Abstract
SARS-CoV-2 is a SARS-like coronavirus of likely zoonotic origin first identified in December 2019 in Wuhan, the capital of China's Hubei province. The virus has since spread globally, resulting in the currently ongoing COVID-19 pandemic. The first whole genome sequence was published on January 5 2020, and thousands of genomes have been sequenced since this date. This resource allows unprecedented insights into the past demography of SARS-CoV-2 but also monitoring of how the virus is adapting to its novel human host, providing information to direct drug and vaccine design. We curated a dataset of 7666 public genome assemblies and analysed the emergence of genomic diversity over time. Our results are in line with previous estimates and point to all sequences sharing a common ancestor towards the end of 2019, supporting this as the period when SARS-CoV-2 jumped into its human host. Due to extensive transmission, the genetic diversity of the virus in several countries recapitulates a large fraction of its worldwide genetic diversity. We identify regions of the SARS-CoV-2 genome that have remained largely invariant to date, and others that have already accumulated diversity. By focusing on mutations which have emerged independently multiple times (homoplasies), we identify 198 filtered recurrent mutations in the SARS-CoV-2 genome. Nearly 80% of the recurrent mutations produced non-synonymous changes at the protein level, suggesting possible ongoing adaptation of SARS-CoV-2. Three sites in Orf1ab in the regions encoding Nsp6, Nsp11, Nsp13, and one in the Spike protein are characterised by a particularly large number of recurrent mutations (>15 events) which may signpost convergent evolution and are of particular interest in the context of adaptation of SARS-CoV-2 to the human host. We additionally provide an interactive user-friendly web-application to query the alignment of the 7666 SARS-CoV-2 genomes.
Keywords: Betacoronavirus, Homoplasies, Mutation, Phylogenetics
Highlights
-
•
Phylogenetic estimates support that the COVID-2 pandemic started sometimes around 6 October 2019 – 11 December 2019.
-
•
The diversity of SARS-CoV-2 strains in many countries recapitulates its entire global diversity.
-
•
198 sites in the SARS-CoV-2 genome appear to have already undergone recurrent, independent mutations.
-
•
Detected recurrent mutations may indicate ongoing adaptation of SARS-CoV-2 to its novel human host.
-
•
Monitoring the build-up of genetic diversity in SARS-CoV-2 has potential to inform on targets for drugs and vaccines.
1. Introduction
On December 31 2019, China notified the World Health Organisation (WHO) about a cluster of pneumonia cases of unknown aetiology in Wuhan, the capital of the Hubei Province. The initial evidence was suggestive of the outbreak being associated with a seafood market in Wuhan, which was closed on January 1 2020. The aetiological agent was characterised as a SARS-like betacoronavirus, later named SARS-CoV-2, and the first whole genome sequence (Wuhan-HU-1) was deposited on NCBI Genbank on January 5 2020 (Wu et al., 2020). Human-to-human transmission was confirmed on January 14 2020, by which time SARS-CoV-2 had already spread to many countries throughout the world. Further extensive global transmission led to the WHO declaring COVID-19 as a pandemic on March 11 2020.
Coronaviridae comprise a large number of lineages that are found in a wide range of mammals and birds (Shaw et al., 2020), including the other human zoonotic pathogens SARS-CoV-1 and MERS-COV. The propensity of Betacoronaviridiae to undergo frequent host jumps supports SARS-CoV-2 also being of zoonotic origin. To date, the genetically closest-known lineage is found in horseshoe bats (BatCoV RaTG13) (Zhou et al., 2020). However, this lineage shares 96% identity with SARS-CoV-2, which is not sufficiently high to implicate it as the immediate ancestor of SARS-CoV-2. The zoonotic source of the virus remains unidentified at the date of writing (April 23 2020).
The analysis of genetic sequence data from pathogens is increasingly recognised as an important tool in infectious disease epidemiology (Rambaut et al., 2008; Grenfell et al., 2004). Genetic sequence data sheds light on key epidemiological parameters such as doubling time of an outbreak/epidemic, reconstruction of transmission routes and the identification of possible sources and animal reservoirs. Additionally, whole-genome sequence data can inform drug and vaccine design. Indeed, genomic data can be used to identify pathogen genes interacting with the host and allows characterisation of the more evolutionary constrained regions of a pathogen genome, which should be preferentially targeted to avoid rapid drug and vaccine escape mutants.
There are thousands of global SARS-CoV-2 whole-genome sequences available on the rapid data sharing service hosted by the Global Initiative on Sharing All Influenza Data (GISAID; https://www.epicov.org) (Elbe and Buckland-Merrett, 2017; Shu and McCauley, 2017). The extraordinary availability of genomic data during the COVID-19 pandemic has been made possible thanks to a tremendous effort by hundreds of researchers globally depositing SARS-CoV-2 assemblies (Table S1) and the proliferation of close to real time data visualisation and analysis tools including NextStrain (https://nextstrain.org) and CoV-GLUE (http://cov-glue.cvr.gla.ac.uk).
In this work we use this data to analyse the genomic diversity that has emerged in the global population of SARS-CoV-2 since the beginning of the COVID-19 pandemic, based on a download of 7710 assemblies. We focus in particular on mutations that have emerged independently multiple times (homoplasies) as these are likely candidates for ongoing adaptation of SARS-CoV-2 to its novel human host. After filtering, we characterise homoplasies at 198 sites in the SARS-CoV-2 genome. We identify a strong signal of recurrent mutation at nucleotide position 11,083 (Codon 3606 Orf1a), together with two further sites in Orf1ab encoding the non-structural proteins Nsp11 and Nsp13. These, together with a mutation in the Spike protein (21,575, Codon 5), comprise the strongest putative regions under selection in our dataset.
