Abstract
There is a critical need for phenotypes with substantial heritability that can be used as endophenotypes in behavioral genetic studies. Activity monitoring, called actimetry, has potential as a means of assessing sleep and circadian rhythm traits that could serve as endophenotypes relevant to a range of psychopathologies. This study examined a range of actimetry traits for heritability using a classic twin design. The sample consisted of 195 subjects from 45 monozygotic (MZ) and 50 dizygotic (DZ) twin pairs aged 16–40 years. Subjects wore both a research-grade actimeter (GENEActiv) and a consumer-oriented device (FitBit) for 2 weeks. Sleep and circadian traits were extracted from GENEActiv data using PennZzz and ChronoSapiens software programs. Sleep statistics for a limited number of FitBit-collected traits were generated by its accompanying mobile app. Broad sense heritability was computed on a set of 33 MZ and 38 DZ twin pairs with complete data using both OpenMX and SOLAR software. These analyses yielded a large number of actimetry-derived traits, 20 of which showed high heritability (h2 > 0.6), seven of which remain significant after Bonferroni correction. These results indicate that actimetry enables assessing a range of phenotypes with substantial heritability that may be useful as endophenotypes for genetic studies.
Keywords: actimetry, circadian, endophenotype, heritability, sleep, twin
1 |. INTRODUCTION
Understanding the genetic basis of psychiatric disorders would significantly improve their treatment. With increasing number of participants, genome-wide association studies have become very successful in identifying common risk variants.1 However, each individual locus or allele identified to date explains only a small proportion of heritability or relatively small changes in risk.2,3 These small effects and likely existence of diagnostic subtypes with genetic heterogeneity have made it difficult to define specific pathways and molecular mechanisms underlying psychiatric disorders, including major depressive and bipolar disorders.4,5 For example, there are multiple pathways to reach a state of depression, that each may have distinct genetic mechanisms.6 A sample of patients with depression will contain individuals with diverse etiologies, making it difficult to find consistent genetic associations.
One approach to address genetic heterogeneity is to improve phenotyping. In conventional genetic studies, phenotyping usually consists of characterizing clinical symptoms, ideally using semi-structured interviews. Self-reports and clinician-assessed phenotypes are often confounded by a range of factors including self-report biases, inter-rater variability and a focus on only perceived behavioral, cognitive or emotional manifestations of the disorder.7 An alternate approach is to use objective, quantifiable endophenotypes that mediate between susceptibility genes and full clinical expression of the disease.8 Gottesman and Gould proposed criteria for defining an endophenotype: (a) an endophenotype is associated with illness, in the population; (b) an endophenotype is heritable; (c) an endophenotype is state-independent; (d) within families, an endopheoptype and illness cosegregate; and (e) an endophenotype identified in probands is found in nonaffected family members at a higher rate than in the general population.8
The initial expectation was that endophenotypes, such as traits obtained by neuroimaging, EEG or performance tasks, would be less complex and more tractable than the disease state.8,9 While endophenotypes may be useful as quantifiable markers of the disease, they do not necessarily exhibit a simpler genetic architecture.10 In terms of the criterion that endophenotypes show heritability, earlier studies found that the heritability of endophenotypes was not greater than that of diagnostic phenotypes.11 Yet, even without showing strong heritability, endophenotypes can provide insight into underlying mechanisms and may serve as biomarkers of treatment response.
Actimetry, the measurement of patterns of rest/activity, has been used for several decades as an assessment of physical activity and to estimate sleep/wake patterns.12 Wrist-worn activity trackers are unobtrusive and can collect quantitative behavioral information over a very long time in the participant’s home environment.13 Actimetry also has a number of advantages over self-report including reduced influence of cognitive biases, substantially higher frequency of data sampling across the day, and no reliance on subjects’ remembering to complete daily entries. Actimetry traits have been used to examine the associations between sleep and health showing, for example, that total sleep time is associated with obesity in older adults.14 Actimetry traits also yield putative endophenotypes that are important dimensions of psychopathology because rest/activity and sleep/wake patterns are disturbed in a range of psychiatric disorders, satisfying the first criterion above.15–17 Activity monitoring also allows cross-species comparisons, thereby enabling the study of disease-related traits in model organisms.
In the context of affective disorders, changes in rest/activity and sleep/wake cycles are among the most prominent aspects of both manic and depressive episodes.18,19 Several groups used actimetry in patients with affective disorders and reported changes in the activity levels, not only in the manic and depressive phases, but also in the euthymic state when compared with controls.20,21 Pagani and col leagues collected actimetry data from 26 bipolar disorder pedigrees (558 individuals) and computed 73 different activity-based phenotypes.22 Forty-nine of the phenotypes showed substantial heritability and 13 were associated with bipolar disorder, with patients having lower activity levels compared with euthymic family members. This paper also showed cosegregation of actimetry traits with illness, meeting criterion four for an endophenotypes.
We sought to build on this approach by utilizing a classic twin design to assess which actimetry traits show high heritability in a population-representative sample (using both scientific and commercial devices, GENEActiv and FitBit, respectively) to determine which might meet the second criterion for endophenotypes. Twin studies can quantify “broad sense” heritability—the degree of genetic influence on a trait as distinct from the effects of the shared family environment and unique environment.23 Our results show that a number of actimetry traits show high heritability and are therefore candidate traits for use in behavioral genetic studies.
2 |. MATERIALS AND METHODS
Pairs of monozygotic (MZ) and dizygotic (DZ) twins were recruited through the Pennsylvania Longitudinal Study of Parents and Children (PALSPAC) twin registry and through advertising in the Philadelphia metropolitan region. The subjects were between 16 and 40 years old. Subjects were required to have sufficient verbal and reading ability in English to be able to sign the consent and respond to interviews and checklist. All procedures were approved by the University of Pennsylvania Institutional Review Board and all subjects provided written informed consent prior to participation in the study.
Subjects were given wrist actimeters to wear on the nondominant wrist for 2 weeks. All subjects in this study wore the GENEActiv device (Activinsights Ltd., Kimbolton, UK). A subset of subjects (n = 97) also wore FitBit Charge or FitBit Flex (FitBit Inc; San Francisco, California). GENEActiv measures body movements and light exposure. It is a tri-axial accelerometer and was set to sample activity at a frequency of 30 Hz. Data were downloaded onto a computer and customized GENEActive software was used to aggregate data into two files with 1-second and 1-minute epochs as these are the epoch lengths used by the sleep scoring algorithms (see below). FitBit devices, worn on the same wrist as GENEActiv, collected data that were synchronized via the FitBit app and stored on FitBit servers. The FitBit app computes a limited number of traits on sleep and physical activity that were then downloaded using the fitabase.com website. During the 2 weeks of actimetry, subjects completed daily sleep diaries by answering questions related to their previous night of sleep (wake time, sleep time, number of awakenings, number of hours asleep, along with other sleep related variables).24
2.1 |. Data processing
Actimetry data were analyzed using two algorithms; PennZzz, an algorithm recently developed by our group for scoring sleep in actimetry data and implemented in Python (McCloskey et al, submitted; source code available at https://github.com/rjmccloskey/PennZZZ), and ChronoSapiens (Chronsulting), a comprehensive tool for analyzing longitudinal actimetry data.25 The raw acceleration data were transformed into a sum vector magnitude (SVM), defined as the sum of acceleration in three dimensions with gravity subtracted from the sum of all readings in the designated 1 second epochs. In PennZzz, tempovascillatory activity (TVA) was computed by finding the SVM-variance within a 2 minutes moving window. The algorithm utilizes the TVA to categorize each 1-second epoch as wake, light sleep, intermediate sleep or deep sleep. In parallel, homothermic states were computed based on computing variance (oscillations) of absolute temperature in a 3 minutes moving window, and temperature below 24°centigrade was considered as nonwear period. The algorithm produced a set of parameters, which fall into four categories: (a) sleep during rest episodes, (b) activity during rest episodes (c) activity during awake and (d) overall activity level. While there are a large number of computed variables, the goal was to cast a wide net in order to identify potentially novel variables that might have utility as endophenotypes.
