Abstract
BackgroundA novel human rhinovirus (HRV) species, HRV-C, was recently discovered, but its clinical features and epidemiology, compared with HRV-A and HRV-B, remains poorly understood, especially in adults
MethodsOne thousand two hundred nasopharyngeal aspirate samples obtained from hospitalized children and adults during a 1-year period were subject to reverse-transcriptase polymerase chain reaction to detect HRV. The clinical and molecular epidemiology of the 3 HRV species was analyzed
ResultsHRVs were detected in 178 (29.7%) of 600 nasopharyngeal aspirate samples from children and 42 (7%) of 600 nasopharyngeal aspirate samples from adults. HRV-A was most prevalent (n=111), followed by HRV-C (n=91) and HRV-B (n=18). Although upper respiratory tract infection was the most common presentation in children, 8 (62%) of the 13 adults with HRV-C infection had pneumonia, compared with 6 (27%) of the 22 adults with HRV-A infection (P<.05). Wheezing episodes were also more common among individuals with HRV-C (37%) and HRV-A (20%) infection than among those with HRV-B (0%) infection (P<.05). Clinical and molecular data analysis revealed HRV-C as a frequent cause of community and institutionalized outbreaks. A diverse set of HRV-C genotypes was circulating throughout the year, among which a potential distinct subgroup of strains was observed
ConclusionHRV-C is associated with pneumonia in adults and outbreaks of respiratory infections requiring hospitalization. A potential novel HRV-C subgroup was identified
Respiratory tract infection is one of the most common and important infectious diseases. However, the etiological agents remain unknown in a significant proportion of cases [1, 2]. The severe acute respiratory syndrome epidemic in 2003 has boosted interest in the discovery of novel respiratory pathogens. Apart from severe acute respiratory syndrome coronavirus [3], a number of novel respiratory viruses have been identified since then, including human coronavirus NL63 [4, 5] human coronavirus HKU1 [6–9], and human bocavirus [9–11]
Human rhinoviruses (HRVs) are the most frequent causes of acute respiratory tract infections. Although they are most often associated with mild upper respiratory tract infections (URTIs), they are increasingly shown to be associated with more severe diseases such as pneumonia, especially in infants and elderly and immunocompromised patients [12–14]. HRVs, which consist of >100 distinct serotypes, have been classified according to several parameters, including receptor specificity, antiviral susceptibility, and nucleotide sequence identities [15]. On the basis of gene sequence analysis, all but 1 HRV serotype were classified into 2 species, HRV-A and HRV-B [16–19]
Recently, several groups have reported the finding of novel HRV genotypes in respiratory tract samples obtained from patients in the United States, Australia, and Hong Kong [20–23]. Analysis of the complete genome sequences suggested that these newly identified HRV genotypes belonged to a potentially novel HRV species, HRV-C, with genome features distinct from HRV-A and HRV-B [22, 24]. Subsequently, HRVs closely related to HRV-C were also identified in more clinical samples worldwide, especially in those obtained from children with respiratory illness [25–28]. However, most of these studies only focused on the molecular identification of HRV-C from clinical samples. No systematic study has been carried out to examine the epidemiology and clinical features of HRV-C, compared with those of HRV-A and HRV-B. In particular, the importance of HRV-C in adult respiratory illness still remains poorly understood. In this study, we describe the clinical and molecular epidemiology of HRV-C in hospitalized children and adults during a 1-year period. The clinical and epidemiological characteristics of HRV-C infection were also compared with those of HRV-A and HRV-B infection
