To the Editor,
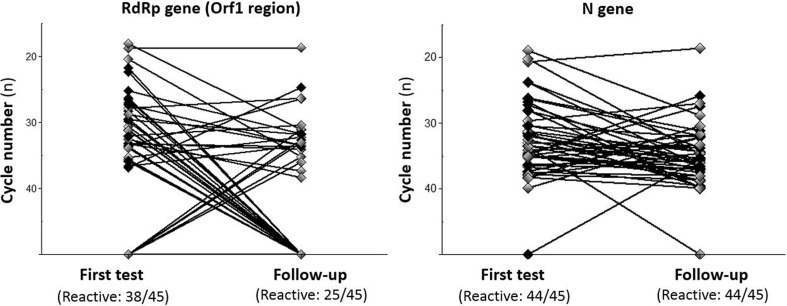
The recent publication by Lv et al. [1] highlighted notable aspects of the PCR-based diagnosis of SARS-CoV-2 infection, namely that the PCR-based detectability of certain loci might differ significantly and change over time. In our practice we also observed that the SARS-CoV-2 PCR positivity in the oropharyngeal swabs of patients might persist for weeks [2], however, the characteristics of positivity (including the target sequences detected) may change in a predictable manner. Although the available test kits offer convenient laboratory workflow and clear instructions for the interpretation of real-time PCR outcome data, critical interpretation of the results may provide additional information. In line with the case presented by Lv et al. we frequently observed a clearly detectable N gene in combination with an absent Orf1a (Open reading frame 1a) or RNA dependent RNA polymerase (RdRp sequence within Orf1 region) in follow-up samples. We aimed to specify this phenomenon. We performed a retrospective analysis of the data of 45 consecutive patients with a positive baseline and at least one positive follow-up test result, both of which we had received using the same PCR assay kit (Allplex™ 2019-nCoV Assay, Seegene Inc., Seoul, Republic of Korea) and thermal cycler (BioRad C1000 Real-time PCR system, BioRad Inc., Hercules, CA, United States). Combined nasal and oropharyngeal swabs were processed, results were analyzed qualitatively. Kyplot 5.0 software (KyensLab Inc., Tokyo, Japan) was used for statistical analysis. The avarage time between baseline and follow-up testing was 6.7 ± 5.1 days. Initial RdRp detectability in 19 patients became undetectable during follow-up. The N gene did not show a similar trend (see Fig. 1 ). A statistically significant difference was observed between the proportions of RdRp positive patient samples at initial and follow-up testing (38/45 vs. 25/45 samples, χ2 p = 0.006). Of note, those becoming RdRp-negative were re-tested after a significantly longer period (9.6 ± 5.2 days vs. 4.6 ± 3.8 in the other 26 patients, T-test p < 0.01). Our results might indicate that the detectability of the viral Orf1a-related RdRp sequence might fade more rapidly during convalescence, only later followed by the N gene. This phenomenon might affect the interpretation of the results, as detecting a lone N gene might suggest a later presentation within the course of the disease. This trend might also be coherent with the gradual expression of genes encoding different virion components. Orf1 (containing RdRp) is among the earliest sequences transcribed and translated, only later followed by the expression of nucleocapsid and coat protein genes [3]. However, certain patients initially presented with an undetectable RdRp, and subsequently became positive. This might raise the possibility of other causes, including a more unstable nature of the Orf1 RNA sequence or even sampling bias, as a lower quantity of virions might result in losing weak signals. Further, large-scale studies are needed to clarify, whether these observations could be diagnostically utilized.
Fig. 1.
The qualitative real-time PCR results of the 45 consecutive patients. Dots indicate real-time PCR cycle numbers, results of the same patients are connected.
Disclosure
The authors have no financial or commercial conflict of interest to disclose.
References
- 1.Lv D., Ying Q., Weng Y., Shen C., Chu J., Kong J., Sun D., Gao X., Weng X., Chen X. Dynamic change process of target genes by RT-PCR testing of SARS-Cov-2 during the course of a Coronavirus Disease patient. Clin. Chim. Acta Int. J. Clin. Chem. 2019;506(2020):172–175. doi: 10.1016/j.cca.2020.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiao A.T., Tong Y.X., Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020 doi: 10.1093/cid/ciaa460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Coronaviruses. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]