Abstract
Marine turtle hybridization is usually sporadic and involves reports of only a few individuals; however, Brazilian populations have high hybridization rates. Here we investigated the presence of hybrids in morphologically identified immature hawksbills (Eretmochelys imbricata) along the South Western Atlantic (SWA). We sequenced one mitochondrial (D-Loop) and three nuclear DNA (RAG1, RAG2, and CMOS) markers to better understand the patterns and characteristics of hybrids. We identified 22 hybrids (n = 270), 11 of them at the extreme South of the SWA. Uruguay had the highest hybrid frequency in the SWA (~37.5%) followed by southern Brazil with 30%. These are common areas for loggerheads (Caretta caretta) but uncommon for hawksbills, and these hybrids may be adopting the behavior of loggerheads. By analyzing nuclear markers, we can infer that 50% of the sampled hybrids are first generation (F1) and 36% are the result of backcrosses between hybrids and pure E. imbricata (> F1). We also report for the first time immature E. imbricata x Lepidochelys olivacea hybrids at the Brazilian coast. Considering the high frequency of hybrids in the SWA, continuous monitoring should be performed to assess the fitness, genetic integrity, and extent of changes in the gene pools of involved populations.
Keywords: Hybridization, Cheloniidae, Genetic markers, Hybrid distribution, Conservation
Introduction
Hybridization can be defined as the production of offspring by the crossbreeding between genetically different populations or species (Harrison, 1990). At least 25% of plant species and 10% of animal species are involved in hybridization processes and potential introgression (Mallet, 2005). According to Rieseberg and Wendell (1993), introgression can be defined as “the incorporation of genes from one set of different populations into another, i.e. the incorporation of external alleles into a new, reproductively integrated population system”. These processes are considered natural in evolution, and continuous events of interspecific and intergeneric hybridization may lead to the appearance of new species (e.g. 50-70% of angiosperms; Whitham et al., 1991). However, hybridization can also be a consequence of anthropogenic factors such as decreasing population sizes, introduction of non-native species and modification of habitats, which may result in the extinction of local species, subspecies and populations (Allendorf et al., 2001).
Hybridization is a common phenomenon in different vertebrate groups, such as birds (Crochet et al., 2003), fish (Rosenfield et al., 2004), and marine mammals - the order Cetacea, for example, has records of hybrids in 20% of species that make up the group (Crossman et al., 2016). In marine turtles, the occurrence of hybrids has already been reported between several species of the Cheloniidae family, especially the closely related species olive ridley (Lepidochelys olivacea), loggerhead (Caretta caretta), and hawksbill (Eretmochelys imbricata) (Bowen and Karl, 2007; Naro-Maciel et al., 2008). It is possible that this interbreeding occurs due to the lack of reproductive barriers (Seminoff et al., 2003) and a chromosomal compatibility between the different species (Karl et al., 1995).
Most data on hybridization in nature comes from morphological evaluation of organisms (Mallet, 2005); however, this alone is insufficient to adequately identify and characterize hybrids, since some hybrids do not present mixed morphology. In marine turtles, the first signs of hybridization were observed in animals with intermediate diagnostic characteristics between two species (Wood et al., 1983), and the first molecular observation of hybrids was done in Brazil by Conceição et al. (1990), using isozymes. With the advance of molecular tools, genetic analyses have been increasingly used to detect and understand this hybridization process (e.g. Vilaça et al., 2012). Mitochondrial (mtDNA) and nuclear (nDNA) markers may therefore aid in the identification of hybrids even when individuals do not have evidence of hybridization observable through morphology.
In most species, mtDNA is maternally inherited. As a result, relying only on mitochondrial information may be misleading when validating morphological observations; for example, if a first-generation (F1) or any subsequent generation (>F1) hybrid shows similar morphological characteristics to the species determined through mtDNA, hybridization cannot be detected. In contrast, nDNA is inherited from the two progenitors, allowing the identification of genes of different species even when the hybrid has the morphological characteristics and mtDNA of only one (Vilaça and Santos, 2013). Thus, the combined use of mtDNA and nDNA markers assists in hybrid identification, allowing a better evaluation of their distribution and frequency. In addition, it is possible to evaluate the number of generations over which hybridization has been occurring, as well as the occurrence of introgression (i.e., whether the individual is the result of a cross between pure parent species (F1 generation), hybrids (F2) or a backcross between a hybrid and one of the pure parent species) (Sunnucks, 2000). Identifying the degree of introgression between species is important in assessing possible losses of locally adapted genes and population fitness (Allendorf et al., 2001). In addition, according to Payseur and Rieseberg (2016), understanding the connection between geographic distribution/gene flow of hybrids and the emergence of new species is fundamental. Considering the hybridization events and species involved, the decision of which individuals and populations should be protected is complex (Wayne and Shaffer 2016), and caution is needed when establishing conservation strategies.
Hybridization events in marine turtles are usually sporadic and involve reports of one or a few individuals (see Table 1); however, Brazilian populations have high rates, which may be due to the endangered status of these animals (IUCN, 2018). At the Bahia state rookery, Lara-Ruiz et al. (2006) observed through mtDNA that, among E. imbricata females analyzed (n = 119), 42% were actually hybrids with C. caretta and 2% hybrids with L. olivacea. When analysing 204 samples of C. caretta nesting females at four rookeries (Rio de Janeiro, Espírito Santo, Bahia and Sergipe states), Reis et al. (2010) observed that 14 out of 51 females from Sergipe were hybrids with L. olivacea, with no record for the other rookeries. Occurrences of C. caretta and E. imbricata immature hybrids have also been reported in Uruguay and Argentina (Alvarez-Varas et al., 2016; Prosdocimi et al., 2014). The occurrence of E. imbricata along the coast of Uruguay is low, with only three individuals registered during twelve years of monitoring by the NGO Karumbé (Vélez-Rubio et al., 2013). However, during these surveys several turtles with inconclusive morphology (i.e. possible hybrids) have been observed (A. Fallabrino, personal communication).
