Abstract
The tRNA isopentenyltransferases (IPTases), which add an isopentenyl group to N6 of A37 (i6A37) of certain tRNAs, are among a minority of enzymes that modify cytosolic and mitochondrial tRNAs. Pathogenic mutations to the human IPTase, TRIT1, that decrease i6A37 levels, cause mitochondrial insufficiency that leads to neurodevelopmental disease. We show that TRIT1 encodes an amino-terminal mitochondrial targeting sequence (MTS) that directs mitochondrial import and modification of mitochondrial-tRNAs. Full understanding of IPTase function must consider the tRNAs selected for modification, which vary among species, and in their cytosol and mitochondria. Selection is principally via recognition of the tRNA A36-A37-A38 sequence. An exception is unmodified tRNATrpCCA-A37-A38 in Saccharomyces cerevisiae, whereas tRNATrpCCA is readily modified in Schizosaccharomyces pombe, indicating variable IPTase recognition systems and suggesting that additional exceptions may account for some of the tRNA-i6A37 paucity in higher eukaryotes. Yet TRIT1 had not been characterized for restrictive type substrate-specific recognition. We used i6A37-dependent tRNA-mediated suppression and i6A37-sensitive northern blotting to examine IPTase activities in S. pombe and S. cerevisiae lacking endogenous IPTases on a diversity of tRNA-A36-A37-A38 substrates. Point mutations to the TRIT1 MTS that decrease human mitochondrial import, decrease modification of mitochondrial but not cytosolic tRNAs in both yeasts. TRIT1 exhibits clear substrate-specific restriction against a cytosolic-tRNATrpCCA-A37-A38. Additional data suggest that position 32 of tRNATrpCCA is a conditional determinant for substrate-specific i6A37 modification by the restrictive IPTases, Mod5 and TRIT1. The cumulative biochemical and phylogenetic sequence analyses provide new insights into IPTase activities and determinants of tRNA-i6A37 profiles in cytosol and mitochondria.
Author summary
tRNA isopentenyltransferases (IPTases) are tRNA modification enzymes that are conserved in bacteria and eukaryotes. They add an isopentenyl group to the Adenosine base at position 37, adjacent to the anticodon of specific subsets of tRNAs that decode codons that begin with Uridine. This modification stabilizes the otherwise weak adjacent codon-anticodon base pair and increases the efficiency of decoding of the corresponding codons of the genetic code. IPTases belong to a group of enzymes that modify both cytoplasmic and mitochondrial tRNAs of eukaryotic cells. Interestingly, during evolution there were changes in the way that IPTases are targeted to mitochondria as well as changes in the relative numbers and identities of IPTase tRNA substrates in the cytoplasm vs. mitochondria. The latter is consistent with phenotypic consequences of IPTase deficiencies in fission and budding yeasts, and mammals. Pathogenic mutations to human IPTase (TRIT1) cause mitochondrial insufficiency and neurodevelopmental disease, principally due to decreased modification of the mt-tRNA substrates. In this study, we identify the way human TRIT1 is targeted to mitochondria. We also show that TRIT1 exhibits a tRNA anticodon identity-specific substrate sensitivity. The work leads to new understanding of the IPTases and the variable anticodon identities of their tRNA substrates found throughout nature.
Introduction
45 different eukaryotic cytoplasmic (cy-) and about half as many mitochondrial (mt-) tRNAs contain unique sets of modifications that are considered in two groups [1–3]; those in the body or core, which contribute to folding and/or stability, and those in the anticodon loop (ACL) which contribute to mRNA decoding by cy- and mt- ribosomes. Modifications in the anticodon stem loop (ASL) are more concentrated and diverse than those in the tRNA body. One of these is isopentenylation of N6 of adenosine, found only on tRNAs that decode UNN codons, at position 37 (i6A37) directly 3' to the anticodon. Bacterial i6A37 is hypermodified to 2-methylthio-N6-isopentenyl-A37 (ms2i6A37), whereas eukaryotic cy-tRNA-i6A37 is not. Also, while the mt-tRNAs of S. cerevisiae and S. pombe are not hypermodified, mammalian mt-tRNAs i6A37 are [4–6] (Table 1).
