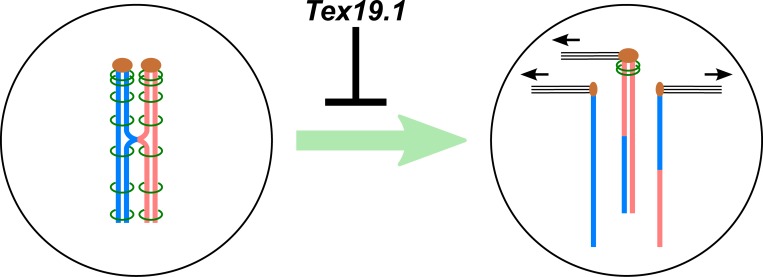
Mammalian oocytes need to maintain sister chromatid cohesion during their prolonged postnatal development to faithfully segregate their chromosomes in meiosis. Reichmann et al. show that Tex19.1 modulates ubiquitin-dependent proteolysis and maintains the cohesive subpopulation of cohesin in postnatal mouse oocytes to prevent oocyte aneuploidy.
Abstract
Age-dependent oocyte aneuploidy, a major cause of Down syndrome, is associated with declining sister chromatid cohesion in postnatal oocytes. Here we show that cohesion in postnatal mouse oocytes is regulated by Tex19.1. We show Tex19.1−/− oocytes have defects maintaining chiasmata, missegregate their chromosomes during meiosis, and transmit aneuploidies to the next generation. Furthermore, we show that mouse Tex19.1 inhibits N-end rule protein degradation mediated by its interacting partner UBR2, and that Ubr2 itself has a previously undescribed role in negatively regulating the acetylated SMC3 subpopulation of cohesin in mitotic somatic cells. Lastly, we show that acetylated SMC3 is associated with meiotic chromosome axes in mouse oocytes, and that this population of cohesin is specifically depleted in the absence of Tex19.1. These findings indicate that Tex19.1 regulates UBR protein activity to maintain acetylated SMC3 and sister chromatid cohesion in postnatal oocytes and prevent aneuploidy from arising in the female germline.
Graphical Abstract
Introduction
Chromosome missegregation in the mammalian germline can cause embryonic lethality or conditions such as Down syndrome in the next generation (Hassold and Hunt, 2001; Nagaoka et al., 2012). In humans, meiotic chromosome segregation errors are prevalent in oocytes, increase dramatically with maternal age, and are associated with reduced chromosome cohesion (Hassold and Hunt, 2001; Nagaoka et al., 2012; Herbert et al., 2015; MacLennan et al., 2015; Gruhn et al., 2019). In mice, loss of chromosome cohesion and increased aneuploidy also occurs in aging oocytes and is accompanied by an age-dependent loss of cohesin proteins from the oocytes’ chromosomes (Chiang et al., 2010; Lister et al., 2010). Cohesin is a complex of four proteins (structural maintenance of chromosomes 1α [SMC1α], SMC3, radiation-sensitive mutant 21 [RAD21], and small tumor antigen 1 [STAG1] or STAG2 in mitotic cells) arranged in a ring-like structure that links DNA molecules and promotes cohesion between sister chromatids (Nasmyth and Haering, 2009). Meiotic cells express additional meiosis-specific versions of some of these cohesin subunits (SMC1β, RAD21 ligand, meiotic recombination 8 [REC8], and STAG3; McNicoll et al., 2013). In mitotic cells, only a small subpopulation of chromosome-associated cohesin is marked by acetylation of SMC3 functions in sister chromatid cohesion (Schmitz et al., 2007; Zhang et al., 2008; Nishiyama et al., 2010, 2013). It is not clear whether sister chromatid cohesion in meiotic chromosomes also relies on an equivalent cohesive subpopulation of cohesin.
In female meiosis, cohesin is loaded onto DNA during fetal development and needs to be maintained during postnatal oocytes’ prolonged meiotic arrest, growth, and maturation (Revenkova et al., 2010; Tachibana-Konwalski et al., 2010; Burkhardt et al., 2016). This fetally loaded cohesin plays a crucial role in meiotic chromosome segregation, as it maintains chiasmata between the arms of homologous chromosomes until metaphase I and persists at centromeres to hold sister chromatids together until metaphase II (Revenkova et al., 2004, 2010; Hodges et al., 2005; Tachibana-Konwalski et al., 2010). Aging mouse oocytes have reduced levels of REC8 associated with their chromosomes (Chiang et al., 2010; Lister et al., 2010), which likely contributes to multiple age-related defects, including reduced cohesion between sister centromeres, fewer and more terminally distributed chiasmata, univalent chromosomes at metaphase I, lagging chromosomes during anaphase I, and fragmented kinetochores (Chiang et al., 2010; Lister et al., 2010; Zielinska et al., 2019). Many of these features are also seen in the oocytes of mice carrying mutations in or depleted for cohesin subunits (Revenkova et al., 2004; Hodges et al., 2005; Zielinska et al., 2019).
Elegant studies have provided significant insight into the molecular mechanisms by which cohesin functions (Nasmyth and Haering, 2009). However, it is possible that mammals possess additional mechanisms to help maintain cohesion during their oocytes’ prolonged postnatal development. Tex19.1 (testis expressed 19.1) was originally identified in a screen for genes expressed in mouse spermatogonia (Wang et al., 2001) but is also expressed in postnatal oocytes (Kuntz et al., 2008). Tex19.1 is a member of the mammal-specific family of TEX19 genes that duplicated during rodent evolution (Kuntz et al., 2008). Mouse Tex19.1 is syntenic with human TEX19, and these genes appear to have similar expression patterns in pluripotent cells and in fetal and postnatal male and female germ cells (Öllinger et al., 2008; Kuntz et al., 2008; Hackett et al., 2012; Planells-Palop et al., 2017). Tex19.2 is expressed in somatic cells in the testis and at more restricted stages of gametogenesis (Kuntz et al., 2008; Celebi et al., 2012; Hackett et al., 2012). Loss of Tex19.2 is reported to not have any major phenotypic consequence in mice, even in a Tex19.1−/− background (Tarabay et al., 2017). In contrast, loss of Tex19.1 causes fertility defects in both male and female mice (Öllinger et al., 2008; Yang et al., 2010). The infertility in Tex19.1−/− male mice is associated with defects in meiotic recombination that lead to chromosome asynapsis and germ cell death (Öllinger et al., 2008; Crichton et al., 2017). Tex19.1−/− female mice are subfertile but do not exhibit equivalent defects in homologous recombination (Öllinger et al., 2008; Crichton et al., 2017). The basis of the fertility defect in Tex19.1−/− female mice is currently not understood.
Mouse Tex19.1 also functions to repress retrotransposons in the germline (Öllinger et al., 2008; Reichmann et al., 2012; MacLennan et al., 2017), although it is not clear whether this function contributes to the fertility defects present in Tex19.1−/− mice (Crichton et al., 2017). Mouse TEX19.1 and human TEX19 physically interact with the E3 ubiquitin ligase UBR2 (ubiquitin-protein ligase E3 component N-recognin 2) and with long interspersed nuclear element 1 (LINE-1) retrotransposon proteins, promote ubiquitin-dependent degradation of LINE-1 protein, and prevent LINE-1 mobilizing to new locations in the genome (Yang et al., 2010; MacLennan et al., 2017). Furthermore, although UBR2 is required for the stability of mouse TEX19.1 and human TEX19 (Yang et al., 2010; MacLennan et al., 2017), it is not clear how TEX19 proteins impact on the ability of UBR2 to carry out its normal cellular roles in N-end rule protein degradation (Kwon et al., 2003; Tasaki et al., 2005). Here we report novel roles for mouse Tex19.1 and human TEX19 in inhibiting the N-end rule degradation and regulating acetylated SMC3-containing cohesin, and we show that Tex19.1 functions to maintain sister chromatid cohesion and prevents aneuploidy in postnatal mouse oocytes.
Results
Subfertility in Tex19.1−/− females is associated with oocyte aneuploidy
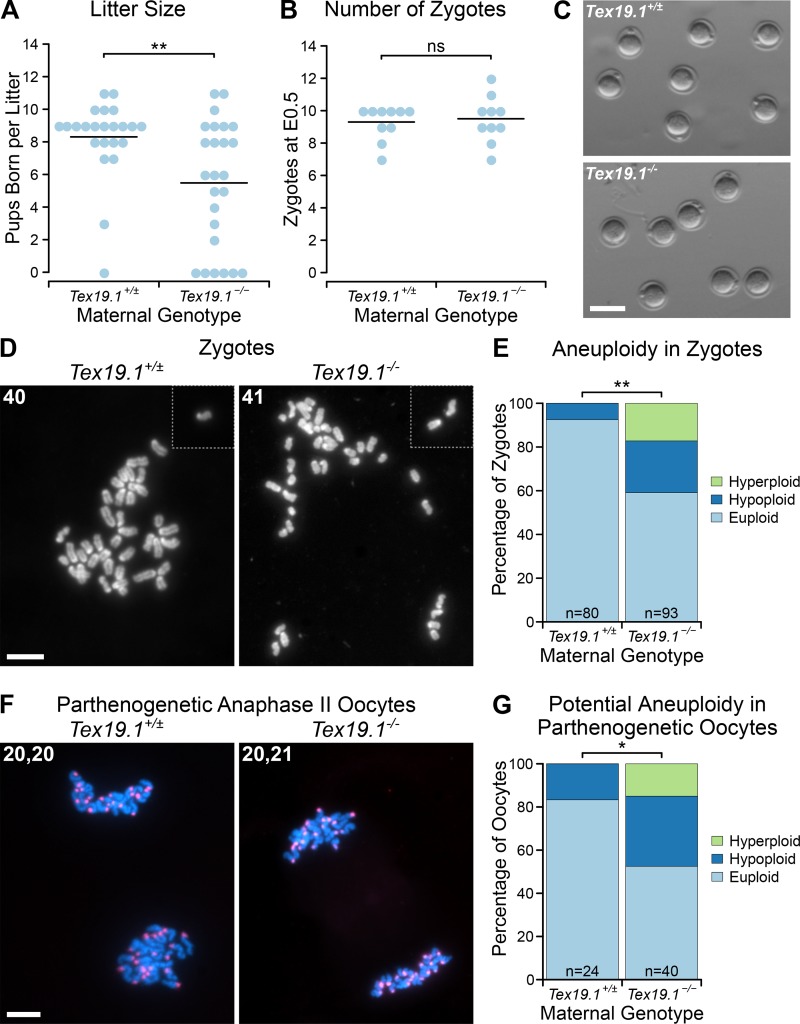
We previously reported that Tex19.1−/− females are subfertile (∼50% reduction in litter size) on a mixed genetic background (Öllinger et al., 2008). In contrast, fertility defects were not detected in Tex19.1−/− females on a C57BL/6 genetic background (Tarabay et al., 2013), although it is not clear if there was sufficient statistical power to detect subfertility in that study. To investigate the mechanistic basis of subfertility in Tex19.1−/− females, we first assessed if this phenotype is present in a C57BL/6 genetic background using an appropriate sample size. Consistent with our previous report on a mixed genetic background (Öllinger et al., 2008), C57BL/6 Tex19.1−/− females have a 33% reduction in litter size when mated to wild-type males (Fig. 1 A). Adult Tex19.1−/− females have normal ovary histology (Fig. S1 A) and contain a normal number of zygotes at embryonic day 0.5 (E0.5; Fig. 1 B) that do not have any gross morphological abnormalities (Fig. 1 C) when mated to wild-type males. However, analysis of chromosome spreads from these zygotes revealed a significant increase in the frequency of aneuploid zygotes from Tex19.1−/− females (41%) compared with Tex19.1+/± controls (7.5%; Fig. 1, D and E). All of the aneuploid zygotes from control Tex19.1+/± females exhibited hypoploidy but never hyperploidy, suggesting this likely represents technical artifacts caused by chromosome loss during preparation of the spreads or clustering that obscures chromosomes during scoring. In contrast, both hypoploidy (24%) and hyperploidy (17%) were observed in zygotes from Tex19.1−/− females (Fig. 1, D and E), which likely represent ∼33.5% of zygotes exhibiting true biological aneuploidy in addition to the ∼7.5% technical hypoploidy. This increased aneuploidy is already present in Tex19.1−/− oocytes before fertilization, as 47.5% of parthenogenetic anaphase II Tex19.1−/− oocytes are potentially aneuploid (32.5% hypoploid and 15% hyperploid) compared with 17% of parthenogenetic anaphase II Tex19.1+/± oocytes (all hypoploid; Fig. 1, E and F; and Fig. S1 B). Again, the hypoploidy without hyperploidy in parthenogenetic anaphase II Tex19.1+/± oocytes likely represents technical artifacts arising from spreading and counting chromosomes. Notably, the increased aneuploidy in Tex19.1−/− anaphase II oocytes (∼31% of oocytes) is comparable to the increased aneuploidy in zygotes (∼33.5% of zygotes) and to the decrease in litter size (33% of pups) from Tex19.1−/− mothers. As aneuploid mouse embryos typically do not develop to term (Yuan et al., 2002), these data indicate that transmission of aneuploidies through the female germline is likely a major contributor to the subfertility in Tex19.1−/− females.
Figure 1.
Subfertility in Tex19.1−/− females is associated with increased oocyte aneuploidy. (A and B) Number of pups born (A) and E0.5 zygotes (B) per litter after mating with wild-type males. Horizontal bars indicate means. Tex19.1+/± and Tex19.1−/− females have litter sizes of 8.3 ± 3.9 and 5.5 ± 2.4 pups born (**, Mann-Whitney U test, P < 0.01; n = 23, 25); and carry 9.3 ± 1.1 and 9.5 ± 1.4 E0.5 zygotes respectively (ns, Mann-Whitney U test, no significant difference; n = 10, 10). Data are from 7 Tex19.1+/± and 7 Tex19.1−/− females (A) and from 10 Tex19.1+/± and 10 Tex19.1−/− females (B). (C) E0.5 zygotes from Tex19.1+/± and Tex19.1−/− females. Scale bar 100 µm. (D and F) Chromosome spreads from E0.5 zygotes (D) and parthenogenetically activated anaphase II oocytes (F). The number of chromosomes is indicated in the top left of each image; dotted lines separate chromosomes from adjacent fields of view. DNA was visualized with DAPI (gray in D, cyan in F) and centromeres by major satellite FISH (red in F). Higher-magnification images of the oocytes shown in F are in Fig. S1 B. Scale bars, 20 µm. (E and G) Quantification of aneuploidy in E0.5 zygotes (E) and potential aneuploidy in parthenogenetically activated anaphase II oocytes (G). Aneuploid zygotes are more frequent in Tex19.1−/− females (24% hypoploid, 17% hyperploid, n = 80) than Tex19.1+/± females (7.5% hypoploid, 0% hyperploid, n = 93; **, Fisher’s exact test, P < 0.01; *, P < 0.05). Data are from eight Tex19.1+/± and eight Tex19.1−/− females. Euploid anaphase II oocytes with unequal numbers of chromosomes in each chromosome mass (e.g., 21, 19) were counted as 0.5 in each of the hypoploid and hyperploid categories (six Tex19.1−/− and no Tex19.1+/± oocytes). Potential aneuploidy in Tex19.1−/− parthenogenetic oocytes (32.5% hypoploid, 15% hyperploid, n = 24) is more frequent than in Tex19.1+/± parthenogenetic oocytes (17% hypoploid, 0% hyperploid, n = 40; *, Fisher’s exact test, P < 0.05). Data are from five Tex19.1+/± and seven Tex19.1−/− females.
Figure S1.