Table 1. Hybridization events reported between Cheloniidae marine turtles, the respective species, number of observed hybrids and country.
| Species A | Species B | No. identified hybrids | Country | References |
|---|---|---|---|---|
| Caretta caretta | Lepidochelys kempii | 1 | USA | Karl et al., 1995 |
| Caretta caretta | Chelonia mydas | 4 | Brazil | Karl et al., 1995 |
| 1 | Canada | James et al., 2004 | ||
| 3 | USA | Komoroske et al., 2019 | ||
| Caretta caretta | Lepidochelys olivacea | 14 | Brazil | Reis et al., 2010 |
| Eretmochelys imbricata | Caretta caretta | 2 | USA | Karl et al., 1995 |
| 50 | Brazil | Lara-Ruiz et al., 2006 | ||
| 1 | Brazil | Conceição et al., 1990 | ||
| 4 | Brazil | Proietti et al., 2012 | ||
| 34 | Brazil | Soares et al., 2017 | ||
| 10 | Brazil | Bass et al., 1996 | ||
| 1 | USA | Komoroske et al., 2019 | ||
| 2 | Argentina | Prosdocimi et al., 2014 | ||
| Eretmochelys imbricata | Chelonia mydas | 1 | Suriname | Karl et al., 1995 |
| 1 | Mexico | Seminoff et al., 2003 | ||
| 1 | Peru | Kelez et al., 2016 | ||
| Eretmochelys imbricata | Lepidochelys olivacea | 2 | Brazil | Lara-Ruiz et al., 2006 |
The Brazilian coast has the highest known rate of hybridization in the world between four sea turtle species. The hybridization pattern involving the most common species in Brazil - E. imbricata, C. caretta and L. olivacea - was investigated by Vilaça et al. (2012) in samples obtained along the coast using 12 nuclear markers. These authors identified L. olivacea hybrids with C. caretta and E. imbricata as being first generation (F1). In contrast, some C. caretta and E. imbricata hybrids showed evidence of backcrosses with pure parent species, indicating a longer process or higher survival of offspring. They also suggested that hybridization events at the region have been occurring for at least 40 years (i.e., around two generations), and may be a result of the historical population decline experienced by both species due to exploitation of eggs and female turtles (Santos et al., 2011).
In Brazil, E. imbricata reproductive areas overlap spatially and temporally with those of C. caretta, with both species reproducing at the Northeast coast and presenting highest nesting concentrations at Bahia and Sergipe states (Marcovaldi et al., 2007; Marcovaldi and Chaloupka, 2007). The C. caretta reproductive season begins in September and ends in February (Santana et al., 2011), and E. imbricata breeding begins in November and extends until March (Marcovaldi et al., 2011). This overlap, together with the population depletion suffered by the species, may contribute to the occurrence of hybridization events. Proietti et al. (2014a) and Vilaça et al. (2012) showed that the hybridization happening in Brazil has a gender bias, and that the encounter of male E. imbricata with female C. caretta would be favored by the larger population size of loggerhead turtles, in conjunction with a temporal overlap at the peak of reproductive season. E. imbricata begins to reproduce near the C. caretta reproductive peak (November-December), leading to the encounter of male E. imbricata with females of both species. In contrast, since the reproductive peak of E. imbricata is after C. caretta, most C. caretta have probably left the area when it presents higher numbers of E. imbricata females, reducing the probability of their reproduction (Soares et al., 2018).
Immature animals resulting from the hybridization process in Brazil were reported for the first time by Proietti et al. (2014a, b), who analyzed 157 E. imbricata along the Brazilian coast and identified four individuals at Cassino beach (Rio Grande do Sul state) with a C. caretta haplotype (CCA4.2). This is an unusual area for E. imbricata, which occupies preferentially tropical regions of the oceans, associated with coral reefs (León and Bjorndal, 2002; Mortimer and Donnelly, 2008); on the other hand, C. caretta is common at temperate latitudes, occurring frequently at the region (Monteiro et al., 2016). The high frequency of immature hybrids found at Cassino shows that the distribution of these animals may present a spatial pattern, with preference for areas used more by C. caretta. Adult hybrids also show differential habitat use: Marcovaldi et al. (2012) tracked pure and hybrid E. imbricata x C. caretta nesting females from Bahia, and observed that they used distinct foraging areas. While pure females migrated to their respective species areas along the Brazilian coast, hybrids migrated predominantly to the northeast Brazilian coast, a C. caretta feeding area, although some hybrids also migrated south to an E. imbricata feeding area (Lima et al., 2013).
The effect of hybridization on marine turtle fitness and reproduction output was investigated for the first time by Soares et al. (2017), who compared factors associated with reproductive success of hybrid and pure E. imbricata and C. caretta females nesting at Bahia. Only 1% of females identified morphologically as C. caretta were hybrids, while more than half of the females identified as E. imbricata presented a C. caretta haplotype. This reaffirms the prevalence of crosses between E. imbricata males and C. caretta females. Based on the analysed reproductive parameters (number of eggs, emergence success, incubation period, number of hatchlings per nest, number of nests per year), hybrid females apparently do not have different reproductive outputs when compared to pure parent species.
Considering the high occurrence of hybrids in the South Western Atlantic (SWA), the potential impacts of this phenomenon on marine turtle populations, and the paucity of studies on the characteristics of hybrid offspring originating from Brazilian populations, the goal of this study was to investigate hybridization in immature turtles along the SWAChange to: SWA, using molecular methods. Through the combined analysis of mtDNA (control region - D-Loop) and nDNA (RAG1, RAG2 and CMOS) markers, we evaluated if: 1) the use of multiple markers enhances the detection of immature hybrids; 2) immature hybrid turtles present preference for certain foraging areas; and (3) the identified immature hybrid turtles are first generation (F1), the result of crossing between hybrids (F2), or of introgression between hybrids and pure parent species (>F1).
Materials and Methods
Sampling
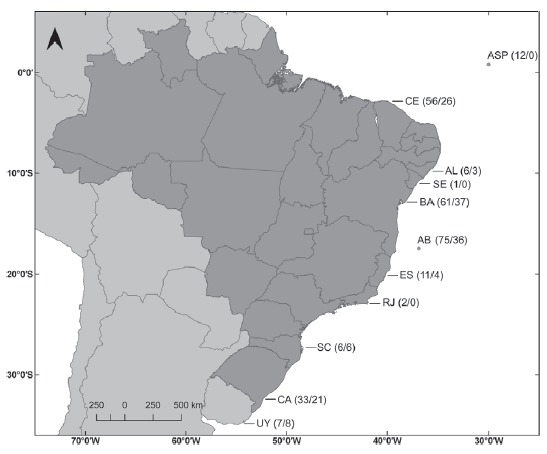
A total of 270 skin and/or muscle samples were obtained from the anterior flippers or inguinal area of immature hawksbills (Curved Carapace Length - CCL - from 13 to 111 cm) incidentally captured by fisheries, stranded on the beach, or intentionally caught in dives along the coast of Brazil and Uruguay. In Brazil, sampling was done at the São Pedro and São Paulo (ASP) and Abrolhos (AB) Archipelagos, along the coast of Alagoas (AL), Bahia (BA), Ceará (CE), Espírito Santo (ES), Santa Catarina (SC) and Sergipe (SE) states, and Cassino beach (Rio Grande do Sul state (Figure 1). Projeto Tamar - Fundação Tamar and Centro Tamar-ICMBio, provided samples from BA, CE, ES and SE; Instituto Biota de Conservação provided samples from AL; and Núcleo de Educação e Monitoramento Ambiental (NEMA) provided samples from Cassino. The NGO Karumbé provided samples from the coast of Uruguay (UY). All samples had their mtDNA characterized (part of which were described in Proietti et al., 2014b); for nDNA 141 samples were analyzed, prioritizing sites where hybrids had been identified through mtDNA.
