Abstract
The recent epidemic outbreak of a novel human coronavirus called SARS-CoV-2 and causing the respiratory tract disease COVID-19 has reached worldwide resonance and a global effort is being undertaken to characterize the molecular features and evolutionary origins of this virus. Therefore, rapid and accurate identification of pathogenic viruses plays a vital role in selecting appropriate treatments, saving people's lives and preventing epidemics. Additionally, general treatments, coronavirus-specific treatments, and antiviral treatments useful in fighting COVID-19 are addressed. This review sets out to shed light on the SARS-CoV-2 and host receptor recognition, a crucial factor for successful virus infection and taking immune-informatics approaches to identify B- and T-cell epitopes for surface glycoprotein of SARS-CoV-2. A variety of improved or new approaches also have been developed. It is anticipated that this will assist researchers and clinicians in developing better techniques for timely and effective detection of coronavirus infection. Moreover, the genomic sequence of the virus responsible for COVID-19, as well as the experimentally determined three-dimensional structure of the Main protease (Mpro) is available. The reported structure of the target Mpro was described in this review to identify potential drugs for COVID-19 using virtual high throughput screening.
Keywords: Coronavirus, SARS-CoV-2, COVID-19, Epitopes, Immune-informatics
Abbreviations: COVID-19, Coronavirus disease 2019; ssRNA, single-stranded RNA; MERS-CoV, Middle East respiratory syndrome coronavirus; SARS-CoV, severe acute respiratory syndrome coronavirus; BtCoV, bat coronavirus; nsp, non-structural proteins; ADE, antibody-dependent enhancement; RNP, ribonucleoprotein; NTD, N-terminal RNA-binding domain; CTD, C-terminal dimerization domain; IBV, infectious bronchitis virus; MHV, mouse hepatitis virus; ACE2, Angiotensin I converting enzyme 2; HIV, human immunodeficiency virus; IVIg, intravenous gammaglobulin; PLP, papain-like protease; LPV, Lopinavir; RTV, ritonavir; RDV, Remdesivir; NO, Nitric oxide; Mpro, Main protease; HLA, human leukocyte antigen
Highlights
-
•
COVID-19 is a highly infectious disease associated with high mortality.
-
•
Rapid and accurate identification of viruses plays a vital role in selecting appropriate treatments.
-
•
The reported structure of the target Mpro was described in this review to identify potential drugs.
-
•
Using virtual high throughput screening will help to identify potential drugs.
1. Introduction
Coronavirus is a type of single-stranded RNA (ssRNA) virus [1] Before the emergence of Sars-CoV-2, there are 6 known human coronaviruses, including the Middle East respiratory syndrome coronavirus (MERS-CoV) and severe acute respiratory syndrome coronavirus (SARS-CoV). The symptoms caused by Sars-CoV-2 infection include acute respiratory distress syndrome (~29%), acute cardiac injury (~12%) or acute kidney injury (~7%) [2], implying that Sars-CoV-2 may infect various human tissues. COVID-19 is a highly infectious disease [3,4] associated with high mortality [5]. SARS-CoV-2, the virus responsible for COVID-19, is a betacoronavirus [6]. The previous name for this virus was Sars-CoV-2. The genome of SARS-CoV-2 has been sequenced [7,8]. The genomic sequence of SARS-CoV-2 has 96% similarity to the bat-coronavirus and 76.5% identity to the SARS-CoV [9]. Although there are no approved drugs or vaccines for COVID-19, some clinical trials are in progress [10]. Lopinavir and Ritonavir were used in preliminary clinical studies [3].
Nevertheless, in the past two decades, a massive amount of work has been done to understand the molecular basis of the coronavirus infection and evolution, develop an effective treatment in forms of both vaccines and antiviral drugs, and propose efficient measures for viral detection and prevention [[11], [12], [13]]. Structures of many individual proteins of SARS, MERS, and related coronaviruses, as well as their biological interactions with other viral and host proteins, have been explored along with the experimental testing of anti-viral properties of small compounds [[14], [15], [16]]. In this context, the use of bioinformatics approaches may help fill the gap of missing data and improve the process of knowledge discovery by elucidating molecular mechanisms behind virus replication, the process of viral attachment to the host cells and the effect on the host molecular pathways [17]. The final understanding of the molecular action of the virus may improve or accelerate the development of anti-viral therapeutic approaches using information and data available for other coronaviruses.
Therefore, understanding the molecular effects of this virus on human proteins play a pivotal role in prioritizing pharmacological strategies [18]. In this review, the crystal structure of the SARS-CoV-2 nucleocapsid N-terminal domain (termed as SARS-CoV-2 N-NTD), as a model for understanding the molecular interactions that govern SARS-CoV-2 N-NTD binding to ribonucleotides is described. and the unique RNA binding site characteristics are discussed, which in turn will aid in the development of new drugs that interfere with viral N protein and viral replication in SARS-CoV-2, and highly related virus SARS-CoV. Furthermore, it is attempted to propose suitable vaccine candidates by identifying B-Cell and T-cell epitopes with computational approaches for drug and vaccine design.
2. Structural genomics of 2019 Wuhan novel coronavirus
Structures of many individual proteins of SARS, MERS, and related coronaviruses, as well as their biological interactions with other viral and host proteins, have been explored along with the experimental testing of anti-viral properties of small compounds [16]. However, experimental data of the same scale for Sars-CoV-2 may take years to obtain by the research community. By leveraging the previously known information on genome sequences as well as protein structure and function, bioinformaticians have been successfully helping the virologists by structurally characterizing proteins of novel viruses, determining the evolutionary trajectories, identifying interactions with host proteins, and providing other important biological insights. In particular, a plethora of results have been achieved through comparative, or homology, modeling principles [19,20]. In addition to the global structural genomics initiatives focusing on determining the 3D structures of proteins on a genome-scale [21] and specific efforts on rapid structural characterization of proteins in emerging viruses [22,23], multiple works have used comparative modeling to predict the structures of protein-protein interaction complexes [[24], [25], [26]], facilitating structure-based drug discovery [20,27,28], inferring protein functions [29], determining the macromolecular interaction network [[30], [31], [32]], and providing molecular insights into viral evolution [[33], [34], [35]].
By providing a comprehensive structural genomics and interactomics road-maps of Sars-CoV-2 and using this information to infer the possible functional differences and similarities with the related SARS coronavirus, Sars-CoV-2 has 16 predicted non-structural proteins constituting a polyprotein, followed by 13 downstream ORFs: Surface, ORF3a, ORF3b, Envelope, Membrane, ORF6, ORF7a, ORF7b, ORF8, Nucleocapsid, ORF9a, ORF9b, and ORF10. The three viral species whose proteins shared the highest similarity were consistently the same: human SARS (SARS- Cov), bat coronavirus (BtCoV), as well as another bat betacoronavirus (BtRf-BetaCoV) [36].
3. RNA binding domain of SARS-CoV-2 nucleocapsid protein
SARS-CoV-2 is a betacoronavirus with single-stranded RNA genomes, like MERS-CoV and SARS-CoV. The first two-thirds of viral 30 kb RNA genome, mainly named as ORF1a/b region, translates into two polyproteins (pp1a and pp1ab) and encodes most of the non-structural proteins (nsp) [37]. The rest parts of the virus genome encode accessory proteins and four essential structural proteins, including spike (S) glycoprotein, small envelope (E) protein, matrix (M) protein, and nucleocapsid (N) protein [37,38] Current antiviral drugs developed to treat coronavirus (CoV) infections primarily target S protein, the 3C-like (3CL) and papain-like (PLP) proteases [39,40]. Because mutant viruses in the S protein is prone to escape the targeted therapeutic with different host-cell receptor binding patterns [39], as well as antibody-dependent enhancement (ADE) effects of S protein antibodies are found in MERS coronavirus [41], there are several limitations on targeting S protein for antiviral approaches. Antiviral protease inhibitors may nonspecifically act on the cellular homologous protease, resulting in host cell toxicity and severe side effects. Therefore, novel antiviral strategies are needed to combat acute respiratory infections caused by this novel coronavirus SARS-CoV-2 (Fig. 1 ).
Fig. 1.
