yunnan Province, over 1500 km from Wuhan [21]. Bats are likely reservoir hosts for SARS-CoV-2; however, whether Bat-CoV RaTG13 directly jumped to humans or transmits to intermediate hosts to facilitate animal-to-human transmission remains inconclusive. No intermediate host sample was obtained by scientists in an initial cluster of infections of the Huanan Seafood and Wildlife Market in Wuhan, where the sale of wild animals may be the source of zoonotic infection. Furthermore, the earliest three patients with symptom onset had no known history of exposure to the Huanan market [1]. Therefore, there may be multiple sources of COVID-19 in the beginning. According to previous studies by metagenomic sequencing for the samples from Malayan pangolins (Manis javanica) in Guangxi and Guangdong, China, it has been suggested that pangolins might be the intermediate hosts between bats and humans because of the similarity of the pangolin coronavirus to SARS-CoV-2 [32,33]. However, the additional phylogenetic analyses effectively trace COVID-19 infection sources. In addition to the zoonotic origins of SARS-CoV-2 by natural evolution, there are still some disputes about the origin of the virus because its spike protein seems to perfectly interact with the human receptor in contributing to human-to-human transmission after evolution in a short period. Nevertheless, more direct evidence is required to clarify the arguments.
Evolution of SARS-COV-2 during the past few months
Replication of RNA viruses could generate mutations due to the low proofreading ability of their RdRP. The genome variations generated by viral RdRP could be beneficial for an emerging virus to adapt to new hosts. However, previous studies have shown that the mutation rates could vary in RNA viruses [34]. The synonymous substitution rate for coronaviruses might be approximately 1 × 10−3/synonymous site/year, which is lower than some other RNA viruses. The mutation rate during coronavirus replication could be partially controlled by the viral exoribonuclease nsp14 [35,36]. Nevertheless, SARS-CoV-2 has been continuously evolving to different groups worldwide during the pandemic.
According to the information of nCoV-19 (SARS-CoV-2) sequences submitted to the GISAID database in January 2020, the virus was first collected in late December 2019 from Wuhan, China. However, those viral sequences varied from the latest submitted sequence collected in early April 2020 from North America. Since the viral sequences continuously change, the construction of a phylogenetic network is crucial to investigate the adaption of the virus in different human populations and environments. Although the virus keeps evolving within humans who could also be susceptible to other human coronaviruses, recombination between SARS-CoV-2 and old human coronaviruses, such as HCoV-229E, OC43, NL63, and HKU1, has not been found. Nevertheless, a recent study claimed that three genetic types of the virus have been circulating globally [37]. The study demonstrated that the genotypes could also correlate to the geographic locations, while the sample size and analysis methods in the study are still being argued in the research field [38]. Therefore, it is still unclear whether the evolution of SARS-CoV-2 could be affected by replication environments, such as genetic and immunological restrictions in different human populations. With evolutionary pressure, the selection of SARS-CoV-2 mutations will be ongoing. The investigation of the geographic patterns of SARS-CoV-2 variations will provide information on vaccine development for different populations.
Conclusions
Human coronaviruses usually cause mild upper respiratory diseases. However, in the past two decades, two coronaviruses transmitted from animals, SARS-CoV and MERS-CoV, have caused severe pneumonia and death in humans. In addition, since late December 2019, the COVID-19 pandemic has spread globally and consequently resulted in at least 772,296 deaths worldwide as of August 18, 2020 [39]. Due to the high sequence homology with a coronavirus isolated from bats, SARS-CoV-2 is considered a zoonotic origin coronavirus. Undoubtedly, SARS-CoV-2 has become the fifth human coronavirus, and it is possible that this virus will continuously circulate in the human population in the future. Because specific antiviral treatments and vaccines are still under development, testing, quarantine, and social distancing are encouraged to prevent virus spread. Nonetheless, since the virus keeps mutating and evolving during the pandemic, studies on viral pathogenicity, treatments and prophylactic vaccines should closely consider the genetic characteristics of the virus.
Conflict of interest statement
The authors declare no conflicts of interest.