Methods
Patients and microbiological methodsAll nasopharyngeal aspirate (NPA) samples in this study were collected from hospitalized patients at 2 acute regional hospitals in Hong Kong during a 1-year period (April 2004 through March 2005) and had negative test results for influenza A and B viruses, parainfluenza viruses types 1, 2, and 3, respiratory syncytial virus and adenovirus by direct immunofluorescence, and human metapneumovirus, human coronavirus 229E, human coronavirus OC43, human coronavirus NL63, and human coronavirus HKU1 by reverse-transcriptase polymerase chain reaction (RT-PCR) [6, 7, 29, 30]. For each month, 50 NPA samples from children aged <18 years and 50 NPA samples from adults were randomly selected and subjected to RT-PCR to detect HRV. The clinical features, laboratory results, and outcome of patients with positive results for HRV were analyzed
RT-PCR for HRV and sequencingViral RNA was extracted from NPA samples with use of the QIAamp Viral RNA Mini Kit (QIAgen). RT was performed using random hexamers and the SuperScript III kit (Invitrogen) as described elsewhere [7, 8]. PCR to detect HRV was performed according to protocols described elsewhere with use of conserved primers targeting the VP4 region [19, 22]. The amplified products were detected by agarose gel electrophoresis. Both strands of PCR products were sequenced twice with an ABI Prism 3700 DNA Analyzer (Applied Biosystems) with use of the PCR primers. The nucleotide sequences of the VP4 regions were compared with those of HRV-A, HRV-B, and HRV-C strains with sequences available in GenBank. Phylogenetic tree construction was performed using neighbor-joining method with GrowTree using Kimura’s 2-parameter correction, with bootstrap values calculated from 1000 trees (Genetics Computer Group)
Statistical analysisComparison between different groups was performed using the χ2 test (SPSS, version 11.5). P<.05 was considered to be statistically significant
Nucleotide sequence accession numberThe VP4 nucleotide sequences of the HRV-C strains have been entered into the GenBank sequence database under accession no. FJ937360–FJ937450
Results
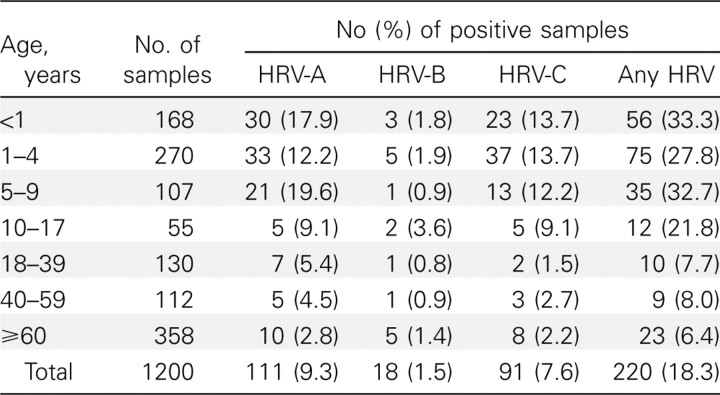
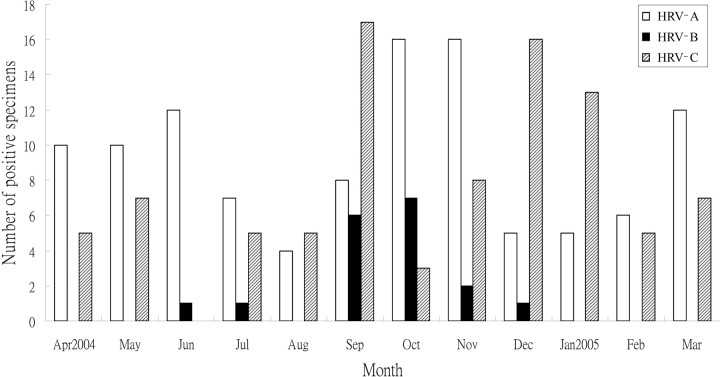
Detection of HRV in NPA samples from hospitalized patientsAmong the 1200 tested NPA samples, 220 (18.3%) had positive results for HRV by RT-PCR. These 220 NPA samples were from 210 patients (male:female ratio, 133:87; median age, 3 years; age range, 19 days to 89 years), and 8 patients had >1 NPA sample with positive results for HRV. HRV was detected in 178 (29.7%) of the 600 tested NPA samples from children and 42 (7%) of the 600 tested NPA samples from adults. Sequencing and phylogenetic analysis of the VP4 genes of viruses isolated from these 220 NPA samples showed that 111 were HRV-A viruses, 18 were HRV-B viruses (data not shown), and 91 were HRV-C viruses. The age distribution of infection due to the 3 HRV species is shown in Table 1. HRV-A and HRV-C had a higher detection rate in NPA samples from children than in those from adults. HRV infection occurred throughout the year during the study period, although a higher incidence was observed during the fall and winter months for all 3 HRV species (Figure 1)
Table 1.
