Epithelial cell integrity and remodeling require proper actomyosin organization at adherens junctions through αE-catenin complexed with β-catenin. Sakakibara et al. show that afadin binds to αE-catenin complexed with β-catenin and enhances its F-actin–binding activity in a novel mechanism.
Abstract
Actomyosin-undercoated adherens junctions are critical for epithelial cell integrity and remodeling. Actomyosin associates with adherens junctions through αE-catenin complexed with β-catenin and E-cadherin in vivo; however, in vitro biochemical studies in solution showed that αE-catenin complexed with β-catenin binds to F-actin less efficiently than αE-catenin that is not complexed with β-catenin. Although a “catch-bond model” partly explains this inconsistency, the mechanism for this inconsistency between the in vivo and in vitro results remains elusive. We herein demonstrate that afadin binds to αE-catenin complexed with β-catenin and enhances its F-actin–binding activity in a novel mechanism, eventually inducing the proper actomyosin organization through αE-catenin complexed with β-catenin and E-cadherin at adherens junctions.
Introduction
Actomyosin-undercoated adherens junctions (AJs) are critical for epithelial cell integrity and remodeling, such as invagination and convergent extension of the epithelial monolayer (Cavey and Lecuit, 2009; Harris and Tepass, 2010; Nishimura and Takeichi, 2009). This actomyosin is present as two types of structures, mesh-like filamentous actomyosin and circumferential actomyosin bundles (Efimova and Svitkina, 2018; Maruthamuthu et al., 2010; Steinbacher and Ebnet, 2018). αE-catenin complexed with β-catenin binds actomyosin in the mesh-like structure and associates the circumferential actomyosin bundles with E-cadherin (Efimova and Svitkina, 2018; Maiden and Hardin, 2011). F-actin in both structures binds myosin II, which regulates the cell adhesion strength and plasticity of AJs (Smutny et al., 2010). However, in vitro biochemical studies have shown that the binding of β-catenin to αE-catenin in solution inhibits its F-actin–binding (FAB) activity (Yamada et al., 2005). The mechanism for this inconsistency between the in vivo and in vitro results has remained elusive.
A “catch-bond model” was recently proposed in which the force exerted to the αE-catenin–β-catenin–E-cadherin complex induces its transient binding to F-actin (Buckley et al., 2014). This model was further supported by a structural analysis showing that the force applied to αE-catenin induces the conformational change and the enhancement of its FAB activity (Ishiyama et al., 2018). However, the mechanism by which this transient binding of the αE-catenin–β-catenin–E-cadherin complex to F-actin is stabilized has remained elusive.
We examined here using biochemical, cell biological, and molecular biological approaches whether afadin plays a role in this stabilization, because mammalian afadin (the Afdn gene product) binds to αE-catenin (Pokutta et al., 2002; Tachibana et al., 2000) and enhances the accumulation of αE-catenin, β-catenin, and E-cadherin at AJs (Sakakibara et al., 2018; Sato et al., 2006), and the Drosophila melanogaster homologue of afadin canoe regulates the proper actomyosin association with AJs in Drosophila during apical constriction (Sawyer et al., 2009), although the precise roles or the modes of action of canoe and afadin in these actions have remained elusive. We showed here that afadin bound to αE-catenin complexed with β-catenin and enhanced its FAB activity in a novel mechanism, eventually inducing the proper actomyosin organization through αE-catenin complexed with β-catenin and E-cadherin at AJs.
Results
Regulation by afadin of the proper actomyosin organization at AJs
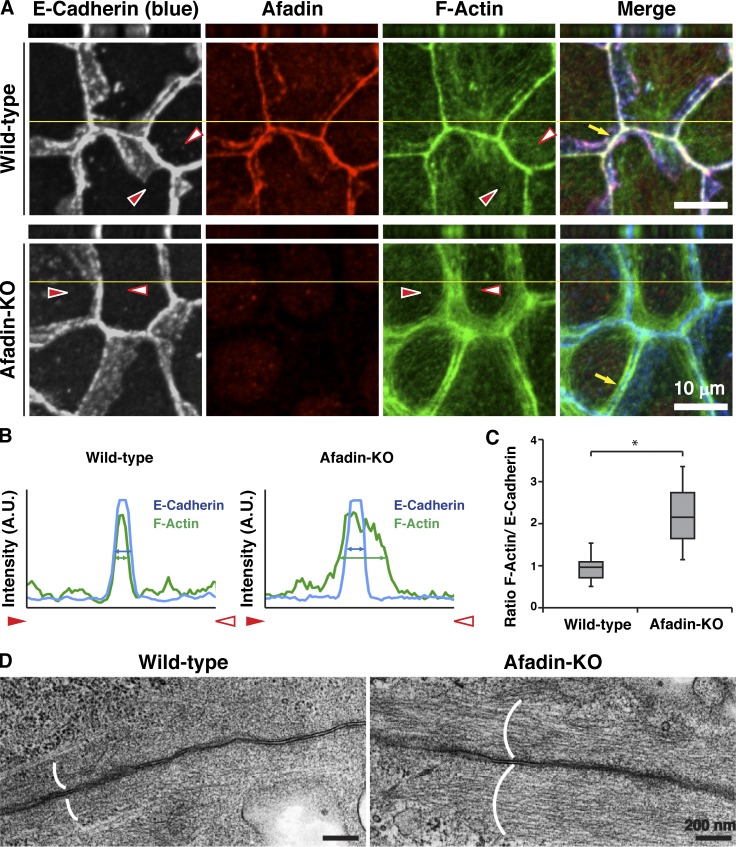
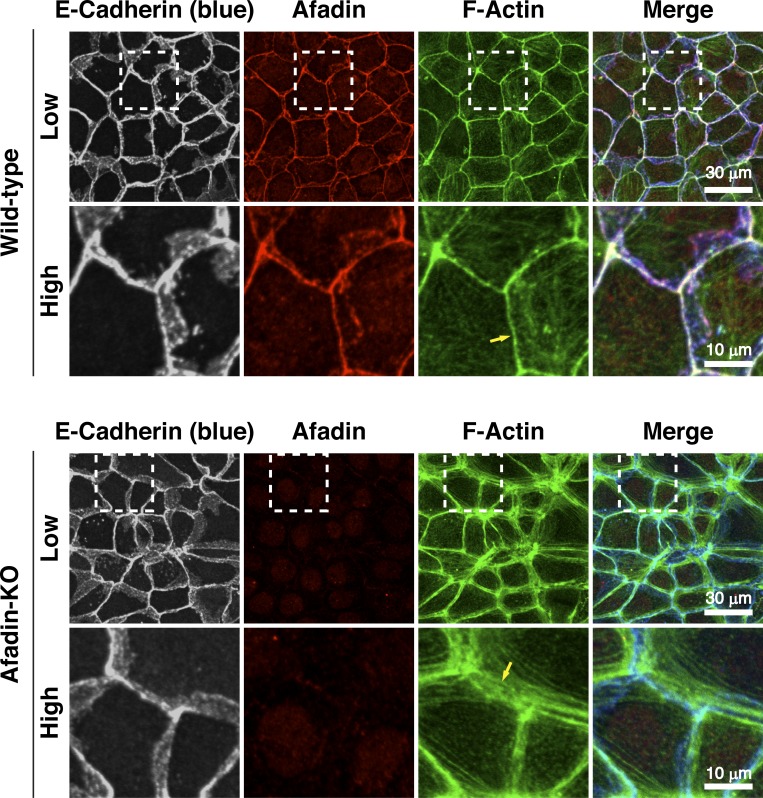
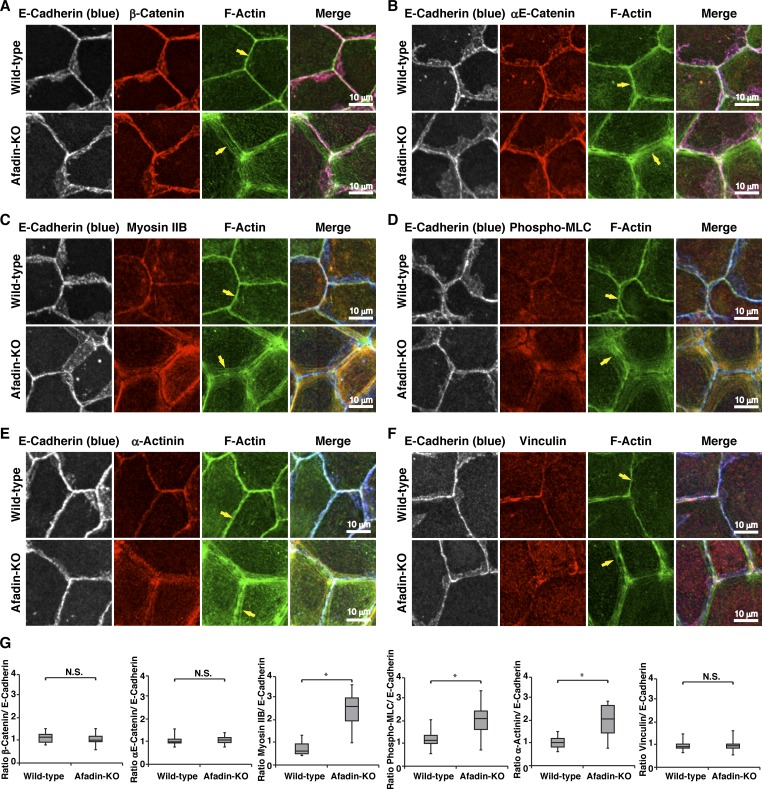
We first examined by immunofluorescence microscopy using EpH4 mouse mammary gland epithelial cells whether afadin is required for the proper actomyosin organization at AJs. E-cadherin localized at bicellular and tricellular junctions similarly in wild-type EpH4 cells and in EpH4 cells in which afadin was knocked out (afadin-KO cells; Sakakibara et al., 2018; Fig. 1 A and Fig. S1). Afadin colocalized with E-cadherin in wild-type cells, but not in afadin-KO cells. β-Catenin and αE-catenin colocalized with E-cadherin in both cells (Fig. S2, A, B, and G). However, at bicellular junctions, F-actin localized differently between wild-type and afadin-KO cells: its signal was observed as a single thick line in wild-type cells, whereas it was observed as several thicker lines and localized in the region farther from the plasma membrane in afadin-KO cells than in wild-type cells (Fig. 1, A–C; and Fig. S1, arrows). It was noted that neither the overall height nor the z-axis position of F-actin relative to that of E-cadherin was apparently different between wild-type and afadin-KO cells as estimated by the lateral side view images. Myosin IIB, phosphorylated myosin light chain II, and α-actinin colocalized with F-actin in both cells but localized differently between wild-type and afadin-KO cells: myosin IIB, phosphorylated myosin light chain II, and α-actinin signals were observed as a single thick line in wild-type cells, whereas they were observed as several thicker lines and localized in the region farther from the plasma membrane in afadin-KO cells than in wild-type cells (Fig. S2, C–E and G). As for vinculin, it localized at bicellular and tricellular junctions in wild-type cells, whereas it did not localize at the bicellular junctions and mainly localized near the tricellular junctions in afadin-KO cells (Fig. S2 F), suggesting that tension at the tricellular junctions is higher than that at the bicellular junctions in afadin-KO cells. In transmission electron microscopy, F-actin bundles accumulated underneath AJs in wild-type cells but accumulated there more broadly in afadin-KO cells (Fig. 1 D). These results indicate that afadin regulates the proper actomyosin organization at AJs.