The current distribution of genomic diversity as well as ongoing allele frequency changes both between isolates and along the SARS-CoV-2 genome are publicly available as an open access and interactive web-resource available here:
2. Material and methods
2.1. Data acquisition
7710 SARS-CoV-2 assemblies flagged as “complete (>29,000 bp)”, “high coverage only”, “low coverage excl” were downloaded from the GISAID Initiative EpiCoV platform as of April 19 2020 (11:30 GMT). A full acknowledgements table of those labs which generated and uploaded data is provided in Table S1. Filtering was performed on the downloaded assemblies to exclude those deriving from animals (bat, pangolin), those with more than 1% missing sites, and otherwise spurious assemblies as also listed by nCov-GLUE (http://cov-glue.cvr.gla.ac.uk/#/excludedSeqs). This left a final dataset of 7666 assemblies for downstream analysis. Sequence metadata was obtained from the NextStrain Github repository (https://github.com/nextstrain/ncov/tree/master/data). While results presented here predominately focus on an analysis of the available assemblies as of April 19 2020, equivalent analyses were performed daily from March 24 2020. This allowed tracking of the emergence of genomic variants in public sequence data as assemblies were uploaded during the course of the pandemic.
2.2. Multi-sequence alignment and maximum likelihood tree
Assemblies were aligned against the Wuhan-Hu-1 reference genome (NC_045512.2, EPI_ISL_402125) using MAFFT (Katoh and Standley, 2013) implemented via the rapid phylodynamic alignment pipeline provided by Augur (https://github.com/nextstrain/augur). Sites in the first 130 bp and last 50 bp of the alignment were masked, as were positions 18,529, 29,849, 29,851 and 29,853, following the protocol also advocated by NextStrain and to account for the fact many putatively artefactual SNPs are located at the beginning and ends of the alignment. Resulting alignments were manually inspected in UGene (http://ugene.net). Subsequently a maximum likelihood phylogenetic tree was built using the Augur tree implementation selecting RAxML as the tree-building method (Kozlov et al., 2019). The resulting phylogeny was viewed and annotated using ggtree (Yu et al., 2017) (Figs. S1-S2). Throughout, site numbering and genome structure are given using Wuhan-Hu-1 (NC_045512.2) as reference.
2.3. Phylogenetic dating
The maximum likelihood phylogenetic tree was tested for the presence of significant molecular evolution over the sampling period using the roottotip() function provided in BactDating (Didelot et al., 2018). After confirmation of a significant regression following 1000 random permutations of sampling dates (Fig. S3), temporal calibration of the phylogeny was performed using TreeDater (Volz and Frost, 2017), assuming a strict clock model of evolution, as we do not expect a significant difference in rate variation across lineages at these time scales (Fig. S4). To obtain confidence intervals around each temporal point estimate we conducted a parametric bootstrapping analysis with 50 replicates on the unmasked alignment, keeping the tree topology constant while generating new branch length estimates using a Poisson distribution and running the same model in TreeDater (Volz and Frost, 2017). We also evaluated all currently available estimates for tip-calibration estimates of the tMRCA of SARS-CoV-2 together with rate estimates for other closely related betacoronaviruses (Table 1 , Table S2).
Table 1.
Estimates of SARS-CoV-2 time to most recent common ancestor (tMRCA). BCI: Bayesian Credible Interval; HPD: Highest Posterior Density; CI: Confidence Interval. Asterix * denotes non-peer reviewed estimate of tMRCA. ‘N.’ denotes the number of whole genomes analysed.
| Reference | N. | Substitution Rate (per site per year) | Estimated tMRCA | Method |
|---|---|---|---|---|
| Li et al. 2020 (Li et al., 2020) | 32 | 1.0 × 10–3 (95% BCI 1.854 × 10–4, 4.0 × 10–3) | October 15, 2019 (95% BCI May 2, 2019; January 17, 2020) | Rate-informed strict clock model (BEAST v1.8.4) |
| Li et al. 2020 (Li et al., 2020) | 32 | 1.8266 × 10–3 (95% BCI 7.5813 × 10–4, 3.0883 × 10–3) | December 6, 2019 (95% BCI November 16, 2019; December 21, 2019) | Rate-estimated relaxed clock model (BEAST v1.8.4) |
| Giovanetti et al. 2020 (Giovanetti et al., 2020) | 54 | 6.58 × 10–3 (95% HPD 5.2 × 10–3, 8.1 × 10–3) | November 25, 2019 (95% CI September 28, 2019; December 21, 2019) | Relaxed clock model (BEAST v1.10.4) |
| Hill & Rambaut 2020*1 | 75 | 0.92 × 10–3 (95% HPD 0.33 × 10–3–1.46 × 10–3) | November 29, 2019 (95% CI October 28, 2019; December 20, 2019) | Unreported clock model (BEAST v1.7.0) |
| Hill & Rambaut 2020*1 | 86 | 0.80 × 10–3 (95% HPD 0.14 × 10–3, 1.31 × 10–3) | November 17, 2019 (95% CI August 27, 2019; December 19, 2019) | Unreported clock model (BEAST v1.7.0) |
| Hill & Rambaut 2020*1 | 116 | 1.04 × 10–3 (95% HPD 0.71 × 10–3, 1.40 × 10–3) | December 3, 2019 (95% CI November 16, 2019; December 17, 2019) | Unreported clock model (BEAST v1.7.0) |
| Lu et al. 2020* (41) | 53 | − | November 29, 2019 (95% HPD November 14, 2019; December 13, 2019) | Strict clock model (BEAST v1.10.0) |
| Duchene et al. 2020*2 | 47 | 1.23 × 10–4(95% HPD 5.63 × 10–4, 1.98 × 10–3) | November 19, 2019 (HPD October 21, 2019; December 11, 2019) | Strict clock model (BEAST v1.10) |
| Duchene et al. 2020*2 | 47 | 1.29 × 10–3 (HPD 5.35 × 10–4, 2.15 × 10–3) | November 12, 2019 (HPD September 26, 2019; December 11, 2019) | Relaxed clock model (BEAST v1.10) |
| Volz et al. 2020*3 | 53 | Model constrained between 7 × 10–4 & 2 × 10–3 | December 8, 2019 (95% CI November 21, 2019; December 20, 2019) | Strict clock model (BEAST v2.6.0) |
| Volz et al. 2020*3 | 53 | Model constrained between 5 × 10–4 & 1.25 × 10–3 | December 5, 2019(95% CI November 6, 2019; December 13, 2019) | Maximum Likelihood regression (treedater R package v0.5.0) |
2.4. Maximum parsimony tree and homoplasy screen
In parallel a Maximum Parsimony tree was built using the fast tree inference and bootstrap approximation offered by MPBoot (Hoang et al., 2018). MPBoot was run on the alignment to reconstruct the Maximum Parsimony tree and to assess branch support following 1000 replicates (−bb 1000). The resulting Maximum Parsimony treefile was used, together with the input alignment, to rapidly identify recurrent mutations (homoplasies) using HomoplasyFinder (Crispell et al., 2019).
