Abstract
Proteases play a pivotal role in several biological processes, from digestion, cell proliferation, and differentiation to fertility. Deregulation of protease metabolism can result in several pathological conditions (i.e., cancer, neurodegenerative disorders, and others). Therefore, monitoring proteolytic activity in real time could have a fundamental role in the early diagnosis of these diseases. Herein, the main approaches used to develop biosensors for monitoring proteolytic activity are reviewed. A comparison of the advantages and disadvantages of each approach is provided along with a discussion of their importance and promising opportunities for the early diagnosis of severe diseases. This new era of biosensors can be characterized by the ability to control and monitor biological processes, ultimately improving the potential of personalized medicine.
Keywords: personalized medicine, biosensors, proteases, proteolytic activity, real time
Highlights
Proteases are widespread in nature and play a pivotal role in many biological processes in life forms and viruses.
Deregulation of protease metabolism can result in several pathological conditions (i.e., cancer, neurodegenerative disorders, and others).
Assays and biosensors that measure proteolytic activity in real time can provide a wealth of information contributing to the rapid evaluation of individual health conditions.
Protease biosensors are mostly based on molecular rulers and usually rely on optical, electrical, or magnetic detection.
Novel and improved protease sensing systems are rapidly emerging with the potential to impact healthcare, drug development, and fundamental studies in biology and biochemistry.
Biosensing in Personalized Medicine: The Emergence of Protease Biomarkers
A deep understanding of the individual characteristics of biological processes and precise control of responses and feedback to therapies are essential in a world that is running towards personalized medicine (see Glossary). Recent reports predict that this market will grow at a compound annual growth rate (CAGR) of over 11% from US$92.4 billion dollars in 2017 to US$194.4 billion in 2024 (https://www.researchandmarkets.com/research/n5kqz7/global?w=5), with oncology responsible for 30% of the revenues generated. Biosensors are one of the underlying technologies of personalized medicine, giving support to the highly informed decisions that are critical to obtain better clinical outcomes and to decrease undesired side-effects. Compared with the personalized medicine market, a similar tendency is observed for the biosensors market where growth from US$18.6 billion dollars in 2018 to US$31.5 billion in 2025 (8% CAGR) is expected (https://www.gminsights.com/industry-analysis/biosensors-market).
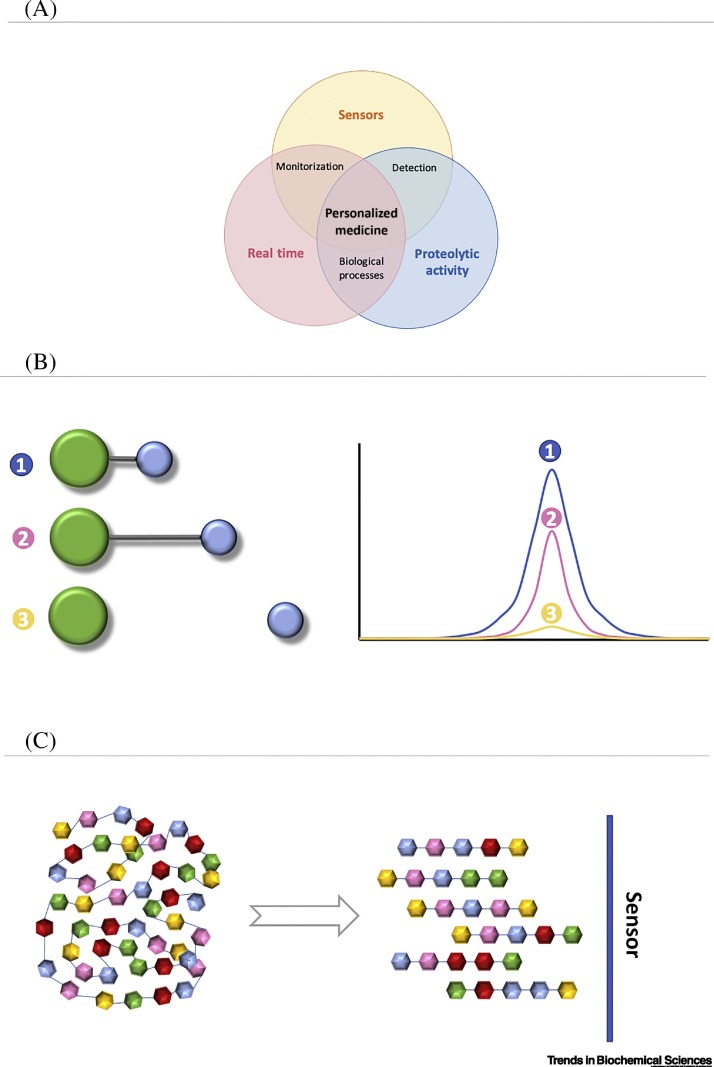
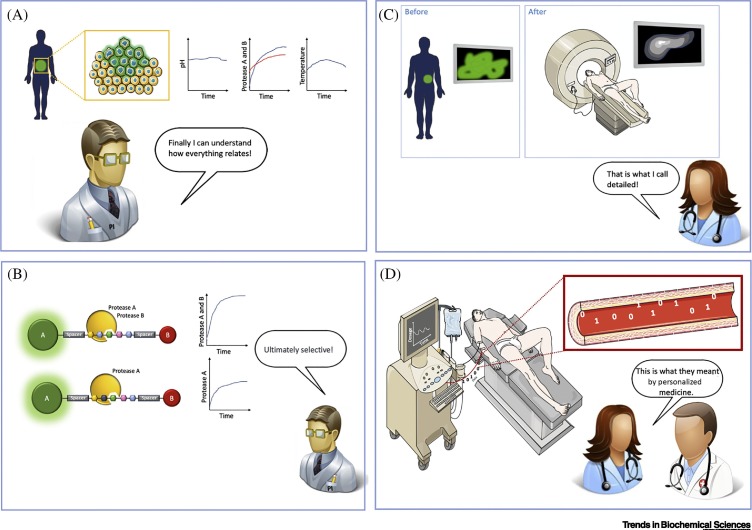
One of the primary goals of a biosensor is to determine the presence/absence or activity of a given biomarker and to correlate these results with pathophysiological conditions. Proteases are emerging as a new category among the wide variety of biomarkers targeted by biosensors due to their key role in health and disease. In this review, we present the main approaches used to measure proteolytic activity in real time – for both in vivo and in vitro applications – and discuss recent developments in and future possibilities for protease biosensors (Figure 1A).
Figure 1.
Real-Time Proteolytic-Activity Biosensors and Personalized Medicine.
(A) Triangulation of proteolytic-activity sensors in real time onto personalized medicine. (B) Generalized scheme illustrating the working principle of a ‘molecular ruler’. Here, the interaction between two or more entities is distance dependent; thus, the signal detected by the sensor is strictly dependent on the distance. In proteolytic sensors, molecular-rulers operate to differentiate the signal of the intact molecule (absence of target protease) and that of a cleaved molecule (presence of target protease) in which the interaction between the entities cannot occur. (C) Schematic of proteolytic sensor based on the fragmentation of an original target molecule. In such systems, the cleavage of the original molecule induces a change of a given property, which will provide a sensor reading that is proportional to the proteolytic activity.
The Roles and Idiosyncrasies of Proteases: Why Monitor Proteolytic Activity?