Age Distribution of Nasopharyngeal Aspirates Positive for Human Rhinoviruses (HRVs)
Figure 1.
Seasonality of the 3 human rhinovirus (HRV) species in the present study
Clinical characteristics of patients with HRV-C infectionThe clinical characteristics of patients with HRV-A, HRV-B, and HRV-C infection are summarized and compared in Table 2. Of the 91 HRV-C isolates, 78 were detected in children, and 13 were detected in adults. Sixty-two (68%) infections occurred in patients with underlying diseases, with asthma or other chronic lung diseases being the most common. Although URTI was the most common diagnosis in children with HRV-C infection (47%), lower respiratory tract infection was particularly common among adults, where 8 (62%) of the 13 adults with HRV-C infection had pneumonia. Fever, cough, and shortness of breath were common symptoms in both children and adults with pneumonia associated with HRV-C infection. Although chest radiographs often showed increased haziness in perihilar or lower zone regions in these children, consolidation and pleural effusion were present in 3 and 2 of the adults, respectively. Wheezing episodes in the form of exacerbations of asthma or chronic obstructive pulmonary disease or febrile wheeze were common among individuals with HRV-C (37%) and HRV-A (20%) infection. In particular, 33 (42%) of the 78 children with HRV-C infection experienced asthmatic exacerbations or febrile wheeze. Acute otitis media and croup each occurred in 1 child with HRV-C infection. Although our study included only NPA samples with negative results for common respiratory viruses, respiratory bacterial coinfecting pathogens were occasionally observed. Gastroenteritis, with or without coinfecting pathogens, occurred in 9% of patients with HRV-A and HRV-C infection and in 17% of patients with HRV-B infection