HomoplasyFinder provides, for each site, the minimum number of state changes required on the tree to explain the observed character states at the tips, as described by Fitch (Fitch, 1971), and measured via the site specific consistency index. For this analysis all ambiguous sites in the alignment were set to ‘N’. To assess whether any particular Open Reading Frame (ORF) showed evidence of more homoplasies than expected given the length of the ORF, an empirical distribution was obtained by sampling, with replacement, equivalent length windows and recording the number of homoplasies detected (Table S3).
HomoplasyFinder identified 1132 homoplasies (1042 excluding masked sites), which were distributed over the SARS-CoV-2 genome (Fig. S5, Table S4). Of these, 40 sites have a derived allele at >1% of the total isolates. However, homoplasies can arise due to convergent evolution (putatively adaptive), recombination, or via errors during the processing of sequence data. The latter is particularly problematic here due to the mix of technologies and methods employed by different contributing research groups. We therefore filtered identified homoplasies using a set of thresholds attempting to circumvent this problem (filtering scripts and figures are available at https://github.com/liampshaw/CoV-homoplasy-filtering).
In summary, for each homoplasy we computed the proportion of isolates with the homoplasy p nn where the nearest neighbouring isolate in the phylogeny also carried the homoplasy (excluding identical sequences). This metric ranges between p nn = 0 (all isolates with the homoplasy present as singletons) and p nn = 1 (no singletons i.e. clustering of isolates with the homoplasy in the phylogeny). We reasoned that artefactual sequencing homoplasies would tend to show up as singletons, so excluded all homoplasies with p nn < 0.1 from further analysis.
To obtain a set of high confidence homoplasies, we then used the following criteria: ≥0.1% isolates in the alignment share the homoplasy (equivalent to >8 isolates), p nn > 0.1, and derived allele found in strains sequenced from >1 originating lab and > 1 submitting lab. We also required the proportion of isolates where the homoplasic site was in close proximity to an ambiguous base (± 5 bp) to be zero. The application of these various filters reduced the number of homoplasies to 198 (Table S5). We also plotted the distributions of cophenetic distances between isolates carrying each homoplasy compared to the distribution for all isolates (Fig. S6), and inspected the distribution of all identified homoplasies in the phylogenies from our own analyses and on the phylogenetic visualisation platform provided by NextStrain. Finally, we examined whether ambiguous bases were seen more often at homoplasic sites than at random bases (excluding masked sites), which was not the case (Fig. S7).
To further validate the homoplasy detection method applied to the alignment of the 7666 SARS-CoV-2 genome assemblies, we took advantage of the genome sequences for which raw reads were available on the Short Read Archive (SRA). A variant calling pipeline (available at https://github.com/DamienFr/CoV-homoplasy) was used to obtain high-confidence alignments for the 348 (out of 889 as of April 19 2020) SRA genomic datasets both meeting our quality criterions and matching GISAID assemblies. The topology of the Maximum Likelihood phylogeny of these 348 samples was compared to that of the corresponding samples from the GISAID genome assemblies using a Mantel test and the Phytools R package (Revell, 2012) (Figs. S8-S9, see Supplementary text).
As discussed, the GISAID dataset comprises assemblies of variable quality, potentially impairing the detection of genuine homoplasies and/or leading to false positive SNPs due to sequencing error or spurious allele assignment during the production of the de novo assembly from raw sequence reads. Therefore, to further assess the detection of homoplasies, we applied HomoplasyFinder to the two datasets comprising the same 348 strains (GISAID and SRA) (Table S6). We detected 19 homoplasies on the dataset originating from the SRA, and 21 on the dataset originating from GISAID assemblies. Of these, 19 were detected in both datasets (Table S7). Using the same filters as for the main dataset (with the exception of the ≥0.1% frequency set to ≥1%), 10 and 11 homoplasies were kept in the SRA dataset and in the GISAID dataset, respectively. Nine sites were detected in both datasets. For sites which failed the filtering thresholds, this was largely due to the low number of studied accessions, which increases the probability of an isolated strain displaying a homoplasy e.g. if n = 2 isolates have a homoplasy, by definition they cannot be nearest neighbours, so p nn = 0.
2.5. Annotation of variant and homoplasic sites
The alignment was translated to amino acid sequences using SeaView V4 (Gouy et al., 2010). Sites were identified as synonymous or non-synonymous and amino acid changes corresponding to these mutations were retrieved via multiple sequence alignment. We assessed the change in hydrophobicity and charge of amino acid residues arising due to homoplastic non-synonymous mutations using the hydrophobicity scale proposed by Janin (Janin, 1979). The ten most hydrophobic residues on this scale were considered hydrophobic and the rest as hydrophilic. In addition, amino acid residues were either classified as positively charged, negatively charged or neutral at pH 7. The charge of each residue can either increase, decrease or remain the same (neutral mutation) due to mutation (Fig. S10).
2.6. Comparison with SARS-CoV-1 and MERS-CoV
SARS-CoV-1 and MERS-CoV are both zoonotic pathogens related to SARS-CoV-2, which underwent a host jump into the human host previously. We investigated whether the major homoplasies we detect in SARS-CoV-2 affect sites which also underwent recurrent mutations in these related viruses as these adapted to their human host. All Coronaviridae assemblies were downloaded (NCBI TaxID:11118) on April 8 2020 and human associated MERS-CoV and SARS-CoV-1 assemblies extracted. This gave a total of 15 assemblies for SARS-CoV-1 and 255 assemblies for MERS-CoV. Following the same protocol (Augur align) as applied to SARS-CoV-2 assemblies, each species was aligned against the respective RefSeq reference genomes: NC_004718.3 for SARS-CoV-1 and NC_019843.3 for MERS-CoV. This produced alignments of 29,751 bp (187 SNPs) and 30,119 bp (1588 SNPs) respectively.