Proteases are a widely explored research topic as illustrated by more than 487 000 entries registered in PUBMED. However, it was only recently that proteases emerged as an analyte of interest for biosensing [68]. These hydrolytic enzymes (EC 3.4) play a pivotal role in several biological processes, such as digestion, cell proliferation, and differentiation [1], as well as apoptosis [2], coagulation [3], immunity [4], and fertility [5]. The deregulation of protease metabolism can result in numerous pathological conditions (i.e., cancers [6], neurodegenerative disorders [7], and others [8]). For this reason, the pharmaceutical industry is actively evaluating the potential of proteases as drug targets or as biomarkers for diagnosis and prognosis. In this regard, thrombin and prostate-specific antigen (PSA) can be highlighted as two of the most common protease targets in clinical biochemistry [9,10]. And, one notable example of a protease-targeted therapy is the administration of HIV-1 protease inhibitors (i.e., lopinavir/ritonavir branded as Kaletra from Abbott Laboratories) in the context of AIDS [11]. Lopinavir is a specific inhibitor of HIV proteases, which prevents maturation of the virus, hindering its ability to infect new cells and replicate. However, lopinavir is quickly degraded within the human body by cytochrome P450 3A4 enzyme (CYP3A4) [12]. To overcome this and increase the lifetime of lopinavir, a potent CYP3A4 inhibitor (ritonavir) is included in the formulation. During the preparation of this manuscript (March 27, 2020), the world is facing an unprecedented pandemic due to the outbreak of the coronavirus disease, COVID-19, caused by the virus SARS-CoV-2. Similar to HIV, proteases are pivotal during viral maturation, affecting the ability of the virus to infect new host cells [13]. Likewise, protease-targeted therapies with protease inhibitors are one of the forefront strategies being pursued to fight the outbreak. Several drugs are undergoing testing and clinical trials (https://www.sciencemag.org/news/2020/03/who-launches-global-megatrial-four-most-promising-coronavirus-treatments#), with mild results [14,15], notably lopinavir/ritonavir and arbidol (https://clinicaltrials.gov/ct2/show/NCT04252885). In this regard, the ability to monitor proteolytic activity in real-time can play a pivotal role in screening potential protease inhibitors for therapeutic purposes [16]. The spread of COVID-19 caught most countries off-guard, with their health-care systems being unable to restrain transmission before adequate testing to allow controlled isolation and tracking [17]. This may due to the fact that the ‘gold-standard’ RT-PCR test is time-consuming and expensive, among other limitations (https://www.fda.gov/media/136151/download; https://www.iaea.org/newscenter/news/how-is-the-covid-19-virus-detected-using-real-time-rt-pcr). Alternative, less expensive and easier to use serological tests also have limitations [17]. Both tests may also provide non-definitive results (https://www.politico.eu/article/spanish-government-under-fire-after-defective-testing-kits-fiasco). Although to our knowledge diagnosis of viral infections by proteolytic activity monitoring is unavailable in health care, experimental studies on viral proteolytic maturation should clarify the potential of proteases as drug targets and as diagnostic tools [18].
In view of its biological and therapeutic relevance, understanding and evaluation of proteolytic activity, both in vitro and in vivo, is extremely informative. However, determining protease concentration in serum, tissues, and other samples without correlation with its activity can give limited or erroneous information due to particular features of these enzymes, as described below. Despite the disadvantages of measuring protease concentration instead of proteolytic activity, this is still performed due to the simple adaptation of gold-standard procedures to these enzymes. The presence of proteases is usually established indirectly by measuring genetic expression via RNA quantification or by using immunoassays. A good example of this approach is the use of reverse transcription PCR (RT-PCR) to infer the presence of matrix metalloproteases (MMPs) in rheumatoid arthritis [19]. However, in these studies immunosensing and mRNA quantification can be misleading because proteases are naturally synthetized in an inactive pro-form, which is activated post-expression. Unfortunately, most immunoassays cannot distinguish between the two forms and the detection of mRNA cannot be taken as a guarantee of the presence of proteolytic activity. Additionally, enzymes are sensitive to disease-related environmental conditions (e.g., pH is often lower in cancerous cells) and thus genetic expression does not necessarily correlate with activity [20,21]. Also, proteases are characterized by a high catalytic constant (108–109 M−1s−1), which means that even a very small variation in genetic expression can have a major outcome difference. Finally, these enzymes are often regulated by endogenous inhibitors, such as tissue inhibitors of metalloproteinase (TIMPs), which are responsible for restraining the activity of MMPs, and a disintegrin and metalloproteinases (ADAMs) within the homeostatic range [22]. For the reasons mentioned above, concentration is not always correlated with activity. Alternatively, by measuring the proteolytic activity one is directly evaluating the function of the protease at physiological conditions and not merely its presence. This creates several opportunities, as described next.
Monitoring Proteolytic Activity in Real Time: What Are the Advantages?
Given the involvement of proteases in many metabolic and cellular processes, the determination of proteolytic activity is important to understand the role of specific proteases and of proteolytic cascades in cellular events/pathways. For example: caspases play essential roles in programmed cell death and inflammation [23]; MMPs are key to the degradation of most extracellular matrix proteins during organogenesis, growth, and tissue turnover [24]; and the HIV-1 viral protease is critical for the budding of virions from host cells [25]. In real-time detection, more data points are obtained, allowing the correlation between an occurring event and its influence on proteolytic activity.
In healthcare, determination of the protease biomarkers thrombin and PSA is considered the gold standard in the diagnosis of blood disorders and prostate cancer, respectively. More specifically, recent studies have shown the importance of prothrombin/thrombin regulation and its involvement in neurological diseases [26,27]. Here, it is critical to understand and correlate inactive/active thrombin to predict potential outcomes [28]. Therefore, measuring proteolytic activity in real time can overcome limitations of standard biomarkers while potentially leading to new protease-focused biomarkers, which will greatly contribute to optimize healthcare procedures.
Most current proteolytic activity assays are neither real-time measurements nor suitable for in vivo studies (i.e., zymography assays or protocols that use azo dye-marked substrates such as casein [29,30]). Changing the paradigm to real-time detection, where the sensor gives an immediate response proportional to the proteolytic activity, would be extremely interesting in drug development, fundamental studies, and healthcare. From the perspective of the development of protease-targeted drugs/therapies, measuring enzyme kinetics over time using the same sample and without the need for suicidal experiments decreases the number of assays needed for each test [31]. Usually, proteases and pharmaceutical compounds, especially those of clinical relevance, can be expensive and, by decreasing the number of performed tests, the development costs can diminish significantly. Additionally, in assays that determine the concentration of a given analyte, usually one molecule produces one output signal. When measuring enzyme activity, however, a single enzyme can be responsible for multiple events of substrate cleavage thus leading to multiple readout signals, which ultimately produce signal amplification with a concomitant increase of the signal-to-noise ratio (SNR).
Looking for Activity in Real Time: How Is It Done?
Proteases are hydrolases responsible for cleaving peptide bonds in proteins/peptides. In general, real-time proteolytic assays exploit the molecular-ruler principle [32], whereby a sensor molecule is used that contains two labels bridged by the target peptide sequence (Figure 1B). The distance between the two components should change only due to the proteolytic activity of the target protease – as result of cleavage of the target sequence – thus producing a signal readout, while in control samples this bridging sequence remains intact and, in this case, no readout is produced. Other real-time sensors, by contrast, explore the fact that the cleavage of the original protein target generates smaller fragments and often induces a change in a given property (impedance, diffusion), which will influence the sensor readings proportionally to the proteolytic activity (Figure 1C).
Different Approaches to Different Needs: Which Designs Are Implemented?
Several strategies are currently being explored in real-time proteolytic sensors, as highlighted in the next section and listed in Table 1 .
Table 1.