While the PennZzz program was used to compute sleep and activity statistics, phase analysis of the data was conducted using the ChronoSapiens program (Figure 1).25 This function in ChronoSapiens is based on the traditional approach of fitting a cosine curve to actimetry data.26 ChronoSapiens models both first and second harmonic fits. In addition, the fit-models’ parameters can be computed on moving averages allowing sensitive detections of changes over time and different conditions. Here, we chose a static cosinor fit for each 24-hour day. The model parameters are used to compute the center of gravity (CoG) for activity (see Figure 1). The parameters generated by ChronoSapiens provide a range of indicators of activity’s timing (represented by the CoG phase for the 1H-fit and by the phases of the potentially two maxima and minima for the 2H-fit) and activity’s amplitude (represented by the max value of the CoG for the 1H-fit or the range of oscillation, RoO = Max − Min for the 2H-fit). Additional parameters are the Munich Rhythmicity Index (MRI), which quantifies the extent of rhythmicity in the time series by taking both the fit’s goodness and amplitude into account27—a high MRI indicates rhythms that are close to 24 hours and high in amplitude; finally, the alpha/rho ratio (Figure 1A,B), which was originally developed to reflect the relative durations of activity (alpha) and rest (rho) within a day. We use it here based on the time when activity values are above vs the time they are below the daily mean.
FIGURE 1.
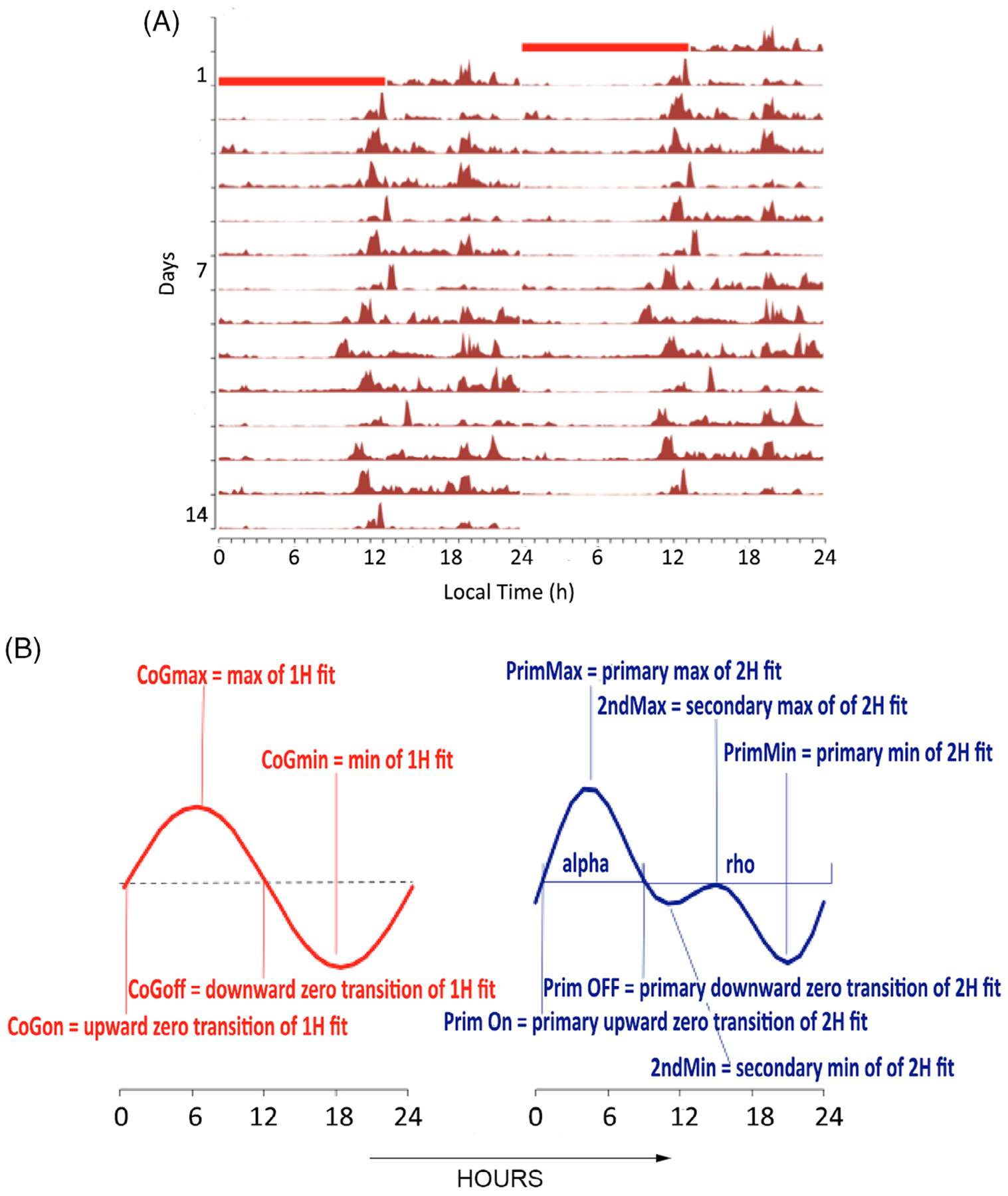
A, Sample figure of14 days actimetry data from a twin with fitted curve from ChronoSapiens. The activity level is indicated in brown picks. Red squares on the top indicate when the watch was off-wrist. B, Parameters generated by ChronoSapiens
We have also imported 1-minute epoch aggregated data from GENEActiv into “nparACT” package for R Core Team28,29 to compute five additional measures not available in PennZzz and ChronoSapiens. These include the interdaily stability, intradaily variability and relative amplitude (RA) of activity and gives the start times and average activity values of M10 (ie, the 10 hours with maximal activity) and L5 (ie, the 5 hours with least activity). The formula for the computation of RA is:
Finally, statistics from the FitBit devices were downloaded through the Fitabase website (https://www.fitabase.com/) and consisted of the following variables: sleep efficiency, minutes after wakeup, minutes asleep, time in bed, awake duration, restless count and restless duration. The definition of each of these variables and the formulas for their computation are not provided by FitBit so their exact meaning is unknown.
For the sleep diaries, the overall mean of each variable was computed. The mean total sleep time from the sleep diary was then compared with that computed from the PennZzz algorithm and from the FitBit using Pearson correlations. This comparison was only made for total sleep time as it was the primary parameter available across all three methods.
2.2 |. Data analysis
Descriptive statistics for each of the actimetry variables were computed. Plots were created to display the distributions of each trait separately for all MZ and all DZ twins in order to visually compare their means and variances. We examined outliers to determine if there were errors in data entry or in computations but all observations were found to be valid and were retained in the analyses. Given the large number of variables computed by the PennZzz program, the correlations among the variables were computed to better understand how the parameters related to each other. Next, the Broad sense heritability of each of the statistics was computed using the open-access program OpenMX (http://openmx.ssri.psu.edu/). We estimated heritability using the basic ACE model where ACE stands for(A) Additive genetic variance, (C) Shared environmental variance from environmental influence shared by family members and (E) Residual variance from environmental influence that are not shared by family members. The program subsequently employs maximum likelihood modeling procedures to determine what combination of A, C and E best fits the observed data. The models were conducted under the default settings that assume a common mean and variance between zygosity groups.