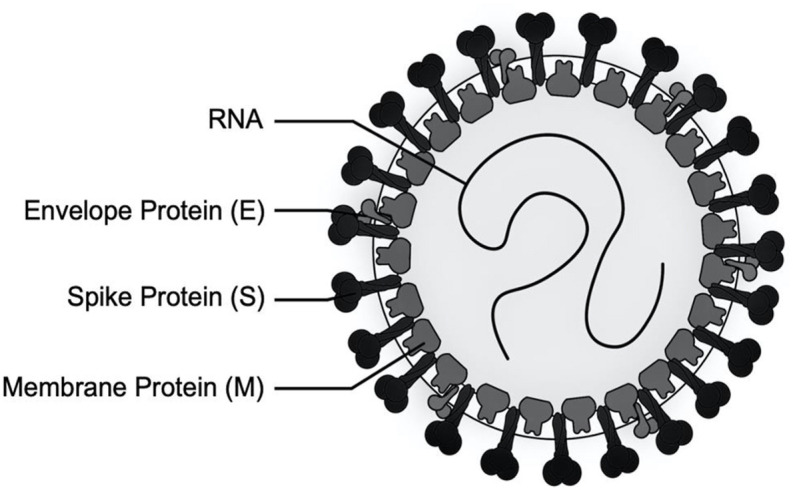
Schematic diagram of the SARS coronavirus structure. Like all coronaviruses, Sars-CoV-2 consist of a minimum of three viral proteins namely spike protein (S), a type of glycoprotein, a membrane protein (M) that spans the membrane and an envelope protein (E), a highly hydrophobic protein that covers the entire structure of the coronavirus. The spike (S) glycoprotein in the coronavirus recognizes the host cell receptors and causes an important role in viral infection.
The CoV N protein is a multifunctional RNA-binding protein necessary for viral RNA transcription and replication [42]. It plays many pivotal roles in forming helical ribonucleoproteins during packaging the RNA genome, regulating viral RNA synthesis during replication, transcription and modulating infected cell metabolism [42,43]. The primary functions of N protein are binding to the viral RNA genome, and packing them into a long helical nucleocapsid structure or ribonucleoprotein (RNP) complex [44,45]. In vitro and in vivo experiments revealed that N protein bound to leader RNA, and was critical for maintaining highly ordered RNA conformation suitable for replicating, and transcribing the viral genome [43,45,46]. More studies implicated that N protein regulated host-pathogen interactions, such as actin reorganization, host cell cycle progression, and apoptosis [47,48]. The N protein is also a highly immunogenic and abundantly expressed protein during infection, capable of inducing protective immune responses against SARS-CoV and SARS-CoV-2 [[49], [50], [51]]. The common domain architectures of coronavirus N protein are consisting of three distinct but highly conserved parts: An N-terminal RNA-binding domain (NTD), a C-terminal dimerization domain (CTD), and intrinsically disordered central Ser/Arg (SR)-rich linker. Previous studies have revealed that the NTD are responsible for RNA binding, CTD for oligomerization, and (SR)-rich linker for primary phosphorylation, respectively [[52], [53], [54]]. The crystal structures of SARS-CoV N-NTD [55], infectious bronchitis virus (IBV) N-NTD [56,57], HCoV-OC43 N-NTD [53] and mouse hepatitis virus (MHV) N-NTD [58] have been solved. The CoVs N-NTD have been found to associate with the 3′ end of the viral RNA genome, possibly through electrostatic interactions. Additionally, several critical residues have been identified for RNA binding and virus infectivity in the N-terminal domain of coronavirus N proteins [[58], [59], [60]]. However, the structural and mechanistic basis for newly emerged novel SARS-CoV-2 N protein remains largely unknown. Understanding these aspects should facilitate the discovery of agents that specifically block the coronavirus replication, transcription and viral assembly [61]. Kang et al. [62] reported the crystal structure of SARS-CoV-2 nucleocapsid N-terminal domain (termed as SARS-CoV-2 N-NTD), as a model for understanding the molecular interactions that govern SARS-CoV-2 N-NTD binding to ribonucleotides. This finding will aid in the development of new drugs that interfere with viral N protein and viral replication in SARS-CoV-2, and highly related virus SARS-CoV [62].
4. Single-cell RNA sequencing of human tissues
Angiotensin I converting enzyme 2 (ACE2), is the host receptor by Sars-CoV-2 to infect human cells. Viruses bind to host receptors on the target cell surface to establish infection. Membrane proteins mediated membrane fusion allowed the entry of enveloped viruses [63]. As recently reported, both nCoV and SARS-CoV could use ACE2 protein to gain entry into the cells [64]. Since the outbreak, many data analysis have shown a wide distribution of ACE2 across human tissues, including lung [65], liver [66], stomach [67], ileum [67], colon [67] and kidney [68], indicating that Sars-CoV-2 may infect multiple organs. However, these data showed that AT2 cells (the main target cell of Sars-CoV-2) in the lung expressed rather low levels of ACE2 [68]. Hence, the nCoVs may depend on co-receptor or other auxiliary membrane proteins to facilitate its infection. It is reported that viruses tend to hijack co-expressed proteins as their host factors [69]. For example, Hoffmann et al. recently showed that Sars-CoV-2-S use ACE2 for entry and depends on the cellular protease TMPRSS2 for priming [70], showing that 2019- nCoV infections also require multiple factors. Understanding the receptors’ usage by the viruses could facilitate the development of intervention strategies. Therefore, identifying the potential co-receptors or auxiliary membrane proteins for Sars-CoV-2 is of great significance.
Although ACE2 is reported to be expressed in the lung, liver, stomach, ileum, kidney, and colon, its expressing levels are rather low, especially in the lung [71]. Sars-CoV-2 may use co-receptors/auxiliary proteins as ACE2 partners to facilitate the virus entry [72]. To identify the potential candidates [73], explored the single-cell gene expression atlas including 119 cell types of 13 human tissues and analyzed the single-cell co-expression spectrum of 51 reported RNA virus receptors and 400 other membrane proteins. Consistent with other recent reports [73], confirmed that ACE2 was mainly expressed in lung AT2, liver cholangiocyte, colon colonocytes, esophagus keratinocytes, ileum ECs, rectum ECs, stomach epithelial cells, and kidney proximal tubules. Intriguingly [73], found that the candidate co-receptors, manifesting the most similar expression patterns with ACE2 across 13 human tissues, are all peptidases, including ANPEP, DPP4, and ENPEP. Among them, ANPEP and DPP4 are the known receptors for human CoVs, suggesting ENPEP as another potential receptor for human CoVs.
5. Receptors of human coronaviruses
The viruses target host cells via binding host receptors before engaging the infection cycle. ACE2 was proved to be the cell receptor of Sars-CoV-2, the same receptor as SARS-CoV [65,70]. The expression profiles of ACE2 across different cell types of different organs will reveal clues of the virus transmission routes and its potential pathogenesis. In previous studies, ACE2 was found to express in the esophagus upper and stratified epithelial cells, absorptive enterocytes from ileum and colon, alveolar type II cells in the lung, liver cholangiocyte and kidney proximal tubules [66,67]. These findings suggested that the clinical symptoms of hepatic failure, respiratory injury, acute kidney injury or diarrhoea may be associated with the pervasive ACE2 expressing cells in these tissues.
However, we and others found that ACE2 is lowly expressed, especially in the lung (the main target organ of nCoVs), raising the possible existence of co-receptors facilitating nCoV infection. It is well recognized that ssRNA viruses tend to have multiple receptors [69]. For example, ACE2, CD209 (Dendritic Cell-Specific ICAM-3-Grabbing Non-Integrin 1), CLEC4G (C-type lectin domain family 4 member G) and CLEC4M (C-type lectin domain family 4 member M) are receptors of SARS-CoV [71,74]. Also, other membrane proteins may also assist virus entry [63]. Since the viral receptors and co-receptors should be co-expressed on the same cell types, single-cell co-expression patterns covering 400 membrane proteins and 51 known viral receptors was analyzed [64]. After calculating gene expression similarity, they found ANPEP, ENPEP and DPP4 are the top three genes correlated with ACE4 (R > 0.8). Interestingly, both ANPEP and DPP4 are viral receptors of human coronaviruses [75], while ENPEP is also a peptidase, despite that its involvement in virus infection is unclear. For mysterious reasons, human coronaviruses use peptidases as their receptors [76]. In addition, Li et al. [77] showed co-expression profiles of these molecules, indicating that different human CoVs target similar cell types across different human tissues.
6. General treatment for viral infection
Currently, there is no registered treatment or vaccine for the disease. In the absence of a specific treatment for this novel virus, there is an urgent need to find an alternative solution to prevent and control the replication and spread of the virus.
