Abstract
In December 2019, a novel human-infecting coronavirus (SARS-CoV-2) was recognized in China. In a few months, SARS-CoV-2 has caused thousands of disease cases and deaths in several countries. Phylogenetic analyses indicated that SARS-CoV-2 clusters with SARS-CoV in the Sarbecovirus subgenus and viruses related to SARS-CoV-2 were identified from bats and pangolins. Coronaviruses have long and complex genomes with high plasticity in terms of gene content. To date, the coding potential of SARS-CoV-2 remains partially unknown. We thus used available sequences of bat and pangolin viruses to determine the selective events that shaped the genome structure of SARS-CoV-2 and to assess its coding potential. By searching for signals of significantly reduced variability at synonymous sites (dS), we identified six genomic regions, one of these corresponding to the programmed −1 ribosomal frameshift. The most prominent signal of dS reduction was observed within the E gene. A genome-wide analysis of conserved RNA structures indicated that this region harbors a putative functional RNA element that is shared with the SARS-CoV lineage. Additional signals of reduced dS indicated the presence of internal ORFs. Whereas the presence ORF9a (internal to N) was previously proposed by homology with a well characterized protein of SARS-CoV, ORF3h (for hypothetical, within ORF3a) was not previously described. The predicted product of ORF3h has 90% identity with the corresponding predicted product of SARS-CoV and displays features suggestive of a viroporin. Finally, analysis of the putative ORF10 revealed high dN/dS (3.82) in SARS-CoV-2 and related coronaviruses. In the SARS-CoV lineage, the ORF is predicted to encode a truncated protein and is neutrally evolving. These data suggest that ORF10 encodes a functional protein in SARS-CoV-2 and that positive selection is driving its evolution. Experimental analyses will be necessary to validate and characterize the coding and non-coding functional elements we identified.
Keywords: SARS-CoV-2, Coronaviruses, Functional RNA elements, Coding potential
Abbreviations: ORF, open reading frame; -1 PRF, programmed -1 ribosomal frameshifting; dS, synonymous substitution rate; dN, nonsynonymous substitution rate; GTR, General Time Reversible; SLAC, single-likelihood ancestor counting; PAML, Phylogenetic Analysis by Maximum Likelihood
Highlights
-
•
We analyzed the coding region of SARS-CoV-2 and related bat/pangolin viruses.
-
•
We identified six regions of significantly low variability at sysnonymous sites.
-
•
One of these corresponds to a conserved RNA structure shared with the SARS-CoV lineage.
-
•
The dS reduction within ORF3a corresponds to a potential ORF encoding a viroporin.
-
•
In SARS-CoV-2 and related viruses, the putative 3′ terminal ORF10 has high dN/dS.
1. Introduction
Human-infecting coronaviruses are potentially dangerous pathogens of zoonotic origin (Cui et al., 2019; Forni et al., 2017). In December 2019, a novel coronavirus, now referred to as SARS-CoV-2 (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, 2020), was described in China. The virus caused respiratory disease in a large number of people and was responsible for thousands of deaths (Zhu et al., 2020). Eventually, SARS-CoV-2 reached other countries: outbreaks in Italy, Japan, South Korea, and other areas are ongoing (www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/). This is reminiscent of the events that, two decades ago, led to the emergence and spread of SARS-CoV (severe acute respiratory syndrome-related coronavirus) and, in 2012, of MERS-CoV (Middle East respiratory syndrome-related Coronavirus) (Cui et al., 2019, Forni et al., 2017).
Coronaviruses (family Coronaviridae, order Nidovirales) are positive-sense, single stranded RNA viruses. Several coronavirus genera and subgenera are recognized (https://talk.ictvonline.org/ictv-reports/). Phylogenetic analyses indicated that SARS-CoV-2 clusters with SARS-CoV and other viruses in the Sarbecovirus subgenus (Betacoronavirus genus) (Zhou et al., 2020).
As most human coronaviruses, both SARS-CoV and MERS-CoV originated in bats and were transmitted to humans via intermediate hosts: small carnivores in the case of SARS-CoV and dromedary camels for MERS-CoV (Cui et al., 2019; Forni et al., 2017). A bat origin seems also to be likely for SARS-CoV-2, as to date its closest relative (BatCoV RaTG13) was sequenced from horseshoe bats (Rhinolophus affinis) (Zhou et al., 2020). Two other bat-derived coronaviruses (bat-SL-CoVZC45 and bat-SL-CoVZXC21) display high levels of similarity (> 70%) to SARS-CoV-2 (Lu et al., 2020; Paraskevis et al., 2020; Wu et al., 2020b). Recently, though, several reports suggested that viruses related to SARS-CoV-2 exist in pangolins (Manis javanica) (Lam et al., 2020; Xiao et al., 2020; Wong et al., 2020; Liu et al., 2020). In particular, striking similarity between pangolin-derived viruses and SARS-CoV-2 was detected in the receptor binding domain (RBD) of the surface spike glycoprotein (Wong et al., 2020), which represents a major determinant of coronavirus host range (Haijema et al., 2003, Kuo et al., 2000,McCray Jr et al., 2007, Moore et al., 2004, Schickli et al., 2004). Overall, these reports suggest that recombination events have shaped the diversity of the spike protein and contributed to the diversification of SARS-CoV-2 and related viruses (Wong et al., 2020; Zhang et al., 2020). Nevertheless, it still remains unclear whether this novel human pathogen emerged as a zoonosis transmitted from bats, pangolins, or other animals. Moreover, determinants located in genomic regions other than the RBD most likely contribute to determine coronavirus host range (Peck et al., 2015). Importantly, several coronavirus proteins modulate immune evasion, tissue tropism, and virulence, eventually playing a role in disease severity and pathogenesis (Cui et al., 2019, Forni et al., 2017). Indeed, coronaviruses have unusually long and complex genomes if compared to those of other RNA viruses. Two thirds of the coronavirus genome are occupied by two large overlapping open reading frames (ORF1a and ORF1b), that are translated into polyproteins. These latter are processed to generate 16 nonstructural proteins (nsp1 to nsp16). The remaining portion of the genome includes ORFs for the structural proteins (spike, envelope, membrane, and nucleoprotein) and a variable number of accessory proteins (Cui et al., 2019, Forni et al., 2017). Gene gains and losses during the evolutionary history of coronaviruses have resulted in a diverse array of accessory ORFs, even among viruses belonging to the same lineage (Forni et al., 2017).