MPBoot (Hoang et al., 2018) was run on both sets of alignments to reconstruct the maximum parsimony tree and to assess branch support following 1000 replicates (−bb 1000). The resulting maximum parsimony treefiles were used, together with the input alignment, to rapidly identify homoplasies using HomoplasyFinder (Crispell et al., 2019). For SARS-CoV-1 we detected six homoplasies and for MERS-CoV we detected 350 homoplasies (pre-filtering) (Fig. S11-S12). The distribution of homoplasies was assessed relative to the Genbank annotation files and in the context of the high confidence homoplasies that we detect in SARS-CoV-2.
3. Results
3.1. Emergence of SARS-CoV-2 genomic diversity over time
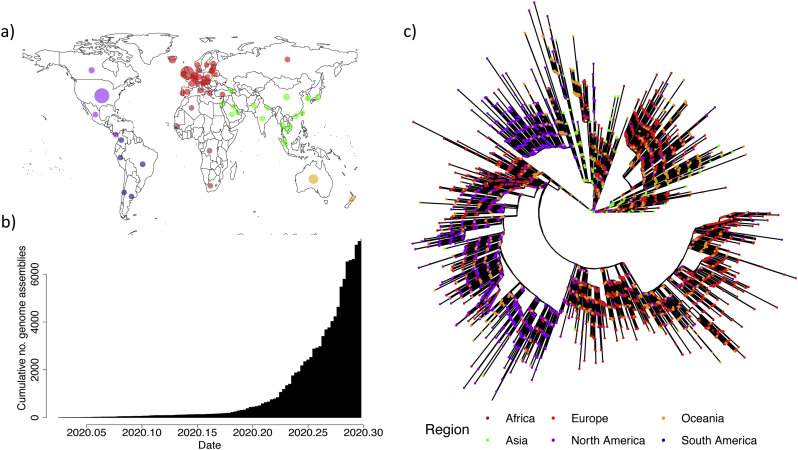
The 7666 SARS-CoV-2 genomes offer an excellent geographical and temporal coverage of the COVID-19 pandemic (Fig. 1a-b). The genomic diversity of the 7666 SARS-CoV-2 genomes is represented as Maximum Likelihood phylogenies in a radial (Fig. 1c) and linear layout (Fig. S1-S2). There is a robust temporal signal in the data, captured by a statistically significant correlation between sampling dates and ‘root-to-tip’ distances for the 7666 SARS-CoV-2 (Fig. S3; R 2 = 0. 20, p < .001). Such positive association between sampling time and evolution is expected to arise in the presence of measurable evolution over the timeframe over which the genetic data was collected. Specifically, more recently sampled strains have accumulated additional mutations in their genome than older ones since their divergence from the Most Recent Common Ancestor (MRCA, root of the tree).
Fig. 1.
Global sequencing efforts have contributed hugely to our understanding of the genomic diversity of SARS-CoV-2. a) Viral assemblies available from global regions as of 19/04/2020. b) Cumulative total of viral assemblies uploaded to GISAID included in our analysis. c) Radial Maximum Likelihood phylogeny for 7666 complete SARS-CoV-2 genomes. Colours represent continents where isolates were collected. Green: Asia; Red: Europe; Purple: North America; Orange: Oceania; Dark blue: South America according to metadata annotations available on NextStrain (https://github.com/nextstrain/ncov/tree/master/data). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The origin of the regression between sampling dates and ‘root-to-tip’ distances (Fig. S3) provides a cursory point estimate for the time to the MRCA (tMRCA) around late 2019. Using TreeDater (Volz and Frost, 2017), we observe an estimated tMRCA, which corresponds to the start of the COVID-19 epidemic, of 6 October 2019–11 December 2019 (95% CIs) (Fig. S4). These dates for the start of the epidemic are in broad agreement with previous estimates performed on smaller subsets of the COVID-19 genomic data using various computational methods (Table 1), though they should still be taken with some caution. Indeed, the sheer size of the dataset precludes the use of some of the more sophisticated inference methods available.
The SARS-CoV-2 global population has accumulated only moderate genetic diversity at this stage of the COVID-19 pandemic with an average pairwise difference of 9.6 SNPs between any two genomes, providing further support for a relatively recent common ancestor. We estimated a mutation rate underlying the global diversity of SARS-Cov-2 of ~6 × 10−4 nucleotides/genome/year (CI: 4 × 10−4–7 × 10−4) obtained following time calibration of the maximum likelihood phylogeny. This rate is largely unremarkable for an RNA virus (Domingo-Calap et al., 2018; Holmes et al., 2016), despite Coronaviridae having the unusual capacity amongst viruses of proofreading during nucleotide replication, thanks to the non-structural protein nsp14 exonuclease, which excises erroneous nucleotides inserted by their main RNA polymerase nsp12 (Snijder et al., 2003; Minskaia et al., 2006).
3.2. Everything is everywhere
Some of the major clades in the maximum likelihood phylogeny (Fig. 1c and Fig. S1) are formed predominantly by strains sampled from the same continent. However, this likely represents a temporal rather than a geographic signal. Indeed, the earliest available strains were collected in Asia, where the COVID-19 pandemic started, followed by extensive genome sequencing efforts first in Europe and then in the USA.