6.1. Nutritional interventions
Vitamin A supplementation reduced morbidity and mortality in different infectious diseases, such as measles, diarrheal disease, measles-related pneumonia, human immunodeficiency virus (HIV) infection, and malaria, as described by Semba et al. [78]. Keil et al. [79] had reported that vitamin B2 and UV light effectively reduced the titer of MERS-CoV in human plasma products. Atherton et al. [80] had reported that vitamin C increased the resistance of chick embryo tracheal organ cultures to avian coronavirus infection. In addition, the decreased vitamin D status in calves had been reported to cause the infection of Bovine coronavirus [81]. Vitamin E deficiency had been reported to intensify the myocardial injury of coxsackievirus B3 (a kind of RNA viruses) infection in mice [82]. Protection D1, the omega-3 PUFA-derived lipid mediator, could markedly attenuate influenza virus replication via RNA export machinery [83]. Dietary selenium deficiency that causes oxidative stress in the host can alter a viral genome so that a normally benign or mildly pathogenic virus can become highly virulent in the deficient host under oxidative stress [84]. The combination of zinc and pyrithione at low concentrations inhibits the replication of SARS coronavirus [85]. Iron is required for both host and pathogen and iron deficiency can impair host immunity, while iron overload can cause oxidative stress to propagate harmful viral mutations [86].
6.2. Immunoenhancers
A combination of interferon-α-2a with ribavirin was administered to patients with severe MERS-CoV infection and the survival of these patients was improved [87]. During the SARS outbreak in 2003, Intravenous gammaglobulin (IVIg) was used extensively in Singapore. However, one-third of critically ill patients developed venous thromboembolism including pulmonary embolism despite the use of low-molecular-weight heparin prophylactic [88]. Thymosin α-1 could increase resistance to glucocorticoid-induced death of the thymocyte [89]. Thymosin α-1 could also be used as an immune enhancer to SARS patients and it was effective in controlling the spread of the disease [90,91]. Thymopentin (TP5, munox), a synthetic pentapeptide corresponding to the active site of thymopoietin, had been shown to restore antibody production in old mice [92]. levamisole can act as either an immunostimulant agent or an immunosuppressive agent depending upon the dosing and the timing [93]. Luo et al. [94] had speculated that nucleocapsid protein (NP) of SARS-CoV played an important role in the process of virus particle assembly and release and it might also bind to human cyclophilin A.
6.3. Coronavirus-specific treatments
Chymotrypsin-like (3C-like) and papain-like protease (PLP) are coronavirus encoded protein. They have an essential function for coronaviral replication and also have an additional function for inhibition of host innate immune responses [95]. Cinanserin, an old drug, is well-known for serotonin receptor antagonists. It could inhibit the 3 chymotrypsin-like (3C-like) protease and was a promising inhibitor of replication of SARS-CoV [96]. Flavonoids are an important class of natural products and have several subgroups, which include chalcones, flavones, flavonols, and isoflavones [97]. Other flavonoids (Herbacetin, isobavachalcone, quercetin 3-β-d-glucoside, and helichrysetin) were also found to be able to block the enzymatic activity of MERS-CoV/3CLpro [98]. Papain-like protease (PLP) of human coronavirus is a novel viral-encoded deubiquitinase and is an IFN antagonist for inhibition of host innate antiviral immune response. Diarylheptanoids is a natural product and is extracted from the stem bark of Alnus japonica. It was able to inhibit papain-like protease of SARS-CoV [95].
Angiotensin-converting enzyme-2 (ACE2) is a type I integral membrane protein which that functions as a carboxypeptidase and is the first human homolog of ACE. Besides, ACE2 is also a functional receptor of SARS-CoV and it mediates virus entry into the cell through binding with spike (S) protein [99,100]. Zhang et al. [101] reported that COVID-19 used ACE2 as a sole receptor for the entry, but did not use other coronavirus receptors, aminopeptidase N and dipeptidyl peptidase, for the entry. Blocking the binding of S protein to ACE2 is important for the treatment of SARS-CoV infection [102]. One recombinant human monoclonal antibody (mAb) (single-chain variable region fragments, scFvs 80R) against the S1 domain of S protein of SARS-CoV from two nonimmune human antibody libraries. The mAb could efficiently neutralize SARS-CoV and inhibit syncytia formation between cells expressing the S protein and those expressing the SARS-CoV receptor ACE2 were found by Sui et al. [103].
Chloroquine has many interesting biochemical properties including antiviral effects. In addition, it had been used against viral infection [104]. Emodin is an anthraquinone compound derived from genus Rheum boosted protease inhibitor in the treatment of HIV infection. Lopinavir (LPV) is usually combined with ritonavir (RTV) to increase and Polygonum and it is also a virucidal agent [105]. Emodin could significantly block the interaction between the S protein of SARS-CoV and ACE2 [106]. Promazine, an anti-psychotic drug, shares a similar structure with emodin. It has been found to exhibit a significant effect in inhibiting the replication of SARS-CoV [107]. As compared to emodin, promazine exhibited potent inhibition of the binding of S protein to ACE2. These findings suggested that emodin and promazine might be able to inhibit SARS-CoV infectivity by blocking the interaction of S protein and ACE2. Therefore, the monoclonal antibody (scFv80R), chloroquine, emodin, and promazine could be used as choices for the treatment of COVID-19. Nicotianamine is an important metal-ligand in plants [108] and it is found a novel angiotensin-converting enzyme-2 inhibitor in soybean [109]. So, it is another potential option to be used to reduce the infection of COVID-19.
6.4. Antiviral treatments
Ribavirin, a broad-spectrum antiviral agent, is routinely used to treat hepatitis C [110]. Morgenstern et al. [111] had reported that ribavirin and interferon-β synergistically inhibited the replication of SARS-associated coronavirus in animal and human cell lines. Given adverse reactions and the lack of in vitro efficacy, the use of ribavirin should be seriously considered for the treatment of COVID-19, even in combination with other antiviral drugs. The combination of lopinavir (LPV) with ritonavir (RTV) is widely used as a boosted protease inhibitor in the treatment of HIV infection [112]. Kim et al. [113] had also reported a successful case of MERS-CoV disease treated with triple combination therapy LPV/RTV, ribavirin, and IFN-α2a. Remdesivir (RDV), a nucleoside analog GS-5734, had been reported to inhibit human and zoonotic coronavirus in vitro and to restrain severe acute respiratory syndrome coronavirus (SARS-CoV) in vivo [114]. Recently, the antiviral activity of RDV and IFN-β was found to be superior to that of LPV/RTV–IFN–β against MERS-CoV in vitro and in vivo [115]. Therefore, the use of RDV with IFN-β could be a better choice for the treatment of COVID-19 comparing with that of the triple combination of LPV/RTV–IFN–β. However, randomized and controlled trials are still needed to determine the safety and efficacy of remdesivir. Yamamoto et al. [116] had found that nelfinavir could strongly inhibit the replication of SARS-CoV.
Arbidol and its derivatives, arbidol mesylate, had been reported to have antiviral activity against the pathogen of SARS in the cell cultures and arbidol mesylate was nearly 5 times as effective as arbidol in reducing the reproduction of SARS virus in the cultured cells [117]. Nitric oxide (NO) is a gas with diverse biological activities and is produced from arginine by NO synthases [118]. Akerström et al. [119] had reported that organic NO donor, S-nitroso- N-acetylpenicillamine, could significantly inhibit the replication cycle of SARS-CoV in a concentration-dependent manner. Therefore, NO inhalation could be also chosen as an option for the treatment of severely COVID-19 infected patients (Table 1 ).
Table 1.
