Abstract
Respiratory infections after air travel are frequent, but epidemiological data are incomplete. Using sensitive polymerase chain reactions, we studied the spectrum of atypical bacteria and respiratory viruses in travelers fulfilling the case definition of severe acute respiratory syndrome. A pathogen was identified in 67 travelers (43.2%). Influenza and parainfluenza viruses were most prevalent, at 14.2% and 15.5%, respectively. Prevalences of adenoviruses, human metapneumovirus, coronaviruses, and rhinoviruses ranged between 2.6% and 4.8%. Human bocavirus, respiratory syncytial virus, and Legionella, Mycoplasma, and Chlamydophila species were absent or appeared at frequencies of <1%. To our knowledge, these are the first specific baseline data for the mentioned agents in the context of air travel.
Acute respiratory infections are frequently experienced after air travel. Because most patients do not see a doctor, current knowledge about incidence and etiology is imprecise and anecdotal [1]. The very few systematic studies available suggested that up to 20% of passengers may develop respiratory infections within 1 week after air travel and that flight attendants show significantly higher incidence rates of respiratory infections than do control groups [2, 3]. Even though antibiotic therapy against common bacterial respiratory disease is often administered, laboratory data on the spectrum of causative agents are actually not available [1, 4, 5]. The epidemic of severe acute respiratory syndrome (SARS) in 2003 involved a period of heightened awareness of respiratory infections after air travel. Samples for laboratory testing were routinely obtained from patients, which provided a unique opportunity for studying their disease etiologies.
After initial characterization of the causative agent of SARS, we acted as a diagnostic reference laboratory for the World Health Organization (WHO) [6]. During the epidemic, we accepted samples for initial and confirmatory testing exclusively from those patients who fulfilled the WHO case definition of suspected or probable SARS. The definition was designed to be sensitive and thus to prevent any possible transmission, but its specificity was low. It covered most respiratory illnesses compatible with viral or atypical bacterial infection.
Respiratory agents are best diagnosed by direct assays, the most sensitive of which is polymerase chain reaction (PCR). Because of the rapid pace at which the technique evolves, we searched the literature for the most up-to-date PCR assays that cover the broadest possible range of genetic diversity for each respective agent. High sensitivities had to be clearly proved in studies. Where assays fulfilling these criteria were not available, we established sensitive real-time PCRs de novo. These assays were used to determine a point prevalence of the full spectrum of respiratory viruses and atypical bacteria in SARS-compatible patients.
Patients and methods. Respiratory samples (n=214) from 172 patients were available. All patients fulfilled the WHO case definition of suspected or probable SARS, which required a combination of fever and lower-respiratory-tract symptoms (e.g., cough or difficulty breathing) plus either a stay in an affected area during the preceding 10 days or close contact with suspected patients. Probable cases additionally had radiological evidence of respiratory distress syndrome without other reason.
Samples were prepared as described elsewhere [6]. Assays included adenovirus (AdV) [7]; rhinovirus (RV) [8]; human bocavirus (hBoV) [9]; human coronaviruses (hCoV) 229E, NL63, and OC43 (table 1); influenza virus (Inf) A (table 1); and a combined assay for Inf A and B, parainfluenzaviruses (PIVs) 1–3, respiratory syncytial virus (RSV), and human metapneumovirus (hMPV) (Hexaplex Plus; Prodesse). The commercial system was chosen because several published respiratory multiplex assays gave instable results in our laboratory. We also tested for the presence of atypical bacteria, including Mycoplasma, Chlamydophila, and Legionella species [10]. for mulations of new assays established for the present study are provided in table 1. Their limits of detection (LODs) were determined using in vitro-transcribed RNA [6]. LODs for hCoVs NL63 and Inf A were <5 copies of synthetic RNA genomes per reaction, corresponding to ∼50 RNA copies/swab sample. LODs for hCoV 229E and OC43 were <20 copies/ reaction, corresponding to ∼200 copies/swab sample.
Table 1.
Formulations of reverse-transcription (RT)-polymerase chain reaction (PCR) assays designed for the study.

>Results. Travel histories were reconstructed by telephone interviews with hospitals, family physicians, or patients themselves. For 164 patients (hereafter called “flight patients”), additional information was retrieved. This included 124 for whom the exact dates and destinations of departure to Germany were known and 22 for whom only destinations could be reconstructed. Seventeen patients had not traveled as initially reported but had been in contact with patients who had suspected SARS. For 8 patients, no additional information could be retrieved. They were included in the cohort because a travel history had been confirmed before samples were received. Only those patients who had reportedly not traveled were evaluated separately. They are hereafter called “contact patients.”
