Fascin is highly dynamic within filopodia and undergoes rapid changes in phosphorylation that dictate localized F-actin bundling activity. Pfisterer et al. show that fascin binds directly to FMNL2, and this controls fascin positioning and entry into filopodia to control filopodia stability and cell-matrix sensing.
Abstract
Filopodia are peripheral F-actin–rich structures that enable cell sensing of the microenvironment. Fascin is an F-actin–bundling protein that plays a key role in stabilizing filopodia to support efficient adhesion and migration. Fascin is also highly up-regulated in human cancers, where it increases invasive cell behavior and correlates with poor patient prognosis. Previous studies have shown that fascin phosphorylation can regulate F-actin bundling, and that this modification can contribute to subcellular fascin localization and function. However, the factors that regulate fascin dynamics within filopodia remain poorly understood. In the current study, we used advanced live-cell imaging techniques and a fascin biosensor to demonstrate that fascin phosphorylation, localization, and binding to F-actin are highly dynamic and dependent on local cytoskeletal architecture in cells in both 2D and 3D environments. Fascin dynamics within filopodia are under the control of formins, and in particular FMNL2, that binds directly to dephosphorylated fascin. Our data provide new insight into control of fascin dynamics at the nanoscale and into the mechanisms governing rapid cytoskeletal adaptation to environmental changes. This filopodia-driven exploration stage may represent an essential regulatory step in the transition from static to migrating cancer cells.
Introduction
Environmental sensing is a key property enabling cancer cells to dynamically adapt to changes in the ECM during migration away from the primary tumor to distant sites in the body. Key players that can effectively fulfil the task of exploring nanostructures in the extracellular microenvironment are filopodia (Albuschies and Vogel, 2013). These highly dynamic finger-like membrane protrusions are stabilized by fascin, a key molecule in controlling parallel F-actin bundling in a range of cancer cell types (Jacquemet et al., 2015). Fascin is low or absent in normal epithelial cells but is significantly up-regulated in numerous human cancers, and this increased expression correlates with poor clinical prognosis and higher incidence of metastasis (Jansen et al., 2011; Jayo et al., 2016; Jayo and Parsons, 2010; Schoumacher et al., 2014; Vignjevic et al., 2007). Fascin is therefore emerging as both a key prognostic marker and a potential therapeutic target for metastatic disease (Chen et al., 2010).
Fascin consists of four β-trefoil domains with two actin-binding sites located at the N- and C-termini that enable bundling of adjacent actin filaments (Jayo and Parsons, 2010; Sedeh et al., 2010). Structural analysis suggest that fascin adopts a compact globular conformation (Sedeh et al., 2010), but possible conformation changes during cycles of actin bundling have been proposed (Yang et al., 2013). Fascin-dependent bundling of F-actin is controlled by PKC-dependent phosphorylation of serine 39 within the N-terminus (Adams et al., 1999; Anilkumar et al., 2003). Phosphorylation at serine 39 (pS39) can be positively regulated by extracellular cues, resulting in a reduction of filopodia due to the loss of F-actin bundling by fascin (Adams et al., 1999; Zhang et al., 2009). We have shown previously that pS39-fascin associates with Nesprin-2 and thereby couples F-actin to the nuclear envelope (Jayo et al., 2016). This interaction is essential for nuclear movement and plasticity in migrating cells and may be a crucial F-actin–bundling independent mechanism used by invading cancer cells. Fascin is also important for focal adhesion dynamics through binding to microtubules and associated adhesion components, contributing to adhesion turnover and cell migration (Elkhatib et al., 2014; Villari et al., 2015). However, despite increasing understanding of fascin functions within the cell, very little is known about how fascin rapidly changes localization or function in response to changing extracellular environments.
Cancer cell migration is dependent on coordination between the physical characteristics of the ECM, cell adhesion, actin-driven contractility, and membrane protrusion. Two main F-actin architectures regulate membrane protrusion: linear formin-dependent and branched actin-related protein 2 (Arp2)/Arp3 complex-dependent F-actin. Linear F-actin–rich membrane protrusions, such as filopodia, act as sensory organs, whereas branched sheet-like F-actin is the predominant structure in protruding lamellipodia at the leading edge during migration and spreading (Faix and Rottner, 2006). Filopodia emerge de novo from the lamellipodium via a cell division cycle 42 (Cdc42)-mediated mechanism or are initiated from precursor forms called microspikes, which are fully embedded in branched F-actin (Faix and Rottner, 2006; Mellor, 2010). After initiation, formins are essential in nucleating and elongating linear F-actin at the barbed end in protruding filopodia, often in a profilin-dependent manner (Chhabra and Higgs, 2007). A recent study has shown that loss of the Cdc42 effector formin proteins, FMNL2 and FMNL3, results in altered lamellipodia ultrastructure with less dense branched F-actin, resulting in impaired protrusive membrane velocities and lamellipodia force generation (Kage et al., 2017). It has been suggested that the homeostasis of branched and linear F-actin could have a direct effect on filopodia initiation and stability. However, it remains unclear whether fascin distribution, positioning, dynamics, and/or function are controlled by local F-actin architecture or the molecular complexes that control this.
A common hallmark of neoplastic disease is the loss of mechanical tissue homeostasis. Emerging evidence suggests that these changes in local ECM stiffness and the subsequent changes to forces exerted on the resident cells play a key role in directly controlling protein function (Lu et al., 2012). Many primary tumors show stiffening of the surrounding ECM or the tumor itself, and this increased rigidity can directly activate biochemical mechanisms enhancing cell invasion, leading to a poorer clinical outcome (Broders-Bondon et al., 2018; Seewaldt, 2014). As cells use filopodia to actively probe the ECM before transition to a promigratory phenotype (Carey et al., 2016) and are guided by increased matrix stiffness (also called durotaxis), this form of ECM sensing represents a significant mechanism in coordinating the escape of cells from the primary tumor (van Helvert et al., 2018; Wong et al., 2014).
In the current study, we used advanced live-cell imaging techniques and implementation of a fascin conformation biosensor to demonstrate that fascin phosphorylation, localization, and binding to F-actin are highly dynamic and dependent on local cytoskeletal architecture in cells in both 2D and 3D environments. Moreover, the dynamics and conformation of fascin are sensitive to changes in substrate stiffness. We show that fascin-actin binding is highest at the base of growing filopodia, and cycles of phosphorylation are required for stabilization of this interaction. Fascin entry into and exit from filopodia are under the control of formins, and in particular FMNL2, which binds directly and preferentially to dephosphorylated fascin. Our data provide new insight into control of fascin dynamics at the nanoscale and into the mechanisms governing rapid cytoskeletal adaptation to environmental changes.
Results
Fascin is highly mobile and exhibits fast molecular kinetics within filopodia
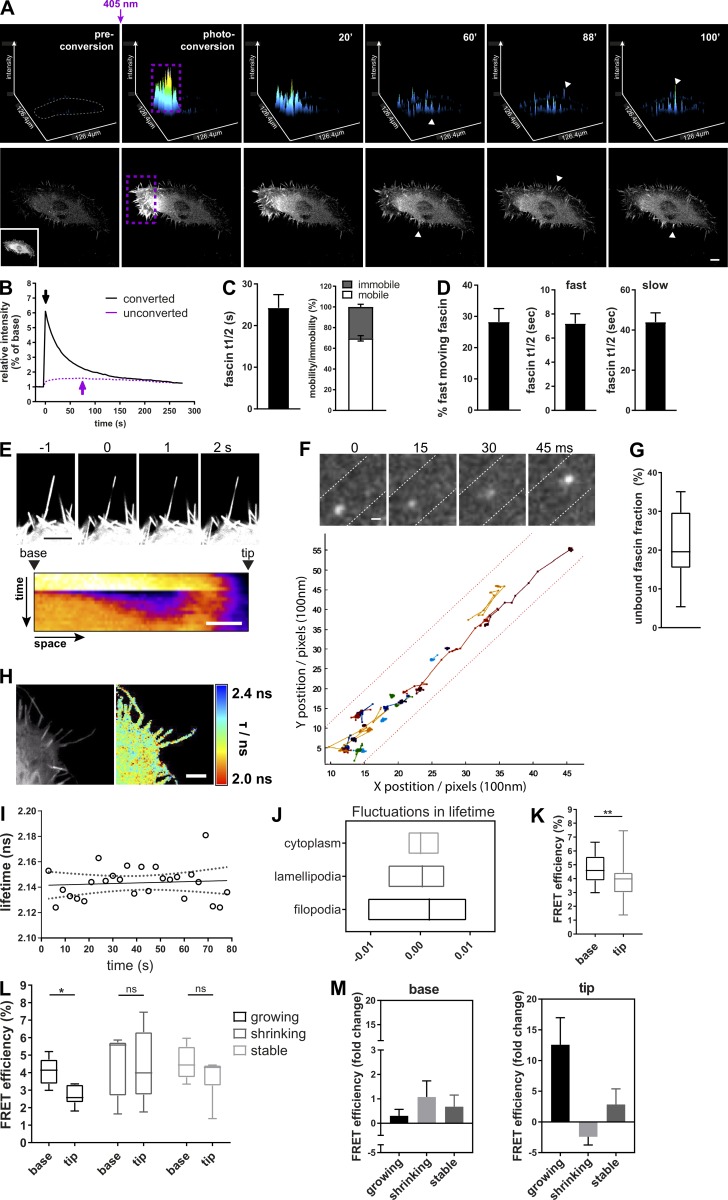
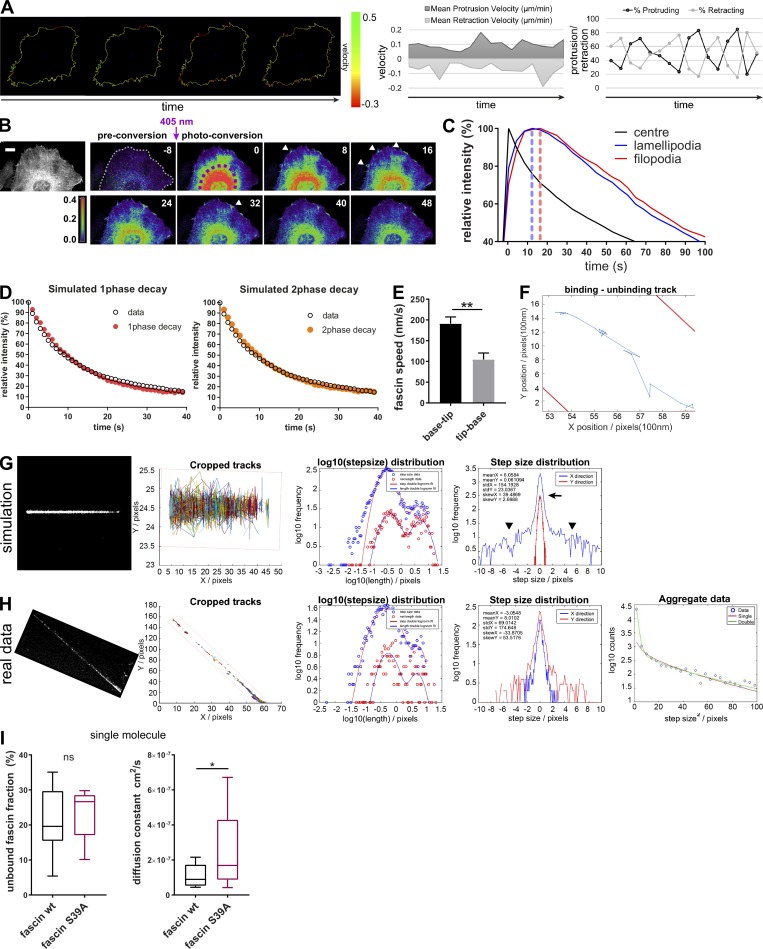
To understand the kinetics of fascin movement across cells, fascin-depleted HeLa cells stably expressing shRNA-resistant mEOS2-fascin at physiological levels (comparable to endogenous fascin; Jayo et al., 2016) were subjected to photoconversion using a short 405-nm laser pulse within multiple filopodia at one side of the cell, followed by tracking of converted fascin movement through the cell over time (Fig. 1 A and Video 1). Photoconverted fascin molecules appeared within filopodia at the opposite side of the cell within 40–60 s of 405-nm illumination (arrowheads). Comparison of fluorescence curves showed the diffusion out of the photoconverted region of interest (ROI) and subsequent increase of photoconverted fascin in the unconverted ROI with a delay of peak maxima of ∼40–50 s (Fig. 1 B, arrow). To determine whether translocation of fascin across the cell was directed toward specific membrane regions, protrusion and retraction of cells was quantified over time and compared with rates of photoconverted fascin appearance. Data demonstrated that cells exhibited random, nondirectional protrusion and retraction cycles with varying velocities (Fig. S1 A) that were not apparently coincident with fascin recruitment, suggesting that fascin is transported in a nondirectional and unbiased manner from filopodia on one side of the cell to the other. To further determine whether perinuclear fascin that links to Nesprin and F-actin at the nuclear membrane (Jayo et al., 2016) exhibited differential kinetics from peripheral fascin, mEOS2-fascin was photoconverted within the perinuclear region, and populations were tracked over time. Notably, the movement of cytoplasmic fascin was so rapid that parts of photoconverted fascin were already outside of the conversion ROI after 30-s illumination with 405 nm (time point 0; Fig. S1 B). Analysis revealed that fascin was efficiently translocated from perinuclear regions, via lamellipodia to filopodia at the cell periphery, with a speed of ∼3 µm/s (Fig. S1 C). These data combined demonstrate that fascin is rapidly mobilized from all sites within the cell to enable fast translocation to new regions.
Figure 1.
Fascin–actin interactions are highly dynamic and spatially regulated. (A) Photoconverted mEOS2-fascin in fascin knockdown cells imaged by confocal microscopy. Top panel shows surface plot visualization (intensity in x,y). Cell border in preconverted frame is highlighted in gray, and photoconversion region, in purple (conversion ROI contained multiple filopodia per cell). Bottom panel shows the respective cell and the intensity of the photoconverted channel (red). Inset picture shows the unconverted cell (green channel). One representative time lapse is shown, scale bar = 10 µm. Arrowheads depict photoconverted fascin in filopodia outside of photoconversion ROI. (B) Representative relative intensities of photoconverted and unconverted regions over time (black and purple, respectively; 4-s frame rate). Peak intensity values (overall maximum intensity) are depicted with arrows. (C) Fascin half-life (t1/2 in seconds, left) showing the time needed for 50% of fascin molecules to exit filopodia after photoconversion and fascin mobile and immobile fractions (right) are shown. Values were extracted from monoexponential curves of photoconverted areas. n = 9 cells (ROIs with multiple filopodia). (D) Percentage of fast moving fascin and t1/2 of fast- and slow-moving fascin. Values were extracted from biexponential curves of photoconverted areas. n = 9 cells (ROIs with multiple filopodia). (E) Small area photobleaching (FRAP) along single filopodium of GFP-fascin expressing cells. Bottom panel shows representative pseudocolored kymograph of fascin recovery. Scale bar, 2 μm. (F) Single molecules of mEOS2-fascin along filopodia (top panel, filopodia borders are outlined as dashed lines, 15 ms frame rate, scale bar = 0.5 µm). Bottom graph: Multiple single-molecule tracks of fascin along the filopodial shaft. The base is located at xy = 0. (G) Unbound fascin fraction estimated by single-molecule tracking analysis as described in Supplemental Methods; average single-molecule tracks of n = 15 filopodia. (H) Interaction of GFP-fascin and RFP-actin measured with multiconfocal FLIM-FRET microscope. Left, intensity image; right, FLIM-FRET data. High interactions are red (low lifetime), whereas low interactions are blue (high lifetime). Scale bar = 2 µm. (I) Fascin–actin interaction measured over time in single filopodium of living cell in 2D with live FLIM-FRET at a 3-s frame rate. Lifetime values are plotted over time (circles) with linear regression (solid line) and 95% confidence interval (dotted line). (J) Fluctuation range of fascin–actin interaction measurement by live FLIM-FRET in different cellular compartments. Lowess residuals were calculated as positive or negative residual value of lifetime data at a certain time point in respect to the fitted Lowess curve of lifetime data. Lowess residuals are plotted as minimum to maximum floating bar graphs for cytoplasm, lamellipodia, and filopodia measurements. The line represents the median. n = 28 cells. (K) FRET efficiency data of GFP-fascin interaction with RFP-actin obtained for the base and the tip of filopodia. n = 29 filopodia, Student’s t test; **, P ≤ 0.01. (L) Fascin–actin interaction at the base versus the tip in growing, shrinking, or stable filopodia measured by live FLIM-FRET. *, P ≤ 0.05. n = 16, 15, or 17 filopodia from two independent experiments. (M) Comparison of live FLIM-FRET measurements of fascin–actin interaction at the base and the tip of actively growing, shrinking, or stable filopodia. Fold change over time (e.g., fold FRET change between the beginning and the end of the growth phase) is shown. n = 16, 14, or 16 filopodia from one of two independent experiments.
Video 1.
2D photoconversion of fascin in filopodia reveals fast redistribution throughout the cell. Filopodia of mEOS2-fascin–expressing HeLa cells were photoconverted from green to red using a 405-nm laser. Frame rate = 4 s.
Figure S1.
Single-molecule analysis of fascin behavior in filopodia. (A) HeLa cell protrusion and retraction (percentage and velocity) was measured with the open source ImageJ plugin ADAPT. Left image sequence shows protrusion and retraction velocity along the cell border. Scale bar, 10 μm. (B) mEOS2-fascin was photoconverted in the cell center (perinuclear region), and diffusion of mEOS2-fascin was monitored over time. Shown is a representative time-lapse series with preconversion on the left, followed by pseudocolored ratios of converted/nonconverted intensities. Scale bar = 5 µm. (C) Relative intensity values (% of maximum for each ROI) of photoconverted mEOS2-fascin fluorescence measured for the photoconversion area, unconverted lamellipodia and filopodia over time. Peak intensity times are highlighted with dashed lines. Data from one representative experiment are shown. Fascin moved at ∼3 µm/s from the center toward the periphery. (D) Simulation of monoexponential (red) and biexponential (orange) decay curve fitting for fascin movement out of filopodia. For biexponential decay curves, fast moving fascin was estimated at ∼20%. Simulated data (color) were overlaid with real experimental data (black). (E) Kymographs of small region FRAP experiments (as in Fig. 1 E) were analyzed for fascin speed (base to tip movement vs. tip-base movement). Student’s t test; **, P ≤ 0.01. (F) Single-molecule track on one fascin molecule along the filopodia shaft (borders in red) with binding and unbinding behavior. (G) Simulation of single-molecule movement. Cropped tracks show simulated single-molecule tracks along hypothetical filopodium (borders in red). Log10 step-size distribution for a bimodal single molecule behavior. Simulation of step size distribution in X (along filopodia shaft) and Y (perpendicular to filopodia shaft) demonstrates localization precision of the method used, with an error peak ∼0 ± 1 pixel (arrow) and real diffusive motion left and right to the peak (arrowheads). (H) Experimental data for one filopodium showing comparative statistics as in G. Number of tracked molecules was limited as visualized in cropped tracks. Log10 stepsize distribution shows bimodal distribution, and step size distribution graph validates model used (with error peak around XY = 0 and free diffusive motion identified around the peak). As tracks were limited for some filopodia, data from separated filopodia were aggregated and fitted to reduce the variation and extract reliable numbers for diffusion and bound/unbound fractions. (I) Unbound fraction (left) and diffusion constant (right) values for WT and fascin-S39A single molecules extracted from individual filopodia (WT n = 15, S39A n = 18) of three independent experiments. Student’s t test; *, P ≤ 0.05; ns, not significant.