Table 1. tRNA-i6A37 identity variation among species, cytoplasm and mitochondria.
| tRNA anticodon (as set by DNA) |
E. coli MiaA |
S. pombe tit1+ |
S. cerevisiae MOD5 |
human or bovine TRIT1 |
|---|---|---|---|---|
| cy-Ser GGA | A37 | |||
| cy-Ser AGA | i6A37 | i6A37 | i6A37 | |
| cy-Ser CGA | ms2i6A37 | i6A37 | i6A37 | i6A37 |
| cy-Ser UGA | ms2i6A37 | i6A37 | i6A37 | i6A37 |
| cy-SerSeC UCA | i6A37 | i6A37 | ||
| cy-Tyr GUA | ms2i6A37 | i6A37 | i6A37 | m1G37 |
| cy-Cys GCA | ms2i6A37 | (G) | i6A37 | m1G37 |
| cy-Trp CCA | ms2i6A37 | i6A37 | A37 | m1G37 |
| cy-Phe GAA | ms2i6A37 | yW37 | yW37 | yW37 |
| cy-Leu CAA | ms2i6A37 | (G) | m1G37 | m1G37 |
| cy-Leu UAA | ms2i6A37 | (G) | m1G37 | m1G37 |
| No. isoacceptors@/ cy-tRNAs-i6A37 | 6/9 | 3/5 | 3/5 | 1/4 |
| #mt-Trp CCA | i6A37 | |||
| #mt-Trp UCA | i6A37 | ms2i6A37 | ||
| mt-Tyr GUA | (G) | i6A37 | ms2i6A37 | |
| mt-Ser UGA | (G) | m1G37 | ms2i6A37 | |
| mt-Phe GAA | (G) | m1G37 | ms2i6A37 | |
| mt-Cys GCA | i6A37 | i6A37 | i6A37 | |
| mt-Leu UAA | (A)* | m1G37 | A* | |
| No. isoacceptors/ mt-tRNAs-i6A37 | 2/2 | 3/3 | 5/5 | |
| Total number of tRNAs-i6A37 | 9 | 7 | 8 | 9 |
@Isoacceptors are tRNAs with different anticodons that are acylated with the same amino acid.
The blank white rectangle spaces denote that the tRNA is not expressed in this species [71].
#Position 34 of S. pombe mt-tRNATrp is encoded as C [84] whereas in S. cerevisiae [50, 104], bovine and human [105], it is encoded to be U, and was determined to be 5-taurinomethyluridine (tm5U) in bovine [4], and cmnm5U in S. cerevisiae [92] (see tRNAmodViz server [83]).
*A designates an A37 that is not in an A36-A37-A38 context, containing C38 [4] instead of A as is found in its E. coli counterpart.
The pattern listed in the human or bovine column is also the same for C. elegans, Drosophila melanogaster, Xenopus tropicalis, Gallus gallus, and Mus musculus [71].
yW = ybutosine.
All entries with subscript 37 have been verified as modified or unmodified; the A37 in the S. cerevisiae MOD5 column indicates an exceptional unmodified A37 in a A36-A37-A38 context (see text).
Entries in parentheses show the DNA-encoded nucleotide but have not been validated as modified or not in RNA [4, 43, 59, 60].
i6A37 increases tRNA affinity for the ribosome, increases anticodon-codon pairing occupancy, can activate mRNA decoding and decrease frameshifting (reviewed in [7, see 8]). ASL modifications contribute to "circuits" [9], in which one is prerequisite for formation of others, and whose disruptions can cause disease [10, 11]. I6A37 is required for m3C32 formation on the tRNAsSer that decode UCN codons [12, 13].