Table 2.
Comparison of Clinical Characteristics of Patients with Human Rhinovirus (HRV) A, HRV-B, and HRV-C Infection
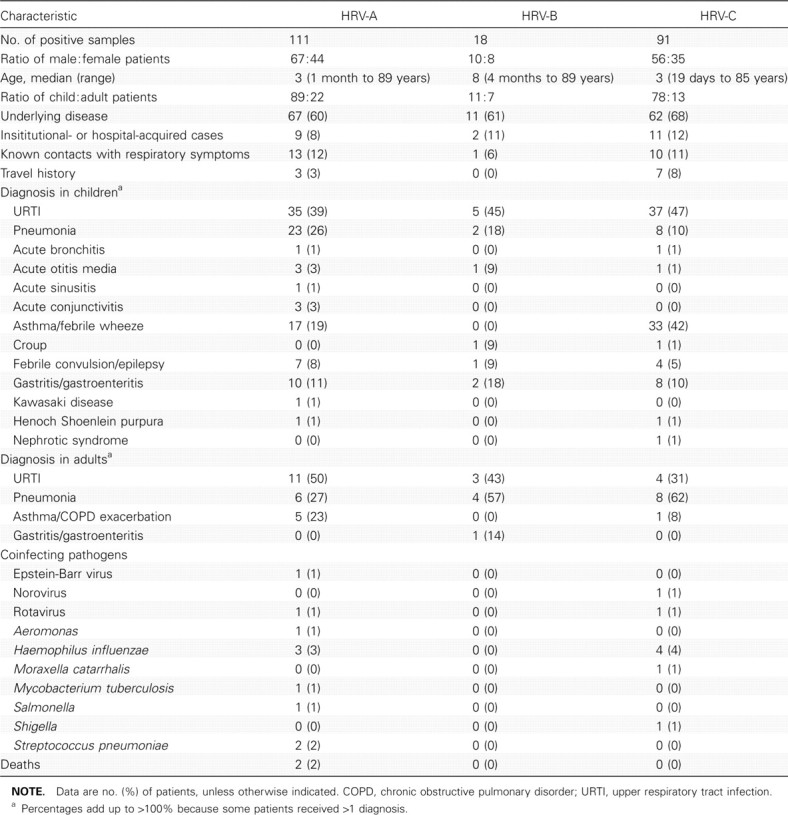
Reports of contacts or household members with respiratory symptoms were not uncommon among patients with HRV infection. A significant proportion of patients (8%–12%) acquired HRV infection at their resident institutions or during prolonged hospital stay. Three patients were institutionalized in a baby care center that reported staff members or other children who had similar illness. Three percent of patients with HRV-A infection and 8% of patients with HRV-C infection had recent travel histories to mainland China, the Philippines, Malaysia, or Pakistan. Although 2 patients with HRV-A infection died (one 70-year-old receiving immunosuppressants with concomitant active pulmonary tuberculosis and one 51-year-old intravenous drug user with severe community-acquired pneumonia complicated by acute renal failure), all patients with HRV-B or HRV-C infection survived
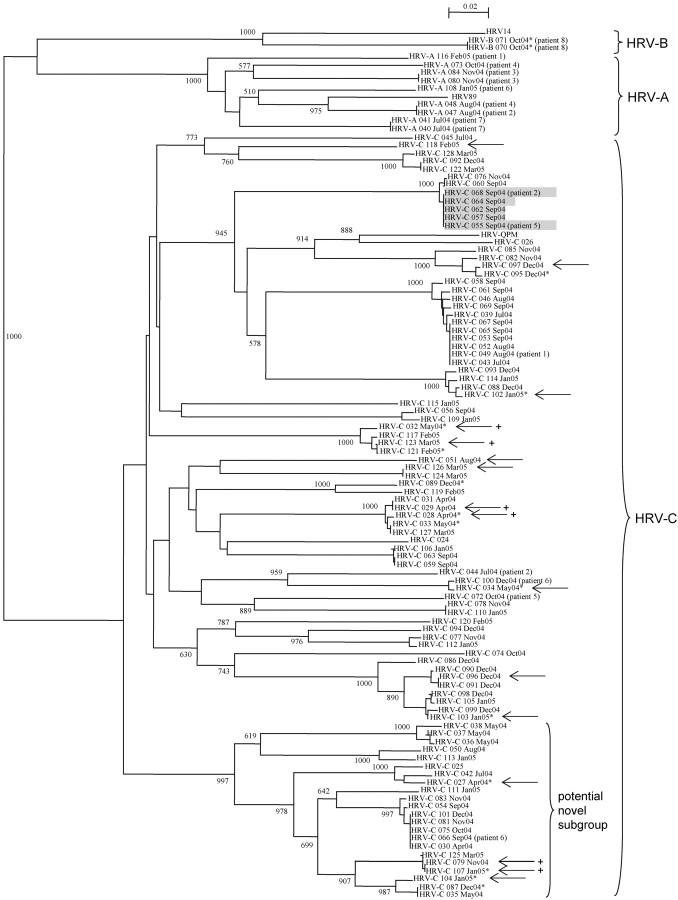
Molecular epidemiology of HRV-C infectionThe VP4 sequences of the HRV-C isolates showed diverse genetic variations, with 67.2%–82.1% nucleotide identity to HRV-C strain 024 (GenBank accession no. NC_009996), suggesting that diverse genotypes among the species were circulating throughout the year (Figure 2). Among these diverse HRV-C genotypes, a subgroup of strains were found to form a distinct cluster with a high bootstrap value of 997 (Figure 2). A similar phenomenon was also observed when VP4 sequences of other HRV-C or related strains available from GenBank were included in the analysis (data not shown). No temporal pattern among the diverse HRV-C genotypes and no apparent genetic difference between isolates from children and adults were observed (Figure 2)
Figure 2.