The heritability of each of the statistics was further verified using Sequential Oligogenic Linkage Analysis Routines (SOLAR) (Almasy and Blangero, 1998). The SOLAR software package https://helix.nih.gov/Documentation/solar-6.6.2-doc/03.chapter.html provides extensive capabilities for analyzing heritability and has been widely used in family and twin studies. We used SOLAR to estimate heritability of traits by creating a pedigree structure of MZ and DZ twin families, and we considered three possible covariates, sex, age and body mass index (BMI), given their known influence on sleep/wake and circadian traits.30–33 The SOLAR “polygenic” command performs a variance component analysis to determine broad sense heritability.
2.2.1 |. Data availability
We deposited accelerometry files to the NIMH Data Archive.
3 |. RESULTS
Complete data were available from 190 subjects from 45 MZ and 50 DZ twin pairs. Descriptive statistics for the sample are provided in Table 1. Data for one MZ twin pair were excluded because they were outliers on multiple phenotypes, drawing into question the validity of their data. Additionally, subjects were required to have seven or more days of complete actigraphy data on twin pairs (both siblings) to be included in analyses (see supplemental Figure S1), resulting in 65 pairs of twins (33 MZ and 38 DZ).
TABLE 1.
Descriptive statistics for the sample overall and by zygosity
| Overall | MZ | DZ | |
|---|---|---|---|
| Age | 21.7 (4.6) | 21.2 (3.9) | 22.2 (5.1) |
| BMI | 24.3 (5.3) | 23.5 (4.9) | 25.2 (5.6) |
| Sex (% female) | 53.8% | 55.1% | 52.6 |
| Ethnicity | |||
| Hispanic | 5.0% | 8.1% | 2.1% |
| Non-Hispanic | 95.0% | 92.0% | 97.9% |
| Race/ethnicity | |||
| White | 90.9% | 92.0% | 87.8% |
| African American | 3.2% | 0.0% | 6.1% |
| Asian | 5.9% | 5.6% | 6.1% |
| Education | |||
| 0–4 years | 1.1% | 0.0% | 2.0% |
| 5–8 years | 0.0% | 0.0% | 0.0% |
| Some high school | 11.8% | 14.8% | 9.1% |
| Completed high school or G.E.D. | 17.7% | 14.8% | 20.2% |
| Business or trade school | 0.5% | 0.0% | 1.0% |
| 1–3 years college | 40.6% | 50.0% | 32.3% |
| Completed college | 14.4% | 10.2% | 18.2% |
| Post-graduate college | 13.9% | 10.2% | 17.2% |
| Employment status | |||
| Working, full time | 18.7% | 12.5% | 24.2% |
| Working, part time | 19.3% | 25.0% | 14.1% |
| Not currently employed | 3.2% | 3.4% | 3.0% |
| Student | 56.7% | 55.7% | 57.6% |
| Retired | 0.0% | 0.0% | 0.0% |
| Disabled | 0.0% | 0.0% | 0.0% |
| Other | 2.1% | 3.4% | 1.0% |
Descriptive statistics for the parameters derived for the GENEActiv (analyzed by PennZzz, ChronoSapiens and traits computed by the nparACT package) and FitBit devices are provided in Tables 2–5. Given the large number of PennZzz statistics, we examined the distributions for each parameter (Supplemental Figure S2A) and the correlations among the parameters to better understand the extent to which they captured unique information (Supplemental Figure S2B). Generally, correlations were high among parameters that fell within the same category. For example, parameters describing aspects of activity levels during rest episodes were all highly correlated with each other (both positively and negatively, for example, number and mean duration of sleep episodes). This suggests that many of the parameters are not contributing much unique information relative to other related parameters and that it should be possible to reduce the number of parameters to a minimal set that captures the same amount of information.
TABLE 2.
Descriptive statistics for GENEActiv parameters derived from the PennZzz algorithm
| GENEActiv traits | SD | Mean | Median | Range |
|---|---|---|---|---|
| Minutes of active wake (MAW) | 961.44 | 970.345 | 1067.07 | 279.26 |
| Minutes of quiet wake (mQW) | 26.24 | 78.98 | 74.67 | 159.82 |
| Minutes of total wake (TW) | 55.39 | 1040.43 | 1048.05 | 339.15 |
| Minutes of intermediate sleep | 23.9 | 120.03 | 120.76 | 137.64 |
| Minutes of light sleep (mLS) | 45.79 | 392.97 | 386.44 | 233.35 |
| Minutes of deep sleep (mDS) | 23.35 | 69.88 | 67.8 | 119.655 |
| Minutes of total sleep (mTS) | 61.72 | 582.53 | 578.71 | 280.53 |
| Wake after sleep onset (WASO) in minutes | 22.2 | 138.25 | 137.02 | 109.95 |
| Rest episodes duration in hours (REDhrs) | 0.871 | 8.69 | 8.65 | 5.15 |
| Sleep efficiency (SE) % | 0.034 | 0.73 | 0.74 | 0.17 |
| Mean activity during main rest episode | 0.187 | 0.66 | 0.62 | 0.95 |
| Number of sleep episodes | 10.14 | 48.54 | 48.73 | 53.37 |
| Mean duration of sleep episodes in minutes | 2.405 | 10.86 | 10.66 | 12.47 |
| Number of wake episodes | 10.252 | 48.05 | 48.25 | 53.77 |
| Mean duration of wake episodes in minutes | 4.046 | 17.01 | 16.5 | 18.37 |
| Sleep during rest episode hours | 0.72 | 6.38 | 6.38 | 3.74 |
| Median activity during rest episodes | 0.206 | 0.47 | 0.39 | 0.86 |
| Mean variance of activity during main rest episode | 0.723 | 1.25 | 1.09 | 4.97 |
| Mean activity during wake | 0.49 | 2.08 | 2 | 2.21 |
| Median activity during wake | 0.277 | 0.842 | 0.81 | 1.48 |
| Variance of activity during wake | 8.73 | 12.61 | 9.125 | 40.28 |
| Number of sleep episodes during main sleep | 7.457 | 30.09 | 30.24 | 33.6 |
| Mean duration of sleep episodes during main sleep in minutes | 4.07 | 14.29 | 13.525 | 18.66 |
| Number of wake episodes during main sleep | 7.477 | 30.09 | 30.32 | 33.6 |
| Mean duration of wake episodes during main sleep in minutes | 0.576 | 3.842 | 3.71 | 3.74 |
| Total time in homothermic state in hours | 0.47 | 1.09 | 1.09 | 3.38 |
| Total time in homothermic state during wake in hours | 0.341 | 0.65 | 0.62 | 2.34 |
| Total time in homothermic state during sleep in hours | 0.183 | 0.671 | 0.67 | 1.15 |
TABLE 5.
Descriptive statistics for FitBit variables
| FitBit traits | SD | Mean | Median | Range |
|---|---|---|---|---|
| Sleep duration (min) | 40.84 | 457.72 | 456.71 | 268.4 |
| Sleep efficiency | 2.26 | 93.94 | 94.27 | 10.57 |
| Minutes after wakeup | 0.77 | 0.95 | 0.772 | 4.28 |
| Minutes asleep | 36.86 | 427.89 | 426.43 | 212.68 |
| Minutes to fall asleep | 3.505 | 0.79 | 0 | 27.75 |
| Awake count | 1.47 | 1.53 | 1.26 | 12.29 |
| Awake duration | 3.40 | 3.49 | 2.69 | 26.39 |
| Restless count | 5.38 | 13.23 | 12.65 | 25.33 |
| Restless duration | 12.24 | 25.32 | 23.77 | 73.28 |
Distributions of ChronoSapiens traits and their inter-correlations are shown in Supplemental Figures S3A and S3B. The correlations among parameters were much lower in magnitude compared to the PennZzz variables, indicating that they each capture unique aspects of rhythmicity and should be retained in future studies. Distributions of rest-activity measures from nparACT are shown in Figure S4. Plots of the distributions of each trait separated by their MZ or DZ status are shown in Figures S5A–S5C.