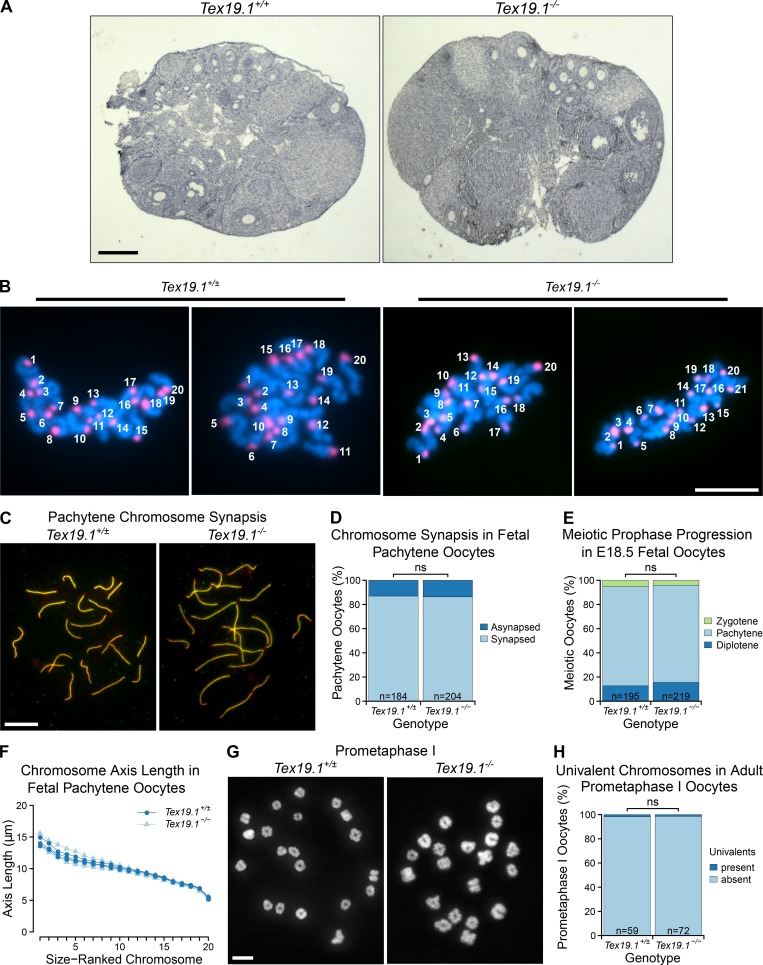
Oogenesis and meiotic prophase proceed normally in Tex19.1−/− female mice. (A) Hematoxylin-stained paraffin sections from Tex19.1+/+ and Tex19.1−/− adult ovaries. No gross abnormalities were evident in Tex19.1−/− ovaries. Primary, secondary, and antral follicles containing growing oocytes were observed in Tex19.1+/+ and Tex19.1−/− ovaries. Scale bar, 1 mm. (B) Example of chromosome counts from parthenogenetically activated anaphase II oocytes. Anaphase II chromosome masses are those shown in Fig. 1 F. Centromeres were visualized by FISH for major satellites (red), and DNA was stained with DAPI (cyan). Scale bar, 20 µm. (C) Immunostained E18.5 chromosome spreads from Tex19.1+/± and Tex19.1−/− oocytes showing chromosome synapsis. Axial elements and transverse filaments of the synaptonemal complex were stained with anti-SYCP3 (red) and anti-SYCP1 (green) antibodies, respectively. Scale bar, 10 µm. (D) Quantification of synapsis in E18.5 pachytene chromosome spreads. Asynapsis was present in 24 of 184 pachytene Tex19.1+/± oocytes and 28 of 204 pachytene Tex19.1−/− oocytes (ns, Fisher’s exact test, no significant difference). Data are derived from five Tex19.1+/± and five Tex19.1−/− fetuses. (E) SYCP3-positive nuclei in E18.5 oocyte chromosome spreads were classified into substages of meiotic prophase based on SYCP3 and SYCP1 immunostaining. The distribution of prophase substages was not significantly different between Tex19.1+/± and Tex19.1−/− oocytes (ns, Fisher’s exact test, no significant difference; n = 195, 219). Data are derived from five Tex19.1+/± and five Tex19.1−/− fetuses. (F) Chromosome axis lengths in pachytene nuclei from E18.5 fetal oocyte chromosome spreads as determined by anti-SYCP3 and anti-SYCP1 immunostaining. Chromosomes are ordered on the basis of size. 20 nuclei were scored for each fetus, and the mean axis length for each chromosome is plotted. Data for three Tex19.1+/± and three Tex19.1−/− fetuses are shown. Tex19.1+/± and Tex19.1−/− axis lengths are not significantly different for any chromosome (t test; n = 3). (G) Chromosome spreads from prometaphase I Tex19.1+/± and Tex19.1−/− oocytes 3 h after GVBD. DNA is stained with DAPI. Scale bar, 10 µm. (H) Quantification of number of prometaphase I oocytes containing univalents. Tex19.1+/± and Tex19.1−/− oocytes had similar frequencies of oocytes containing univalents (1/59 and 1/72 respectively; ns, Fisher’s exact test, no significant difference). All 59 Tex19.1+/± and 72 Tex19.1−/− oocytes had 40 chromosomes; therefore Tex19.1−/− oocytes are not already aneuploid before the first meiotic division. Data are derived from three Tex19.1+/± and three Tex19.1−/− female mice.
Tex19.1 prevents homologue missegregation and premature sister chromatid separation during oocyte meiosis I
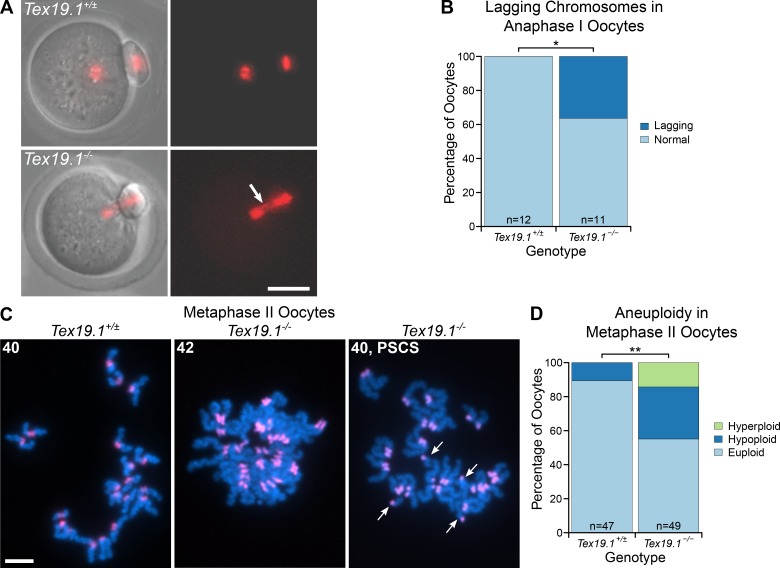
We next investigated why aneuploidy arises in Tex19.1−/− oocytes. In contrast to Tex19.1−/− spermatocytes (Öllinger et al., 2008; Crichton et al., 2017, 2018), Tex19.1−/− oocytes showed no detectable changes in progression through the first stages of meiotic prophase or the frequency of asynapsis or chromosome axis length at pachytene (Fig. S1, C–F). Furthermore, no aneuploidy or elevated frequency of univalent chromosomes was evident in Tex19.1−/− prometaphase I oocytes 3 h after germinal vesicle breakdown (GVBD; Fig. S1, G and H). Thus, the aneuploidy in Tex19.1−/− oocytes does not appear to be a consequence of defects in homologous chromosome synapsis or the establishment of bivalents during meiotic prophase.
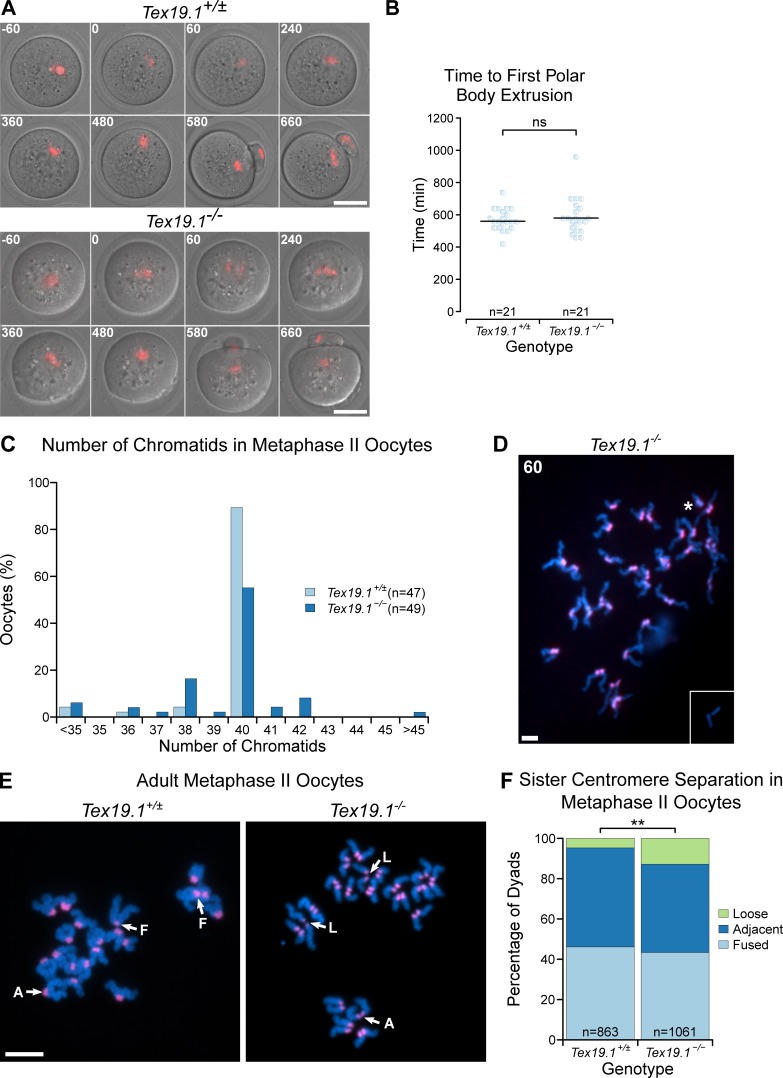
We next determined if errors in meiosis I chromosome segregation could be causing the aneuploidy in Tex19.1−/− oocytes. Live imaging of oocytes microinjected with histone H2B-RFP RNA at the germinal vesicle (GV) stage showed that the interval between GVBD and extrusion of the first polar body is similar between Tex19.1+/± and Tex19.1−/− oocytes (Fig. S2, A and B) and suggests that the spindle assembly checkpoint is not defective in these oocytes (Homer et al., 2005; McGuinness et al., 2009; Touati et al., 2015). However, lagging chromosomes were observed during anaphase I in approximately one third of Tex19.1−/− but not control oocytes (Fig. 2, A and B), indicating that meiosis I chromosome segregation may be perturbed. Furthermore, approximately one third of metaphase II Tex19.1−/− oocytes are aneuploid, with 17% of metaphase II Tex19.1−/− oocytes possessing at least one isolated sister chromatid (Fig. 2, C and D; and Fig. S2, C and D). This suggests that premature sister chromatid separation is contributing to the aneuploidy in Tex19.1−/− oocytes. Sister centromeres within intact dyads also displayed increased separation in Tex19.1−/− metaphase II oocytes (Fig. S2, E and F). However, five of the seven hyperploid Tex19.1−/− metaphase II oocytes had ≥21 dyads where sister chromatid cohesion appeared to be intact, indicating that missegregation of homologues had occurred during meiosis I in these oocytes (Fig. 2, C and D; and Fig. S2, C and D). Taken together, these data indicate that both premature sister chromatid separation and homologue missegregation during meiosis I are contributing to the aneuploidy in Tex19.1−/− oocytes.
Figure S2.
Meiotic chromosome segregation in adult Tex19.1−/− oocytes. (A) Live imaging of meiosis I in Tex19.1+/± and Tex19.1−/− oocytes. Chromatin was visualized with histone H2B-RFP (red). Time relative to GVBD in minutes is indicated in the top left corner of each image. Data are derived from six Tex19.1+/± and three Tex19.1−/− females across seven microinjection and imaging sessions. Scale bar, 50 µm. (B) Beeswarm plot showing the time to polar body extrusion relative to GVBD in Tex19.1+/± and Tex19.1−/− oocytes. Median values are indicated with a horizontal line (ns, Mann-Whitney U test, no significant difference; n = 21, 21). (C) Histogram showing the percentage of Tex19.1+/± and Tex19.1−/− metaphase II oocytes analyzed in Fig. 2 (C and D) that contained the indicated number of chromatids. Note the presence of aneuploid oocytes with odd numbers of chromatids, indicating potential premature segregation of sister chromatids. Of the seven hyperploid Tex19.1−/− oocytes, two exhibited cytologically detectable premature separation of sister chromatids; four had ≥21 dyads exhibiting intact sister chromatid cohesion, indicating missegregation of homologues had occurred; and one hyperploid Tex19.1−/− oocyte exhibited both these traits. (D) Chromosome spreads from an extreme hyperploid Tex19.1−/− metaphase II oocyte containing 60 chromatids. This oocyte contains >20 dyads indicating that homologous chromosomes missegregated without undergoing premature separation of sister centromeres during meiosis I in this oocyte. Note that a pair of broken chromatids lacking centromeric FISH signals (inset) was located just outside the field of view in this spread. These could potentially represent sister chromatids that broke away from their centromeres during preparation of the chromosome spreads, possibly from the centromere pair indicated with an asterisk. Scale bar, 10 µm. (E) Chromosome spreads from Tex19.1+/± and Tex19.1−/− metaphase II oocytes. DNA was visualized with DAPI (cyan), and centromeres were detected by FISH for major satellites (red). Sister centromere separation was scored as fused (F, one continuous centromeric domain between two chromatids), adjacent (A, two separate distinct centromeric domains separated by less than half the width of the chromatid arm), or loose (L, two separate distinct centromeric domains separated by more than half the width of the chromatid arm). Examples of these configurations are annotated. Scale bar, 10 µm. (F) Percentage of metaphase II dyads with different levels of sister centromere separation. 46% of dyads in 863 Tex19.1+/± oocytes have a fused configuration, 49% adjacent, and 5% loose. 43% of dyads in 1061 Tex19.1−/− oocytes have a fused configuration, 44% adjacent, and 13% loose (**, Fisher’s exact test, P < 0.01). Data are derived from 8 Tex19.1+/± and 12 Tex19.1−/− females.
Figure 2.
Tex19.1−/− oocytes missegregate homologous chromosomes and prematurely separate sister chromatids during meiosis I. (A) Live imaging of meiosis I in Tex19.1+/± and Tex19.1−/− oocytes. Chromatin was visualized with histone H2B-RFP (red). Lagging chromosomes are indicated with an arrow. Scale bar, 50 µm. (B) 36% of Tex19.1−/− anaphase I oocytes (n = 12) but no Tex19.1+/± anaphase I oocytes (n = 11) contained lagging chromosomes (*, Fisher’s exact test, P < 0.05). Data are from six Tex19.1+/± and three Tex19.1−/− females. (C) Chromosome spreads from metaphase II oocytes. DNA was visualized with DAPI (cyan) and centromeres by major satellite FISH (red). The number of chromatids is indicated. An aneuploid Tex19.1−/− oocyte with 42 chromatids but no overt premature sister chromatid separation (PSCS) and a euploid Tex19.1−/− oocyte with 40 chromatids and PSCS (arrows) are shown. Scale bar, 10 µm. (D) 31% of metaphase II Tex19.1−/− oocytes were hypoploid and 14% hyperploid compared with 11% hypoploid and 0% hyperploid for metaphase II Tex19.1+/± oocytes (**, Fisher’s exact test, P < 0.01; n = 47, 49). Data are from 8 Tex19.1+/± and 12 Tex19.1−/− females.