Figure 1.
Requirement of afadin for the proper actin organization at AJs. (A–C) Localization of E-cadherin and F-actin in wild-type and afadin-KO cells. (A) Wild-type and afadin-KO cells were fixed. The samples were stained with the indicated Abs and phalloidin, followed by immunofluorescence microscopic analysis. Projected xy views (lower panels) as well as z-sectional xz views (upper panels) are presented. Planes of orthogonal sections are indicated by yellow lines. Arrows indicate the F-actin signal at bicellular AJs. Arrowheads indicate both ends of the areas where the line-scans were performed in B. (B) Quantification by line-scans of the immunofluorescence images of wild-type and afadin-KO cells in A. Line-scans were performed between the indicated arrowheads in the immunofluorescence images of A. The pixel intensities for E-cadherin (blue) and F-actin (green) are shown. The blue and green double-ended arrows indicate the full width at half maximum (FWHM) of E-cadherin and F-actin pixel intensities, respectively. (C) Statistical analysis of B. The ratios of FWHM of F-actin to FWHM of E-cadherin are indicated. Data are expressed as a box-and-whisker plot of three independent experiments (n = 10). The whiskers indicate the maximum and minimum values, and the box corresponds to the 75th percentile, median, and 25th percentile values. *, P < 0.01 (two-tailed, unpaired Student’s t test). (D) Ultrastructural images of F-actin bundles at AJs. Wild-type and afadin-KO cells were fixed. The samples were subjected to transmission electron microscopic analysis. Parentheses indicate the F-actin bundles at AJs. A.U., arbitrary units.
Figure S1.
Requirement of afadin for the proper actin organization at AJs in low and high magnifications. Localization of E-cadherin and F-actin in wild-type and afadin-KO cells. Wild-type and afadin-KO cells were fixed. The samples were stained with the indicated Abs and phalloidin, followed by immunofluorescence microscopic analysis. The boxed regions in the upper panels are magnified in the lower panels. Arrows indicate the F-actin signal at bicellular AJs. The results are representative of three independent experiments.
Figure S2.
Requirement of afadin for the proper localization of AJ proteins. (A–F) Localization of AJ proteins, myosin IIB, phosphorylated myosin light chain II, and FAB proteins in wild-type and afadin-KO cells. Wild-type and afadin-KO cells were fixed. The samples were stained with the indicated Abs and phalloidin, followed by immunofluorescence microscopic analysis. (A) E-cadherin and β-catenin; (B) E-cadherin and αE-catenin; (C) E-cadherin and myosin IIB; (D) E-cadherin and phosphorylated myosin light chain II (phospho-MLC); (E) E-cadherin and α-actinin; and (F) E-cadherin and vinculin. Arrows indicate the F-actin signal at bicellular AJs. The results are representative of three independent experiments. (G) Quantification by line-scans of the immunofluorescence images of wild-type and afadin-KO cells in A–F. Statistical analysis was performed as described in the legend to Fig. 1. The ratios of FWHM of proteins of interest to FWHM of E-cadherin are indicated. Data are expressed as a box-and-whisker plot of three independent experiments (n = 10). The whiskers indicate the maximum and minimum values, and the box corresponds to the 75th percentile, median, and 25th percentile values. *, P < 0.01 (two-tailed, unpaired Student’s t test). N.S., not significant.
Involvement of the coiled-coil (CC) region–mediated binding of afadin to αE-catenin in the proper actomyosin organization at AJs
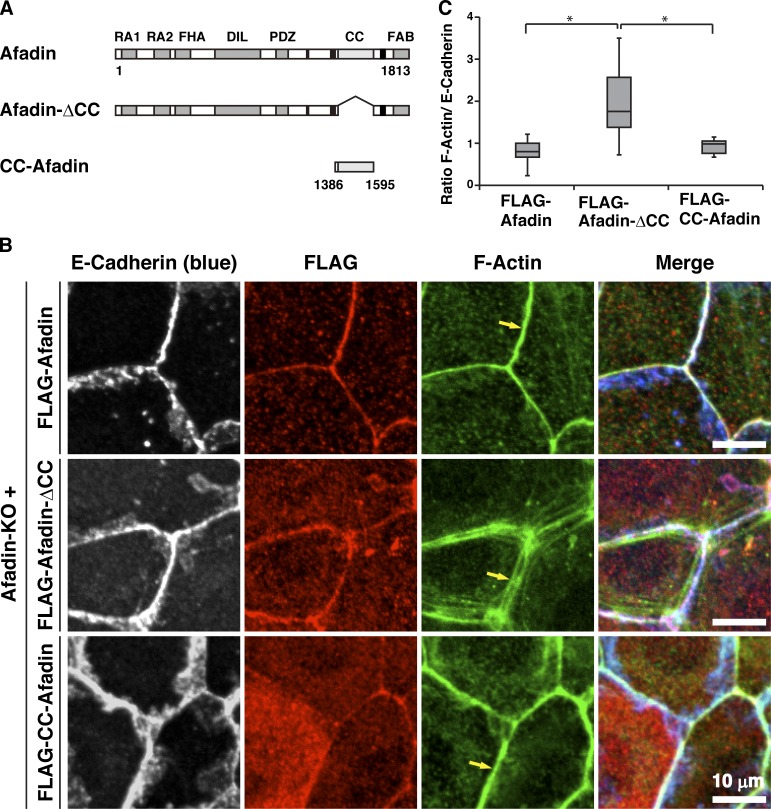
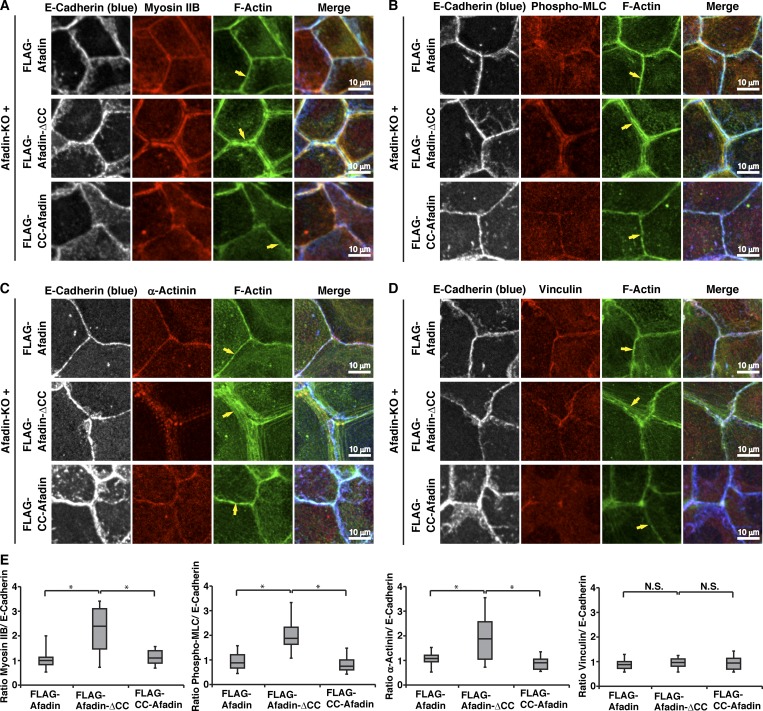
We previously showed that the CC region of afadin (CC-afadin) is necessary and sufficient for its binding to αE-catenin (Maruo et al., 2018). To determine the role of the CC region–mediated binding of afadin to αE-catenin in the proper actomyosin organization at AJs, various afadin fragments shown in Fig. 2 A were expressed in afadin-KO cells to generate EpH4 cells stably expressing each FLAG-afadin fragment. The reexpression of full-length (FL) afadin in afadin-KO cells (FLAG-afadin cells) restored the proper localization patterns of F-actin, myosin IIB, phosphorylated myosin light chain II, α-actinin, and vinculin, but the expression of the afadin mutant in which the αE-catenin–binding CC region was deleted in afadin-KO cells (FLAG-afadin-ΔCC cells) did not restore the proper localization patterns of these molecules with the exception of vinculin (Fig. 2, B and C; and Fig. S3, A–D, arrows, and E). Vinculin localized at the bicellular and tricellular junctions in FLAG-afadin-ΔCC cells, similar to wild-type cells. The expression of CC-afadin in afadin-KO cells (FLAG-CC-afadin cells) restored the proper localization patterns of these molecules with the exception of vinculin. Vinculin did not localize at the bicellular junctions and mainly localized near the tricellular junctions in FLAG-CC-afadin cells, similar to afadin-KO cells. These results indicate that the αE-catenin–binding CC region of afadin is necessary and sufficient for the proper actomyosin organization at AJs, implying that the CC region–mediated binding of afadin to αE-catenin is involved in regulating the proper actomyosin organization at AJs.
Figure 2.