Highlights of Proteolytic Activity in Real-Time Biosensors and Their Features
| Type of biosensor | Target protease | Method of detection | Limit of detection | Suitable for in vivo | Advantage | Drawback | Refs |
|---|---|---|---|---|---|---|---|
| Optical | MMP-14 | FRET | ND | ✔ | Versatile | May have misleading results | [59]a |
| Spectral properties and brightness | ↓ In-depth penetration imaging | ||||||
| MMP-14 | ND | ✔ | ↓ Background | Genetic modification is needed | [60] | ||
| Micro PA + MMP-9 | ND | ✔ | Multiplexing measurements | ↓ In-depth penetration imaging | [40]a | ||
| Caspase-9 | ND | ✔ | ↑ SNR | ↑ Photobleaching | [39]a | ||
| Caspase-3/8/9 | ND | ✔ | ↓ False-positive interference | [41]a | |||
| Trypsin, chymotrypsin | Double FRET | 1 nM | ✘ | Double validation | Limited by spectra overlapping | [56]a | |
| Multiplexing measurements | |||||||
| Can be used as logic gates | |||||||
| MMP-2 | FRET, NIR-II optical imaging | ND | ✔ | Versatile to use for several proteases | May have misleading results | [35]a | |
| ↑ Spatial resolution through deep tissues | Limited for long-term studies | ||||||
| Photostability and brightness | Laborious manufacture | ||||||
| Caspase-3/8/9 | Bioluminescence resonance ET | 12.5 pM | ✔ | Does not need an excitation light source | May have misleading results | [61] | |
| Easy manufacture | Conditioned by surrounding medium | ||||||
| Compared with FRET, ↓ photobleaching and ↓ light scattering | |||||||
| Caspase-3 | Nanosurface ET | 12.0 pM | ✔ | ↑ Sensitivity | ↓ In-depth penetration imaging | [37] | |
| ↓ Photobleaching | |||||||
| ↑ SNR | |||||||
| Trypsin | Surface-enhanced Raman spectroscopy (SERS) | 8.6 nM | ✘ | Multiplexing measurements | Cannot be used for a wide range of proteases | [62] | |
| Complex equipment needed | |||||||
| Complex manufacture of the nanodomes | |||||||
| PSA | Nanoplasmonic resonator, Raman | 6.0 pM | ✘ | ↑ Sensitivity | Complex equipment needed | [63] | |
| Multiplexing measurements | |||||||
| MMP-2 | Multispectral photoacoustic imaging | ND | ✔ | Spatial resolution | ↑ Scattering level | [64] | |
| Optical contrast without ionizing radiation | ↓ In-depth penetration imaging | ||||||
| Photoacoustic imaging | 0.52 ng/ml | ✔ | Noninvasive | ↑ Scattering level | [65] | ||
| ↑ Depth imaging | ↓ In-depth penetration imaging | ||||||
| ↑ Spatial resolution | |||||||
| Caspase-3 | Optical imaging of plasmon rulers | ND | ✔ | ↓ Photobleaching | Complex equipment is needed | [36]a | |
| Stable over time | Prone to interference | ||||||
| ↑ Enhanced signal intensity | |||||||
| MMP-2/9 | Evanescent wave spectroscopy | 32 nM/256 nM | ✘ | Cheap design | Difficult to adapt | [66] | |
| Portable | ↓ Sensitivity | ||||||
| Constant measurements | ↓ Detection limits | ||||||
| Combined techniques | Thrombin | NIR Fluorescence/ PET | ND | ✔ | ↑ Spatial resolution | [67] | |
| ↑ Sensitivity | Complex equipment is needed | ||||||
| MMP-2/9/13 | ND | ✔ | ↑ Accurate results | Complexity | [45] | ||
| Whole-body imaging | |||||||
| Cathepsin B | NIR fluorescence/ CT | ND | ✔ | ↑ Spatial resolution | ↓ In depth | [42] | |
| Some interference | |||||||
| Rapid imaging | |||||||
| MMP-2/9 | FRET/single-photon emission CT (SPECT) | 4.8 ng/ml | ✔ | Nonradioactive probe | Complex equipment is needed | [43] | |
| Caspase-3/7 | FRET/PET | ND | ✔ | ↑ Spatial resolution | Unstable PET probe | [44] | |
| MMP-2 | FRET/ MRI | 0.64 pM | ✔ | ↑ Time/spatial resolution | Complex equipment is needed | [46]a | |
| ↑ In-depth imaging | |||||||
| ↑ SNR | |||||||
| Non-optical | MMP-9 | IDAMs | 10.0 pM to 10.0 nM | ✘ | ↑ Versatility | Limited to ‘2D systems’ | [49]a |
| Suitable for complex Cause/effect studies | |||||||
| HIV-1 protease | Nanopore | 47.0 pM | ✘ | Stable over time | Steric hindrance | [47]a | |
| Multiplexing | Specific equipment is required | ||||||
| Unambiguous response | Limited to 2D systems | ||||||
| ↑ Sensitive | |||||||
| Caspase-3/7 | MRI | ND | ✔ | ↑ Time/spatial resolution | Complex equipment is needed | [48]a | |
| ↑ In-depth imaging | Very expensive | ||||||
| ↑ SNR | Multiplexing can be difficult |
These examples are detailed in the main text.
Optical Biosensors: From Organic Molecules to Inorganic Particles
Energy transfer (ET)-based biosensors are by far the most commonly used for proteolytic activity determination in real time. These systems are wash free, allow low-depth in vivo imaging, and are very sensitive, and the readout is easily obtained. In most designs, ET occurs through long-range dipole–dipole interactions [Förster mechanism or FRET], in which one entity works as an energy donor and another as an energy acceptor. Herein, the fluorophore properties critically affect the detection limit and the dynamic range of the method; a good fluorophore should possess a high molar absorption coefficient, high quantum yield (QY), chemical and photochemical stability, and, ideally, no or low toxicity [33]. The advantage of organic fluorophores is illustrated by the fact that tailormade proteolytic sensors using cyanine and Atto dyes are commercially available [34]. Furthermore, being molecular-sized entities, these organic dye labels exhibit almost no steric effects, which can be critical in enzymatic assays. Despite these advantages, organic fluorophores, especially near-IR (NIR) dyes, have limiting photophysical properties, such as photobleaching and low QY, which hampers their potential for in vivo applications [34].
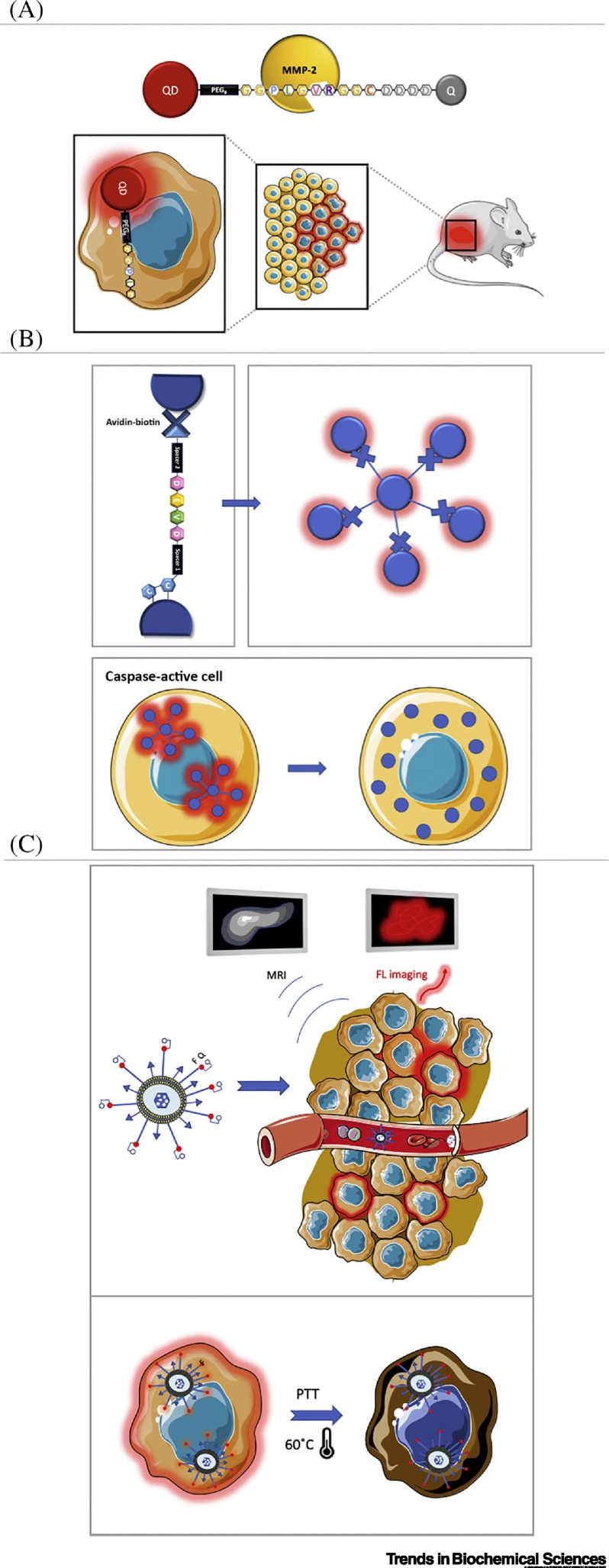
Inorganic emitters such as quantum dots (QDs) can be an alternative to surpass such limitations. These nanocrystals have enhanced optical properties such as high photostability, spectral tunability, and a broad excitation wavelength range, while allowing surface functionalization for biomolecule immobilization [34]. For example, Jeong et al. developed a QD-based ET biosensor in the second NIR region to determine the proteolytic activities of MMP-2 and -9 in real time (Figure 2A) [35]. In this work, a multifunctional peptide was used to bridge a QD to an acceptor that quenches its emission in the absence of MMPs. On breaking the peptide bond, the acceptor is released and the donor brightness increases proportionally to the presence of the active protease. Compared with other NIR dyes, this QD-based sensor displayed better performance for in-depth in vivo imaging. Despite these improvements, the application of QDs in these systems can still be hindered by toxicity, photoblinking phenomena, and size-related issues.