Figure 1. Sampling sites in the South Western Atlantic, indicating the number of individuals analyzed for mtDNA/nDNA. ASP São Pedro and São Paulo Archipelago; AB Abrolhos Archipelago; AL Alagoas; CA Cassino; CE Ceará, BA Bahia; ES Espírito Santo; RJ Rio de Janeiro; SC Santa Catarina; SE Sergipe; UY Uruguay.
Molecular analyses
Genomic DNA was extracted using a commercial extraction kit (Qiagen DNEasy Extraction Kit), according to the manufacturer’s protocol. Fragments of the mtDNA control region (~850 bp) were amplified via Polymerase Chain Reaction (PCR) using primers LCM15382 and H950 (Abreu-Grobois et al., 2006), under the following conditions: 5’ at 94 °C; 36 cycles of 30 s at 94 °C, 30 s at 50 °C 1min at 72 °C; and a final extension of 10 min at 72 °C. Fragments of nDNA were amplified using primers previously described by Vilaça et al. (2012), for three different genes: oocyte maturation factor (CMOS - 601 bp and 13 polymorphic sites), and two somatic recombination activating genes (RAG1 - 368 bp and 10 polymorphic sites; RAG2 - 620 bp and 8 polymorphic sites). These markers were shown to be species-specific and effectively differentiate various sea turtle species and their hybrids (Vilaça et al., 2012). PCR cycle conditions were: 5’ denaturation at 94 °C; 35 cycles consisting of 30 s at 94 °C; 1 min under specific annealing temperatures (62.5 °C for RAG1 and 67 °C for RAG2 and CMOS), 1 min at 72 °C; and a final extension of 10 min at 72 °C. Each nDNA marker was amplified for a distinct number of samples, since it was not possible to amplify the three markers for all analysed individuals.
Amplified products were purified with purification kits (GE Healthcare Illustra GFX Purification kit) and quantified by spectrophotometry using a BioDrop μLITE. The purified products were then sequenced at Macrogen (http://dna.macrogen.com/eng/). Quality analysis of the sequences obtained for both mtDNA and nDNA was performed with Chromas 2.6.5 software (https://technelysium.com.au). The mtDNA fragments were aligned using the Clustal W tool (Larkin et al., 2007) implemented in BioEdit 7.0.9 (Hall, 1999). Sequences were cropped to 740 bp and classified according to GenBank (https://www.ncbi.nlm.nih.gov/) and the Archie Carr Center for Sea Turtle Research database (http://accstr.ufl.edu/resources/mtdna-sequences/). Since all individuals sequenced were morphologically identified as E. imbricata, those that had mtDNA of other species were considered as hybrids based on this marker. For nDNA, sequence analysis was performed using the PHASE tool of DNAsp software (Librado and Rozas, 2009) to identify the haplotype of each allele of the analysed markers. The nDNA haplotypes previously described by Vilaça et al. (2012; Table S1) were used as prior information.
To characterize hybridization and introgression, we followed the considerations presented by Vilaça et al. (2012): F1 hybrids exhibit for all loci alleles derived from different species, e.g., a C. caretta x E. imbricata F1 hybrid shows for all loci one C. caretta and one E. imbricata allele; introgressed animals (>F1) show for one or more loci two alleles of the same species, e.g. for RAG1 the hybrid individual presents two alleles exclusive to C. caretta.
Data analyses
To update the frequency of occurrence and distribution of immature hybrids based on mtDNA, we grouped the sequences of the 112 samples analyzed in this work with the 158 presented by Proietti et al. (2014b). Based on the haplotype identified for each sample, a haplotype network was built using PopArt (Leigh and Bryant, 2015), with the Median-Joining method (Bandelt et al., 1999).
Bayesian clustering methods were used to detect the level of introgression through STRUCTURE (Pritchard et al., 2000) and NewHybrids (Anderson and Thompson, 2002), based on the nDNA markers. These programs require information on the characteristic haplotypes of each species (coded as bi-allelic genotypes) to infer the ancestry/admixture of the individuals analyzed, and we therefore used as input the database generated by Vilaça et al. (2012). STRUCTURE analysis was performed assuming non-correlated allele frequencies in the admixture model, with a burn-in of 100,000 and 1,000,000 randomizations collected via Markov Chain Monte Carlo (MCMC), with a K value ranging from 1 to 10, with 5 independent iterations. The best K was chosen using the online tool CLUMPAK (Kopelman et al., 2015), according to the Evanno method (Evanno et al., 2005).
The NewHybrids analysis was implemented considering six classes for identification: two classes for pure species (Ei and Cc), and four for hybrids - first generation (F1), second generation (F2), and backcrosses with pure species (F1xEi and F1xCc) - since no hybrid above F2 could be statistically detected by this method (Anderson and Thompson, 2002). This analysis was performed with a burn-in of 10,000 and 1,000,000 randomizations collected via the Markov Chain (MCMC). NewHybrids uses a model that considers only two pure parent species, so in this case we used samples identified by Vilaça et al. (2012) as being of E. imbricata and C. caretta, excluding from the analysis the other species of the database. Thus, samples “CE32" and ”CE43" were not considered in this analysis, since they possessed mtDNA of L. olivacea.
Results
mtDNA
We analysed mtDNA from a total of 270 individuals distributed along the 11 collection sites, based on the 740bp fragments of the control (D-Loop) region. Fifteen haplotypes were identified (Figure 2), with eleven being characteristic of E. imbricata and four of other species. The E. imbricata haplotype distribution (Figure 3) showed a predominance of haplotype EiA01, found in all areas and with a total frequency of 78%. The less frequent haplotypes were EiA62 (6%), EiA32 (4%), EiA09 (1%), and rare haplotypes, with only one occurrence each, were EiA11, EiA23, EiA24, EiA28, EiA61, EiA76 and EiA92.
Figure 2. mtDNA (D-Loop) haplotype network constructed based on the Median-Joining method. Circles represent each of the identified haplotypes, size corresponds to the frequency of occurrence of the haplotype, and color represents the sampling location. Dashes between haplotypes represent the number of distinct bases between them.
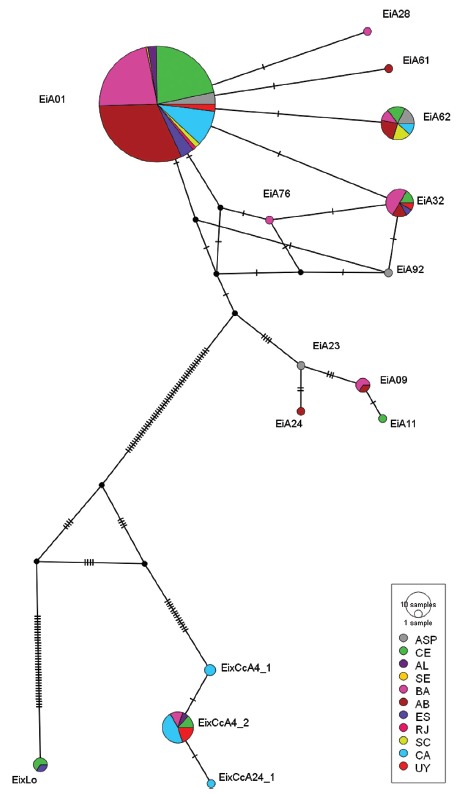
Figure 3. Geographic distribution of D-Loop haplotypes found in the mtDNA analysis along the South Western Atlantic. ASP São Pedro and São Paulo Archipelago; (n = 12); AB Abrolhos Archipelago (n = 75); AL - Alagoas (n = 6); CA - Cassino (n = 33); CE - Ceará (n = 56), BA - Bahia (n = 61); ES - Espírito Santo (n = 11); RJ Rio de Janeiro (n = 2); SC Santa Catarina (n = 6); SE - Sergipe (n = 1); UY - Uruguay (n = 7).