To further explore the dynamic behavior of fascin, monoexponential (one-phase) and biexponential (two-phase) decay curves were simulated assuming that fascin movement is represented by (1) one population only (monoexponential) or (2) two different populations with differing speeds (biexponential). Comparison of these hypothetical dynamics with real data derived from photoconverted single filopodia showed that fascin dynamics could be fitted to both models (Fig. S1 D). Monoexponential curve fitting of multiple filopodia revealed a fascin t1/2 of ∼25 s and ∼70% mobile fraction (Fig. 1 C). Biexponential fitting demonstrated that 20–30% of fascin was faster moving, with a mean t1/2 of ∼7 s, and the remaining slower-moving population exhibiting a t1/2 of 40 s (Fig. 1 D). FRAP analysis of a small region of GFP-fascin within a single filopodia further revealed that fascin moved rapidly into filopodia from the base (at ∼200 nm/s), whereas retrograde movement from the tip to cell body was significantly slower (∼100 nm/s; Figs. 1 E and S1 E). These data demonstrate that populations of fascin display distinct molecular speeds depending on the directional movement into or out of filopodia, potentially as a result of variations in the local microenvironment within a filopodium.
To more accurately define fascin dynamics within filopodia at the single-molecule level, mEOS2-fascin–expressing cells were imaged using a customized two-color stochastic optical reconstruction microscopy (STORM)/photoactivated localization microscopy (PALM) microscope (Marsh et al., 2018). This enabled detection of unconverted, bulk fluorescence in the 488-nm channel simultaneous with photoconverted single molecules in the 560-nm channel at a frame rate of 15 ms. Tracking of subsequent datasets revealed that fascin within filopodia exhibited bimodal behavior, with a fast, freely diffusing motion and a near-static (on the time scale of the camera frame exposure time) population. Molecules in the bound state move only a few nanometers in 1 frame, which is lower than the localization precision of single molecules; as such, they appeared as spots in the graph (tracks in Fig. 1 F). At the single-molecule level, frequent transitions between the two types of motion were observed, with fascin spending ∼20% of the time in the fast-moving state (Fig. S1 F). This fast motion was observed throughout the length of the filopodia and without apparent directional bias, with a maximum molecular speed of 40 µm/s and diffusion constant of 7.6 × 10−8 cm2/s (Figs. 1 G and S1, G–H; and Videos 2 and 3). Notably, the low signal-to-noise of these experiments precluded us from determining whether this fast motion was composed of two or more populations with differing diffusion constants. As pS39 is known to inhibit fascin-dependent F-actin bundling (Ono et al., 1997; Yamakita et al., 1996), a S39A phosphodead mutant of fascin was analyzed to determine whether single-molecule dynamics were altered when fascin-actin binding was enhanced. Surprisingly, analysis showed that the diffusion constant of fascin-S39A was significantly higher than WT fascin, but with no change in the percentage of unbound fascin (Fig. S1 I). These datasets combined suggest that fascin undergoes rapid F-actin binding/unbinding events within filopodia, and that nonpS39-fascin is more dynamic in this context, suggesting that actin binding may positively regulate fascin movement.
Video 2.
Rapid forward and backward jumps of fascin single molecule. mEOS2-fascin single molecules imaged on a dual color PALM microscope. mEOS-fascin was photoconverted with a 405-nm laser. Frame rate = 20 ms; filopodia borders are depicted in white. Scale bar = 0.5 µm.
Video 3.
Unbinding–moving–binding motion of fascin single molecule. mEOS2-fascin single molecules imaged on a dual color PALM microscope. mEOS-fascin was photoconverted with a 405-nm laser. Frame rate = 20 ms; filopodia borders are depicted in white. Scale bar = 0.5 µm.
Fascin–actin interactions are highly dynamic and spatially regulated
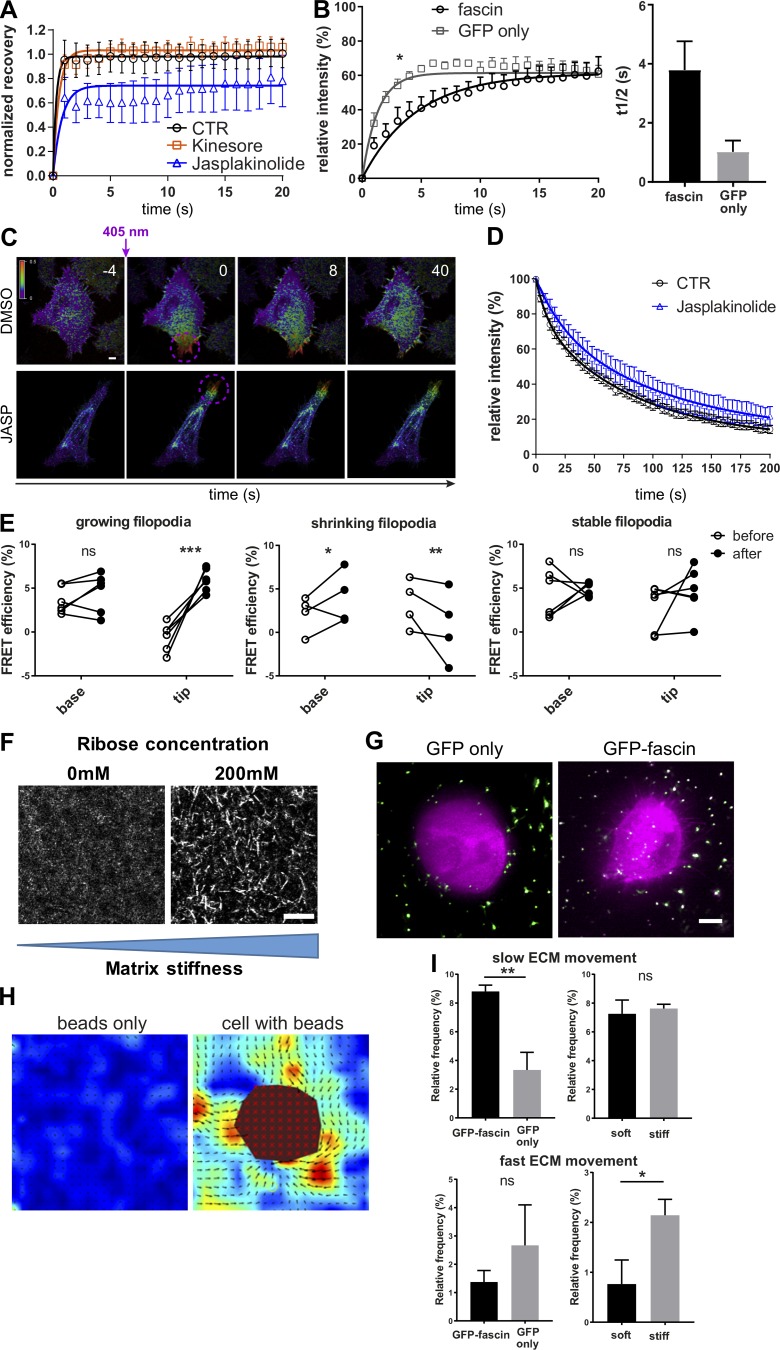
Fascin binds and bundles parallel F-actin fibers but can also interact with microtubules (Villari et al., 2015). To determine whether integrity of actin and/or tubulin cytoskeletal networks contributes to fascin dynamics, FRAP analysis was performed on GFP-fascin–expressing fascin knockdown cells. Treatment of cells with Kinesore, a modulator of Kinesin-1–based microtubule transport (Randall et al., 2017), did not affect fascin recovery into filopodia after photobleaching, whereas reducing F-actin dynamics by treatment with Jasplakinolide led to decreased fascin entry into filopodia compared with control cells (Fig. S2 A). Notably, fascin recovery curves were distinct from that of GFP alone, further suggesting that regulated transport rather than passive diffusion is required for fascin translocation into filopodia (Fig. S2 B). Jasplakinolide had no effect on movement out of filopodia (Fig. S2, C and D), suggesting that fascin movement into and not out of filopodia depends on F-actin turnover but not microtubule-based transport.
Figure S2.
Fascin dynamics and actin binding are dependent on F-actin turnover, and fascin contributes to local force transmission. (A) Filopodia FRAP on confocal microscope for evaluation of the influence of cytoskeleton on fascin movement without (CTR, black) or with Kinesore (orange) or Jasplakinolide (blue); n = 9 cells for each; data were normalized to the mean value of control filopodia to allow comparison between the four independent experiments. (B) Comparison of FRAP recovery curves of GFP-fascin or GFP alone showed early influx retention of fascin, suggesting active transport or regulated entry and not passive diffusion. FRAP curves were compared by two-way ANOVA, and t1/2 values, using Student’s t test; *, P ≤ 0.05. (C) Photoconversion of filopodia of mEOS2-fascin expressing HeLa cells that were left untreated or treated with Jasplakinolide. Representative time-lapse experiment is shown. Scale bar, 2 μm. (D) Mean relative intensity data of photoconversion experiment (as in B) for control and Jasplakinolide-treated cells. n = 7 cells from one of two independent experiments. (E) Fascin–actin interaction was evaluated over time using live FLIM-FRET in growing, shrinking, and stable filopodia. Graphs show FRET efficiency (calculated relative to GFP only lifetime) at the base and the tip of filopodia at the beginning (before) and at the end (after) of the respective process. Groups were compared using two-way ANOVA followed by a Bonferroni posttest, n = 14 for each condition and representative of three independent experiments; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant. (F) Soft and stiff 3D collagen matrix visualized by confocal reflection microscopy. Collagen fibers were bundled and stiffened using 200 mM ribose. Scale bar = 10 µm. (G) HeLa cells expressing GFP only (left) and GFP-fascin in soft 3D collagen matrix with embedded 200-nm beads. Images were captured using LLSM. Scale bar = 5 µm. (H) Analysis of beads only or cells with beads in 3D ECM using PIVlab identifies pulling and pushing forces exerted by the cell on the ECM. Matrix deformation is pseudocolored depending on matrix velocity. Vector arrows indicate the direction of matrix movement, and the size corresponds to the velocity magnitude. The cell body is excluded in the analysis (red mask). Scale bar, 5 μm. (I) Velocity data were separated into slow (1–1.5 µm/s, top panel) and fast (2–2.5 µm/s, bottom panel) ECM movement, and percentages are shown on bar graphs for GFP only and GFP-fascin–expressing cells in soft matrix (left) and GFP-fascin–expressing cells in soft and stiff matrix (right). n = 15 for each condition and representative of three independent experiments; *, P ≤ 0.05; **, P ≤ 0.01.
To further define the dynamic interaction between fascin and actin in more detail, direct binding between GFP-fascin and RFP-actin was quantified in live cells using a custom-built multiconfocal high-speed fluorescence lifetime imaging microscope (FLIM) to measure fluorescence resonance energy transfer (FRET; Fig. 1 H). This technique enables direct analysis of inherent fluorescence lifetimes of the GFP donor, and upon direct interaction (<10 nm) with the acceptor fluorophore, GFP lifetime values decrease because of energy transfer. Analysis of data demonstrated that the fascin–actin interaction was highly dynamic over time, as seen by the fluctuations in lifetime over the time course of imaging (Fig. 1 I). This was particularly pronounced in filopodia that exhibited significantly greater fluctuations in fascin-actin FRET over time compared with those within the lamellipodia and cytoplasm (Fig. 1 J).
More detailed spatial quantification of fascin-actin FRET revealed that fascin–actin interactions were higher at the base of growing filopodia compared with the tip. However, analysis in shrinking and stable filopodia showed similar levels of fascin-actin FRET in both the tip and base of filopodia (Fig. 1, K–M). Moreover, in growing filopodia, fascin-actin FRET was maintained at a similar level at the base but increased over time within the tip as the filopodia stabilized, and this behavior was not seen in shrinking or stable filopodia (Figs. 1 M and S2 E). These data demonstrate that fascin–actin interactions are highly dynamic within filopodia, and that the degree and positioning of interaction correspond to filopodia growth.
Fascin dynamics are sensitive to the extracellular mechanical environment
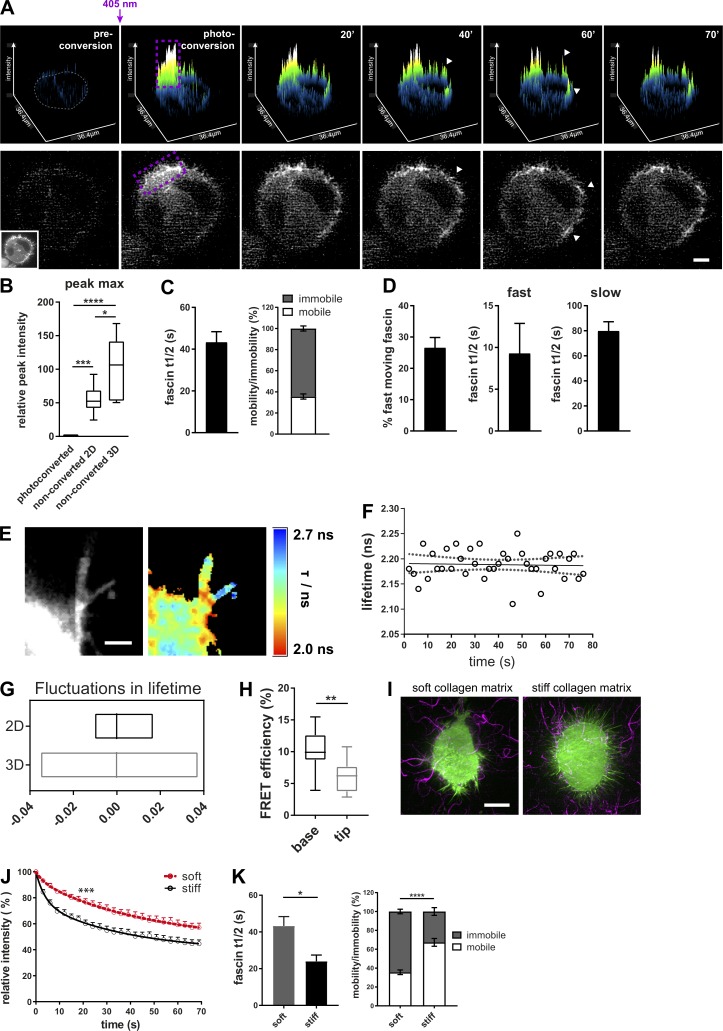
Filopodia are thought to play a crucial role in enabling cells to explore the extracellular environment (Jacquemet et al., 2015). The role of filopodia in physiologically relevant 3D environments is far less well studied than on 2D surfaces, but in the context of fascin-dependent invasion, determining dynamics in cells in 3D represents an important challenge to address and provide insight into fascin function in metastatic disease. To determine whether fascin dynamics differ on 2D versus within 3D environments, mEOS2-fascin–expressing HeLa cells were seeded in native 3D collagen matrices, and photoconversion analyses were performed using lattice light sheet microscopy (LLSM; Chen et al., 2014). Cells within 3D matrices adopted a rounded morphology and formed numerous filopodia (Video 4). After photoconversion, fascin was detected in nonconverted filopodia within 30–40 s (Fig. 2 A), similar to that seen in fully spread cells on 2D surfaces (Fig. 1 A). However, when we normalized fascin velocity to the cell area, we found that the time for fascin to reach peak intensities in unconverted areas in cells in 3D was 100 s, compared with 50 s for cells on 2D surfaces, suggesting that fascin speed is reduced in cells in 3D environments (Fig. 2 B). Indeed, the half-life for fascin movement out of the photoconversion area of cells in 3D was almost double that in 2D. Moreover, the proportion of mobile fascin in cells within 3D matrices was ∼30% (Fig. 2 C), compared with ∼70% seen in cells on 2D (Fig. 1 C). Biexponential fitting of the data further revealed that the percentage and t1/2 values of fast-moving fascin were the same between 2D and 3D, but that t1/2 values of slow-moving fascin doubled (Fig. 2 D) compared with cells in 2D. This suggests that the slower-moving, actin-bound fascin population is sensitive to the dimensionality of the cell microenvironment.
Video 4.
GFP-fascin–expressing HeLa cells assemble numerous filopodia in 3D collagen matrix. 3D volume reconstruction of GFP-fascin–expressing fascin knockdown HeLa cell (green) embedded in 3D collagen matrix and imaged with LLSM. Collagen fibers were prestained with Cy5 dye (magenta). Images were acquired at 1-µm intervals in the Z plane. 3D rendering was performed with IMARIS.
Figure 2.
Fascin dynamics are sensitive to the extracellular mechanical environment. (A) Photoconversion of mEOS2-fascin expressed in cells in 3D collagen matrix imaged at an LLSM. Top panel: Surface plot visualization as shown in Fig. 1. Cell border in preconverted frame is highlighted in gray, and photoconversion region, in purple. Arrowheads depict photoconverted fascin in filopodia outside of conversion area. Bottom panel shows the respective cell and the intensity of the photoconverted channel. Inset picture shows the unconverted cell (green channel). Scale bar = 10 µm. (B) Relative peak intensity values (intensity maxima as highlighted in Fig. 1 B, arrows) of photoconverted and unconverted ROIs of cells in 2D and 3D (photoconverted values were set to 1). 2D and 3D data were normalized to cell size. n = 9 cells for 2D and n = 5 cells for 3D. One-way ANOVA with Bonferroni posttest; *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001. (C) Extracted curve-fitting values (fascin t1/2 values and fascin mobility) for monoexponential curves of photoconverted areas. n = 30 cells (ROIs with multiple filopodia per cell). (D) Percentage of fast moving fascin and t1/2 values for fast- and slow-moving fascin were extracted from biexponential fitted curves of photoconverted areas. n = 30 cells (ROIs with multiple filopodia). (E) Interaction of GFP-fascin and RFP-actin of live HeLa cells in 3D collagen matrix measured with multiconfocal FLIM-FRET microscope. Left, intensity image; right, FLIM-FRET data. High interactions are red (low lifetime), whereas low interactions are blue (high lifetime). Scale bar = 2 µm. (F) Fascin–actin interaction measured over time in single filopodium of living cell in 3D collagen matrix at a 3-s frame rate. Plotted are lifetime values over time (circles) with linear regression (solid line) and 95% confidence interval (dotted line). (G) Fluctuation range of fascin–actin interaction measurements by live FLIM-FRET in 2D and 3D. Lowess residuals were calculated as positive or negative residual value of lifetime data at a certain time point in respect to the fitted Lowess curve. Shown are minimum to maximum floating bar graphs for filopodia data of cells in 2D and 3D, and the line represents the median. n = 23 filopodia of two independent experiments. (H) FLIM-FRET measurement of GFP-fascin and RFP-actin in 3D filopodia. Data for base and tip were extracted from single filopodia. n = 14 filopodia (one to two filopodia per cell) of three independent experiments, one-way ANOVA; **, P ≤ 0.01. (I) GFP-fascin–expressing HeLa cells (green) in soft and stiff 3D collagen matrix. Collagen was stained with Cy5 for visualization (magenta). Scale bar = 10 µm. (J) Fascin movement out of filopodia in soft and stiff ECM. Intensity of photoconverted mEOS-fascin in filopodia of HeLa cells embedded in soft (red) and stiff (black) 3D collagen matrix over time. n = 8 cells for soft and n = 13 cells for stiff matrix from one of two independent experiments, two-way ANOVA; ***, P ≤ 0.001. (K) Fascin t1/2 and mobility data of cells in soft and stiff 3D matrix were extracted from monoexponential fitting curves of photoconverted fascin in filopodia (as in J). n = 30 cells for soft and n = 13 cells for stiff; Student’s t test and two-way ANOVA; *, P ≤ 0.05; ****, P ≤ 0.0001.