Treatment options available for COVID‐19.
| Compounds | Virus target | Functions | Representative References |
|---|---|---|---|
| General treatments for viral infection | |||
| Nutritional interventions | |||
| Vitamin A | Measles virus, avian coronavirus | Prevention of lung infection. | (Semba 1999) |
| B vitamins | MERS‐CoV; ventilator‐induced lung injury | Enhance their immune system | (Keil, Bowen et al., 2016) |
| Vitamin C | Avian coronavirus; lower respiratory tract infections | Act as an antioxidant | (Atherton, Kratzing et al., 1978) |
| Vitamin D | Bovine coronavirus | Maintaining bone integrity | (Nonnecke, McGill et al., 2014) |
| Vitamin E | Coxsackievirus, bovine coronavirus | Reducing oxidative stress | (Beck, Kolbeck et al., 1994) |
| PUFAa | Influenza virus, human immunodeficiency virus | Anti‐inflammatory and pro‐inflammatory effects. | (Morita, Kuba et al., 2013) |
| Selenium | Influenza virus, avian coronavirus; viral mutations | Defense against infectious diseases | (Rayman 2012) |
| Zinc | Measles virus, SARS‐CoV | Maintaining immune system | (Te Velthuis, van den Worm et al., 2010) |
| Iron | Viral mutations | Development of recurrent acute respiratory infections | (Wessling-Resnick 2018) |
| Immunoenhancers | |||
| Interferons | SARS‐CoV, MERS‐CoV | Immune response to virus infection. | (Momattin, Al-Ali et al., 2018) |
| Intravenous gammaglobulin | SARS‐CoV | Increase of viscosity in hypercoagulable states | (Lew, Kwek et al., 2003) |
| Thymosin α‐1 | SARS‐CoV | Increase resistance to glucocorticoid‐induced death | (Baumann, Badamchian et al., 1997) |
| Thymopentin | hepatitis B | Restore antibody production | (Duchateau, Servais et al., 1985) |
| Levamisole | SARS‐CoV | Immunostimulant agent or immunosuppressive agent | (Joffe, Sukha et al., 1983) |
| Cyclosporine A | SARS‐CoV, avian infectious bronchitis virus | Treatment of autoimmune disorders | (Luo, Luo et al., 2004) |
| Coronavirus‐specific treatments | |||
| Coronavirus protease inhibitors | |||
| Chymotrypsin‐like (3C‐like) inhibitors | |||
| Cinanserin | SARS‐CoV | Serotonin receptor antagonist | (Chen, Gui et al., 2005) |
| Flavonoids | SARS‐CoV/MERS‐CoV | Antioxidant effects/antiviral abilities | (Diwan, Ninawe et al., 2017) |
| Papain‐like protease (PLP) inhibitors | |||
| Diarylheptanoids | SARS‐CoV | Anti-inflammatory, antioxidant, antitumor | (Park, Jeong et al., 2012) |
| Spike (S) protein‐angiotensin‐converting enzyme‐2 (ACE2) blockers | |||
| Human monoclonal antibody | SARS‐CoV | Treatment of many solid tumors | (Sui, Li et al., 2004) |
| Chloroquine | SARS‐CoV | Prevention of malaria in adults | (Savarino, Boelaert et al., 2003) |
| Emodin | SARS‐CoV | Pancreatic disease, inflammatory, and diabetes | (Vickers 2017) |
| Promazine | SARS‐CoV | Using in paranoid and manic-depressive conditions, | (Cauwenberghs, Feijge et al., 2006) |
| Nicotianamine | SARS‐CoV | To reduce the infection | (Cauwenberghs, Feijge et al., 2006) |
| Antiviral treatments | |||
| Ribavirin | SARS‐CoV | Treatment of hepatitis C | (Ksiazek, Erdman et al., 2003) |
| (LPV)b/(RTV)c (Kaletra) | MERS‐CoV | Treatment of HIV infection | (Tsang and Zhong 2003) |
| Remdesivir | SARS‐CoV/MERS‐CoV | Treatment of Ebola virus disease and Marburg virus | (Yamamoto, Yang et al., 2004) |
| Nelfinavir | SARS‐CoV | Treatment of HIV | (Mohanasundaram and Sekhar 2018) |
| Arbidol | SARS‐CoV | Treatment for influenza infection | (Khamitov, Loginova et al., 2008) |
| Nitric oxide | SARS‐CoV | Treatment of inflammatory airway disease | (Robbins and Grisham 1997) |
Omega‐3 polyunsaturated fatty acid.
Lopinavir.
Ritonavir.
7. Prediction of potential drugs for COVID-19
7.1. Virtual high throughput screening
Computational methods can be utilized for the design and engineering of drugs [120,121]. The low time requirements of computational methods are conducive for high throughput screening of available drugs to identify potential drugs for novel diseases as well as to predict the adverse effects of novel drugs [122]. Development of novel drugs is a time-consuming process and generally, several years of work are required for clinical approval [121]. Drug repositioning, also known as repurposing, is an effective strategy to combat novel diseases caused by infectious agents that spread rapidly [123]. Drugs that have been approved for some disease, are safe for human use, and only their effectiveness against the disease of interest needs to be established [124]. In life-threatening cases, where there is no alternative medicine or vaccine, such a drug repurposing strategy is particularly attractive. However, clinical trials are necessary to ensure that such treatment is better than a placebo [125].
Lopinavir and Ritonavir were identified in earlier studies to target the Main protease (Mpro) of SARS virus. The protein sequences of COVID-19 Main protease (Sars-CoV-2 Mpro) and SARS-CoV Mpro are 96% identical [126]. In several early studies, the similarities in the sequence of a potential target for COVID-19 to that of the SARS Mpro were utilized to build a model for the structure of SARS- CoV-2 Mpro [126]. Homology based models were utilized to screen a library of compounds to predict that Nelfinavir, an approved antiviral protease inhibitor, is a potential drug for COVID-19 [122]. The sequence similarity of the SARS-CoV-2 Mpro to the SARS Mpro is sufficiently high to build a good model for the structure of SARS-CoV-2 Mpro [123,127]. However, the predictions of virtual screening studies and binding energy calculations are generally more accurate if a high-resolution experimental structure of the target is available.
The recent availability of the genomic sequence of the virus responsible for COVID-19, as well as the experimentally determined three-dimensional structure of the Mpro was utilized as the target for virtual high throughput screening [120], indicating that the results confirm earlier preliminary reports based on studies of homologs that some of the drugs approved for the treatment of other viral infections also have the potential for the treatment of COVID-19. Therefore, approved anti-viral drugs that target proteases were ranked for potential effectiveness against COVID-19 and novel candidates for drug repurposing were identified.
7.2. Drug and vaccine design against novel coronavirus spike protein
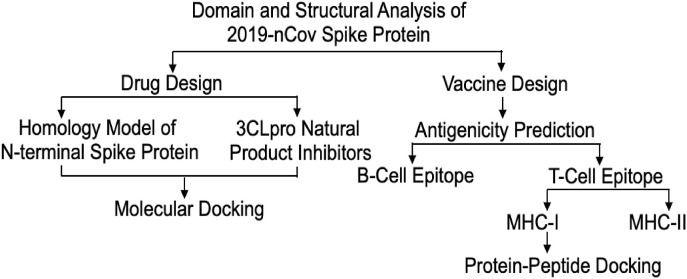
There is an urgent need for the development of anti-viral drugs and vaccines against the 2019- nCov virus due to the high mortality rate of patients. During the epidemic and pandemic outbreak of new viral pathogens, the conventional method of development of drugs and vaccination is not possible to control as it is a time-consuming process [128,129]. Consequently, the rapid approach based on in-silico informatics has become very popular with recent advances in the sequencing of many pathogen genomes and protein sequence databases [[130], [131], [132]]. The continuous increase of patients and a high mortality rate of Sars-CoV-2 infection highlight the urgent need for the development of a safe and effective vaccine [133]. The aim of this is to use to computational approach to design both anti-viral drug and vaccine candidates. The spike protein in the novel coronavirus sequence is used to design both anti-viral drug and vaccine candidates. For anti-viral drug design, receptor-binding protein of novel coronavirus present in the N-terminal of spike protein is homology modelled and used as a protein receptor [134]. 3C like proteinase (3CLpro) plays a role in viral pathogenicity and replication by clevaging the polyprotein. The inhibitors of 3CLpro can block the clevage reaction thereby controlling viral replication and pathogenicity [135]. The natural inhibitors were used as ligands and docked with homology modelled coronavirus receptor binding protein [136]. For vaccine design, the full sequence of the spike protein of novel coronavirus is used to predict B-cell and T-cell epitopes. The select best T-cell epitopes based on antigenicity used as peptides and docked with human allele protein.
The computational approach is proposed for drug and vaccine design [120]. To explore suitable natural inhibitors for the N-terminal receptor-binding domain of spike protein, the spike protein sequences were collected from a protein database. And also they were analyzed with various bioinformatics tools, attempting to identify suitable vaccine candidates by identifying B-Cell and T-cell epitopes [137]. In the drug design, the tanshinone Iia and methyl Tanshinonate were identified as natural inhibitors based on the docking score [138]. In the vaccine design, B-cell epitope VLLPLVSSQCVNLTTRTQLPPAYTN was found to have the highest antigenicity. FVFLVLLPL of MHC class-I allele and FVFLVLLPL of MHC class-II allele were identified as best peptides based on several alleles and antigenicity scores [139]. Therefore, identifies natural inhibitors and putative antigenic epitopes might be useful as effective drug and vaccine candidates for the eradication of novel coronavirus.