Requirement of the binding of afadin to αE-catenin for the proper actomyosin organization at AJs. (A) Schematic representation of afadin, afadin-ΔCC, and CC-afadin. (B) Afadin-KO cells were reexpressed with FLAG-tagged afadin, FLAG-tagged afadin-ΔCC, or FLAG-tagged CC-afadin. The cells were fixed and stained with the indicated Abs and phalloidin, followed by immunofluorescence microscopic analysis. Arrows indicate the F-actin signal at bicellular AJs. (C) Quantification by line-scans of the immunofluorescence images of FLAG-afadin, FLAG-afadin-ΔCC, and FLAG-CC-afadin cells in B. Statistical analysis was performed as described in the legend to Fig. 1. The ratios of FWHM of F-actin to FWHM of E-cadherin are indicated. Data are expressed as a box-and-whisker plot of three independent experiments (n = 10). The whiskers indicate the maximum and minimum values, and the box corresponds to the 75th percentile, median, and 25th percentile values. *, P < 0.01 (two-tailed, unpaired Student’s t test).
Figure S3.
Requirement of the binding of afadin to αE-catenin for the proper localization of AJ proteins. (A–D) Localization of myosin IIB, phosphorylated myosin light chain II, and FAB proteins in afadin-KO cells expressing FLAG-tagged afadin, FLAG-tagged afadin-ΔCC, or FLAG-tagged CC-afadin. Afadin-KO cells were expressed with FLAG-tagged afadin, FLAG-tagged afadin-ΔCC, or FLAG-tagged CC-afadin. The cells were fixed and stained with the indicated Abs and phalloidin, followed by immunofluorescence microscopic analysis. (A) E-cadherin and myosin IIB; (B) E-cadherin and phosphorylated myosin light chain II (phospho-MLC); (C) E-cadherin and α-actinin; and (D) E-cadherin and vinculin. Arrows indicate the F-actin signal at bicellular AJs. The results are representative of three independent experiments. (E) Quantification by line-scans of the immunofluorescence images of FLAG-afadin, FLAG-afadin-ΔCC, and FLAG-CC-afadin cells in A–D. Statistical analysis was performed as described in the legend to Fig. 1. The ratios of FWHM of proteins of interest to FWHM of E-cadherin are indicated. Data are expressed as a box-and-whisker plot of three independent experiments (n = 10). The whiskers indicate the maximum and minimum values, and the box corresponds to the 75th percentile, median, and 25th percentile values. *, P < 0.01 (two-tailed, unpaired Student’s t test). N.S., not significant.
No requirement of afadin for the formation of the αE-catenin–β-catenin–E-cadherin complex at AJs
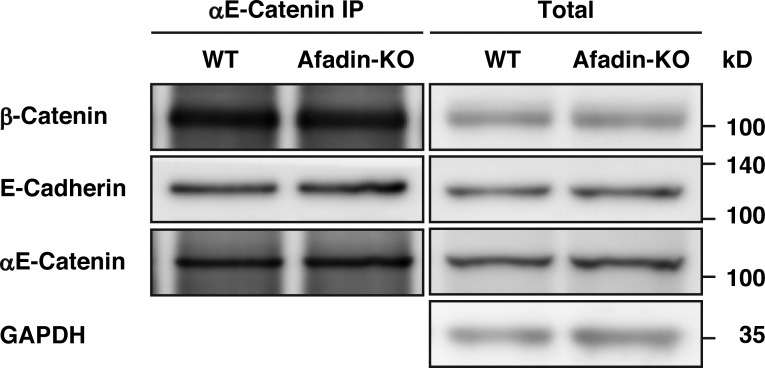
We next examined by an immunoprecipitation assay whether afadin is required for the formation of the αE-catenin–β-catenin–E-cadherin complex at AJs. When αE-catenin was immunoprecipitated using the anti–αE-catenin polyclonal antibody (pAb) from the lysates of wild-type and afadin-KO cells, β-catenin and E-cadherin were coimmunoprecipitated with αE-catenin to similar extents (Fig. 3). These results indicate that the binding of afadin to αE-catenin is not required for the localization or the formation of the αE-catenin–β-catenin–E-cadherin complex at AJs.
Figure 3.
No requirement of afadin for the formation of the αE-catenin–β-catenin–E-cadherin complex. αE-catenin was immunoprecipitated with the anti–αE-catenin pAb from the lysates of wild-type or afadin-KO cells, and the immunoprecipitates were subjected to Western blotting with the indicated Abs. The results are representative of three independent experiments.
Regulation of the proper actomyosin association with AJs by the binding of afadin to αE-catenin
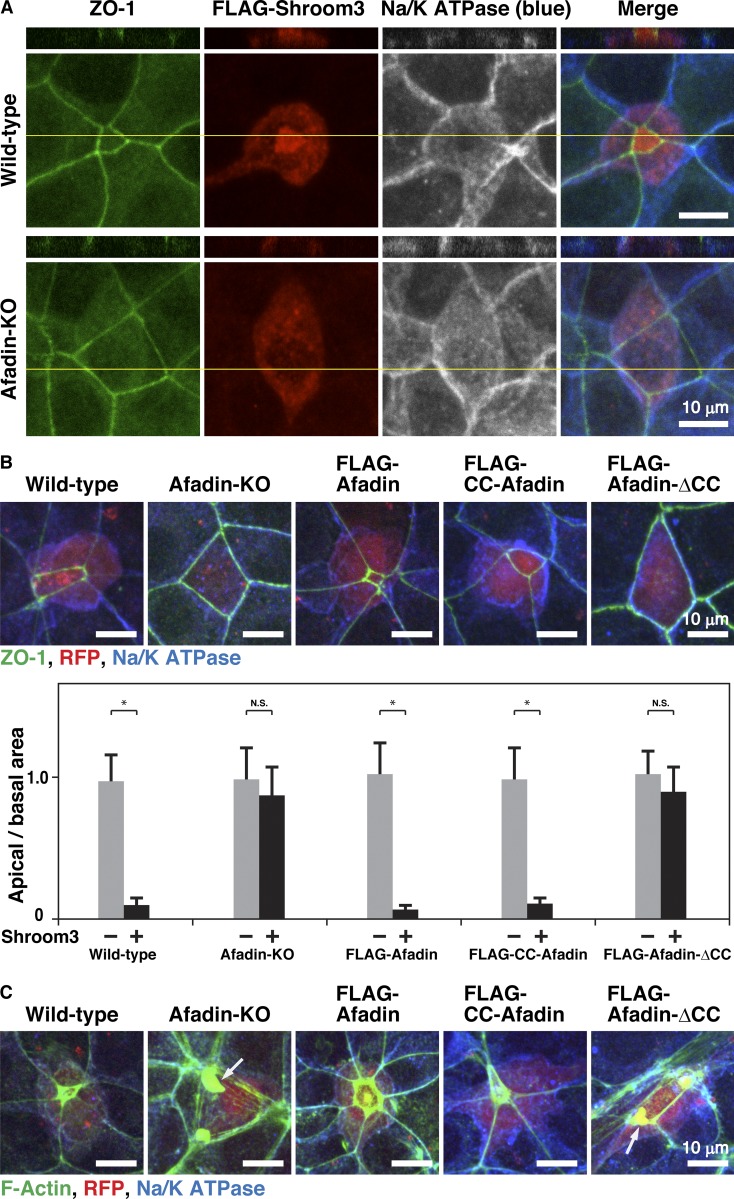
It was previously shown that the Drosophila homologue of afadin canoe regulates the proper actomyosin association with AJs in Drosophila during apical constriction (Sawyer et al., 2009). Shroom3 is the Rho kinase–binding protein that regulates apical constriction in epithelial cells (Hildebrand and Soriano, 1999). Overexpression of shroom3 induces apical constriction in epithelial cells along with actomyosin accumulation at AJs (Haigo et al., 2003; Hildebrand, 2005). To first examine whether afadin is required for the proper apical constriction, we overexpressed FLAG-tagged shroom3 (FLAG-shroom3) in wild-type and afadin-KO cells and analyzed the shapes of the FLAG-shroom3–overexpressing cells by staining ZO-1 and Na/K ATPase as markers for the apical complex and the basolateral membrane, respectively (McNeill et al., 1990; Stevenson et al., 1986). The overexpression of FLAG-shroom3 in afadin-KO cells did not induce apical constriction under the condition where the overexpression of FLAG-shroom3 in wild-type cells induced apical constriction (Fig. 4 A). It was noted that overall height was not apparently different between FLAG-shroom3–overexpressing wild-type and afadin-KO cells as estimated by the lateral side view images. We next examined by overexpressing FLAG-shroom3 in FLAG-afadin, FLAG-CC-afadin, or FLAG-afadin-ΔCC cells in addition to wild-type and afadin-KO cells whether the CC region–mediated binding of afadin to αE-catenin is required for the proper apical constriction. In this series of experiments, wild-type and afadin-KO cells and the cells expressing each FLAG-tagged afadin FL or mutant were further overexpressed with FLAG-shroom3 and expressed with RFP to identify FLAG-shroom3–overexpressing cells. Consistent with the above results, the overexpression of FLAG-shroom3 in afadin-KO cells did not induce apical constriction under the condition where the overexpression of FLAG-shroom3 in wild-type cells induced apical constriction (Fig. 4 B). The overexpression of FLAG-shroom3 in FLAG-afadin or FLAG-CC-afadin cells, but not FLAG-afadin-ΔCC cells, induced apical constriction (Fig. 4 B). We further examined the localization of F-actin in these cells during the FLAG-shroom3–induced apical constriction. F-actin accumulated at AJs in wild-type, FLAG-afadin, and FLAG-CC-afadin cells (Fig. 4 C), whereas it was separated from the plasma membrane at AJs in afadin-KO and FLAG-afadin-ΔCC cells (Fig. 4 C, arrows). These results indicate that the αE-catenin–binding CC region of afadin is necessary and sufficient for the shroom3-induced apical constriction, implying that the CC-region–mediated binding of afadin to αE-catenin regulates the proper actomyosin association with the β-catenin–E-cadherin complex through αE-catenin during the shroom3-induced apical constriction.
Figure 4.