Figure 2.
Optically Based Sensors.
(A) Matrix metalloprotease (MMP)-2/9 detection using a near-IR (NIR-II)-emitting quantum dot (QD) and a quencher (Q). When the enzymes are active, fluorescence (FL) increases and nearby cells are marked. This system was able to detect colon cancer in mouse models. Adapted from [35]. (B) Crown-shaped assembly of 40-nm plasmonic gold nanoparticles bridged by a peptide sequence for Caspase-3/7 detection. Scattered light from the plasmonic assembly is proportional to proteolytic activity, and particles can freely diffuse within the cell after cleavage. Adapted from [36]. (C) Multifunctional nanohybrid comprising Gd-doped CuS nanodisks, a fluorescent probe for MMP-2 activity determination and tumor cell-targeting peptide. After proteolytic cleavage of the activatable probe, tumor margins are determined by FL imaging. Thus, the nanohybrids accumulate on tumor cells allowing MRI with increased spatiotemporal resolution and subsequent photothermal therapy (PTT). Adapted from [46].
As an alternative to luminescence-based sensors, a system based on the scattering of light from plasmonic nanoparticles was develop by Jun et al. [36] to detect Caspase-3 and -7 inside cells using an adenocarcinoma (SW 620) cell line. In their design, the authors used a crown nanoparticle assembly comprising a core 40-nm plasmonic gold particle coupled to several gold nanoparticles by a target sequence for caspase detection (Figure 2B). Using this plasmon ruler, the authors were able to optically probe caspase activity by measuring the scattered light from the nanoassembly and to follow cell-signaling pathways in vivo at a single-molecule level during a period of over 2 h (continuous measuring). The crown nanoassemblies were crucial to obtain a good SNR in such a highly scattering environment, because two or more particles display a scattering signal 44× greater than individual particles. Additionally, the nanoassembly was functionalized with a cell-penetrating peptide (TAT) allowing intracellular/cytosolic sensing of Caspase-3 and -7 activity in real time. The movement of the particles was minimal during experiments due to constriction from the network of cytoskeleton filaments, although after cleavage the resulting particles were able to freely diffuse away from the region of interest. This system proved to be a powerful tool to determine single proteolytic events in real time inside living cells and allowed the researchers to characterize biochemical parameters, such as time lag, and to determine cell heterogeneity in the same sample. The main disadvantage of the strategy appeared to be the difficulty in preparing the nanocrown systems and their inherent heterogeneity.
As shown in several reported works [37,38], gold nanoparticles are valuable optical labels in the field of proteolytic sensors. However, most of the strategies used to conjugate gold nanoparticles and peptides rely on the establishment of sulfur–gold bonds, which can be compromised by interference from other thiol-containing molecules that may be present in the medium. This may result in the effective displacement of the thiol-bonded peptide and thus lead to false-positive results. As a strategy to overcome this problem, Tang and coworkers evaluated the stability of selenol (–SeH)-containing peptides as functionalizing groups in cell cultures [39]. The authors demonstrated that R–SeH-functionalized particles exhibit higher performance and increased stability when exposed to high concentrations of glutathione (GSH) (5 mM) and to high temperatures, relative to their R–SH counterparts. The selenium-functionalized systems showed increased SNR during in vitro tests in MCF-7 cells and serum, showing the potential of this strategy in the development of highly stable and reproducible gold-based nanosensors. Furthermore, the authors developed a multiplexing sensing system to study protease cascades in vitro that was able to discriminate between the activity of Caspase-3 (executioner) and that of -8 and -9 (initiators) with minimal background and nonspecific signal in real time [40,41].
Combined Techniques: What Can Be Done to Surpass Optical Sensors’ Limitations?
A next generation of proteolytic sensors is emerging with the expansion to multiplex-modality systems with activatable probes for in vivo detection/imaging, where optically based sensors are conjugated with other techniques, such as photoacoustic imaging, to improve imaging resolution. So far, the performance of fluorescence-based sensors have been improved and tested using tumor models. For example, Matthias et al. [42] and Yin et al. [43] have combined optical detection with CT. Also, Elvas et al. [44] and Lee et al. [45] have developed probe systems for, respectively, photoacoustic imaging and positron emission tomography (PET) to expand the information provided by their proteolytic sensors.
One notable example is the system recently reported by Shi et al., which includes various techniques to enhance the diagnosis and to kill gastric tumor cells in vivo (Figure 2C) [46]. The authors developed a tumor-targeted and MMP-2-activatable nanoprobe, denoted T-MAN, through an engineered nanoparticle that combined fluorescence/magnetic resonance (MR) bimodal imaging with photothermal therapy (PTT) capabilities. Succinctly, tumor margins are determined using fluorescence imaging activated by the proteolytic activity of MMP-2. After cleavage from MMP-2, a ligand (cRGD) becomes accessible and leads to preferential accumulation and internalization of T-MAN on tumor cells. Consequently, an MR image with enhanced contrast can be acquired due to the presence of Gd-doped CuS nanoparticles in the T-MAN nanohybrids. Additionally, CuS nanodisks exhibit high photothermal conversion efficiency under NIR irradiation, which allows the killing of gastric tumor cells in vitro and in vivo in a mouse model. The potential of these nanoprobes for theranostics of gastric tumors was demonstrated and it was proposed they may also be easily adaptable for other malignant tumors [46].
In the examples above, a map of a metabolic process (proteolytic activity) was acquired using fluorescence imaging and complementary techniques with increased spatial resolution and/or in-depth imaging. Moreover, as demonstrated by the last example, therapeutic agents can be included in the formulation to render theranostic features. For these reasons, we believe that multimodal imaging systems complemented with drug delivery mechanisms are a pathway full of potential to be exploited in the future.