7.3. Epitope-based peptide vaccine design and target site characterization against novel coronavirus
Structural proteins are important targets for vaccine and anti-viral drug development due to their indispensable function to fuse and enter into the host cell [140]. SARS-CoV-2 utilizes glycosylated spike (S) protein to gain entry into host cells. The S protein is a trimeric class I fusion protein and exists in a metastable prefusion conformation that undergoes a dramatic structural rearrangement to fuse the viral membrane with the host-cell membrane [141,142]. The S protein includes the receptor binding S1-subunit and the membrane fusion S2-subunit. The S1 subunit receptor-binding domain (RDB) is specifically recognized by the host receptor. When the S1 subunit binds to a host-cell receptor, the prefusion trimer is destabilized, resulting in the shedding of the S1 subunit, and the state transition of the S2 subunit to a stable postfusion conformation [143]. The critical function of the S protein can be a breakthrough in vaccine design and development.
Great efforts are being made for the discovery of antiviral drugs, but there are no licensed therapeutic or vaccine for the treatment of SARS-CoV-2 infection available in the market. Developing an effective treatment for SARS-CoV-2 is, therefore, a research priority. It is time-consuming and expensive to design novel vaccines against viruses by the use of kits and related antibodies [144]. Thus, choosing the method of immune-informatics is more efficient and more applicable for deep analysis of viral antigens, B- and T-cell linear epitope prediction, and evaluation of immunogenicity and virulence of pathogens. Among those can be analyzed, B-cell can recognize and activate defense responses against viral infection, T-cell and antibody reactions may recover extreme respiratory infection (Fig. 2 ).
Fig. 2.
The overall workflow of computational drug and vaccine design by using a novel coronavirus spike protein.
The immune-informatics approaches to identify B- and T-cell epitopes for surface glycoprotein (S) of SARS-CoV-2, followed by estimating their antigenicity and interactions with the human leukocyte antigen (HLA) alleles was taken by Li et al. [145]. Four B cell epitopes, two MHC class-I and nine MHC class-II binding T-cell epitopes, which showed highly antigenic features were identified. Allergenicity, toxicity and physiochemical properties analysis confirmed the specificity and selectivity of epitopes. The stability and safety of epitopes were confirmed by digestion analysis. No mutations were observed in all the selected B- and T-cell epitopes across all isolates from different locations worldwide. Epitopes were thus identified and some of them can be potential candidates for vaccine development, as described by Li et al. [145].
8. Conclusion
As noted before, COVID-19 has become a global concern, due to widespread outbreaks and lack of treatment. Therefore, it is necessary to find and evaluate treatment methods more quickly in this case computer methods are very effective and helpful. The predicted binding and ranking of drugs will also be useful to interpret the results of ongoing clinical trials that are testing existing drugs for effectiveness against COVID-19. Notwithstanding the limitations, it has been described several diseases and traits which may be causally related to ACE2 expression the lung, which in turn may mediate susceptibility to Sars-CoV-2 infection. In addition, the proteome-wide MR analysis revealed proteins that could lead to changes in ACE2 expression. Subsequent drug repositioning analysis highlighted several candidates that may warrant further investigations. We stress that most of the findings require replications and validation in further studies, especially the part on drug repositioning. Nevertheless, it is believed that this work is of value given the urgency to address the outbreak of Sars-CoV-2.
Funding
This study was supported by Tabriz University of Medical Sciences with grant number 65174.
Declaration of competing interestCOI
None to declare.
Acknowledgment
We thank all staff of Imam Reza Hospital for all they did during Covid-19 pandemic and in memorial of all our colleagues lost their life during this pandemic.
Contributor Information
Ehsaneh Khodadadi, Email: ehsaneh.khodadadi@gmail.com.
Parham Maroufi, Email: Dr.parhammaroufi@yahoo.com.
Ehsan Khodadadi, Email: E.khodadadi@yahoo.com.
Isabella Esposito, Email: Isabellaesposito@libero.it.
Khudaverdi Ganbarov, Email: Khuda1949@mail.ru.
Silvano Espsoito, Email: silvanoesposito@libero.it.
Mehdi Yousefi, Email: yousefime@tbzmed.ac.ir.
Elham Zeinalzadeh, Email: Elham_leyenda@yahoo.com.
Hossein Samadi Kafil, Email: Kafilhs@tbzmed.ac.ir.
References
- 1.Ahlquist P., Noueiry A.O., Lee W.-M., Kushner D.B., Dye B.T. Host factors in positive-strand RNA virus genome replication. J. Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao S., Lin Q., Ran J., Musa S.S., Yang G., Wang W. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int. J. Infect. Dis. 2020;92:214–217. doi: 10.1016/j.ijid.2020.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ozma M.A., Maroufi P., Khodadadi E., Köse Ş., Esposito I., Ganbarov K. Clinical manifestation, diagnosis, prevention and control of SARS-CoV-2 (Covid-19) during the outbreak period. Inf. Med. 2020;28 [PubMed] [Google Scholar]
- 5.Hui D.S., I Azhar E., Madani T.A., Ntoumi F., Kock R., Dar O. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2020;7:1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.F.-W., Kok K.-H., Zhu Z., Chu H., To K.K.-W., Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microb. Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J. Pathogenicity and transmissibility of 2019-nCoV—a quick overview and comparison with other emerging viruses. Microb. Infect. 2020;22(2):69–71. doi: 10.1016/j.micinf.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV) Biosci. Trends. 2020;14:69–71. doi: 10.5582/bst.2020.01020. [DOI] [PubMed] [Google Scholar]
- 11.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotten M., Watson S.J., Kellam P., Al-Rabeeah A.A., Makhdoom H.Q., Assiri A. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: a descriptive genomic study. Lancet. 2013;382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fathizadeh H., Maroufi P., Momen-Heravi M., Dao S., Köse Ş., Ganbarov K. Protection and disinfection policies against SARS-CoV-2 (COVID-19) Inf. Med. 2020;28 [PubMed] [Google Scholar]
- 14.Killerby M.E., Biggs H.M., Midgley C.M., Gerber S.I., Watson J.T. Middle East respiratory syndrome coronavirus transmission. Emerg. Infect. Dis. 2020;26:191. doi: 10.3201/eid2602.190697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho C.-C., Lin M.-H., Chuang C.-Y., Hsu C.-H. Macro domain from Middle East respiratory syndrome coronavirus (MERS-CoV) is an efficient ADP-ribose binding module crystal structure and biochemical studies. J. Biol. Chem. 2016;291:4894–4902. doi: 10.1074/jbc.M115.700542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang N., Rosen O., Wang L., Turner H.L., Stevens L.J., Corbett K.S. Structural definition of a neutralization-sensitive epitope on the MERS-CoV S1-NTD. Cell Rep. 2019;28:3395–3405. e6. doi: 10.1016/j.celrep.2019.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang R., Zhang X., Irwin D.M., Shen Y. Emergence of SARS-like Coronavirus poses new challenge in China. J. Infect. 2020;80:350–371. doi: 10.1016/j.jinf.2020.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzzi P.H., Mercatelli D., Ceraolo C., Giorgi F.M. Master regulator analysis of the SARS-CoV-2/human interactome. bioRxiv. 2020;9(4):982. doi: 10.3390/jcm9040982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martí-Renom M.A., Stuart A.C., Fiser A., Sánchez R., Melo F., Šali A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 20.Cavasotto C.N., Phatak S.S. Homology modeling in drug discovery: current trends and applications. Drug Discov. Today. 2009;14:676–683. doi: 10.1016/j.drudis.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 21.Burley S.K., Almo S.C., Bonanno J.B., Capel M., Chance M.R., Gaasterland T. Structural genomics: beyond the human genome project. Nat. Genet. 1999;23:151–157. doi: 10.1038/13783. [DOI] [PubMed] [Google Scholar]