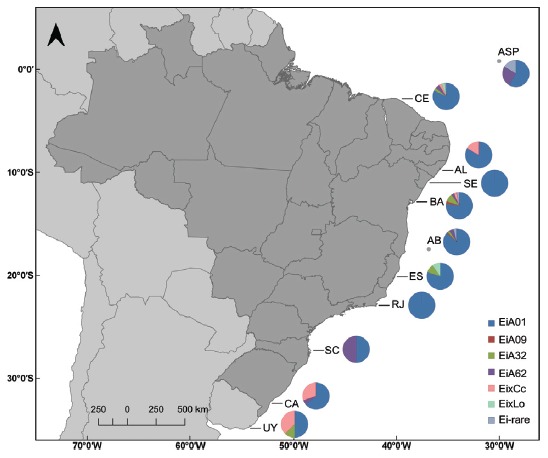
Of the haplotypes of other species, three are specific to C. caretta (CC-A4.2 - 5.6%, CC-A4.1 - 0.7% and CC-A24.1 - 0.4%), and one to L. olivacea (Haplotype F - 1.1%), all of which were previously shown to occur in Brazil’s nesting grounds (Bowen et al., 1998; Shamblin et al., 2014). Two hybrid turtles with L. olivacea haplotypes were found in Espírito Santo and one in Ceará, and all three hybrids had haplotype F. Of the 17 hybrids with C. caretta, observed at five sites, 10 were found in Cassino, one in Alagoas, two in Bahia, two in Ceará, and two in Uruguay. These hybrids showed a predominance of the CC-A4.2 haplotype, with one occurring in Alagoas (out of a total of 6), two in Bahia (total n = 61) and two in Ceará (total n = 56). Interestingly, Cassino showed a high frequency of hybrids with three distinct C. caretta haplotypes - CC-A4.2 (21%), CC-A4.1 (6%) and CC-A24.1 (3%) - and the presence of the most frequent E. imbricata haplotypes - EiA01 (64%) and EiA62 (6%) (total n = 33). Uruguay presented two E. imbricata haplotypes - EiA01 (50%) and EiA32 (12.5%) - and displayed only one C. caretta haplotype, CCA4.2 (37.5%) (total n = 8).
nDNA
Six haplotypes were found for RAG1 (n = 126): Hap3, species-specific of E. imbricata; Hap1 and Hap4, shared between E. imbricata and L. olivacea; Hap2, species-specific of C. caretta; Hap8, species-specific of C. mydas; and a previously unidentified haplotype (named Hap10, Table S1). For RAG2 (n = 89), four haplotypes were found: Hap5, species-specific of E. imbricata; Hap2, species-specific of C. caretta; Hap6, species-specific of C. mydas; and a previously unidentified haplotype (named Hap7, Table S1). For CMOS (n = 35), six haplotypes were found: Hap3, Hap5, Hap9 and Hap10, species-specific of E. imbricata; and Hap1 and Hap2, species-specific of C. caretta.
In general, most samples were homozygotes (RAG1 = 83%, RAG2 = 94%, CMOS = 63%), while heterozygotes were in most cases hybrid individuals that had alleles of different species. The exception was RAG1, in which 11 of the 21 heterozygotes had a haplotype that can be found in both E. imbricata and L. olivacea, and therefore it was not possible to determine if they were hybrids or pure E. imbricata. The CMOS marker was the only one to identify eleven heterozygotes among the pure individuals, i.e., individuals that presented two distinct but species-specific E. imbricata haplotypes in each of the alleles.
In STRUCTURE, the best K (K = 4) separated E. imbricata (Ei), C. caretta (Cc), L. olivacea (Lo) and C. mydas (Cm) in different groups (Figure 4), and hybrids that had alleles of two or more distinct species were also identified. Of the 17 E. imbricata x C. caretta hybrids identified through mtDNA, for five samples it was not possible to amplify any of the nuclear markers used in this work. Therefore, these five samples were not included in the subsequent analysis. Clustering results showed that E. imbricata x C. caretta (Ei x Cc) hybrids presented different degrees of hybridization. Surprisingly, two individuals (CA23 from Cassino and PF06 from Bahia), classified as E. imbricata through morphological analysis and mtDNA, presented alleles of C. caretta and therefore appear in the ‘Ei’ group with ‘Cc’ components. These hybrids have the particularity of having mtDNA and morphology of E. imbricata, a condition observed at low frequency (1%) by Vilaça et al. (2012). Therefore, these two individuals might be: 1) the result of the cross between a female E. imbricata and a male C. caretta (rare occurrence), and are therefore F1; or 2) a cross between a female E. imbricata female and a male Ei x Cc hybrid, hence generation >F1. Based only on the available data, it was not possible to distinguish between the two possibilities.
Figure 4. Cluster analysis performed in STRUCTURE for three nDNA markers (RAG1, RAG2 and CMOS). X-axis represents each of the individuals analysed, y-axis is the estimated mixture ratio of each of the parent species in the composition of these individuals. Species abbreviation and colors: blue, E. imbricata (Ei); orange, C. caretta (Cc); purple, L. olivacea (Lo); green, C. mydas (Cm).

For the three individuals identified as hybrids with L. olivacea (Ei x Lo) through mtDNA, it was possible to amplify only one nDNA marker (RAG1) in two individuals, and both presented Hap1, shared between E. imbricata and L. olivacea. Therefore, inference on the degree of hybridization of these individuals was not possible since these samples were not included in the STRUCTURE analysis, and they were categorized as hybrids based only on mtDNA. In addition to the E. imbricata x C. caretta hybrids previously identified by mtDNA, it was possible to find two additional hybrids (CA23 and PF06) that presented mtDNA of E. imbricata and nDNA with one allele of each of the pure parent species, for all amplified markers.
NewHybrids correctly identified all pure individuals as E. imbricata (p > 0.98, Figure 5). Seven hybrids (AP56, CA24, CA25, CA31, CA36, CE04, CE51) were classified as F1 (p > 0.90), and had in all loci a species-specific haplotype of the parent species with no evidence of introgression. Five other hybrids (CA12, CA14, CA22, UY01, and UY05) were not classified as hybrids since they had species-specific E. imbricata haplotypes in all loci, despite having C. caretta mtDNA, thus demonstrating signs of introgression with E. imbricata. An information summary for all hybrids identified at the South Western Atlantic, with mtDNA haplotype, parental species involved, and inference on generations, can be seen in Table 2.