To date, the coding potential of SARS-CoV-2 remains partially unknown, and distinct studies have provided different genome annotations (Zhou et al., 2020; Wu et al., 2020b; Wu et al., 2020a; Chan et al., 2020). We thus used available sequences of bat and pangolin viruses related to SARS-CoV-2 to determine the selective events that shaped the genome structures of these viruses and to assess their coding potential.
2. Materials and methods
2.1. Sequences, alignments, and recombination
Viral genome sequences from pangolins were retrieved from the GISAID (https://www.gisaid.org) database, whereas all other genome sequences were downloaded from the NCBI (National Center for Biotechnology Information) database (http://www.ncbi.nlm.nih.gov/) (Supplementary Table S1).
All alignments were generated using MAFFT (Katoh and Standley, 2013), setting sequence type as codons or nucleotide, as appropriate.
In order to analyze interspecies diversity, pairwise identity scores were calculated for pangolin viruses. Scores were calculated as 1-(M/L) where M is the number of mismatching nucleotides and L is the total number of positions along the alignment at which neither sequence has a gap or a N character, as previously suggested (Muhire et al., 2014). We considered genomes with a identity score less than 0.90 and with the highest genome coverage (Supplementary Fig. S1). We thus randomly selected Guangxi/P1E among Guangxi/P1E, Guangxi/P5E, Guangxi/P5L, Guangxi/P4L, Guangxi/P3B, and Guangxi/P2V. We also selected Guandong/1 instead of Guangdong/P2S based on their genomic coverage (Supplementary Fig. S1).
Recombination was evaluated using RDP4. Specifically, we applied four different methods (RDP, GENECONV, MaxChi, and Chimera) (Martin et al., 2017; Sawyer, 1989; Martin and Rybicki, 2000; Posada and Crandall, 2001; Smith, 1992) and only recombination events with a p value < .05 for at least two methods were considered as significant.
2.2. Detection of synonymous substitution reduction
Synonymous substitution (dS) reduction was calculated using synplot2 program (Firth, 2014). This tool is designed to identify overlapping functional elements along coding sequence alignments. A peculiar signature of these elements is a reduction of dS variability. We used windows of 25 codons, as suggested (Firth, 2014), and a p-value threshold calculated on the basis of the number of windows (i.e., 0.05/number of windows).
2.3. Detection of conserved RNA structures
RNA secondary structures were searched for using RNAz v2.1 (Washietl et al., 2005). This software predicts conserved structures in a multiple sequence alignment. RNAz was run using default parameters, with sliding windows of 120 nucleotides moving with a step of 40. We accepted only RNA structures with a mean z-score < −4, a structure conservation index ≥ mean pairwise identity, and an SVM RNA-class probability > .95. A visual representation of the secondary structures was obtained with RNAalifold (Bernhart et al., 2008).
2.4. Phylogenetic trees and positive selection analyses
Phylogenetic trees were generated using the phyML software using a General Time Reversible (GTR) model with gamma-distributed rates, 4 substitution rate categories, and estimation of transition/transversion ratio and proportion of invariable sites (Guindon et al., 2009).
The ORF10 average nonsynonymous substitution (dN)/synonymous substitution (dS) rate was calculated using the single-likelihood ancestor counting (SLAC) method(Kosakovsky Pond and Frost, 2005).
Evidence of positive selection was searched for using the codon-based codeml program implemented in the PAML (Phylogenetic Analysis by Maximum Likelihood) suite (Yang, 2007). We applied different site (NSsite) models. M7 and M8a are null models, M8 is a positive selection model. M7 assumes that 0 < dN/dS < 1 and is beta distributed among sites; M8 is the same as M7 but also includes a category of sites with dN/dS > 1, and M8a is the same as M8, but allows only neutral evolution. The models were run with the F3x4 codon frequency model, an initial value of ω = 0.4, and a phylogenetic tree generated for each ORF analyzed, after accounting for recombination events. To assess statistical significance, twice the difference of the likelihood (ΔlnL) for the two models is compared to a χ2 distribution (M7 vs M8, degrees of freedom = 2; (M8a vs M8, degrees of freedom = 1).