To determine whether the altered dynamic behavior of fascin in 3D corresponded to changes in F-actin binding, FLIM analysis was performed in cells coexpressing GFP-fascin and RFP-actin in 3D (Fig. 2 E). Lifetime values of fascin–actin interaction along filopodia as a function of time showed fluctuations over time (Fig. 2 F) similar to those seen in cells in 2D, but direct comparison revealed increased fluctuations in fascin–actin binding in cells in 3D compared with 2D (Fig. 2 G). Moreover, detailed analysis of spatial FRET signals further showed that filopodia in 3D exhibited the same topographical distribution of fascin–actin interaction, with higher FRET at the base and lower FRET at the tip (Fig. 2 H). These data demonstrate that the percentage and the respective speed of the fast-moving fascin population are intrinsic, constant, and independent of the environment. Moreover, the localizations of fascin–actin interactions are similar in cells within 2D and 3D environments. However, the dynamics of fascin–actin binding are dependent on the extracellular environment.
2D surfaces are known to be orders of magnitude stiffer (approximately >1 GPa) than those found within normal connective tissue (∼0.2–5 kPa; Butcher et al., 2009). To determine whether fascin dynamics were responsive to stiffness of the microenvironment, photoconverted fascin was tracked in cells within native 3D matrices or those that were cross-linked with ribose to increase matrix stiffness from ∼200 to ∼800 Pa without altering the number of ligand-binding sites for cell attachment (Mason et al., 2013; Fig. S2 F). Collagen fibers were also labeled with a far-red fluorescent dye (as shown in Video 4) to enable cell–matrix interactions to be visualized (Fig. 2 I and Videos 5 and 6). Analysis of photoconverted fascin demonstrated that the speed of fascin movement increased when cells were embedded in a stiffer matrix compared with the non–cross-linked counterparts (Fig. 2 J). The average t1/2 of fascin movement was significantly lower in stiffer gels, which corresponded to a significant increase in the percentage of fast-moving fascin (Fig. 2 K) up to levels seen for cells in 2D (compare with Fig. 1 C). As force sensing between cells and their environment is bidirectional (Butcher et al., 2009; Ringer et al., 2017), we next aimed to determine whether fascin was required for cells to exert force on the surrounding matrix. Fascin-depleted cells expressing GFP or GFP-fascin were seeded in soft and stiff matrix containing 200-nm microbeads, and bead movement was tracked and quantified over time using particle imaging velocimetry (PIV; Fig. S2, G and H). Tracking revealed two populations of bead movement: slow (1–1.5 nm/s) and fast (2–2.5 nm/s) that correspond to different levels of cell-dependent traction forces applied to the matrix. GFP-expressing, fascin-depleted cells did not assemble filopodia and were less able to exert slow 3D ECM movement in close vicinity to the cell compared with GFP-fascin–expressing cells (Fig. S2 I, top left panel, soft matrix), whereas fast bead movement was not fascin dependent (Fig. S2 I, bottom left panel, soft matrix). Slow bead movement was unaffected when GFP-fascin cells were grown in soft or stiff 3D ECM (Fig. S2 I, top right panel), but the frequency of fast bead movement was significantly increased in cells within stiff matrices compared with non–cross-linked ECM (Fig. S2 I, bottom right panel). Combined, these data suggest that stiffening of 3D collagen matrices leads to increased, faster fascin movement out of filopodia, and this is coupled with a higher frequency of matrix deformation.
Video 5.
HeLa cell in soft 3D collagen matrix. GFP-fascin–expressing fascin knockdown HeLa cell (green) embedded in 3D collagen matrix and imaged at a frame rate of 7 s with LLSM. Collagen fibers were prestained with Cy5 dye (magenta).
Video 6.
HeLa cell in stiff 3D collagen matrix. GFP-fascin–expressing fascin knockdown HeLa cell (green) embedded into ribose-stiffened collagen matrix and imaged at a frame rate of 7 s with LLSM. Collagen fibers were prestained with Cy5 dye (magenta).
Local F-actin organization regulates fascin movement into and out of filopodia
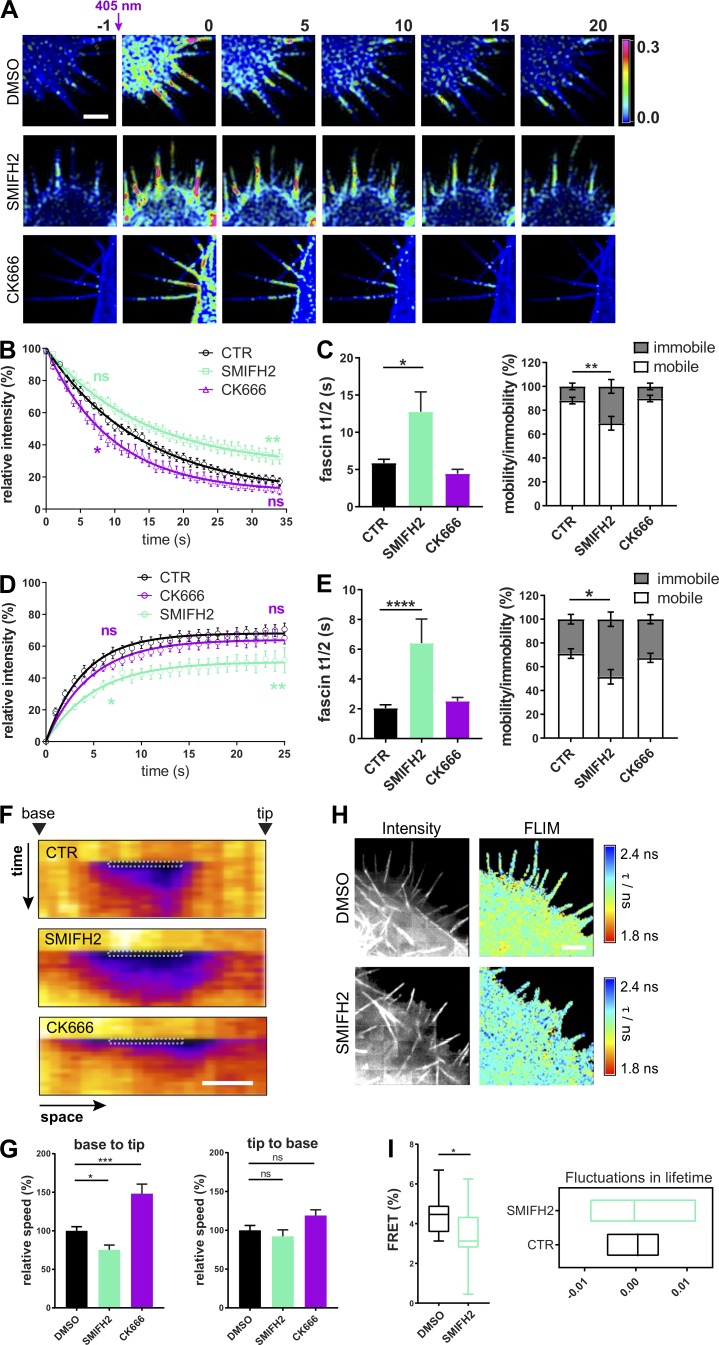
Fascin is known to bundle F-actin within filopodia and associate with actin both at the nuclear envelope and within adhesions (Elkhatib et al., 2014; Jayo et al., 2016; Villari et al., 2015). However, the potential impact of the local organization of F-actin structures on fascin mobility remains unknown. To determine whether the architecture of F-actin contributes to fascin behavior, cells were treated with inhibitors of formins (SMIFH2) or the Arp2/3 complex (CK666; Nolen et al., 2009; Rizvi et al., 2009) that broadly contribute to linear and branched F-actin assembly, respectively. Live cells were first assessed for filopodia formation after inhibitor treatment. Filopodia of untreated cells were ∼5 µm long and had an average lifetime of 12–13 s (Fig. S3 A). For comparison of inhibitor experiments, data were normalized and are displayed as relative values. Quantification demonstrated that treatment with SMIFH2 for 4 h resulted in a significant decrease of filopodia length and number, whereas both parameters were increased following Arp2/3 inhibition by CK666 (Fig. S3 B, left panels, 4-h treatment). Shorter treatment times had negligible effects on filopodia (Fig. S3 B, right panels, 1-h treatment). This time period therefore provided a window to enable analysis of the dependence of fascin dynamics on formins and Arp2/3 while maintaining filopodia assembly. mEOS2-fascin–expressing HeLa cells were preincubated for 1 h with DMSO, SMIFH2, or CK666 before photoconversion of single filopodia (Fig. 3 A). Analysis of subsequent recovery curves revealed that SMIFH2 significantly reduced fascin movement out of filopodia, and this was coincident with increased fascin t1/2 and decreased mobility by ∼20%, whereas CK666 had only a marginal effect (Fig. 3, B and C). Both inhibitors had no effect on the percentage of faster-moving fascin (Fig. S3 C), further indicating that this population represents an intrinsic property of the dynamic behavior of this molecule.
Figure S3.
Disturbing the organization of F-actin alters filopodia and fascin movement. (A) Filopodia length in micrometers (left) and filopodia lifetime in seconds (right) of fascin knockdown HeLa cells expressing GFP-fascin. n = 22 cells, three independent experiments. (B) Filopodia length (normalized to DMSO control) and coverage (relative filopodia number/perimeter) was evaluated after long-time (left, n = 22, 28, or 25 cells) or short-time (right, n = 51, 28, or 22 cells) treatment with SMIFH2 and CK666. One-way ANOVA; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant. (C) Percentage of fast-moving fascin was extracted from filopodia photoconversion data from HeLa cells treated with DMSO, SMIFH2, or CK666. Shown are data from three independent experiments (n = 16, 18, or 11 filopodia respectively), one-way ANOVA. (D) Morphology of GFP-fascin–expressing HeLa cells embedded in soft 3D ECM after inhibition of formins (SMIFH2) and Arp2/3 complex (CK666). Images were captured using LLSM. Scale bar = 10 µm. (E) Inhibition of formins decreases pulling forces on ECM. Velocity magnitude of cell-induced bead displacement in 3D collagen matrix without (n = 220 bead velocity vectors) and with inhibitor treatment. Analysis was performed as described in Fig. S2, G and H (SMIFH2, n = 140; CK666, n = 120; Jasplakinolide, n = 137; beads only, n = 60). **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. (F) Evaluation of fascin-S39A speed in comparison to WT fascin by FRAP. Filopodia were bleached in RFP-fascin WT (black) and RFP-fascin-S39A (green) expressing fascin knockdown HeLa cells, and FRAP intensities were determined over time. Representative of three independent experiments. n = 17 filopodia, two-way ANOVA with Bonferroni posttest; **, P ≤ 0.01. (G) Photoconversion of mEOS2-fascin in HeLa cells on 2D was performed to compare fascin recovery in lamellipodia and filopodia. n = 3 ROI measurements per cell area from 18 cells from three experiments. (H) Retention of fascin in lamellipodia is dependent on F-actin density. Small lamellipodial regions of DMSO control (black) and CK666-treated (magenta) GFP-fascin–expressing HeLa cells were photobleached, and FRAP was analyzed over time; n = 10 from one of three experiments. (I–K) Characterization of the CS-fascin. Filopodia length (I) and number (filopodia/perimeter; J) were measured for GFP-fascin–expressing cells and CS-fascin expressing cells. CS-fascin (tagged with both GFP and mScarlet) or GFP-fascin and mScarlet-fascin (K) were coexpressed in fascin knockdown HeLa cells; cells were fixed, and lifetime was measured by FLIM-FRET. Shown are data from two independent experiments (n = 15, 16, or 12 cells). One-way ANOVA with Bonferroni posttest; ***, P ≤ 0.001; ****, P ≤ 0.0001. (L) CS-fascin–expressing cells were treated with the Arp2/3 inhibitor CK666 and imaged on a confocal microscope to measure ratiometric FRET. Bar graph shows FRET/donor (in %) for control (n = 40 cells) and CK666 treatment (n = 15 cells). One-way ANOVA with uncorrected Fisher posttest. (M) FRET signal of HeLa cells expressing CS-fascin from time binned datasets. Binning of time-lapse images shows increased FRET signal of CS-fascin both in filopodia and at the base of filopodia after combining 50 time-lapse frames. High FRET is shown in yellow/green, and low FRET is shown in dark blue/black. Imaging was performed by SIM; representative example is shown; scale bar = 5 µm. (N) Representative image of the GFP-fascin donor-only control for 3D FRET experiment shows the lack of filopodia and no FRET signal. Scale bar = 2 µm.
Figure 3.
Local F-actin organization regulates fascin movement into and out of filopodia. (A) Filopodia of mEOS2-fascin–expressing HeLa cells were photoconverted, and exit of photoconverted fascin was monitored over time using a confocal microscope at a 1-s frame rate. Upper panel, control cells (DMSO treated cells); middle panel, formin inhibition (SMIFH2-treated cells); bottom panel, Arp 2/3 complex inhibition (CK666-treated cells). Shown are pseudocolored ratios of photoconverted/unconverted intensities before (first column, −1 s) and after (shown are every 5 s) conversion. Scale bar = 5 µm. (B) Fascin movement out of filopodia. Photoconversion intensities plotted versus time for control and SMIFH2- and CK666-treated cells. Early and late phases were compared using two-way ANOVA with uncorrected Fisher’s posttest; shown are data (n = 15, 24, or 17 cells, one ROI per cell, and two to four filopodia per ROI) from three independent experiments; *, P ≤ 0.05; **, P ≤ 0.01; ns, not significant. (C) Fascin t1/2 values and mobility versus immobility (in %) extracted from monoexponential single-curve photoconversion fitting (n = 15, 24, or 17 cells, one ROI per cell, and two to four filopodia per ROI). One-way ANOVA; *, P ≤ 0.05; **, P ≤ 0.01. (D) Fascin movement into filopodia. Filopodia of GFP-fascin–expressing HeLa were bleached (FRAP), and recovery intensities were plotted over time for control and SMIFH2- and CK666-treated cells. Early and late phases were compared using two-way ANOVA with uncorrected Fisher’s posttest; shown are data (n = 18, 21, or 17 cells) from three independent experiments; *, P ≤ 0.05; **, P ≤ 0.01. (E) Fascin t1/2 values and mobile versus immobile fractions extracted from monoexponential single-curve FRAP fitting (n = 18, 21, or 17 cells). One-way ANOVA; *, P ≤ 0.05; ****, P ≤ 0.0001. (F) Small regions within single filopodia were bleached in control and SMIFH2- and CK666-treated cells and displayed as pseudocolored kymographs (space vs. time). Arrowheads point to the base or the tip of the filopodium. Scale bar, 1 μm. (G) Analysis of kymographs shown in F. Relative base to tip (left) and tip to base (right) speeds are shown. Data are from three independent experiments (n = 18, 21, or 17 single filopodia from ≥15 individual cells per condition), one-way ANOVA; *, P ≤ 0.05; ***, P ≤ 0.001. (H) Interaction of GFP-fascin and RFP-actin measured in live cells by multiconfocal FLIM-FRET imaging. Examples of control cells (top panel) and SMIFH2 treated cells (bottom panel) are shown. Intensity images on the left and pseudocolored FLIM image on the right. Warm colors indicate FRET (low lifetime) and cool colors indicate no FRET (high lifetime). Scale bar = 2 µm. (I) Mean FRET efficiency (left, n = 16) and lifetime Lowess residuals (right, n = 28) of fascin–actin interaction of single filopodia measured by live FLIM in cells with and without SMIFH2 treatment. Student’s t test; *, P ≤ 0.05.
FRAP analysis of GFP-fascin was then performed under identical conditions to determine rates of fascin recruitment into filopodia. Data revealed that the speed of fascin influx into filopodia was also significantly impaired by SMIFH2 treatment, whereas CK666 had no effect (Fig. 3, D and E). Analysis of GFP-fascin recovery into small regions within filopodia further confirmed that SMIFH2 reduced recovery speeds of fascin into filopodia; however, interestingly, CK666 treatment increased this rate (Fig. 3, F and G, left panel). Neither drug had an effect on fascin movement from the filopodia tip to the cell body (Fig. 3 G, right panel). To determine whether SMIFH2-dependent effects on fascin dynamics also altered fascin–actin interactions, live FLIM analysis was performed on cells coexpressing GFP-fascin and RFP-actin in the presence of the drug (Fig. 3 H). Quantification of resulting FRET efficiencies demonstrated that SMIFH2 treatment resulted in significantly reduced fascin-actin binding within filopodia (Fig. 3 I, left panel), with increased fluctuations in lifetime (Fig. 3 I, right panel), suggesting that destabilization of the interaction through inhibition of formins results in lower fascin mobility and speed. These data together demonstrate that a balance of branched and linear F-actin is required for efficient fascin movement into and out of filopodia, as well as for stability of fascin-actin binding.
To determine whether formin inhibition also altered fascin-dependent interactions between cells and soft 3D matrices, beads within collagen gels containing GFP-fascin–expressing inhibitor-treated cells were imaged using LLSM. Analysis of resulting tracks revealed that SMIFH2 treatment significantly reduced bead displacement by cells pulling on the matrix, whereas CK666 had no effect on this process (Fig. S3, D and E). Treatment of cells with Jaskplakinolide to stabilize F-actin filaments and reduce treadmilling also significantly reduced bead displacement in this context (Fig. S3 E). These experiments demonstrate that formins are required for fascin-dependent cell–ECM interactions and the ability to exert forces on 3D microenvironments.