Phylogenetic tree of the VP4 region of the 91 human rhinovirus (HRV) C strains and HRV strains detected in patients with >1 NPA samples with positive results. Two hundred one nucleotide positions in each VP4 region were included in the analysis. All field strains in the present study are indicated with dates of detection. Strains detected from adults are marked with asterisks. Strains associated with pneumonia are marked with arrows. The 5 strains with the same nucleotide differences detected from 5 patients within 3 days are shaded. The 6 strains of 3 different clusters (2 closely related strains from each cluster) associated with pneumonia are marked with a plus sign. The scale bar indicates the estimated number of substitutions per 50 bases. The HRV-QPM strain was from Queensland. The GenBank accession numbers of the previously published sequences are as follows: HRV-QPM, EF186077; HRV89, A10937; HRV14, NC_001490; HRV-C 024, NC_009996; HRV-C 025, EF582386; and HRV-C 026, EF582387
Analysis of clinical data for HRV-C isolates with identical VP4 sequences revealed a cluster of 5 community-acquired cases admitted to the hospital within 3 days in infants or young children presenting with respiratory illness (Figure 2). One of the patients was a 5-month-old infant who was institutionalized in a baby care center with staff members who also reported respiratory illness. Two similar small clusters of HRV-C isolates with identical sequences were also observed, though they were detected over a longer period of time, one from July to September and the other from April to December (Figure 2)
Eight patients with HRV infection had >1 NPA sample positive for HRV, of whom 3 patients (patient 3 and patient 7 with HRV-A infection and patient 8 with HRV-B infection) had 2 separate NPA samples with positive results for the same strain during the same episode of illness (Figure 2). The other 5 patients had recurrent HRV infection due to different strains of the same species or different species during the same year. All 5 patients were young children with underlying illnesses. For example, patient 6, an infant with underlying congenital heart disease and laryngomalacia, was infected by 2 different HRV-C strains 3 months apart and subsequently an HRV-A strain 1 month later
Discussion
We report the first clinical and molecular epidemiology study of HRV-C infection in comparison to HRV-A and HRV-B infection in both children and adults. Because laboratory diagnosis of HRV infection is generally not available in clinical virology laboratories, their impact on health is often ignored. Although recent reports have implicated HRVs in more severe URTIs and lower respiratory tract infections, especially in children, the elderly, and immunocompromised adults [12–14], the relative importance and disease severity associated with the 2 pre-existing species, HRV-A and HRV-B, and the newly described species, HRV-C, are not well understood. In this study, HRVs, especially HRV-A and HRV-C, are shown to be important causes of hospitalizations due to respiratory illness in our locality. In other studies that described the detection of HRV-A and HRV-B in addition to the new genotypes, the results were often limited by either the small number of samples with positive results or a lack of clinical and epidemiological data [20, 23, 27, 28, 32]. In our previous study, HRVs were detected in 26 (12.8%) of 203 NPA samples obtained from hospitalized children, among which 21 were HRV-C and 5 were HRV-A viruses [22]. In this study, 18.3% of the tested NPA samples had positive results for HRV, with HRV-A being the most prevalent species (111 isolates), followed by HRV-C (91 isolates) and HRV-B (18 isolates). These results suggest that HRV-A and HRV-C may be more prevalent than HRV-B in our population or that they are associated with higher hospitalization rates. Interestingly, HRV-A and HRV-C appeared to alternate as the most common HRV species at different times during the peak seasons (Figure 1). Alternate disease activity has been reported in human parainfluenza virus infection, where human parainfluenza virus 3 activity also appeared to be greater during years when human parainfluenza virus 1 was not circulating, a phenomenon believed to be attributable to viral interference or cross-serological protection [31]. It would be interesting to investigate possible viral interference between HRV-A and HRV-C