Test for relaxation of selective pressure, we used the RELAX tool (Wertheim et al., 2015). RELAX evaluates if selection on the test branches is relaxed (k < 1) or intensified (k > 1) compared to background branches. In particular, we masked the stop codon in SARS-CoV lineage strains and we analyzed the full potential coding sequence both in SARS-CoV lineage and SARS-CoV-2 lineage viruses. We set as test branches all the branches in the phylogenetic tree that led to SARS-CoV-2 lineage species, and as background branches all the branches leading to the SARS-CoV lineage. SLAC and RELAX were performed using the datamonkey webtool (https://www.datamonkey.org/).
2.5. Protein domain prediction
Transmembrane domains were predicted using different methods: TMpred (https://embnet.vital-it.ch/software/TMPRED_form.html), Phobius (Kall et al., 2007), and the MEMSAT-SVM (Nugent and Jones, 2010) tool from PSIPRED (Buchan and Jones, 2019). MEMSAT-SVM was also used for the prediction of pore-lining residues.
3. Results
3.1. Sequence conservation and recombination
As mentioned above, in addition to three bat viruses, eight available pangolin viruses display high sequence similarity to SARS-CoV-2 (Supplementary Table S1) (Zhou et al., 2020; Lam et al., 2020; Xiao et al., 2020). The genomes of these viruses were obtained using different approaches and display different levels of coverage. We thus calculated pairwise sequences identities among these genomes. Because our aim was to perform an analysis of interspecies diversity and conservation, we included in our analysis two pangolin virus genomes with less that 0.90 identity and with the highest coverage (Supplementary Fig. S1 and Fig. 1a). Thus, the final genome dataset included SARS-CoV-2 (reference genome, Wuhan-Hu-1), three bat viruses (BatCoV RaTG13, bat-SL-CoVZC45 and bat-SL-CoVZXC21), and the two pangolin viruses (Fig. 1a).
Fig. 1.
Sequence conservation, coding potential, and RNA structure prediction in the SARS-CoV-2 lineage. (a) Phylogenetic tree of SARS-CoV-2 and related animal viruses. The maximum-likelihood tree was calculated using the ORF1a non-recombining region (see panel b). Colored dots indicate viral sequences. (b) Schematic representation of recombination events. Each bar represents the recombinant sequence with the corresponding name and the genomic location of the recombination event. Colors indicate the major and the minor parental sequences (color coding as in panel a); gray color indicates an unknown parental sequence. The location of the spike protein, its receptor-binding domain (RBD), and the receptor-binding motif (RBM) are also shown. (c) Distribution of synonymous site variation. Plot of synonymous sites substitution (dS) along SARS-CoV-2 lineage coding sequences. The brown line indicates relative dS variability calculated as the ratio of the observed over the expected values of dS in a sliding window of 25 codons. The red line shows the corresponding p-value and the dashed line represents the p-value cutoff. A schematic representation of SARS-CoV-2 ORFs is reported; ORFs that are not annotated in the SARS-CoV-2 reference genome (NC_045512.2) are in red (genomic positions 25,457–25,582, 28,284–28,577, and 28,734–28,955 of the reference SARS-CoV-2 genome correspond to ORF3h, ORF9a, and ORF9b). Gray boxes indicate recombination events that were masked in the analysis. The location of the three predicted conserved RNA secondary structures is shown. Structures were rendered using RNAalifold. The color scheme reflects the mutational pattern with respect to the structure. Thus, colors indicate conserved base pairs: from red (conservation of only one base-pair type) to purple (all six base-pair types are found); from dark (all sequences contain this base pair) to light colors (1 or 2 sequences are unable to form this base pair). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Recombination is known to affect evolutionary inference (Martin et al., 2011). We thus screened the genome alignment using four methods implemented in the RDP4 software (Martin et al., 2017). In particular, RDP, GENECONV, MaxChi, and Chimera (Martin et al., 2017; Sawyer, 1989; Martin and Rybicki, 2000; Posada and Crandall, 2001; Smith, 1992) were used due to their good power in previous simulation analyses (Posada and Crandall, 2001; Bay and Bielawski, 2011). Because our aim was to account for recombination signals, we used relatively relaxed criteria to define recombination events (i.e., p value cutoff = 0.05 and detection by at least two methods). Five recombination events were detected (Fig. 1b). One of these involves the RBD domain of the BatCoV RaTG13, BetaCoV/pangolin/Gd/1, and BetaCoV/pangolin/Gx/P1E. We thus split the genomic alignments into three large non-recombining regions, whereas shorter regions involved in recombination events were not analyzed further.
We first used synplot2 to analyze the genomic alignments (Firth, 2014). This program was devised to detect functional elements that overlap with viral coding sequences. To identify such elements, which include ORFs with alternative reading frames and RNA structural elements, synplot2 searches for regions with a statistically significant reduction in the rate of substitution at synonymous sites (dS). The analysis identified six regions of significantly low dS (Fig. 1c). One of these corresponds to the region of programmed −1 ribosomal frameshifting (−1 PRF) between ORFs 1a and 1b. A similar dS reduction was previously reported in this region for SARS-CoV (using an alignment of betacoronaviruses) and indicates the presence of an RNA pseudoknot structure (Firth, 2014; Plant et al., 2005). One signal was present in the nsp3 region within ORF1a, but no potential ORF was found in the corresponding region, in line with the fact that alternative proteins encoded from ORF1a or ORF1b have not been described in coronaviruses. Likewise, no alternative reading frame was detected in the prominent signal within the E (envelope) gene. Notably, a reduction of dS was previously noted in the E gene for other betacoronaviruses, including SARS-CoV (Firth, 2014). Conversely, the dS reduction within ORF3a corresponded to the presence of a potential ORF (we refer to this ORF as ORF3h, for hypothetical, see below) (Fig. 1c and Fig. 2 ). Likewise, the signals in the N gene, some of which did not pass the p value cutoff, might be accounted for by the presence of two additional ORFs, as previously described on the basis of similarity to SARS-CoV (Wu et al., 2020b; Wu et al., 2020a). Failure to reach statistical significance might be due to low power resulting from the small number of sequences.