Tex19.1−/− oocytes have defects in maintaining the number and position of chiasmata during postnatal development
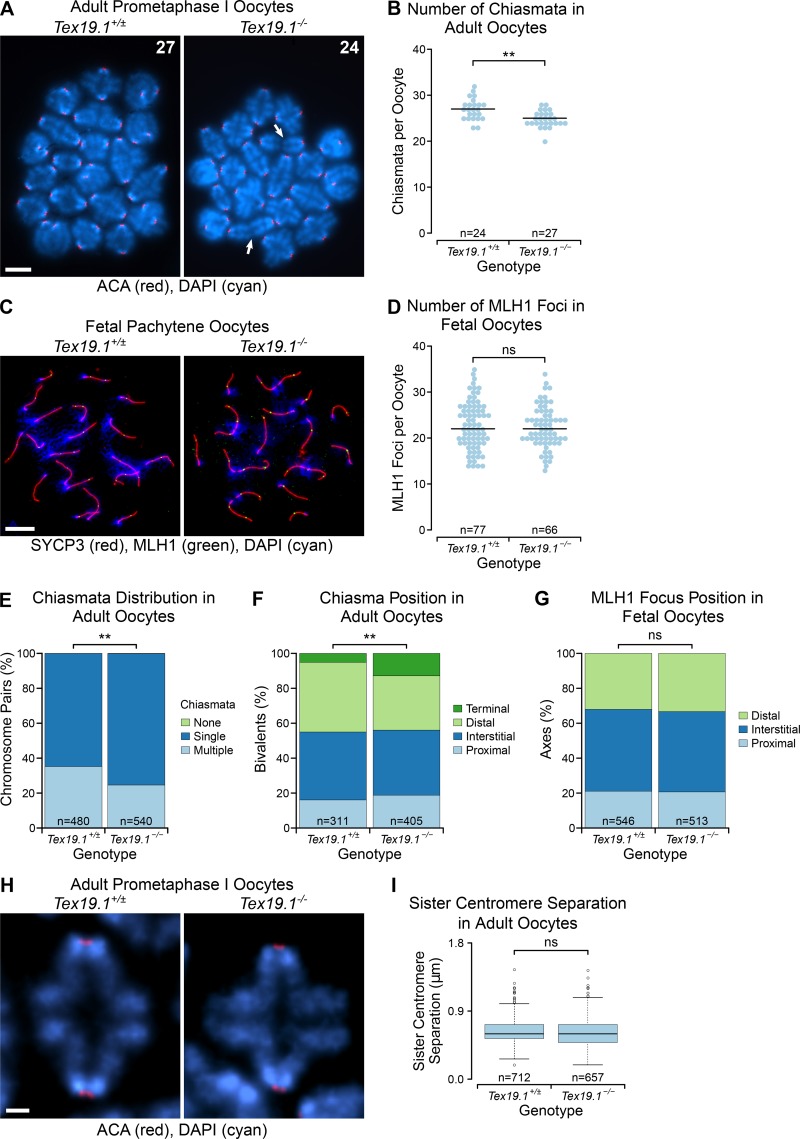
Homologue missegregation and premature sister chromatid separation during meiosis I are suggestive of defects in sister chromatid cohesion. As defects in cohesin function in oocytes can also cause reduced numbers and increased terminalization of chiasmata (Hodges et al., 2005), we analyzed chiasmata in Tex19.1−/− oocytes. Interestingly, the number of chiasmata in Tex19.1−/− prometaphase I oocytes 5 h after GVBD was significantly lower than in Tex19.1+/± controls (Fig. 3, A and B). This reduction in chiasmata frequency primarily reflects Tex19.1−/− oocytes having fewer bivalents with multiple chiasmata. (Fig. 3 E). To determine whether the reduction in chiasmata in adult Tex19.1−/− oocytes arises from defects in the establishment of crossovers during fetal development, we immunostained fetal oocyte chromosome spreads for mutL homolog 1 (MLH1), which marks ∼90% of crossovers (Baker et al., 1996; Holloway et al., 2008). The number of MLH1 foci in fetal Tex19.1−/− oocytes was not significantly different from controls (Fig. 3, C and D), suggesting that either Tex19.1 primarily affects the generation of MLH1-independent crossovers (Holloway et al., 2008), or meiotic crossovers are established correctly in fetal Tex19.1−/− oocytes but are not maintained during postnatal oocyte development.
Figure 3.
Tex19.1−/− oocytes have impaired maintenance of arm cohesion. (A) Chromosome spreads from adult prometaphase I oocytes. Centromeres are labeled with anti-centromere antibodies (ACA, red), DNA is stained with DAPI (cyan). Arrows highlight bivalents linked by terminal chiasmata, and the total number of chiasmata is indicated. Scale bar, 10 µm. (B) Tex19.1−/− prometaphase I oocytes have 24.9 ± 1.7 chiasmata, fewer than the 27.0 ± 2.3 in Tex19.1+/± prometaphase I oocytes (**, Mann-Whitney U test, P < 0.01; n = 24, 27). Horizontal lines indicate medians. (C) Chromosome spreads from E18.5 fetal pachytene oocytes. Synaptonemal complex is labeled with anti-SYCP3 antibodies (red), late recombination foci with anti-MLH1 antibodies (green), and DNA with DAPI (blue). Scale bar, 10 µm. (D) E18.5 fetal pachytene Tex19.1+/± oocytes possess 22.9 ± 5.2 MLH1 foci, similar to the 22.4 ± 4.5 in E18.5 fetal pachytene Tex19.1−/− oocytes (ns, Mann Whitney U test, no significant difference; n = 77, 66). Data are from three Tex19.1+/± and three Tex19.1−/− fetuses. Horizontal lines indicate medians. (E) The proportion of univalent chromosome pairs (no chiasmata) is not significantly different (0/480 for Tex19.1+/±, 1/540 for Tex19.1−/−; no significant difference, Fisher’s exact test), but adult Tex19.1−/− oocytes have fewer bivalents with multiple chiasmata (169/480 for Tex19.1+/±, 133/540 for Tex19.1−/−; **, Fisher’s exact test, P < 0.01). Data are from seven Tex19.1+/± and five Tex19.1−/− females. (F and G) Chiasma and MLH1 focus position relative to the centromere. Only bivalents/axes with a single chiasma/MLH1 focus were scored. MLH1 focus position is similar in Tex19.1+/± and Tex19.1−/− fetal oocytes (ns, Fisher’s exact test, no significant difference; n = 546, 513), there are more bivalents with terminal chiasmata (arrows in A) in Tex19.1−/− adult oocytes (13%) than in Tex19.1+/± controls (5%; **, Fisher’s exact test, P < 0.01; n = 311, 405). (H) High-magnification prometaphase I chromosomes labeled with ACA (red) to visualize centromeres and DAPI (cyan) to visualize DNA. The brightest point projections after deconvolution are shown. Scale bar, 1 µm. (I) Mean sister centromere separation at prometaphase I is 0.637 ± 0.174 µm in Tex19.1+/± oocytes (n = 712) and 0.626 ± 0.191 µm in Tex19.1−/− oocytes (n = 657; ns, Mann-Whitney U test, not significantly different).
We next analyzed whether loss of Tex19.1 might cause terminalization of chiasmata, as reported in aging oocytes (Henderson and Edwards, 1968) and Smc1β−/− female mice (Hodges et al., 2005). The position of chiasmata on the chromosome arms in prometaphase I bivalents with a single chiasma were classified as being proximal, interstitial, or distal relative to the centromeres (Hodges et al., 2005), and bivalents in an end-to-end configuration with no outward inflection of the arms (Fig. 3 A, arrows) were classified as having terminal chiasmata (Henderson and Edwards, 1968). Loss of Tex19.1 resulted in a significant increase in the proportion of terminal chiasmata (Fig. 3 F), which resembles the chiasmata terminalization reported in oocytes from aging and Smc1β−/− mice (Henderson and Edwards, 1968; Hodges et al., 2005). However, we cannot exclude that at least some of the bivalents arranged in an end-to-end configuration in prometaphase I Tex19.1−/− oocytes are achiasmate and remain associated through a different type of linkage. In contrast, phenotypes present in fetal Smc1β−/− oocytes, such as fewer MLH1 foci and altered chromosome axis length, were not detected in Tex19.1−/− fetal oocytes (Fig. 3, G and F); furthermore, neither was sister centromere separation detectably altered in adult prometaphase I Tex19.1−/− oocytes (Fig. 3, H and I). These data suggest that Tex19.1 has a role in the maintenance of arm cohesion in postnatal oocytes. Weakened arm cohesion can potentially result in precocious resolution of bivalents as they interact with the spindle during prometaphase I (Sakakibara et al., 2015; Zielinska et al., 2015), and subsequent biorientation or monoorientation of those univalents on the meiosis I spindle can then cause premature sister chromatid separation or homologue missegregation, respectively (LeMaire-Adkins et al., 1997; Kouznetsova et al., 2007). Thus, defects in maintaining arm cohesion could be sufficient to cause the patterns of aneuploidy seen in in Tex19.1−/− oocytes.
Ectopic expression of TEX19 promotes sister chromatid cohesion in mitotic somatic cells
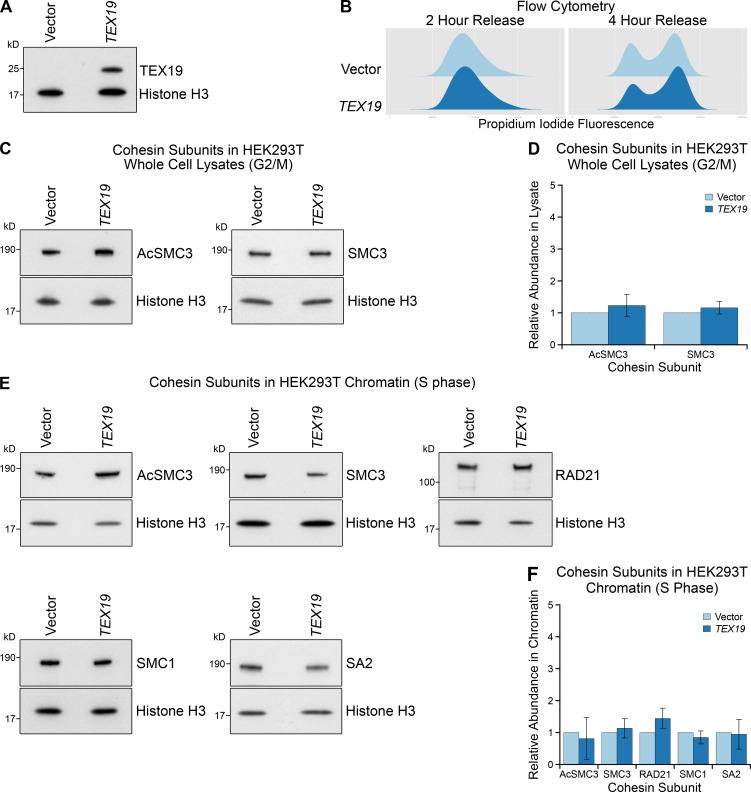
Although the Tex19.1−/− phenotype indicates that Tex19.1 plays a role in maintaining arm cohesion in oocytes, the biochemical function of TEX19.1 is poorly understood. Both mouse Tex19.1 and human TEX19 are functional in some nonmeiotic somatic cell types that naturally express these genes (Reichmann et al., 2013; Tarabay et al., 2013; Planells-Palop et al., 2017). Therefore, to investigate whether the ability of mouse Tex19.1 to regulate sister chromatid cohesion is conserved in human TEX19 and extends from meiosis to mitosis, we expressed human TEX19 in human HEK293T cells that do not normally express this gene (Fig. S3 A). Cohesins are loaded onto chromatin during S phase and removed during M phase in mitotic somatic cells (Waizenegger et al., 2000; Hauf et al., 2001); therefore, we enriched HEK293T cells in G2/M using a double thymidine block and release. Interestingly, ectopic expression of TEX19 in these cells reduces sister chromatid separation in G2/M (Fig. 4, A and B), suggesting that arm cohesion could be enhanced. Flow cytometry suggests that this effect on sister chromatid separation is not a consequence of human TEX19 perturbing cell cycle progression (Fig. S3 B). Thus, the Tex19.1-dependent mechanism promoting maintenance of cohesion in postnatal meiotic oocytes in mice appears to be reconstituted to some extent by ectopically expressing human TEX19 in mitotic somatic cells.
Figure S3.
Ectopic expression of TEX19 in HEK293T cells does not alter total cohesin levels in G2/M or chromatin-associated cohesin in S phase. (A) Western blot showing that transfection of TEX19 expression constructs into HEK293T cells results in detectable expression of TEX19 protein. Results are representative of three independent transfections. (B) Flow cytometry showing the DNA content (propidium iodide fluorescence) in HEK293T cell populations transfected with either empty vector or TEX19, synchronized with a double thymidine block, and released for either 2 or 4 h into fresh media to enrich for S phase and G2/M populations, respectively. (C and D) Representative Western blots (C) and quantification (D) of three replicates determining the abundance of AcSMC3 and SMC3 cohesin subunits in whole-cell lysates from HEK293T cells transfected with TEX19 or empty vector. Cells were synchronized with a double thymidine block and released for 4 h to enrich for G2/M cells. Histone H3 was used as a loading control. Cohesin abundance was normalized to histone H3 and quantified relative to empty vector transfections (means ± SD are indicated). AcSMC3 and SMC3 abundance is not significantly different between cells transfected with TEX19 and controls (t test; n = 3). (E and F) Representative Western blots (E) and quantification (F) of three replicates determining the abundance of cohesin subunits (AcSMC3, SMC3, RAD21, SMC1, SA2) in chromatin from HEK293T cells transfected with TEX19 or empty vector. Cells were synchronized with a double thymidine block and released for 2 h to enrich for S phase cells. Histone H3 was used as a loading control. Cohesin abundance was normalized to histone H3 and quantified relative to empty vector transfections (means ± SD are indicated). The abundance of chromatin-associated cohesin subunits is not significantly different between cells transfected with TEX19 and controls (t test; n = 3).
Figure 4.
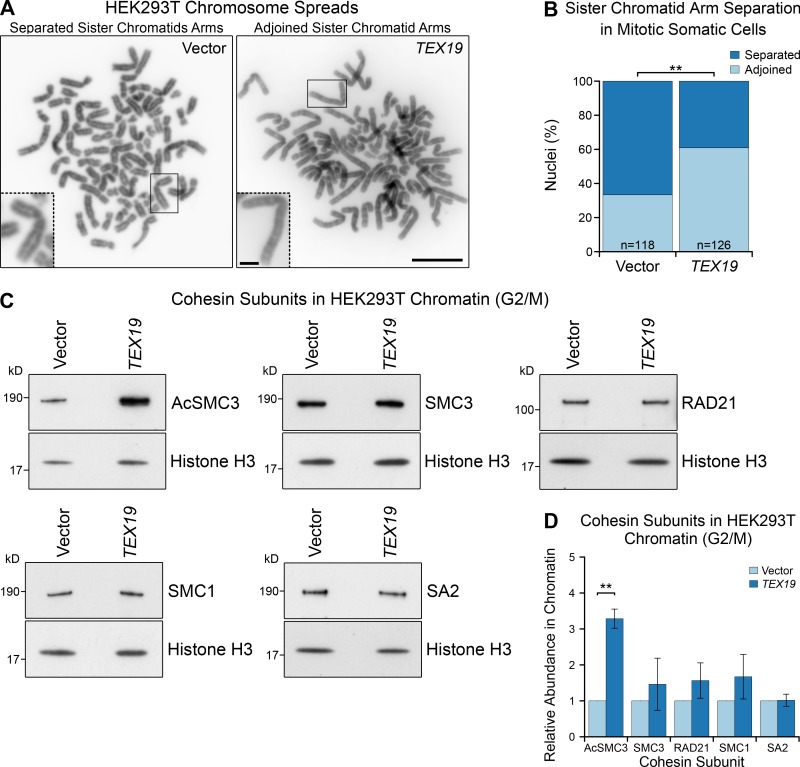
Ectopic expression of TEX19 promotes sister chromatid cohesion in mitotic somatic cells. (A and B) Photographs (A) and quantification (B) of sister chromatid separation in HEK293T cells. Chromosome spreads from HEK293T cells transfected with human TEX19 or empty vector were classed as having separated sister chromatids if most chromosomes had a visible gap between chromosome arms. Scale bar, 10 µm; inset scale bar, 2 µm. Quantification of four independent experiments; n indicates the total number of chromosome spreads. 33% (n = 126) of metaphase spreads from cells transfected with TEX19 had separated sister chromatids compared with 67% (n = 118) of controls (**, Fisher’s exact test, P < 0.01). (C and D) Representative Western blots (C) and quantification (D) of cohesin subunits in chromatin from HEK293T cells transfected with either human TEX19 or empty vector, synchronized to enrich for cells in G2/M. Cohesin abundance was normalized to histone H3 and quantified relative to empty vector transfections. Means ± SD are indicated. Expression of TEX19 induces a 3.3-fold increase in chromatin-associated AcSMC3 (**, t test, P < 0.01; n = 3). Each pair of histone H3 and cohesin bands are from the same gel lane; the high concentration of histones in chromatin causes the sample to spread laterally at low molecular weights (Fig. S5 C).