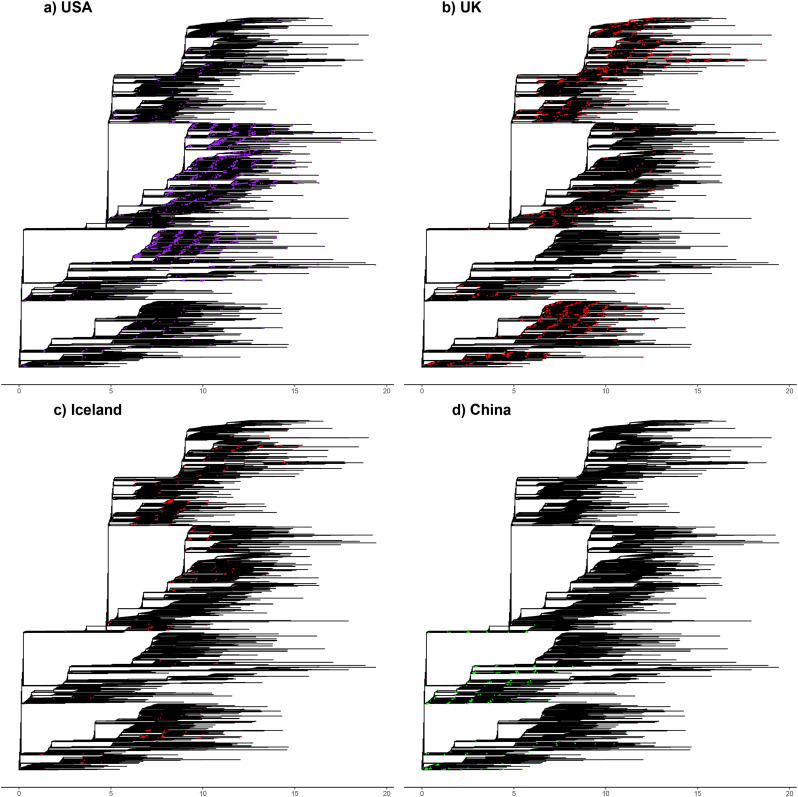
The SARS-CoV-2 genomic diversity found in most countries (with sufficient sequences) essentially recapitulates the global diversity of COVID-19 from the 7666-genome dataset. Fig. 2 highlights the proportion of the global genetic diversity found in the UK, the USA, Iceland and China. In the UK, the USA and Iceland, the majority of the global genetic diversity of SARS-CoV-2 is recapitulated, with representatives of all major clades present in each of the countries (Fig. 2A-C). The same is true for other countries such as Australia (Fig. S2a).
Fig. 2.
Genomic diversity of SARSCoV-2 in the USA, UK, Iceland and China. Strains collected from all four countries are highlighted on the global phylogenetic tree. a) Strains collected in the USA shown in purple. b) Strains from the UK shown in red. c) Strains collected in Iceland shown in red. d) Strains collected in China shown in green. Regional colours match to the global phylogeny shown in Fig. 1c. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
This genetic diversity of SARS-CoV-2 populations circulating in different countries points to each of these local epidemics having been seeded by a large number of independent introductions of the virus. The main exception to this pattern is China, the source of the initial outbreak, where only a fraction of the global diversity is present (Fig. 2d). This is also to an extent the case for Italy (Fig. S2b), which was an early focus of the COVID-19 pandemic. However, this global dataset includes only 35 SARS-CoV-2 genomes from Italy, so some of the genetic diversity of SARS-CoV-2 strains in circulation likely remains unsampled. The genomic diversity of the global SARS-CoV-2 population being recapitulated in multiple countries points to extensive worldwide transmission of COVID-19, likely from extremely early on in the pandemic.
3.3. Genetic diversity along the genome alignment and recurrent mutations (homoplasies)
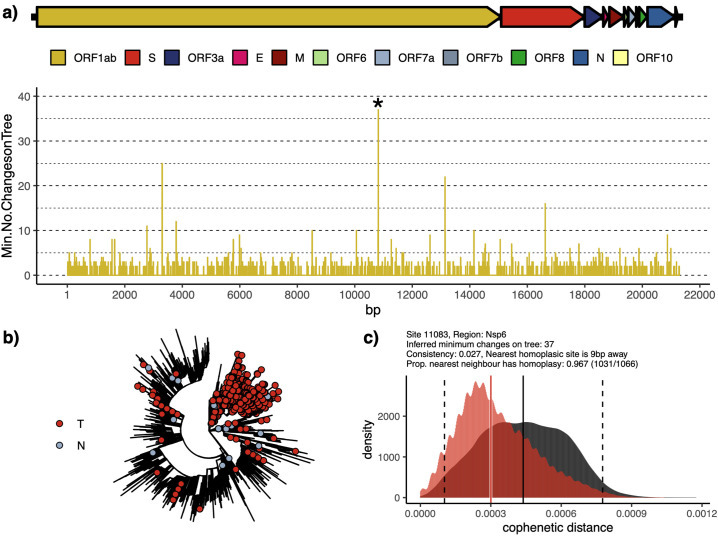
The SARS-CoV-2 alignment can be considered as broken into a large two-part Open Reading Frame (ORF) encoding non-structural proteins, four structure proteins: spike (S), envelope (E), membrane (M) and nucleocapsid (N), and a set of small accessory factors (Fig. 3a). There is variation in genetic diversity across the alignment, with polymorphisms often found in neighbouring clusters (Fig. S5). A simple permutation resampling approach suggests that both Orf3a and N exhibit SNPs which fall in the 95th percentile of the empirical distribution (Table S3). However, not all of these sites can be confirmed as true variant positions, due to the lack of accompanying sequence read data. However, we closely inspected those sites that appear to have arisen multiple times following a maximum parsimony tree building step. We identified a large number of putative homoplasies (n = 1042 excluding masked regions), which were filtered to a high confidence cohort of 198 positions (see Methods).
Fig. 3.