Fig. 2.
ORF3 Transmembrane regions. Schematic representation of the protein product of ORF3a, with the predicted location of transmembrane (TM) regions. The potential alternative protein encoded by ORF3h is also shown, along with an amino acid alignment of SARS-CoV-2 and related animal viruses. PSIPRED predicted the boxed region to be transmembrane (dark orange) and pore-lining (blue profile). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As in the case of the −1 PRF, where an RNA pseudoknot was described (Plant et al., 2005), a dS reduction might indicate the presence of RNA secondary structures. We thus used RNAz (Washietl et al., 2005) to analyze the genome alignments for the presence of conserved RNA structures. As expected, the RNA pseudoknot was not predicted, as RNAz is not devised to predict this kind of structures (Hamada, 2015). Using very conservative criteria (see methods), we detected three conserved secondary structures (Fig. 1c, Supplementary Table S2). The first is located within ORF1a, at the 3′ boundary of one of the recombination events, the second is within ORF3a, and the last one covers almost entirely the E gene. Clearly, structure conservation might be secondary to sequence conservation (Washietl et al., 2005). We thus repeated the RNAz analysis on a genome alignment that included more divergent coronaviruses (SARS-CoV lineage, Supplementary Table S2). Using the same criteria as above, the only highly conserved RNA structure we detected was the one overlapping with the E gene. Together with the strong reduction of dS we observed in the region, this result supports the presence of a conserved functional RNA element.
3.2. Coding potential of SARS-CoV-2
The coding potential of SARS-CoV-2 has mainly been explored by comparison with SARS-CoV. At the amino acid level, the percentage of identity between proteins encoded by the two viruses ranges from ~69% (ORF6) to ~96% (ORF1b and E), with the exclusion of ORF8 (Zhou et al., 2020). SARS-CoV strains of the early epidemic phase acquired a 29-nucleotide deletion which split ORF8 in two functional ORFs (ORF8a and ORF8b) (Chinese SARS Molecular Epidemiology Consortium, 2004). SARS-CoV-2 potentially encodes an ORF8 protein of similar length as the full-length ORF8 of early SARS-Cov isolates, however identity (32%) is definitely lower than for the other proteins (Zhou et al., 2020).
Different annotations exist for the internal ORFs of SARS-CoV-2 (Zhou et al., 2020; Wu et al., 2020b; Wu et al., 2020a; Chan et al., 2020). The potential alternative protein encoded by ORF3 (ORF3h) that we predicted based on the significantly low dS value is 41 amino acid long and does not correspond to those previously reported (23 and 57 amino acids in size) (Wu et al., 2020a, Chan et al., 2020). Inspection of the SARS-CoV genome indicated that protein 3h is potentially encoded by this virus, as well, and the identity with the SARS-CoV-2 protein is 90%. Analysis of ORF3h protein domains indicated the presence of a predicted transmembrane helix possibly involved in pore formation (see methods).
As for the two putative ORFs within the N gene, the results of synplot2 were not conclusive, as multiple signals were observed, but most did not reach statistical significance. Two ORFs (9a and 9b) of 97 and 73 amino acids are predicted from the genome sequence of SARS-CoV-2 and related viruses (Wu et al., 2020b). synplot2 results provide stronger support for ORF9a. The predicted protein has 72% identity with the corresponding product encoded by SARS-CoV.
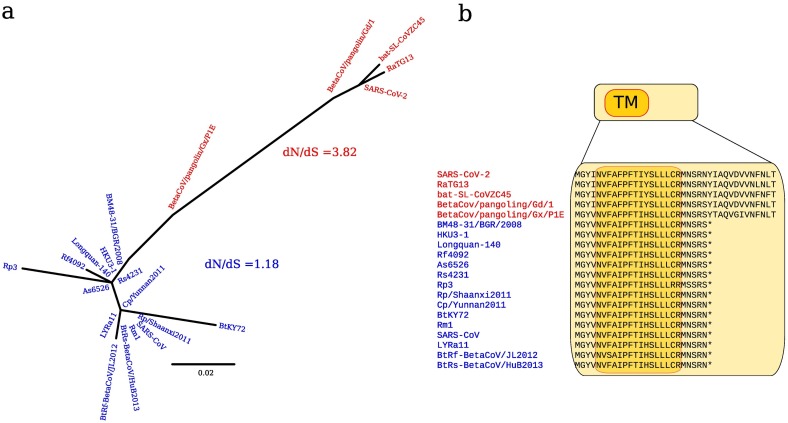
Compared to SARS-CoV, the genome of SARS-CoV-2 may encode an additional ORF at the genomic 3′ end (designated as ORF10) (Wu et al., 2020b) (Fig. 1c). The predicted protein product of ORF10 is 38 amino acids long and is potentially encoded by the pangolin and bat viruses we analyzed, with the exception of bat-SL-CoVZXC21, in which a one nucleotide deletion alters the reading frame. It is presently impossible to determine whether this is the result of a sequencing error or of a mutation event in this virus. Analysis of more distantly related coronaviruses (SARS-CoV lineage) indicated that the reading frame of ORF10 is interrupted by a stop codon (Fig. 3 ). To assess the potential functional relevance of ORF10, we calculated the nonsynonymous over synonymous substitution rate (dN/dS). For viruses potentially encoding the full length protein, dN/dS resulted equal to 3.82, whereas a value close to selective neutrality (dN/dS = 1.18) was observed for viruses carrying a truncated ORF10 protein (Fig. 3). High values of dN/dS may be due to either positive selection or to relaxed purifying selection. To disentangle these possibilities, we applied the RELAX method, which evaluates if selection on the test branches (encoding full-length protein) is relaxed (k < 1) compared to background branches (encoding truncated protein) (see methods) (Wertheim et al., 2015). We obtained a k = 5.65, which excludes the hypothesis of relaxed constraint. Overall, this suggests that ORF10 is evolving under positive selection in viruses related to SARS-CoV-2. Analysis of the potential ORF10 protein indicated the presence of a predicted transmembrane domain.