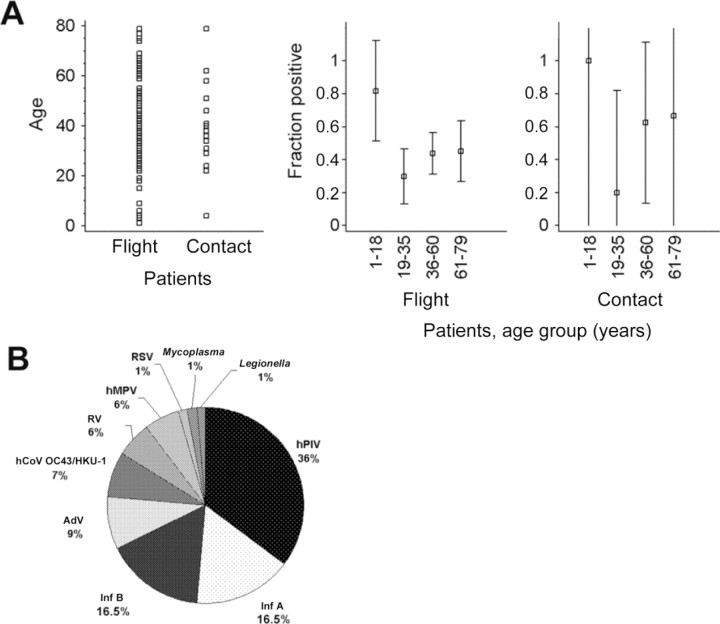
The average age of flight patients was 42.2 years (range, 1–79 years). The average age of contact patients was 41.8 years (range, 4–79 years). Neither the average or median ages nor the SDs in both groups were significantly different. Figure 1 shows the age distributions in both groups. Some 71% of flight patients were between 19 and 60 years old, as were 76% of contact patients (P=.57). Only 12 patients were <19 years old, 11 of whom were flight patients. They contributed 13.4% of all positive findings in flight patients, which was significantly more than the mean positivity rate in all age groups (P<.012, Student's t test).
Figure 1.
A, Age distribution of patients (left) and rates of detection of any tested agent in 4 different age groups (middle and right). The age of 15 of 172 patients could not be recorded because these were coded requests forwarded to us by other laboratories. Range lines in the middle and right panels depict 95% confidence intervals (CIs). Note that large CIs in the contacts panel are due to the small no. of patients in this group (n=17). Sample sizes in the different age np17 groups were as follows. In the middle panel (flight patients), 1–18 years, 8 patients; 19–35 years, 37 patients; 36–60 years, 64 patients; and 61–79 years, 31 patients. In the right panel (contact patients), 1–18 years, 1 patient; 19–35 years, 5 patients; 36–60 years, 8 patients; and 61–79 years, 3 patients. B, Relative detection rates of agents in travelers. AdV, adenovirus; BoV, bocavirus; hCoV, human coronavirus; hMPV, human metapneumovirus; hPIV, human parainfluenza virus; Inf, influenza virus; RSV, respiratory syncytial virus; RV, rhinovirus.
At least 1 pathogen was detected in 67 flight patients (43.2%) and in 8 contact patients (47%) (P=.7, Student's t test). table 2 shows the global pathogen detection rates by age group, as well as the relative and absolute prevalences of pathogens in flight patients. In contact patients (n=17), AdV, human PIV, Inf B, and hMPV were detected in 2 patients each and Inf A in 1 patient. Double infections occurred in only 2 patients: RV/ PIV in a flight patient and AdV/PIV in a contact patient.
Table 2.
Absolute detection rates of agents in flight patients.
Inf and PIV were clearly the most prevalent agents in flight patients, at 14.2% and 15.5%, respectively, without significant differences between age groups (1-way analysis of variance [ANOVA], 95% significance level). Equal distribution of these viruses was also seen in contact patients. CoVs were more frequent than one would expect in a mostly adult cohort. Detection rates did not differ between age groups at the 95% significance level. RSV and hMPV appeared to be significantly more frequent in flight patients <18 years old than in the other age groups (P=.006 and .01, F test). For hMPV, this was also seen in contact patients (P=.02, F test). The novel hBoV was not detected in any patient, which indicates that this agent may be restricted to children. Indeed, all data available so far about BoV have been derived from cohorts of young infants [9, 11, 12]. A complete absence was also observed of Chlamydophila species, but it cannot be derived from our data what fraction of patients had already received preemptive antibiotic treatment at the time of sampling.