HRV-C is likely a frequent cause of both community and institutionalized outbreaks of respiratory illness. In the present study, 12% of cases of HRV-C infection were acquired during institutionalization or hospitalization. Most of them were in children with prolonged stay in pediatric wards or a baby care center, whereas one was in an adult hospitalized in a psychiatric ward for management of schizophrenia. For 1 infant, staff members from the same baby care center were noted to have similar illness. These suggest that HRV-C frequently circulates among institutions. On the other hand, clusters of cases due to the same strains of HRV-C and frequent reports of contacts with similar illness were also observed in the community-acquired cases, suggesting that these viruses are also common causes of community outbreaks that lead to significant morbidity and hospital admissions. Because 8% of patients reported recent travel history to other Asian countries, some infections may have also been acquired during travel
In contrast to children, in whom URTI was the predominant diagnosis, hospitalized adults with HRV-C infection most commonly presented with pneumonia. Although the detection rate of HRV-C infection was higher among children in the present study, adults with HRV-C infection appeared to present with more-severe illness. HRV-C infection has been mostly described in infants and young children with acute respiratory illness. In line with our results reported elsewhere [22], children with HRV-C infection in the present study commonly present with URTI, febrile wheeze, and asthmatic exacerbations. One child also presented with acute otitis media, which concurred with the detection of HRV-C in middle ear fluid samples from patients with acute otitis media in another report [25]. On the other hand, relatively little is known about the significance of HRV-C infection in the adult population. In a study from Australia, 2 of the 17 identified strains were from adults, of whom 1 experienced an exacerbation of chronic obstructive pulmonary disease [21]. In 2 other studies that mentioned the detection of novel HRVs, which corresponded to HRV-C, in respiratory tract samples from adults, no further clinical details were provided [23, 28]. In the present study, 8 (62%) of the 13 adults with HRV-C infection had pneumonia, compared with 6 (27%) of the 22 adults with HRV-A infection (P<.05) (Table 2). Although most of the adults with HRV-C infection had underlying diseases or were of advanced age, 1 of the 8 patients with pneumonia was a 45-year-old healthy man who presented with left upper lobe consolidation without identifiable coinfecting pathogens. Among the 16 cases of HRV-C–associated pneumonia, 6 were caused by viruses of 3 different clusters (2 closely related strains from each cluster), respectively (Figure 2). Wheezing episodes were also more common in patients with HRV-C (37%) and HRV-A (20%) infection, compared with that in patients with HRV-B (0%) infection (P<.05). Although children with HRV-C infection most often presented with URTI, the disease course was not uncommonly complicated by asthmatic exacerbations or febrile wheeze triggered by the virus infection. Further clinical studies that involve more patients with HRV infection are required to investigate whether certain HRV species or strains may be more virulent than others
Recurrent infections in the same patient by different HRV strains within a short period of time suggested that these young children may be particularly susceptible to recurrent HRV infection. This may be attributable to low cross-serological protection between different strains of HRV-C and between different HRV species, especially in the setting of a suboptimal level of neutralizing antibodies from previous infection in patients with underlying diseases. Together with the genetic diversity observed in VP4 sequence analysis throughout the year, HRV-C probably represents a clade of diverse HRV genotypes that can easily evade immune protection. However, successful isolation of different HRV-C genotypes would be required to study their cross-serological reactivity