We next analyzed whether expression of TEX19 in HEK293T cells might affect cohesin. Surprisingly, the reduction in sister chromatid separation in these cells is not accompanied by a detectable statistically significant increase in any of the four core cohesin subunits (Fig. 4, C and D). This finding bears some resemblance to knocking down sororin, which also impairs sister chromatid cohesion without detectably altering the bulk population of cohesin associated with chromatin (Schmitz et al., 2007). Sororin regulates sister chromatid cohesion through protecting the small subpopulation of chromatin-associated cohesin that mediates sister chromatid cohesion (Schmitz et al., 2007; Zhang et al., 2008; Nishiyama et al., 2010, 2013; Ladurner et al., 2016). We therefore tested whether this cohesive subpopulation of cohesin, which is marked by acetylation of SMC3, might be regulated by TEX19. Interestingly, Western blots using anti-AcSMC3 antibodies (Nishiyama et al., 2010) showed that the abundance of AcSMC3 is elevated approximately threefold in G2/M chromatin in response to TEX19 expression (Fig. 4, C and D). This effect appears to be restricted to chromatin-associated AcSMC3, as TEX19 does not detectably change the total amount of AcSMC3 in these cells (Fig. S3, C and D). SMC3 is acetylated during S phase by ESCO1 (establishment of sister chromatid cohesion N-acetyltransferase 1) and/or ESCO2 to establish sister chromatid cohesion (Zhang et al., 2008; Minamino et al., 2015; Ladurner et al., 2016). In contrast to G2/M cells, expression of TEX19 does not detectably affect the amount of chromatin-associated AcSMC3 in cells enriched for S phase (Fig. S3, E and F), suggesting that TEX19 is not strongly influencing establishment of SMC3 acetylation (Zhang et al., 2008; Ladurner et al., 2016). Taken together, these data suggest that expression of TEX19 promotes maintenance of a chromatin-associated subpopulation of cohesin marked by AcSMC3 during G2/M phases of the cell cycle.
Mouse Tex19.1 and human TEX19 inhibit N-end rule degradation
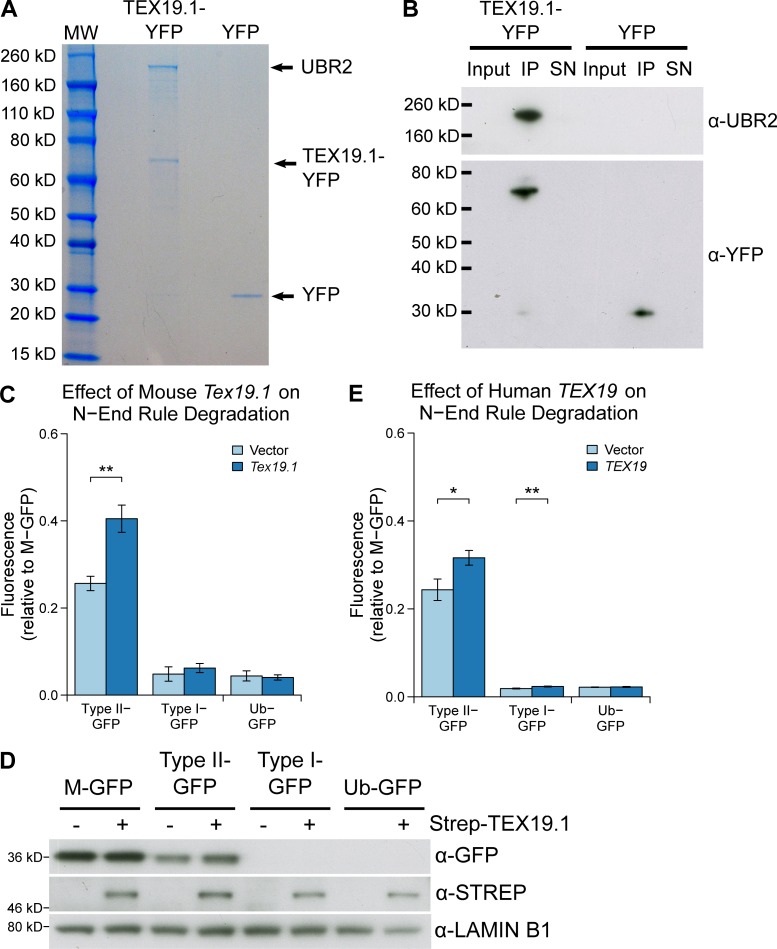
To investigate how Tex19.1/TEX19 genes might regulate AcSMC3 in this experimental context, we next identified proteins that interact with mouse TEX19.1 by performing mass spectrometry on TEX19.1-YFP protein complexes isolated from stable TEX19.1-YFP–expressing HEK293 cells. TEX19.1-YFP is present in anti-YFP immunoprecipitates in stoichiometric amounts with an ∼220-kD protein that was identified by mass spectrometry as the E3 ubiquitin ligase UBR2 (Fig. 5 A). UBR2 has previously been identified as coimmunoprecipitating with TEX19.1 from testes (Yang et al., 2010) and from embryonic stem cells (MacLennan et al., 2017), suggesting that this interaction reflects the behavior of endogenously expressed proteins. Western blotting confirmed that endogenous UBR2 is present in TEX19.1-YFP immunoprecipitates (Fig. 5 B). The stoichiometry of UBR2 and TEX19.1-YFP in these immunoprecipitates suggests that TEX19.1 could represent a regulatory subunit rather than a substrate of UBR2.
Figure 5.
TEX19.1 inhibits the activity of the E3 ubiquitin ligase UBR2 toward type II N-end rule substrates. (A) Colloidal blue-stained anti-YFP immunoprecipitates from cytoplasmic lysates of HEK293T cells stably expressing mouse TEX19.1-YFP or YFP alone. The ∼220-kD band coimmunoprecipitating stoichiometrically with TEX19.1-YFP was identified by mass spectrometry as UBR2 (44 matching peptides covering 25% of UBR2, probability of random match <10−25). (B) Anti-YFP immunoprecipitates as described for A, Western blotted for YFP and endogenous UBR2. (C and E) N-end rule reporters assays. Stable Flp-In-293 cell lines expressing different ubiquitin-GFP fusions were transiently transfected with mouse Tex19.1 or empty vector (C) or with human TEX19 or empty vector (E). GFP reporter fluorescence was assayed by flow cytometry relative to the M-GFP Flp-In-293 cell line. Mouse Tex19.1 increases stability of the type II N-end rule reporter by 58% (**, t test, P < 0.01; n = 3); human TEX19 increases stability of the type II N-end rule reporter by 30% (*, t test, P < 0.05; n = 3) and the type I N-end rule reporter by 24% (**, t test, P < 0.01; n = 3). Graphs indicate mean ± SD. (D) As in C, except STREP-tagged TEX19.1 expression plasmids were used, and GFP reporter stability was assayed by Western blotting against GFP using lamin B1 as a loading control.
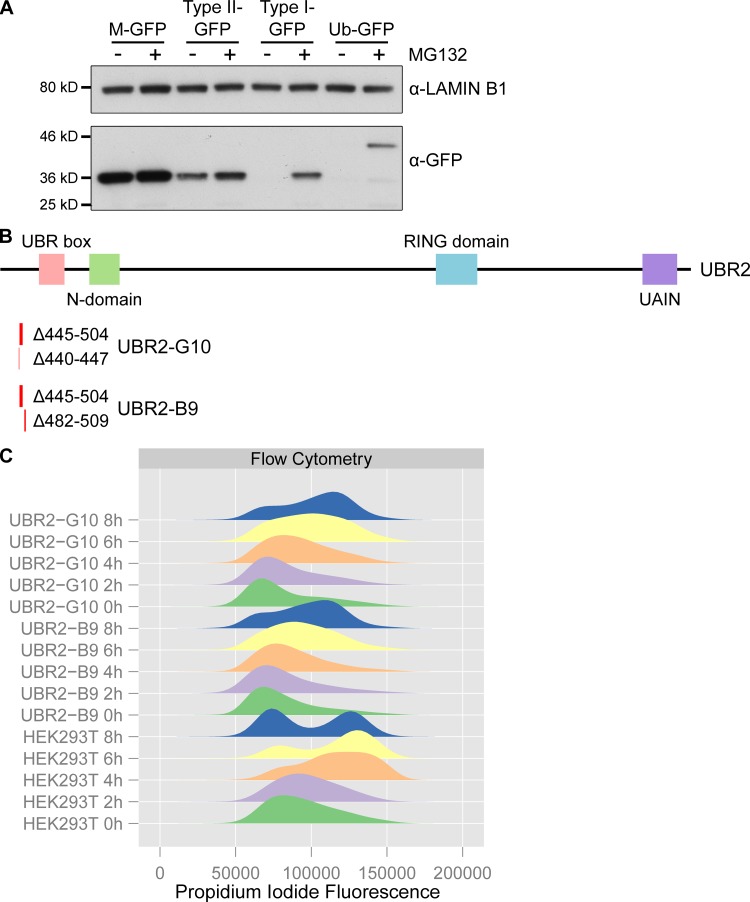
UBR2 is a ubiquitously expressed protein functioning in the N-end rule pathway that degrades proteins depending on their N-terminal amino acid. Proteins with a basic (type I) residue or a large hydrophobic (type II) residue at their N-terminus are degraded by the N-end rule pathway (Tasaki et al., 2005). We therefore used ubiquitin-GFP fusion proteins that are processed by ubiquitin hydrolyases to generate GFP moieties possessing N-terminal methionine (M-GFP), N-terminal arginine (R-GFP, type I), N-terminal leucine (L-GFP, type II), or a noncleavable ubiquitin fusion degradation signal control (Ub-GFP) to test the effect of Tex19.1 on the N-end rule pathway (Dantuma et al., 2000). We confirmed that the abundance of GFP in Flp-In-293 cell lines stably expressing these constructs from the same chromosomal locus was determined by its N-terminal amino acid and was sensitive to the proteasome inhibitor MG132 (Fig. 5, C and D; and Fig. S4 A). Transient transfection of mouse Tex19.1 into these N-end rule reporter cell lines resulted in a ∼50% increase in L-GFP fluorescence but did not affect the R-GFP reporter or the Ub-GFP control substrate (Fig. 5, C and D). This Tex19.1-dependent increase in L-GFP fluorescence represents increased abundance of L-GFP protein (Fig. 5 D). The specificity of Tex19.1 for type II rather than type I N-end rule substrates is consistent with UBR2 primarily binding to type II substrates in vivo (Tasaki et al., 2005). Taken together, these data indicate that TEX19.1 potentially assembles into a stable stoichiometric complex with UBR2 and inhibits UBR2 from targeting type II N-end rule substrates for ubiquitin-dependent degradation.
Figure S4.
Validation of N-end rule reporter and UBR2 mutant cell lines. (A) N-end rule GFP reporter cell lines are sensitive to proteasome inhibition. Ubiquitin fusion constructs that generate GFPs possessing N-end rule degrons (leucine for type II-GFP, arginine for type I-GFP) or methionine (M-GFP) at their N-termini, or a noncleavable Ub-GFP fusion construct, were stably integrated into Flp-In-293 cells. The abundance of the GFP reporters in these cell lines cultured in the presence or absence of the proteasome inhibitor MG132 was assessed by Western blotting using anti-GFP antibodies. Anti-lamin B1 antibodies were used as a loading control. (B) Location of UBR2 deletions recovered in compound heterozygous HEK293T clones generated by CRISPR/Cas9 genome editing. Locations and nucleotides deleted are indicated below a schematic showing the domain structure of UBR2 (Tasaki et al., 2005). Clone B9 has a 28-bp frameshift deletion (Δ482–509) in one UBR2 allele and a 60-bp in-frame deletion (Δ445–504) in another (numbering is relative to UBR2 cDNA NM_015255.2). Clone G10 has one allele with an 8-bp frameshift deletion (Δ440–447) in one UBR2 allele and the same 60 bp in-frame deletion (Δ445–504) present in clone B9 in the other. We did not recover any wild-type UBR2 sequences from these clones, or any CRISPR/Cas9-edited clones that only contained frameshift mutations in UBR2. (C) Clones B9 and G10 are slow growing and have altered cell cycle kinetics. Clones B9 and G10 exhibit cell death during a double thymidine block and release; therefore flow cytometry data represents a single thymidine block and release. Clones B9 and G10 have altered cell cycle distribution in the thymidine block (0 h) and are delayed in progression through to G2/M during the release by 2–4 h relative to the parental HEK293T cells.
The interaction between mouse TEX19.1 and UBR2 is conserved in human TEX19 (MacLennan et al., 2017). To test if inhibition of N-end rule degradation is also conserved in human TEX19, we expressed human TEX19 in the N-end rule reporter cell lines. Like mouse Tex19.1, human TEX19 increased abundance of L-GFP in this assay, but also had a minor effect on R-GFP abundance (Fig. 5 E). Therefore, the ability of mouse Tex19.1 to inhibit N-end rule degradation is conserved in human TEX19. Taken together, the data in this study and in MacLennan et al. (2017) suggest that mouse TEX19.1 and human TEX19 proteins function at least in part by altering the substrate specificity of UBR2 to direct it away from its endogenous N-end rule substrates and toward retrotransposon-encoded proteins.
Ubr2 negatively regulates levels of chromatin-associated AcSMC3
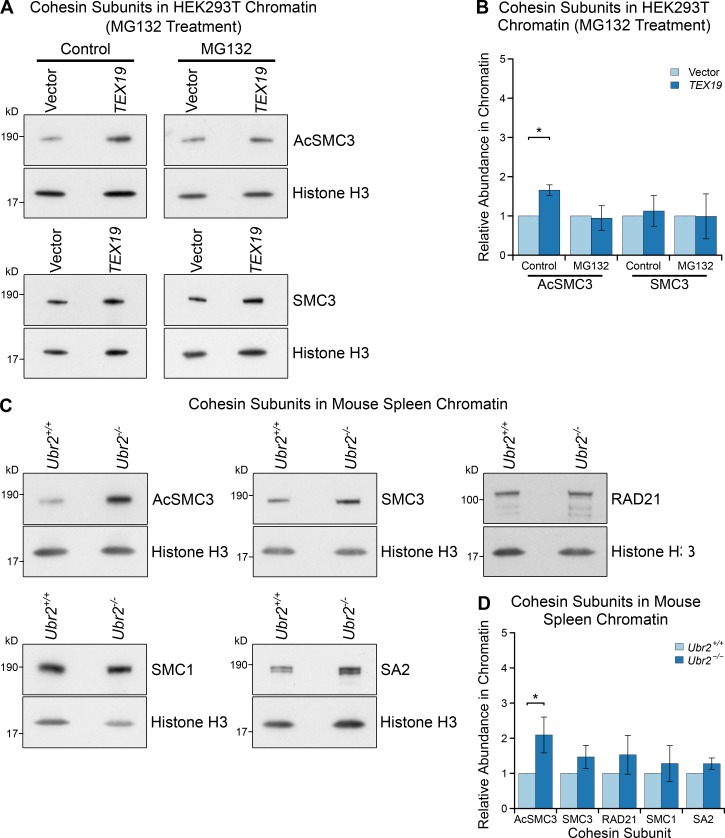
Mouse Tex19.1 and human TEX19 could function at least in part through inhibiting the activity of UBR2 toward some of its endogenous cellular substrates. We therefore tested whether TEX19 requires a functional proteasome to regulate chromatin-associated AcSMC3. HEK293T cells transiently transfected with TEX19 were treated with the proteasome inhibitor MG132, which arrests cells in M phase with high levels of sister chromatid cohesion (Nakajima et al., 2007). MG132 treatment abolishes the ability of TEX19 to increase the amount of chromatin-associated AcSMC3 (Fig. 6, A and B), suggesting that a functional proteasome is required for TEX19 to regulate AcSMC3 cohesin. It is possible that MG132 is abolishing the activity of TEX19 in this assay by arresting cells in a phase of the cell cycle where TEX19-dependent regulation of cohesin is not active; however, TEX19 does appear to regulate cohesin during the G2/M phases of the cell cycle rather than S phase (Fig. 4, C and D; and Fig. S3, E and F). Thus, the biochemical function that we have identified for TEX19 in regulating UBR2 and ubiquitin-dependent proteolysis could be mechanistically relevant for the regulation of AcSMC3.
Figure 6.