Among the numerous enzymes that modify cy- and mt-tRNAs, there are three that recognize the ACLs of their substrates and whose mutations lead to neurodevelopmental disorders. These are TRIT1 (tRNA isopentenyl transferase-1)[14, 15], THG1L (tRNA-histidine guanylyltransferase-1) [16, 17], and TRMT5 (tRNA methyltransferase-5)[18–20]. Thg1 recognizes the anticodon identity element to distinguish its substrate from other tRNAs, even though it modifies a distant site on the tRNA [21]. The Trm5 and IPTase enzymes differentially modify position 37 of numerous tRNAs, using distinct recognition mechanisms [22–24]. As detailed below, it is unclear how IPTases recognize and distinguish among some of their substrates.
IPTases have been conserved through evolution. tRNA i6A37 modification can be functionally complemented in S. cerevisiae lacking its IPTase, Mod5, by cDNAs encoding IPTases from human (TRIT1), silkworm (GRO-1), S. pombe (Tit1), plants or even moss that produce prokaryote-type IPTase [25–29]. Complementation is often shown using tRNA-mediated suppression (TMS) which reflects decoding of an mRNA premature stop codon by a suppressor-tRNATyrUUA that requires i6A37 for activity. A comparable TMS system in S. pombe is mediated by suppressor-tRNASerUCA [30–33]. In both systems, TMS can be readily monitored by a red-white colony assay. Full length TRIT1 (1–467) cloned from human cDNA is active for TMS in S. cerevisiae [25]. Human TRIT1 also complements tRNASerUCA-i6A37-mediated TMS in S. pombe although the TRIT1-R323Q mutation associated with neurodevelopmental disease is largely inactive in the same assay [14].
The single gene-encoded Mod5 localizes to nuclei, nucleoli, cytoplasm and mitochondria [34–36]. Nuclear Mod5 is involved in tRNA gene-mediated silencing [37, 38]; its localization [36] is consistent with this and early tRNA processing [39, 40]. For some tRNAs, A36-A37-A38 is interrupted by an intron, and in yeast where tRNA splicing occurs on the surface of mitochondria, tRNA nuclear export is prerequisite to i6A37 formation [2, 41, also see 42]. A Mod5 nuclear localization signal [36] is C-terminal to the eukaryote-specific IPTase Zn-finger motif [7] that binds the tRNA backbone [24], and is required for the activity of the S. Pombe IPTase Tit1 [43].
I6A37 modification of mt-tRNA in the mitochondrial matrix is a critical TRIT1 function [14]. Pathophysiology of TRIT1-R323Q-associated neurodevelopmental disease is attributable to deficient mt-tRNA modification and consequent impairment of mitochondrial mRNA translation, even though cy-tRNAsSer are also hypomodified [14]. Though S. cerevisiae and C. elegans use alternative translation initiation to target their IPTases to mitochondria [34, 35, 44], the mechanism used by TRIT1 would appear to differ (Fig 1) yet had not been subjected to experimental analysis. In the first part of this study we examine mitochondrial targeting of TRIT1 in human cells. We then extend this to TRIT1 mitochondrial function in S. pombe and S. cerevisiae model systems. Later, we examine modification of a diversity of tRNA substrates by TRIT1, Mod5 and Tit1, and the extent to which substrate-specific negative determinants may contribute to tRNAs-i6A37 profile variation (below).
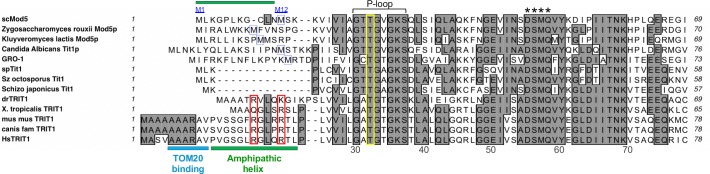
Fig 1. N-termini sequence alignment of representatives of phylogenetic groups of IPTases.