Non-optical Biosensors
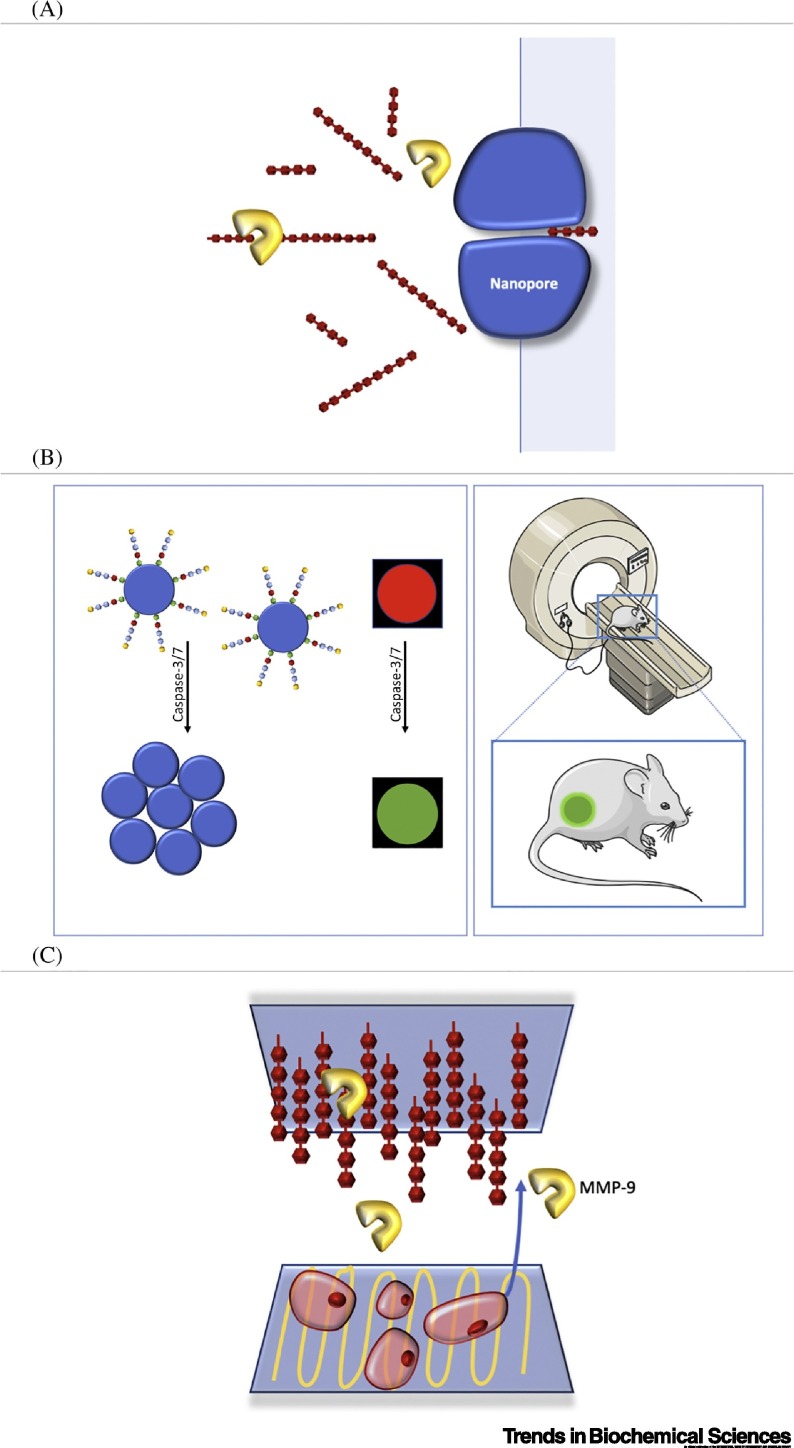
Nanopore-based sensors are an interesting alternative to optical sensors since they allow an immediate clear readout and have become less expensive in recent years. In work by Wang et al. [47], HIV-1 protease activity is monitored in vitro by the translocation of substrate degradation products through a nanopore sensor in real time and in a label-free fashion (Figure 3A). In this design, the peptide substrate that crosses the nanopore-containing membrane produces only one major type of current modulation event. When the target protease is present or active, however, the substrate is cleaved in two differently sized fragments, producing two new types of blockage events with smaller residence times and/or amplitudes relative to the original substrate. Given the high sensitivity of the method and its potential to discriminate the target protease from false positives, this strategy is potentially useful in diagnosis, prognosis, and the development of drugs. The major disadvantage of nanopore systems is that these are not suitable to be used in vivo and in complex media.
Figure 3.
Non-optical Biosensors.
(A) Real-time detection of HIV-1 protease activity using a nanopore. Here, a molecule crossing through the nanopore induces current modulation events. The cleaved peptide rises in two differently sized fragments, which results in different residence times and amplitudes. Adapted from [47]. (B) MRI detection of proteolytic activity from Caspase-3/7 in real time. Peptide-stabilized iron oxide (Fe3O4) nanoparticles lose their colloidal stability and aggregate near apoptotic cells. T2-weighted images were obtained from subcutaneously xenografted HepG2 tumors in nude mice. Adapted from [48]. (C) Simultaneous detection of matrix metalloprotease (MMP)-9 activity and evaluation of cellular behavior using interdigitated-array microelectrodes (IDAMs). Here, the bottom layer detects the cells’ morphology change by differences in resistance while the upper layer detects the presence of secreted MMP-2/9 by the change in electrode capacitance. Adapted from [49].
MRI is a noninvasive and well-established technique for in vivo analysis/diagnosis that allows time and spatial resolution with reasonably high sensitivity and in-depth tissue penetration. In view of these advantages, Yuan et al. [48] developed an MRI proteolytic sensor in real time based on iron oxide (Fe3O4) nanoparticles, which aggregate after proteolytic activity in a process induced by Caspase-3 and/or -7 and concomitantly increase T2 contrast (Figure 3B). This design was tested in nude mice with subcutaneously xenografted HepG2 tumor. These in vivo tumor MRI measurements suggested specificity for T2-enhanced imaging in tumor apoptosis, demonstrating that MRI can be efficiently applied to evaluate chemotherapeutic efficiency, or other treatments, in routine preclinical studies.
Electrochemical-based sensors provide time resolution and render clear readouts with multiplexing capabilities. For example, Tran et al. [49] developed an on-chip dual-sensing device for the detection of cell-secreted MMP-9 that simultaneously monitors cellular resistance and correlates it with the secretome and morphological analysis. The device comprises two independent sensing platforms featuring interdigitated-array microelectrodes (IDAMs) in one common fluidic chamber (Figure 3C). The goal of the device is to detect MMP-9 activity and understand cellular behavior during motility. On migration, cells tend to change morphology by extending their shape (i.e., lamellipodium formation) [50] and that increases resistance to the flow of electrical current between electrodes. The faster cells extend, the higher the value of the electrical resistance measured. During the migration process, MMP-9 was simultaneously secreted and diffused upwards from the cell binding site [49]. The substrate peptides at the upper site were proteolytically cleaved, causing a change in electrode capacitance. We believe that this dual-sensing method is interesting since it affords high versatility and could be applicable to a wide range of cell-secreted proteases by choosing the appropriate target proteins or peptides. Despite not being suitable for in vivo studies, this approach could deliver promising insights on the relation between the cellular secretome and morphological analysis in real time; for example, in the context of bioengineering approaches used for cellular differentiation.
From the Bench to Clinical Trials
Currently, there are at least two clinical trials registered on the National Institutes of Health (NIH) ClinicalTrials.gov database involving the use of in vivo sensors to determine proteolytic activity in real time [51,52]. These sensors were developed to determine the activity of MMPs (AVB-620) and cathepsins (LUM015) in breast cancer. In breast surgery, surgeons rely on physical examination, tissue texture, and correlation with radiographic data to determine tumor margins. By using such sensors, surgeons can precisely differentiate between tumorous tissue and nontumorous with precision with no tissue destruction or processing [53].