Proteasome-dependent and Ubr2-dependent pathways regulate AcSMC3-containing cohesin in mammalian somatic cells. (A and B) Representative Western blots (A) and quantification (B) determining the abundance of AcSMC3 and SMC3 cohesin subunits in chromatin from HEK293T cells treated with the proteasome inhibitor MG132. Cells were transfected with either TEX19 or empty vector before treatment with either MG132 or DMSO as a vehicle control. Cohesin abundance was normalized to histone H3 and quantified relative to empty vector transfections. Means ± SD are indicated. Expression of TEX19 induces a 1.6-fold increase in chromatin-associated AcSMC3 (*, t test, P < 0.05; n = 3), but this effect is abolished in the presence of MG132. (C and D) Representative Western blots (C) and quantification (D) from Ubr2+/+ and Ubr2−/− mice determining the abundance of cohesin subunits in spleen chromatin. Cohesin abundance was normalized to histone H3 and quantified relative to Ubr2+/+ mice. Means ± SD are indicated. Ubr2−/− spleens have a 2.1-fold increase in the amount of chromatin-associated AcSMC3 (*, t test, P < 0.05; n = 4).
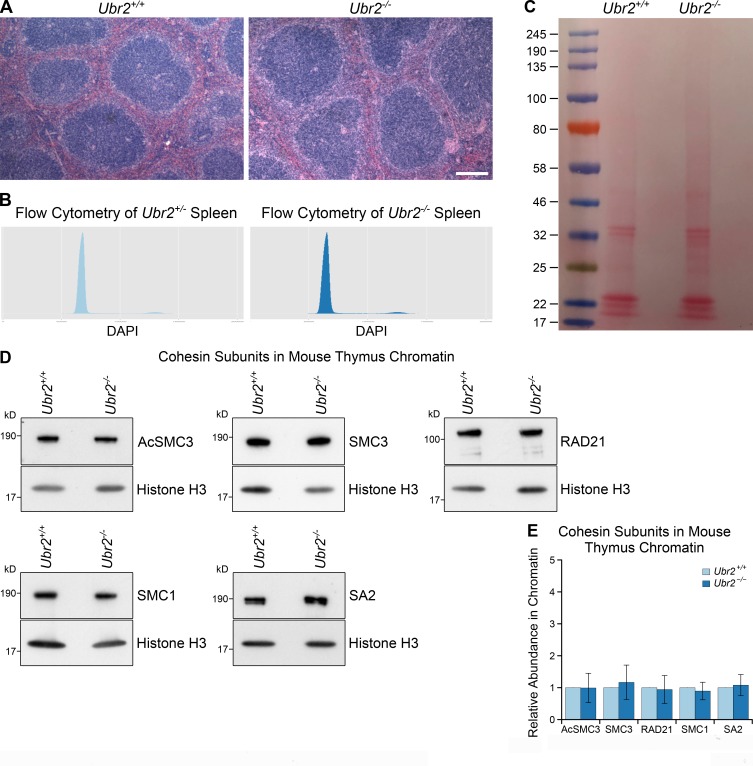
We next tested if UBR2 might have a previously undescribed role in regulating AcSMC3 that TEX19 genes could be modulating. Ubr2 is required to stabilize mouse TEX19.1 (Yang et al., 2010), and in Tex19.1-expressing germ cells, this function of Ubr2 has been difficult to dissect from its role in promoting protein ubiquitylation (Yang et al., 2010; Crichton et al., 2017). We therefore tried to investigate whether UBR2 has TEX19-independent roles in regulating AcSMC3 in HEK293T somatic cells. However, hypomorphic UBR2 mutant HEK293T cells generated by CRISPR/Cas9-mediated genome editing displayed abnormal proliferation and cell cycle kinetics that interfered with analyzing any effects on AcSMC3 in G2/M (Fig. S4, B and C). In contrast, although Ubr2−/− mutant mice exhibit female embryonic lethality and defects in spermatogenesis, somatic tissues are relatively unperturbed in adult Ubr2−/− males (Kwon et al., 2003). Therefore, we investigated whether rapidly proliferating adult mouse tissues might have altered AcSMC3 levels in Ubr2−/− adult male mice. Histology and flow cytometry of adult mouse spleen showed no obvious differences in cell composition or cell cycle distribution in the absence of Ubr2 (Fig. S5, A and B). However, the amount of chromatin-associated AcSMC3 was approximately twofold higher in spleen from Ubr2−/− mice relative to controls (Fig. 6, C and D; and Fig. S5 C). This effect was primarily restricted to AcSMC3: other cohesin subunits were not affected by loss of Ubr2 (Fig. 6, C and D). Thus, loss of Ubr2 in this proliferating somatic tissue has a similar effect on chromatin-associated AcSMC3 as ectopically expressing TEX19 in cultured HEK293T cells. The amount of chromatin-associated AcSMC3 is not detectably affected by loss of Ubr2 in the thymus (Fig. S5, D and E), suggesting that the relative contribution of Ubr2 to AcSMC3 regulation varies in different tissues, potentially reflecting redundancy between different UBR proteins (Tasaki et al., 2005). Nevertheless, Ubr2 appears to have a previously undescribed role in regulating AcSMC3 cohesin in somatic tissues, and it is possible that the reduced sister chromatid cohesion seen in Tex19.1−/− oocytes represents defects in inhibiting the activity of UBR proteins toward AcSMC3 in the germline.
Figure S5.
Additional data from Ubr2−/− mice. (A) Hematoxylin and eosin–stained paraffin sections of Ubr2+/+ and Ubr2−/− spleen. Loss of Ubr2 does not dramatically alter the histological appearance or cell type composition of the spleen. Scale bar, 100 µm. (B) Flow cytometry of Ubr2+/− and Ubr2−/− spleens. Loss of Ubr2 does not grossly perturb the cell cycle distribution in this tissue. (C) Ponceau S stained membrane of chromatin preparations from Ubr2+/+ and Ubr2−/− spleen before Western blotting. Chromatin preparations have high concentrations of histone proteins, which cause the gel lanes to broaden in the low molecular weight (15–25-kD) region. This phenomenon means that some bands visualized with anti-histone H3 antibodies have different widths from bands visualized with antibodies against higher molecular weight cohesins (e.g., anti-SMC3, anti-acetylated SMC3) in this study, despite originating from the same membrane. (D and E) Representative Western blots (D) and quantification (E) from three pairs of Ubr2+/+ and Ubr2−/− mice for the abundance of cohesin subunits (AcSMC3, SMC3, RAD21, SMC1, SA2) in thymus chromatin. Histone H3 was used as a loading control. Cohesin abundance was normalized to histone H3 and quantified relative to Ubr2+/+ mice (graph indicates mean ± SD). The abundance of cohesin subunits in the thymus of Ubr2−/− mice was not significantly different from Ubr2+/+ controls (t test; n = 4).
Tex19.1−/− oocytes have reduced levels of chromosome-associated acetylated SMC3 cohesin
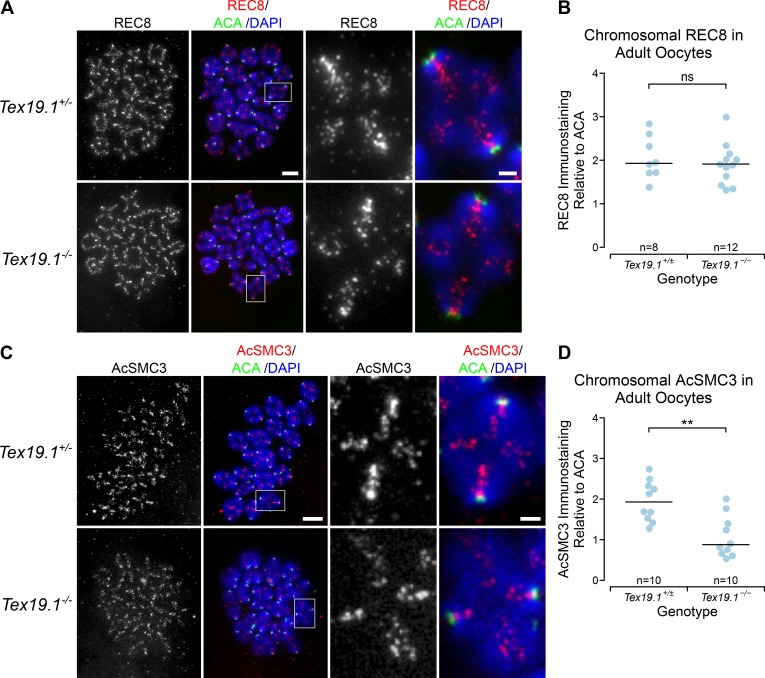
We next tested whether the defects in arm cohesion seen in postnatal Tex19.1−/− oocytes reflect reduced levels of chromatin-associated AcSMC3 in these cells. Although the existence and cohesive function of an AcSMC3-marked subpopulation of cohesin is well established in mitotic cells, it is not clear whether this subpopulation of cohesin exists in meiotic oocyte chromosomes. We performed immunostaining for REC8, a meiotic kleisin subunit of cohesin, and for AcSMC3 in prometaphase I chromosomes from Tex19.1+/± and Tex19.1−/− oocytes. As previously reported (Lister et al., 2010), anti-REC8 staining is primarily located on chromosome axes between sister chromatids in prometaphase I oocyte chromosomes from control mice (Fig. 7 A). However, we could not detect any change in the abundance or distribution of anti-REC8 immunostaining in chromosomes from Tex19.1−/− oocytes (Fig. 7, A and B). This finding is consistent with there being no detectable effect of TEX19 expression on the amount of the RAD21 mitotic kleisin subunit in HEK293T cells (Fig. 4, C and D). Moreover, analogous to mitotic cells (Schmitz et al., 2007), this finding suggests that arm cohesion in meiotic oocyte chromosomes is mediated by a small subpopulation of cohesin.
Figure 7.
Tex19.1−/− oocytes have reduced levels of chromatin-associated AcSMC3 cohesin. (A and C) Tex19.1+/± and Tex19.1−/− prometaphase I chromosome spreads immunostained with anti-centromere antibodies (ACA; green), DAPI (blue), and either anti-REC8 (A, red) or anti-AcSMC3 (C, red) antibodies to visualize cohesin subunits. Example individual bivalents (boxes) are magnified and shown in the righthand panels. Single-channel images of anti-AcSMC3 and anti-REC8 are also shown in grayscale. Scale bars 10 µm; inset scale bars 2 µm. (B and D) Quantification of anti-REC8 (B) and anti-AcSMC3 (D) immunostaining in prometaphase I oocyte chromosomes. Individual bivalents were distinguished by DAPI staining, and total cohesin immunostaining on each bivalent was measured relative to ACA. The median of the ratios for each oocyte is plotted; horizontal lines indicate median ratios for each genotype. REC8 immunostaining is not significantly different in Tex19.1−/− oocytes (ns, Mann-Whitney U test, not significantly different; n = 8, 12), but AcSMC3 is significantly reduced to 45% of the level detected in Tex19.1+/± controls (**, Mann-Whitney U test, P < 0.01; n = 10, 10). Data are from seven Tex19.1+/± and five Tex19.1−/− females for REC8 and from four Tex19.1+/± and four Tex19.1−/− females for AcSMC3.
Consistent with immunostaining for other cohesin subunits (Hodges et al., 2005; Chiang et al., 2010; Lister et al., 2010), anti-AcSMC3 immunostaining is located along chromosome axes between sister chromatids in prometaphase I oocyte chromosomes (Fig. 7 C). However, in contrast to REC8, anti-AcSMC3 immunostaining showed a significant, approximately twofold reduction in prometaphase I chromosomes isolated from Tex19.1−/− oocytes (Fig. 7, C and D). Thus, loss of Tex19.1 primarily affects a specific subpopulation of cohesin marked by AcSMC3. Furthermore, the reduced arm cohesion in Tex19.1−/− oocytes correlates better with anti-AcSMC3 than bulk anti-REC8 immunostaining, suggesting that, as in mitotic cells, arm cohesion is mediated by an AcSMC3-marked subpopulation of cohesin in meiotic oocytes. Together, the phenotypic analyses in this study suggest that Tex19.1 plays a role in maintaining this AcSMC3-marked subpopulation of cohesin and arm cohesion in postnatal mouse oocytes to prevent aneuploidy from arising in the female germline.
Discussion
Maintenance of cohesin in postnatal oocytes
The data in this study (a) suggest that postnatal mouse oocytes maintain sister chromatid cohesion and prevent aneuploidy through a mechanism that depends on Tex19.1 and (b) implicate a subpopulation of cohesin marked by AcSMC3 in this process. Meiotic sister chromatid cohesion is established during fetal development in females, then maintained postnatally in the absence of detectable de novo incorporation of REC8 protein molecules (Revenkova et al., 2010; Tachibana-Konwalski et al., 2010; Burkhardt et al., 2016). It is not clear whether AcSMC3 behaves similarly to REC8 in being established in fetal oocytes and then maintained postnatally without detectable renewal or replacement. However, in mitotic cells, ESCO1 can acetylate SMC3 independently of DNA replication (Minamino et al., 2015), and it is possible that AcSMC3 is more dynamic than REC8 in postnatal oocytes. Further work is needed to assess AcSMC3 dynamics in meiotic oocytes, and how this might be affected by loss of Tex19.1.
Age-dependent loss of cohesion and chromosome-associated REC8 from chromosome arms and centromeres potentially contributes to age-dependent aneuploidy in mouse oocytes (Chiang et al., 2010; Lister et al., 2010). Although Tex19.1 expression declines in aging GV-stage oocytes (Pan et al., 2008), age-dependent changes in Tex19.1 activity would not fully explain the age-dependent loss of cohesion in aging mouse oocytes, as loss of Tex19.1 had no detectable effect on sister centromere separation at prometaphase I. While we cannot exclude that there are defects at meiosis I centromeres or kinetochores that we have not detected in Tex19.1−/− oocytes, other factors are presumably involved in mediating the changes in sister centromere separation seen in aging oocytes. Furthermore, loss of Tex19.1 affects the AcSMC3-marked subpopulation of cohesin rather than the bulk chromosome-associated REC8 that changes in aging oocytes. The differential behavior of AcSMC3 and REC8 in Tex19.1−/− oocytes could reflect deacetylation of AcSMC3 and chromosome-associated REC8-containing cohesin becoming noncohesive in these cells. Or, as in mitotic cells, perhaps only a small proportion of all the cohesin associated with chromosomes is functionally cohesive, and this population is marked by AcSMC3 (Schmitz et al., 2007; Deardorff et al., 2012). There are similarities between the Tex19.1−/− meiotic oocyte phenotype and the effects of depleting sororin, which binds to AcSMC3, in mitotic cells (Schmitz et al., 2007). The Tex19.1−/− phenotypic data suggest that, at least in some situations, chromosome-associated AcSMC3 might be more closely linked to sister chromatid cohesion in meiosis than chromosome-associated REC8. It will be of interest to determine the effect of oocyte aging on AcSMC3 in meiotic chromosomes and to evaluate the contribution of human TEX19 to aging in human oocytes (Sakakibara et al., 2015; Zielinska et al., 2015; Ottolini et al., 2015; Gruhn et al., 2019).
Roles for Tex19.1 and Ubr2 in regulating AcSMC3
The data presented in this study suggests that Tex19.1 and Ubr2 have previously uncharacterized roles in regulating AcSMC3-marked cohesin (Fig. 8). Tex19.1 has not been previously linked to cohesin regulation, but the budding yeast orthologue of Ubr2, UBR1, stimulates degradation of the C-terminal fragment of Rad21 generated by separase cleavage during mitosis (Rao et al., 2001). However, degradation of analogous separase cleavage fragments in mitotic cells in mammals may not be occurring in the same way (Liu et al., 2016), and the effects of Tex19.1 and Ubr2 that we describe here appear to preferentially affect AcSMC3-containing cohesin. It is possible that Ubr2, and its orthologues, might regulate cohesin in multiple ways and that different mechanisms of regulation might be differentially important in different organisms and/or cell types.