Flight destinations were analyzed next. Destinations of departure to Germany included Beijing (n=24), Hong Kong (n=19), Bangkok (n=17), Shanghai (n=14), Hanoi (n=13), Singapore (n=13), Toronto (n=6), Taiwan (n=3), and Kuala Lumpur (n=2); single patients traveled from other airports in China, Southeast Asia, and the United States. Flights were distributed over 69 different days. For 12 of 146 flights evaluated, there was at least 1 other patient who had departed from the same city on the same day. Because flight numbers were not available, results from all patients (n=25) who had traveled on nonunique flight dates were compared. Only in 2 cases did 2 patients have the same diagnosis (hPIV in both). There was no association between the airport of departure and the detection of pathogens in general (P<.3, 1-way ANOVA) or of any pathogen in particular.
To identify any possible influence of sampling on detection rates, we analyzed common categories of clinical samples separately for both flight and contact patients. Samples were categorized as follows: upper-respiratory-tract samples (category 1), throat-wash fluids (category 2), and lower-respiratory-tract samples (sputum and bronchoalveolar lavage [BAL] fluids; category 3). A comparison of age versus sample type yielded a significant lack of lower-respiratory-tract samples in patients <19 years old (1-way ANOVA, 95% significance level). Because a significantly higher detection rate of viruses had been identified in these patients, they were eliminated from the analysis, to avoid a bias. After elimination, no significant age differences in the 3 categories of samples remained (P=.3, F test). The average ages for categories 1, 2, and 3 were 42.1, 43.9, and 47.0 years, respectively.
Upper-respiratory-tract samples analyzed included 39 pharyngeal, nasal, and nasopharyngeal swabs and 99 throat-wash fluids. Lower-respiratory-tract samples included 50 sputum specimens and 4 BAL fluids. The detection frequencies of every pathogen in the 3 categories were compared by separate ANOVAs. Global detection frequencies did not differ significantly between categories (any pathogen: 51.3%, 34.3%, and 46.3% in category 1, 2, and 3, respectively). Only for Inf was the detection frequency in swabs was significantly higher than that for other samples (category 1, 30.1%; category 2, 10.1%; category 3, 14.8%; P=.01, F test). The same was observed for Inf A only (21%, 5%, and 9%, respectively; P=.01) but not for Inf B alone.
Discussion. Baseline data on the prevalence of respiratory viruses and atypical bacteria after air travel are not currently available. These patients constitute a specific subcohort of patients with community-acquired respiratory disease. Unlike general cohorts, children and elderly persons are underrepresented, and adults of working age constitute the majority of patients (71% in our study). Normally, respiratory infections in adults are mild and infrequent, but this may change in the context of travel. There is evidence that air travel increases the incidence of respiratory disease in general and that significant transmission in modern aircraft has occurred of SARS, Inf, and other agents of respiratory infection [2, 4]. Because morbidity during working life is economically relevant and new thera peutic options are at hand, investigation into the etiology of travel-associated respiratory disease seems to be well justified.
The criteria of the WHO SARS case definition, including fever, possibly caused an underrepresentation of milder infections in our cohort (RSV and hMPV). However, this made our patients representative of those who are likely to be absent from work. We used up-to-date diagnostic assays to determine the spectrum of viral or atypical agents in these patients. The rate of resolved etiologies was markedly higher than that in studies of community-acquired respiratory infections, including recent collective analyses and some of the latest original studies [5, 13–15]. This may be due to the extended spectrum of highly sensitive assays applied.
Our main finding is that the spectrum of agents in returning travelers is broad. Almost one-half of all patients were infected with respiratory viruses. Agents with treatment options, such as influenza or atypical bacteria, were present in only 15.4% of flight patients and 15.7% of all patients, making even upto- date diagnostics unrewarding. In this context, it is interesting that lower-respiratory-tract samples did not yield increased detection rates. The general assumption that nasopharyngeal washes are more sensitive than swabs in patients with community- acquired respiratory disease could not be confirmed in our study [5]. Good detection rates and a low risk of aerosolization suggest that swabs should be the preferred in this type of patients.
It cannot be told whether the high prevalence and diversity of respiratory viruses seen in our study is specific to patients with recent intercontinental air travel. Because otherwise healthy adults of working age are usually not tested for respiratory viruses, comparison data from similar cohorts are not available. Our contact patients provided a small control group, and the observed prevalence of viruses confirms the findings seen in flight patients. However, it should be noted that all contact patients fulfilled the case definition of SARS, requiring recent contact with a suspected patient [6]. In Germany, this means that all of them most likely had contact with a recent intercontinental traveler.