Figure 5. Assignment probability model for the 12 E. imbricata x C. caretta hybrids identified by mtDNA analysis (AP56 UY05) and two additional hybrids identified only through nDNA (CA23 and PF06). The y-axis represents the probability that each individual belongs to each of the six categories presented (Ei, Cc, F1, F2, F1xEi e F1xCc).
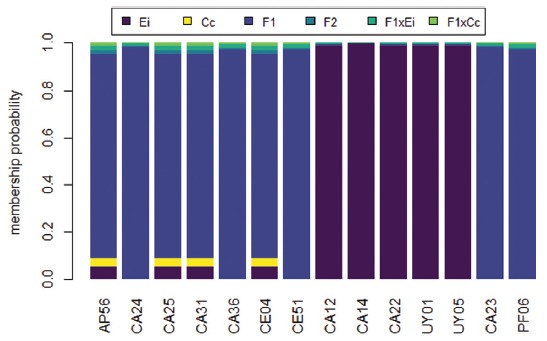
Table 2. Immature sea turtle hybridsImmature sea turtle hybrids identified at the South Western Atlantic, with sampling area, mtDNA haplotype, species involved, and inference on hybrid classes. AL - Alagoas, BA - Bahia, CA - Cassino, CE - Ceará, ES - Espirito Santo, UY - Uruguay.
| Sample | Area | Haplotype | mtDNA | nDNA | F - M* | Classes |
|---|---|---|---|---|---|---|
| AL02 | AL | CCA4.2 | EixCc | - | Cc - Ei | - |
| AP56 | BA | CCA4.2 | EixCc | EixCc | Cc - Ei | F1 |
| PF06 | BA | EiA01 | Ei | EixCc | Ei - Cc or Ei - EixCc | F1 or F1xEi |
| SA10 | BA | CCA4.2 | EixCc | - | Cc - Ei | - |
| CA04 | CA | CCA4.2 | EixCc | - | Cc - Ei | - |
| CA12 | CA | CCA4.2 | EixCc | Ei | EixCc - Ei | F1xEi |
| CA14 | CA | CCA4.2 | EixCc | Ei | EixCc - Ei | F1xEi |
| CA22 | CA | CCA4.2 | EixCc | Ei | EixCc - Ei | F1xEi |
| CA23 | CA | EiA01 | Ei | EixCc | Ei - Cc or Ei - EixCc | F1 or F1xEi |
| CA24 | CA | CCA4.2 | EixCc | EixCc | Cc - Ei | F1 |
| CA25 | CA | CCA4.2 | EixCc | EixCc | Cc - Ei | F1 |
| CA31 | CA | CCA4.2 | EixCc | EixCc | Cc - Ei | F1 |
| CA32 | CA | CCA24.1 | EixCc | - | Cc - Ei | - |
| CA33 | CA | CCA4.1 | EixCc | - | Cc - Ei | - |
| CA36 | CA | CCA4.1 | EixCc | EixCc | Cc - Ei | F1 |
| CE04 | CE | CCA4.2 | EixCc | EixCc | Cc - Ei | F1 |
| CE32 | CE | Hap F | EixLo | Ei/Lo | Lo - Ei | - |
| CE43 | CE | Hap F | EixLo | Ei/Lo | Lo - Ei | - |
| CE51 | CE | CCA4.2 | EixCc | EixCc | Cc - Ei | F1 |
| ES05 | ES | Hap F | EixLo | - | Lo - Ei | - |
| UY01 | UY | CCA4.2 | EixCc | Ei | EixCc - Ei | F1xEi |
| UY05 | UY | CCA4.2 | EixCc | Ei | EixCc - Ei | F1xEi |
| *F: female – M: male. |
Discussion
In this work, we used mtDNA (D-Loop) and nDNA (RAG1, RAG2 and CMOS) markers to investigate hybridization in immature marine turtles in the South Western Atlantic, and identified 22 hybrid turtles in 270 samples (8.1%), with 60% of them occurring at the extreme south of their distribution (South Brazil and Uruguay). A previous study conducted along the Brazilian coast with immature turtles identified as E. imbricata found four hybrids with C. caretta through mtDNA analysis (Proietti et al., 2014b).
With the increase in geographical coverage and sample number, we identified 17 hybrid Ei x Cc on the coast of the South Western Atlantic, based on mtDNA. The highest frequency of these hybrids occurred in Uruguay, which presented three hybrids in eight samples (37.5%), followed by Cassino, where 10 out of the 33 turtles analysed (30%) had C. caretta haplotypes. Proietti et al. (2014a) hypothesized that the high occurrence of hybrid turtles in temperate areas could be due to the adoption of the behavior of C. caretta, which occupies colder regions (Wallace et al., 2010) than E. imbricata, which prefers tropical areas (León and Bjorndal, 2002; Mortimer and Donnelly, 2008). Our results corroborate this hypothesis, and we suggest that other methods such as telemetry or diet analysis should be used to confirm the differential behavior of hybrids.
Previous Ei x Cc records reported the occurrence of only one mtDNA haplotype, CCA4.2, the most common haplotype of females at Brazilian nesting grounds (Shamblin et al., 2014). In the present work, two other haplotypes (CCA4.1 and CCA24.1) were identified for the first time in hybrids. These haplotypes are present only at nesting grounds in Brazil, but at lower frequency (Reis et al., 2010). Based on haplotype frequencies, Shamblin et al. (2014) suggested a possible regionalization of C. caretta populations at the Brazilian coast, into two Regional Management Units (RUMs): 1) Sergipe and Bahia 2) and Espírito Santo and Rio de Janeiro. The hybrids we identified presented haplotypes most frequently found in the first RMU, and therefore we can consider that they most likely originate from the Bahia/Sergipe rookeries, reinforcing the observation that this is the main region where C. caretta and E. imbricata crossbreed.
We found three E. imbricata x L. olivacea hybrids, a type of cross that had not yet been reported for immature marine turtles along the South Western Atlantic coast. These hybrids presented the F haplotype, described by Bowen et al. (1998) as characteristic of L. olivacea and originating from nesting areas of the Atlantic Ocean (in descending order of frequency: Suriname, Brazil and Guinea Bissau). Rookeries for this species in Brazil are concentrated between Bahia and Alagoas, with higher density in Sergipe state (Castilhos et al., 2011). Lara-Ruiz et al. (2006) observed two hybrids with haplotype F when analyzing the mtDNA of 119 adult E. imbricata samples from the north coast of Bahia. Vilaça et al. (2012) reported two occurrences of Ei x Lo at the coast of Bahia among 121 female E. imbricata, and concluded that all were the result of the crossing between a male E. imbricata and a female L. olivacea (F1). In addition, the authors state that these individuals showed no signs of introgression; therefore, they may be infertile or generated from rare hybridization events. The rarity of E. imbricata and L. olivacea hybrids can be explained by both species presenting their highest reproductive population densities in different areas: olive ridleys nest mostly in Sergipe state, reducing the probability of crossings between them. Sanches and Bellini (1999) observed that morphology may also influence this reproduction, since adult L. olivacea are smaller (mean CCL 73 cm; Silva et al., 2007) than E. imbricata (mean CCL 97 cm; Marcovaldi et al., 1999), which is in turn more similar to C. caretta (mean CCL 103 cm; Marcovaldi and Chaloupka, 2007).