Fig. 3.
(a) ORF10 phylogenetic tree for the SARS-CoV-2 and the SARS-CoV lineages. Viral sequences belonging to the SARS-CoV-2 lineage are colored in red, viruses belonging to the SARS-CoV lineage are colored in blue. Their corresponding dN/dS values, calculated using SLAC, are also reported. (b) ORF10 protein alignment. An amino acid alignment of the same viral sequences is shown, along with transmembrane region (TM) prediction (dark orange) by PSIPRED. Asterisks indicate stop codons. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To formally test if positive selection has shaped the diversity of the coding sequences of SARS-CoV-2 and related viruses, we applied the likelihood ratio tests (LRT) implemented in the PAML package (Yang, 2007; Yang, 1997). Specifically, we used the codeml program to compare a models of gene evolution that allow (NSsite model M8, positive selection models) or disallow (NSsite models M7 and M8a, null models) a class of codons to evolve with dN/dS >1. The analysis was performed for ORF1a, 1b, M, 7a, and 8; for ORFs N and 3a, only the portion that does not overlap with the internal reading frames was analyzed. Other ORFs (6, 7b), including ORF10, were not analyzed as the power of the LRTs for short ORFs and few sequences is extremely low. The E ORF was not analyzed because of its constraints on dS. No evidence of positive selection was detected for any ORF (not shown).
4. Discussion
Coronaviruses have long and complex genomes with high plasticity in terms of gene content. This feature is thought to contribute to their ability to adapt to specific hosts and to facilitate host shifts (Cui et al., 2019; Forni et al., 2017). It is therefore essential to characterize coronavirus genomes in terms of coding potential and functional elements. In the case of SARS-CoV-2, the little we know about its genetic make up mainly derives from comparison with SARS-CoV, which has been extensively studied since its emergence as a human pathogen in 2002. However, the coding potential of SARS-CoV-2 is still uncertain and the presence of non-coding functional elements is poorly explored, even for SARS-CoV. By allowing the detection of evolutionary constrained sequences, comparative genomic approaches can provide clues about the presence of functional elements, either coding or non-coding. We thus focused on viruses that are closely related to SARS-CoV-2 and we used evolutionary inference to characterize their genomes. By searching for signals of significantly low dS, we identified six genomic regions, one of these associated with the −1 PRF. The most prominent signal of dS reduction was observed within the E gene and corresponded to the presence of a conserved RNA structure, which is shared among SARS-CoV-2 related viruses and the SARS-CoV lineage. RNA secondary structures are increasingly recognized as functional elements within RNA virus genomes. For coronaviruses, the best characterized RNA secondary structure elements are those located in the genomic 5′ and 3′ ends, which play essential roles in viral transcription and replication (Yang and Leibowitz, 2015). We did not detect these elements as we only focused on coding regions, where dS reduction can be calculated. In addition to the UTRs, the role of an RNA pseudoknot in programmed −1 frameshifting was mentioned above and is a conserved feature of coronaviruses (Firth, 2014; Plant et al., 2005; Irigoyen et al., 2016). However, recent experimental data indicated that structured RNA elements are pervasive throughout the genomes of RNA viruses (Boerneke et al., 2019). For instance, extensive probing of several human-infecting viruses (e.g. HCV, HIV, ZIKV, DENV) revealed that RNA elements have diverse functions and often modulate viral fitness by regulation of viral transcription, protein synthesis, and replication, as well as by favoring the evasion of the host immune responses (Boerneke et al., 2019). For instance, HCV adopts secondary RNA structures that minimize cleavage by RNAse L and recognition by protein kinase R, two major components of the innate immune response (Mauger et al., 2015). Whereas some RNA structures in viral genomes were found to be conserved across species or even genera, others are specific to single species and even strains. Emblematic is the case of an RNA element which is only present in the epidemic Asian ZIKV lineage and absent in the African lineage (Li et al., 2018). This element engages into long-range intramolecular interactions eventually modulating virus infectivity (Li et al., 2018).