Inspection of a major homoplastic site in Orf1ab of SARS-CoV-2 genome (position 11,083). Panel A shows a colour-coded schematic of the SARS-CoV-2 genome annotated as per NC_045512.2 and a plot of all potential homoplastic sites in Orf1ab measured as minimal number of character-state changes on a Maximum Parsimony tree (see Methods). Exemplar homoplasy (denoted with *) has been shown on the radial ML phylogenetic tree in panel B. Panel C shows the distribution of cophenetic distances between isolates carrying the identified homoplasy (red) and the distribution for all isolates (grey), showing that isolates with the homoplasy tend to cluster in the phylogeny. Equivalent figures for other filtered homoplasies are generated as part of the filtering method (see Methods). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
These 198 positions in the SARS-CoV-2 genome alignment (0.67% of all sites) were associated with 290 amino acid changes across all 7666 genomes. Of these amino acid changes, 232 comprised non-synonymous and 58 comprised synonymous mutations. Two non-synonymous mutations involved the introduction or removal of stop codons were found (*13402Y, *26152G). 53 of the remaining 101 non-synonymous mutations involved neutral hydrophobicity changes (Fig. S10a). In addition, 79 of the remaining 101 non-synonymous mutations involved neutral changes (Fig. S10b). Both Orf1ab and N had a four-fold higher frequency of hydrophilic → hydrophobic mutations than hydrophobic → hydrophilic mutations (Fig. S10). In addition, neutral hydrophobic changes were clearly favoured in the S protein. Lastly, 87 of the remaining 110 non-synonymous mutations involved neutral charge changes.
Amongst the strongest filtered homoplasic sites (>15 change points on the tree), three are found within Orf1ab (nucleotide positions 11,083, 13,402, 16,887) and S (21575). We exemplify the strongest signal and our approach using position 11,083 in Fig. 3 and provide a full list of homoplasic sites, both filtered and unfiltered, in Tables S4–5. The strongest hit in terms of the inferred minimum number of changes required (Fig. 3b-c) at Orf1ab (11,083, Codon 3606) falls over a region encoding the non-structural protein, Nsp6, and is also observed in our analyses of the SRA dataset (Table S7).
We note that some of the hits also overlap with positions identified as putatively under selection using other approaches (http://virological.org/t/selection-analysis-of-gisaid-sars-cov-2-data/448/3, accessed April 23 2020), with Orf1ab consistently identified as a region comprising several candidates for non-neutral evolution. Orf1ab is an orthologous gene with other human-associated betacoronaviruses, in particular SARS-CoV-1 and MERS-CoV which both underwent host jumps into humans from likely bat reservoirs (Lau et al., 2005; Memish et al., 2013). We performed an equivalent analysis on human-associated virus assemblies available on the NCBI Virus platform. We identified six putative homoplasic sites within SARS-CoV-1, two occurring within the 3c-like proteinase just upstream of Nsp6 (10,384, 10,793) and a further two homoplasies within Orf1ab at Nsp9 and Nsp13 (Fig. S11). In addition, one homoplasy was identified in the spike protein and one in the membrane protein ORFs.
For MERS-CoV, multiple unfiltered homoplasies were detected, consistent with previous observations of high recombination in this species (Dudas and Rambaut, 2016), though only one invoked more than a minimum number of 10 changes on the maximum parsimony tree (Fig. S12). This corresponded to a a further homoplasy identified in Orf1ab Nsp6 (position 11,631). It is of note that this genomic region coincides with the strongest homoplasy in SARS-CoV-2 which also occurs in the Nsp6 encoding region of Orf1ab. Codon 3606 of Orf1ab shares a leucine residue in MERS-CoV and SARS-CoV-2, though a valine in SARS-CoV. The exact role of these and other homoplasic mutations in human associated betacoronaviruses represents an important area of future work, although it appears that the Orf1ab region may exhibit multiple putatively adapted variants across human betacoronavirus lineages.
The genome alignment of the 7666 SARS-CoV-2 genomes can be queried through an open access, interactive web-application (https://macman123.shinyapps.io/ugi-scov2-alignment-screen/). It provides users with information on every SNP and homoplasy detected across our global SARS-CoV-2 alignment and allows visual inspection both within the sequence alignment and across the maximum likelihood tree phylogeny. Fig. 3 illustrates some of the functionalities of the web application using position 11083 in the alignment as an example. This particular homoplasy was observed 1126 times across the genomes and requires a minimum of 37 character-site changes to become congruent with the observed SARS-CoV-2 phylogeny (Fig. 3a and b).
4. Discussion
Pandemics have been affecting humanity for millennia (Balloux and van Dorp, 2017). Over the last century alone, several global epidemics have claimed millions of lives, including the 1957/58 influenza A (H2N2) pandemic, the sixth (1899–1923) and seventh ‘El Tor’ cholera pandemic (1961–1975), as well as the HIV/AIDS pandemic (1981-today). COVID-19 acts as an unwelcome reminder of the major threat that infectious diseases represent in terms of deaths and disruption.
One positive aspect of the current situation, relative to previous pandemics, is the unprecedented availability of scientific and technological means to face COVID-19. In particular, the rapid development of drugs and vaccines has already begun. Modern drug and vaccine development are largely based on genetic engineering and an understanding of host-pathogen interactions at a molecular level. The mobilisation to address the COVID-19 pandemic by scientists worldwide has been remarkable. This includes the feat of the global scientific community who has already produced and publicly shared well over 11,000 complete SARS-CoV-2 genome sequences at the time of writing (April 23 2020), which we have used here with gratitude. Further initatives in the United Kingdom (https://www.cogconsortium.uk/data/) have already to date produced over 10,000 genomes, some of which overlap with those already available on GISAID.
To put these numbers of SARS-CoV-2 genomes in context, it is interesting to consider parallels with the 2009 H1N1pdm influenza pandemic, the first epidemic for which genetic sequence data was generated in near-real time (Fraser et al., 2009; Smith et al., 2009). The genetic data available at the time looks staggeringly small in comparison to the amount that has already been generated for SARS-CoV-2 during the early stages of the COVID-19 pandemic. For example, Fraser et al. considered 11 partial hemagglutinin gene sequences two months after the WHO had declared 2009 H1N1pdm influenza a pandemic (Fraser et al., 2009).
This unprecedented genomic resource has already provided strong conclusions about the pandemic. For example, analyses by multiple independent groups place the start of the COVID-19 pandemic towards the end of 2019 (Table 1). This rules out any scenario that assumes SARS-CoV-2 may have been in circulation long before it was identified, and hence have already infected large proportions of the population.