Regulation of the proper actomyosin association with AJs by the binding of afadin to αE-catenin. (A) Requirement of afadin for the shroom3-induced apical constriction. 4 h after cell seeding, wild-type and afadin-KO cells were transfected with FLAG-shroom3 and incubated for 20 h. The cells were then fixed. The samples were stained with an anti–ZO-1 mAb (green), an anti-FLAG mAb (red), and an anti-Na/K ATPase pAb (blue), followed by immunofluorescence microscopic analysis. Projected xy views (lower panels) as well as z-sectional xz views (upper panels) are presented. Planes of orthogonal sections are indicated by yellow lines. (B) Requirement of the binding of afadin to αE-catenin for the shroom3-induced apical constriction. 4 h after cell seeding, wild-type, afadin-KO, FLAG-afadin, FLAG-CC-afadin, and FLAG-afadin-ΔCC cells were cotransfected with FLAG-shroom3 and RFP, and incubated for 20 h. The cells were then fixed. The samples were stained with the anti–ZO-1 mAb (green) and the anti-Na/K ATPase pAb (blue), followed by immunofluorescence microscopic analysis. Quantitative analysis is shown in the lower panel. The ratios of apical area and basal area are shown. Data are expressed as the means ± SD of three independent experiments (n = 10). *, P < 0.01 (two-tailed, unpaired Student’s t test). N.S., not significant. (C) Requirement of the binding of afadin to αE-catenin for the proper actomyosin association with AJs during the shroom3-induced apical constriction. 4 h after cell seeding, wild-type, afadin-KO, FLAG-afadin, FLAG-CC-afadin, and FLAG-afadin-ΔCC cells were cotransfected with FLAG-shroom3 and RFP, and incubated for 20 h. The cells were then fixed. The samples were stained with phalloidin (green) and the anti-Na/K ATPase pAb (blue), followed by immunofluorescence microscopic analysis. Arrows indicate the F-actin signal in the cytoplasm.
Binding of afadin to the conformationally open form of αE-catenin in solution in vitro
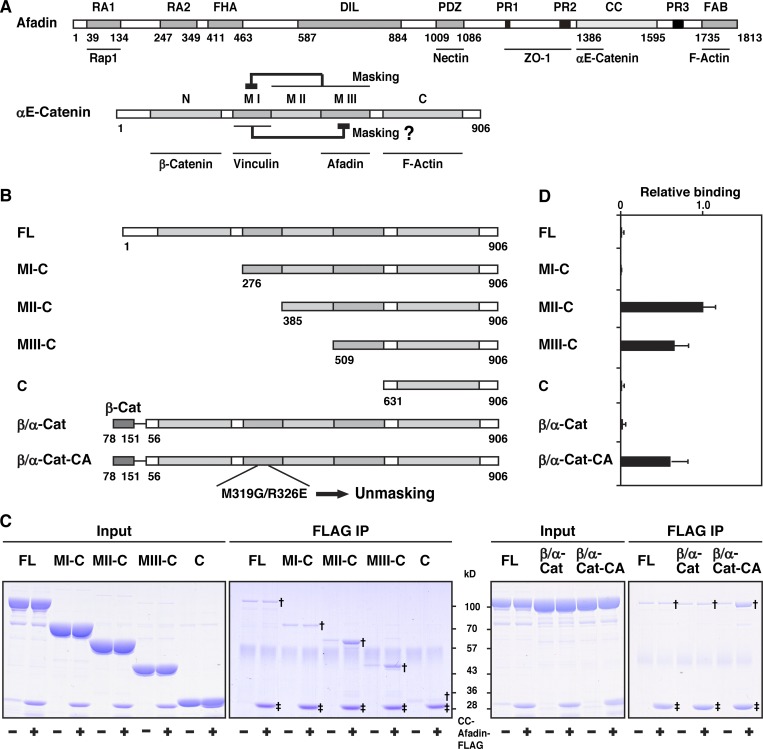
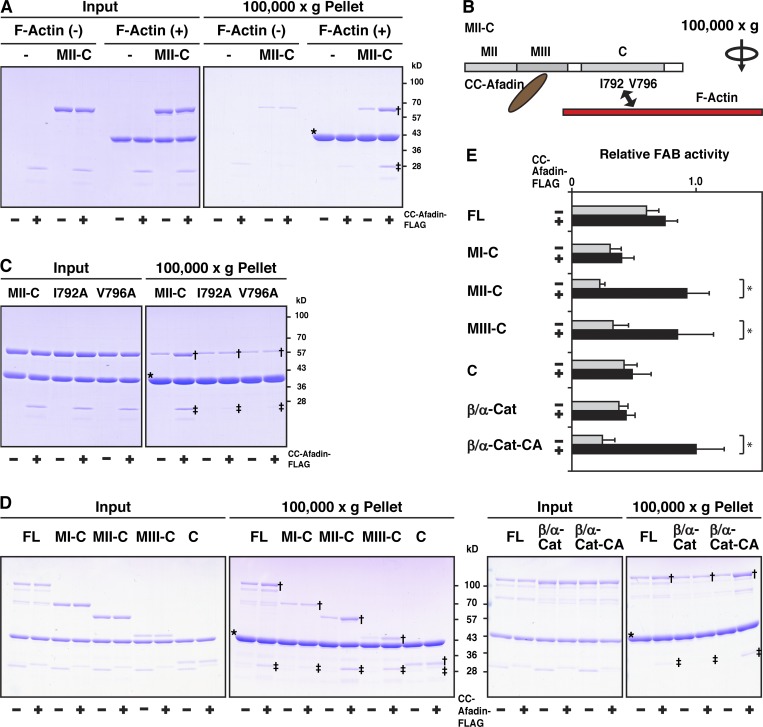
It was previously shown that αE-catenin complexed with β-catenin shows a weak FAB activity in solution in vitro (Yamada et al., 2005). αE-catenin is present as two forms in solution, a monomer and a dimer, with the monomer showing less of a FAB activity than the dimer (Drees et al., 2005). The domain structure of αE-catenin is shown in Fig. 5 A. The N domain of αE-catenin has a strong dimerization activity, but the binding of β-catenin to this domain inhibits its dimerization and FAB activities (Drees et al., 2005). The β-catenin–fused αE-catenin (β/α-cat) lacks the N domain–mediated dimerization activity and shows only a weak FAB activity, as well as that of the β-catenin–αE-catenin complex (Bianchini et al., 2015; Yamada et al., 2005). αE-catenin exists as a conformationally closed form, in which the MI subdomain is stabilized and thereby masked by the subdomains containing the MII and MIII subdomains and the binding of vinculin to the MI subdomain of the closed form is inhibited, unless force is exerted (Hirano et al., 2018; Maki et al., 2016; Matsuzawa et al., 2018; Yonemura et al., 2010). Point mutations in the MI subdomain (M319G/R326E) destabilize the MI subdomain to make αE-catenin conformationally open, enhancing its binding of vinculin (Maki et al., 2016; Matsuzawa et al., 2018). We first examined the binding of afadin to αE-catenin by an immunoprecipitation assay using the purified proteins of CC-afadin, FL αE-catenin, β/α-cat, β-catenin–fused αE-catenin with the M319G/R326E mutations (β/α-cat-CA), and each of αE-catenin fragments that have various regions in addition to the C domain (Fig. 5 B). CC-afadin bound to the fragment containing the MIII subdomain and the C domain (MIII-C) and the fragment containing the MII and MIII subdomains and the C domain (MII-C), but not to the fragment containing the MI, MII, and MIII subdomains and the C domain (MI-C) (Fig. 5, C and D). CC-afadin bound to β/α-cat-CA, but not to FL αE-catenin or β/α-cat (Fig. 5, C and D). These results indicate that the MI subdomain masks the MIII subdomain, inhibiting the binding of afadin to the MIII subdomain, and that afadin binds to only the conformationally open form of αE-catenin.
Figure 5.
Binding of afadin to the αE-catenin–β-catenin complex in solution in vitro. (A) Schematic representation of the domain/region structures and the binding partners of afadin and αE-catenin. The following domains of afadin are shown: RA1, RA2, Ras-associated domain-1 and -2; FHA, forkhead-associated domain; DIL, dilute domain; PDZ, PDZ domain; PR1, PR2, PR3, proline-rich domain-1 to -3; CC, CC region; and FAB, FAB domain. The following domains of αE-catenin are shown: N, N domain; MI, MI subdomain; MII, MII subdomain; MIII, MIII subdomain; and C, C domain. (B) Schematic representation of FL αE-catenin, its mutants, and fusion proteins. (C) Binding of afadin to the αE-catenin–β-catenin complex. Each of the indicated proteins was incubated with the FLAG-tagged fragment of CC-afadin and immunoprecipitated (IP) with the anti-FLAG mAb. The samples were analyzed by SDS-PAGE with CBB staining. Single and double daggers indicate the bands of αE-catenin fragments coimmunoprecipitated with CC-afadin and those of immunoprecipitated CC-afadin, respectively. (D) Quantitative analysis of C. The band intensity of immunoprecipitated MII-C in the presence of CC-afadin is normalized to 1.0.
Enhancement of the FAB activity of the αE-catenin–β-catenin complex by the binding of afadin to αE-catenin in solution in vitro
We then examined the effect of the binding of afadin to αE-catenin on its FAB activity by an F-actin cosedimentation assay using the purified proteins. The FAB activity of MII-C in the presence of CC-afadin was higher than that in the absence of CC-afadin, indicating that CC-afadin enhances the FAB activity of MII-C (Fig. 6 A). However, CC-afadin did not enhance the FAB activity of two MII-C constructs with a point mutation at its FAB interface in the C domain (I792 and V796 in Fig. 6 B), which shows a weak FAB activity (Fig. 6 C; Chen et al., 2015; Ishiyama et al., 2018). These results indicate that the CC region of afadin binds to the MIII subdomain of αE-catenin and enhances its FAB activity.
Figure 6.