Fascin dynamics are reduced by S39 phosphorylation
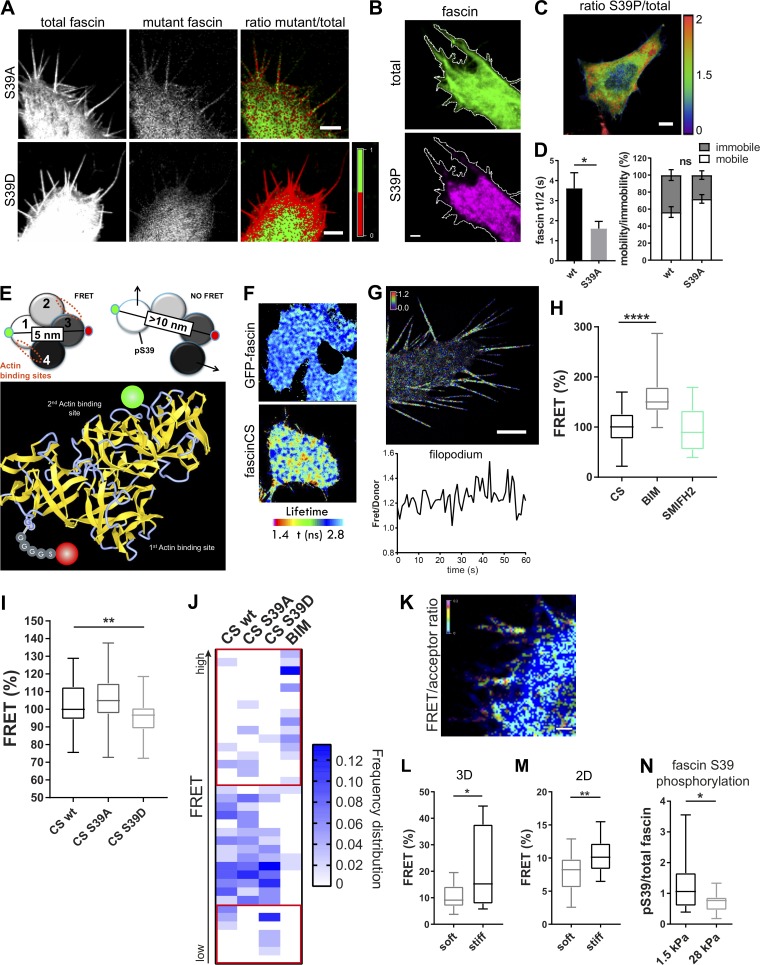
pS39 prevents F-actin bundling by fascin, resulting in cells with very few, highly unstable filopodia (Aratyn et al., 2007); however, the dynamics and spatial localization of fascin phosphorylation cycles remain unknown. To first confirm that fascin phosphorylated at S39 was unable to localize to filopodia when formed, WT GFP-fascin was coexpressed with RFP-fascin-S39A or RFP-fascin-S39D, and localization of coexpressed variants was analyzed (Fig. 4 A). Images demonstrated that both WT and fascin-S39A were strongly recruited to filopodia, whereas fascin-S39D was excluded from filopodia containing WT fascin (Fig. 4 A). Analysis of cells stained for endogenous pS39-fascin demonstrated accumulation at the cell periphery close to the base of filopodia (Fig. 4, B and C), suggesting that this form of fascin may be associated with regions of high F-actin density within lamellipodia. FRAP curves from WT and fascin-S39A–containing filopodia showed that the nonphosphorylated mutant of fascin moved into filopodia with significantly increased speed compared with WT (Figs. 4 D and S3 F), further confirming data in Fig. S1 showing faster single-molecule speed of this mutant. These data suggest a strong correlation between fascin phosphorylation status, molecular speed, and movement into filopodia. To further determine whether organization of F-actin at the plasma membrane correlates with fascin dynamics, fascin movement out of lamellipodia was compared with movement out of filopodia. Data demonstrated that dynamics of photoconverted fascin was significantly slower in lamellipodia regions compared with filopodia (Fig. S3 G), and this speed of movement could be enhanced by treatment of cells with CK666, which depletes branched F-actin at the cell periphery (Fig. S3 H). This supports the notion that molecular retention of fascin within cross-linked F-actin networks could be a regulatory factor in controlling fascin movement into and out of filopodia.
Figure 4.
A biosensor reveals that fascin phosphorylation is mechanosensitive. (A) Fascin knockdown HeLa cells were cotransfected with WT GFP-fascin and either RFP-fascin-S39A (top panel) or RFP-fascin-S39D (bottom panel), and fascin distribution was analyzed by confocal microscopy. Total WT fascin fluorescence is shown on the left, the mutant in the middle, and mutant/total ratio on the right. In the ratio view, a mutant to total fascin ratio of 1:1 (equal distribution) is colored in green; <50% of mutant fascin is colored in red (lack of mutant is red). Scale bar = 5 µm. (B and C) GFP-fascin–expressing cells stained for endogenous pS39-fascin and imaged by confocal microscopy. Representative image (B) of single channels of total fascin (green) and pS39-fascin (magenta) are shown with the respective cell border in white. Scale bar = 5 µm; ratio of pS39/total fascin in the whole cell (C), showing pS39-fascin retention at cell periphery (high pS39-fascin is red, low pS39 is blue), scale bar = 10 µm. (D) Evaluation of fascin-S39A speed and recovery in comparison with WT fascin by FRAP (FRAP curves shown in Fig. S3 F). Statistical analysis of fascin t1/2 and mobility extracted from monoexponential fitting of single curves. n = 7 cells with one ROI per cell and one to two filopodia per ROI; Student’s t test; *, P ≤ 0.05; ns, not significant. (E) Schematic view of CS-fascin in a potential high-FRET and low-FRET conformation (based on crystal structure). Approximate distances of fluorophores (GFP and mScarlet) are indicated (top). Bottom: Ribbon diagram of WT fascin (PDB: 3P53), with actin binding sites and fluorophores on surface exposed loops indicated. (F) FLIM-FRET analysis of CS-fascin in fixed HeLa cells. Top panel shows pseudocolored FLIM data from GFP-fascin only (donor control), and bottom panel shows CS-fascin (donor and acceptor). Scale bar, 5 μm. (G) SIM image of CS-fascin ratiometric view of FRET/donor of one representative live cell. Scale bar = 5 µm. Bottom graph shows fluctuating FRET/donor values for a representative filopodium over time. (H) FRET/donor ratios (FRET %, normalized to mean CS values without treatment) of HeLa cells expressing CS-fascin imaged live on a confocal microscope with and without treatment with PKC inhibitor (BIM) and formin inhibitor (SMIFH2). Shown are data from three independent experiments (n = 40, 53, or 26 cells), one-way ANOVA with Bonferroni posttest; ****, P ≤ 0.0001. (I) FRET/donor ratios of WT, CS-fascin-S39A–, and CS-fascin-S39D–expressing cells (FRET %, normalized to mean CS WT values). Data are from three independent experiments (n = 46, 52, or 52 cells), one-way ANOVA with Bonferroni posttest; **, P ≤ 0.01. (J) Frequency distribution graph of data shown in I showing relative FRET frequency (bin range = 0.8–1.5, bin width = 0.02). Red squares highlight main differences between fascin mutants at the lower and higher FRET value ranges. (K) Representative image of cell expressing CS-fascin and embedded in 3D collagen matrix imaged by LLSM. FRET/acceptor ratio is shown in pseudocolor with high FRET in red/pink and low FRET in blue. Scale bar, 1 μm. (L and M) CS-fascin displays different FRET levels depending on the stiffness of the surrounding substrate. (L) FRET for CS-fascin–expressing cells in soft and stiff 3D matrix (FRET/acceptor). n = 14 cells, Student’s t test; *, P ≤ 0.05. (M) FRET values (FRET/donor) for CS-fascin–expressing HeLa cells on soft and stiff collagen-coated 2D slides (soft: elastically supported surface of 1.5-kPa stiffness, stiff: glass). Data pooled from three independent experiments, with n = 10 cells per experiment. Student’s t test; **, P ≤ 0.01. (N) Fascin pS39 was stained in GFP-fascin–expressing fascin knockdown cells plated on soft and stiff 2D surfaces (1.5 and 28 kPa, respectively). The ratio of pS39 (AF560) and total fascin (GFP) intensities was calculated. Student’s t test; *, P ≤ 0.05, n = 25 and 19 cells for 1.5 and 28 kPa, respectively.
Fascin phosphorylation is mechanosensitive
To directly probe the conformation and phosphorylation status of fascin in live cells, we developed an intramolecular FRET-based fascin conformation sensor (CS-fascin) comprising GFP on the N-terminus and mScarlet positioned within domain 3 (Fig. 4 E). The previously reported structures for fascin suggest that the protein adopts a globular state when associated with F-actin (Chen et al., 2010; Hashimoto et al., 2005). Based on molecular modeling, the N-terminus and domain 3 are within 5-nm distance in this globular state, which would allow for energy transfer from the donor (GFP) to the acceptor (mScarlet). A conformational change would result in an increased distance and a subsequent reduction in FRET. CS-fascin construct expression in fascin-depleted cells showed full rescue of filopodia formation, consistent with that seen in GFP-fascin–expressing cells (Fig. S3, I and J). FLIM-FRET analysis of CS-fascin–expressing cells showed both low and high FRET populations across the cell (Fig. 4 F), with a significant increase in FRET at the periphery in live cells (Fig. 4 G). Coexpression of GFP-fascin and mScarlet-fascin did not result in a FRET signal above the donor-only control (GFP-fascin), eliminating the possibility of intermolecular FRET between two neighboring fascin molecules and confirming that fascin is monomeric, as previously reported (Yang et al., 2013; Fig. S3 K).
Further detailed analysis of CS-fascin activity in live cells demonstrated that FRET fluctuated in filopodia over time (Fig. 4 G, bottom panel, and Videos 7, 8, and 9), suggesting that fascin undergoes fast conformational changes in filopodia. Crystallographic analysis has previously suggested that pS39 may induce conformational changes in fascin (Yang et al., 2013). Indeed, treatment of cells with bisindolylmaleimide I (BIM I), which inhibits class I PKC family members and thus would be expected to block fascin S39 phosphorylation, led to a significant increase in FRET efficiency, suggesting a shift toward a more globular-like fascin conformation upon S39 dephosphorylation (Fig. 4 H). Conversely, SMIFH2 and CK666 did not change CS-fascin FRET (Figs. 4 H and S3 L), indicating that formins and Arp2/3 do not act upstream to control fascin phosphorylation or conformation. To directly determine whether fascin CS FRET correlated with S39 phosphorylation, we generated CS-fascin-S39D (phosphomimic) and S39A (phosphodead) mutants. FRET analysis demonstrated that CS-fascin-S39D showed significantly reduced FRET compared with CS-fascin-WT, whereas CS-fascin-S39A showed no change (Fig. 4 I), in agreement with previous suggested structural changes in response to S39 phosphorylation (Yang et al., 2013). Further analysis of FRET distributions across multiple populations of cells demonstrated that CS-fascin-S39A exhibited increased high-FRET populations and reduced lower-FRET populations, whereas CS-fascin-S39D showed a significant increase in the low-FRET population (Fig. 4 J, highlighted as red squares). Notably BIM treatment led to markedly increased higher-FRET populations compared with those seen in CS-fascin-S39A. BIM may also play additional roles in the control of fascin conformation either through indirect dephosphorylation of other residues (for example, S274 within the second actin binding site, for which the kinase has not yet been identified) or regulation of additional fascin-binding partners. We speculate that the further reduction in CS FRET seen with the BIM treatment compared with the S39A fascin mutant may be due to these additional contributing factors. Moreover, binning of time-lapse series FRET images from 10 or 50 frames demonstrated that low FRET is seen within the cytoplasm, whereas high FRET accumulates predominantly within filopodia, with some bias toward the base of the filopodia (Fig. S3 M). This would agree with the notion that a transition from pS39- to non–pS39-fascin occurs upon entry of fascin into the filopodial shaft. These data combined suggest that fascin changes its conformation in response to phosphorylation of S39, with a more globular high-FRET conformation corresponding to the non-pS39 form and the phosphorylated form adopting a low-FRET conformation, which separates fascin domains 1 and 3.
Video 7.
CS-fascin reveals rapid conformational change of fascin within filopodia. SIM time-lapse video of fascin knockdown HeLa cell expressing fascinCS FRET sensor. Ratiometric view of FRET/donor (high FRET is red and low FRET is blue) of one representative cell. Scale bar = 1 µm.
Video 8.
CS-fascin FRET is highly dynamic over time. CS-fascin was expressed in fascin knockdown HeLa cells, the donor (GFP) was excited with a 488-nm laser, and emission was collected for donor and acceptor (GFP and mScarlet) over time. Shown is acceptor fluorescence without donor signal, which represents only the fascin FRET population. To display spatial protein distribution of FRET, intensities were pseudocolored in cyan, with high intensities shown in yellow/green and low intensities in blue. Scale bar = 5 µm; frame rate = 1 s.
Video 9.
CS-fascin FRET varies spatially in growing filopodia. SIM time-lapse video of fascin knockdown HeLa cell expressing CS-fascin FRET sensor. FRET donor (GFP) was excited with a 488-nm laser, and emission was collected for donor and acceptor (GFP and mScarlet) over time. Ratiometric view of FRET/donor (high FRET is red and low FRET is blue) of one representative cell. Low FRET CS-fascin is at the tip of growing filopodia, whereas high FRET CS-fascin is more abundant at the base.
We next explored fascin phosphorylation dynamics in cells within soft or stiff 3D collagen matrices. Images from these experiments revealed that fascin adopted both high- and low-FRET states, as seen for cells on 2D surfaces. (Fig. 4 K, and respective GFP-fascin only control in Fig. S3 N). Again, high-FRET CS-fascin was enriched in the lower part of filopodia that is closest to the cell body. Interestingly, cells within stiffer 3D matrices showed a significant increase in CS-fascin FRET (Fig. 4 L), indicating that increased mechanical stiffness may lead to lower phosphorylation of S39 fascin and therefore increased potential to bundle F-actin. To determine whether our observations were specific to 3D environments, the same experiments were performed on cells plated on 2D surfaces of differing stiffness (soft, 1.5 kPa; stiff, 1 GPa). Data demonstrated that CS-fascin FRET was also significantly increased in cells plated on stiffer 2D surfaces compared with those on softer ones (Fig. 4 M). Further, we stained pS39-fascin in GFP-fascin–expressing cells plated on soft and stiff collagen-coated 2D surfaces (1.5 and 28 kPa, respectively) and found that high stiffness resulted in reduced S39 phosphorylation of fascin (Fig. 4 N). Taken together, these data demonstrate that fascin conformation is dependent on its phosphorylation status at S39, and that this conformation is sensitive to extracellular stiffness.
FMNL2 regulates fascin movement into and out of filopodia in 2D
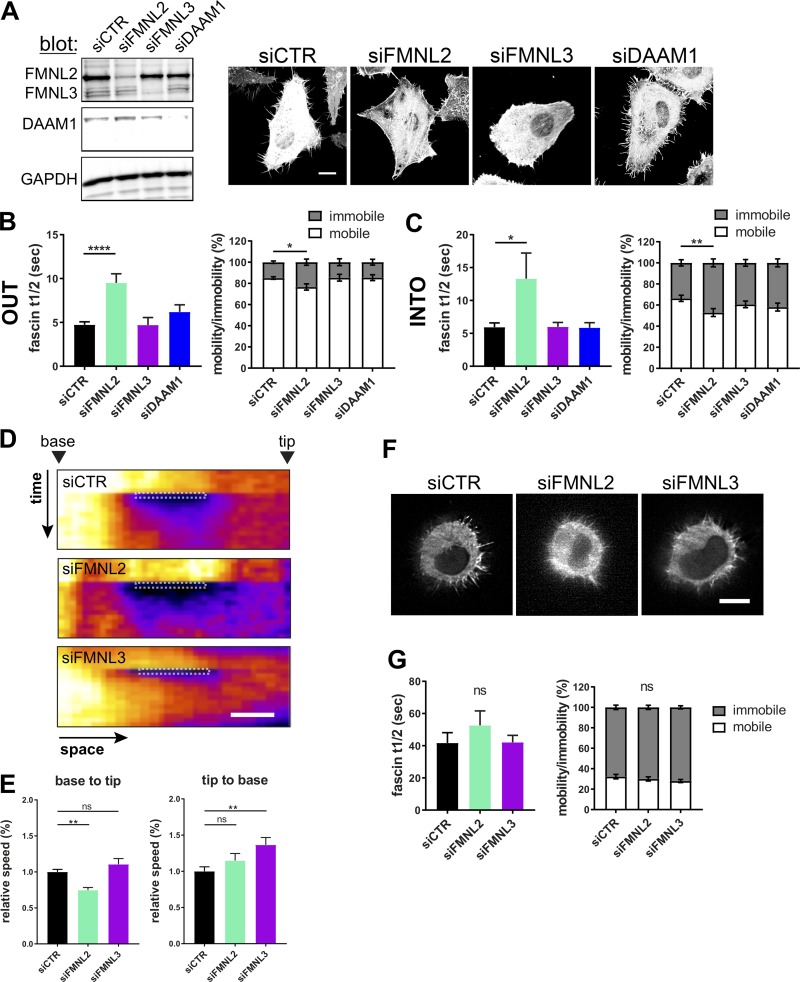
Our data thus far suggested that formins play a key role in controlling fascin dynamics within filopodia. FMNL2, FMNL3, and DAAM1 have all been shown to localize to filopodia (Jacquemet et al., 2019; Jaiswal et al., 2013) and are involved in filopodia assembly (Harris et al., 2010). FMNL2 and FMNL3 have also been linked to poor prognosis in cancer patients through increase of cell migration and metastasis (Gardberg et al., 2016), and DAAM1 has previously been suggested to bind to fascin (Jaiswal et al., 2013). To test whether these formins may contribute to fascin dynamics in filopodia, cells were treated with siRNA pools specifically targeting each of these formins (Fig. 5 A). Knockdown of FMNL2 and FMNL3 led to cells with significantly reduced filopodia length and number, whereas DAAM1 depletion increased filopodia lifetime without changing number and length (Figs. 5 A and S4 A). When we analyzed fascin movement out of filopodia using the mEOS2 photoconversion assay, we found that FMNL2 depletion significantly increased fascin recovery t1/2 values, whereas FMNL3 and DAAM1 depletion had no effect (Fig. 5 B, left graph, and Fig. S4 B). FMNL2 depletion also significantly reduced the mobile fraction of fascin, whereas silencing of FMNL3 and DAAM1 had no effect (Fig. 5 B, right graph). Similarly, FMNL2 depletion also significantly reduced the speed of fascin entry into filopodia and resulted in a significant reduction in the fascin mobile fraction, whereas FMNL3 and DAAM1 silencing had no effect on fascin dynamics (Fig. 5 C). Fascin recruitment from the cell body into small regions within filopodia was also reduced in FMNL2-depleted cells, whereas FMNL3 knockdown increased fascin movement from tip to cell body (Fig. 5, D and E). FMNL2 has been shown to drive actin polymerization in vitro in concert with profilin (Block et al., 2012). Depletion of both isoforms of profilin (1 and 2) had no significant effect on filopodia formation (Fig. S4 C); however, fascin movement out of filopodia was significantly slower in profilin-2 knockdown cells, whereas profilin-1 silencing had no effect on fascin motility (Fig. S4 D). Both profilin-1 and -2 had no effect on filopodia morphology (Fig. S4 E). Interestingly, detailed analysis of filopodia curvature further revealed that silencing FMNL2 led to increased bending of filopodia, whereas FMNL3 knockdown had no effect on this parameter (Fig. S4 F), suggesting that FMNL2 may act to stabilize filopodial structure potentially through promoting fascin-actin binding, and this could play a potential role in filopodia exploring the ECM. To determine whether fascin dynamics in 3D matrices were also FMNL2 regulated, photoconversion experiments were conducted in formin-depleted cells within collagen gels. However, fascin t1/2 and mobility fractions were unchanged in cells cultured in soft 3D gels (Fig. 5, F and G), suggesting an environment-dependent effect of FMNL2 on fascin dynamics. Taken together, these data demonstrate that FMNL2 plays a key role in controlling fascin movement within filopodia.