It is unclear why and how patients acquire viral respiratory disease in the context of air travel [2, 4]. In our study, it was interesting that significant clusters of patients with the same diagnosis, who could have been on the same flight, did not exist. Similarly, no association was detected between any pathogen and a particular airport. It would thus be likely that viruses are picked up during prior travel rather than being acquired in flight. In any case, data from this unique cohort suggest that the prevalence of known, emerging, and potentially novel respiratory viruses in adults must be carefully studied. Understanding the spectrum and the etiological contribution of viruses in adult respiratory disease will be of growing importance in the future. With new antiviral drugs at hand, more consultations will occur for respiratory diseases. Targeted therapy will require broad-spectrum pathogen detection with rapid results, most likely by PCR. Given the large range of assays required for this study and considering current reagent costs, it is obvious that developing more efficient diagnostic assays is as essential as developing drugs themselves. Today, diagnostic technology is far from attaining this aim.
Acknowledgments
We are grateful to Laurent Kaiser, Eric Claas, and Harald Kessler for providing detailed bench protocols for the published polymerase chain reaction assays used in the study.
Footnotes
Potential conflicts of interest: none reported.
Financial support: German Ministry of Health (as part of funding for the National Reference Centre for Tropical Infections, Bernhard Nocht Institute). L.K.S.L. is a fellow of the Conselho Nacional de Desenvolvimento Cientifico e Tecnologico, Brasil.
References
- 1.Leder K, Newman D. Respiratory infections during air travel. Intern Med J. 2005;35:50–5. doi: 10.1111/j.1445-5994.2004.00696.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zitter JN, Mazonson PD, Miller DP, Hulley SB, Balmes JR. Aircraft cabin air recirculation and symptoms of the common cold. JAMA. 2002;288:483–6. doi: 10.1001/jama.288.4.483. [DOI] [PubMed] [Google Scholar]
- 3.Whelan EA, Lawson CC, Grajewski B, et al. Prevalence of respiratory symptoms among female flight attendants and teachers. Occup Environ Med. 2003;60:929–34. doi: 10.1136/oem.60.12.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangili A, Gendreau MA. Transmission of infectious diseases during commercial air travel. Lancet. 2005;365:989–96. doi: 10.1016/S0140-6736(05)71089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.File TM. Community-acquired pneumonia. Lancet. 2003;362:1991–2001. doi: 10.1016/S0140-6736(03)15021-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drosten C, Gunther S, Preiser W, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348:1967–76. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 7.Claas EC, Schilham MW, de Brouwer CS, et al. Internally controlled real-time PCR monitoring of adenovirus DNA load in serum or plasma of transplant recipients. J Clin Microbiol. 2005;43:1738–44. doi: 10.1128/JCM.43.4.1738-1744.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deffernez C, Wunderli W, Thomas Y, Yerly S, Perrin L, Kaiser L. Amplicon sequencing and improved detection of human rhinovirus in respiratory samples. J Clin Microbiol. 2004;42:3212–8. doi: 10.1128/JCM.42.7.3212-3218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–6. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raggam RB, Leitner E, Berg J, Muhlbauer G, Marth E, Kessler HH. Single-run, parallel detection of DNA from three pneumonia-producing bacteria by real-time polymerase chain reaction. J Mol Diagn. 2005;7:133–8. doi: 10.1016/S1525-1578(10)60019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, Mackay IM. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma X, Endo R, Ishiguro N, et al. Detection of human bocavirus in Japanese children with lower respiratory tract infections. J Clin Microbiol. 2006;44:1132–4. doi: 10.1128/JCM.44.3.1132-1134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee BE, Robinson JL, Khurana V, Pang XL, Preiksaitis JK, Fox JD. Enhanced identification of viral and atypical bacterial pathogens in lower respiratory tract samples with nucleic acid amplification tests. J Med Virol. 2006;78:702–10. doi: 10.1002/jmv.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaye M, Skidmore S, Osman H, Weinbren M, Warren R. Surveillance of respiratory virus infections in adult hospital admissions using rapid methods. Epidemiol Infect. 2006;134:792–8. doi: 10.1017/S0950268805005364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louie JK, Hacker JK, Gonzales R, et al. Characterization of viral agents causing acute respiratory infection in a San Francisco University Medical Center Clinic during the influenza season. Clin Infect Dis. 2005;41:822–8. doi: 10.1086/432800. [DOI] [PMC free article] [PubMed] [Google Scholar]