Enhancement of the FAB activity of the αE-catenin-β-catenin complex by the binding to afadin in solution in vitro. (A) Enhancement of the FAB activity of αE-catenin by the binding of afadin. MII-C was incubated with F-actin in the presence or absence of CC-afadin and subjected to the F-actin cosedimentation assay. The samples were separated by SDS-PAGE and analyzed by CBB staining. (B) Schematic representation of MII-C, CC-afadin, and F-actin. I792 and V796 are the FAB interface. (C) Inability of enhancement of the FAB activity of αE-catenin mutants by binding of CC-afadin. MII-C or its mutants were incubated with F-actin in the presence or absence of CC-afadin and subjected to the F-actin cosedimentation assay. The samples were separated by SDS-PAGE and analyzed by CBB staining. (D) Enhancement of the FAB activity of the αE-catenin–β-catenin complex by the binding of afadin. Each of the indicated proteins was incubated with F-actin in the presence or absence of CC-afadin and subjected to the F-actin cosedimentation assay. The samples were separated by SDS-PAGE and analyzed by CBB staining. The arrows indicate the bands of αE-catenin fragments cosedimented with F-actin in the presence of CC-afadin. Single and double daggers and asterisks in A, C, and D indicate the bands of αE-catenin fragments cosedimented with F-actin in the presence of CC-afadin, those of CC-afadin cosedimented with F-actin, and those of pelleted F-actin, respectively. (E) Quantitative analysis of D. The band intensity of β/αE-cat-CA cosedimented with F-actin in the presence of CC-afadin is normalized to 1.0. Data are expressed as the means ± SD of three independent experiments. *, P < 0.05 (two-tailed, unpaired Student’s t test).
CC-afadin enhanced the FAB activity of MIII-C as well as MII-C, but not that of MI-C (Fig. 6, D and E). The FAB activity of MII-C or MIII-C in the presence of CC-afadin was not significantly different from that of FL αE-catenin in the presence or absence of CC-afadin. β/α-Cat showed less of a FAB activity than FL αE-catenin, and CC-afadin did not enhance the FAB activity of β/α-cat. However, CC-afadin enhanced the FAB activity of β/α-cat-CA, and the FAB activity of β/α-cat-CA in the presence of CC-afadin was not significantly different from that of FL αE-catenin in the presence or absence of CC-afadin. It was noted that MII-C, MIII-C, and β/α-cat-CA, all of which were able to bind CC-afadin (Fig. 5, C and D), showed the marked CC-afadin–enhanced FAB activities. Collectively, these results indicate that the binding of afadin to the αE-catenin–β-catenin complex through the CC region of afadin and the MIII subdomain of αE-catenin enhances the FAB activity of this complex.
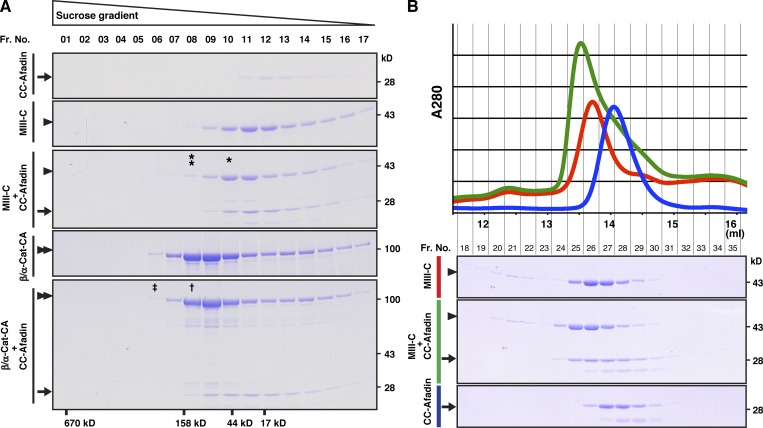
Formation of the afadin–αE-catenin–β-catenin complex in solution in vitro
It was previously shown that αE-catenin complexed with β-catenin shows less of a FAB activity than the αE-catenin dimer in solution in vitro (Drees et al., 2005; Yamada et al., 2005). The in solution in vitro results that the binding of afadin to the αE-catenin–β-catenin complex enhances its FAB activity raised the possibility that the binding of afadin to αE-catenin causes its dimerization. To examine this possibility, we first performed a sucrose gradient centrifugation analysis using the purified proteins of CC-afadin, MIII-C, and β/α-cat-CA. These proteins were present as a monomer (Fig. 7 A). When CC-afadin and MIII-C or β/α-cat-CA were mixed, CC-afadin bound to both proteins, and each complex was present as a heterodimer, but not a heterotetramer (Fig. 7 A, single asterisk and dagger). We further confirmed this result using a gel filtration column chromatography. The essentially same results were obtained, and the possibility that the CC region of afadin and the αE-catenin fragment form a heterotetramer was excluded, although the molecular weight of the complex could not be accurately evaluated in this analysis (Fig. 7 B). These results indicate that CC-afadin binds to αE-catenin complexed with β-catenin to form the ternary complex without its dimerization.
Figure 7.
Formation of the afadin–αE-catenin–β-catenin complex in solution in vitro. (A) Sucrose density gradient centrifugation analysis. MIII-C or β/αE-cat-CA was incubated with CC-afadin and subjected to a sucrose density gradient centrifugation analysis. Each fraction was separated by SDS-PAGE and analyzed by CBB staining. The single asterisk indicates the position of the 1:1 complex of CC-afadin and MIII-C. The double asterisk indicates the position of the 2:2 complex. The dagger indicates the position of the 1:1 complex of CC-afadin and β/αE-cat-CA. The double dagger indicates that of the 2:2 complex. The results shown are representative of three independent experiments. (B) Gel filtration analysis. The fragment of MIII-C was incubated with the fragment of CC-afadin and subjected to a gel filtration analysis. Each fraction was separated by SDS-PAGE and analyzed by CBB staining. Fr. No., fraction number; arrows, CC-afadin; arrowheads, MIII-C; and double arrowheads, β/αE-cat-CA.
Discussion
The proper actomyosin organization is regulated by many αE-catenin–binding proteins, including vinculin, α-actinin, formin-1, ZO-1, afadin, and EPLIN, all of which are FAB proteins (Maiden and Hardin, 2011). We previously showed that afadin enhances AJ formation and that the FAB activity of afadin is required for this activity (Sakakibara et al., 2018; Sato et al., 2006). However, the role of afadin after AJ formation has remained unknown. We showed here using wild-type and afadin-KO EpH4 cells for the first time that afadin was essential for the proper actomyosin organization after AJ formation. We further showed here using afadin-KO cells stably expressing various domain/region deletion mutants of afadin that the αE-catenin–binding CC region of afadin is necessary and sufficient for the binding of afadin to αE-catenin and for the proper actomyosin organization at AJs, and that the FAB domain is not required for this activity of afadin. These results are consistent with the previous observations that the Drosophila homologue of afadin canoe regulates the proper actomyosin association with AJs in Drosophila during apical constriction (Sawyer et al., 2009) and indicate that the CC region of afadin regulates the proper actomyosin organization through αE-catenin complexed with β-catenin and E-cadherin at AJs.
Actomyosin is associated with the β-catenin–E-cadherin complex through αE-catenin, strengthening E-cadherin–mediated cell adhesion (Cavey and Lecuit, 2009). Apical constriction of epithelial cells, which is especially important for epithelial invagination to form the neural cleft and tube during embryo developmental stages, is induced by the contraction of actomyosin properly associating with the β-catenin–E-cadherin complex through αE-catenin (Nishimura and Takeichi, 2009; Takeichi, 2014). However, when actomyosin is not properly associated with the β-catenin–E-cadherin complex through αE-catenin, E-cadherin–mediated cell adhesion is not properly coupled with the actomyosin contraction, and apical constriction is not induced. This apical constriction is regulated by the Rho kinase–binding protein shroom3: it directly binds to Rho kinase, which inhibits a myosin light chain phosphatase activity and thereby enhances myosin light chain phosphorylation and subsequent myosin II ATPase activation, eventually causing actomyosin contraction (Kimura et al., 1996; Uehata et al., 1997). We showed here that the CC region–mediated binding of afadin to αE-catenin was essential for the shroom3-induced apical constriction in EpH4 cells. These results indicate that the binding of afadin to αE-catenin is critical for the proper actomyosin association with the β-catenin–E-cadherin complex through αE-catenin. Furthermore, these results, together with the previous observations that the actomyosin undercoating AJs strengthens E-cadherin–mediated cell adhesion (Angres et al., 1996; Chu et al., 2004; Imamura et al., 1999), raised the possibility that the proper actomyosin association with AJs by the CC-region–mediated binding of afadin to αE-catenin regulates cell adhesion strength mainly mediated by E-cadherin in EpH4 cells.
We then examined here the regulatory mechanism for the binding of afadin to αE-catenin to reveal the role of the CC region of afadin in the FAB activity of αE-catenin. It was previously shown that αE-catenin is present as the conformationally closed form in solution in vitro (Maki et al., 2016; Yonemura et al., 2010). In the conformationally closed form of αE-catenin, the MI subdomain is stabilized and thereby masked by the subdomains containing the MII and MIII subdomains, causing the inhibition of the binding of vinculin to the MI subdomain (Hirano et al., 2018; Maki et al., 2016; Yonemura et al., 2010). The amino acid residues at the interface among the MI, MII, and MIII subdomains stabilize the conformationally closed form to inhibit the binding of vinculin to the MI subdomain of αE-catenin (Ishiyama et al., 2013). This conformationally closed form is opened by force, exposing the vinculin-binding region in the MI subdomain (Ishiyama et al., 2013; Maki et al., 2016; Yonemura et al., 2010). Point mutations in these residues (M319G/R326E) destabilize the conformationally closed form and convert it to the conformationally open form to cause the binding of vinculin to the MI subdomain of αE-catenin (Maki et al., 2016; Matsuzawa et al., 2018). We showed here using various domain/deletion mutants and the conformationally open form mutant (M319G/R326E) of αE-catenin that the MI subdomain conversely masked the MIII subdomain and inhibited the binding of afadin to the MIII subdomain, indicating that afadin binds to only the conformationally open form of αE-catenin. Previous magnetic tweezers measurements revealed that the force required to expose the vinculin-binding region in the MI subdomain is less than that required to fully stretch all the three MI, MII, and MIII subdomains (Maki et al., 2016; Yao et al., 2014), but the force required to expose the afadin-binding region in the MIII subdomain remains unexplored in this study. In addition to force, there may be other possible regulatory mechanisms for the conformational changes of αE-catenin for its binding of vinculin and afadin. For instance, vinculin and afadin may mutually regulate their respective binding to αE-catenin. It was shown that ponsin binds to both vinculin and afadin (Mandai et al., 1999) and that αE-catenin is phosphorylated by casein kinase (Escobar et al., 2015). Therefore, ponsin and the phosphorylation of αE-catenin could regulate the conformational changes of αE-catenin for its binding of vinculin and afadin. Thus, it is important to investigate how the conformationally closed form is destabilized to expose the vinculin-binding region in the MI subdomain and the afadin-binding region in the MIII subdomain for our better understanding of the respective roles of vinculin and afadin in the proper actomyosin association with αE-catenin complexed with β-catenin and E-cadherin.