For almost all parameters an AE model provided the best fit. The only exceptions to this pattern were intermediate sleep, primary max value, primary mas phase and center of gravity max phase, for which an ACE model provided a better fit. For intermediate sleep, C was equal to A in magnitude but for the other variables C ws much less than A.
Ten traits among the 29 PennZzz variables showed high heritability (using a threshold of h2 > 0.6; Tables 6 and S1; Figure 2; SOLAR results are in Figure S6). High heritability was seen for the number of minutes of active wake, number of sleep episodes, number of wake episodes, overall mean activity, overall variance in activity, mean activity during wake, variance of activity during wake, number of sleep episodes during the main sleep period, number of wake episodes during the main sleep period, total time in homothermic state and total time in homothermic state during wake. Variables related to the number of sleep or wake episodes were highly correlated with each other, as were those related to homothermic state. Heritability estimates for parameters from ChronoSapiens, nparACT and FitBit are shown in Figures 3–5 (SOLAR results are in Figures S7–S9 for ChronoSapiens, nparACT and FitBit, respectively). For ChronoSapiens, six of the traits showed high heritability: CoG phase, Primary max value, Primary offset, Primary min phase, RoO and MRI (see Table 2 for variable definitions). Only one variable from nparACT showed high heritability, the 10 period with maximal activity. The heritability of FitBit statistics were moderate-to-high, with 5 out of 10 variables showing heritability >0.6.
TABLE 6.
Heritability estimates and associated statistics from Mx
| Pennzzz_trait | H2 using covariate age & sex | P_value | Cl lower bound | Cl upper bound | C using covariate age & sex | SE | H2 using covariate BMI & sex | P_value | Cl lower bound | Cl upper bound | C using covariate BMI & sex | SE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Active wake | 0.46 | 0.0004 | 0.24 | 0.651 | 0 | 0.13 | 0.47 | 0.1 | 0.28 | 0.65 | 0 | 0.13 |
| Light sleep | 0.41 | 0.0002 | 0.15 | 0.606 | 0 | 0.11 | 0.4 | 0.001 | 0.16 | 0.61 | 0 | 0.12 |
| Deep sleep | 0.21 | 0.1 | 0 | 0.4741 | 0 | 0.08 | 0.2 | 0.1 | 0 | 0.4673 | 0 | 0.08 |
| Total sleep | 0.49 | 0.2 | 0.24 | 0.67 | 0 | 0.15 | 0.33 | 0.07 | 0 | 0.6766 | 0.16 | 0.1 |
| No of sleep episodes | 0.64 | 0.003 | 0.33 | 0.814 | 0 | 0.05 | 0.68 | 0.002 | 0.4 | 0.83 | 0 | 0.05 |
| Mean duration of sleep episodes | 0.44 | 0.05 | 0.13 | 0.672 | 0 | 0.08 | 0.44 | 0.05 | 0.131 | 0.674 | 0 | 0.08 |
| No of wake episodes | 0.64 | 0.003 | 0.32 | 0.811 | 0 | 0.05 | 0.67 | 0.002 | 0.382 | 0.83 | 0 | 0.05 |
| Mean duration of wake episodes | 0.47 | 0.04 | 0.19 | 0.680 | 0 | 0.09 | 0.49 | 0.05 | 0 | 0.69 | 0 | 0.1 |
| Mean activity during wake | 0.69 | 0.003 | 0.16 | 0.788 | 0 | 0.1 | 0.65 | 0.02 | 0.08 | 0.788 | 0 | 0.12 |
| Variance of activity during wake | 0.66 | 2.00E-02 | 0.46 | 0.793 | 0 | 0.13 | 0.66 | 0.02 | 0.08 | 0.796 | 0 | 0.13 |
| Number of sleep episodes during deep sleep | 0.61 | 0.01 | 0.3 | 0.788 | 0 | 0.06 | 0.63 | 0.01 | 0.33 | 0.81 | 0 | 0.07 |
| Mean duration of sleep episodes during deep sleep | 0.6 | 0.01 | 0.33 | 0.767 | 0 | 0.08 | 0.6 | 0.01 | 0.32 | 0.769 | 0 | 0.08 |
| Number of wake episodes during deep sleep | 0.63 | 0.01 | 0.31 | 0.794 | 0 | 0.06 | 0.64 | 0.01 | 0.34 | 0.806 | 0 | 0.07 |
| Mean duration of wake episodes during deep sleep | 0.37 | 0.08 | 0 | 0.6434 | 0 | 0.07 | 0.36 | 0.09 | 0 | 0.64 | 0 | 0.07 |
| Wake after sleep onset | 0.26 | 0.1 | 0 | 0.536 | 0 | 0.09 | 0.27 | 0.1 | 0 | 0.5525 | 0 | 0.09 |
| Total time in homothermic state during wake | 0.64 | 0.005 | 0.27 | 0.793 | 0 | 0.07 | 0.64 | 0.005 | 0.26 | 0.793 | 0 | 0.07 |
| Sleep efficiency | 0.58 | 0.05 | 0 | 0.749 | 0 | 0.14 | 0.58 | 0.07 | 0 | 0.7504 | 0 | 0.14 |
| ChronoSapiens_traits | ||||||||||||
| Center of gravity MAX value | 0.58 | 0.05 | 0.738 | 0 | 0 | 0.12 | 0.6 | 0.06 | 0.75 | 0 | 0 | 0.14 |
| Center of gravity MAX phase | 0.55 | 0.06 | 0.81 | 0 | 0.12 | 0.14 | 0.52 | 0.08 | 0.81 | 0 | 0.14 | 0.15 |
| Primary MAX value | 0.42 | 0.1 | 0.75 | 0 | 0.19 | 0.15 | 0.4 | 0.2 | 0.76 | 0 | 0.23 | 0.17 |
| Primary MAX phase | 0.41 | 0.2 | 0.71 | 0 | 0.11 | 0.14 | 0.38 | 0.2 | 0.7 | 0 | 0.12 | 0.14 |
| Primary MIN value | 0.45 | 0.1 | 0.64 | 0.22 | 0 | 0.13 | 0.4 | 0.2 | 0.68 | 0 | 0.11 | 0.13 |
| Primary MIN phase | 0.88 | 0 | 0.940 | 0.74 | 0 | 0.03 | 0.91 | 1.28E-05 | 0.95 | 0.8 | 0 | 0.03 |
| Primary_OFF_phase | 0.35 | 0.2 | 0.732 | 0 | 0.23 | 0.16 | 0.31 | 0.2 | 0.72 | 0 | 0.24 | 0.16 |
| Rhythm of oscillation | 0.28 | 0.1 | 0.73 | 0 | 0.5 | 0.18 | 0.28 | 0.1 | 0.73 | 0 | 0.5 | 0.18 |
| Munich Rythmicity Index | 0.46 | 0.09 | 0.810 | 0 | 0.23 | 0.16 | 0.43 | 0.1 | 0.8 | 0 | 0.26 | 0.17 |
| Alpha_rho | 0.22 | 0.5 | 0.610 | 0 | 0.17 | 0.13 | 0.24 | 0.5 | 0.62 | 0 | 0.16 | 0.13 |
| Rest-activity_traits | ||||||||||||
| Interdaily stability | 0.3 | 0.4 | 0 | 0.6863732 | 0.17 | 0.14 | 0.24 | 0.5 | 0.31 | 0.77 | 0.25 | 0.15 |
| Relative amplitude | 0.57 | 0.09 | 0 | 0.7344203 | 0 | 0.14 | 0.6 | 0.1 | 0 | 0.75 | 0.07 | 0.15 |
| L5 (5 hours with minimal activity) | 0.45 | 0.1 | 0.07 | 0.7078336 | 0 | 0.09 | 0.47 | 0.1 | 0.1 | 0.71 | 0 | 0.1 |
| M10 (10 hours with maximal activity) | 0.64 | 0.0007 | 0.28 | 0.9165123 | 0.22 | 0.14 | 0.61 | 0.001 | 0.25 | 0.91 | 0.25 | 0.15 |
| Fitbitjraits | ||||||||||||
| Alpha/rho | 0.56 | 0.1 | 0 | 0.7828844 | 0 | 0.12 | 0.56 | 0.1 | 0 | 0.78 | 0 | 0.13 |
| Efficiency | 0.88 | 0.0001 | 0.48 | 0.94 | 0 | 0.1 | 0.89 | 0.0001 | 0.47 | 0.94 | 0 | 0.11 |
| Minutes after wakeup | 0.56 | 0.1 | 0 | 0.79 | 0 | 0.13 | 0.56 | 0.1 | 0 | 0.78 | 0 | 0.13 |
| Minutes asleep | 0.51 | 0.2 | 0.13 | 0.74 | 0 | 0.1 | 0.5 | 0.009 | 0.14 | 0.74 | 0 | 0.1 |
| Time in bed in minutes | 0.5 | 0.3 | 0.16 | 0.73 | 0 | 0.1 | 0.51 | 0.3 | 0 | 0.74 | 0 | 0.1 |
| Restless count | 0.92 | 0 | 0.82 | 0.96 | 0 | 0.05 | 0.92 | 3.63E-06 | 0.81 | 0.96 | 0 | 0.05 |
| Restless duration | 0.81 | 0.005 | 0.91 | 0.48 | 0 | 0.08 | 0.8 | 0.005 | 0.9 | 0.6 | 0 | 0.08 |
FIGURE 2.