HRV-C has been proposed as a new HRV species within the genus Enterovirus by the International Committee on Taxonomy of Viruses (http://talk.ictvonline.org/media/p/1035.aspx), which may contain a distinct subgroup of strains. Since the discovery of this new clade of HRV, related viruses have been identified from different countries from Africa, Asia, Australia, Europe, and North America [20–28, 32]. Although these viruses were referred to as novel genotypes or HRV-X in some studies, they are now believed to represent a distinct species, HRV-C [22, 24–26, 28, 32, 33]. First, these viruses formed a distinct cluster separate from HRV-A and HRV-B on phylogenetic analysis of the 5′ noncoding regions and predicted proteins [22, 24]. Second, they possessed genome features distinct from HRV-A and HRV-B, namely, a different putative cleavage site between VP4 and VP2 and major differences within the VP1 region. In particular, only a few of the conserved amino acid residues within VP1 that determine receptor binding of HRV-A and HRV-B were found [22]. With use of structural homology modeling, the VP1 of HRV-C was also found to possess structural disparities within sites for receptor binding, compared with HRV-A and HRV-B [24]. These suggested that HRV-C may use a different cellular receptor, which may reflect the inability to successfully isolate HRV-C in traditional cell cultures. By completing the genome sequences for all known HRV serotypes, a recent study also confirmed species-specific sequence and RNA structure elements that differentiate HRV-A and HRV-B from HRV-C [33]. The study also identified a potential novel clade, clade D, which forms a separate cluster within HRV-A. Interestingly, we also identified a potential distinct cluster of strains of HRV-C during phylogenetic analysis in the present study. In fact, we found that a similar separate cluster of strains was also present in our and several other studies of HRV-C strains, although bootstrap values were not shown in some studies [20, 22, 25, 27, 28]. When all available VP4 sequences of HRV-C isolates were included in analysis, these strains were clustered with high bootstrap values, suggesting a possible distinct subgroup within HRV-C (analysis was performed using available sequences published up to 15 March 2009; data not shown). The same tree topology was obtained when a different method, UPGMA, was used for tree construction (data not shown). However, the present data are limited by the fact that only VP4 sequences were available for analysis. Although additional studies are required to elucidate the receptor(s) for HRV-C, the existing data support the classification of HRV-C as a separate species. Longer gene sequences or complete genome sequences of the diverse HRV-C genotypes should allow better delineation of their genetic variability and allow ascertainment of the existence of distinct subgroups within the species
Acknowledgments
We are grateful for the generous support of Mr. Hui Hoy and Mr. Hui Ming in the genomic sequencing platform
Footnotes
Potential conflicts of interest: none reported
Financial support: Research Grant Council Grant (HKU 7687/09M); University Development Fund, HKU Special Research Achievement Award and Outstanding Young Researcher Award, The University of Hong Kong; the Croucher Senior Medical Research Fellowship 2006–2007; the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare, and Food Bureau; and Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for Department of Health of the HKSAR
S.K.P.L. and C.C.Y.Y. contributed equally to the manuscript
References
- 1.Macfarlane JT, Colville A, Guion A, Macfarlane RM, Rose DH. Prospective study of aetiology and outcome of adult lower-respiratory-tract infections in the community. Lancet. 341:511–4. doi: 10.1016/0140-6736(93)90275-l. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz M, Ewig S, Marcos MA, et al. Etiology of community-acquired pneumonia: impact of age, comorbidity, and severity. Am J Respir Crit Care Med. 160:397–405. doi: 10.1164/ajrccm.160.2.9808045. [DOI] [PubMed] [Google Scholar]
- 3.Peiris JS, Lai ST, Poon LL, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fouchier RA, Hartwig NG, Bestebroer TM, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 101:6212–6. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Hoek L, Pyrc K, Jebbink MF, et al. Identification of a new human coronavirus. Nat Med. 10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lau SK, Woo PC, Yip CC, et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 44:2063–71. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo PC, Lau SK, Chu CM, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 79:884–95. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Woo PC, Lau SK, Tsoi HW, et al. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J Infect Dis. 191:1898–907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. 102:12891–6. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau SKP, Yip CCY, Que TL, et al. Clinical and molecular epidemiology of human bocavirus in respiratory and fecal samples from children in Hong Kong. J Infect Dis. 196:986–93. doi: 10.1086/521310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaiser L, Aubert JD, Pache JC, et al. Chronic rhinoviral infection in lung transplant recipients. Am J Respir Crit Care Med. 174:1392–9. doi: 10.1164/rccm.200604-489OC. [DOI] [PubMed] [Google Scholar]