It was noted that actomyosin was not completely separated from the plasma membrane at AJs in afadin-KO cells unless shroom3 was overexpressed and that vinculin localized near the tricellular junctions, which are the hot spots of epithelial tension (Higashi and Miller, 2017), in afadin-KO cells. These results raised the possibility that another unidentified afadin-independent mechanism is present for maintaining the proper actomyosin association with αE-catenin complexed with β-catenin and E-cadherin. Therefore, two mechanisms, afadin-dependent and afadin-independent ones, may be present for maintaining the proper actomyosin association with αE-catenin complexed with β-catenin and E-cadherin.
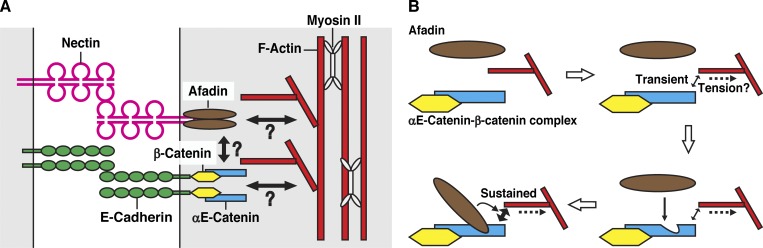
The previous studies performed in solution in vitro showed that the dimer of αE-catenin shows a stronger FAB activity than the monomer and that the dimerization of αE-catenin is inhibited by its binding of β-catenin (Drees et al., 2005; Yamada et al., 2005). However, the present results indicate that the CC region of afadin allosterically enhanced the FAB activity of αE-catenin when it bound to the conformationally open form of αE-catenin and that the dimerization of αE-catenin was not absolutely essential for its increased FAB activity as far as tested in our solution in vitro assays. A “catch-bond model” was recently proposed, in which the force exerted to the αE-catenin–β-catenin–E-cadherin complex induces its transient binding to F-actin (Buckley et al., 2014). Although it is not clear whether force is required to expose the afadin-binding region in the MIII subdomain of αE-catenin, it is possible that the force required to induce the transient binding of the αE-catenin–β-catenin–E-cadherin complex to F-actin also exposes the afadin-binding region in the MIII subdomain, resulting in the CC-afadin–mediated sustained binding of the αE-catenin–β-catenin–E-cadherin complex to F-actin (Fig. 8). This novel role of afadin explains at least a part of the mechanism by which the transient binding of the αE-catenin–β-catenin–E-cadherin complex to F-actin is stabilized. The previous structure-function analyses reported that some αE-catenin mutations affect the FAB activity, in which a linker region between the MIII subdomain and the C domain (Escobar et al., 2015) or an α helix in the C domain is mutated (Ishiyama et al., 2018). The binding of CC region of afadin to αE-catenin may alter the structure of these regions and regulate the FAB activity of αE-catenin.
Figure 8.
Schematic representation of actomyosin-undercoated AJs and the afadin-regulated FAB activity of αE-catenin. (A) The molecular compositions of AJs. α-Actinin and vinculin are not shown. (B) A model for the mode of action of afadin in the FAB activity of αE-catenin.
Actomyosin shows plasticity and undergoes rapid dissociation even after AJ formation, given that AJs are dynamically remodeled in morphogenesis and tissue regeneration (Harris and Tepass, 2010; Maruthamuthu et al., 2010). Previous studies on afadin-KO mice showed developmental defects at stages during and after gastrulation, including disorganization of the ectoderm, impaired migration of the mesoderm, and loss of somites and other structures derived from both the ectoderm and the mesoderm (Ikeda et al., 1999; Zhadanov et al., 1999). The defects of afadin-KO mice are quite diverse depending on tissues and developmental stages. The tissue-specific KO of afadin in the intestine showed apparently normal phenotypes, including actomyosin organization (Tanaka-Okamoto et al., 2011). The studies in Drosophila showed that canoe regulates the maintenance of AJs in some morphogenetically active cells, but not in steady-state cells (Sawyer et al., 2009). In addition, recent studies revealed that afadin or canoe plays critical roles for the dynamic regulation of AJs (Choi et al., 2016; Manning et al., 2019). Therefore, afadin may play especially important roles in the maintenance of AJs and the proper actomyosin organization at AJs where the cell dynamics and force in tissue are elevated. Consistent with these previous observations, the present results suggest that the association and dissociation among afadin, αE-catenin, and F-actin play an important role in not only cell adhesion strength but also AJ plasticity. However, further studies are needed to elucidate the dynamic regulation of the proper actomyosin organization at AJs and the involvement of protein–protein interaction networks at AJs.
Materials and methods
Cell culture and transfection
Wild-type and afadin-KO cells were obtained as described previously (Sakakibara et al., 2018). The cells were maintained in DMEM with 10% FBS and 1% penicillin/streptomycin and were cultured under 5% CO2 at 37°C. Lipofectamine 3000 (Invitrogen) was used as a reagent for plasmid transfection.
Plasmids
An expression vector for FLAG-tagged shroom3 (pCA-FLAG-shroom3; Nishimura and Takeichi, 2008) and the cDNA for mouse αE-catenin were kind gifts from M. Takeichi (Center for Biosystems Dynamics Research, RIKEN, Kobe, Japan). The cDNA for mouse afadin was described previously (Sakakibara et al., 2018). For the generation of the cDNAs for β/α-cat, the cDNA for the β-catenin fragment (aa 78–151) and the αE-catenin fragment (aa 56–906) were fused by In-Fusion PCR cloning (Takara Bio USA, Inc.). The cDNAs for β/α-cat-CA, MII-C I792A, and MII-C V796A were constructed by site-directed mutagenesis. The cDNAs for FL αE-catenin (aa 1–906), αE-catenin fragments of MI-C (aa 276–906), MII-C (aa 385–906), MIII-C (aa 509–906), C (aa 631–906), β/α-cat, β/α-cat-CA, MII-C I792A, and MII-C V796A were subcloned into pGEX-6P-1 (GE Healthcare). The cDNAs for CC-afadin (aa 1386–1595) and GFP were subcloned into pGEX-6P-1-FLAG, which was constructed by subcloning the insert encoding the FLAG epitope into pGEX-6P-1. The cDNAs for FL afadin, afadin-ΔCC (aa 1–1385 and 1596–1813), and CC-afadin were subcloned into the pCAG-FLAG-IRESpuro vector, which was constructed by subcloning the CAG promoter, the IRES puro resistance cassette, and the insert encoding the FLAG epitope into pEGFP-C1 (Takara Bio USA, Inc.).
Stable cell lines
For the generation of stable cell lines expressing FLAG-tagged afadin, FLAG-tagged afadin-ΔCC, or FLAG-tagged CC-afadin, afadin-KO cells were transfected with pCAG-FLAG-Afadin-IRESpuro, pCAG-FLAG-Afadin-ΔCC-IRESpuro, or pCAG-FLAG-CC-Afadin-IRESpuro, respectively. After the transfected cells were selected using 2.5 µg/ml puromycin (Sigma-Aldrich), single cell–derived clones were established.
Antibodies (Abs)
A rabbit anti–l-afadin pAb was prepared as described previously (Mandai et al., 1997). The Abs listed below were purchased from commercial sources: rabbit anti–α-catenin pAb (C2081; Sigma-Aldrich); mouse anti–α-actinin mAb (A5044; Sigma-Aldrich); rabbit anti–β-catenin pAb (C2206; Sigma-Aldrich); mouse anti-FLAG mAb (F3165; Sigma-Aldrich); rabbit anti-GAPDH mAb (14C10; Cell Signaling Technology); rabbit anti-myosin light chain (phospho S20) pAb (ab2480; Abcam); rabbit anti-sodium potassium ATPase pAb (ab76020; Abcam); rabbit anti-nonmuscle myosin heavy chain II-B pAb (PRB-445P; BioLegend); mouse anti-vinculin mAb (V9264; Sigma-Aldrich); and rat anti–ZO-1 mAb (sc-33725; Santa Cruz Biotechnology). A rat anti–E-cadherin mAb (ECCD2) was a kind gift from M. Takeichi. The HRP-conjugated secondary Abs used for Western blotting were purchased from GE Healthcare. The Alexa Fluor–conjugated secondary Abs used for immunocytochemistry were purchased from Thermo Fisher Scientific.
Immunocytochemistry
The cells to be examined were seeded in a 24-well plate (1 × 105 cells per well), cultured under normal conditions for 24 h, and fixed with a fixative containing 2% PFA, 4% sucrose, 1 mM sodium pyruvate, HBSS containing 1 mM CaCl2 and 1 mM MgCl2 (Thermo Fisher Scientific), and 10 mM Hepes, pH 7.4, at 37°C for 15 min. The fixed cells were permeabilized with 0.25% Triton X-100 in PBS for 10 min, and then blocked in PBS containing 10% goat serum at room temperature for 20 min. The cells were then incubated with primary Abs in PBS containing 20% BlockAce (DS Pharma Biomedical) at 4°C overnight. After three washes in PBS at room temperature, the cells were incubated with Alexa Fluor–conjugated secondary Abs (Thermo Fisher Scientific) at room temperature for 45 min and then washed three times with PBS. To visualize F-actin, Alexa Fluor 488–conjugated phalloidin (Thermo Fisher Scientific, A12379) was used. The samples were then mounted in a FluorSave reagent (Merck Millipore). The images were acquired at a 0.4-µm step along the z axis using a Nikon C2 confocal system (Nikon, Inc.) with a Plan Apo 60×/1.2 numerical aperture water immersion objective lens (Nikon, Inc.). Maximum signal intensity projection images were obtained using the ImageJ software program (Schneider et al., 2012).