The analysis of both mtDNA and nDNA increased the number of hybrid sea turtle detections at the South Western Atlantic coast. Based on mtDNA alone 20 hybrids were identified, and nDNA analysis revealed two additional hybrids not identified by morphology or mtDNA. In addition, with the analysis of three nDNA markers, it was possible to infer that 50% of these hybrids were first generation and 36% were backcrossed between hybrids and pure E. imbricata (>F1). The generations of two hybrids (CA23 and PF06) could not be directly determined, since they had morphology and mtDNA of the same species (E. imbricata) but nDNA with alleles of two different species (Ei x Cc). With these characteristics the individuals could be: F1, result of the unusual crossing between female E. imbricata and male C. caretta; or >F1, the result of a backcross between a hybrid male (Ei x Cc) and an E. imbricata female.
As mentioned above, most first generation hybrids (F1) result from mating between C. caretta females and E. imbricata males (Vilaça et al., 2012). Crossings between F1 females and both pure parent species also occur, but crossings of pure females and hybrid males are apparently less frequent (Vilaça et al., 2012, Soares et al., 2018). Female E. imbricata, C. caretta and Ei x Cc hybrids have different nesting peaks, with pure C. caretta nesting earlier than hybrids, which nest earlier than pure E. imbricata (Soares et al., 2017). Considering this, along with the gender bias of the reproductive groups, it is unlikely that the CA23 and PF06 individuals are the result of a cross between a female E. imbricata (pure) and a male C. caretta (F1). Since the E. imbricata nesting peak in Bahia occurs when most C. caretta males have already left the area, the likelihood of them reproducing is reduced. Indeed, this type of hybrid cross was observed in only one of the 82 females from Bahia analysed by Soares et al. (2017). This hybrid was also identified as C. caretta through morphological analysis, which did not occur in the individuals found in this study. Hybrid males (Ei x Cc) may show an intermediate reproductive period between pure parent species, as observed for hybrid females. In this case, they would encounter more E. imbricata females than the C. caretta males. Additionally, considering that backcrosses between Ei x Cc and pure E. imbricata have been observed more frequently than introgression with C. caretta, it is likely that CA23 and PF06 are a result of the cross between E. imbricata and hybrid males. Given our findings, we recommend that future studies sequence at least one nuclear loci in conjunction with the mtDNA to identify hybrids and potential introgression.
Conclusions
Our study shows the importance of the combined use of mtDNA and nDNA markers in the evaluation of hybridization among marine turtles. Although mtDNA analysis is of paramount importance in the study of hybrids, nDNA analysis is also crucial for identifying generations/parental species, as well as for detecting hybrids in cases where mtDNA cannot (e.g. hybrid backcrosses with pure E. imbricata). This is confirmed by our observation of two hybrids that presented E. imbricata morphology and mtDNA, which could only be identified as Ei x Cc through nDNA. This observation may have been underestimated since we were not able to amplify the three nDNA markers for all samples. Another limitation was that the only nuclear marker amplified for all animals (RAG1) presents two shared haplotypes between E. imbricata and L. olivacea, and therefore it is not possible to infer conclusively about Ei x Lo hybrids with nDNA analysis. The hybridization events between E. imbricata and L. olivacea seems to be rare, but additional studies are necessary to identify species-specific haplotypes, allowing us to understand the generation and parental species of these hybrids.
Considering the high frequency of hybrids found in the South Western Atlantic, studies on the behavior, distribution, feeding strategy, migration and demography of hybrid turtles should be performed. Satellite tracking, stable isotope analysis and more comprehensive molecular tools such as genomics (e.g. SNP analysis), are techniques that would aid in understanding the process and better determining the ecological role of these hybrids, as well as their influence on marine turtle populations. According to Bohling (2016), the greatest concern regarding hybridization events is the extinction of genetic, phenotypic and/or evolutionary units due to a continuous hybridization process or hybrid vigor. Schwartz et al. (2007) suggest that this process be biologically and ecologically monitored in a continuous manner, providing a framework for determining and tracking the extent of hybridization, trends over time, and results of management strategies.
There are currently no guidelines for the management of hybrids and hybrid zones in Brazil, and most existing programs are focused on areas where the event has anthropogenic causes such as habitat change (Crispo et al., 2011) and overexploitation (Bohling and Waits, 2015). To avoid the genetic extinction of pure species in the presence of a hybrid swarm, hybrid animals can be eliminated from the reproductive stock by eradication or sterilization. This type of action has already been carried out to control the hybridization beween the estuarine fish Cyprinodon variegatus and Cyprinodon bovinus, which was successful due to the limited geographic extension of the hybrid group (Echelle and Echelle, 1997). Considering the current endangered status of marine turtles and the high frequency of hybrids along the SWA coast, continuous monitoring should be carried out to assess the fitness, genetic integrity, and to detect the extent of changes in the gene pools of the involved populations. This is fundamental to evaluate if management of hybrid individuals should be considered, and ensure the conservation of SWA marine turtle populations.
Acknowledgments
C. Brito was a graduate student of the Programa de Pós-graduação em Oceanografia Biológica, and received a scholarship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), which is suffering serious budget cuts by the current Brazilian government. CNPq also provided a research fellowship to M.C.P. (312470/2018-5). Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES Finance code 001) provided access to the Portal de Periódicos and financial support through Programa de Excelência Acadêmica (PROEX). This study was funded by The Rufford Foundation and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). We thank Luis F. Marins and Luiz Felipe Cestari Dumont, as well as anonymous reviewers, for suggestions on this manuscript. We thank Projeto Tamar - Fundação Tamar and Centro Tamar-ICMBio, Núcleo de Educação e Monitoramento Ambiental (NEMA), Instituto Biota de Conservação, Associação Civil Karumbé, Parque Nacional Marinho dos Abrolhos and Reserva Biológica Marinha Arvoredo for providing samples and field assistance. We also thank the Berlin Center for Genomics in Biodiversity Research - BeGenDiv - for support in laboratorial analyses. This is a contribution of the research group “ECOMEGA - Ecologia da Megafauna Marinha”.
Supplementary material.