Figure 5.
Formins regulate fascin movement into and out of filopodia in 2D. (A) Representative Western blots showing specific silencing of FMNL2, FMNL3, or DAAM1 using siRNA (left) and morphology of control and knockdown cells (right). Scale bar = 10 µm. (B) Analysis of mEOS2-fascin movement out of filopodia in formin silenced cells. Fascin t1/2 values and mobile versus immobile fractions from monoexponential single-curve fitting (n = 53, 12, 8, or 11 cells, one ROI per cell and one to two filopodia per ROI). One- and two-way ANOVA; *, P ≤ 0.05; ****, P ≤ 0.0001. (C) FRAP of filopodia to analyze GFP-fascin movement into filopodia in formin-silenced cells. Fascin t1/2 values and mobile versus immobile fractions extracted from monoexponential single-curve FRAP fitting (n = 34, 30, 26, or 24 cells). One- and two-way ANOVA; *, P ≤ 0.05; **, P ≤ 0.01. (D) Small regions within single filopodia were bleached in control and formin knockdown cells and displayed as pseudocolored kymographs (space vs. time). Arrowhead depicts the base or the tip of the filopodium. Scale bar = 1 µm. (E) Analysis of kymographs shown in D. Relative base to tip (left) and tip to base (right) speeds are shown from three independent experiments (n = 25, 23, or 23), one-way ANOVA; **, P ≤ 0.01; ns, not significant. (F) Morphology of control, FMNL2, and FMNL3 knockdown cells in soft 3D collagen matrix imaged using LLSM. Scale bar = 10 µm. (G) mEOS2-fascin photoconversion and monitoring over time in cells in 3D matrices. Data from monoexponential (fascin t1/2 and mobility) analysis of photoconversion experiments. n = 23 cells (siCTR), 22 cells (siFMNL2), and 25 cells (siFMNL3), 5–10 filopodia per cell, one- and two-way ANOVA.
Figure S4.
FMNL2, FMNL3, DAAM1, and profilin differentially contribute to filopodia formation and stability. (A) Filopodia length (normalized to siCTR), coverage (relative filopodia number/perimeter), and lifetime (normalized to siCTR) was evaluated after knockdown of FMNL2, FMNL3, and DAAM1. One-way ANOVA with uncorrected Fisher posttest (n = 62, control cells; 62, siFMNL2 cells; 36, siFMNL3 cells; 39, siDAAM1 cells); *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. (B) Western blot (left) and filopodia FRAP experiments of siCTR cells (black) and siFMNL2 cells silenced with two different single siRNAs (turquoise). FRAP curves were compared by two-way ANOVA, t1/2 values using Student’s t test, and fascin mobility using two-way ANOVA; data were pooled from two independent experiments; n = 26 filopodia for siCTR and 27 and 28 for siFMNL2 1 and 2 (single), respectively; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. (C) Evaluation of profilin-1 and -2 siRNA knockdown efficiency and specificity via Western blot (left). GAPDH and HSP70 served as loading control. Representative images of HeLa cells after knockdown of the indicated profilins are shown in right. Scale bar = 10 µm. (D) Fascin movement out of filopodia after profilin knockdown. mEOS2-fascin was photoconverted and monitored over time. Early and late phases were compared using two-way ANOVA with uncorrected Fisher posttest; shown are data from three independent experiments; *, P ≤ 0.05. Fascin t1/2 values and mobility versus immobility (in %) extracted from monoexponential single-curve fitting (n = 53, 58, or 30 filopodia). One- and two-way ANOVA; ****, P ≤ 0.0001; ns, not significant. (E) Filopodia length, coverage, and lifetime was evaluated after control siRNA or combined knockdown of profilin-1 and -2 (n = 63 and 35 cells, respectively). Student’s t test; data are representative of three or more independent experiments. (F) Filopodia flexibility depends on FMNL2. Filopodia of siCTR, siFMNL2, and siFMNL3 GFP-fascin HeLa cells were analyzed for the potential to bend by dividing the area of the traced filopodium with the convex area as outlined in the schematic illustration (left). Straight filopodia have a ratio close to 1; lower ratios indicate more filopodia bending. One-way ANOVA; *, P ≤ 0.05, n = 84, 75, and 54 single filopodia of ≥20 siCTR, siFMNL2, and siFMNL3 cells, respectively, from four independent experiments.
Active FMNL2 increases fascin movement into filopodia
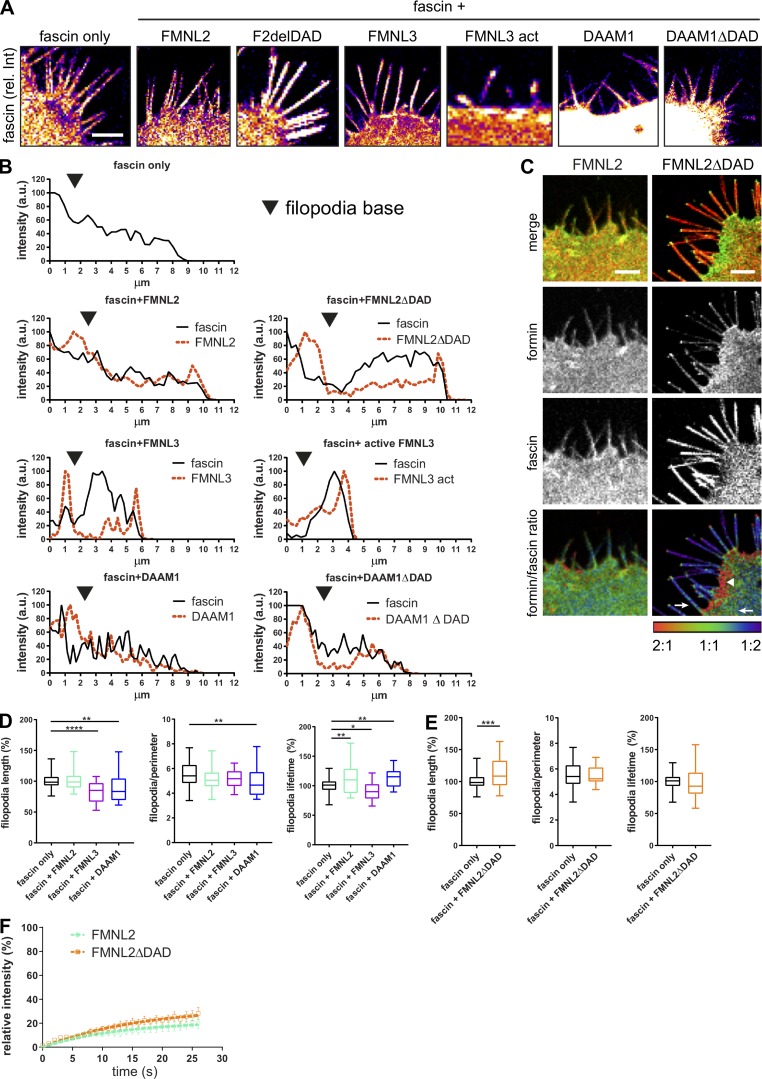
Formins are regulated through GTPase binding, which relieves autoinhibition afforded through the DAD domain and promotes activation and binding to other effector proteins, including actin (Kühn and Geyer, 2014). To further analyze the colocation of different formins with fascin and determine whether formin activation altered fascin localization, GFP-tagged WT or constitutively active FMNL2, FMNL3, or DAAM1 were coexpressed with RFP-fascin in fascin-depleted cells, followed by live-cell confocal imaging. Images demonstrated WT FMNL2 was enriched in the lamellipodia and along the filopodial shaft, whereas active FMNL2 was predominantly at the plasma membrane and the tips of filopodia (Fig. 6 A), confirming previous reports (Gardberg et al., 2010; Kühn et al., 2015; Péladeau et al., 2016). Active FMNL2 also promoted fascin enrichment within filopodia, leading to broader fascin-positive filopodia in these cells (Fig. S5, A and B). WT FMNL3 was also localized at the plasma membrane and in filopodia tips (Fig. 5 A). Overexpression of WT and constitutively active FMNL3 resulted in fascin enrichment in filopodia tips (Fig. S5, A and B). Conversely, DAAM1 was highly abundant within the cytoplasm and also along the filopodia shafts (Fig. 6 A), and overexpression of WT and active DAAM1 did not alter fascin localization within filopodia (Fig. S5, A and B). Line scan analysis of fascin and formin intensities within filopodia revealed high overlap between FMNL2 and DAAM1 with fascin, whereas FMNL3 and fascin occupied distinct regions of the filopodia (Fig. S5 B). Analysis of the ratio of fascin:formin signal in FMNL2-overexpressing cells further demonstrated a high accumulation of both WT and active FMNL2 at the base of filopodia, coincident with fascin enrichment within the filopodial shaft (Fig. S5 C). To determine whether these distinct localizations of each formin led to altered filopodia behavior, live-cell imaging and filopodia tracking were performed. Data revealed that overexpression of either FMNL3 or DAAM1 decreased filopodia length (Fig. S5 D), whereas FMNL2 overexpression significantly increased filopodia stability, and constitutively active FMNL2 increased filopodia length (Fig. S5, D and E). These data combined suggest that FMNL2 positively contributes to fascin enrichment within filopodia, resulting in enhanced filopodia stability.
Figure 6.
Active FMNL2 increases fascin movement into filopodia. (A) Representative confocal images of HeLa cells coexpressing RFP-fascin (red) and FMNL2-GFP (WT and active; ΔDAD), FMNL3-GFP, or DAAM1-GFP (green). Scale bar = 5 µm. (B and C) Analysis of fascin recovery by FRAP in filopodia (one ROI per cell/filopodia) in cells coexpressing specified formins. Fascin t1/2 values (B) and relative mobility (C; normalized to mean fascin only) extracted from monoexponential single FRAP curve fitting. One-way ANOVA; *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001, data from at least three independent experiments are shown, n = 101 (fascin only), 31 (+FMNL2), 27 (+FMNL2ΔDAD Nt), 33 (+FMNL3), 13 (+FMNL3 act), 16 (+DAAM1), and 14 (+DAAM1ΔDAD) cells. (D) FLIM-FRET analysis of HeLa cells expressing RFP-fascin and FMNL2-GFP, FMNL3-GFP, or GFP-DAAM1. Intensity images of GFP fluorescence (top panel) and FLIM data in pseudocolor (bottom panel) are shown. Warm colors show high FRET (low GFP lifetime), and cool colors indicate low FRET (high lifetime) GFP served as donor-only control and was used for calculation of FRET efficiency shown in E. Scale = 5 µm. (E) Calculated FRET efficiency (relative to GFP only) for interaction between fascin and FMNL2, FMNL3, and DAAM1 (n = 26, 35, and 11 cells). One-way ANOVA; ***, P ≤ 0.001. (F) Representative example image of fascin and active FMNL2 (FMNL2ΔDAD) FRET. Scale bar = 2 µm. (G) Interaction of FMNL2 with WT fascin, fascin-S39A (phospho-dead), or fascin-S39D (phospho-mimic) measured by FLIM-FRET in HeLa cells. FRET efficiency was calculated based on FMNL2-GFP only (donor-only control). Shown are data from three independent experiments (n = 26, 29, and 16 cells, respectively). One-way ANOVA; *, P ≤ 0.05; **, P ≤ 0.01. (H) Interaction of FMNL2 and fascin before or after disassembly of F-actin with Cytochalasin D measured by FLIM-FRET. Two-way ANOVA; ***, P ≤ 0.001; ****, P ≤ 0.0001, n = 10 and 15 cells for FMNL2 only (+ and – CytD), 20 and 28 cells for FMNL2 + fascin (+ and – CytD) from three independent experiments. Scale bar, 5 μm.
Figure S5.
WT and active FMNL2, FMNL3, and DAAM1 alter fascin distribution at the cell periphery. (A) Fascin distribution was evaluated with and without overexpression of WT and active FMNL2, FMNL3, and DAAM1 using a confocal microscope. Fluorescence intensity values of fascin are shown in pseudocolor to highlight high (white) and low (purple) intensity. Scale bar = 5 µm. (B) Line scans along filopodia in HeLa cells expressing RFP-fascin only (upper panel) or RFP-fascin with coexpression of WT and active FMNL2 (second panel), FMNL3 (third panel), and DAAM1 (fourth panel). Intensity plots versus micrometers show the intensity in arbitrary units for fascin (black, solid line) and the respective formin (red, dashed line) for comparison. (C) Colocalization analysis of FMNL2 (WT and active form, second panel) and fascin (third panel) in HeLa cells upon overexpression of formins. Top panel, merged image; bottom panel, formin/fascin ratio; areas of higher abundance of formin and lower abundance of fascin illustrated in red (2:1 ratio), colocalization in green (1:1 ratio), and high abundance of fascin with low formin in blue (1:2 ratio). Images show high colocalization of FMNL2 and fascin throughout the cell and filopodia and more specific colocalization of active FMNL2ΔDAD with fascin at the filopodia base and a few microns into the cytoplasm (arrow), whereas fascin appears excluded from the lamellipodium close to the plasma membrane (arrowhead). Scale bar = 5 µm. (D and E) Filopodia length (normalized to fascin only), coverage (relative filopodia no/perimeter), and lifetime (normalized to fascin only) was evaluated after overexpression of FMNL2, FMNL3, and DAAM1 (D) and after overexpression of active FMNL2 (FMNLΔDAD; E). One-way ANOVA with uncorrected Fisher posttest or Student’s t test, data from three or more experiments are shown; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. (F) Filopodia FRAP of GFP-FMNL2– and active GFP-FMNL2ΔDAD–expressing cells with recovery intensities plotted over time. n = 9 cells from one of three independent experiments.
To further determine whether formin overexpression altered fascin dynamics, FRAP analysis was performed on cells coexpressing GFP-formins and RFP-fascin. Resulting data demonstrated that overexpression of WT or constitutively active FMNL2 decreased the speed of fascin recruitment into filopodia, although active FMNL2 increased the mobile fraction of fascin (Fig. 6, B and C). Overexpression of both WT and active FMNL3 significantly decreased both fascin recruitment speeds and mobile fractions, whereas DAAM1 overexpression had no effect on either parameter (Fig. 6, B and C). Recovery of all formins was also monitored simultaneously with fascin during the FRAP analysis, and interestingly, the recovery rates of all formins were not coupled with fascin influx into filopodia, further suggesting that these proteins do not enter filopodia together (Fig. S5 F). Taken together, these data show that active FMNL2 promotes fascin entry into filopodia, resulting in more stable filopodia, whereas FMNL3 overexpression leads to reduced fascin entry into filopodia, resulting in reduced filopodia length. Conversely, DAAM1 overexpression resulted in shorter and fewer filopodia over time, but with no effect on fascin mobility and speed.
FMNL2 directly interacts with fascin
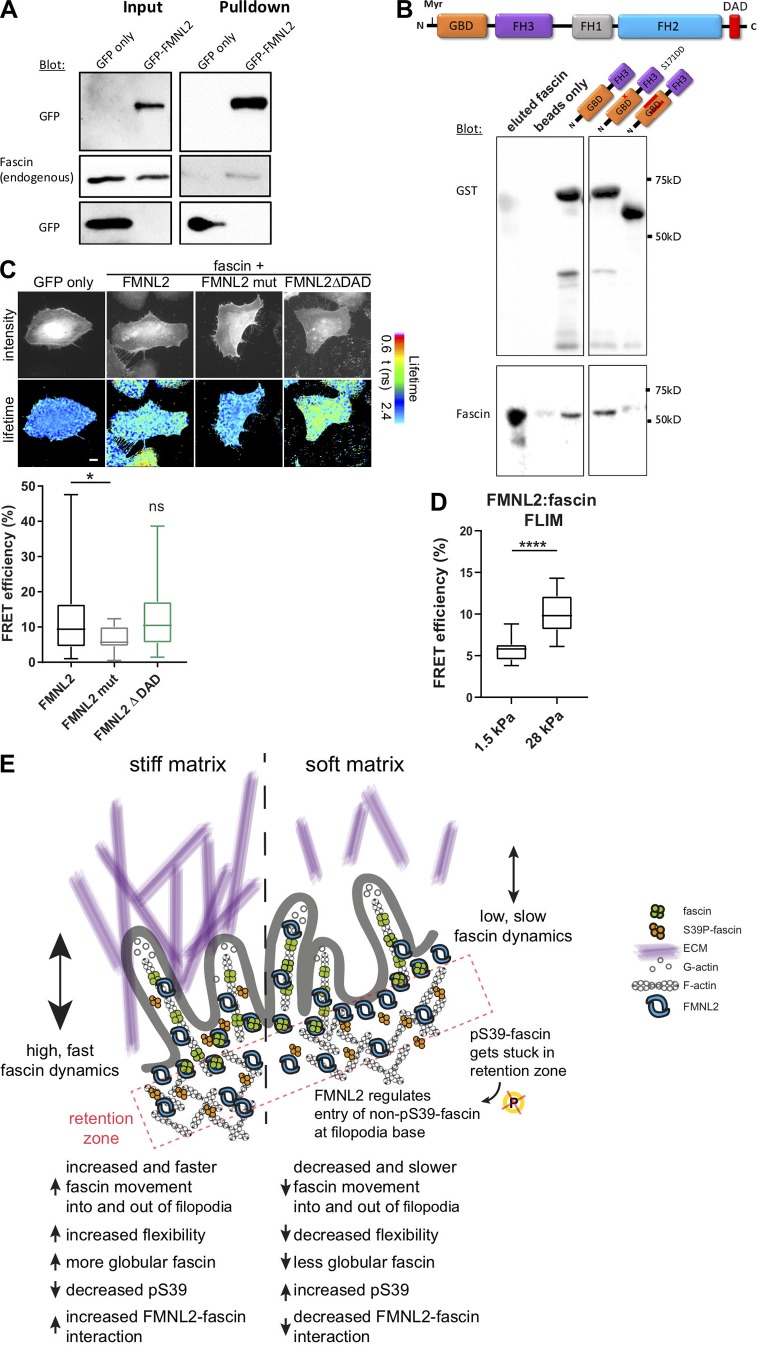
DAAM1 has previously been shown to interact with fascin (Jaiswal et al., 2013), but this study proposed a role for fascin in DAAM1 recruitment to filopodia rather than vice versa. To determine whether fascin could also interact with FMNL2 to explain the reliance of fascin dynamics on this formin, GFP-formins were coexpressed with mScarlet-fascin, and FLIM-FRET analysis was performed (Fig. 6 D). Data demonstrated a direct interaction between fascin and both FMNL2 and DAAM1, but not FMNL3 (Fig. 6, D and E). More detailed analysis of filopodia in active FMNL2-overexpressing cells confirmed high levels of FRET, particularly at the base of filopodia (Fig. 6 F). To determine whether fascin–FMNL2 interactions were regulated by fascin pS39, FRET between FMNL2-GFP and RFP-fascin-S39A or -S39D was quantified. Resulting data demonstrated FMNL2–fascin interactions were significantly increased in fascin-S39A–expressing cells compared with WT, whereas mutation to S39D significantly impaired fascin-FMNL2 binding (Fig. 6 G). To determine whether F-actin may act as a molecular bridge to mediate the association between fascin and FMNL2, FRET analysis between FMNL2 and WT fascin was repeated in cells treated with Cytochalasin D, which disrupts F-actin filaments. Data demonstrated that disruption of F-actin did not significantly affect the established fascin–FMNL2 complexes (Fig. 6 H), suggesting this is indeed a direct interaction, and that maintenance of the complex does not rely on actin as a scaffold.