Acknowledgements
This work was financially supported by the Research Center for Emerging Viral Infections from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education in Taiwan and the Ministry of Science and Technology, Taiwan (MOST 108-3017-F-182-001).
Footnotes
Peer review under responsibility of Chang Gung University.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coronaviridae Study Group of the International Committee on Taxonomy of V The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson N.P., Mueller J. Updating the accounts: global mortality of the 1918-1920 "Spanish" influenza pandemic. Bull Hist Med. 2002;76:105–115. doi: 10.1353/bhm.2002.0022. [DOI] [PubMed] [Google Scholar]
- 7.Kain T., Fowler R. Preparing intensive care for the next pandemic influenza. Crit Care. 2019;23:337. doi: 10.1186/s13054-019-2616-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonsen L., Clarke M.J., Schonberger L.B., Arden N.H., Cox N.J., Fukuda K. Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J Infect Dis. 1998;178:53–60. doi: 10.1086/515616. [DOI] [PubMed] [Google Scholar]
- 9.Viboud C., Simonsen L., Fuentes R., Flores J., Miller M.A., Chowell G. Global mortality impact of the 1957-1959 influenza pandemic. J Infect Dis. 2016;213:738–745. doi: 10.1093/infdis/jiv534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microb Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan J.F., To K.K., Tse H., Jin D.Y., Yuen K.Y. Interspecies transmission and emergence of novel viruses: lessons from bats and birds. Trends Microbiol. 2013;21:544–555. doi: 10.1016/j.tim.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28:465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin Y., Wunderink R.G. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130–137. doi: 10.1111/resp.13196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jevsnik M., Ursic T., Zigon N., Lusa L., Krivec U., Petrovec M. Coronavirus infections in hospitalized pediatric patients with acute respiratory tract disease. BMC Infect Dis. 2012;12:365. doi: 10.1186/1471-2334-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 18.Reusken C.B., Haagmans B.L., Muller M.A., Gutierrez C., Godeke G.J., Meyer B. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 20.Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020 doi: 10.1126/science.abb7498. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020 doi: 10.1016/j.cell.2020.03.045. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181 doi: 10.1016/j.cell.2020.02.058. 281–92.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao Y., Shi M., Chommanard C., Queen K., Zhang J., Markotter W. Surveillance of bat coronaviruses in Kenya identifies relatives of human coronaviruses NL63 and 229E and their recombination history. J Virol. 2017;91 doi: 10.1128/JVI.01953-16. e01953-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo P.C., Lau S.K., Lam C.S., Lau C.C., Tsang A.K., Lau J.H. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- 31.Haagmans B.L., Al Dhahiry S.H., Reusken C.B., Raj V.S., Galiano M., Myers R. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lam T.T., Shum M.H., Zhu H.C., Tong Y.G., Ni X.B., Liao Y.S. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. in press. [DOI] [PubMed] [Google Scholar]
- 33.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr Biol. 2020;30:1578. doi: 10.1016/j.cub.2020.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanada K., Suzuki Y., Gojobori T. A large variation in the rates of synonymous substitution for RNA viruses and its relationship to a diversity of viral infection and transmission modes. Mol Biol Evol. 2004;21:1074–1080. doi: 10.1093/molbev/msh109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8:270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eckerle L.D., Lu X., Sperry S.M., Choi L., Denison M.R. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J Virol. 2007;81:12135–12144. doi: 10.1128/JVI.01296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forster P., Forster L., Renfrew C., Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci USA. 2020;117:9241–9243. doi: 10.1073/pnas.2004999117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mavian C., Pond S.K., Marini S., Magalis B.R., Vandamme A.M., Dellicour S. Sampling bias and incorrect rootingmake phylogenetic network tracing of SARS-COV-2 infections unreliable. Proc Natl Acad Sci USA. 2020 doi: 10.1073/pnas.2007295117. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.JHU . 2020. COVID-19 dashboard by the center for systems science and engineering (CSSE) at johns hopkins university (JHU)https://coronavirus.jhu.edu/map.html [Google Scholar]