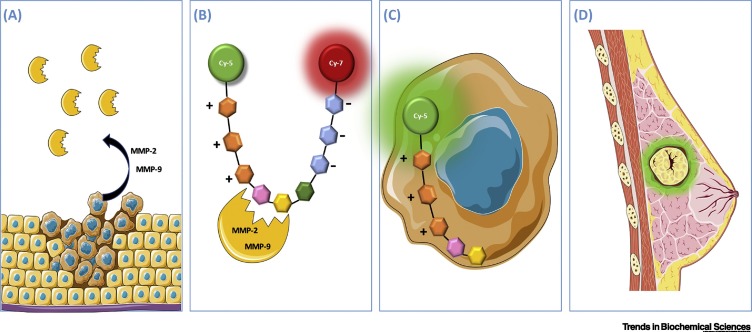
Although the target proteases differ, the working principles underlying AVB-620 and LUM015 are similar. For this reason, only AVB-620 is detailed herein. AVB-620 is a fluorescence-based sensor that is currently undergoing Phase III trials for breast cancer and entering Phase I trials for multiple cancers (Figure 4 ) [54]. In this hairpin sensor design, two fluorophores (Cy-5 and Cy-7) are in close proximity due to electrostatic interactions between two oppositely charged domains of the sensor molecule, which are bridged by a specifically targeted peptide sequence. In the absence of tumor-associated enzymes such as MMP-2 and -9, the two fluorophores are close to each other, with Cy-5 acting as an energy donor to Cy-7, which then emits brightly. In the presence of MMP-2/9, the target sequence is cleaved and the two fluorophores are separated, leading to a decrease in Cy-7 emission that is dependent on the protease activity as detected by fluorescence cameras, allowing intraoperative imaging in real time. Additionally, the presence of an activated cell penetration peptide (positively charged) in the resulting Cy-5-containing peptide fragment allows the fluorophore to be internalized by nearby cells, precisely delimiting tumor margins. The system did not exhibit significant side effects and showed improved performance in defining tumor margins. If the AVB-620 sensor is able to pass through the whole validation process, it will potentially encourage the development of protease-based theranostic systems.
Figure 4.
Workflow for AVB-620 Detection System.
(A) Breast cancer cells producing matrix metalloprotease (MMP)-2/9. (B) The AVB-620 molecule is U-shaped due to electrostatic interactions, which increases the proximity between the Förster resonance energy transfer (FRET) pair (Cy-5 and Cy-7). (C) After cleavage, the pair dissociates and the positively charged side of the peptide is internalized by nearby cells. Consequently, Cy-5 emission increases preferentially in breast cancer cells. (D) Doctors are able to determine tumor margins with high precision.
Concluding Remarks
Throughout this review, we highlighted the advantages of measuring proteolytic activity in real time and the reasons why they can provide a wealth of information about biological processes. Despite the fast pace of development of novel and improved detection systems, the field has plenty of space to grow. In our perspective, there are several opportunities and challenges that can and should be addressed in the future. For instance, to the best of our knowledge, multiplexing systems are so far based on optical methods only. New strategies for multiplexing can overcome the current limitations, increase diagnostic precision, and be helpful in correlating metabolic information between different enzymes. Within the same rational multiparametric systems, the work of Zhao et al. [38], in which proteolytic activity and pH are simultaneously evaluated in real time, also seems promising (Figure 5A).
Figure 5.
Real-Time Biosensors in the Foreseeable Future.
(A) Multiplex proteolytic activity and multiparametric sensors in real time. (B) Specific target peptide sequence for single protease detection. (C) Imaging of ‘in-depth’ tissues with high resolution. (D) Programmable medicine representation comprising a real-time feedback system based on proteolytic-activity logic gates.
Another important issue is specificity. Despite most proteases tested as potential biomarkers being specific towards a target peptide sequence, often it is not possible to discriminate between two or more family-related proteases (i.e., MMP-2/9 and Cas-3/7). One possible strategy to achieve this goal was developed by Bainbridge et al. [55], who exchanged L-amino acids for their D- counterparts in the peptide structure when designing a fibroblast activation protein (FAP)-specific substrate. Although the kinetic parameters of the target protease were slightly influenced in a negative way, a specific target peptide without dipeptidyl peptidase-4 (DPPIV) interference was obtained (Figure 5B).
The lack of in-depth imaging is one of the major pitfalls of optical sensors. Magnetic sensors, however, can reliably provide good 3D reconstruction and high in-depth abilities with almost no background signal (Figure 5C). Another interesting vision for proteolytic sensors in real time suggests their integration in logic gates [56]. Also, Kwong et al. [57,58] used nonreal-time proteolytic activity as biologic bits and integrated them in the field of programmable medicine. By merging these concepts, one can imagine a machine analyzing the readout in real time and giving an immediate and proper response – for example, to stop/start the administration of a drug – in a feedback scheme and without the constant supervision of a healthcare professional. Also, this approach has the potential to decrease the amount of drug administered and to avoid drug spikes in the patient’s body, since the drug can be delivered through microdosing (Figure 5D).
In summary, a wealth of research is being dedicated to the field of protease sensing with a diversity of purposes, such as drug development, tumor imaging, and others. Although these types of biosensors are already under development, we strongly believe that there are plenty of unexplored concepts that can arise in the future (see Outstanding Questions). We cannot predict the pathways that are going to be pursued henceforth in real-time proteolytic-activity biosensors. Nevertheless, we hope that platforms for general use and the accessibility of these sensors will accelerate protease-based research and intensively propel the field. Undoubtedly, optimized systems to monitor and control biological processes will play a role in improving healthcare and moving towards a better version of personalized medicine.
Outstanding Questions.
Protease activity is sensitive to environmental conditions such as temperature, pH, and other. What challenges arise from the integration of proteolytic activity onto multiple parametric sensors?
As we are currently experiencing due to Coronavirus and globalization, infectious diseases can quickly spread, ultimately resulting in a pandemic. Can proteolytic sensors be an ally in drug development and/or diagnosis to mitigate the outcome of a potential pandemic event?
Proteolytic sensors have shown great potential to be used as diagnostic tools, although the addition of therapeutic features to these sensors is still almost unexplored. Which new designs can be explored for theranostic purposes?
Is increased selectivity and sensitivity a requirement for real-life applications of proteolytic-activity sensors?
Optically based biosensors have limited in-depth capabilities and low photophysical stability in complex media, which can be critical for in vivo imaging. Will future technologies be able to overcome this limitation?
MRI sensors allow high in-depth imaging with spatiotemporal resolution. However, MRI machines are expensive and have limited access compared with other technologies. Can magnetic sensors become accessible and enable high-throughput analysis?
In translation from academia to the clinical environment, how far along are real-time proteolytic-activity biosensors?
Can artificial intelligence and programmable medicine be incorporated with such systems to create a new era in drug development, therapy, and other applications?
Alt-text: Outstanding Questions
Acknowledgments
R.O-S. acknowledges Dr Peter Zijlstra and Dr Fábio Trindade for all the fruitful discussions and support. The authors also acknowledge the FCT – Portuguese Foundation for Science and Technology, grant numbers UIDB/04565/2020, UIDP/50011/2020, and UID/QUI/00100/2019, project CICECO - Aveiro Institute of Materials (grant nos UIDB/50011/2020 & UIDP/50011/2020) and Programa Operacional Regional de Lisboa 2020, grant number 007317. R.O-S. and D.B. acknowledge the FCT program BIOTECnico fellowships PD/BD/116850/2016 and PD/BD/113630/2015, respectively. M.S-J. acknowledges the FCT project number 29460 (POCI-01-0145-FEDER-29460).
Glossary
- Biosensors
analytical devices used for detection and/or measurement of specific analyte, biometric parameters, and body conditions.
- Caspases
a family of intracellular cysteine-aspartic proteases that are pivotal in programmed cell death, such as apoptosis, pyroptosis, and necrosis.
- Compound annual growth rate (CAGR)
rate of return required for an investment to grow from its beginning balance to its ending balance, assuming the profits are reinvested at the end of each year of the investment’s lifespan.
- Energy transfer (ET) based
systems in which ET can occur from an electronically excited state of a donor to an acceptor. In biosensors the efficiency of this process can be influenced by a target analyte, thus signaling its presence by means of a spectroscopic observable (emission, intensity or lifetime).
- Logic gate
a device that performs logical operations to convert the signal from the sensor to a binary output (0 or 1). Logic gates based on proteolytic activity operate by differentiating ‘low-activity’ from ‘high-activity’ states and further address them as 0 and 1, respectively.