To further delineate potential domains within FMNL2 that mediate fascin binding, we performed biochemical experiments. Immunoprecipitation experiments further confirmed that FMNL2-GFP forms a complex with endogenous fascin biochemically (Fig. 7 A). We then tested direct protein–protein interactions between in vitro purified His-tagged WT fascin and GST-tagged N-terminal truncated versions of FMNL2 to investigate whether this region of FMNL2 was sufficient to support fascin binding. The truncated form of FMNL2 consists of the myristoylation site, the GBD-binding domain, and parts of the FH3 domain (Fig. 7 B). We tested 2 mutants of this construct, one introducing the S171DD point mutation that has previously been shown to stabilize FMNL2-Cdc42 binding, and one lacking part of the GBD that interferes with GTPase binding (Kühn et al., 2015). Resulting data demonstrated that fascin bound to the longest FMNL2 protein, but not the version lacking part of the GBD (Fig. 7 B). This suggests that fascin binds to the FMNL2 N-terminal region. Analysis of fascin interactions with FMNL2 mutant forms using FLIM-FRET further demonstrated that fascin interacted with the WT and constitutively active (ΔDAD) version of FMNL2 (Fig. 7 C). However, no FRET was seen between fascin and a proposed constitutively active mutant of FMNL2 (mut, A272E; Kage et al., 2017), suggesting either that fascin requires this residue for binding or that this mutant adopts a conformation that is no longer permissive for fascin binding (Fig. 7 C, bottom panel). Finally, as our previous data showed that higher-stiffness substrates led to reduced fascin phosphorylation, and that S39A-fascin bound more strongly to FMNL2, we performed FLIM analysis on cells coexpressing fascin and FMNL2 in cells on 2D surfaces of differing stiffness. Data demonstrated a significant increase in fascin-FMNL2 binding in cells on higher-stiffness (28-kPa) surfaces compared with lower equivalents (1.5-kPa; Fig. 7 D). These data combined support the notion that fascin is mechanosensitive and that stiffer environments trigger a shift to dephosphorylated fascin that has higher binding to both FMNL2 and F-actin, facilitating filopodia assembly.
Figure 7.
FMNL2 directly interacts with fascin. (A) GFP trap analysis to evaluate complex formation between FMNL2-GFP and endogenous fascin. FMNL2-GFP was overexpressed in HeLa cells, and GFP trap was performed from cell lysates. Levels of GFP and fascin were evaluated in lysates (input) and pulldowns via Western blot. Representative Western blot of three independent experiments is shown. (B) Direct protein–protein interaction between full-length fascin and FMNL2. Top: Schematic view of full-length FMNL2 with GBD, FH3, FH1, FH2, and DAD domains. Bottom: GST-pulldown experiments of fascin and N-terminal FMNL2 truncations (GBD-FH3, GBD-FH3 with point mutation S171DD) and GBD-FH3 with deletion in GBD domain). GST-tagged FMNL2 constructs and fascin levels were evaluated using Western blot. Representative experiment of three independent experiments is shown. (C) FLIM-FRET lifetime measurement of fixed HeLa cells expressing RFP-fascin and GFP-tagged WT FMNL2, mutant FMNL2 (A272E), and active FMNL2ΔDAD. Intensity images of GFP fluorescence (top panel) and FLIM data in pseudocolor (bottom panel) are shown. Warm colors show high FRET (low GFP lifetime), whereas cool colors indicate low FRET (high lifetime). GFP-fascin alone served as donor-only control and for calculations of FRET efficiency (%). Scale = 5 µm. Calculated FRET efficiency (relative to GFP-fascin alone) for fascin:formin interaction (n = 26, 18, or 21 cells respectively). One-way ANOVA; *, P ≤ 0.05; ns, not significant. (D) FMNL2–fascin interaction on soft (1.5-kPa) and stiff (28-kPa) 2D surfaces were measured with FLIM-FRET. Student t test; ****, P ≤ 0.0001, n = 18 and 17 cells for 1.5 and 28 kPa, respectively. (E) Proposed model. pS39-fascin accumulates at the cell periphery and is subsequently dephosphorylated before it interacts with FMNL2. FMNL2 acts as gatekeeper to control fascin movement into and out of filopodia to allow flexibility and adaptation to the stiffness of the ECM.
Discussion
Here we show that fascin phosphorylation, localization, and binding to F-actin are highly dynamic and dependent on local cytoskeletal architecture in cells in both 2D and 3D environments. We provide evidence that fascin pS39 induces a conformational change in agreement with previous suggestions based on structural data (Jansen et al., 2011; Yang et al., 2013). We further show that fascin S39 phosphorylation is highly dynamic, suggesting that the cyclical actions of PKC and phosphatases on fascin are correlated with local positioning of fascin within the cell, which in turn is dependent on the local F-actin architecture. Previous in vitro studies demonstrated that intrinsic properties of fascin allow it to segregate to appropriate F-actin structures (Winkelman et al., 2016), and our data show that binding to FMNL2 can control localized fascin dynamics and interactions with actin. Indeed, activation-dependent redistribution of both PKCα and FMNL2 to the plasma membrane (Wang et al., 2015) is also likely to contribute to this local phosphorylation switching in fascin subpopulations at the membrane, enabling both FMNL2 binding and fascin entry into filopodia.
The density of branched F-actin structures at the cell periphery is not solely dependent on Arp2/3; FMNL2 and FMNL3 also play important roles in the regulation of the actin architecture at these sites. Both of these formins are localized within lamellipodia and regulate protrusion stability and forces (Kage et al., 2017). We show that S39 dephosphorylated fascin preferentially interacts with FMNL2, which actively controls non–pS39-fascin entry into filopodia. Although the exact mechanisms that define spatial binding of these two proteins remain to be defined, our data support a model wherein the conformational change of fascin following S39 dephosphorylation results in the detachment of fascin from branched F-actin and an increased affinity for parallel linear F-actin structures. Increased lamellipodia density through elevated polymerization against a counterforce such as the plasma membrane can increase intracellular forces powered by actomyosin networks (Mueller et al., 2017). This positive net forward force coupled with localized complex formation with active FMNL2 may promote the active recruitment of dephosphorylated fascin into filopodia. In 3D environments that better mimic the topographical and mechanical features of tissues, stiffening of the ECM also induces higher intracellular forces (Ahmadzadeh et al., 2017), and this triggers filopodia assembly before the cell starts to migrate to a new site (Huang et al., 2015; Jacquemet et al., 2016; Wong et al., 2014). A further report has shown that stretching cells decreases overall S39 phosphorylation levels of fascin (Kliewe et al., 2017). Our own data indicate that fascin undergoes changes in S39 phosphorylation, conformation, and dynamics in response to altered 3D extracellular stiffness, and fascin-dependent filopodia are subsequently required to increase local matrix deformation. This coupled with our demonstration of FMNL2-dependent recruitment of fascin to filopodia favors a model in which cytoskeletal regulators, such as formins, can directly rapidly feed back changes to external forces to mobilize and control fascin activity and function (Fig. 7 E).
Our data provide evidence at both bulk and single-molecule levels that fascin in filopodia is highly mobile and not stably bound to F-actin, as has also been previously suggested (Aratyn et al., 2007). Our single-molecule imaging identified at least two distinct motions of fascin within filopodia. The fast movement of fascin was ∼10 times higher than previously reported speeds for this molecule using FRAP analysis (Aratyn et al., 2007). Our single-molecule and FLIM data strongly suggest rapid on/off rates of the interaction of fascin and F-actin in filopodia in a nondirectional manner, which suggests a rather flexible nature of this interaction. This highly dynamic behavior could enable rapid adaptation of filopodia to environment cues while ensuring stability and strength through differential fascin-dependent F-actin bundling within specific sites along the filopodia shaft, as also shown by our FRET data. It is possible that the rapid change in fascin conformational state we report here may also contribute to the fluctuations in fascin-actin FRET through alteration of the orientation of the fluorophores with respect to each other. Moreover, the highly confined 2D nature of filopodia also likely contributes to the high molecular speeds of fascin within these structures. Importantly our data show that confinement is not the only regulating factor, as pS39 status alters fascin molecular movement. Thus, our data reveal that F-actin bundling by fascin is not a stable event as previously hypothesized, but rather a highly dynamic interaction that fluctuates an order of magnitude faster than the growth rate of the filopodia itself. This raises interesting questions around the dynamic properties of other F-actin bundling and cross-linking molecules at the nanoscale that warrant further investigation in future.
We have shown that fascin directly interacts with the N-terminal domain of FMNL2 and that FMNL2 promotes fascin translocation into filopodia. Although we cannot rule out the possibility that actin binding provides an initial scaffold for complex assembly, our data show that F-actin is not required for complex stability. This may provide means for active FMNL2 to bind non–pS39-fascin at the cell periphery and position it within the lamellipodia for rapid entry into filopodia. FMNL2 has recently been shown to control lamellipodial actin organization and force production in melanoma cells (Kage et al., 2017), as well as localizing to filopodial tips (Gardberg et al., 2010). Interestingly, FMNL2, while widely expressed, is particularly abundant in neuronal and epithelial tissues (Gardberg et al., 2010), corresponding to tissues in which fascin is also highly expressed. Moreover, both fascin and FMNL2 are up-regulated in many types of cancer and correlate with metastasis formation and poor patient prognosis (Gardberg et al., 2016; Hashimoto et al., 2005; Jayo and Parsons, 2010; Péladeau et al., 2016; Schoumacher et al., 2014; Zhong et al., 2018; Zhu et al., 2008). The high stiffness of human tumors and the resulting destabilization of adult tissue homeostasis is an emerging hallmark of tumor cell evasion and metastatic progression (Seewaldt, 2014). The potential role for FMNL2-dependent fascin dynamics, under specific mechanical environments, might enable cancer cells to exploit increased stiffness of the ECM and leave primary tumors under these permissive conditions.
Fascin has also previously been shown to directly interact with DAAM1 in mouse cells (Jaiswal et al., 2013), and our data confirm this interaction in human cells. Both FMNL2 and DAAM1 are localized to the filopodia shaft; however, our data demonstrate that the interaction of DAAM1 and fascin primarily occurs within the cytoplasm. Moreover, DAAM1 overexpression or knockdown does not result in altered fascin dynamics within filopodia, or a change in filopodia number, suggesting that the interaction between these two proteins may regulate an alternative function for fascin. The difference in our findings, compared with the previous study of DAAM1-fascin cooperation (Jaiswal et al., 2013), may be the result of different experimental approaches. Much of the proposed cooperative behavior in the previous study was demonstrated using in vitro approaches or in fixed cells. As filopodia are highly sensitive to fixation, we used only live cells throughout the present study, and our analysis is restricted to filopodia that clearly protrude from the cell membrane and that were not embedded into the lamellipodium, otherwise known as microspikes. Microspikes are surrounded by highly branched Arp2/3-dependent F-actin structures and are suggested as precursors to filopodia (Faix and Rottner, 2006; Mellor, 2010). Fascin appears at the tips of microspikes, which resembles the base of a cell-surface protruding, free-moving filopodium. It is therefore possible that DAAM1 plays a role in regulating fascin within microspikes; analyzing cooperativity between formins and fascin within other parallel F-actin structures will be interesting avenues to pursue in future.
Taken together, we have identified a regulatory role of the formin FMNL2 in the spatiotemporal regulation of fascin. We propose a model (Fig. 7 E) whereby pS39-fascin accumulates within dense branched F-actin regions at the plasma membrane. Following dephosphorylation, fascin interacts with FMNL2, which acts as a gatekeeper to control fascin movement into the filopodia base. The molecular speed and subcellular distribution of fascin is regulated by S39 phosphorylation and sensitive to the stiffness of the ECM, as demonstrated by the fact that dephosphorylated fascin enters filopodia. High extracellular stiffness decreases S39 phosphorylation, increases the interaction of fascin and FMNL2 and fascin mobility, and results in increased filopodia flexibility compared with softer substrates. Fast conformational changes in fascin and its highly dynamic interaction with F-actin within filopodia enable cancer cells to rapidly adapt to the extracellular environment. Future analysis will shed further light on the specific roles of formins and their interaction with fascin to potentially provide new modes of action for therapeutic targeting.
Materials and methods
Reagents, dyes, antibodies, and plasmids
The following antibodies were used: anti-fascin mouse monoclonal antibody (Dako), anti-fascin pS39 (Abcam), anti-GFP mouse antibody (Roche), anti-FMNL2/3 (Abcam), anti-Profilin1 and anti-Profilin2 (Cell Signaling), and anti-DAAM1 (Santa Cruz), anti-GAPDH (Chemicon). Anti-mouse and anti-rabbit HRP-conjugated antibodies were purchased from Dako, and AF555- and AF647-coupled secondary antibodies were from Life Technologies. The following compounds were used: CK666 and Jasplakinolide (Santa Cruz), SMIFH2 (Tocris), Cytochalasin D (Sigma-Aldrich), BIM (Sigma-Aldrich), phorbol 12,13-dibutyrate (Sigma-Aldrich), tetradecanoyl phorbol acetate (Sigma-Aldrich), and Kinesore (kindly provided by Mark Dodding, University of Bristol, UK). Cy5 monoreactive dye and the dialysis kit were from GE Healthcare, rat tail collagen I was purchased from Corning, and ribose was from AppliChem. AF647 carboxylate-modified microbeads (0.2-µm diameter) were kindly provided by the Advanced Image Center Team (Janelia Research Campus). To generate stable fascin knockdown HeLa cells, we used the shRNA 5′-GCCTGAAGAAGAAGCAGATCT-3′, cloned into pLKO (RNAi consortium), and used cells for either transient transfection or stable viral transduction of GFP-fascin and mEOS2-fascin as described previously (Jayo et al., 2016). Effectene (Qiagen) was used for DNA transfections and Dharmafect (Dharmacon) for siRNA transfections. The following plasmids were used for transient expression: GFP-fascin WT, RFP-fascin WT, RFP-fascin-S39A, RFP-fascin-S39D, CS-fascin, CS-fascin-S39A, CS-fascin-S39D, mScarlet-lifeact, RFP-actin, mEOS2-fascin, FMNL2-GFP (Block et al., 2012; Kage et al., 2017), FMNL2ΔDAD-GFP (generated by subcloning FMNL2ΔDAD from Block et al. [2012]), FMNL2-A272E-GFP (Woodham et al., 2017), FMNL3-GFP, FMNL3act-GFP (Kage et al., 2017), GFP-DAAM1, and GFP-DAAM1ΔDAD (Liu et al., 2008). GST FMNL2 constructs were described in Kühn et al. (2015). To generate the CS-fascin, GFP-fascin in pEGFP-C1 was mutated using the Q5 mutagenesis kit (NEB) to create restriction sites within an unconserved loop in domain 3 of fascin, predicted to face to the outside of the molecule in a closed, globular conformation. On-TARGET siRNA pools directed against FMNL2, FMNL3, DAAM1, Profilin-1, and Profilin-2 or control siRNAs were from Dharmacon. Single control or siRNAs against FMNL2 were from Sigma-Aldrich (SASI_Hs01_00144237 and SASI_Hs02_00361655).
Cell culture and transfections
HeLa cells were cultured in high-glucose DMEM (Sigma-Aldrich) supplemented with 10% FCS (Thermo Fisher Scientific), 1% glutamine, and 1% penicillin/streptomycin (Sigma-Aldrich) and maintained under standard conditions (37°C, 5% CO2, and 95% humidity). Fascin knockdown cells were selected and maintained using puromycin, and cells were subcultured using 0.05% trypsin-EDTA in PBS. For expression of fascin (transient or stable), we exclusively used fascin knockdown HeLa cells and reexpressed fascin at physiological levels. mEOS2-fascin–expressing cells were maintained in medium without phenol-red, to decrease background fluorescence. For fascin, actin, or formin cDNA, cells were transfected with Effectene according to the manufacturer’s instructions and used for imaging after 24 h. 50 nM siRNA was transfected into cells using Dharmafect, and cells were lysed for Western blot or used for live imaging after 72 h.
Western blotting
Western blotting was performed as described previously (Jayo et al., 2016). Briefly, cells were lysed in sample buffer, and proteins were separated under reducing conditions by SDS-PAGE, blotted onto polyvinylidene fluoride membranes using a semidry method, blocked with 5% BSA, and probed for specified proteins.