- 22.Wichapong K., Pianwanit S., Sippl W., Kokpol S. Homology modeling and molecular dynamics simulations of Dengue virus NS2B/NS3 protease: insight into molecular interaction. J. Mol. Recogn.: Interdiscipl. J. 2010;23:283–300. doi: 10.1002/jmr.977. [DOI] [PubMed] [Google Scholar]
- 23.Ekins S., Liebler J., Neves B.J., Lewis W.G., Coffee M., Bienstock R. Illustrating and homology modeling the proteins of the Zika virus. F1000Research. 2016;5 doi: 10.12688/f1000research.8213.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis F.P., Barkan D.T., Eswar N., McKerrow J.H., Sali A. Host–pathogen protein interactions predicted by comparative modeling. Protein Sci. 2007;16:2585–2596. doi: 10.1110/ps.073228407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell R.B., Alber F., Aloy P., Davis F.P., Korkin D., Pichaud M. A structural perspective on protein–protein interactions. Curr. Opin. Struct. Biol. 2004;14:313–324. doi: 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Q.C., Petrey D., Deng L., Qiang L., Shi Y., Thu C.A. Structure-based prediction of protein–protein interactions on a genome-wide scale. Nature. 2012;490:556–560. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang S.-Q., Du Q.-S., Chou K.-C. Study of drug resistance of chicken influenza A virus (H5N1) from homology-modeled 3D structures of neuraminidases. Biochem. Biophys. Res. Commun. 2007;354:634–640. doi: 10.1016/j.bbrc.2006.12.235. [DOI] [PubMed] [Google Scholar]
- 28.Cavasotto C.N., Orry A.J., Murgolo N.J., Czarniecki M.F., Kocsi S.A., Hawes B.E. Discovery of novel chemotypes to a G-protein-coupled receptor through ligand-steered homology modeling and structure-based virtual screening. J. Med. Chem. 2008;51:581–588. doi: 10.1021/jm070759m. [DOI] [PubMed] [Google Scholar]
- 29.Loewenstein Y., Raimondo D., Redfern O.C., Watson J., Frishman D., Linial M. Protein function annotation by homology-based inference. Genome Biol. 2009;10:207. doi: 10.1186/gb-2009-10-2-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vidalain P.-O., Tangy F. Virus-host protein interactions in RNA viruses. Microb. Infect. 2010;12:1134–1143. doi: 10.1016/j.micinf.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Zhang A., He L., Wang Y. Prediction of GCRV virus-host protein interactome based on structural motif-domain interactions. BMC Bioinf. 2017;18:145. doi: 10.1186/s12859-017-1500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durmuş S., Ülgen K.Ö. Comparative interactomics for virus–human protein–protein interactions: DNA viruses versus RNA viruses. FEBS Open Biol. 2017;7:96–107. doi: 10.1002/2211-5463.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Warren S., Wan X.-F., Conant G., Korkin D. Extreme evolutionary conservation of functionally important regions in H1N1 influenza proteome. PloS One. 2013;8 doi: 10.1371/journal.pone.0081027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Valderramos S.G., Wu A., Ouyang S., Li C., Brasil P. From mosquitos to humans: genetic evolution of Zika virus. Cell Host Microbe. 2016;19:561–565. doi: 10.1016/j.chom.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel H., Kukol A. Prediction of ligands to universally conserved binding sites of the influenza a virus nuclear export protein. Virology. 2019;537:97–103. doi: 10.1016/j.virol.2019.08.013. [DOI] [PubMed] [Google Scholar]
- 36.Srinivasan S., Cui H., Gao Z., Liu M., Lu S., Mkandawire W., Narykov O., Sun M., Korkin D. Structural Genomics of SARS-CoV-2 Indicates Evolutionary Conserved Functional Regions of Viral Proteins. Viruses. 2020;12:360. doi: 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawicki S.G. Viral Genome Replication. Springer; 2009. Coronavirus genome replication; pp. 25–39. [Google Scholar]
- 39.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramajayam R., Tan K.-P., Liang P.-H. Portland Press Ltd.; 2011. Recent Development of 3C and 3CL Protease Inhibitors for Anti-coronavirus and Anti-picornavirus Drug Discovery. [DOI] [PubMed] [Google Scholar]
- 41.Shang J., Wan Y., Liu C., Yount B., Gully K., Yang Y. Structure of mouse coronavirus spike protein complexed with receptor reveals mechanism for viral entry. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson G.W., Stohlman S.A., Tahara S.M. High affinity interaction between nucleocapsid protein and leader/intergenic sequence of mouse hepatitis virus RNA. J. Gen. Virol. 2000;81:181–188. doi: 10.1099/0022-1317-81-1-181. [DOI] [PubMed] [Google Scholar]
- 43.Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94 doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masters P.S., Sturman L.S. Coronaviruses and Their Diseases: Springer. 1990. Background paper functions of the coronavirus nucleocapsid protein; pp. 235–238. [DOI] [PubMed] [Google Scholar]
- 45.McBride R., Van Zyl M., Fielding B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses. 2014;6:2991–3018. doi: 10.3390/v6082991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang T.K., Wu M.P.J., Chen S.T., Hou M.H., Hong M.H., Pan F.M. Biochemical and immunological studies of nucleocapsid proteins of severe acute respiratory syndrome and 229E human coronaviruses. Proteomics. 2005;5:925–937. doi: 10.1002/pmic.200401204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Du L., Zhao G., Lin Y., Chan C., He Y., Jiang S. Priming with rAAV encoding RBD of SARS-CoV S protein and boosting with RBD-specific peptides for T cell epitopes elevated humoral and cellular immune responses against SARS-CoV infection. Vaccine. 2008;26:1644–1651. doi: 10.1016/j.vaccine.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hsieh P.-K., Chang S.C., Huang C.-C., Lee T.-T., Hsiao C.-W., Kou Y.-H. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005;79:13848–13855. doi: 10.1128/JVI.79.22.13848-13855.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12:254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu S.-J., Leng C.-H., Lien S-p, Chi H.-Y., Huang C.-Y., Lin C.-L. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24:3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shang B., Wang X.-Y., Yuan J.-W., Vabret A., Wu X.-D., Yang R.-F. Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus. Biochem. Biophys. Res. Commun. 2005;336:110–117. doi: 10.1016/j.bbrc.2005.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lo Y.-S., Lin S.-Y., Wang S.-M., Wang C.-T., Chiu Y.-L., Huang T.-H. Oligomerization of the carboxyl terminal domain of the human coronavirus 229E nucleocapsid protein. FEBS Lett. 2013;587:120–127. doi: 10.1016/j.febslet.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen I.-J., Yuann J.-M.P., Chang Y.-M., Lin S.-Y., Zhao J., Perlman S. Crystal structure-based exploration of the important role of Arg106 in the RNA-binding domain of human coronavirus OC43 nucleocapsid protein. Biochim. Biophys. Acta Protein Proteonomics. 2013;1834:1054–1062. doi: 10.1016/j.bbapap.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chang C-k, Chen C.-M.M., Chiang M-h, Hsu Y-l, Huang T-h. Transient oligomerization of the SARS-CoV N protein–implication for virus ribonucleoprotein packaging. PloS One. 2013;8 doi: 10.1371/journal.pone.0065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saikatendu K.S., Joseph J.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C. Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. J. Virol. 2007;81:3913–3921. doi: 10.1128/JVI.02236-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jayaram H., Fan H., Bowman B.R., Ooi A., Jayaram J., Collisson E.W. X-ray structures of the N-and C-terminal domains of a coronavirus nucleocapsid protein: implications for nucleocapsid formation. J. Virol. 2006;80:6612–6620. doi: 10.1128/JVI.00157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fan H., Ooi A., Tan Y.W., Wang S., Fang S., Liu D.X. The nucleocapsid protein of coronavirus infectious bronchitis virus: crystal structure of its N-terminal domain and multimerization properties. Structure. 2005;13:1859–1868. doi: 10.1016/j.str.2005.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grossoehme N.E., Li L., Keane S.C., Liu P., Dann C.E., III, Leibowitz J.L. Coronavirus N protein N-terminal domain (NTD) specifically binds the transcriptional regulatory sequence (TRS) and melts TRS-cTRS RNA duplexes. J. Mol. Biol. 2009;394:544–557. doi: 10.1016/j.jmb.2009.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan Y.W., Fang S., Fan H., Lescar J., Liu D. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keane S.C., Liu P., Leibowitz J.L., Giedroc D.P. Functional transcriptional regulatory sequence (TRS) RNA binding and helix destabilizing determinants of murine hepatitis virus (MHV) nucleocapsid (N) protein. J. Biol. Chem. 2012;287:7063–7073. doi: 10.1074/jbc.M111.287763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lin S.-M., Lin S.-C., Hsu J.-N., Chang C-k, Chien C.-M., Wang Y.-S. Structure-based stabilization of non-native protein-protein interactions of coronavirus nucleocapsid proteins in antiviral drug design. J. Med. Chem. 2020;63(6):3131–3141. doi: 10.1021/acs.jmedchem.9b01913. [DOI] [PubMed] [Google Scholar]