Electron microscopy
The cells to be examined were seeded in a 24-well plate (1 × 105 cells per well), cultured under normal conditions for 24 h, and fixed with a fixative containing 2% PFA, 2.5% glutaraldehyde, and 50 mM Hepes, pH 7.5, at room temperature for 2 h. After the samples were washed with the same buffer, they were post-fixed with 1% OsO4 in 50 mM Hepes, pH 7.5, at 4°C for 30 min. The samples were then embedded in resin (epon 812; TAAB). Ultrathin sections were stained with uranyl acetate and lead citrate before observation with an electron microscope (JEM-1400 Plus; JEOL).
Immunoprecipitation assay
The cells to be examined were washed with ice-cold PBS and then lysed with a lysis buffer (20 mM Tris-HCl, pH 7.5, 1% Triton X-100, 10% glycerol, 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1 mM NaF, and 1 mM DTT). The cell lysates were obtained by centrifugation at 20,000 g for 15 min. The cell lysates were incubated with the rabbit anti–α-catenin pAb-conjugated protein A–sepharose at 4°C for 3 h. After the beads were extensively washed with the lysis buffer, proteins bound to the beads were eluted using an SDS sample buffer (60 mM Tris-HCl at pH 6.7, 3% SDS, 2% 2-mercaptoethanol, and 5% glycerol) and boiled for 5 min. The samples were separated by SDS-PAGE, followed by Western blotting with the indicated Abs.
Western blotting
The samples separated by SDS-PAGE were transferred to polyvinylidene difluoride membranes (Merck Millipore). After the membranes were blocked with 5% skim milk in TBS plus 0.05% Tween 20, they were incubated with the indicated Abs. After the membranes were washed with TBS plus 0.05% Tween 20 three times, they were incubated with HRP-conjugated anti-rabbit or anti-mouse IgG Abs. The signals for the proteins were detected using Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore) and a LAS-4000 luminescent image analyzer (Fujifilm).
Shroom3 overexpression assay
4 h after cell seeding, the cells to be examined were transfected with pCA-FLAG-shroom3 and incubated for 20 h. The cells were then fixed and stained with the indicated Abs and phalloidin.
Recombinant protein expression and purification
Various fragments of αE-catenin, β/α-cat, and β/α-cat-CA and the FLAG-tagged fragment of CC-afadin and GFP were expressed as GST-fused proteins, which contained the recognition sequence for site-specific cleavage by PreScission Protease between the GST domain and the proteins of interest, in Escherichia coli BL21-CodonPlus (DE3)-RIPL cells. The E. coli cells were grown to an A600 of 0.8 at 37°C and induced with 0.2 mM IPTG at 30°C for 3 h. The E. coli cells were sonicated in buffer A (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1 mM DTT), followed by centrifugation at 100,000 g at 4°C for 20 min. The supernatant was then incubated with glutathione-sepharose 4B beads (GE Healthcare) at 4°C for 1 h to immobilize GST-fused proteins. After the beads were washed with buffer A, the proteins of interest were removed from GST by cleavage with PreScission Protease (GE Healthcare) at 4°C for 4 h and stored at –80°C.
Immunoprecipitation assay of recombinant proteins
Various fragments of αE-catenin, β/α-cat, and β/α-cat-CA were incubated with the FLAG-tagged fragment of CC-afadin at a final concentration of 4 µM in buffer A at room temperature for 1 h, and immunoprecipitated with an anti-FLAG mAb. The samples were mixed with the SDS sample buffer, separated by SDS-PAGE, and analyzed by Coomassie Brilliant Blue (CBB) staining. The gel images were scanned with an EPSON GT-X750 scanner (Seiko Epson, Inc.). The intensity of the gel bands was measured using the ImageJ software program.
F-actin cosedimentation assay
Human platelet nonmuscle G-actin (Cytoskeleton, Inc.) was incubated in an actin polymerization buffer (20 mM Hepes, pH 7.5, 100 mM KCl, 2 mM MgCl2, 0.5 mM ATP, and 1 mM EGTA) at room temperature for 1 h. αE-catenin fragments, CC-afadin, and GFP were dialyzed against an F-actin cosedimentation buffer (20 mM Hepes, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.5 mM ATP, 1 mM EGTA, and 1 mM DTT), diluted to the final concentration of 2 µM, and incubated with 8 µM F-actin at room temperature for 1 h. The samples were centrifuged at 100,000 g at 4°C for 20 min. The pellets were resuspended in the F-actin cosedimentation buffer. The samples were mixed with the SDS sample buffer, separated by SDS-PAGE, and analyzed by CBB staining. The gel images were scanned with an EPSON GT-X750 scanner. The intensity of the gel bands was measured using the ImageJ software program.
Sucrose density gradient centrifugation
20 μM αE-catenin fragments in buffer A with or without 20 µM CC-afadin were incubated at 4°C overnight. Then, 200 µl of these samples were layered onto a 4.8-ml linear 10–40% sucrose density gradient in buffer A, followed by centrifugation at 200,000 g at 4°C for 20 h with a swing rotor (P55ST2; Hitachi Ltd.). Fractions (300 µl each) were collected. An aliquot of each fraction was mixed with the SDS sample buffer, separated by SDS-PAGE, and analyzed by CBB staining.
Size exclusion chromatography
10 μM MIII-C in buffer A with or without 10 µM CC-afadin were incubated at 25°C overnight. Then, 100 µl of these samples were applied to an analytical Superdex 200 size exclusion column maintained at 4°C. An aliquot of each fraction was mixed with the SDS sample buffer, separated by SDS-PAGE, and analyzed by CBB staining.
Online supplemental material
Fig. S1 and Fig. S2 include additional data showing the role of afadin in the proper actomyosin organization at AJs. Fig. S3 includes additional data showing the role of the binding of afadin to αE-catenin in the proper actomyosin organization at AJs.
Acknowledgments
The authors thank Dr. Masatoshi Takeichi for providing the rat anti–E-cadherin mAb (ECCD2) and pCA-FLAG-shroom3 and for his useful discussions; Dr. Yasunori Yamamoto for assistance with the sucrose density gradient centrifugation analysis; Dr. Tomohiko Maruo for his useful discussions; and Dr. Brian Quinn of Japan Medical Communication for careful editing of the manuscript.
This study was supported by MEXT KAKENHI (Ministry of Education, Culture, Sports, Science, and Technology Grants-in-Aid for Scientific Research) grant JP26114007 (Y. Takai), AMED (Japan Agency for Medical Research and Development) grant JP19cm0106111 (Y. Takai), JSPS KAKENHI (Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research) grants JP16J06236 (S. Sakakibara), JP17K08636 (A. Sakane), and JP18H02617 (S. Yonemura), and JST CREST (Japan Science and Technology Agency Core Research for Evolutional Science and Technology) grant JPMJCR13W4 (S. Yonemura).
The authors declare no competing financial interests.
Author contributions: Y. Takai conceived the research project. S. Sakakibara, K. Mizutani, and Y. Takai designed the experiments. S. Sakakibara, K. Mizutani, A. Sugiura, A. Sakane, T. Sasaki, and S. Yonemura performed the experiments. S. Sakakibara, K. Mizutani, A. Sugiura, A. Sakane, T. Sasaki, S. Yonemura, and Y. Takai analyzed the data and wrote the manuscript.
References
- Angres B., Barth A., and Nelson W.J.. 1996. Mechanism for transition from initial to stable cell-cell adhesion: kinetic analysis of E-cadherin-mediated adhesion using a quantitative adhesion assay. J. Cell Biol. 134:549–557. 10.1083/jcb.134.2.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini J.M., Kitt K.N., Gloerich M., Pokutta S., Weis W.I., and Nelson W.J.. 2015. Reevaluating αE-catenin monomer and homodimer functions by characterizing E-cadherin/αE-catenin chimeras. J. Cell Biol. 210:1065–1074. 10.1083/jcb.201411080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley C.D., Tan J., Anderson K.L., Hanein D., Volkmann N., Weis W.I., Nelson W.J., and Dunn A.R.. 2014. Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science. 346:1254211 10.1126/science.1254211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavey M., and Lecuit T.. 2009. Molecular bases of cell-cell junctions stability and dynamics. Cold Spring Harb. Perspect. Biol. 1:a002998 10.1101/cshperspect.a002998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.S., Hong S., Indra I., Sergeeva A.P., Troyanovsky R.B., Shapiro L., Honig B., and Troyanovsky S.M.. 2015. α-Catenin-mediated cadherin clustering couples cadherin and actin dynamics. J. Cell Biol. 210:647–661. 10.1083/jcb.201412064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W., Acharya B.R., Peyret G., Fardin M.A., Mège R.M., Ladoux B., Yap A.S., Fanning A.S., and Peifer M.. 2016. Remodeling the zonula adherens in response to tension and the role of afadin in this response. J. Cell Biol. 213:243–260. 10.1083/jcb.201506115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y.-S., Thomas W.A., Eder O., Pincet F., Perez E., Thiery J.P., and Dufour S.. 2004. Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J. Cell Biol. 167:1183–1194. 10.1083/jcb.200403043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees F., Pokutta S., Yamada S., Nelson W.J., and Weis W.I.. 2005. α-catenin is a molecular switch that binds E-cadherin-β-catenin and regulates actin-filament assembly. Cell. 123:903–915. 10.1016/j.cell.2005.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimova N., and Svitkina T.M.. 2018. Branched actin networks push against each other at adherens junctions to maintain cell-cell adhesion. J. Cell Biol. 217:1827–1845. 10.1083/jcb.201708103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escobar D.J., Desai R., Ishiyama N., Folmsbee S.S., Novak M.N., Flozak A.S., Daugherty R.L., Mo R., Nanavati D., Sarpal R., et al. 2015. α-Catenin phosphorylation promotes intercellular adhesion through a dual-kinase mechanism. J. Cell Sci. 128:1150–1165. 10.1242/jcs.163824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigo S.L., Hildebrand J.D., Harland R.M., and Wallingford J.B.. 2003. Shroom induces apical constriction and is required for hingepoint formation during neural tube closure. Curr. Biol. 13:2125–2137. 10.1016/j.cub.2003.11.054 [DOI] [PubMed] [Google Scholar]