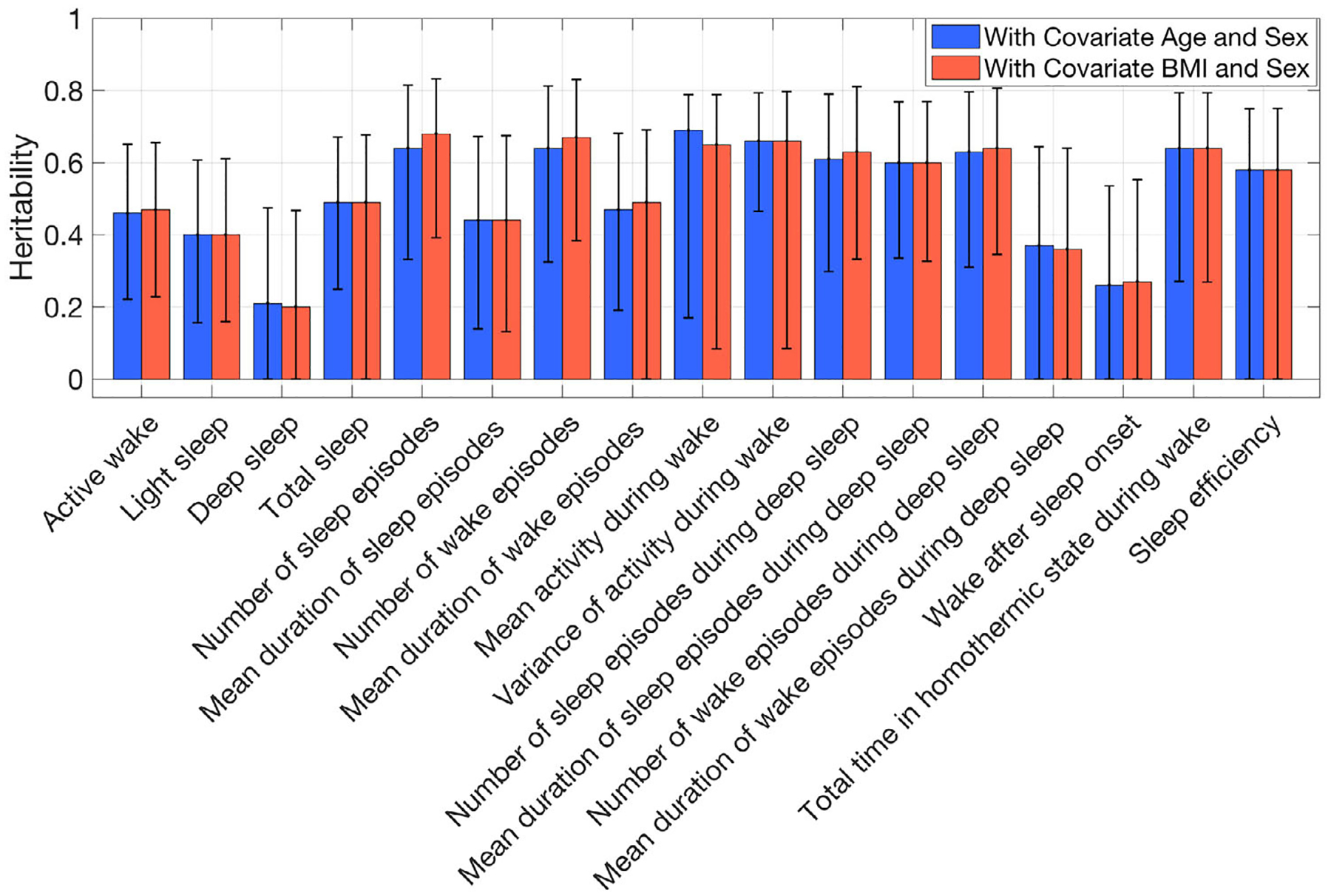
Heritability of PennZzz-detected traits computed in OpenMX by using covariates age and body mass index
FIGURE 3.
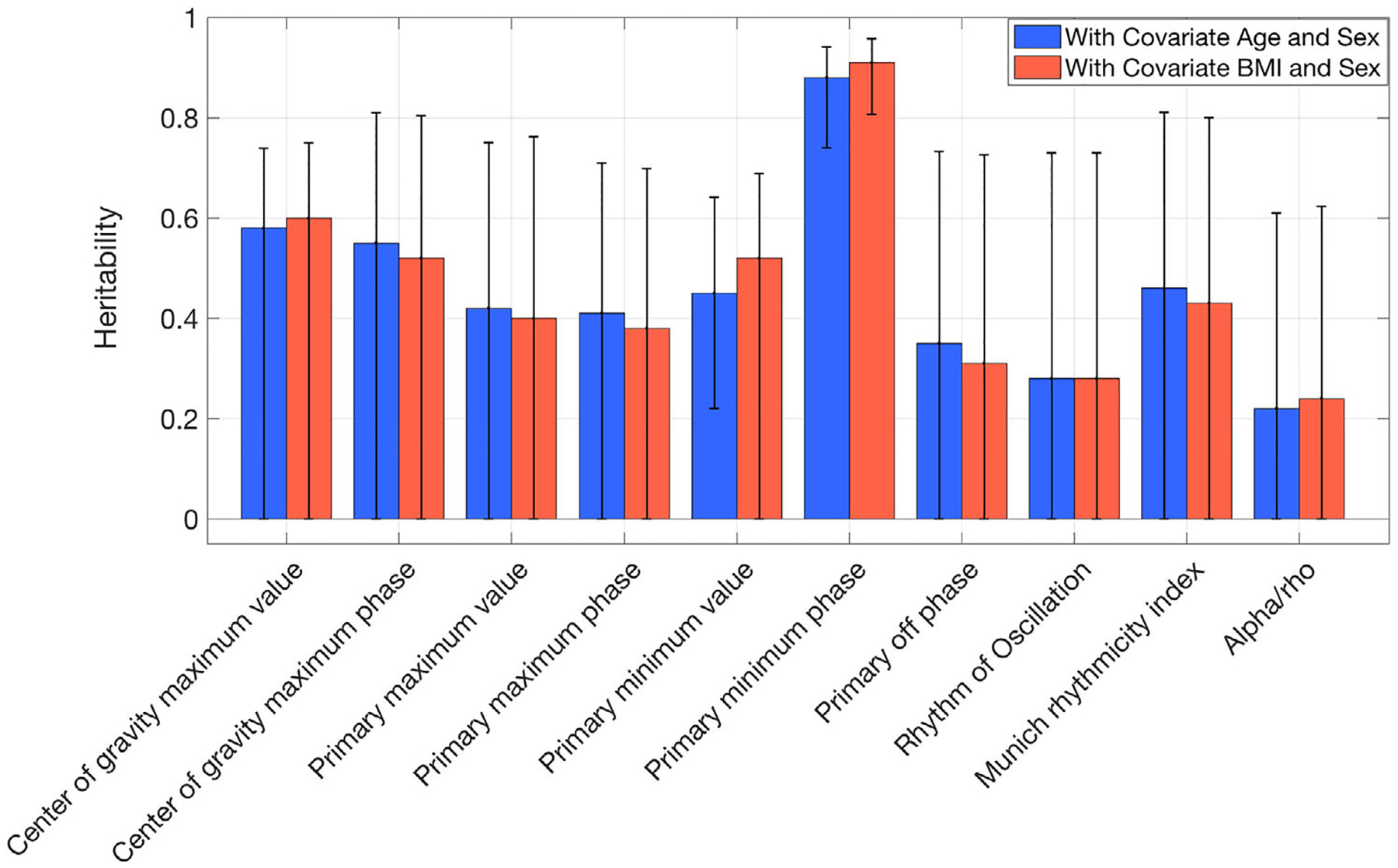
Heritability of ChronoSapiens-detected traits computed in OpenMX by using covariates age and body mass index
FIGURE 5.