The following online material is available for this article:
References
- Abreu-Grobois FA, Horrocks JA, Formia A, Dutton PH, LeRoux RA, Velez-Zuazo X, Soares LS, Meylan AB. In: Frick M, Panagopoulou A, Rees AF, Williams K, editors. New mtDNA D-loop primers which work for a variety of marine turtle species may increase the resolution of mixed stock analysis; 26thAnnual Symposium on Sea Turtle Biology; Crete. 2006. pp. 179–179. Proceedings. [Google Scholar]
- Allendorf FW, Leary RF, Spruell P, Wenburg JK. The problems with hybrids: Setting conservation guidelines. Trends Ecol Evol. 2001;16:613–622. [Google Scholar]
- Alvarez-Varas R, Berzins R, Bilo K, Chevalier J, Chevallier D, Thoisy BDE, Fallabrino A, Cruz MG, Kelez S, Lopez-Mendilaharsu M, et al. Sea Turtles of South America. SWOT Rep. 2016;11:14–27. [Google Scholar]
- Anderson EC, Thompson EA. A model-based method for identifying species hybrids using multilocus data. Genetics. 2002;160:1217–1229. doi: 10.1093/genetics/160.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-Joining Networks for inferring intraspecific phylogenies. Mol Biol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Bohling JH. Strategies to address the conservation threats posed by hybridization and genetic introgression. Biol Conserv. 2016;203:321–327. [Google Scholar]
- Bohling JH, Waits LP. Factors influencing red wolfcoyote hybridization in eastern North Carolina, USA. Biol Conserv. 2015;184:108–116. [Google Scholar]
- Bowen BW, Clark AM, Abreu-Grobois FA, Chaves A, Reichart HA, Ferl RJ. Global phylogeography of the Ridley sea turtles (Lepidochelys spp.) as inferred from mitochondrial DNA sequences. Genetica. 1998;101:179–189. doi: 10.1023/a:1018382415005. [DOI] [PubMed] [Google Scholar]
- Bowen BW, Karl SA. Population genetics and phylogeography of sea turtles. Mol Ecol. 2007;16:4886–4907. doi: 10.1111/j.1365-294X.2007.03542.x. [DOI] [PubMed] [Google Scholar]
- Castilhos JC De, Coelho CA, Argolo JF, Allan E. Avaliação do estado de conservação da tartaruga marinha Lepidochelys olivacea (Eschscholtz, 1829) no Brasil. Biodiversidade Bras. 2011;1:26–34. [Google Scholar]
- Conceição MB, Levy JA, Marins LF, Marcovaldi MA. Electrophoretic characterization of a hybrid between Eretmochelys imbricata and Caretta caretta (Cheloniidae) Comp Biochem Physiol. 1990;97:275–278. [Google Scholar]
- Crispo E, Moore JS, Lee-Yaw JA, Gray SM, Haller BC. Broken barriers: Human-induced changes to gene flow and introgression in animals: An examination of the ways in which humans increase genetic exchange among populations and species and the consequences for biodiversity. BioEssays. 2011;33:508–518. doi: 10.1002/bies.201000154. [DOI] [PubMed] [Google Scholar]
- Crochet PA, Chen JZ, Pons JM, Lebreton JD, Hebert PDN, Bonhomme F. Genetic differentiation at nuclear and mitochondrial loci among large white-headed gulls: Sexbiased interspecific gene flow? Evolution. 2003;57:2865–2878. doi: 10.1111/j.0014-3820.2003.tb01527.x. [DOI] [PubMed] [Google Scholar]
- Crossman CA, Taylor EB, Barrett-Lennard LG. Hybridization in the Cetacea: Widespread occurrence and associated morphological, behavioral, and ecological factors. Ecol Evol. 2016;6:1293–1303. doi: 10.1002/ece3.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva ACCD, de Castilhos JC, Lopez GG, Barata PCR. Nesting biology and conservation of the olive Ridley sea turtle (Lepidochelys olivacea) in Brazil, 1991/1992 to 2002/2003. J Mar Biol Assoc UK. 2007;87:1047–1047. [Google Scholar]
- Echelle AA, Echelle AF. Genetic introgression of endemic taxa by non-natives: a case study with Leon Springs Pupfish and Sheepshead Minnow. Conserv Biol. 1997;11:153–161. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Harrison RG. Hybrid zones: windows on evolutionary process. In: Futuyma D, Antonovics J, editors. Oxford Surveys in Evolutionary Biology. Oxford University Press; Oxford: 1990. pp. 69–128. [Google Scholar]
- Karl SA, Bowen BW, Avise JC. Hybridization among the ancient mariners: characterization of marine turtle hybrids with molecular genetic assays. J Hered. 1995;86:262–268. doi: 10.1093/oxfordjournals.jhered.a111579. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 2015;15:1179–1191. doi: 10.1111/1755-0998.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Ruiz P, Lopez GG, Santos FR, Soares LS. Extensive hybridization in hawksbill turtles (Eretmochelys imbricata) nesting in Brazil revealed by mtDNA analyses. Conserv Genet. 2006;7:773–781. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez RI, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Leigh JW, Bryant D. POPART: Full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. [Google Scholar]
- León YM, Bjorndal KA. Selective feeding in the hawksbill turtle, an important predator in coral reef ecosystems. Mar Ecol Prog Ser. 2002;245:249–258. [Google Scholar]
- Librado P, Roza J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lima EHSM, Melo MTD, Godfrey MH, Barata PCR. Sea turtle in the waters of Almofala, Ceará, in Northeastern Brazil, 2001-2010. Mar Turt Newsl. 2013;137:5–9. [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends Ecol Evol. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Marcovaldi MA, Lopez GG, Soares L, Santos AJB, Bellini C, Barata PCR. Fifteen years of Hawksbill Sea Turtle (Eretmochelys imbricata) nesting in Northern Brazil. Chelonian Conserv Biol. 2007;6:223–223. [Google Scholar]
- Marcovaldi M, Chaloupka M. Conservation status of the loggerhead sea turtle in Brazil: an encouraging outlook. Endanger Species Res. 2007;3:133–143. [Google Scholar]
- Marcovaldi MÂ, Vieitas CF, Godfrey MH. Nesting and conservation management of hawksbill turtles (Eretmochelys imbricata) in Northern Bahia, Brazil. Chelonian Conserv Biol. 1999;3:301–307. [Google Scholar]
- Marcovaldi MÂ, Lopez GG, Soares LS, Santos AJB, Bellini C, Santos AS, Lopez M. Avaliação do estado de conservação da tartaruga marinha Eretmochelys imbricata (Linnaeus, 1766) no Brasil. Biodiversidade Bras. 2011;1:20–27. [Google Scholar]
- Marcovaldi M, Lopez G, Soares L, López-Mendilaharsu M. Satellite tracking of hawksbill turtles (Eretmochelys imbricata) nesting in northern Bahia, Brazil: turtle movements and foraging destinations. Endanger Species Res. 2012;17:123–132. [Google Scholar]