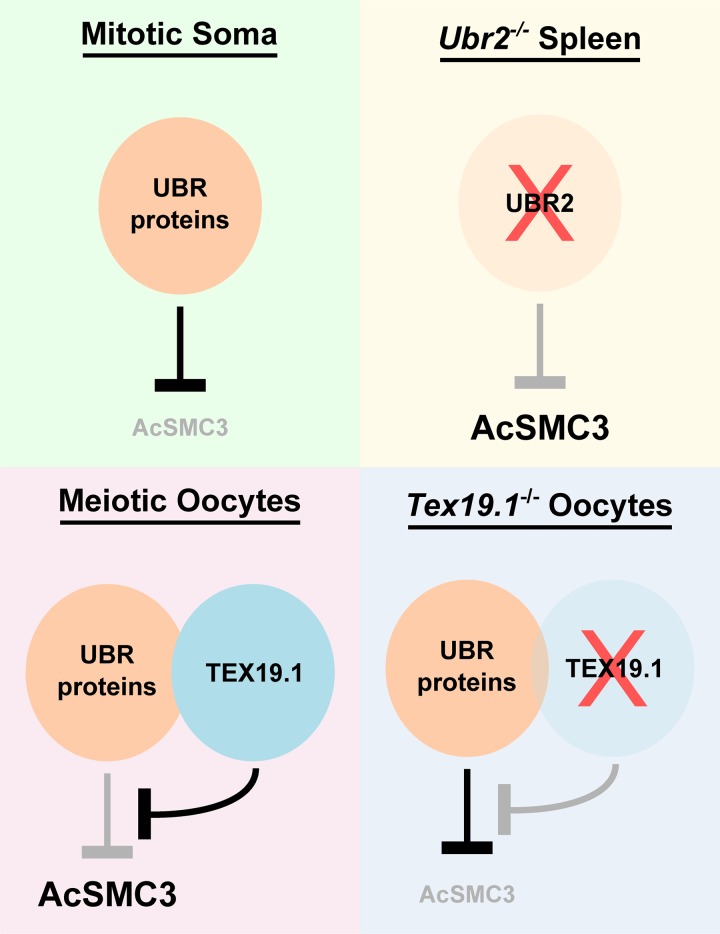
Figure 8.
Regulation of AcSMC3 By TEX19.1 and UBR proteins. Model outlining how TEX19.1 and UBR proteins might influence chromosome-associated AcSMC3. In mitotic somatic cells, UBR proteins such as UBR2 negatively regulate AcSMC3 directly or indirectly. In Ubr2−/− spleen, this negative regulation is reduced and AcSMC3 abundance increases. Expression of Tex19.1, for example, when ectopically expressed in HEK293T cells or endogenously expressed in meiotic oocytes, results in the formation of TEX19.1-UBR protein complexes (e.g., TEX19.1-UBR2) that inhibit the activity of UBR proteins toward some substrates (e.g., N-end rule substrates) and result in increased AcSMC3 abundance. In Tex19.1−/− oocytes, the inhibitory effect of TEX19.1 on UBR proteins is lost, which could allow negative regulation of AcSMC3 by UBR proteins to contribute to the reduced levels of AcSMC3 in prometaphase I Tex19.1−/− oocyte chromosomes. Although Tex19.1 appears to function through Ubr2 in mouse spermatocytes (Yang et al., 2010; Crichton et al., 2017), redundancy between UBR proteins for stabilizing TEX19.1 and/or regulating AcSMC3 in other cell types cannot be excluded.
Although we have shown that Ubr2 negatively regulates AcSMC3 in mouse spleen, the substrates and pathways involved are currently not clear. UBR2 could directly target AcSMC3-marked cohesin for ubiquitin-dependent proteolysis through a potential Met-Φ motif (Kim et al., 2014) at the N-terminus of SMC3. Alternatively, UBR2 could regulate the AcSMC3-marked population of cohesin indirectly through regulating sororin, which protects AcSMC3-containing cohesin from wings apart-like (WAPL)-dependent removal (Nishiyama et al., 2010), through regulating the acetylation or deacetylation of SMC3 (Zhang et al., 2008; Deardorff et al., 2012; Minamino et al., 2015), or through regulating other cohesin subunits in the AcSMC3-containing cohesin complexes. Although regulation of AcSMC3 by UBR proteins is unlikely to be a major pathway regulating removal of sister chromatid cohesion during mitosis, oocytes experience a prolonged dictyate arrest and spend much longer in prometaphase than mitotic cells. It might therefore be important to regulate this pathway in meiotic oocytes where there could be sufficient time for gradual UBR protein-dependent depletion of cohesin to reach physiologically critical levels.
Reconciling cohesin and retrotransposon regulatory functions for Tex19.1
Tex19.1 regulates retrotransposons at multiple levels in vivo (Öllinger et al., 2008; Yang et al., 2010; Reichmann et al., 2012, 2013; Tarabay et al., 2013; MacLennan et al., 2017). In testes, Tex19.1 represses MMERVK10C retrotransposons transcriptionally (Öllinger et al., 2008; Yang et al., 2010; Crichton et al., 2017), loss of Tex19.1 de-represses multiple retrotransposon RNAs in placenta (Reichmann et al., 2013; Tarabay et al., 2013), and TEX19.1/TEX19 proteins interact with UBR2 to promote degradation of LINE-1 retrotransposon proteins in multiple cell types (Yang et al., 2010; MacLennan et al., 2017). TEX19.1 has also been reported to interact with components of the piRNA pathway present in testes (Tarabay et al., 2017). We show in this study that both mouse Tex19.1 and human TEX19 inhibit degradation of some N-end rule substrates and regulate AcSMC3-containing cohesin. As Ubr2 can regulate AcSMC3 independently of Tex19.1 in somatic cells, the cohesin-regulatory function of Tex19.1/TEX19 genes likely relates to these proteins physically interacting with UBR2 (Yang et al., 2010; MacLennan et al., 2017). Budding yeast Ubr1 has multiple substrate binding sites, and peptides binding to the N-end rule binding site in Ubr1 inhibit other substrates binding to Ubr1 via internal degrons (Du et al., 2002; Xia et al., 2008). It is possible that mammalian TEX19 proteins effect, at least in part, a similar mechanism on UBR2 to promote binding and ubiquitylation of some substrates (e.g., LINE-1 ORF1p, internal degron) at the expense of others (e.g., type II N-end rule substrates).
Retrotransposons evolve rapidly between mammalian species (Crichton et al., 2014); therefore, the parts of TEX19 that physically interact with retrotransposons will also need to evolve rapidly to conserve this mechanism (van Valen, 1973). In contrast, parts of TEX19 that interact with the UBR2 are more conserved (Yang et al., 2010). Mutating mammalian TEX19 genes could therefore potentially result in two distinct sets of consequences. One set could result from increased abundance and activity of retrotransposon proteins, which could be relatively species specific but similar between TEX19 and UBR2 mutants, and one set could result from deregulation of UBR2 toward N-end rule or other nonretrotransposon substrates, which might be more conserved but potentially occurring in opposite directions in TEX19 and UBR2 mutants. Regulation of AcSMC3 by mouse Tex19.1/Ubr2 appears to belong to the latter category. The reduced levels of AcSMC3 in Tex19.1−/− postnatal oocytes could potentially reflect increased activity of UBR2 toward nonretrotransposon substrates in these cells. However, there is redundancy between UBR proteins in the N-end rule pathway (Tasaki et al., 2005), and peptides for two additional N-end rule UBR proteins, UBR4 and UBR5, are also present in TEX19.1-YFP immunoprecipitates from mouse embryonic stem cells (MacLennan et al., 2017). It would be of interest to determine the contribution of different UBR proteins to the regulation of AcSMC3 and sister chromatid cohesion in postnatal oocytes, and whether these proteins have a role in age-dependent oocyte aneuploidy.
Materials and methods
Mice
Tex19.1−/− mice on a C57BL/6 genetic background were bred from heterozygous crosses and genotyped using primers 5′-CTTCAGGAGGTCTGATGCCCTCT-3′ and 5′-GAGTGTTGTGTGGTGGGTGTTATGG-3′ to detect the wild-type allele and 5′-CACCGCCTGTGCTCTAGTAGCTT-3′ and 5′-CTTCAGGAGGTCTGATGCCCTCT-3′ to detect the mutant allele (Öllinger et al., 2008). The entire Tex19.1 gene is replaced by a neomycin resistance cassette in these mice. For embryonic stages, the day the vaginal plug was found was designated E0.5. Tex19.1−/− females were analyzed at 6–14 wk old alongside either Tex19.1+/+ or Tex19.1+/− age-matched control animals from the same breeding colony. Tex19.1+/− control females have normal fertility, and data from these and Tex19.1+/+ females were combined as Tex19.1+/± controls. Ubr2−/− mice generated by CRISPR/Cas9 double nickase-mediated genome editing in zygotes were genotyped using primers 5′-TCTGAGGTTGCAAGAGAATGT-3′ and 5′-GGCCACAGATCAGCTAAACC-3′, followed by restriction digestion to detect the XbaI site incorporated into the mutant allele (Crichton et al., 2017). These Ubr2−/− mice carry a premature stop codon at cysteine-121 within the UBR domain of UBR2 (Uniprot Q6WKZ8-1). Heterozygous founder pups were backcrossed to C57BL/6 and then interbred. Ubr2−/− mice were phenotypically grossly normal except for small testes and an almost complete absence of epididymal sperm, as previously reported (Kwon et al., 2003). Animal experiments were performed under UK Home Office Project Licenses PPL 60/4424 and P93007F29 in accordance with local ethical guidelines.
Oocyte collection, culture, and imaging
For hormone injections, mice were injected intraperitoneally with 5 IU pregnant mare serum, followed by 5 IU human chorionic gonadotrophin (hCG) 46–48 h later (Nagy et al., 2003). For meiosis I, GV-stage oocytes were isolated from ovaries 42 h after pregnant mare serum injection by pricking with a needle in M2 (Sigma-Aldrich), separated from cumulus cells by pipetting, then cultured in M16 (Sigma-Aldrich) at 37°C in 5% CO2. Oocytes that underwent GVBD within 2 h were cultured for an additional 3 or 5 h to obtain prometaphase I oocytes. Ovulated metaphase II oocytes (16–18 h after hCG injection) and zygotes were recovered from the oviduct in flushing-holding medium (FHM; Millipore) and separated from cumulus cells by treating with 0.5 mg/ml hyaluronidase in FHM for 2–5 min (Nagy et al., 2003). Metaphase II oocytes were parthenogenetically activated by culturing in potassium-supplemented simplex optimized medium (KSOM; Millipore) containing 5 mM SrCl2 and 2 mM EGTA at 37°C in 5% CO2 for 2 h (Kishigami and Wakayama, 2007). For chromosome spreads, zygotes were cultured overnight in KSOM containing 0.1 µg/ml colcemid (Life Technologies) at 37°C in 5% CO2.
Fluorescence imaging was performed using an Orca AG charge-coupled device (CCD; Hamamatsu Photonics) or Coolsnap HQ2 CCD (Photometrics) camera, Zeiss Axioplan II fluorescence microscope with Plan-neofluar objectives, a 100-W Hg source (Carl Zeiss), and either a Chroma #83000 triple bandpass filter set (Chroma Technology) with the excitation filters installed in a motorized filter wheel (Prior Scientific Instruments) or Chroma #89014ET three-color or Chroma #89000ET four-color filter set with excitation and emission filters installed in motorized filter wheels. Image capture was performed using iVision or IPLab software (BioVision Technologies). Three-color RGB images were constructed from single-channel images using ImageJ (National Institutes of Health; Schindelin et al., 2012) or GIMP (https://www.gimp.org/). For two-color experiments that used DAPI to visualize DNA, 50% of the DAPI image was passed to the green channel and 100% to the blue channel to aid visualization, with the experimental image passed to the red channel.
For live imaging, GV-stage oocytes were maintained in M16 containing 100 µM 3-isobutyl-1-methylxanthine at 37°C for 2 h during transportation between Edinburgh and Newcastle, then microinjected using a pressure injector (Narishige) on a Nikon Diaphot microscope. Oocytes were microinjected with mRNA encoding Histone H2-RFP, placed in G-IVF culture medium (Vitrolife), and imaged for 14–20 h on a Nikon Ti inverted microscope fitted with a stage-mounted incubator at 37°C in 7% CO2. Bright-field and fluorescence images were acquired every 20 min on five 0.75-µm planes using a Photometrics CoolSnapHQ interline cooled CCD camera (Roper Scientific). Hardware control was performed using MetaMorph (Molecular Devices), and images were analyzed and processed using Fiji software (Schindelin et al., 2012) using maximum-intensity projections. Only oocytes in which chromosomes remained in the imaging plane throughout nuclear division were used for analysis of lagging chromosomes.
Chromosome spreads
Chromosome spreads were performed by incubating zygotes or postnatal oocytes in 1% trisodium citrate for 15–20 min; the individual zygotes or oocytes were transferred to a 50–100 µl drop of 3:1 methanol:acetic acid and allowed to dry (Yuan et al., 2002). Slides were mounted in Vectashield hard set mounting medium containing DAPI (Vector Labs). FISH for major satellite DNA was performed on methanol:acetic acid–fixed chromosome spreads as described previously (Boyle et al., 2001). Briefly, chromosome spreads were treated with 100 µg/ml RNaseA in 2× SSC for 1 h at 37°C, dehydrated through an ethanol series, denatured in 70% formamide and 2× SSC at 70°C for 75 s, dehydrated through an ethanol series again, and air dried. 100 ng biotin-labeled mouse major satellite FISH probe prepared by nick translation using bio-16-dUTP (Roche), DNaseI (Roche), and DNA polymerase I (Invitrogen) according to manufacturer’s instructions was resuspended in hybridization mix (50% formamide, 4× SSC, and 10% dextran sulfate), denatured at 70°C for 5 min, preannealed at 37°C for 15 min, and then hybridized to prewarmed slides overnight at 37°C. Slides were washed four times for 3 min each in 2× SSC at 45°C, four times for 3 min each in 0.1× SSC at 60°C, and then in 4× SSC and 0.1% Tween-20 at room temperature. Slides were incubated in blocking buffer (4× SSC and 5% nonfat skimmed milk powder) for 5 min at room temperature, incubated with 2 µg/ml FITC-conjugated avidin (Vector Laboratories) for 60 min at 37°C, and washed three times in 4× SSC and 0.1% Tween for 2 min each. Slides were stained with 5 µg/ml biotinylated anti-avidin (Vector Laboratories) then with 2 µg/ml FITC-conjugated avidin (Vector Laboratories) as described for the first round of FITC-conjugated avidin staining. Slides were then incubated in 0.5 µg/ml DAPI in PBS for 3 min, washed, and mounted underneath a glass coverslip with Vectashield hard set mounting medium containing DAPI (Vector Laboratories).
Preparation and analysis of pachytene spreads from E18.5 oocytes were performed as described (Bolcun-Filas et al., 2009). Two fetal ovaries were incubated for 15–30 min at room temperature in hypotonic extraction buffer (30 mM Tris, 50 mM sucrose, 17 mM trisodium citrate dihydrate, 5 mM EDTA, 0.5 mM DTT, and 0.5 mM PMSF, pH 8.2), before transferring to 20 µl of 100 mM sucrose and repeatedly piercing with a needle to release cells from the tissue. 10 µl of the cell suspension was applied to a glass microscope slide previously dipped in fixative (1% PFA and 0.15% Triton-X-100, pH 9.2), incubated in a humid chamber overnight, then air dried. Slides were washed in PBS; blocked for 1 h with PBS containing 0.15% BSA, 0.1% Tween-20, and 5% goat or donkey serum; and incubated with primary antibodies diluted in block solution in a humid chamber for up to 3 h at room temperature or overnight at 4°C. Slides were washed three times with PBS, incubated with secondary antibodies diluted in block solution, washed a further three times with PBS, and mounted under a glass coverslip with 90% glycerol, 10% PBS, and 0.1% p-phenylenediamine. To assess the position of MLH1 foci, centromeric ends of chromosome axes were identified by DAPI-dense pericentromeric heterochromatin. Primary antibodies were 1:200 mouse anti-SYCP3 (Santa Cruz, sc-74569), 1:250 rabbit anti-SYCP1 (Abcam, ab15090), 1:50 mouse anti-MLH1 (BD PharMingen, 51-1327GR), and 1:500 rabbit anti-SYCP3 (LSBio, LS-B175). Texas Red– and FITC-conjugated secondary antibodies (Jackson ImmunoResearch) and Alexa Fluor–conjugated secondary antibodies (Invitrogen) were used at 1:500, and DAPI (Sigma-Aldrich) was used at 0.02 µg/ml.