- Harris T.J., and Tepass U.. 2010. Adherens junctions: from molecules to morphogenesis. Nat. Rev. Mol. Cell Biol. 11:502–514. 10.1038/nrm2927 [DOI] [PubMed] [Google Scholar]
- Higashi T., and Miller A.L.. 2017. Tricellular junctions: how to build junctions at the TRICkiest points of epithelial cells. Mol. Biol. Cell. 28:2023–2034. 10.1091/mbc.e16-10-0697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand J.D. 2005. Shroom regulates epithelial cell shape via the apical positioning of an actomyosin network. J. Cell Sci. 118:5191–5203. 10.1242/jcs.02626 [DOI] [PubMed] [Google Scholar]
- Hildebrand J.D., and Soriano P.. 1999. Shroom, a PDZ domain-containing actin-binding protein, is required for neural tube morphogenesis in mice. Cell. 99:485–497. 10.1016/S0092-8674(00)81537-8 [DOI] [PubMed] [Google Scholar]
- Hirano Y., Amano Y., Yonemura S., and Hakoshima T.. 2018. The force-sensing device region of α-catenin is an intrinsically disordered segment in the absence of intramolecular stabilization of the autoinhibitory form. Genes Cells. 23:370–385. 10.1111/gtc.12578 [DOI] [PubMed] [Google Scholar]
- Ikeda W., Nakanishi H., Miyoshi J., Mandai K., Ishizaki H., Tanaka M., Togawa A., Takahashi K., Nishioka H., Yoshida H., et al. 1999. Afadin: A key molecule essential for structural organization of cell-cell junctions of polarized epithelia during embryogenesis. J. Cell Biol. 146:1117–1132. 10.1083/jcb.146.5.1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura Y., Itoh M., Maeno Y., Tsukita S., and Nagafuchi A.. 1999. Functional domains of α-catenin required for the strong state of cadherin-based cell adhesion. J. Cell Biol. 144:1311–1322. 10.1083/jcb.144.6.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama N., Tanaka N., Abe K., Yang Y.J., Abbas Y.M., Umitsu M., Nagar B., Bueler S.A., Rubinstein J.L., Takeichi M., and Ikura M.. 2013. An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J. Biol. Chem. 288:15913–15925. 10.1074/jbc.M113.453928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiyama N., Sarpal R., Wood M.N., Barrick S.K., Nishikawa T., Hayashi H., Kobb A.B., Flozak A.S., Yemelyanov A., Fernandez-Gonzalez R., et al. 2018. Force-dependent allostery of the α-catenin actin-binding domain controls adherens junction dynamics and functions. Nat. Commun. 9:5121 10.1038/s41467-018-07481-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Ito M., Amano M., Chihara K., Fukata Y., Nakafuku M., Yamamori B., Feng J., Nakano T., Okawa K., et al. 1996. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science. 273:245–248. 10.1126/science.273.5272.245 [DOI] [PubMed] [Google Scholar]
- Maiden S.L., and Hardin J.. 2011. The secret life of α-catenin: moonlighting in morphogenesis. J. Cell Biol. 195:543–552. 10.1083/jcb.201103106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki K., Han S.W., Hirano Y., Yonemura S., Hakoshima T., and Adachi T.. 2016. Mechano-adaptive sensory mechanism of α-catenin under tension. Sci. Rep. 6:24878 10.1038/srep24878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai K., Nakanishi H., Satoh A., Obaishi H., Wada M., Nishioka H., Itoh M., Mizoguchi A., Aoki T., Fujimoto T., et al. 1997. Afadin: A novel actin filament-binding protein with one PDZ domain localized at cadherin-based cell-to-cell adherens junction. J. Cell Biol. 139:517–528. 10.1083/jcb.139.2.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandai K., Nakanishi H., Satoh A., Takahashi K., Satoh K., Nishioka H., Mizoguchi A., and Takai Y.. 1999. Ponsin/SH3P12: an l-afadin- and vinculin-binding protein localized at cell-cell and cell-matrix adherens junctions. J. Cell Biol. 144:1001–1017. 10.1083/jcb.144.5.1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning L.A., Perez-Vale K.Z., Schaefer K.N., Sewell M.T., and Peifer M.. 2019. The Drosophila Afadin and ZO-1 homologues Canoe and Polychaetoid act in parallel to maintain epithelial integrity when challenged by adherens junction remodeling. Mol. Biol. Cell. 30:1938–1960. 10.1091/mbc.E19-04-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruo T., Sakakibara S., Miyata M., Itoh Y., Kurita S., Mandai K., Sasaki T., and Takai Y.. 2018. Involvement of l-afadin, but not s-afadin, in the formation of puncta adherentia junctions of hippocampal synapses. Mol. Cell. Neurosci. 92:40–49. 10.1016/j.mcn.2018.06.006 [DOI] [PubMed] [Google Scholar]
- Maruthamuthu V., Aratyn-Schaus Y., and Gardel M.L.. 2010. Conserved F-actin dynamics and force transmission at cell adhesions. Curr. Opin. Cell Biol. 22:583–588. 10.1016/j.ceb.2010.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa K., Himoto T., Mochizuki Y., and Ikenouchi J.. 2018. α-Catenin controls the anisotropy of force distribution at cell-cell junctions during collective cell migration. Cell Rep. 23:3447–3456. 10.1016/j.celrep.2018.05.070 [DOI] [PubMed] [Google Scholar]
- McNeill H., Ozawa M., Kemler R., and Nelson W.J.. 1990. Novel function of the cell adhesion molecule uvomorulin as an inducer of cell surface polarity. Cell. 62:309–316. 10.1016/0092-8674(90)90368-O [DOI] [PubMed] [Google Scholar]
- Nishimura T., and Takeichi M.. 2008. Shroom3-mediated recruitment of Rho kinases to the apical cell junctions regulates epithelial and neuroepithelial planar remodeling. Development. 135:1493–1502. 10.1242/dev.019646 [DOI] [PubMed] [Google Scholar]
- Nishimura T., and Takeichi M.. 2009. Remodeling of the adherens junctions during morphogenesis. Curr. Top. Dev. Biol. 89:33–54. 10.1016/S0070-2153(09)89002-9 [DOI] [PubMed] [Google Scholar]
- Pokutta S., Drees F., Takai Y., Nelson W.J., and Weis W.I.. 2002. Biochemical and structural definition of the l-afadin- and actin-binding sites of α-catenin. J. Biol. Chem. 277:18868–18874. 10.1074/jbc.M201463200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara S., Maruo T., Miyata M., Mizutani K., and Takai Y.. 2018. Requirement of the F-actin-binding activity of l-afadin for enhancing the formation of adherens and tight junctions. Genes Cells. 23:185–199. 10.1111/gtc.12566 [DOI] [PubMed] [Google Scholar]
- Sato T., Fujita N., Yamada A., Ooshio T., Okamoto R., Irie K., and Takai Y.. 2006. Regulation of the assembly and adhesion activity of E-cadherin by nectin and afadin for the formation of adherens junctions in Madin-Darby canine kidney cells. J. Biol. Chem. 281:5288–5299. 10.1074/jbc.M510070200 [DOI] [PubMed] [Google Scholar]
- Sawyer J.K., Harris N.J., Slep K.C., Gaul U., and Peifer M.. 2009. The Drosophila afadin homologue Canoe regulates linkage of the actin cytoskeleton to adherens junctions during apical constriction. J. Cell Biol. 186:57–73. 10.1083/jcb.200904001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smutny M., Cox H.L., Leerberg J.M., Kovacs E.M., Conti M.A., Ferguson C., Hamilton N.A., Parton R.G., Adelstein R.S., and Yap A.S.. 2010. Myosin II isoforms identify distinct functional modules that support integrity of the epithelial zonula adherens. Nat. Cell Biol. 12:696–702. 10.1038/ncb2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher T., and Ebnet K.. 2018. The regulation of junctional actin dynamics by cell adhesion receptors. Histochem. Cell Biol. 150:341–350. 10.1007/s00418-018-1691-8 [DOI] [PubMed] [Google Scholar]
- Stevenson B.R., Siliciano J.D., Mooseker M.S., and Goodenough D.A.. 1986. Identification of ZO-1: a high molecular weight polypeptide associated with the tight junction (zonula occludens) in a variety of epithelia. J. Cell Biol. 103:755–766. 10.1083/jcb.103.3.755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana K., Nakanishi H., Mandai K., Ozaki K., Ikeda W., Yamamoto Y., Nagafuchi A., Tsukita S., and Takai Y.. 2000. Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J. Cell Biol. 150:1161–1176. 10.1083/jcb.150.5.1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeichi M. 2014. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15:397–410. 10.1038/nrm3802 [DOI] [PubMed] [Google Scholar]
- Tanaka-Okamoto M., Hori K., Ishizaki H., Itoh Y., Onishi S., Yonemura S., Takai Y., and Miyoshi J.. 2011. Involvement of afadin in barrier function and homeostasis of mouse intestinal epithelia. J. Cell Sci. 124:2231–2240. 10.1242/jcs.081000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehata M., Ishizaki T., Satoh H., Ono T., Kawahara T., Morishita T., Tamakawa H., Yamagami K., Inui J., Maekawa M., and Narumiya S.. 1997. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 389:990–994. 10.1038/40187 [DOI] [PubMed] [Google Scholar]
- Yamada S., Pokutta S., Drees F., Weis W.I., and Nelson W.J.. 2005. Deconstructing the cadherin-catenin-actin complex. Cell. 123:889–901. 10.1016/j.cell.2005.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M., Qiu W., Liu R., Efremov A.K., Cong P., Seddiki R., Payre M., Lim C.T., Ladoux B., Mège R.M., and Yan J.. 2014. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun. 5:4525 10.1038/ncomms5525 [DOI] [PubMed] [Google Scholar]
- Yonemura S., Wada Y., Watanabe T., Nagafuchi A., and Shibata M.. 2010. α-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12:533–542. 10.1038/ncb2055 [DOI] [PubMed] [Google Scholar]
- Zhadanov A.B., Provance D.W. Jr., Speer C.A., Coffin J.D., Goss D., Blixt J.A., Reichert C.M., and Mercer J.A.. 1999. Absence of the tight junctional protein AF-6 disrupts epithelial cell-cell junctions and cell polarity during mouse development. Curr. Biol. 9:880–888. 10.1016/S0960-9822(99)80392-3 [DOI] [PubMed] [Google Scholar]