The two additional conserved RNA structure elements we detected were not associated with a statistically significant reduction in the degree of variability at synonymous sites. This might indicate that they are less strongly constrained or that we had insufficient power to detect a dS reduction in the corresponding regions. One of these structures was located at the 3′ end of one of the recombination events we detected. Notably, RNA secondary structures were proposed to play a role in recombination in coronaviruses and other positive sense RNA viruses, although the mechanisms and effective role of structural elements is still matter of debate (Bentley and Evans, 2018; Rowe et al., 1997). In general, only experimental analysis will shed light on the functional role of the RNA secondary structures we detected and on their relevance for SARS-CoV-2 fitness.
We also detected two regions of reduced variability at sysnonymous sites (one in the nsp3 region and the other in ORF6) which were not associated with the presence of alternative reading frames nor with prediction of secondary structure elements. However, we run RNAz with a recommended window size of 120 nucleotides, which might fail to detect short structural motifs. Thus, these unexplained signals might represent additional conserved RNA structures. Further analysis, possibly with a larger number of SARS-CoV-2-related genomes will clarify this point. Conversely, two regions of reduced dS corresponded to alternative reading frames (ORF9a and ORF3h). The presence ORF9a was previously proposed (Wu et al., 2020b) and the encoded predicted protein shares homology with a multifunctional, well characterized protein (9b) of SARS-CoV (Xu et al., 2009; Meier et al., 2006). The 9b protein encoded by SARS-CoV is incorporated into mature virions (Xu et al., 2009) but is dispensable for viral replication in vitro and in vivo ( von Brunn et al., 2007 ; DeDiego et al., 2008 ). When retained in the nucleus, protein 9b can induce caspase-dependent apoptosis, but its role in SARS-CoV pathogenesis and virulence is unknown (Sharma et al., 2011). Conversely, ORF3h was not previously described. The predicted product of ORF3h, a 40 amino acid protein with a single transmembrane domain, shares high homology with the corresponding predicted product encoded by SARS-CoV and, interestingly, displays features suggestive of a viroporin. Coronaviruses are known to encode diverse viroporins, which were acquired through distinct processes and often independently of each other (Forni et al., 2017). Whereas some of these proteins are multi-pass membrane proteins, other are short, with a single transmembrane domain. For instance, the 8a protein encoded by SARS-CoV is a 39 amino acid long protein with a single transmembrane domain. The protein oligomerizes to form ion channels and localizes to the mitochondria, where it can induce apoptosis by depolarization of the membrane potential (Chen et al., 2011; Chen et al., 2007). Several coronaviruses encode two viroporins, whereas SARS-CoV encodes three proteins with channel-forming activity (3a, E, and 8a) (Castano-Rodriguez et al., 2018), suggesting that multiple viroporins are beneficial for the virus. Because viroporins can modulate virulence and are regarded as possible targets for antivirals (Forni et al., 2017; Royle et al., 2015), it will be important to determine whether the 3h protein is translated and if it displays channel-forming activity. It is also worth noting that SARS-CoV encodes a 3b protein (154 amino acids in size) that is translated in a different frame than 3a and inhibits interferon (IFN) production and signaling (Kopecky-Bromberg et al., 2007). Based on the synplot2 results, the 3b protein was not predicted to be encoded by SARS-CoV-2, as no dS reduction was observed other than the putative ORF3h. In line with this observation, analysis of ORF3 revealed that the reading frame for the 3b protein is interrupted by a STOP codon at position 23, indicating that SARS-CoV-2 lacks this IFN antagonist. However, as previously reported, the function of the 3b protein of SARS-CoV is redundant with those of ORF6 (with an effect on both IFN production and signaling) and N (which only affects IFN production), suggesting that distinct, although related coronaviruses display different arrays of immunomodulatory proteins (Kopecky-Bromberg et al., 2007).
Finally, we investigated the evolutionary history of the putative ORF10, which, in its full-length form, appears to be specific for SARS-CoV-2 and related coronaviruses. Calculation of dN/dS revealed a value of 3.82 in these viruses, whereas in the SARS-CoV lineage the ORF encoding a truncated protein appears to be neutrally evolving. These data suggest that ORF10 might encode a functional protein in SARS-CoV-2 and that positive selection is driving its evolution. Again, additional data, including genomes of other coronaviruses encoding a full-length protein, as well as experimental analyses, will be required to test this hypothesis. Indeed, a clear limitation of our study lies in the quality and paucity of genomes from viruses related to SARS-CoV-2. Available sequences were obtained using different methods: they most likely contain errors and display missing information in several regions (especially the genomes deriving from pangolins). Whereas this is unlikely to strongly affect most results, especially analyses that were performed over sliding windows and thus rely signals from relatively long regions, the availability of additional genomes will indubitably increase the power to detect selective events and the confidence with which evolutionary patterns are inferred. For instance, the lack of evidence of positive selection should not be regarded as conclusive, but might be due to the low statistical power of the LRT with as few as six sequences.
The following are the supplementary data related to this article.
Pairwise identity matrix for pangolin coronaviruses.
List of analyzed strains.
Detection of RNA structures.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgments
Acknowledgments
We gratefully acknowledge the authors, originating and submitting laboratories of the sequences from GISAID's EpiCoV™ database on which this research is based. The list is in supplementary Table S1.