Confocal imaging, FRET, FRAP, and photoconversion in 2D
For live- and fixed-cell imaging, HeLa cells were seeded in glass-bottom imaging chambers (Lab-Tek, Nunc) the day before imaging. After one washing step, medium was exchanged with imaging medium, containing DMEM (phenol-red-free), 10% FCS, and 20 mM Hepes for live-cell imaging. For pS39-fascin detection, GFP-fascin–expressing HeLa cells were fixed with prewarmed 4% PFA, washed with PBS, and permeabilized with 0.1% Triton X-100 in PBS. After blocking in 3% BSA, cells were stained with primary anti-pS39-fascin antibody for 2 h at RT, washed, and incubated with AF 555- or AF647-labeled secondary antibody for 1 h, followed by three washes and mounting or covering with PBS. Confocal microscopy was performed using a Nikon A1R inverted confocal laser scanning microscope with a 60× oil objective. For ratiometric FRET imaging, CS-fascin was excited using the 488-nm wavelength, and emission was simultaneously collected at 525 nm (GFP) and 595 nm (mScarlet). Data were background corrected and analyzed using NIS Elements Advanced Research imaging software (Nikon). FRET/donor ratios were used for presentation of FRET (in %) and illustrated in pseudocolor. For comparison of stiffness in 2D, cells were plated on either collagen-coated glass dishes or 35-mm imaging dishes with an elastically supported surface of 1.5-kPa stiffness (IBIDI). For measuring the change in pS39 phosphorylation depending on surface stiffness, GFP-fascin–expressing cells were plated on collagen-coated 35-mm imaging dishes with an elastically supported surface of 1.5- or 28-kPa stiffness, fixed, stained, and imaged. For FRAP experiments, ROIs were manually defined for each cell (multiple filopodia, single filopodia, or small patches along single filopodia). After baseline measurement, fluorophores were photobleached with 70–100% laser power for 0.5 s, and recovery was measured in 1-s intervals. FRAP data were background and photobleaching corrected in NIS Elements and exported to Excel. Data were normalized (0–100%) in GraphPad Prism and fitted using mono- and biexponential functions (1- and 2-phase decay). For inhibitor, siRNA, and overexpression experiments, the mobile fraction (plateau) was set to experimental control values (60%) for comparable t1/2 value calculations. For photoconversion experiments, fascin-depleted HeLa cells that constitutively express silencing-resistant mEOS2-fascin were converted from green to red with a 405-nm laser for 0.5 s with 70% laser power. The frame rate for tracking photoconverted mEOS-fascin = 1 s. Data were background corrected in NIS Elements, and corrected time-lapse intensity data were exported to Excel. Data were normalized (% of maximum) in GraphPad Prism and fitted using mono- and biexponential functions. For 3D, inhibitor, overexpression, and siRNA experiments, the immobile fraction was set to control 2D values (30%) for comparable t1/2 value calculations.
Structured illumination microscopy (SIM)
Cells were seeded in 35-mm round glass-bottom dishes (Greiner), and medium was changed to Hepes containing imaging medium. iSIM imaging of CS-fascin was performed at the VT-iSIM (VisiTech, Sunderland, UK) with a spatial resolution of 125 nm laterally and 350 nm axially. Images and time lapses were taken using a 100× oil objective (SR Apo total internal reflection fluorescence [TIRF] AC) and a frame rate of 1 s. For CS-fascin imaging, ratiometric FRET was measured upon donor excitation at 488 nm and simultaneous emission collection at 538 nm (GFP) and 594 nm (mScarlet). FRET/donor ratios are shown in pseudocolor. Richardson–Lucy deconvolution (20 iterations) was done using the Nikon software.
FLIM
FLIM experiments were performed using a custom microscope design based around an inverted microscope frame (Ti Eclipse, Nikon). A frequency-doubled Ti:Sapphire laser (Chameleon II, Coherent) delivered pulses of 488-nm light at a repetition rate of 80 MHz. The laser light was incident on a spatial light modulator (LCOS-SLM, P512, Meadowlark) that was used to generate an 8 × 8 array of beamlets. The beamlet array was reflected off a dichroic beamsplitter (FF495-Di03-25 × 36, Semrock) and raster scanned across the sample using a tip/tilt piezo mirror (S334.2SD, Physik Instrumente) through a 100×, NA 1.45 objective lens (Nikon Instruments). The interbeamlet spacing at the sample was 2 µm, and each beamlet scanned 32 × 32 points, giving a total field of view of 16 × 16 µm, with 256 × 256 voxels per image. The fluorescence was descanned, passed through the tdichroic beamsplitter, and focused using a 4× NA 0.13 objective lens (Nikon Instruments) onto a single-photon avalanche diode array detector with on-board 10-bit timing electronics (Richardson, J., R. Walker, L. Grant, D. Stoppa, F. Borghetti, E. Charbon, M. Gersbach, and R.K. Henderson. 2009. A 32x32 50ps Resolution 10 bit Time to Digital Converter Array in 130 nm complementary metal–oxide–semiconductor for Time Correlated Imaging. Proceedings of the IEEE 2009 Custom Integrated Circuits Conference). LabVIEW Virtual Instruments were used for hardware control and data acquisition, and the time-resolved data were processed using either a center of mass method (Poland et al., 2016) or time-correlated single-photon counting to determine the fluorescence lifetimes. Center of mass method reads were intensity corrected for differences in array beamlets and processed with Labview. The time-correlated single-photon counting data were fitted using a monoexponential decay model in the TRI2 analysis package (Barber et al., 2009) to generate FLIM images. FLIM images were acquired with an acquisition time of 3 s per frame.
For fixed FLIM-FRET imaging, cells were fixed with 4% PFA for 10 min at RT, rehydrated with PBS, and permeabilized with 0.1% Triton X-100 in PBS for 10 min before quenching with freshly prepared 1 mg/ml sodium borohydride in PBS for 10 min at RT. Cells were washed 3× with PBS and either mounted in hardening mounting medium (Dabco, Sigma-Aldrich) or kept in PBS until imaging. For measuring the interaction of FMNL2 (GFP) and fascin (RFP) in response to surface stiffness, cells were plated on collagen-coated 35-mm imaging dishes with an elastically supported surface of 1.5- or 28-kPa stiffness, fixed, and treated with sodium borohydride before imaging. Imaging was performed on a confocal microscope (Ti Eclipse, Nikon) with a 40× objective throughout (Plan Fluor NA 1.3; differential interference contrast [DIC] H, WD 0.2; Nikon), with fluorescence lifetime acquired by time-correlated single-photon counting electronics (SPC-830) on a DCC-100 control (both Becker & Hickl). Acquisition was performed at 900-nm laser excitation (MaiTai, DeepSee; Spectra-Physics) for 2–3 min to collect sufficient photons for fitting, while avoiding pulse pile-up or significant photobleaching. Corresponding wide-field images were taken for GFP and RFP/mScarlet (DS-Qi1Mc camera; Nikon). Lifetime raw data were analyzed with TRI2 (Paul Barber), and monoexponential fitting was done, unless stated otherwise. Histogram data are plotted as mean FRET efficiency (calculated on the basis of GFP lifetime without acceptor) from the indicated number of cells per sample. FLIM images are presented in reverse rainbow pseudocolor, with red representing low lifetime and high FRET/interaction and blue representing high lifetime and low FRET/interaction.
Dual-color PALM single-molecule microscopy
For single-molecule experiments, fascin knockdown HeLa cells stably expressing mEOS2-fascin that were maintained in phenol-red free DMEM were plated in 35-mm round dishes (Greiner) the day before imaging. Medium was changed to phenol red–free imaging medium containing 20 mM Hepes 30 min before imaging. All single-molecule experiments were performed on a custom PALM/STORM microscope. This consisted of a DMi8 microscopy body fitted with a SuMo ultra-high-stability stage (Leica Microsystems). A 1.43-NA, 160× TIRF objective (Leica Microsystems) was mounted to the underside of the stage via a piezo drive (PI), which was used to provide focus control. This combination allows very low sample drift. Illumination was provided by diode lasers of 561 nm (Oxxius), 473 nm (Dragon Laser), and 405 nm (Vortran). The output of each laser was coupled through an optical fiber (ThorLabs), which depolarized it before being combined, apertured, and expanded using various optics (ThorLabs). The beams were focused and displaced in the back aperture of the objective to provide TIRF illumination. Fluorescence was split according to wavelength by an image splitter (Photometrics Dual-view) and imaged simultaneously side by side on a fast electron-multiplying charge-coupled device camera (Photometrics Evolve). The filter windows used were 500–530 nm for the unconverted mEOS2 channel and 575–630 nm for the photoconverted channel. Low-intensity 473-nm illumination was used to excite the unconverted mEos2, which constituted the vast majority of the total mEOS2-fascin population. This effectively provided a wide-field image of the cell body and filopodia. Simultaneous strong 561-nm illumination (∼20–30 mW in the back aperture) excited a very small proportion of mEOS2-fascin photoconverted to the long wavelength emitting state by the weak 473-nm illumination. Emission from individual mEOS2-fascin molecules was then imaged in the photoconverted channel. If the density of single emitters in the photoconverted channel was insufficient, it was increased with periodic low-intensity 405-nm illumination. By comparing the two simultaneously acquired channels, the density of single-molecule emitters specific to filopodia could be discerned from the cell as a whole. Acquisitions typically consisted of 10,000 or 15,000 frames of 20- or 25-ms exposure time. The image pixel size was 100 nm.
Single-molecule tracking analysis
The raw image sequence was spatially split in half separating the two imaging channels. To improve signal to noise, the unconverted channel was temporally downsampled by averaging 100 frame blocks of the original sequence. This gives a precise record of the location, movement growth, and retraction of the filopodia with a time resolution of ∼2 s. Using this information, cells were selected for analysis that possessed one or more filopodia that were fairly straight, maintained a fairly fixed orientation, and did not cross any other filopodia for a large proportion of the acquired image sequence. An additional constraint was that the filopodia of interest remained close to the coverslip and therefore in focus. A large majority of candidate filopodia would repeatedly move into and out of the focal TIRF plane. For each successful candidate, a start and end frame in the original image sequence was identified for which all these criteria were met. To aid the single-molecule tracking program in correctly identifying molecular trajectories constrained to the target filopodia and reject spurious background molecules, a dynamic ROI around the filopodia was used. This was implemented using a custom Matlab (MathWorks) script. At the previously identified start frame, several analysis reference points positioned from the base to the tip of the filopodia separated by a few hundred nanometers are identified. A line connecting these points marks the approximate center line of the filopodia. In each subsequent frame, an intensity profile along a line perpendicular to this line is fitted to a Gaussian profile. The reference point is then shifted sideways to match the maxima of the fit. An additional sensing point is located ahead of the tip of the filopodia by extrapolation of the connecting line; if the magnitude of the fitted profile is above a threshold, this point is added to the line of reference points. However, if the last point in the list is below the threshold, it is removed from the list. In this manner, the reference line tracked movement and lengthening/shortening of the filopodium from frame to frame. An ROI was then drawn in each frame using these reference points, which is the length of the filopodium and ∼200–400 nm wide. Each ROI was then transferred to the corresponding 100-frame block of the converted channel sequence. All pixel values outside the ROI were replaced with noise equivalent to the mean background level. The resulting sequence contained only single-molecule images or fascin located in the target filopodium, eliminating other cell structures and most of the background molecules. The width of the ROI allowed for small errors in registration between the two channels.
Single-molecule tracking analysis was performed on the processed image sequences using the MosaicSuite plugin for ImageJ (Sbalzarini and Koumoutsakos, 2005). The list of identified molecular tracks were then further analyzed in Matlab. Overlaying all the individual trajectories reconstructed the central line of the filopodia. Any molecular tracks that showed sharp tangential steps were judged to result from inclusion of background localizations and were excluded. Histograms of the size of individual molecular steps from frame to frame showed dynamics over very different time scales. For all filopodia, these were found to fit better to a triple Gaussian distribution than a double (Fig. S1, F and G). We attribute these three modes of motion to the following. The broad large step-size distribution results from free pseudo-1D diffusion of fascin along the filopodium. The other two much slower (small step-size) distributions could result from fascin bound to F-actin. The width of these distributions reflects a combination of the localization precision and localization errors (such as mistaking two overlapping emitters as one). This interpretation is supported by two observations, the first of which is made by examining individual molecular tracks. The typical ON time of the mEOS2 before photobleaching was quite short, ∼175 ms averaged over all filopodia, corresponding to only seven to nine camera frames, depending on exposure time. For the small proportion of tracks that contained a much larger number of tracks, long periods of very small randomly oriented steps were interspersed with shorter periods of much larger 1D motion. The number of molecules with sufficiently long tracks was too small for further analysis. Second, the downsampled unconverted channel visually shows mEOS2-fascin density fluctuations that persist over many frames, moving slowly with a retrograde motion toward the cell body, yet fascin is not depleted from the tip of the filopodium. This is consistent with a bound fascin population following the normal actin treadmilling and an unbound diffusing population, with slow exchange between the two. Following this interpretation, the diffusion coefficient of mEOS2-fascin was extracted from the large scale motion using
where σf is the SD of the Gaussian fit to the fast motion and tex is the camera exposure time. The proportion of fascin in the unbound state is the area of the fit to the fast motion compared with the total area.
Filopodia quantification
Filopodia were quantified using the Matlab plugin CellGeo (Tsygankov et al., 2014). Briefly, confocal images or time-lapse image sequences were thresholded using MovThresh, followed by BisectoGraph, which maps arbitrarily a polygon on a tree graph required for FiloTrack, which identifies filopodia and tracks their length, number, and lifetime over time.
To measure the flexibility of filopodia, filopodia bending was analyzed in siControl (siCTR), siFMNL2, and siFMNL3 GFP-fascin HeLa cells. Briefly, single frames from time-lapse videos of live cells were imported into ImageJ, and an ROI was drawn along the outline of single filopodia (polygon selection). The area of the traced filopodium was divided by the convex hull drawn for each object. The shape description
was used as a parameter to estimate the potential of filopodia to be flexible enough to bend. Straight filopodia have a ratio close to 1, whereas bent filopodia have a ratio <1.
3D collagen I matrix preparation for 3D cell imaging
Precooled HeLa cells were mixed on ice with a collagen solution containing 20 mM Hepes, 20 µg/ml fibronectin, 0.3% sodium bicarbonate, and 1 M NaOH to neutralize 0.02 N collagen in acetic acid to obtain a final collagen concentration of 2 mg/ml and cell density of 5–8 × 105 cells/ml (∼200 Pa; Mason et al., 2013). For stiff collagen matrix (∼800 Pa), collagen stock solution was preincubated for 30 min on ice with ribose (200 mM) and thereafter used for preparation. For visualization of collagen fibers, collagen was stained with Cy5 in 0.1 M sodium bicarbonate and dialyzed overnight in 0.1% acetic acid. For measurement of forces exerted on the collagen matrix, beads were embedded into the 3D matrix. Briefly, AF647-carboxylate–modified microbeads (0.2-µm diameter; Molecular Probes) were sedimented at 3,000 g for 20 min, resuspended in medium by vortexing, and sonicated for 60 min. Beads were mixed at 1.8 × 109 beads/ml final concentration with collagen-cell solution on ice to prevent polymerization. 10-20 µl of collagen-cell mix (with or without beads) were pipetted on collagen-coated 5-mm round glass coverslips (CS-5R, Warner Instruments), left to polymerize at 37°C for 30 min, and imaged within 36 h.
LLSM, FRET, and photoconversion in 3D
The LLSM instrument used in these experiments is housed in the Advanced Image Center at the Howard Hughes Medical Institute Janelia research campus. The system is configured and operated as previously described (Chen et al., 2014). 3D collagen samples were illuminated using a 2D optical lattice generated by a spatial light modulator (Fourth Dimension Displays). The sample is excited by 488-, 560-, or 641-nm diode lasers (MPB Communications) at 30-, 75-, and 50-mW initial box power through an excitation objective (Special Optics, 0.65 NA, 3.74-mm WD). Fluorescent emission was collected by detection objective (Nikon, CFI Apo LWD 25XW, 1.1 NA), and detected by an scientific complementary metal–oxide–semiconductor camera (Hamamatsu Orca Flash 4.0 v2). Acquired data were deskewed as previously described (Chen et al., 2014) and deconvolved using an iterative Richardson–Lucy algorithm. Point-spread functions for deconvolution were experimentally measured using 200-nm tetraspeck beads adhered to 5-mm glass coverslips (Invitrogen, T7280) for each excitation wavelength. Photomanipulation in these experiments was performed with a 405-nm laser (2% laser power) controlled by a pair of galvanic mirrors and necessary optics (Firefly system, Rapp-Opto Electronics) that was custom fit to the LLSM. ROIs for photomanipulation were kept at a constant size and manually defined for each cell.
PIV analysis of bead displacement in 3D gels
For bead displacement measurements in 3D collagen gels, beads were tracked, and time-resolved PIV was analyzed using the Matlab plugin PIVlab (Thielicke, 2014). Briefly, cell areas were excluded, whereas the cell surroundings (proximal and distal) were analyzed. Interrogation areas to step values were set to 160–80, 90–45, and 40–20 pixels, and velocity limits were set to 1.5 × 1.5 pixels/frame. Velocity maps are shown in Jet pseudocolor. When necessary, automatic drift correction was performed using the StackReg plugin in Fiji before analysis in PIVlab.
Protein production
His-tagged fascin was produced as described before (Zanet et al., 2012). GST-tagged FMNL2 constructs were transformed into Escherichia coli BL21 strains and expressed overnight at 18°C with 400 µM isopropyl β-d-1-thiogalactopyranoside. Pellets were resuspended in GST lysis buffer (50 mM Tris HCl, pH 8.0, 500 mM NaCl, 10% glycerol, 2 mM β-mercaptoethanol, and protease inhibitor), sonicated, and cleared by centrifugation. GST-FMNL2 constructs were isolated using glutathione Sepharose beads as described before (Jayo et al., 2016).
Pulldown experiments
For GST pulldown experiments, His-fascin bound to Sepharose beads was eluted using 50 mM NaH2PO4, pH 8.0, 300 mM NaCl, and 250 mM imidazole and dialyzed against GST lysis buffer. GST-FMNL2 beads were incubated for 2 h with eluted fascin at 4°C, washed 4× with radioimmunoprecipitation assay buffer (1% Triton X-100, 1% sodium deoxycolate, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 50 mM Tris-HCl, pH 8, and protease inhibitor) and analyzed by Western blot for GST and fascin. For GFP trap experiments, GFP only or FMNL2-GFP was expressed in WT HeLa cells, cells were lysed (1% NP-40, 50 mM Tris-HCl, 150 mM NaCl, pH 7.5, and protease inhibitor), and cleared lysates were incubated with GFP trap agarose beads (ChromoTek) for 2 h at 4°C. Beads were washed 4× with lysis buffer and resuspended with Laemmli buffer for Western blot analysis of endogenous fascin and GFP.
Statistical analysis
Statistical tests were performed using Student’s t tests and one- or two-way ANOVA, followed by a posttest (uncorrected Fisher’s least significant difference or Bonferroni as indicated) using Prism8 (GraphPad). In general, box plots display the median (line), and whiskers show 2.5–97.5th percentile borders, unless stated differently. Potential outliers were identified using the robust regression and outlier removal (ROUT) method (Q = 10%). A P value <0.05 was considered significant. All experiments were performed at least three times unless stated otherwise, and data are expressed as mean ± SEM (or mean ± SD, when indicated).
Online supplemental material
Fig. S1 shows analysis of photoconverted mEOS2-fascin in whole cells and by single-molecule analysis and that S39Afascin shows faster dynamics than WTfascin. Fig. S2 shows that fascin dynamics and actin binding are dependent on F-actin turnover and that fascin contributes to local force transmission using traction for microscopy in 3D collagen gels. Fig. S3 shows that disturbing the organization of F-actin alters filopodia formation and fascin movement, and further provides characterization of the CSfascin biosensor. Fig. S4 shows that FMNL2, FMNL3, DAAM1, and profilin differentially contribute to filopodia formation and stability, and that FMNL2 plays the dominant role in these phenotypes. Fig. S5 shows that overexpressed FMNL2, FMNL3, and DAAM1 have distinct localization within filopodia, and FMNL2 alters fascin distribution. Video 1 shows the movement of mEOS-fascin in cells on 2D surfaces. Videos 2 and 3 show single-molecule dynamics of fascin in filopodia. Video 4 shows GFP-fascin in filopodia within 3D matrices imaged using LLSM. Videos 5 and 6 show cells expressing GFP-fascin in soft (Video 5) or stiff (Video 6) collagen gels. Videos 7, 8, and 9 show dynamics of the CSfascin FRET biosensor within filopodia.