Extensive genomic resources for SARS-CoV-1 should in principle also be key to informing on optimal drug and vaccine design, particularly when coupled with knowledge of human proteome and immune interactions (Gordon et al., 2020). Ideally, drugs and vaccines should target relatively invariant, strongly constrained regions of the SARS-CoV-2 genome, to avoid drug resistance and vaccine evasion. Therefore ongoing monitoring of genomic changes in the virus will be essential to gain a better understanding of fundamental host-pathogen interactions that can inform drug and vaccine design.
As most (but not all) pathogens capable of causing epidemic at a pandemic scale, SARS-CoV-2 is in all likelihood of zoonotic origin. This implies that SARS-CoV-2 may not be fine-tuned to its novel human host. However, it is near-impossible to predict future trajectories for the virulence and transmissibility of horizontally transmitted pathogens (Anderson and May, 1991). It is also possible that the population of SARS-CoV-2 will evolve into different lineages characterised by variable levels of virulence and transmissibility. However, despite existing phylogenetic structure (Rambaut et al., 2020), it is important to stress that there is no evidence for the evolution of distinct phenotypes in SARS-CoV-2 at this stage.
The vast majority of mutations observed so far in SARS-CoV-2 circulating in humans are likely neutral (Cagliani et al., 2020; Dearlove et al., 2020) or even deleterious (Nielsen et al., 2020). Homoplasies, such as those we detect here, can arise by product of neutral evolution or as a result of ongoing selection. Of the 198 homoplasies we detect (after applying stringent filters), some proportion are very likely genuine targets of positive selection which signpost to ongoing adaptation of SARS-CoV-2 to its new human host. Indeed, we do observe an enrichment for non-synonymous changes (80%) in our filtered sites. As such, our provided list (Table S5) contains candidates for mutations which may affect the phenotype of SARS-CoV-2 and virus-host interactions and which require ongoing monitoring. Conversely, the finding that 78% of the homoplasic mutations involve no polarity change could still reflect strong evolutionary constraints at these positions (Hughes, 2007; Yampolsky et al., 2005). The remaining non-neutral changes to amino acid properties at homoplasic sites may be enriched in candidates for functionally relevant adaptation and could warrant further experimental investigation.
One of the strongest homoplasies lies at site 11,083 in the SARS-CoV-2 genome in a region of Orf1a encoding Nsp6. This site passed our stringent filtering cirteria and was also present in our analysis of the SRA dataset (Table S7). Interestingly, this region overlaps a putative immunogenic peptide predicted to result in both CD4+ and CD8+ T-cell reactivity (Grifoni et al., 2020). More minor homoplasies amongst our top candidates, identified within Orf3a (Table S5), also map to a predicted CD4 T cell epitope. While the immune response to SARS-CoV-2 is poorly understood at this point, key roles for CD4 T cells, which activate B cells for antibody production, and cytotoxic CD8 T cells, which kill virus-infected cells, are known to be important in mediating clearance in respiratory viral infections (Kohlmeier and Woodland, 2009). Of note, we also identify a strong recurrent mutation in nucleotide position 21,575, corresponding to the SARS-CoV-2 spike protein (codon 5). While the spike protein is the known mediator of host-cell entry, our detected homoplasy falls outside of the N-terminal and receptor binding domains.
Our analyses presented here provide a snapshot in time of a rapidly changing situation based on available data. Although we have attempted to filter out homoplasies caused by sequencing error with stringent thresholds, and also used available short-read data to validate a subset of homoplasic sites in a smaller dataset, our analysis nevertheless remains reliant on the underlying quality of the publicly available assemblies. As such, it is possible that some results might be artefactual, and further investigation will be warranted as additional raw sequencing data becomes available.
However, given the crucial importance of identifying potential signatures of adaptation in SARS-CoV-2 for guiding ongoing development of vaccines and treatments, we have suggested what we believe to be a plausible approach and initial list in order to facilitate future work and interpretation of the observed patterns. More data continues to be made available, which will allow ongoing investigation by ourselves and others. We believe it is important to continue to monitor SARS-CoV-2 evolution in this way and to make the results available to the scientific community. In this context, we hope that the interactive web-application we provide will help identify key recurrent mutations in SARS-CoV-2 as they emerge and spread.
Author contributions
L.v.D., and F.B. conceived and designed the study; L.v.D., M.A, D.R L.P.S., C.E.F., L.O., C.J.O., J.P., C.C.S.T., F.A.T.B., and A.T.O analysed data and performed computational analyses; L.v.D., and F.B. wrote the paper with inputs from all co-authors.
Acknowledgments and funding
L.v.D and F.B. acknowledge financial support from the Newton Fund UK-China NSFC initiative (grant MR/P007597/1) and the BBSRC (equipment grant BB/R01356X/1). Computational analyses were performed on UCL Computer Science cluster and the South Green bioinformatics platform hosted on the CIRAD HPC cluster. We thank Jaspal Puri for insights and assistance on the development of the alignment visualisation tool and Nicholas McGranahan and Rachel Rosenthal for their comments on the manuscript. We additionally wish to acknowledge the very large number of scientists in originating and submitting labs who have readily made available SARS-CoV-2 assemblies to the research community.
Declaration of Competing Interest
The authors have no competing interests to declare.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2020.104351.
Contributor Information
Lucy van Dorp, Email: lucy.dorp.12@ucl.ac.uk.
François Balloux, Email: f.balloux@ucl.ac.uk.