Immunostaining on prometaphase I oocyte chromosomes was performed as described (Susiarjo et al., 2009), with minor modifications. Zona pellucidae were removed from oocytes by incubating in a 50–100 µl drop of acid Tyrode’s for 5–10 s until the zona pellucidae were no longer visible, then washed through six 20–50 µl drops of M2 culture medium (Sigma-Aldrich) and incubated for 2 min in a 50–100 µl drop of 0.5% trisodium citrate. Individual oocytes were transferred to a glass slide dipped in fixative (1% PFA, 0.15% Triton-X-100, and 3 mM DTT, pH 9.2), and the slides were incubated in a humid chamber overnight then allowed to air dry. Slides were washed with 0.1% Tween-20 before blocking and incubation with antibodies as described for fetal ovary spreads. Primary antibodies were 1:50 human anti-centromere antibodies (Antibodies Inc.), 1:50 affinity-purified guinea pig anti-REC8 (Kouznetsova et al., 2005), and 1 µg/ml mouse anti-acetylated SMC3 (Nishiyama et al., 2010). Images from chromosome spreads were scored blindly for aneuploidy, chiasmata frequency, and chiasmata position by coding filenames via computer script before scoring. Immunostaining was quantified using Fiji (Schindelin et al., 2012). The immunofluorescence signal above background was measured within each bivalent’s DAPI area for each antibody, and the ratio of anti-cohesin:anti-centromere antibody staining was calculated for each bivalent. The median bivalent ratio was then determined for each oocyte (≥16 bivalents per oocyte). Sister centromere separation in metaphase II was measured from anti-centromeric antibody–stained chromosome spreads imaged in three dimensions using an Axioplan II fluorescence microscope (Zeiss) fitted with a piezoelectrically driven objective mount and deconvolved with Volocity (PerkinElmer).
Analysis of cohesin and cohesion in HEK293T cells
HEK293-derived cells were grown in DMEM (Invitrogen) containing 10% FCS, 2 mM l-glutamine, and 1% penicillin-streptomycin at 37°C in 5% CO2. HEK293T cells were transfected with pCMV-TEX19 vector expressing human TEX19 or empty pCMV vector using Lipofectamine 2000 (Invitrogen). To synchronize cells with a double thymidine block, HEK293T cells 8 h after transfection were incubated in medium containing 1.25 mM thymidine for 16 h, in fresh medium for 8 h, and then in medium containing 1.25 mM thymidine for 16 h. Cells were then released into fresh medium for 0, 2, or 4 h to obtain populations enriched for cells in G1/S, S, or G2/M phases of the cell cycle, respectively. Cells were fixed for flow cytometry in ice-cold 70% ethanol, and then incubated in 50 µg/ml propidium iodide and 100 µg/ml RnaseA in PBS for 1 h. DNA content was analyzed using a BD LSRFortessa flow cytometer (BD Biosciences). Chromatin was isolated from HEK293T cells as described (Méndez and Stillman, 2000) by washing cells in PBS then resuspending in buffer A (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, 1 mM DTT, 5 mM sodium butyrate, and complete protease inhibitors [Roche]). Triton X-100 was then added to a final concentration of 0.1%, and cells were lysed on ice for 5 min. Nuclei were pelleted (1,300 g, 4 min, 4°C), washed once in buffer A, and lysed in buffer B (3 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 5 mM sodium butyrate, and complete protease inhibitors) on ice for 30 min. Chromatin was pelleted (1,700 g, 4 min, 4°C), washed once in buffer B, resuspended in Laemmli sample buffer (Sigma-Aldrich), sonicated for 20 cycles (30 s on, 30 s off), and boiled for 5 min before analysis by SDS-PAGE. Chromatin was analyzed by SDS-PAGE/Western blotting using the following primary antibodies: 1:1,000 mouse anti-AcSmc3 (Nishiyama et al., 2010), 1:1,000 rabbit anti-Smc3 (Abcam, ab128919), 1:1,000 rabbit anti-Smc1 (Abcam, ab9262), 1:500 rabbit anti-SA2 (Abcam, ab155081), 1:100 rabbit anti-Rad21 (Abcam, ab992), and 1:25,000 rabbit anti-histone H3 (Abcam, ab1791). HRP-conjugated goat anti-mouse (Bio-Rad) and goat anti-rabbit (NEB) secondary antibodies were used at 1:5,000 and developed using Supersignal West Pico Chemiluminescent Substrate (Invitrogen). Membranes were cut before antibody incubation to allow anti-cohesin and anti-histone H3 signals for a sample to be quantified from the same membrane (Fig. S5 C). X-ray films were scanned, and ImageJ (Schindelin et al., 2012) was used to determine the density of signal in specific bands over background.
For MG132 experiments, transfected HEK293T cells were treated 8 h after transfection with 20 mM MG132 or DMSO for 18 h, and chromatin was isolated for analysis by Western blotting. 1:2,000 rabbit anti-TEX19 (Abcam, ab185507) was used to confirm expression of human TEX19 in transfected cells. Chromosome spreads from HEK293T cells were prepared by resuspending cells in hypotonic solution (0.5% sodium citrate and 0.56% potassium chloride) and fixing in 3:1 methanol:acetic acid, and images were scored blindly for cohesion between chromosome arms after coding filenames by computer script. Two different slides were scored for each condition in each experiment to assess any variation between slides originating from the same chromosome preparation.
CRISPR/Cas9 gene editing
UBR2 mutant HEK293T cells were generated by annealing guide RNAs targeting exon 2 of UBR2 (5′-CACCGGGGACCCCTGCAGTAGATTT-3′ and 5′-AAACAAATCTACTGCAGGGGTCCCC-3′; 5′-CACCGGCTGGCACAGCATGTTTTGT-3′ and 5′-AAACACAAAACATGCTGTGCCAGCC-3′) and then cloning them into plasmid PX461 (Ran et al., 2013). HEK293T cells were treated with 200 ng/ml nocodazole for 17 h (Lin et al., 2014) to enhance homology-directed repair and then transfected with the resulting guide RNA plasmids and a repair template oligonucleotide (5′-GCCCACTATGTACCCAAAATCTACTGCAGGGGTCCCAACCCTTTTCCACAGAAATAAGACATGATGGCACAGCATGTTTTGTTGGGACCAATGGAATGGTACCTTTGTGGTGAA-3′) using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were cultured for 24 h before GFP-positive cells were isolated using a BD Jazz FACS sorter (BD Biosciences) and allowed to grow, and individual colonies were picked, cultured, and genotyped.
GFP-Trap and mass spectrometry
The CMV promoter in pEYFP-N1 (Clontech) was replaced with the CAG promoter (Niwa et al., 1991), then the mouse Tex19.1 open reading frame was subcloned in-frame with EYFP. HEK293 cells were transfected with pCAG-TEX19.1-YFP and pCAG-YFP, and stable cell lines expressing similar levels of YFP fluorescence were isolated by flow cytometry. Cytoplasmic lysates were prepared by Dounce homogenizing cells in buffer A (10 mM Hepes, pH 7.6, 15 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, and complete protease inhibitors) and then adding 1/10th volume buffer B (50 mM Hepes, pH 7.6, 1 M KCl, 30 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, and complete protease inhibitors). Nuclei were removed by centrifugation at 3,400 g for 15 min at 4°C, and glycerol was added to the supernatant to 10%. YFP-containing protein complexes were isolated from cytoplasmic lysates using GFP-Trap agarose beads (Chromotek) according to the supplier’s instructions. Coimmunoprecipitating proteins were analyzed by SDS-PAGE and visualized by colloidal blue staining, and prominent bands were excised. In-gel digestion with trypsin and mass spectrometry using a 4800 MALDI TOF/TOF Analyser (ABSciex) equipped with an Nd:YAG 355nm laser were performed at St. Andrews University Mass Spectrometry and Proteomics Facility. Data were analyzed using the Mascot search engine (Matrix Science) to interrogate the NCBInr database using tolerances of ±0.2 D for peptide and fragment masses, allowing for one missed trypsin cleavage, fixed cysteine carbamidomethylation, and variable methionine oxidation. Protein identities were confirmed by SDS-PAGE/Western blotting using mouse anti-GFP (Roche, 1:2,000 dilution) and mouse anti-UBR2 (Abcam, 1:1,000 dilution) antibodies.
N-end rule reporter assays
Ubiquitin fusion proteins that generate M-GFP, L-GFP, R-GFP, and UbG76V-GFP (Ub-GFP) reporters (Dantuma et al., 2000) were subcloned into pcDNA5/FRT (Invitrogen) and integrated into Flp-In-293 cells (Invitrogen) according to the supplier’s instructions. The resulting stable cell lines were transiently transfected with a 1:3 ratio of mCherry expression plasmid and either an empty expression vector (pMONO-zeo, Invitrogen) or pMONO-zeo-TEX19.1–expressing mouse Tex19.1, using Lipofectamine 2000 (Invitrogen) as instructed by the manufacturer. Cells were analyzed by flow cytometry using a BD FACSAria II cell sorter (BD Biosciences) 48 h after transfection, and the amount of GFP fluorescence in the mCherry-positive population was measured. For assays with human TEX19, cell lines were transiently transfected with either pCMV or pCMV-TEX19 without mCherry cotransfection, and GFP fluorescence was measured in the total population 24 h after transfection. For MG132 treatment, these stable cell lines were incubated in culture medium containing 25 µM MG132 (Cayman Chemicals) for 7 h. To assess GFP protein abundance, the stable cell lines were transiently transfected with pEXPR-IBA105 (IBA Life Sciences) or pEXPR-IBA105-TEX19.1–expressing Strep-tagged mouse Tex19.1, lysed in radioimmunoprecipitation assay buffer 48 h after transfection, and analyzed by SDS-PAGE/Western blotting. Mouse anti-GFP antibodies (Roche, 11814460001) were used at 1:6,000 dilution, rabbit anti-lamin B1 antibodies (Abcam, ab16048) at 1:10,000, and rabbit anti-Strep Tag II (Abcam, ab76950) at 1:6,000.
Statistical analyses
Two-sided Mann–Whitney U tests were used as a nonparametric test for differences between two populations. For statistical testing between experiments with small numbers of replicates (n < 5), data distribution was assumed to be normal, and two-sided unequal variance t test was used. Two-sided Fisher’s exact test was used to detect differences in categorical data. P < 0.05 was used as a threshold for statistical significance (ns, not significant; *, P < 0.05; **, P < 0.01). Means are reported ±SD. Statistical analysis was performed in R (R Core Team, 2017).
Online supplemental material
Fig. S1 shows additional phenotyping data from fetal and adult Tex19.1−/− oocytes during meiotic prophase and prometaphase I. Fig. S2 shows additional phenotyping data showing meiotic chromosome segregation defects in Tex19.1−/− adult oocytes. Fig. S3 shows the effects of expressing human TEX19 on the total amount of cohesin in HEK293T cells and on chromosome-associated cohesin in S phase. Fig. S4 shows characterization of the N-end rule reporter and UBR2 mutant cell lines generated in this study. Fig. S5 shows additional phenotyping data from rapidly proliferating tissues in Ubr2−/− mice.
Acknowledgments
We thank Christer Höög and Nico Dantuma (Karolinska Institutet, Stockholm, Sweden) for providing anti-REC8 antibodies and Ub-L-GFP constructs, respectively. We are grateful to Sally Shirran and Catherine Botting (Mass Spectrometry and Proteomics Facility, University of St. Andrews, St. Andrews, UK) for performing mass spectrometry on anti-YFP immunoprecipitates; to imaging services at Medical Research Council Human Genetics Unit; and to bioresearch and veterinary services at University of Edinburgh. We thank Wendy Bickmore and Nick Hastie (both Medical Research Council Human Genetics Unit), Jan Ellenberg (European Molecular Biology Laboratory, Heidelberg, Germany), and Mariana Coelho Correia Da Silva (Research Institute of Molecular Pathology, Vienna, Austria) for support, advice, and helpful suggestions, and Javier Caceres (Medical Research Council Human Genetics Unit) for comments on the manuscript.
Funding: J. Reichmann and E.S. Raymond were supported by Medical Research Council Doctoral Training Awards. K. Dobie, J.H. Crichton, D. Best, M. MacLennan, D. Read, C.-C. Hung, S. Boyle, H.J. Cooke, and I.R. Adams were supported by Medical Research Council Intramural Program Grants MC_PC_U127580973 and MC_UU_00007/6. K. Shirahige was supported by Japan Science and Technology Agency CREST grant JPMJCR18S5. L.M. Lister and M. Herbert were supported by funding from the European Union’s Horizon 2020 - Research and Innovation Framework Programme under grant agreement no. 634113 (GermAge) and funding from the Medical Research Council (MR/J003603/1).
The authors declare no competing financial interests.
Author contributions: J. Reichmann, J.H. Crichton, D. Read, S. Boyle, E.S. Raymond, and I.R. Adams performed phenotypic analysis of Tex19.1−/− mice; L.M. Lister and M. Herbert performed live imaging of oocytes; K. Dobie and D. Read performed phenotypic analysis of Ubr2−/− mice; K. Dobie, D. Best, and M. MacLennan performed cell culture experiments; C.-C. Hung, K. Shirahige, and H.J. Cooke provided reagents. I.R. Adams coordinated the study and wrote the manuscript with contributions from all authors.