- 62.Kang S., Yang M., Hong Z., Zhang L., Huang Z., Chen X. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharmaceutica Sinica B. 2020 doi: 10.1016/j.apsb.2020.04.009. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chazal N., Gerlier D. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 2003;67:226–237. doi: 10.1128/MMBR.67.2.226-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the putative receptor of Wuhan 2019-nCov. BioRxiv. 2020 doi: 10.1164/rccm.202001-0179LE. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chai X., Hu L., Zhang Y., Han W., Lu Z., Ke A. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection. bioRxiv. 2020 Preprint. [Google Scholar]
- 67.Zhang H., Kang Z., Gong H. Digestive system is a potential route of COVID-19: an analysis of single-cell coexpression pattern of key proteins in viral entry process. Gut. 2020 doi: 10.1136/gutjnl-2020-320953. [DOI] [Google Scholar]
- 68.Zou X., Chen K., Zou J., Han P., Hao J., Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front. Med. 2020:1–8. doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Z., Zhu Z., Chen W., Cai Z., Xu B., Tan Z. Cell membrane proteins with high n-glycosylation, high expression and multiple interaction partners are preferred by mammalian viruses as receptors. Bioinformatics. 2019;35:723–728. doi: 10.1093/bioinformatics/bty694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoffmann M., Kleine-Weber H., Krüger N., Mueller M.A., Drosten C., Pöhlmann S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv. 2020 Preprint. [Google Scholar]
- 71.Wang S., Guo F., Liu K., Wang H., Rao S., Yang P. Endocytosis of the receptor-binding domain of SARS-CoV spike protein together with virus receptor ACE2. Virus Res. 2008;136:8–15. doi: 10.1016/j.virusres.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu Y., Cheng Y., Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol. Sin. 2020:1–6. doi: 10.1007/s12250-020-00207-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526(1):135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marzi A., Gramberg T., Simmons G., Möller P., Rennekamp A.J., Krumbiegel M. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 2004;78:12090–12095. doi: 10.1128/JVI.78.21.12090-12095.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Peck K.M., Scobey T., Swanstrom J., Jensen K.L., Burch C.L., Baric R.S. Permissivity of dipeptidyl peptidase 4 orthologs to Middle East respiratory syndrome coronavirus is governed by glycosylation and other complex determinants. J. Virol. 2017;91 doi: 10.1128/JVI.00534-17. e00534-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li F. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J. Virol. 2015;89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Semba R.D. Vitamin A and immunity to viral, bacterial and protozoan infections. Proc. Nutr. Soc. 1999;58:719–727. doi: 10.1017/s0029665199000944. [DOI] [PubMed] [Google Scholar]
- 79.Keil S.D., Bowen R., Marschner S. Inactivation of M iddle E ast respiratory syndrome coronavirus (MERS‐C o V) in plasma products using a riboflavin‐based and ultraviolet light‐based photochemical treatment. Transfusion. 2016;56:2948–2952. doi: 10.1111/trf.13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Atherton J., Kratzing C., Fisher A. The effect of ascorbic acid on infection of chick-embryo ciliated tracheal organ cultures by coronavirus. Arch. Virol. 1978;56:195–199. doi: 10.1007/BF01317848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nonnecke B., McGill J., Ridpath J., Sacco R., Lippolis J., Reinhardt T. Acute phase response elicited by experimental bovine diarrhea virus (BVDV) infection is associated with decreased vitamin D and E status of vitamin-replete preruminant calves. J. Dairy Sci. 2014;97:5566–5579. doi: 10.3168/jds.2014-8293. [DOI] [PubMed] [Google Scholar]
- 82.Beck M.A., Kolbeck P.C., Rohr L.H., Shi Q., Morris V.C., Levander O.A. Vitamin E deficiency intensifies the myocardial injury of coxsackievirus B3 infection of mice. J. Nutr. 1994;124:345–358. doi: 10.1093/jn/124.3.345. [DOI] [PubMed] [Google Scholar]
- 83.Morita M., Kuba K., Ichikawa A., Nakayama M., Katahira J., Iwamoto R. The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell. 2013;153:112–125. doi: 10.1016/j.cell.2013.02.027. [DOI] [PubMed] [Google Scholar]
- 84.Rayman M.P. Selenium and human health. Lancet. 2012;379:1256–1268. doi: 10.1016/S0140-6736(11)61452-9. [DOI] [PubMed] [Google Scholar]
- 85.Te Velthuis A.J., van den Worm S.H., Sims A.C., Baric R.S., Snijder E.J., van Hemert M.J. Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wessling-Resnick M. Crossing the iron gate: why and how transferrin receptors mediate viral entry. Annu. Rev. Nutr. 2018;38:431–458. doi: 10.1146/annurev-nutr-082117-051749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Momattin H., Al-Ali A.Y., Mohammed K., Al-Tawfiq J.A. Benchmarking of antibiotic usage: an adjustment to reflect antibiotic stewardship program outcome in a hospital in Saudi Arabia. J. Infect. Publ. Health. 2018;11:310–313. doi: 10.1016/j.jiph.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lew T.W., Kwek T.-K., Tai D., Earnest A., Loo S., Singh K. Acute respiratory distress syndrome in critically ill patients with severe acute respiratory syndrome. JAMA. 2003;290:374–380. doi: 10.1001/jama.290.3.374. [DOI] [PubMed] [Google Scholar]
- 89.Baumann C.A., Badamchian M., Goldstein A.L. Thymosin α1 antagonizes dexamethasone and CD3-induced apoptosis of CD4+ CD8+ thymocytes through the activation of cAMP and protein kinase C dependent second messenger pathways. Mech. Ageing Dev. 1997;94:85–101. doi: 10.1016/s0047-6374(96)01860-x. [DOI] [PubMed] [Google Scholar]
- 90.Gao Z., Zhu J., Sun Y., Ding X., Ma J., Cui Y. Clinical investigation of outbreak of nosocomial severe acute respiratory syndrome. Protection and disinfection policies against SARS-CoV-2 (COVID-19) Chin. Crit. Care Med. 2003;15:332–335. [PubMed] [Google Scholar]
- 91.Kumar V., Jung Y.-S., Liang P.-H. Anti-SARS coronavirus agents: a patent review (2008–present) Expert Opin. Ther. Pat. 2013;23:1337–1348. doi: 10.1517/13543776.2013.823159. [DOI] [PubMed] [Google Scholar]
- 92.Duchateau J., Servais G., Vreyens R., Delespesse G., Bolla K. Modulation of immune response in aged humans through different administration modes of thymopentin. Surv. Immunol. Res. 1985;4:94. [PubMed] [Google Scholar]
- 93.Joffe M., Sukha N., Rabson A. Lymphocyte subsets in measles. Depressed helper/inducer subpopulation reversed by in vitro treatment with levamisole and ascorbic acid. J. Clin. Invest. 1983;72:971–980. doi: 10.1172/JCI111069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Luo C., Luo H., Zheng S., Gui C., Yue L., Yu C. Nucleocapsid protein of SARS coronavirus tightly binds to human cyclophilin A. Biochem. Biophys. Res. Commun. 2004;321:557–565. doi: 10.1016/j.bbrc.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Park J.-Y., Jeong H.J., Kim J.H., Kim Y.M., Park S.-J., Kim D. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012;35(11):2036–2042. doi: 10.1248/bpb.b12-00623. [DOI] [PubMed] [Google Scholar]
- 96.Chen L., Gui C., Luo X., Yang Q., Günther S., Scandella E. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Diwan A., Ninawe A., Harke S. Gene editing (CRISPR-Cas) technology and fisheries sector. Can. J. Biotechnol. 2017;1:65–72. [Google Scholar]