Supplementary Material
Acknowledgments
We thank Clemens Rottner (Helmholtz Centre for Infection Research, Braunschweig, Germany), Harry Mellor (University of Bristol, Bristol, UK), Matthias Geyer (Max Planck Institute of Molecular Physiology, Dortmund, Germany), and Ray Habas (Temple University, Philadelphia, PA) for providing plasmids for use in this study. We also thank Ben Goult for helpful discussions on fascin conformation. We also gratefully acknowledge assistance with imaging from Daniel Matthews in the Nikon Imaging Centre at King’s College London.
Funding for this work was provided by the Medical Research Council (MR/K015664/1). K. Pfisterer was further funded by the Company of Biologists and the Biochemical Society for travel grants to visit the Janelia Research Campus. The LLSM imaging experiments were performed at the Advanced Imaging Center at Howard Hughes Medical Institute Janelia Research Campus. The Advanced Imaging Center is a jointly funded venture of the Gordon and Betty Moore Foundation and the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
Author contributions: K. Pfisterer conducted all experiments and analysis, with help from J. Levitt, C.D. Lawson and R.J. Marsh for FLIM, FRAP/S39 fascin analysis, and single-molecule analysis respectively. S. Ameer-Beg assisted with design and development of FLIM system and experimental approaches. S. Cox cobuilt the single-molecule instrument and assisted with experimental design and data interpretation. RJM performed single-molecule data analysis. J.M. Heddleston and E. Wait assisted with data acquisition and analysis for 3D LLSM datasets. M. Parsons conceived the study and designed experiments with K. Pfisterer. M. Parsons and K. Pfisterer wrote the manuscript with input from all authors.
References
- Adams J.C., Clelland J.D., Collett G.D., Matsumura F., Yamashiro S., and Zhang L.. 1999. Cell-matrix adhesions differentially regulate fascin phosphorylation. Mol. Biol. Cell. 10:4177–4190. 10.1091/mbc.10.12.4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadzadeh H., Webster M.R., Behera R., Jimenez Valencia A.M., Wirtz D., Weeraratna A.T., and Shenoy V.B.. 2017. Modeling the two-way feedback between contractility and matrix realignment reveals a nonlinear mode of cancer cell invasion. Proc. Natl. Acad. Sci. USA. 114:E1617–E1626. 10.1073/pnas.1617037114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuschies J., and Vogel V.. 2013. The role of filopodia in the recognition of nanotopographies. Sci. Rep. 3:1658 10.1038/srep01658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anilkumar N., Parsons M., Monk R., Ng T., and Adams J.C.. 2003. Interaction of fascin and protein kinase Calpha: a novel intersection in cell adhesion and motility. EMBO J. 22:5390–5402. 10.1093/emboj/cdg521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aratyn Y.S., Schaus T.E., Taylor E.W., and Borisy G.G.. 2007. Intrinsic dynamic behavior of fascin in filopodia. Mol. Biol. Cell. 18:3928–3940. 10.1091/mbc.e07-04-0346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber P.R., Ameer-Beg S.M., Gilbey J., Carlin L.M., Keppler M., Ng T.C., and Vojnovic B.. 2009. Multiphoton time-domain fluorescence lifetime imaging microscopy: practical application to protein-protein interactions using global analysis. J. R. Soc. Interface. 6(suppl 1):S93–S105. 10.1098/rsif.2008.0451.focus [DOI] [Google Scholar]
- Block J., Breitsprecher D., Kühn S., Winterhoff M., Kage F., Geffers R., Duwe P., Rohn J.L., Baum B., Brakebusch C., et al. 2012. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr. Biol. 22:1005–1012. 10.1016/j.cub.2012.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broders-Bondon F., Nguyen Ho-Bouldoires T.H., Fernandez-Sanchez M.-E., and Farge E.. 2018. Mechanotransduction in tumor progression: The dark side of the force. J. Cell Biol. 217:1571–1587. 10.1083/jcb.201701039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher D.T., Alliston T., and Weaver V.M.. 2009. A tense situation: forcing tumour progression. Nat. Rev. Cancer. 9:108–122. 10.1038/nrc2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey S.P., Goldblatt Z.E., Martin K.E., Romero B., Williams R.M., and Reinhart-King C.A.. 2016. Local extracellular matrix alignment directs cellular protrusion dynamics and migration through Rac1 and FAK. Integr. Biol. 8:821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B.C., Legant W.R., Wang K., Shao L., Milkie D.E., Davidson M.W., Janetopoulos C., Wu X.S., Hammer J.A. III, Liu Z., et al. 2014. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 346:1257998 10.1126/science.1257998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Yang S., Jakoncic J., Zhang J.J., and Huang X.Y.. 2010. Migrastatin analogues target fascin to block tumour metastasis. Nature. 464:1062–1066. 10.1038/nature08978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra E.S., and Higgs H.N.. 2007. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9:1110–1121. 10.1038/ncb1007-1110 [DOI] [PubMed] [Google Scholar]
- Elkhatib N., Neu M.B., Zensen C., Schmoller K.M., Louvard D., Bausch A.R., Betz T., and Vignjevic D.M.. 2014. Fascin plays a role in stress fiber organization and focal adhesion disassembly. Curr. Biol. 24:1492–1499. 10.1016/j.cub.2014.05.023 [DOI] [PubMed] [Google Scholar]
- Faix J., and Rottner K.. 2006. The making of filopodia. Curr. Opin. Cell Biol. 18:18–25. 10.1016/j.ceb.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Gardberg M., Heuser V.D., Koskivuo I., Koivisto M., and Carpén O.. 2016. FMNL2/FMNL3 formins are linked with oncogenic pathways and predict melanoma outcome. J. Pathol. Clin. Res. 2:41–52. 10.1002/cjp2.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardberg M., Talvinen K., Kaipio K., Iljin K., Kampf C., Uhlen M., and Carpén O.. 2010. Characterization of Diaphanous-related formin FMNL2 in human tissues. BMC Cell Biol. 11:55 10.1186/1471-2121-11-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E.S., Gauvin T.J., Heimsath E.G., and Higgs H.N.. 2010. Assembly of filopodia by the formin FRL2 (FMNL3). Cytoskeleton (Hoboken). 67:755–772. 10.1002/cm.20485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y., Skacel M., and Adams J.C.. 2005. Roles of fascin in human carcinoma motility and signaling: prospects for a novel biomarker? Int. J. Biochem. Cell Biol. 37:1787–1804. 10.1016/j.biocel.2005.05.004 [DOI] [PubMed] [Google Scholar]
- Huang F.K., Han S., Xing B., Huang J., Liu B., Bordeleau F., Reinhart-King C.A., Zhang J.J., and Huang X.Y.. 2015. Targeted inhibition of fascin function blocks tumour invasion and metastatic colonization. Nat. Commun. 6:7465 10.1038/ncomms8465 [DOI] [PubMed] [Google Scholar]
- Jacquemet G., Baghirov H., Georgiadou M., Sihto H., Peuhu E., Cettour-Janet P., He T., Perälä M., Kronqvist P., Joensuu H., and Ivaska J.. 2016. L-type calcium channels regulate filopodia stability and cancer cell invasion downstream of integrin signalling. Nat. Commun. 7:13297 10.1038/ncomms13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemet G., Hamidi H., and Ivaska J.. 2015. Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr. Opin. Cell Biol. 36:23–31. 10.1016/j.ceb.2015.06.007 [DOI] [PubMed] [Google Scholar]
- Jacquemet G., Stubb A., Saup R., Miihkinen M., Kremneva E., Hamidi H., and Ivaska J.. 2019. Filopodome Mapping Identifies p130Cas as a Mechanosensitive Regulator of Filopodia Stability. Curr. Biol. 29:202–216.e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal R., Breitsprecher D., Collins A., Corrêa I.R. Jr., Xu M.Q., and Goode B.L.. 2013. The formin Daam1 and fascin directly collaborate to promote filopodia formation. Curr. Biol. 23:1373–1379. 10.1016/j.cub.2013.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen S., Collins A., Yang C., Rebowski G., Svitkina T., and Dominguez R.. 2011. Mechanism of actin filament bundling by fascin. J. Biol. Chem. 286:30087–30096. 10.1074/jbc.M111.251439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayo A., and Parsons M.. 2010. Fascin: a key regulator of cytoskeletal dynamics. Int. J. Biochem. Cell Biol. 42:1614–1617. 10.1016/j.biocel.2010.06.019 [DOI] [PubMed] [Google Scholar]
- Jayo A., Malboubi M., Antoku S., Chang W., Ortiz-Zapater E., Groen C., Pfisterer K., Tootle T., Charras G., Gundersen G.G., and Parsons M.. 2016. Fascin Regulates Nuclear Movement and Deformation in Migrating Cells. Dev. Cell. 38:371–383. 10.1016/j.devcel.2016.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kage F., Winterhoff M., Dimchev V., Mueller J., Thalheim T., Freise A., Brühmann S., Kollasser J., Block J., Dimchev G., et al. 2017. FMNL formins boost lamellipodial force generation. Nat. Commun. 8:14832 10.1038/ncomms14832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewe F., Scharf C., Rogge H., Darm K., Lindenmeyer M.T., Amann K., Cohen C.D., Endlich K., and Endlich N.. 2017. Studying the role of fascin-1 in mechanically stressed podocytes. Sci. Rep. 7:9916 10.1038/s41598-017-10116-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn S., and Geyer M.. 2014. Formins as effector proteins of Rho GTPases. Small GTPases. 5:e29513 10.4161/sgtp.29513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn S., Erdmann C., Kage F., Block J., Schwenkmezger L., Steffen A., Rottner K., and Geyer M.. 2015. The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation. Nat. Commun. 6:7088 10.1038/ncomms8088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Sato A., Khadka D., Bharti R., Diaz H., Runnels L.W., and Habas R.. 2008. Mechanism of activation of the Formin protein Daam1. Proc. Natl. Acad. Sci. USA. 105:210–215. 10.1073/pnas.0707277105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu P., Weaver V.M., and Werb Z.. 2012. The extracellular matrix: a dynamic niche in cancer progression. J. Cell Biol. 196:395–406. 10.1083/jcb.201102147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh R.J., Pfisterer K., Bennett P., Hirvonen L.M., Gautel M., Jones G.E., and Cox S.. 2018. Artifact-free high-density localization microscopy analysis. Nat. Methods. 15:689–692. 10.1038/s41592-018-0072-5 [DOI] [PubMed] [Google Scholar]
- Mason B.N., Starchenko A., Williams R.M., Bonassar L.J., and Reinhart-King C.A.. 2013. Tuning three-dimensional collagen matrix stiffness independently of collagen concentration modulates endothelial cell behavior. Acta Biomater. 9:4635–4644. 10.1016/j.actbio.2012.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor H. 2010. The role of formins in filopodia formation. Biochim. Biophys. Acta. 1803:191–200. [DOI] [PubMed] [Google Scholar]
- Mueller J., Szep G., Nemethova M., de Vries I., Lieber A.D., Winkler C., Kruse K., Small J.V., Schmeiser C., Keren K., et al. 2017. Load Adaptation of Lamellipodial Actin Networks. Cell. 171:188–200.e16. 10.1016/j.cell.2017.07.051 [DOI] [PubMed] [Google Scholar]
- Nolen B.J., Tomasevic N., Russell A., Pierce D.W., Jia Z., McCormick C.D., Hartman J., Sakowicz R., and Pollard T.D.. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 460:1031–1034. 10.1038/nature08231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono S., Yamakita Y., Yamashiro S., Matsudaira P.T., Gnarra J.R., Obinata T., and Matsumura F.. 1997. Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J. Biol. Chem. 272:2527–2533. 10.1074/jbc.272.4.2527 [DOI] [PubMed] [Google Scholar]
- Péladeau C., Heibein A., Maltez M.T., Copeland S.J., and Copeland J.W.. 2016. A specific FMNL2 isoform is up-regulated in invasive cells. BMC Cell Biol. 17:32 10.1186/s12860-016-0110-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland S.P., Erdogan A.T., Krstajić N., Levitt J., Devauges V., Walker R.J., Li D.D.-U., Ameer-Beg S.M., and Henderson R.K.. 2016. New high-speed centre of mass method incorporating background subtraction for accurate determination of fluorescence lifetime. Opt. Express. 24:6899–6915. 10.1364/OE.24.006899 [DOI] [PubMed] [Google Scholar]
- Randall T.S., Yip Y.Y., Wallock-Richards D.J., Pfisterer K., Sanger A., Ficek W., Steiner R.A., Beavil A.J., Parsons M., and Dodding M.P.. 2017. A small-molecule activator of kinesin-1 drives remodeling of the microtubule network. Proc. Natl. Acad. Sci. USA. 114:13738–13743. 10.1073/pnas.1715115115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringer P., Colo G., Fassler R., and Grashoff C.. 2017. Sensing the mechano-chemical properties of the extracellular matrix. Matrix Biol. 644:6–16. [DOI] [PubMed] [Google Scholar]
- Rizvi S.A., Neidt E.M., Cui J., Feiger Z., Skau C.T., Gardel M.L., Kozmin S.A., and Kovar D.R.. 2009. Identification and characterization of a small molecule inhibitor of formin-mediated actin assembly. Chem. Biol. 16:1158–1168. 10.1016/j.chembiol.2009.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbalzarini I.F., and Koumoutsakos P.. 2005. Feature point tracking and trajectory analysis for video imaging in cell biology. J. Struct. Biol. 151:182–195. 10.1016/j.jsb.2005.06.002 [DOI] [PubMed] [Google Scholar]
- Schoumacher M., El-Marjou F., Laé M., Kambou N., Louvard D., Robine S., and Vignjevic D.M.. 2014. Conditional expression of fascin increases tumor progression in a mouse model of intestinal cancer. Eur. J. Cell Biol. 93:388–395. 10.1016/j.ejcb.2014.08.002 [DOI] [PubMed] [Google Scholar]
- Sedeh R.S., Fedorov A.A., Fedorov E.V., Ono S., Matsumura F., Almo S.C., and Bathe M.. 2010. Structure, evolutionary conservation, and conformational dynamics of Homo sapiens fascin-1, an F-actin crosslinking protein. J. Mol. Biol. 400:589–604. 10.1016/j.jmb.2010.04.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seewaldt V. 2014. ECM stiffness paves the way for tumor cells. Nat. Med. 20:332–333. 10.1038/nm.3523 [DOI] [PubMed] [Google Scholar]
- Thielicke W.S. 2014. PIVlab – Towards User-friendly, Affordable and Accurate Digital Particle Image Velocimetry in MATLAB. J. Open Res. Softw. 2:e30 10.5334/jors.bl [DOI] [Google Scholar]
- Tsygankov D., Bilancia C.G., Vitriol E.A., Hahn K.M., Peifer M., and Elston T.C.. 2014. CellGeo: a computational platform for the analysis of shape changes in cells with complex geometries. J. Cell Biol. 204:443–460. 10.1083/jcb.201306067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Helvert S., Storm C., and Friedl P.. 2018. Mechanoreciprocity in cell migration. Nat. Cell Biol. 20:8–20. 10.1038/s41556-017-0012-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignjevic D., Schoumacher M., Gavert N., Janssen K.P., Jih G., Laé M., Louvard D., Ben-Ze’ev A., and Robine S.. 2007. Fascin, a novel target of beta-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Res. 67:6844–6853. 10.1158/0008-5472.CAN-07-0929 [DOI] [PubMed] [Google Scholar]
- Villari G., Jayo A., Zanet J., Fitch B., Serrels B., Frame M., Stramer B.M., Goult B.T., and Parsons M.. 2015. A direct interaction between fascin and microtubules contributes to adhesion dynamics and cell migration. J. Cell Sci. 128:4601–4614. 10.1242/jcs.175760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Arjonen A., Pouwels J., Ta H., Pausch P., Bange G., Engel U., Pan X., Fackler O.T., Ivaska J., and Grosse R.. 2015. Formin-like 2 Promotes β1-Integrin Trafficking and Invasive Motility Downstream of PKCα. Dev. Cell. 34:475–483. 10.1016/j.devcel.2015.06.015 [DOI] [PubMed] [Google Scholar]
- Winkelman J.D., Suarez C., Hocky G.M., Harker A.J., Morganthaler A.N., Christensen J.R., Voth G.A., Bartles J.R., and Kovar D.R.. 2016. Fascin- and α-Actinin-Bundled Networks Contain Intrinsic Structural Features that Drive Protein Sorting. Curr. Biol. 26:2697–2706. 10.1016/j.cub.2016.07.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S., Guo W.-H., and Wang Y.-L.. 2014. Fibroblasts probe substrate rigidity with filopodia extensions before occupying an area. Proc. Natl. Acad. Sci. USA. 111:17176–17181. 10.1073/pnas.1412285111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodham E.F., Paul N.R., Tyrrell B., Spence H.J., Swaminathan K., Scribner M.R., Giampazolias E., Hedley A., Clark W., Kage F., et al. 2017. Coordination by Cdc42 of Actin, Contractility, and Adhesion for Melanoblast Movement in Mouse Skin. Curr. Biol. 27:624–637. 10.1016/j.cub.2017.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakita Y., Ono S., Matsumura F., and Yamashiro S.. 1996. Phosphorylation of human fascin inhibits its actin binding and bundling activities. J. Biol. Chem. 271:12632–12638. 10.1074/jbc.271.21.12632 [DOI] [PubMed] [Google Scholar]
- Yang S., Huang F.K., Huang J., Chen S., Jakoncic J., Leo-Macias A., Diaz-Avalos R., Chen L., Zhang J.J., and Huang X.Y.. 2013. Molecular mechanism of fascin function in filopodial formation. J. Biol. Chem. 288:274–284. 10.1074/jbc.M112.427971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J., Jayo A., Plaza S., Millard T., Parsons M., and Stramer B.. 2012. Fascin promotes filopodia formation independent of its role in actin bundling. J. Cell Biol. 197:477–486. 10.1083/jcb.201110135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fonovic M., Suyama K., Bogyo M., and Scott M.P.. 2009. Rab35 controls actin bundling by recruiting fascin as an effector protein. Science. 325:1250–1254. 10.1126/science.1174921 [DOI] [PubMed] [Google Scholar]
- Zhong B., Wang K., Xu H., and Kong F.. 2018. Silencing Formin-like 2 inhibits growth and metastasis of gastric cancer cells through suppressing internalization of integrins. Cancer Cell Int. 18:79 10.1186/s12935-018-0576-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.L., Liang L., and Ding Y.Q.. 2008. Overexpression of FMNL2 is closely related to metastasis of colorectal cancer. Int. J. Colorectal Dis. 23:1041–1047. 10.1007/s00384-008-0520-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.