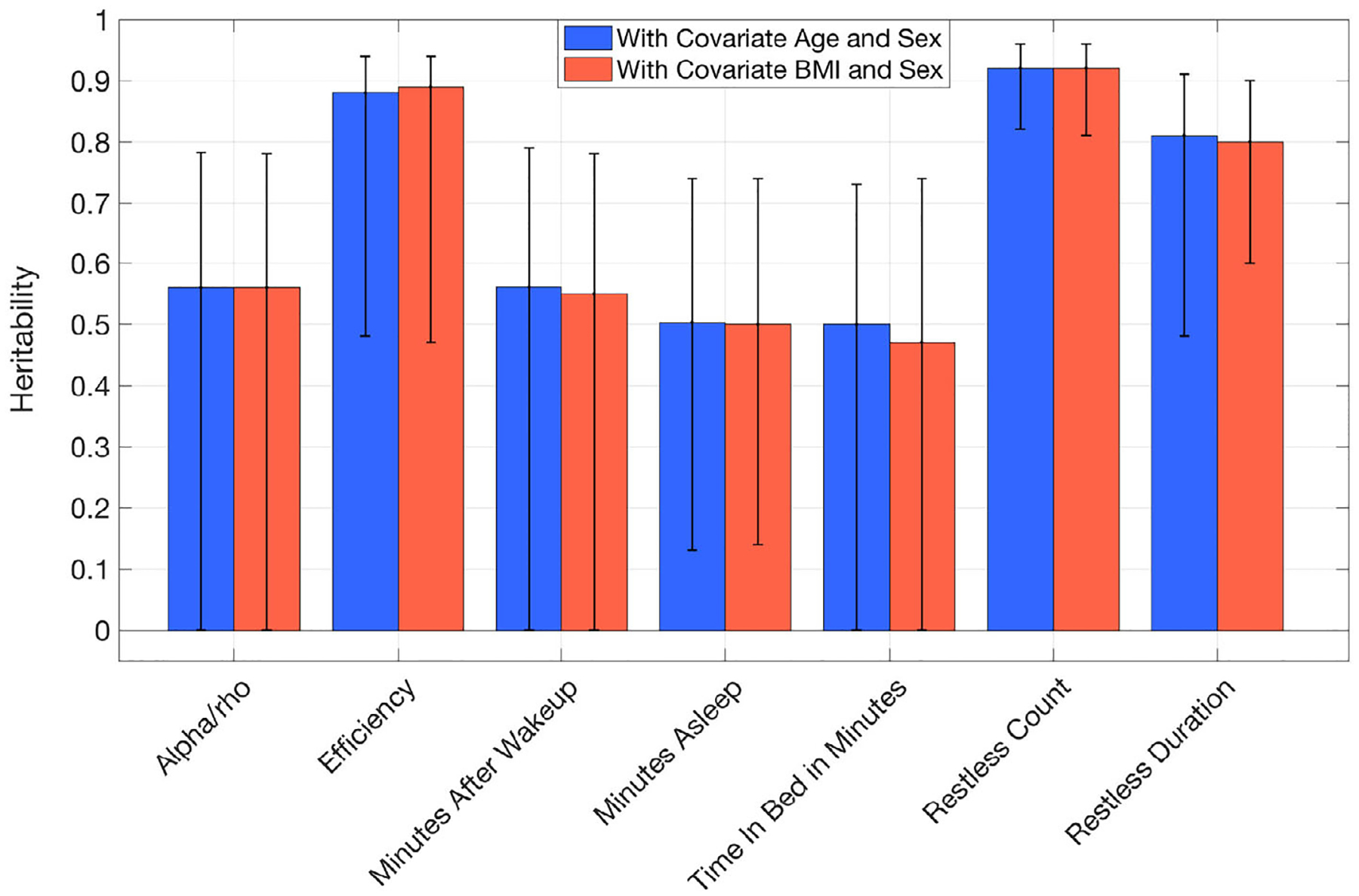
Heritability of FitBit traits computed in OpenMX by using covariates age and body mass index
The mean total sleep time derived from each method was 442.5 minutes for the sleep diary, 469.1 minutes for the PennZzz analysis of the GENEActiv, and 427.9 minutes for the FitBit. Sleep diary total sleep time was significantly correlated with that from GENEActiv (r = 0.41, P = 0.0001) and FitBit (r = 0.39, P = 0.0004) but the actimetry methods were not correlated with each other (r = 0.003, P = 0.987).
4 |. DISCUSSION
The goal of this study was to identify actimetry traits as potential endophenotypes for behavioral genetic studies. A large number of parameters were generated using software to compute statistics on sleep and activity (PennZzz) and phase analysis (ChronoSapiens and nparACT) from a research grade actimeter. A limited set of sleep traits were collected with FitBit, a widely used consumer-oriented device. By examining a large number of parameters, we identified several traits with high heritability using a classical twin design. The high heritability of these traits suggest that they may be appropriate for use in behavioral genetic studies.
The traits with the highest heritability estimates covered a number of different sleep and rhythmicity features. For PennZzz, these traits included measures of variability in activity, fragmentation of sleep and wake and time spent in periods of stable temperature. Several traits related to measures of phase and range of oscillation emerged from ChronoSapiens. It is noteworthy that phase of activity and rest was more dominant than activity levels. Phase relates to temporal behavior, which is controlled to a large extent by the circadian clock. The dominance of phase in heritability of complex behavior was to be expected because the circadian system is known for its strong genetic basis. Only one trait from nparACT was highly heritable. The FitBit traits with the strongest heritability were related primarily to periods of restless sleep, although overall sleep efficiency was also prominent. This shows that a wide range of sleep and rhythmicity traits are strongly influenced by genetic factors. These traits represent a range of characteristics of activity. Parameters describing overall levels of activity or sleep were less likely to stand out compared with traits related to variability and sleep quality (eg, fragmentation and restless sleep). Of note, estimates of heritability are limited by measurement error and, consequently, the phenotypes with higher heritabilities may be those that are more reliably measured.
The sleep diaries and actimetry methods were compared in their estimates of mean total sleep time. While the estimates were generally in the same ballpark, there were still substantial differences with the estimates from GENEActiv and FitBit differing by 33.1 minutes. It is noteworthy that the actimetry methods were more highly correlated with the sleep diary estimate than they were with each other. This suggests that the actimetry methods may not be directly comparable although it is not clear whether this is because of differences in hardware or scoring algorithms.