- 98.Jo S., Kim H., Kim S., Shin D.H., Kim M.S. Characteristics of flavonoids as potent MERS‐CoV 3C‐like protease inhibitors. Chem. Biol. Drug Des. 2019;94:2023–2030. doi: 10.1111/cbdd.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dimitrov D.S. The secret life of ACE2 as a receptor for the SARS virus. Cell. 2003;115:652–653. doi: 10.1016/S0092-8674(03)00976-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang L., Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 2020;92(5):479–490. doi: 10.1002/jmv.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yeung K.S., Yamanaka G.A., Meanwell N.A. Severe acute respiratory syndrome coronavirus entry into host cells: opportunities for therapeutic intervention. Med. Res. Rev. 2006;26:414–433. doi: 10.1002/med.20055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sui J., Li W., Murakami A., Tamin A., Matthews L.J., Wong S.K. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. Unit. States Am. 2004;101:2536–2541. doi: 10.1073/pnas.0307140101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today's diseases. Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alves D.S., Pérez-Fons L., Estepa A., Micol V. Membrane-related effects underlying the biological activity of the anthraquinones emodin and barbaloin. Biochem. Pharmacol. 2004;68:549–561. doi: 10.1016/j.bcp.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 106.Vickers N.J. Animal communication: when I'm calling you, will you answer too? Curr. Biol. 2017;27:R713–R715. doi: 10.1016/j.cub.2017.05.064. [DOI] [PubMed] [Google Scholar]
- 107.Zhang X.W., Yap Y.L. Old drugs as lead compounds for a new disease? Binding analysis of SARS coronavirus main proteinase with HIV, psychotic and parasite drugs. Bioorg. Med. Chem. 2004;12:2517–2521. doi: 10.1016/j.bmc.2004.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cauwenberghs S., Feijge M.A., Harper A.G., Sage S.O., Curvers J., Heemskerk J.W. Shedding of procoagulant microparticles from unstimulated platelets by integrin‐mediated destabilization of actin cytoskeleton. FEBS Lett. 2006;580:5313–5320. doi: 10.1016/j.febslet.2006.08.082. [DOI] [PubMed] [Google Scholar]
- 109.Takahashi S., Yoshiya T., Yoshizawa-Kumagaye K., Sugiyama T. Nicotianamine is a novel angiotensin-converting enzyme 2 inhibitor in soybean. Biomed. Res. 2015;36:219–224. doi: 10.2220/biomedres.36.219. [DOI] [PubMed] [Google Scholar]
- 110.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 111.Morgenstern B., Michaelis M., Baer P.C., Doerr H.W., Cinatl J., Jr. Ribavirin and interferon-β synergistically inhibit SARS-associated coronavirus replication in animal and human cell lines. Biochem. Biophys. Res. Commun. 2005;326:905–908. doi: 10.1016/j.bbrc.2004.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Tsang K., Zhong N.S. SARS: pharmacotherapy. Respirology. 2003;8:S25–S30. doi: 10.1046/j.1440-1843.2003.00525.x. [DOI] [PubMed] [Google Scholar]
- 113.Kim U.J., Won E.-J., Kee S.-J., Jung S.-I., Jang H.-C. Case report Combination therapy with lopinavir/ritonavir, ribavirin and interferon-α for Middle East respiratory syndrome. Antivir. Ther. 2016;21:455–459. doi: 10.3851/IMP3002. [DOI] [PubMed] [Google Scholar]
- 114.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. e00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yamamoto N., Yang R., Yoshinaka Y., Amari S., Nakano T., Cinatl J. HIV protease inhibitor nelfinavir inhibits replication of SARS-associated coronavirus. Biochem. Biophys. Res. Commun. 2004;318:719–725. doi: 10.1016/j.bbrc.2004.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Khamitov R., Loginova S., Shchukina V., Borisevich S., Maksimov V., Shuster A. Antiviral activity of arbidol and its derivatives against the pathogen of severe acute respiratory syndrome in the cell cultures. Vopr. Virusol. 2008;53:9–13. [PubMed] [Google Scholar]
- 118.Robbins R.A., Grisham M.B. Nitric oxide. Int. J. Biochem. Cell Biol. 1997;29:857–860. doi: 10.1016/s1357-2725(96)00167-7. [DOI] [PubMed] [Google Scholar]
- 119.Åkerström S., Mousavi-Jazi M., Klingström J., Leijon M., Å Lundkvist, Mirazimi A. Nitric oxide inhibits the replication cycle of severe acute respiratory syndrome coronavirus. J. Virol. 2005;79:1966–1969. doi: 10.1128/JVI.79.3.1966-1969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Talluri S. 2020. Virtual High Throughput Screening Based Prediction of Potential Drugs for COVID-19. [DOI] [PubMed] [Google Scholar]
- 121.Macalino S.J.Y., Gosu V., Hong S., Choi S. Role of computer-aided drug design in modern drug discovery. Arch Pharm. Res. (Seoul) 2015;38:1686–1701. doi: 10.1007/s12272-015-0640-5. [DOI] [PubMed] [Google Scholar]
- 122.Mohanasundaram N., Sekhar T. Computational studies of molecular targets regarding the adverse effects of isoniazid drug for tuberculosis. Curr. Pharmacogenom. Pers. Med. (Former. Curr. Pharmacogenom.) 2018;16:210–218. [Google Scholar]
- 123.Li J., Zheng S., Chen B., Butte A.J., Swamidass S.J., Lu Z. A survey of current trends in computational drug repositioning. Bioinf. 2016;17:2–12. doi: 10.1093/bib/bbv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hurle M., Yang L., Xie Q., Rajpal D., Sanseau P., Agarwal P. Computational drug repositioning: from data to therapeutics. Clin. Pharmacol. Therapeut. 2013;93:335–341. doi: 10.1038/clpt.2013.1. [DOI] [PubMed] [Google Scholar]
- 125.Novac N. Challenges and opportunities of drug repositioning. Trends Pharmacol. Sci. 2013;34:267–272. doi: 10.1016/j.tips.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 126.Liu X., Zhang B., Jin Z., Yang H., Rao Z. RCSB Protein Data Bank; 2020. The Crystal Structure of 2019-NCoV Main Protease in Complex with an Inhibitor N3. [Google Scholar]
- 127.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92(4):418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 129.Hughes J.P., Rees S., Kalindjian S.B., Philpott K.L. Principles of early drug discovery. Br. J. Pharmacol. 2011;162:1239–1249. doi: 10.1111/j.1476-5381.2010.01127.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang H.-J., Yu H.W., Chen C.-Y., Hsu C.-H., Chen H.-Y., Lee K.-J. Current developments of computer-aided drug design. J. Taiwan Inst. Chem. Eng. 2010;41:623–635. [Google Scholar]
- 131.Kapetanovic I. Computer-aided drug discovery and development (CADDD): in silico-chemico-biological approach. Chem. Biol. Interact. 2008;171:165–176. doi: 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kumar S. Computational identification and binding analysis of orphan human cytochrome P450 4X1 enzyme with substrates. BMC Res. Notes. 2015;8:9. doi: 10.1186/s13104-015-0976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Van Drie J.H. Computer-aided drug design: the next 20 years. J. Comput. Aided Mol. Des. 2007;21:591–601. doi: 10.1007/s10822-007-9142-y. [DOI] [PubMed] [Google Scholar]
- 134.Veselovsky A., Ivanov A. Strategy of computer-aided drug design. Curr. Drug Targets - Infect. Disord. 2003;3:33–40. doi: 10.2174/1568005033342145. [DOI] [PubMed] [Google Scholar]
- 135.Coordinators N.R. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2017;45:D12–D17. doi: 10.1093/nar/gkw1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Marchler-Bauer A., Lu S., Anderson J., Chitsaz F., Derbyshire M., DeWeese-Scott C. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chou K.-C., Shen H.-B. Cell-PLoc 2.0: an improved package of web-servers for predicting subcellular localization of proteins in various organisms. Nat. Sci. 2010;2:1090. doi: 10.1038/nprot.2007.494. [DOI] [PubMed] [Google Scholar]
- 138.Kelley L., Mezulis S., Yates C., Wass M., Sternberg M. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kumar S. 2020. Drug and Vaccine Design against Novel Coronavirus (2019-nCoV) Spike Protein through Computational Approach. [Google Scholar]
- 140.Lindenbach B.D., Rice C.M. Molecular biology of flaviviruses. Adv. Virus Res. 2003;59:23–62. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 141.Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Walls A.C., Tortorici M.A., Snijder J., Xiong X., Bosch B.-J., Rey F.A. Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc. Natl. Acad. Sci. Unit. States Am. 2017;114:11157–11162. doi: 10.1073/pnas.1708727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.ul Qamar M.T., Bari A., Adeel M.M., Maryam A., Ashfaq U.A., Du X. Peptide vaccine against chikungunya virus: immuno-informatics combined with molecular docking approach. J. Transl. Med. 2018;16:298. doi: 10.1186/s12967-018-1672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li L., Sun T., He Y., Li W., Fan Y., Zhang J. Epitope-based peptide vaccines predicted against novel coronavirus disease caused by SARS-CoV-2. BioRxiv. 2020 doi: 10.1016/j.virusres.2020.198082. Preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]