References
- Baker S.M., Plug A.W., Prolla T.A., Bronner C.E., Harris A.C., Yao X., Christie D.M., Monell C., Arnheim N., Bradley A., et al. 1996. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat. Genet. 13:336–342. 10.1038/ng0796-336 [DOI] [PubMed] [Google Scholar]
- Bolcun-Filas E., Hall E., Speed R., Taggart M., Grey C., de Massy B., Benavente R., and Cooke H.J.. 2009. Mutation of the mouse Syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLoS Genet. 5:e1000393 10.1371/journal.pgen.1000393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle S., Gilchrist S., Bridger J.M., Mahy N.L., Ellis J.A., and Bickmore W.A.. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211–219. 10.1093/hmg/10.3.211 [DOI] [PubMed] [Google Scholar]
- Burkhardt S., Borsos M., Szydlowska A., Godwin J., Williams S.A., Cohen P.E., Hirota T., Saitou M., and Tachibana-Konwalski K.. 2016. Chromosome Cohesion Established by Rec8-Cohesin in Fetal Oocytes Is Maintained without Detectable Turnover in Oocytes Arrested for Months in Mice. Curr. Biol. 26:678–685. 10.1016/j.cub.2015.12.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celebi C., van Montfoort A., Skory V., Kieffer E., Kuntz S., Mark M., and Viville S.. 2012. Tex 19 paralogs exhibit a gonad and placenta-specific expression in the mouse. J. Reprod. Dev. 58:360–365. 10.1262/jrd.11-047K [DOI] [PubMed] [Google Scholar]
- Chiang T., Duncan F.E., Schindler K., Schultz R.M., and Lampson M.A.. 2010. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr. Biol. 20:1522–1528. 10.1016/j.cub.2010.06.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton J.H., Dunican D.S., Maclennan M., Meehan R.R., and Adams I.R.. 2014. Defending the genome from the enemy within: mechanisms of retrotransposon suppression in the mouse germline. Cell. Mol. Life Sci. 71:1581–1605. 10.1007/s00018-013-1468-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton J.H., Playfoot C.J., MacLennan M., Read D., Cooke H.J., and Adams I.R.. 2017. Tex19.1 promotes Spo11-dependent meiotic recombination in mouse spermatocytes. PLoS Genet. 13:e1006904 10.1371/journal.pgen.1006904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crichton J.H., Read D., and Adams I.R.. 2018. Defects in meiotic recombination delay progression through pachytene in Tex19.1−/− mouse spermatocytes. Chromosoma. 127:437–459. 10.1007/s00412-018-0674-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma N.P., Lindsten K., Glas R., Jellne M., and Masucci M.G.. 2000. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18:538–543. 10.1038/75406 [DOI] [PubMed] [Google Scholar]
- Deardorff M.A., Bando M., Nakato R., Watrin E., Itoh T., Minamino M., Saitoh K., Komata M., Katou Y., Clark D., et al. 2012. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature. 489:313–317. 10.1038/nature11316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du F., Navarro-Garcia F., Xia Z., Tasaki T., and Varshavsky A.. 2002. Pairs of dipeptides synergistically activate the binding of substrate by ubiquitin ligase through dissociation of its autoinhibitory domain. Proc. Natl. Acad. Sci. USA. 99:14110–14115. 10.1073/pnas.172527399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn J.R., Zielinska A.P., Shukla V., Blanshard R., Capalbo A., Cimadomo D., Nikiforov D., Chan A.C.-H., Newnham L.J., Vogel I., et al. 2019. Chromosome errors in human eggs shape natural fertility over reproductive life span. Science. 365:1466–1469. 10.1126/science.aav7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J.A., Reddington J.P., Nestor C.E., Dunican D.S., Branco M.R., Reichmann J., Reik W., Surani M.A., Adams I.R., and Meehan R.R.. 2012. Promoter DNA methylation couples genome-defence mechanisms to epigenetic reprogramming in the mouse germline. Development. 139:3623–3632. 10.1242/dev.081661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T., and Hunt P.. 2001. To err (meiotically) is human: the genesis of human aneuploidy. Nat. Rev. Genet. 2:280–291. 10.1038/35066065 [DOI] [PubMed] [Google Scholar]
- Hauf S., Waizenegger I.C., and Peters J.M.. 2001. Cohesin cleavage by separase required for anaphase and cytokinesis in human cells. Science. 293:1320–1323. 10.1126/science.1061376 [DOI] [PubMed] [Google Scholar]
- Henderson S.A., and Edwards R.G.. 1968. Chiasma frequency and maternal age in mammals. Nature. 218:22–28. 10.1038/218022a0 [DOI] [PubMed] [Google Scholar]
- Herbert M., Kalleas D., Cooney D., Lamb M., and Lister L.. 2015. Meiosis and maternal aging: insights from aneuploid oocytes and trisomy births. Cold Spring Harb. Perspect. Biol. 7:a017970 10.1101/cshperspect.a017970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C.A., Revenkova E., Jessberger R., Hassold T.J., and Hunt P.A.. 2005. SMC1beta-deficient female mice provide evidence that cohesins are a missing link in age-related nondisjunction. Nat. Genet. 37:1351–1355. 10.1038/ng1672 [DOI] [PubMed] [Google Scholar]
- Holloway J.K., Booth J., Edelmann W., McGowan C.H., and Cohen P.E.. 2008. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4:e1000186 10.1371/journal.pgen.1000186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer H.A., McDougall A., Levasseur M., Yallop K., Murdoch A.P., and Herbert M.. 2005. Mad2 prevents aneuploidy and premature proteolysis of cyclin B and securin during meiosis I in mouse oocytes. Genes Dev. 19:202–207. 10.1101/gad.328105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.-K., Kim R.-R., Oh J.-H., Cho H., Varshavsky A., and Hwang C.-S.. 2014. The N-terminal methionine of cellular proteins as a degradation signal. Cell. 156:158–169. 10.1016/j.cell.2013.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishigami S., and Wakayama T.. 2007. Efficient strontium-induced activation of mouse oocytes in standard culture media by chelating calcium. J. Reprod. Dev. 53:1207–1215. 10.1262/jrd.19067 [DOI] [PubMed] [Google Scholar]
- Kouznetsova A., Lister L., Nordenskjöld M., Herbert M., and Höög C.. 2007. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nat. Genet. 39:966–968. 10.1038/ng2065 [DOI] [PubMed] [Google Scholar]
- Kouznetsova A., Novak I., Jessberger R., and Höög C.. 2005. SYCP2 and SYCP3 are required for cohesin core integrity at diplotene but not for centromere cohesion at the first meiotic division. J. Cell Sci. 118:2271–2278. 10.1242/jcs.02362 [DOI] [PubMed] [Google Scholar]
- Kuntz S., Kieffer E., Bianchetti L., Lamoureux N., Fuhrmann G., and Viville S.. 2008. Tex19, a mammalian-specific protein with a restricted expression in pluripotent stem cells and germ line. Stem Cells. 26:734–744. 10.1634/stemcells.2007-0772 [DOI] [PubMed] [Google Scholar]
- Kwon Y.T., Xia Z., An J.Y., Tasaki T., Davydov I.V., Seo J.W., Sheng J., Xie Y., and Varshavsky A.. 2003. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol. Cell. Biol. 23:8255–8271. 10.1128/MCB.23.22.8255-8271.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladurner R., Kreidl E., Ivanov M.P., Ekker H., Idarraga-Amado M.H., Busslinger G.A., Wutz G., Cisneros D.A., and Peters J.-M.. 2016. Sororin actively maintains sister chromatid cohesion. EMBO J. 35:635–653. 10.15252/embj.201592532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMaire-Adkins R., Radke K., and Hunt P.A.. 1997. Lack of checkpoint control at the metaphase/anaphase transition: a mechanism of meiotic nondisjunction in mammalian females. J. Cell Biol. 139:1611–1619. 10.1083/jcb.139.7.1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S., Staahl B.T., Alla R.K., and Doudna J.A.. 2014. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 3:e04766 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister L.M., Kouznetsova A., Hyslop L.A., Kalleas D., Pace S.L., Barel J.C., Nathan A., Floros V., Adelfalk C., Watanabe Y., et al. 2010. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr. Biol. 20:1511–1521. 10.1016/j.cub.2010.08.023 [DOI] [PubMed] [Google Scholar]
- Liu Y.-J., Liu C., Chang Z., Wadas B., Brower C.S., Song Z.-H., Xu Z.-L., Shang Y.-L., Liu W.-X., Wang L.-N., et al. 2016. Degradation of the Separase-cleaved Rec8, a Meiotic Cohesin Subunit, by the N-end Rule Pathway. J. Biol. Chem. 291:7426–7438. 10.1074/jbc.M116.714964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan M., Crichton J.H., Playfoot C.J., and Adams I.R.. 2015. Oocyte development, meiosis and aneuploidy. Semin. Cell Dev. Biol. 45:68–76. 10.1016/j.semcdb.2015.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLennan M., García-Cañadas M., Reichmann J., Khazina E., Wagner G., Playfoot C.J., Salvador-Palomeque C., Mann A.R., Peressini P., Sanchez L., et al. 2017. Mobilization of LINE-1 retrotransposons is restricted by Tex19.1 in mouse embryonic stem cells. eLife. 6:e26152 10.7554/eLife.26152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuinness B.E., Anger M., Kouznetsova A., Gil-Bernabé A.M., Helmhart W., Kudo N.R., Wuensche A., Taylor S., Hoog C., Novak B., and Nasmyth K.. 2009. Regulation of APC/C activity in oocytes by a Bub1-dependent spindle assembly checkpoint. Curr. Biol. 19:369–380. 10.1016/j.cub.2009.01.064 [DOI] [PubMed] [Google Scholar]
- McNicoll F., Stevense M., and Jessberger R.. 2013. Cohesin in gametogenesis. Curr. Top. Dev. Biol. 102:1–34. 10.1016/B978-0-12-416024-8.00001-5 [DOI] [PubMed] [Google Scholar]
- Méndez J., and Stillman B.. 2000. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20:8602–8612. 10.1128/MCB.20.22.8602-8612.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino M., Ishibashi M., Nakato R., Akiyama K., Tanaka H., Kato Y., Negishi L., Hirota T., Sutani T., Bando M., and Shirahige K.. 2015. Esco1 Acetylates Cohesin via a Mechanism Different from That of Esco2. Curr. Biol. 25:1694–1706. 10.1016/j.cub.2015.05.017 [DOI] [PubMed] [Google Scholar]
- Nagaoka S.I., Hassold T.J., and Hunt P.A.. 2012. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13:493–504. 10.1038/nrg3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A., Gertsenstein M., Vintersten K., and Behringer R.. 2003. Manipulating the Mouse Embryo: A Laboratory Manual. Third edition Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Nakajima M., Kumada K., Hatakeyama K., Noda T., Peters J.-M., and Hirota T.. 2007. The complete removal of cohesin from chromosome arms depends on separase. J. Cell Sci. 120:4188–4196. 10.1242/jcs.011528 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., and Haering C.H.. 2009. Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43:525–558. 10.1146/annurev-genet-102108-134233 [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Ladurner R., Schmitz J., Kreidl E., Schleiffer A., Bhaskara V., Bando M., Shirahige K., Hyman A.A., Mechtler K., and Peters J.-M.. 2010. Sororin mediates sister chromatid cohesion by antagonizing Wapl. Cell. 143:737–749. 10.1016/j.cell.2010.10.031 [DOI] [PubMed] [Google Scholar]
- Nishiyama T., Sykora M.M., Huis in ‘t Veld P.J., Mechtler K., and Peters J.-M.. 2013. Aurora B and Cdk1 mediate Wapl activation and release of acetylated cohesin from chromosomes by phosphorylating Sororin. Proc. Natl. Acad. Sci. USA. 110:13404–13409. 10.1073/pnas.1305020110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., and Miyazaki J.. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 108:193–199. 10.1016/0378-1119(91)90434-D [DOI] [PubMed] [Google Scholar]
- Öllinger R., Childs A.J., Burgess H.M., Speed R.M., Lundegaard P.R., Reynolds N., Gray N.K., Cooke H.J., and Adams I.R.. 2008. Deletion of the pluripotency-associated Tex19.1 gene causes activation of endogenous retroviruses and defective spermatogenesis in mice. PLoS Genet. 4:e1000199 10.1371/journal.pgen.1000199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottolini C.S., Newnham L., Capalbo A., Natesan S.A., Joshi H.A., Cimadomo D., Griffin D.K., Sage K., Summers M.C., Thornhill A.R., et al. 2015. Genome-wide maps of recombination and chromosome segregation in human oocytes and embryos show selection for maternal recombination rates. Nat. Genet. 47:727–735. 10.1038/ng.3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Ma P., Zhu W., and Schultz R.M.. 2008. Age-associated increase in aneuploidy and changes in gene expression in mouse eggs. Dev. Biol. 316:397–407. 10.1016/j.ydbio.2008.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planells-Palop V., Hazazi A., Feichtinger J., Jezkova J., Thallinger G., Alsiwiehri N.O., Almutairi M., Parry L., Wakeman J.A., and McFarlane R.J.. 2017. Human germ/stem cell-specific gene TEX19 influences cancer cell proliferation and cancer prognosis. Mol. Cancer. 16:84 10.1186/s12943-017-0653-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2017. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H., Uhlmann F., Nasmyth K., and Varshavsky A.. 2001. Degradation of a cohesin subunit by the N-end rule pathway is essential for chromosome stability. Nature. 410:955–959. 10.1038/35073627 [DOI] [PubMed] [Google Scholar]
- Reichmann J., Crichton J.H., Madej M.J., Taggart M., Gautier P., Garcia-Perez J.L., Meehan R.R., and Adams I.R.. 2012. Microarray analysis of LTR retrotransposon silencing identifies Hdac1 as a regulator of retrotransposon expression in mouse embryonic stem cells. PLOS Comput. Biol. 8:e1002486 10.1371/journal.pcbi.1002486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichmann J., Reddington J.P., Best D., Read D., Öllinger R., Meehan R.R., and Adams I.R.. 2013. The genome-defence gene Tex19.1 suppresses LINE-1 retrotransposons in the placenta and prevents intra-uterine growth retardation in mice. Hum. Mol. Genet. 22:1791–1806. 10.1093/hmg/ddt029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenkova E., Eijpe M., Heyting C., Hodges C.A., Hunt P.A., Liebe B., Scherthan H., and Jessberger R.. 2004. Cohesin SMC1 beta is required for meiotic chromosome dynamics, sister chromatid cohesion and DNA recombination. Nat. Cell Biol. 6:555–562. 10.1038/ncb1135 [DOI] [PubMed] [Google Scholar]
- Revenkova E., Herrmann K., Adelfalk C., and Jessberger R.. 2010. Oocyte cohesin expression restricted to predictyate stages provides full fertility and prevents aneuploidy. Curr. Biol. 20:1529–1533. 10.1016/j.cub.2010.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara Y., Hashimoto S., Nakaoka Y., Kouznetsova A., Höög C., and Kitajima T.S.. 2015. Bivalent separation into univalents precedes age-related meiosis I errors in oocytes. Nat. Commun. 6:7550 10.1038/ncomms8550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Watrin E., Lénárt P., Mechtler K., and Peters J.-M.. 2007. Sororin is required for stable binding of cohesin to chromatin and for sister chromatid cohesion in interphase. Curr. Biol. 17:630–636. 10.1016/j.cub.2007.02.029 [DOI] [PubMed] [Google Scholar]
- Susiarjo M., Rubio C., and Hunt P.. 2009. Analyzing mammalian female meiosis. Methods Mol. Biol. 558:339–354. 10.1007/978-1-60761-103-5_20 [DOI] [PubMed] [Google Scholar]
- Tachibana-Konwalski K., Godwin J., van der Weyden L., Champion L., Kudo N.R., Adams D.J., and Nasmyth K.. 2010. Rec8-containing cohesin maintains bivalents without turnover during the growing phase of mouse oocytes. Genes Dev. 24:2505–2516. 10.1101/gad.605910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarabay Y., Achour M., Teletin M., Ye T., Teissandier A., Mark M., Bourc’his D., and Viville S.. 2017. Tex19 paralogs are new members of the piRNA pathway controlling retrotransposon suppression. J. Cell Sci. 130:1463–1474. 10.1242/jcs.188763 [DOI] [PubMed] [Google Scholar]
- Tarabay Y., Kieffer E., Teletin M., Celebi C., Van Montfoort A., Zamudio N., Achour M., El Ramy R., Gazdag E., Tropel P., et al. 2013. The mammalian-specific Tex19.1 gene plays an essential role in spermatogenesis and placenta-supported development. Hum. Reprod. 28:2201–2214. 10.1093/humrep/det129 [DOI] [PubMed] [Google Scholar]
- Tasaki T., Mulder L.C.F., Iwamatsu A., Lee M.J., Davydov I.V., Varshavsky A., Muesing M., and Kwon Y.T.. 2005. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 25:7120–7136. 10.1128/MCB.25.16.7120-7136.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati S.A., Buffin E., Cladière D., Hached K., Rachez C., van Deursen J.M., and Wassmann K.. 2015. Mouse oocytes depend on BubR1 for proper chromosome segregation but not for prophase I arrest. Nat. Commun. 6:6946 10.1038/ncomms7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Valen L. 1973. A new evolutionary law. Evol. Theory. 1:1–30. [Google Scholar]
- Waizenegger I.C., Hauf S., Meinke A., and Peters J.M.. 2000. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 103:399–410. 10.1016/S0092-8674(00)00132-X [DOI] [PubMed] [Google Scholar]
- Wang P.J., McCarrey J.R., Yang F., and Page D.C.. 2001. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 27:422–426. 10.1038/86927 [DOI] [PubMed] [Google Scholar]
- Xia Z., Webster A., Du F., Piatkov K., Ghislain M., and Varshavsky A.. 2008. Substrate-binding sites of UBR1, the ubiquitin ligase of the N-end rule pathway. J. Biol. Chem. 283:24011–24028. 10.1074/jbc.M802583200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Cheng Y., An J.Y., Kwon Y.T., Eckardt S., Leu N.A., McLaughlin K.J., and Wang P.J.. 2010. The ubiquitin ligase Ubr2, a recognition E3 component of the N-end rule pathway, stabilizes Tex19.1 during spermatogenesis. PLoS One. 5:e14017 10.1371/journal.pone.0014017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Liu J.-G., Hoja M.-R., Wilbertz J., Nordqvist K., and Höög C.. 2002. Female germ cell aneuploidy and embryo death in mice lacking the meiosis-specific protein SCP3. Science. 296:1115–1118. 10.1126/science.1070594 [DOI] [PubMed] [Google Scholar]
- Zhang J., Shi X., Li Y., Kim B.-J., Jia J., Huang Z., Yang T., Fu X., Jung S.Y., Wang Y., et al. 2008. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol. Cell. 31:143–151. 10.1016/j.molcel.2008.06.006 [DOI] [PubMed] [Google Scholar]
- Zielinska A.P., Bellou E., Sharma N., Frombach A.-S., Seres K.B., Gruhn J.R., Blayney M., Eckel H., Moltrecht R., Elder K., et al. 2019. Meiotic Kinetochores Fragment into Multiple Lobes upon Cohesin Loss in Aging Eggs. Curr. Biol. 29:3749–3765.e7. 10.1016/j.cub.2019.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska A.P., Holubcova Z., Blayney M., Elder K., and Schuh M.. 2015. Sister kinetochore splitting and precocious disintegration of bivalents could explain the maternal age effect. eLife. 4:e11389 10.7554/eLife.11389 [DOI] [PMC free article] [PubMed] [Google Scholar]