Most prior actimetry studies relied on a small number of standard sleep/wake or circadian statistics. The study by Pagani and colleagues22 was unique in its comprehensive assessment of a wider range of traits, however, they limited their analyses to previously reported statistics. While future studies are needed to determine the utility of the parameters in this study, the heritability analyses provide initial support of biological plausibility for a number of them.
The twin design provides a solid framework for estimating heritability that has advantages over the family design used previously.22 The twins in the current study were not selected with regard to any biomedical phenotype and are generally healthy individuals, whereas the families studied by Pagani were selected for a high density of individuals with bipolar disorder. Thus the present study shows that actimetry traits are heritable beyond the context of bipolar disorder, supporting its use as a general endophenotype. Estimation of heritability through twin and family studies relies on different assumptions regarding shared environment. As such, it is valuable to compare heritabilities estimated via the two methods. Unless shared environment is specifically modeled, family studies have an implicit assumption that correlations among family members are due only to the additive effect of genes. Studies of nuclear families, where all pairs of individuals have equal genetic sharing and potentially similar environmental sharing, may have inflated estimates of heritability because of confounding with shared environment. Twin studies account for shared environment but implicitly assume that environmental sharing is equivalent for MZ and DZ pairs. Another consideration is the estimation of narrow sense heritability in family studies vs broad sense heritability in twin studies. Thus, to the extent that heritabilities are higher in family studies than twin studies we might posit effects of shared environment and to the extent that they are higher in twins than in families we might posit non-additive genetic components. It should also be noted that heritability is population-specific. Thus the present study also has value in that it focuses on a different population from the previously published Pagani study.
The large quantity of parameters generated in this study, while a strength, is also a weakness in that heritability was computed for a large number of traits and may have led to an inflated Type I error rate. However, if a Bonferroni Type I error correction had been applied seven of the traits would have survived multiple testing. The novel statistics computed are also as yet unproven for utility and relevance. However, this reflects the exploratory nature of this study. We plan to further examine and refine this list of candidate endophenotypes in subsequent studies. Data loss was also a limitation of this study, with quite a few sets of twins not having useable data for the GENEActiv, FitBit or both. There was also variability in the amount of data subjects provided, with some wearing the devices for only one of the 2 weeks, which could add variability across subjects in the robustness of parameter estimates. We did not observe any sex differences, nor were there considerable effects of other covariates (age, BMI), which may reflect the relative homogeneity of the sample. These results will need to be examined in more diverse samples in the future.
In summary, our results show the usefulness of actimetry for behavioral genetic research. They provide the means to collect objective, high-frequency data unobtrusively to generate statistics with substantial heritability, satisfying one of the proposed criteria for endophenotypes. Actimetry can be used in large samples, as has been showed in the UK Biobank, which collected 1 week recordings in >100 000 subjects.34 Future studies will need to build on these results to determine which traits are of the most value and satisfy other endophenotype criteria and then include them in genetic studies in order understand their underlying biology. The results of genetic studies will be particularly informative as to whether these endophenotypes do indeed have simpler genetic architecture as opposed to clinical symptoms or syndromes. It will also be important to examine these traits longitudinally given the known developmental changes that occur in sleep. Speficially, a set of different actimetry traits may be more or less useful at certain stages of development.
Supplementary Material
FIGURE 4.
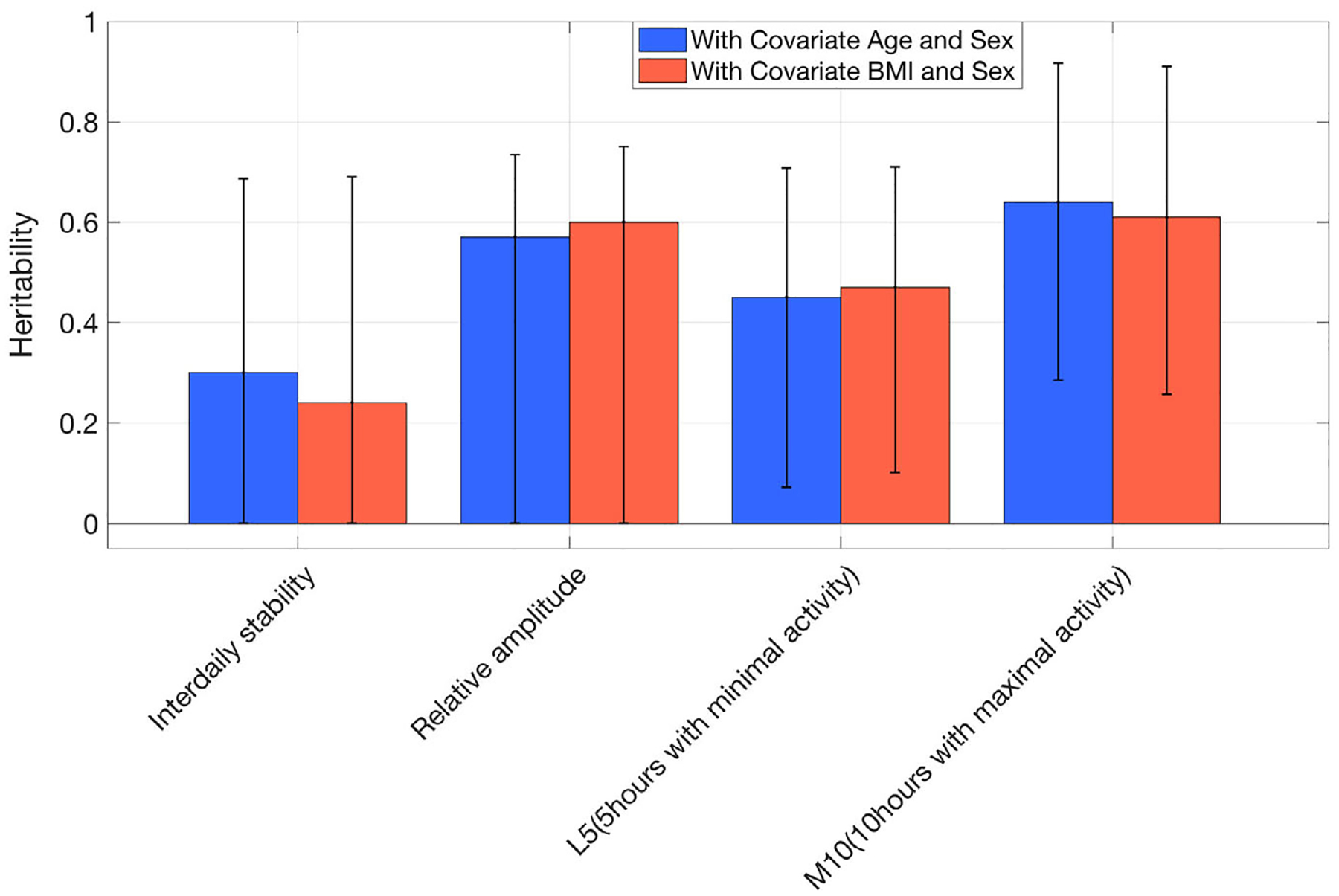
Heritability of rest-activity measures from nparACT computed in OpenMX by using covariates age and body mass index
TABLE 3.