Appendix A. Supplementary data
Supplementary Table S1
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Supplementary Figures and Tables
References
- Anderson R.M., May R.M. Oxford University Press; Oxford: 1991. Infectious Diseases of Humans. Dynamics and Control. [Google Scholar]
- Balloux F., van Dorp L. Q&a: what are pathogens, and what have they done to and for us? BMC Biol. 2017;15:6. doi: 10.1186/s12915-017-0433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R., Forni D., Clerici M., Sironi M. Computational inference of selection underlying the evolution of the novel coronavirus, SARS-CoV-2. J. Virol. 2020 doi: 10.1128/JVI.00411-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crispell J., Balaz D., Gordon S.V. HomoplasyFinder: a simple tool to identify homoplasies on a phylogeny. Microbial Genom. 2019;5(1):10. doi: 10.1099/mgen.0.000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearlove B.L., Lewitus E., Bai H., Li Y., Reeves D.B., Joyce M.G. A SARS-CoV-2 vaccine candidate would likely match all currently circulating strains. bioRxiv. 2020 doi: 10.1073/pnas.2008281117. 2020.04.27.064774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X., Croucher N.J., Bentley S.D., Harris S.R., Wilson D.J. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res. 2018;46(22):11. doi: 10.1093/nar/gky783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Calap P., Schubert B., Joly M., Solis M., Untrau M., Carapito R. An unusually high substitution rate in transplant-associated BK polyomavirus in vivo is further concentrated in HLA-C-bound viral peptides. PLoS Pathog. 2018;14(10):18. doi: 10.1371/journal.ppat.1007368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudas G., Rambaut A. MERS-CoV recombination: implications about the reservoir and potential for adaptation. Virus Evol. 2016;2(1):11. doi: 10.1093/ve/vev023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Chall. 2017;1(1):33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch W.M. Toward defining course of evolution - minimum change for a specific tree topology. Syst. Zool. 1971;20(4):406–416. [Google Scholar]
- Fraser C., Donnelly C.A., Cauchemez S., Hanage W.P., Van Kerkhove M.D., Hollingsworth T.D. Pandemic potential of a strain of influenza A (H1N1) : early findings. Science. 2009:1557–1561. doi: 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanetti M., Benvenuto D., Angeletti S., Ciccozzi M. The first two cases of 2019-nCoV in Italy: where they come from? J. Med. Virol. 2020;92(5):518–521. doi: 10.1002/jmv.25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., O’Meara M.J. A SARS-CoV-2-human protein-protein interaction map reveals drug targets and potential drug-repurposing. Nature. 2020 doi: 10.1038/s41586-020-2286-9. 2020.03.22.002386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user Interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Grenfell B.T., Pybus O.G., Gog J.R., Wood J.L.N., Daly J.M., Mumford J.A. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303(5656):327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(4):671–680. doi: 10.1016/j.chom.2020.03.002. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang D.T., Vinh L.S., Flouri T., Stamatakis A., von Haeseler A., Minh B.Q. MPBoot: fast phylogenetic maximum parsimony tree inference and bootstrap approximation. BMC Evol. Biol. 2018;18:11. doi: 10.1186/s12862-018-1131-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes E.C., Dudas G., Rambaut A., Andersen K.G. The evolution of Ebola virus: insights from the 2013-2016 epidemic. Nature. 2016;538(7624):193–200. doi: 10.1038/nature19790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L. Looking for Darwin in all the wrong places: the misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99(4):364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- Janin J. Surface and inside volumes in globular proteins. Nature. 1979;277(5696):491–492. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmeier J.E., Woodland D.L. Immunity to respiratory viruses. Annu. Rev. Immunol. 2009;27:61–82. doi: 10.1146/annurev.immunol.021908.132625. [DOI] [PubMed] [Google Scholar]
- Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35(21):4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K.P., Woo P.C.Y., Li K.S.M., Huang Y., Tsoi H.W., Wong B.H.L. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.G., Wang W., Zhao X.F., Zai J.J., Zhao Q., Li Y. Transmission dynamics and evolutionary history of 2019-nCoV. J. Med. Virol. 2020;92(5):501–511. doi: 10.1002/jmv.25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memish Z.A., Mishra N., Olival K.J., Fagbo S.F., Kapoor V., Epstein J.H. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013;19(11):1819–1823. doi: 10.3201/eid1911.131172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minskaia E., Hertzig T., Gorbalenya A.E., Campanacci V., Cambillau C., Canard B. Discovery of an RNA virus 3 ’-> 5 ’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A. 2006;103(13):5108–5113. doi: 10.1073/pnas.0508200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Wang H., Pipes L. Synonymous mutations and the molecular evolution of SARS-Cov-2 origins. bioRxiv. 2020 doi: 10.1093/ve/veaa098. 2020.04.20.052019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Pybus O.G., Nelson M.I., Viboud C., Taubenberger J.K., Holmes E.C. The genomic and epidemiological dynamics of human influenza a virus. Nature. 2008;453(7195):615–U2. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Holmes E.C., Hill V., O’Toole Á., McCrone J., Ruis C. A dynamic nomenclature proposal for SARS-CoV-2 to assist genomic epidemiology. bioRxiv. 2020 doi: 10.1038/s41564-020-0770-5. 2020.04.17.046086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell L.J. Phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol. Evol. 2012;3(2):217–223. [Google Scholar]
- Shaw L.P., Wang A.D., Dylus D., Meier M., Pogacnik G., Dessimoz C. The phylogenetic range of bacterial and viral pathogens of vertebrates. bioRxiv. 2020:670315. doi: 10.1111/mec.15463. [DOI] [PubMed] [Google Scholar]
- Shu Y.L., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Eurosurveillance. 2017;22(13):2–4. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G.J.D., Vijaykrishna D., Bahl J., Lycett S.J., Worobey M., Pybus O.G. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–U107. doi: 10.1038/nature08182. [DOI] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L.M. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E.M., Frost S.D.W. Scalable relaxed clock phylogenetic dating. Virus Evol. 2017;3(2):9. [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798) doi: 10.1038/s41586-020-2008-3. 265−+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yampolsky L.Y., Kondrashov F.A., Kondrashov A.S. Distribution of the strength of selection against amino acid replacements in human proteins. Hum. Mol. Genet. 2005;14(21):3191–3201. doi: 10.1093/hmg/ddi350. [DOI] [PubMed] [Google Scholar]
- Yu G.C., Smith D.K., Zhu H.C., Guan Y., Lam T.T.Y. GGTREE: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol. Evol. 2017;8(1):28–36. [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798) doi: 10.1038/s41586-020-2012-7. 270−+. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1
Supplementary Table S4
Supplementary Table S5
Supplementary Table S6
Supplementary Figures and Tables