Sequences of the N-termini of IPTases. scMod5: Saccharomyces cerevisiae; GRO-1: IPTase from C. elegans; spTit1: Schizosaccharomyces (Sz) pombe; Sz octosporus; Sz japonicus; drTRIT1: D. rerio: Danio rerio; mus mus: Mus musculus; canis fam: Canis familiaris; X. tropicalis: Xenopus tropicalis; Hs: Homo sapiens. Numerals below the alignment show human amino acid numbering. The region between M1 and M12 of Mod5 that is required for mitochondrial targeting is indicated by a green rectangle above [34, 35]. The second methionines that may be used as alternative translation start sites for cytoplasmic localization are boxed in blue. The invariant Threonine within the conserved P-loop that functions in catalysis is in the yellow rectangle. Asterisks indicate the conserved DSMQ sequence that forms a network of contacts that mediate A37 recognition; the D of which acts as a general base for catalysis [24] and the M of which is M57 of TRIT1(57–467). As predicted by MitoFates [53], the TOM20-binding site of human TRIT1 is underlined with a blue bar and the amphipathic helix of TRIT1 and Mod5 are indicated with green bars. The two key basic residues in the conserved amphipathic helix of mammalian MTS that were mutated in human TRIT1 are in red rectangles.
A conserved positive determinant of all known IPTase substrates is the A36-A37-A38 recognition sequence required for i6A37 formation. Structures of bacterial and S. cerevisiae IPTases bound to their substrates and biochemical studies show that they directly recognize the ACLs of their target tRNAs and mediate i6A37 formation using dimethylallyl-pyrophosphate (DMAPP) as the isopentenyl donor [24, 45–48]. Intriguingly, despite these conserved features and effects of i6A37 on translation, variation exists in the tRNAs-i6A37 profiles among species (Table 1). Although i6A37 occurs widely on bacterial tRNAs for UNN codons, fewer cy-tRNAs bear i6A37 in S. pombe, S. cerevisiae and human (Table 1). tRNA-i6A37 identities also vary between mitochondria and cytosol, within and among species. Better understanding of a basis of this variation is a goal of our study.
Table 1 emphasizes that while a single gene encodes the cytoplasmic and mitochondrial IPTases of the species listed, the cy- and mt- tRNA-i6A37 profiles differ. Specifically, while TRIT1 modifies only tRNAsSer in cytosol, it modifies five tRNA types in mitochondria (Table 1).
Table 1 represents a phylogenetic analysis that reveals substitution of A37 with G as a major source of variability among cy-tRNAs-i6A37, especially eukaryotes. tRNASerGGA is only one of ten A36-A37-A38 tRNAs in E. coli that lack i6A37; weak ASL base pairs decrease its substrate activity for MiaA [49]. Lack of i6A37 on S. cerevisiae cy-tRNATrpCCA-A37-A38 is attributable to its CCA ACL as determined in vivo and in vitro, the latter with recombinant Mod5 on cell-derived cy-tRNATrp and synthetic ASL substrates [43]. By contrast, S. pombe Tit1, which has an extended anticodon recognition loop relative to Mod5, readily modifies S. pombe cy-tRNATrpCCA and the same ASL substrates [43]. Hence, Mod5 is denoted as a restrictive, and Tit1 as a nonrestrictive type eukaryotic IPTase. It is notable that while A37 of cy-tRNATrpCCA-A37-A38 is unmodified in S. cerevisiae, the A37 of mt-tRNATrpUCA-A37-A38 was reported as modified with i6A [50, see 51, Table 1, below]. Accordingly, the S. cerevisiae tRNA-i6A37 identity profile is defined in part by the A36-A37-A38 motif and in part by a distinct characteristic of its IPTase. Yet, the significance of the cy-tRNATrpCCA exception in which the AC is a negative determinant in the presence of A36-A37-A38 was enigmatic and deserved further consideration.