Funding sources
This work was supported by the Italian Ministry of Health (“Ricerca Corrente 2019-2020” to MS, “Ricerca Corrente 2018-2020” to DF).
References
- Bay R.A., Bielawski J.P. Recombination detection under evolutionary scenarios relevant to functional divergence. J. Mol. Evol. 2011;73:273–286. doi: 10.1007/s00239-011-9473-0. [DOI] [PubMed] [Google Scholar]
- Bentley K., Evans D.J. Mechanisms and consequences of positive-strand RNA virus recombination. J. Gen. Virol. 2018;99:1345–1356. doi: 10.1099/jgv.0.001142. [DOI] [PubMed] [Google Scholar]
- Bernhart S.H., Hofacker I.L., Will S., Gruber A.R., Stadler P.F. RNAalifold: improved consensus structure prediction for RNA alignments. BMC Bioinformatics. 2008;9 doi: 10.1186/1471-2105-9-474. 474-2105-9-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerneke M.A., Ehrhardt J.E., Weeks K.M. Physical and functional analysis of viral RNA genomes by SHAPE. Annu. Rev. Virol. 2019;6:93–117. doi: 10.1146/annurev-virology-092917-043315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Brunn A., Teepe C., Simpson J.C., Pepperkok R., Friedel C.C., Zimmer R., Roberts R., Baric R., Haas J. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PLoS One. 2007;2 doi: 10.1371/journal.pone.0000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan D.W.A., Jones D.T. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019;47:W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castano-Rodriguez C., Honrubia J.M., Gutierrez-Alvarez J., DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeno J.M., Regla-Nava J.A., Fernandez-Delgado R., Verdia-Baguena C., Queralt-Martin M., Kochan G., Perlman S., Aguilella V.M., Sola I., Enjuanes L. Role of severe acute respiratory syndrome coronavirus Viroporins E, 3a, and 8a in replication and pathogenesis. mBio. 2018;9:e02325–17. doi: 10.1128/mBio.02325-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F., Kok K.H., Zhu Z., Chu H., To, K.K, Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.C., Kruger J., Sramala I., Hsu H.J., Henklein P., Chen Y.M., Fischer W.B. ORF8a of SARS-CoV forms an ion channel: experiments and molecular dynamics simulations. Biochim. Biophys. Acta. 2011;1808:572–579. doi: 10.1016/j.bbamem.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.Y., Ping Y.H., Lee H.C., Chen K.H., Lee Y.M., Chan Y.J., Lien T.C., Jap T.S., Lin C.H., Kao L.S., Chen Y.M. Open reading frame 8a of the human severe acute respiratory syndrome coronavirus not only promotes viral replication but also induces apoptosis. J. Infect. Dis. 2007;196:405–415. doi: 10.1086/519166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinese SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303:1666–1669. doi: 10.1126/science.1092002. [DOI] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dediego M.L., Pewe L., Alvarez E., Rejas M.T., Perlman S., Enjuanes L. Pathogenicity of severe acute respiratory coronavirus deletion mutants in hACE-2 transgenic mice. Virology. 2008;376:379–389. doi: 10.1016/j.virol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth A.E. Mapping overlapping functional elements embedded within the protein-coding regions of RNA viruses. Nucleic Acids Res. 2014;42:12425–12439. doi: 10.1093/nar/gku981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Delsuc F., Dufayard J.F., Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol. Biol. 2009;537:113–137. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- Haijema B.J., Volders H., Rottier P.J. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 2003;77:4528–4538. doi: 10.1128/JVI.77.8.4528-4538.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada M. RNA secondary structure prediction from multi-aligned sequences. Methods Mol. Biol. 2015;1269:17–38. doi: 10.1007/978-1-4939-2291-8_2. [DOI] [PubMed] [Google Scholar]
- Irigoyen N., Firth A.E., Jones J.D., Chung B.Y., Siddell S.G., Brierley I. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall L., Krogh A., Sonnhammer E.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg S.A., Martinez-Sobrido L., Frieman M., Baric R.A., Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007;81:548–557. doi: 10.1128/JVI.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond S.L., Frost S.D. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kuo L., Godeke G.J., Raamsman M.J., Masters P.S., Rottier P.J. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 2000;74:1393–1406. doi: 10.1128/jvi.74.3.1393-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam T.T., Shum M.H., Zhu H., Tong Y., Ni X., Liao Y., Wei W., Cheung W.Y., Li W., Li L., Leung G.M., Holmes E.C., Hu Y., Guan Y. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature. 2020 doi: 10.1038/s41586-020-2169-0. [DOI] [PubMed] [Google Scholar]
- Li P., Wei Y., Mei M., Tang L., Sun L., Huang W., Zhou J., Zou C., Zhang S., Qin C.F., Jiang T., Dai J., Tan X., Zhang Q.C. Integrative analysis of Zika virus genome RNA structure reveals critical determinants of viral infectivity. Cell Host Microbe. 2018;24:875–886. doi: 10.1016/j.chom.2018.10.011. e5. [DOI] [PubMed] [Google Scholar]