- Matrix metalloproteases (MMPs)
metal-dependent endopeptidases found in the extracellular matrix, which are responsible for degrading several proteins and bioactive products.
- Multiplex-modality systems
combine two or more complementary techniques that enable imaging of metabolic processes, such as proteolytic activity, with increased resolution compared with their individual counterparts.
- Personalized medicine
a new paradigm in medicine that views patients not as members of a group, but rather as unique individuals with intrinsic characteristics that must be diagnosed, evaluated, and used to inform therapeutic decisions.
- Photothermal therapy (PTT)
uses electromagnetic radiation – commonly IR wavelengths – which is absorbed by a molecule/particle (photosensitizer). This activation brings the sensitizer to an excited state further releasing heat. The resulting heat increases the temperature locally, killing the targeted cells.
- Plasmonic nanoparticles
display strong interactions with light – scattering and absorption– critically dependent on the particle’s material, size, and geometry and interparticle distances.
- Positron emission tomography (PET)
a nuclear medical imaging technique based on the detection of radioactivity emitted by a radioactive tracer delivered to an individual to monitor physiological functions.
- Programmable medicine
current medical practice, based on analysis of the symptomatology and medical examination results. Programmable medicine does not consider the phenotypic and genotypic characterization, sociodemographic and environmental factors, or lifestyle of an individual and does not include disease preventive strategies.
- Real-time proteolytic assays
(in the authors’ definition) assays that have an immediate response that is proportional to proteolytic activity and can be rapidly read by a sensor, but not necessarily as a continuous measurement.
- Signal-to-noise ratio (SNR)
defined as the ratio between the intensity of a specific signal generated during the measurement and the intensity or standard deviation of a signal generated from unspecific sources (noise). A high SNR corresponds to a bigger difference between the signal generated from an occurring event and the background noise, increasing the confidence/sensitivity.
- Suicidal experiments
experiments that can be used only once to determine a specific parameter at a given time. Therefore, an experiment needs to be done for each desired time point.
References
- 1.Scher W. The role of extracellular proteases in cell proliferation and differentiation. Lab. Investig. 1987;57:607–633. [PubMed] [Google Scholar]
- 2.Li J., Yuan J. Caspases in apoptosis and beyond. Oncogene. 2008;27:6194–6206. doi: 10.1038/onc.2008.297. [DOI] [PubMed] [Google Scholar]
- 3.Narayanan S. Multifunctional roles of thrombin. Ann. Clin. Lab. Sci. 1999;29:275–280. [PubMed] [Google Scholar]
- 4.Heutinck K.M. Serine proteases of the human immune system in health and disease. Mol. Immunol. 2010;47:1943–1955. doi: 10.1016/j.molimm.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 5.Haley S.A., Wessel G.M. Regulated proteolysis by cortical granule serine protease 1 at fertilization. Mol. Biol. Cell. 2004;15:2084–2092. doi: 10.1091/mbc.E03-11-0843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koblinski J.E. Unraveling the role of proteases in cancer. Clin. Chim. Acta. 2000;291:113–135. doi: 10.1016/s0009-8981(99)00224-7. [DOI] [PubMed] [Google Scholar]
- 7.Krenzlin H. The importance of thrombin in cerebral injury and disease. Int. J. Mol. Sci. 2016;17:84. doi: 10.3390/ijms17010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao B. Kallikrein-related peptidase 8 is expressed in myocardium and induces cardiac hypertrophy. Sci. Rep. 2016;6:20024. doi: 10.1038/srep20024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danforth C.M. Defining the boundaries of normal thrombin generation: investigations into hemostasis. PLoS One. 2012;7:e30385. doi: 10.1371/journal.pone.0030385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cabarkapa S. Prostate cancer screening with prostate-specific antigen: a guide to the guidelines. Prostate Int. 2016;4:125–129. doi: 10.1016/j.prnil.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lv Z. HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV. AIDS (Auckl). 2015;7:95–104. doi: 10.2147/HIV.S79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mangum E.M., Graham K.K. Lopinavir-ritonavir: a new protease inhibitor. Pharmacother. J. Hum. Pharmacol. Drug Ther. 2001;21:1352–1363. doi: 10.1592/phco.21.17.1352.34419. [DOI] [PubMed] [Google Scholar]
- 13.Liu C. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baden L.R., Rubin E.J. Covid-19 — the search for effective therapy. N. Engl. J. Med. 2020 doi: 10.1056/NEJMe2005477. Published online March 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao B. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2001282. Published online March 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science (80-. ) 2020 doi: 10.1126/science.abb3405. Published online March 20, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheridan Cormac. Fast, portable tests come online to curb coronavirus pandemic. Nat. Biotechnol. 2020 doi: 10.1038/d41587-020-00010-2. Published online March 20, 2020. [DOI] [PubMed] [Google Scholar]
- 18.Nair P. QnAs with Sangeeta N. Bhatia. Proc. Natl. Acad. Sci. 2020;117:1243–1245. doi: 10.1073/pnas.1922176117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burrage P.S. Matrix metalloproteinases: role in arthritis. Front. Biosci. 2006;11:529–543. doi: 10.2741/1817. [DOI] [PubMed] [Google Scholar]
- 20.Salganik M. Evidence for pH-dependent protease activity in the adeno-associated virus capsid. J. Virol. 2012;86:11877–11885. doi: 10.1128/JVI.01717-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mason S.D., Joyce J.A. Proteolytic networks in cancer. Trends Cell Biol. 2011;21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brew K., Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim. Biophys. Acta. 2010;1803:55–71. doi: 10.1016/j.bbamcr.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McIlwain D.R. Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Almalki S.G., Agrawal D.K. Effects of matrix metalloproteinases on the fate of mesenchymal stem cells. Stem Cell Res Ther. 2016;7:129. doi: 10.1186/s13287-016-0393-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanne J. Stimulated emission depletion nanoscopy reveals time-course of human immunodeficiency virus proteolytic maturation. ACS Nano. 2016;10:8215–8222. doi: 10.1021/acsnano.6b03850. [DOI] [PubMed] [Google Scholar]
- 26.Lorenzano S. Editorial: Role of coagulation pathways in neurological diseases. Front. Neurol. 2019;10:791. doi: 10.3389/fneur.2019.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ebrahimi S. Role of thrombin in the pathogenesis of central nervous system inflammatory diseases. J. Cell. Physiol. 2017;232:482–485. doi: 10.1002/jcp.25501. [DOI] [PubMed] [Google Scholar]
- 28.Ta H.T. Activatable magnetic resonance nanosensor as a potential imaging agent for detecting and discriminating thrombosis. Nanoscale. 2018;10:15103–15115. doi: 10.1039/c8nr05095c. [DOI] [PubMed] [Google Scholar]
- 29.Leber T.M., Balkwill F.R. Zymography: a single-step staining method for quantitation of proteolytic activity on substrate gels. Anal. Biochem. 1997;249:24–28. doi: 10.1006/abio.1997.2170. [DOI] [PubMed] [Google Scholar]