Thus, it might appear that tRNA-i6A37 variation can be accounted for by a simple two component model, i) direct IPTase recognition of A36-A37-A38, ii) A37G substitution in tRNAs that decode UNN codons, and an exception, the unmodified A37 of cy-tRNATrpCCA-A37-A38 in S. cerevisiae. The exception is intriguing because its proposed mechanism involves the AC recognition loop of Mod5, mutagenesis of which altered the hierarchical substrate preference for tRNASer, tRNATyr, and tRNACys [43]. By this model, cy-tRNATrpCCA would be a very poor competitive substrate for the restrictive Mod5. However, this model also raises the possibility that paucity of cy-tRNA-i6A37 in higher eukaryotes reflects an IPTase type hyper-restriction that expanded to additional cy-tRNA isoacceptors. In this case, ASLs with A36-A37-A38 that could not have been modified may have undergone A37G substitution during evolution (Table 1). For example, TRIT1 might be restrictive against human cy-tRNATyrGUA-G37-A38 even if it had A37 because it bears other ASL determinants that negatively impact activity. In such a model, TRIT1 would modify the human mt-tRNAs-i6A37 because they do not bear negative determinants. An attempt to experimentally test a prediction of this model has not been performed. In addition, a restrictive vs. nonrestrictive type analysis had not been extended to a higher eukaryotic IPTase.
Here, we first establish that TRIT1 uses an N-terminal MTS in human cells, and that its function is required for modification of mt-tRNAs, but not cy-tRNAs, in S. pombe and S. cerevisiae. In addition, new phylogenetic and experimental analyses advance our understanding of tRNA-i6A37 identity determinants in eukaryotes. Presence of G37 rather than A in the tRNAs for UNN codons is the major determinant of the progressive paucity of cy-tRNAs-i6A37 in eukaryotes including human. A role for IPTases independent of A36-A37-A38 appears influential but limited to substrate-specific restriction against tRNATrpCCA-A37-A38. We show that the generally nonrestrictive Mod5 and TRIT1 are restrictive against tRNATrpCCA-A37-A38, largely dependent on the source of the tRNA, S. cerevisiae or S. pombe which are different in their ACLs only at position 32. Additional data support that C32 is part of a tRNATrpCCA substrate-specific inhibitory circuit for Mod5 (and TRIT1). The collective data advance knowledge of how tRNA sequence and IPTase activities contribute to cy- and mt- tRNA-i6A37 profiles.
Results
TRIT1 has a mitochondrial targeting sequence (MTS) but is not expected to undergo cleavage
Mitochondrial targeting of human TRIT1 was predicted by the online tool MitoProt II [14] which also predicted a proteolytic cleavage site after amino acid 47 [52]. However, such cleavage would remove the conserved P-loop involved in DMAPP binding and catalysis (Fig 1, and below). We used the prediction algorithm MitoFates which was designed as an improved method for predicting cleavable N-terminal MTSs, referred to as presequences, and their cleavage sites [53]. MitoFates predicted a MTS comprised of a TOM20 (translocase of the outer mitochondrial membrane-20) receptor binding site at amino acids 5–10 (AAARAV, Fig 1) followed by an amphipathic helix at position 11–23 (PVGSGLRGLQRTL), but with the low probability of cleavage of 0.106, well below the default cutoff of 0.5. We note that the cNLS mapper [54] predicted overlapping mono- and bi-partite importin-dependent nuclear localization signals at amino acids 420–453 of TRIT1.