- Liu P., Jiang J., Wan X., Hua Y., Wang X., Hou F., Chen J., Zou J., Chen J. Are pangolins the intermediate host of the 2019 novel coronavirus (2019-nCoV) ? bioRxiv. 2020 doi: 10.1371/journal.ppat.1008421. 2020.02.18.954628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D., Rybicki E. RDP: detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Martin D.P., Lemey P., Posada D. Analysing recombination in nucleotide sequences. Mol. Ecol. Resour. 2011;11:943–955. doi: 10.1111/j.1755-0998.2011.03026.x. [DOI] [PubMed] [Google Scholar]
- Martin D.P., Murrell B., Khoosal A., Muhire B. Detecting and Analyzing genetic recombination using RDP4. Methods Mol. Biol. 2017;1525:433–460. doi: 10.1007/978-1-4939-6622-6_17. [DOI] [PubMed] [Google Scholar]
- Mauger D.M., Golden M., Yamane D., Williford S., Lemon S.M., Martin D.P., Weeks K.M. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc. Natl. Acad. Sci. U. S. A. 2015;112:3692–3697. doi: 10.1073/pnas.1416266112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., Meyerholz D.K., Kirby P., Look D.C., Perlman S. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier C., Aricescu A.R., Assenberg R., Aplin R.T., Gilbert R.J., Grimes J.M., Stuart D.I. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore M.J., Dorfman T., Li W., Wong S.K., Li Y., Kuhn J.H., Coderre J., Vasilieva N., Han Z., Greenough T.C., Farzan M., Choe H. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 2004;78:10628–10635. doi: 10.1128/JVI.78.19.10628-10635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhire B.M., Varsani A., Martin D.P. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent T., Jones D.T. Predicting transmembrane helix packing arrangements using residue contacts and a force-directed algorithm. PLoS Comput. Biol. 2010;6 doi: 10.1371/journal.pcbi.1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79:104212. doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck K.M., Burch C.L., Heise M.T., Baric R.S. Coronavirus host range expansion and Middle East respiratory syndrome coronavirus emergence: biochemical mechanisms and evolutionary perspectives. Annu. Rev. Virol. 2015;2:95–117. doi: 10.1146/annurev-virology-100114-055029. [DOI] [PubMed] [Google Scholar]
- Plant E.P., Perez-Alvarado G.C., Jacobs J.L., Mukhopadhyay B., Hennig M., Dinman J.D. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D., Crandall K.A. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe C.L., Fleming J.O., Nathan M.J., Sgro J.Y., Palmenberg A.C., Baker S.C. Generation of coronavirus spike deletion variants by high-frequency recombination at regions of predicted RNA secondary structure. J. Virol. 1997;71:6183–6190. doi: 10.1128/jvi.71.8.6183-6190.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle J., Dobson S.J., Muller M., Macdonald A. Emerging roles of Viroporins encoded by DNA viruses: novel targets for antivirals? Viruses. 2015;7:5375–5387. doi: 10.3390/v7102880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S. Statistical tests for detecting gene conversion. Mol. Biol. Evol. 1989;6:526–538. doi: 10.1093/oxfordjournals.molbev.a040567. [DOI] [PubMed] [Google Scholar]
- Schickli J.H., Thackray L.B., Sawicki S.G., Holmes K.V. The N-terminal region of the murine coronavirus spike glycoprotein is associated with the extended host range of viruses from persistently infected murine cells. J. Virol. 2004;78:9073–9083. doi: 10.1128/JVI.78.17.9073-9083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Akerstrom S., Sharma A.K., Chow V.T., Teow S., Abrenica B., Booth S.A., Booth T.F., Mirazimi A., Lal S.K. SARS-CoV 9b protein diffuses into nucleus, undergoes active Crm1 mediated nucleocytoplasmic export and triggers apoptosis when retained in the nucleus. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Washietl S., Hofacker I.L., Stadler P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim J.O., Murrell B., Smith M.D., Kosakovsky Pond S.L., Scheffler K. RELAX: detecting relaxed selection in a phylogenetic framework. Mol. Biol. Evol. 2015;32:820–832. doi: 10.1093/molbev/msu400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong M.C., Javornik Cregeen S.J., Ajami N.J., Petrosino J.F. Evidence of recombination in coronaviruses implicating pangolin origins of nCoV-2019. bioRxiv. 2020 2020.02.07.939207. [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020 doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K., Zhai J., Feng Y., Zhou N., Zhang X., Zou J., Li N., Guo Y., Li X., Shen X., Zhang Z., Shu F., Huang W., Li Y., Zhang Z., Chen R., Wu Y., Peng S., Huang M., Xie W., Cai Q., Hou F., Liu Y., Chen W., Xiao L., Shen Y. Isolation and characterization of 2019-nCoV-like coronavirus from Malayan pangolins. bioRxiv. 2020 2020.02.17.951335. [Google Scholar]
- Xu K., Zheng B.J., Zeng R., Lu W., Lin Y.P., Xue L., Li L., Yang L.L., Xu C., Dai J., Wang F., Li Q., Dong Q.X., Yang R.F., Wu J.R., Sun B. Severe acute respiratory syndrome coronavirus accessory protein 9b is a virion-associated protein. Virology. 2009;388:279–285. doi: 10.1016/j.virol.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Leibowitz J.L. The structure and functions of coronavirus genomic 3′ and 5′ ends. Virus Res. 2015;206:120–133. doi: 10.1016/j.virusres.2015.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhang T., Wu Q., Zhang Z. Pangolin homology associated with 2019-nCoV. bioRxiv. 2020 2020.02.19.950253. [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pairwise identity matrix for pangolin coronaviruses.
List of analyzed strains.
Detection of RNA structures.