- 13.Miller EK, Lu X, Erdman DD, et al. Rhinovirus-associated hospitalizations in young children. J Infect Dis. 195:773–81. doi: 10.1086/511821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner RB. Rhinovirus: more than just a common cold virus. J Infect Dis. 195:765–6. doi: 10.1086/511829. [DOI] [PubMed] [Google Scholar]
- 15.Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 73:1941–8. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laine P, Savolainen C, Blomqvist S, Hovi T. Phylogenetic analysis of human rhinovirus capsid protein VP1 and 2A protease coding sequences confirms shared genus-like relationships with human enteroviruses. J Gen Virol. 86:697–706. doi: 10.1099/vir.0.80445-0. [DOI] [PubMed] [Google Scholar]
- 17.Ledford RM, Patel NR, Demenczuk TM, et al. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J Virol. 78:3663–74. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savolainen C, Laine P, Mulders MN, Hovi T. Sequence analysis of human rhinoviruses in the RNA-dependent RNA polymerase coding region reveals large within-species variation. J Gen Virol. 85:2271–7. doi: 10.1099/vir.0.79897-0. [DOI] [PubMed] [Google Scholar]
- 19.Savolainen C, Blomqvist S, Mulders MN, Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J Gen Virol. 83:333–40. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- 20.Lamson D, Renwick N, Kapoor V, et al. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 194:1398–402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McErlean P, Shackelton LA, Lambert SB, Nissen MD, Sloots TP. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau SK, Yip CC, Tsoi HW, et al. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, human rhinovirus-C, associated with acute respiratory illness in children. J Clin Microbiol. 45:3655–64. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kistler A, Avila PC, Rouskin S, et al. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 196:817–25. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McErlean P, Shackelton LA, Andrews E, et al. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C) PLoS ONE. 3:e1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savolainen-Kopra C, Blomqvist S, Kilpi T, Roivainen M, Hovi T. Novel species of human rhinoviruses in acute otitis media. Pediatr Infect Dis J. 28:59–61. doi: 10.1097/INF.0b013e318182c90a. [DOI] [PubMed] [Google Scholar]
- 26.Dominguez SR, Briese T, Palacios G. Multiplex MassTag-PCR for respiratory pathogens in pediatric nasopharyngeal washes negative by conventional diagnostic testing shows a high prevalence of viruses belonging to a newly recognized rhinovirus clade. J Clin Virol. 43:219–22. doi: 10.1016/j.jcv.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renwick N, Schweiger B, Kapoor V, et al. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J Infect Dis. 196:1754–60. doi: 10.1086/524312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Briese T, Renwick N, Venter M, et al. Global distribution of novel rhinovirus genotype. Emerg Infect Dis. 14:944–7. doi: 10.3201/eid1406.080271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peiris JS, Tang WH, Chan KH, et al. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis. 9:628–33. doi: 10.3201/eid0906.030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woo PCY, Chiu SSS, Seto WH, Peiris M. Cost-effectiveness of rapid diagnosis of viral respiratory tract infections in pediatric patients. J Clin Microbiol. 35:1579–81. doi: 10.1128/jcm.35.6.1579-1581.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fry AM, Curns AT, Harbour K, Hutwagner L, Holman RC. Seasonal trends of human parainfluenza viral infections: United States, 1990–2004. Clin Infect Dis. 43:1016–22. doi: 10.1086/507638. [DOI] [PubMed] [Google Scholar]
- 32.Lee WM, Kiesner C, Pappas T, et al. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE. 2:e966. doi: 10.1371/journal.pone.0000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmenberg AC, Spiro D, Kuzmickas R, et al. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 324:55–9. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]