Descriptive statistics for GENEActiv parameters derived from ChronoSapiens software
| ChronoSapiens_traits | SD | Mean | Median | Range |
|---|---|---|---|---|
| Maximum value of the fitted first harmonic (CoG_MAX-value) | 0.14 | 1.69 | 1.69 | 1.95 |
| Timing of the peak of the fitted first harmonic (CoG_MAX-phase) | 1.72 | 15.7 | 15.7 | 9.60 |
| Maximum value of the fitted second harmonic (Prim MAX-value) | 0.26 | 1.95 | 1.69 | 1.64 |
| Timing of the peak of the fitted second harmonic (Prim MAX-phase) | 2.05 | −8.31 | −8.3 | 11.30 |
| Time of the primary downward zero transition of the fitted second harmonic (Prim Offset) | 1.76 | −2.79 | −2.85 | 11.3 |
| Minimum value of the fitted second harmonic (Prim MIN-value) | 14.28 | 9.48 | 3.7 | 94.23 |
| Timing of the nadir of the fitted second harmonic (Prim MIN-phase) | 2.63 | 3.59 48.05 |
3.70 48.25 |
30.20 53.77 |
| Range of Oscilation (RoO) | 83.37 | 165.68 | 143.56 | 519.26 |
| Munich Rhythmicity Index (MRI) | 43.53 | 71.5 | 61.81 | 302.71 |
| Alpha_Rho | 0.16 | 1.05 | 1.02 | 0.78 |
TABLE 4.
Descriptive statistics for rest-activity measures from nparACT
| Rest-activity traits | Mean | SD | Median | Range |
|---|---|---|---|---|
| Interdaily stability | 0.22 | 0.19 | 0.11 | 0.68 |
| Intradaily variability | 0.87 | 0.83 | 0.28 | 1.35 |
| Relative amplitude | 0.54 | 0.55 | 0.13 | 0.67 |
| L5 (5 hours with minimal activity) | 31.4 | 28.42 | 12.27 | 86.14 |
| M10 (10 hours with maximal activity) | 108.54 | 99.18 | 50.12 | 425.86 |
ACKNOWLEDGMENTS
We would like to thank the participants in the Pennsylvania Twin Registry for their collaboration on this project and B. Cleveland,J. Neiderhiser and A. Ramos for help with the recruitment of twins. This work was supported by the US National Institute of Health (NIMH) grant R21 MH103963.
Funding informationNational Institute of Mental Health, Grant/Award Number: R21 MH103963
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Goes FS. Genetics of bipolar disorder: recent update and future directions. Psychiatr Clin North Am. 2016;39(1):139–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scott LJ, Muglia P, Kong XQ, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A. 2009;106(18):7501–7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Craddock N, Sklar P. Genetics of bipolar disorder: successful start to a long journey. Trends Genet. 2009;25(2):99–105. [DOI] [PubMed] [Google Scholar]
- 4.Flint J, Kendler KS. The genetics of major depression. Neuron. 2014; 81(3):484–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Georgi B, Craig D, Kember RL, et al. Genomic view of bipolar disorder revealed by whole genome sequencing in a genetic isolate. PLoS Genet. 2014;10(3):e1004229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunugi H, Hori H, Ogawa S. Biochemical markers subtyping major depressive disorder. Psychiatry Clin Neurosci. 2015;69(10):597–608. [DOI] [PubMed] [Google Scholar]
- 7.Jobe JB. Cognitive psychology and self-reports: models and methods. Qual Life Res. 2003;12(3):219–227. [DOI] [PubMed] [Google Scholar]
- 8.Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160(4):636–645. [DOI] [PubMed] [Google Scholar]
- 9.Lander ES. Splitting schizophrenia. Nature. 1988;336(6195):105–106. [DOI] [PubMed] [Google Scholar]
- 10.Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007;37(2):163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glahn DC, Knowles EE, McKay DR, et al. Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am J Med Genet B Neuropsychiatr Genet. 2014;165B(2):122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tryon W. Issues of validity in actigraphic sleep assessment. Sleep. 2004;27:158–165. [DOI] [PubMed] [Google Scholar]
- 13.Morgenthaler T, Alessi C, Friedman L, et al. Practice parameters for the use of actigraphy in the assessment of sleep and sleep disorders: an update for 2007. Sleep. 2007;30(4):519–529. [DOI] [PubMed] [Google Scholar]
- 14.Patel SR, Hayes AL, Blackwell T, et al. The association between sleep patterns and obesity in older adults. Int J Obes (Lond). 2014;38(9): 1159–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuthbert BN, Insel TR. Toward the future of psychiatric diagnosis: the seven pillars of RDoC. BMC Med. 2013;11:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickie IB, Naismith SL, Robillard R, Scott EM, Hermens DF. Manipulating the sleep-wake cycle and circadian rhythms to improve clinical management of major depression. BMC Med. 2013;11:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones SG, Benca RM. Circadian disruption in psychiatric disorders. Sleep Med Clin. 2015;10(4):481–493. [DOI] [PubMed] [Google Scholar]
- 18.McClung CA. Circadian genes, rhythms and the biology of mood disorders. Pharmacol Ther. 2007;114(2):222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jagannath A, Peirson SN, Foster RG. Sleep and circadian rhythm disruption in neuropsychiatric illness. Curr Opin Neurobiol. 2013;23(5): 888–894. [DOI] [PubMed] [Google Scholar]
- 20.Wolff EA 3rd, Putnam FW, Post RM. Motor activity and affective illness. The relationship of amplitude and temporal distribution to changes in affective state. Arch Gen Psychiatry. 1985;42(3): 288–294. [DOI] [PubMed] [Google Scholar]
- 21.Jones SH, Hare DJ, Evershed K. Actigraphic assessment of circadian activity and sleep patterns in bipolar disorder. Bipolar Disord. 2005;7(2):176–186. [DOI] [PubMed] [Google Scholar]
- 22.Pagani L, St Clair PA, Teshiba TM, et al. Genetic contributions to circadian activity rhythm and sleep pattern phenotypes in pedigrees segregating for severe bipolar disorder. Proc Natl Acad Sci U S A. 2016;113(6):E754–E761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boomsma D, Busjahn A, Peltonen L. Classical twin studies and beyond. Nat Rev Genet. 2002;3(11):872–882. [DOI] [PubMed] [Google Scholar]
- 24.Carney CE, Buysse DJ, Ancoli-Israel S, et al. The consensus sleep diary: standardizing prospective sleep self-monitoring. Sleep. 2012;35(2):287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roenneberg T, Keller LK, Fischer D, Matera JL, Vetter C, Winnebeck EC. Human activity and rest in situ. Methods Enzymol. 2015;552:257–283. [DOI] [PubMed] [Google Scholar]
- 26.Box G, Jenkins G. Time Series Analysis: Forecasting and Control. San Francisco, CA: Holden-Day; 1970. [Google Scholar]
- 27.Blautzik J, Vetter C, Peres I, et al. Classifying fMRI-derived resting-state connectivity patterns according to their daily rhythmicity. Neuroimage. 2013;71:298–306. [DOI] [PubMed] [Google Scholar]
- 28.Blume C, Santhi N, Schabus M. nparACT’ package for R: A free software tool for the non-parametric analysis of actigraphy data. MethodsX. 2016;3:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Someren EJ, Lijzenga C, Mirmiran M, Swaab DF. Long-term fitness training improves the circadian rest-activity rhythm in healthy elderly males. J Biol Rhythms. 1997;12(2):146–156. [DOI] [PubMed] [Google Scholar]
- 30.Pengo MF, Won CH, Bourjeily G. Sleep in women across the life span. Chest. 2018;154(1):196–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Vitiello MV, Gooneratne NS. Sleep in normal aging. Sleep Med Clin. 2018;13(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan WS, Levsen MP, McCrae CS. A meta-analysis of associations between obesity and insomnia diagnosis and symptoms. Sleep Med Rev. 2018;40:170–182. [DOI] [PubMed] [Google Scholar]
- 33.Merikangas KR, Swendsen J, Hickie IB, et al. Real-time mobile monitoring of the dynamic associations among motor activity, energy, mood, and sleep in adults with bipolar disorder. JAMA Psychiatry. 2019;76(2):190–198. 10.1001/jamapsychiatry.2018.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sudlow C, Gallacher J, Allen N, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We deposited accelerometry files to the NIMH Data Archive.