- Monteiro DS, Estima SC, Gandra TBR, Silva AP, Bugoni L, Swimmer Y, Seminoff JA, Secchi ER. Long-term spatial and temporal patterns of sea turtle strandings in southern Brazil. Mar Biol. 2016;163:247–247. [Google Scholar]
- Mortimer JA, Donnelly M. Eretmochelys imbricata. The IUCN Red List of Threatened Species e.T8005A12881238. 2008. [DOI]
- Naro-Maciel E, Le M, FitzSimmons NN, Amato G. Evolutionary relationships of marine turtles: A molecular phylogeny based on nuclear and mitochondrial genes. Mol Phylogenet Evol. 2008;49:659–662. doi: 10.1016/j.ympev.2008.08.004. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Rieseberg LH. A genomic perspective on hybridization and speciation. Mol Ecol. 2016;25:2337–2360. doi: 10.1111/mec.13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietti MC, Reisser J, Marins LF, Marcovaldi MA, Soares LS, Monteiro DS, Wijeratne S, Pattiaratchi C, Secchi ER. Hawksbill loggerhead sea turtle hybrids at Bahia, Brazil: where do their offspring go? Peer J. 2014;2:e255. doi: 10.7717/peerj.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proietti MC, Reisser J, Marins LF, Rodriguez-Zarate C, Marcovaldi MA, Monteiro DS, Pattiaratchi C, Secchi ER. Genetic structure and natal origins of immature hawksbill turtles (Eretmochelys imbricata) in Brazilian waters. PLoS One. 2014;9:e88746. doi: 10.1371/journal.pone.0088746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosdocimi L, Bruno I, Diaz L, Carman VG, Albareda DA, Remis MI. Southernmost reports of the hawksbill sea turtle, Eretmochelys imbricata, in temperate waters of Argentina and evidence of a hybrid origin supported by mitochondrial DNA analysis. Herpetol Rev. 2014;45:1–5. [Google Scholar]
- Reis EC, Soares LS, Vargas SM, Santos FR, Young RJ, Bjorndal KA, Bolten AB, Lôbo-Hajdu G. Genetic composition, population structure and phylogeography of the loggerhead sea turtle: Colonization hypothesis for the Brazilian rookeries. Conserv Genet. 2010;11:1467–1477. [Google Scholar]
- Rieseberg L, Wendell JF. Gene flow and its consequences in plants, Hybrid zones and the evolutionary process. Annu Rev Ecol Evol Syst. 1993;23:237–261. [Google Scholar]
- Rosenfield JA, Nolasco S, Lindauer S, Sandoval C, Kodric-Brown A. The role of hybrid vigor in the replacement of pecos pupfish by its hybrids with sheepshead minnow. Conserv Biol. 2004;18:1589–1598. [Google Scholar]
- Sanches TM, Bellini C. Juvenile Eretmochelys imbricata and Chelonia mydas in the Archipelago of Fernando de Noronha, Brazil. Chelonian Conserv Biol. 1999;3:308–311. [Google Scholar]
- Santana A, Soares L, Marcovaldi MÂ, Monteiro S. Avaliação do estado de conservação da tartaruga marinha Caretta caretta Linnaeus, 1758. Biodiversidade Bras. 2011;1:3–11. [Google Scholar]
- Santos AS, Almeida AP, Santos AJB, Giffoni B, Gallo B, Baptistotte C, Coelho CA, Lima EHSM, Sales G, Lopez GG, et al. Plano de ação nacional para conservação das tartarugas marinhas. Instituto Chico Mendes de Conservação da Biodiversidade; Brasília: 2011. 122 p. (Série Espécies Ameaçadas nº 25) [Google Scholar]
- Schwartz MK, Luikart G, Waples RS. Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol. 2007;22:25–33. doi: 10.1016/j.tree.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Seminoff JA, Karl SA, Schwartz TS, Resendiz A. Hybridization of the green turtle (Chelonia mydas) and hawksbill turtle (Eretmochelys imbricata) in the Pacific Ocean: Indication of an absence of gender bias in the directionality of crosses. Bull Mar Sci. 2003;73:643–652. [Google Scholar]
- Shamblin BM, Bolten AB, Abreu-Grobois FA, Bjorndal KA, Cardona L, Carreras C, Clusa M, Monzón-Argüello C, Nairn CJ, Nielsen JT, et al. Geographic patterns of genetic variation in a broadly distributed marine vertebrate: New insights into loggerhead turtle stock structure from expanded mitochondrial DNA sequences. PLoS One. 2014;9:e85956. doi: 10.1371/journal.pone.0085956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares LS, Bolten AB, Wayne ML, Vilaça ST, Santos FR, Marcovaldi MAG, Bjorndal KA. Comparison of reproductive output of hybrid sea turtles and parental species. Mar Biol. 2017;164:9–9. [Google Scholar]
- Soares LS, Bjorndal KA, Bolten AB, Marcovaldi MAG, Luz PB, Machado R, Lo R, McDaniel SF, Payton AC, Waltzek TB, et al. Effects of hybridization on sea turtle fitness. Conserv Genet. 2018;19:1311–1322. [Google Scholar]
- Sunnucks P. Efficient genetic markers for population biology. Trends Ecol Evol. 2000;15:199–203. doi: 10.1016/s0169-5347(00)01825-5. [DOI] [PubMed] [Google Scholar]
- Vélez-Rubio GM, Estrades A, Fallabrino A, Tomás J. Marine turtle threats in Uruguayan waters: Insights from 12 years of stranding data. Mar Biol. 2013;160:2797–2811. [Google Scholar]
- Vilaça ST, Santos FR. Molecular data for the sea turtle population in Brazil. Dataset Pap Sci. 2013;2013:1–7. [Google Scholar]
- Vilaça ST, Vargas SM, Lara-Ruiz P, Molfetti É, Reis EC, Lôbo-Hajdu G, Soares LS, Santos FR. Nuclear markers reveal a complex introgression pattern among marine turtle species on the Brazilian coast. Mol Ecol. 2012;21:4300–4312. doi: 10.1111/j.1365-294X.2012.05685.x. [DOI] [PubMed] [Google Scholar]
- Wallace BP, DiMatteo AD, Hurley BJ, Finkbeiner EM, Bolten AB, Chaloupka MY, Hutchinson BJ, Abreu-Grobois AF, Amorocho D, Bjorndal KA, et al. Regional management units for marine turtles: A novel framework for prioritizing conservation and research across multiple scales. PLoS One. 2010;5:1–11. doi: 10.1371/journal.pone.0015465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne RK, Shaffer HB. Hybridization and endangered species protection in the molecular era. Mol Ecol. 2016;25:2680–2689. doi: 10.1111/mec.13642. [DOI] [PubMed] [Google Scholar]
- Whitham T, Morrow P, Potts B. Conservation of hybrid plants. Science. 1991;254:779–780. doi: 10.1126/science.254.5033.779-b. [DOI] [PubMed] [Google Scholar]
- Wood J, Wood F, Critchley K. Hybridization of Chelonia mydas and Eretmochelys imbricata. Copeia. 1983;1983:839–842. [Google Scholar]
Internet Resources
- IUCN . The IUCN Red List of Threatened Species. 2018. [accessed 14 February 2018]. IUCN (2018) The IUCN Red List of Threatened Species, www.iucnredlist.org. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