- 30.Caldas C. Purification and characterization of an extracellular protease from Xenorhabdus nematophila involved in insect immunosuppression. Appl. Environ. Microbiol. 2002;68:1297–1304. doi: 10.1128/AEM.68.3.1297-1304.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Son J. Customizing morphology, size, and response kinetics of matrix metalloproteinase-responsive nanostructures by systematic peptide design. ACS Nano. 2019;13:1555–1562. doi: 10.1021/acsnano.8b07401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathew-Fenn R.S. A molecular ruler for measuring quantitative distance distributions. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lavis L.D., Raines R.T. Bright ideas for chemical biology. ACS Chem. Biol. 2008;3:142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Resch-Genger U. Quantum dots versus organic dyes as fluorescent labels. Nat. Methods. 2008;5:763–775. doi: 10.1038/nmeth.1248. [DOI] [PubMed] [Google Scholar]
- 35.Jeong S. Cancer-microenvironment-sensitive activatable quantum dot probe in the second near-infrared window. Nano Lett. 2017;17:1378–1386. doi: 10.1021/acs.nanolett.6b04261. [DOI] [PubMed] [Google Scholar]
- 36.Jun Y. Continuous imaging of plasmon rulers in live cells reveals early-stage caspase-3 activation at the single-molecule level. Proc. Natl. Acad. Sci. 2009;106:17735–17740. doi: 10.1073/pnas.0907367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y. multifunctional gold nanoclusters-based nanosurface energy transfer probe for real-time monitoring of cell apoptosis and self-evaluating of pro-apoptotic theranostics. Anal. Chem. 2016;88:11184–11192. doi: 10.1021/acs.analchem.6b03389. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X. Dual-stimuli responsive and reversibly activatable theranostic nanoprobe for precision tumor-targeting and fluorescence-guided photothermal therapy. Nat. Commun. 2017;8:14998. doi: 10.1038/ncomms14998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu B. Avoiding thiol compound interference: a nanoplatform based on high-fidelity Au–Se bonds for biological applications. Angew. Chem. Int. Ed. 2018;57:5306–5309. doi: 10.1002/anie.201712921. [DOI] [PubMed] [Google Scholar]
- 40.Luan M. A gold–selenium-bonded nanoprobe for real-time in situ imaging of the upstream and downstream relationship between uPA and MMP-9 in cancer cells. Chem. Commun. 2019;55:5817–5820. doi: 10.1039/c9cc01454c. [DOI] [PubMed] [Google Scholar]
- 41.Liu X. Real-time in situ visualizing of the sequential activation of caspase cascade using a multicolor gold–selenium bonding fluorescent nanoprobe. Anal. Chem. 2019;91:5994–6002. doi: 10.1021/acs.analchem.9b00452. [DOI] [PubMed] [Google Scholar]
- 42.Matthias N. Hybrid in vivo FMT-CT imaging of protease activity in atherosclerosis with customized nanosensors. Arterioscler. Thromb. Vasc. Biol. 2009;29:1444–1451. doi: 10.1161/ATVBAHA.109.193086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin L. Rational design and synthesis of a metalloproteinase-activatable probe for dual-modality imaging of metastatic lymph nodes in vivo. J. Org. Chem. 2019;84:6126–6133. doi: 10.1021/acs.joc.9b00331. [DOI] [PubMed] [Google Scholar]
- 44.Elvas F. Caspase-3 probes for PET imaging of apoptotic tumor response to anticancer therapy. Org. Biomol. Chem. 2019;17:4801–4824. doi: 10.1039/c9ob00657e. [DOI] [PubMed] [Google Scholar]
- 45.Lee S. Tumor-homing glycol chitosan-based optical/PET dual imaging nanoprobe for cancer diagnosis. Bioconjug. Chem. 2014;25:601–610. doi: 10.1021/bc500020g. [DOI] [PubMed] [Google Scholar]
- 46.Shi H. Magnetic semiconductor Gd-doping CuS nanoparticles as activatable nanoprobes for bimodal imaging and targeted photothermal therapy of gastric tumors. Nano Lett. 2019;19:937–947. doi: 10.1021/acs.nanolett.8b04179. [DOI] [PubMed] [Google Scholar]
- 47.Wang L. Real-time label-free measurement of HIV-1 protease activity by nanopore analysis. Biosens. Bioelectron. 2014;62:158–162. doi: 10.1016/j.bios.2014.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan Y. Casp3/7-instructed intracellular aggregation of Fe3O4 nanoparticles enhances T2 MR imaging of tumor apoptosis. Nano Lett. 2016;16:2686–2691. doi: 10.1021/acs.nanolett.6b00331. [DOI] [PubMed] [Google Scholar]
- 49.Tran T.B. Electrical dual-sensing method for real-time quantitative monitoring of cell-secreted MMP-9 and cellular morphology during migration process. Biosens. Bioelectron. 2016;77:631–637. doi: 10.1016/j.bios.2015.10.030. [DOI] [PubMed] [Google Scholar]
- 50.Innocenti M. New insights into the formation and the function of lamellipodia and ruffles in mesenchymal cell migration. Cell Adhes. Migr. 2018;12:401–416. doi: 10.1080/19336918.2018.1448352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Avelas Biosciences and Inc Trial of AVB-620 in women with primary, non-recurrent breast cancer undergoing surgery. 2017. https://clinicaltrials.gov/ct2/show/NCT03113825 Published online April 14, 2017.
- 52.Kirsch D. Cathepsin activatable fluorescent probe (LUM015) 2012. https://clinicaltrials.gov/ct2/show/NCT01626066 Published online June 22, 2012.
- 53.Unkart J.T. intraoperative tumor detection using a ratiometric activatable fluorescent peptide: a first-in-human phase 1 study. Ann. Surg. Oncol. 2017;24:3167–3173. doi: 10.1245/s10434-017-5991-3. [DOI] [PubMed] [Google Scholar]
- 54.Avelas Biosciences, Inc https://www.avelasbio.com/
- 55.Bainbridge T.W. Selective homogeneous assay for circulating endopeptidase fibroblast activation protein (FAP) Sci. Rep. 2017;7:12524. doi: 10.1038/s41598-017-12900-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bui H. Transducing protease activity into DNA output for developing smart bionanosensors. Small. 2019;15:1805384. doi: 10.1002/smll.201805384. [DOI] [PubMed] [Google Scholar]
- 57.Holt B.A., Kwong G.A. Proteases as biological bits for programmable medicine. bioRxiv. 2019 doi: 10.1101/607895. Published online April 12, 2019. [DOI] [Google Scholar]
- 58.Mac Q.D. Non-invasive early detection of acute transplant rejection via nanosensors of granzyme B activity. Nat. Biomed. Eng. 2019;3:281–291. doi: 10.1038/s41551-019-0358-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung E.Y. Activatable and cell-penetrable multiplex FRET nanosensor for profiling MT1-MMP activity in single cancer cells. Nano Lett. 2015;15:5025–5032. doi: 10.1021/acs.nanolett.5b01047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Braun A. A cell surface display fluorescent biosensor for measuring MMP14 activity in real-time. Sci. Rep. 2018;8:5916. doi: 10.1038/s41598-018-24080-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.den Hamer A. Bright bioluminescent BRET sensor proteins for measuring intracellular caspase activity. ACS Sens. 2017;2:729–734. doi: 10.1021/acssensors.7b00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wuytens P.C. Gold nanodome SERS platform for label-free detection of protease activity. Faraday Discuss. 2017;205:345–361. doi: 10.1039/c7fd00124j. [DOI] [PubMed] [Google Scholar]
- 63.Sun C. Time-resolved single-step protease activity quantification using nanoplasmonic resonator sensors. ACS Nano. 2010;4:978–984. doi: 10.1021/nn900757p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu C. Multispectral photoacoustic imaging of tumor protease activity with a gold nanocage-based activatable probe. Mol. Imaging Biol. 2018;20:919–929. doi: 10.1007/s11307-018-1203-1. [DOI] [PubMed] [Google Scholar]
- 65.Yin L. Quantitatively visualizing tumor-related protease activity in vivo using a ratiometric photoacoustic probe. J. Am. Chem. Soc. 2019;141:3265–3273. doi: 10.1021/jacs.8b13628. [DOI] [PubMed] [Google Scholar]
- 66.Schyrr B. Fiber-optic protease sensor based on the degradation of thin gelatin films. Sens. Bio-Sensing Res. 2015;3:65–73. [Google Scholar]
- 67.Page M.J. Non-invasive imaging and cellular tracking of pulmonary emboli by near-infrared fluorescence and positron-emission tomography. Nat. Commun. 2015;6:8448. doi: 10.1038/ncomms9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.López-Otín C., Bond J.S. Proteases: Multifunctional Enzymes in Life and Disease. J. Biol. Chem. 2008;283:30433–30437. doi: 10.1074/jbc.R800035200. [DOI] [PMC free article] [PubMed] [Google Scholar]