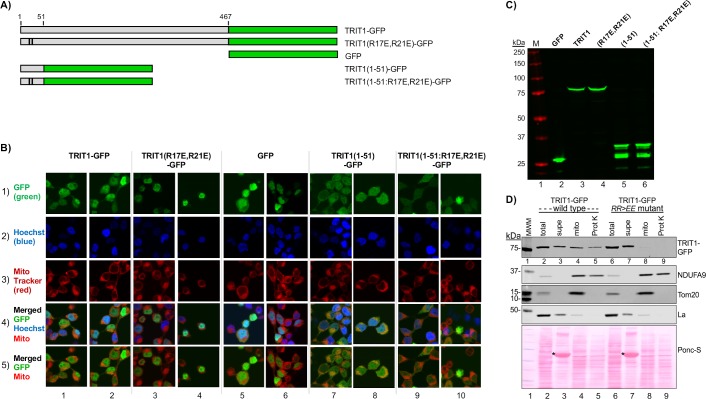
We created multiple GFP fusion constructs to test for mitochondrial localization by confocal microscopy of human embryonic kidney (HEK)-293 cells (Fig 2A and 2B). To test for MTS activity of the TRIT1 N-terminal region we fused only its first 51 amino acids to GFP, as in TRIT1(1–51)-GFP, and examined it for mitochondrial targeting. Two key point mutations, R17E, R21E, in the basic amphipathic helix component of the predicted MTS (Fig 1) were made in two contexts, the N-terminal fusion 1–51 construct, TRIT1(1–51:R17E/R21E)-GFP, and full length TRIT1(R17E/R21E)-GFP. These and other constructs were transfected into HEK293 cells. After 48 hr. the cells were exposed to MitoTracker to stain mitochondria red, fixed and treated with Hoechst to stain nuclei blue (Fig 2B). Cell lysates were examined for protein expression by western blot using anti-GFP antibody (Fig 2C). Because a significant fraction of TRIT1 localizes to nuclei, it was helpful to visualize the patterns in which the Hoechst stained nuclei were and were not included in the merged images as in rows 4 and 5 of Fig 2B respectively. Full length TRIT1-GFP was found localized to nuclei and cytosol with some evidence of mitochondrial localization reflected by overlap of GFP and MitoTracker as a stippling yellow-orange pattern in the merged panels (Fig 2B, columns 1 and 2, lower two panels). This pattern was not observed with full length TRIT1(R17E/R21E)-GFP containing the MTS mutations, which instead accumulated mostly in nuclei and was not found to localize to mitochondria (Fig 2B, columns 3 and 4). Even the least bright TRIT1-GFP expressing cells in the lower left corner of column 1, row 1 show orange stippling in column 1 row 5, whereas cells of comparable GFP brightness in column 3, row 1, show more red stippling in column 3 row 5.
Fig 2. Human TRIT1 contains an N-terminal mitochondrial targeting sequence (MTS).
A) Cartoon of the GFP-fusion constructs. Numbering represents amino acids of TRIT1; black vertical lines indicate positions of two mutated residues in the R17E/R21E constructs. B) Representative confocal microscopic images from HEK293 cells transfected 48 hours prior with the constructs in A and stained with Hoechst and MitoTracker. C) Western blot from cells transfected with constructs in A) using anti-GFP antibody (Ab). D) Western blot subcellular fractionation analysis. Transfected HEK293 cells were fractionated 48-hour later into supe (cytosol), mitochondria (mito) and mitochondria treated with Proteinase K (ProtK), the latter to digest proteins on the outer membrane of the mitochondria. GFP-TRIT1 WT and RR>EE mutant was detected using anti-GFP Ab. NDUFA9 is a mitochondrial inner membrane protein, La is a nuclear and cytosolic protein, TOMM20 is on the mitochondrial outer membrane. Total protein staining with Ponceau S is shown in the lower panel. Asterisks indicate BSA that was added during the mitochondrial isolation protocol (see text).
Additional confocal evidence of mitochondrial localization by the TRIT1 MTS came from the TRIT1(1–51)-GFP construct. GFP alone produced a diffuse pattern with nuclei generally more intense than cytoplasm (columns 5 and 6). By contrast, TRIT1(1–51)-GFP (columns 7 and 8) was generally more intense in the cytoplasm and with distinct punctate foci that colocalized with MitoTracker in the transfected cells. Importantly, colocalized punctate foci were not observed with the MTS mutant, TRIT1(1–51:R17E/R21E)-GFP (Fig 2B, columns 9 and 10).
Mitochondrial fractionation shows that the R17E, R21E mutations debilitate the TRIT1 MTS
We next performed subcellular fractionation on HEK293 cells after transfection of full length TRIT1-GFP and MTS mutant, TRIT1(R17E/R21E)-GFP. Western blots of total cell extracts, protein contents of their mitochondrial subfractions and supernatants, as well as of mitochondria after proteinase K-treatment, are shown in Fig 2D. TRIT1-GFP was detectable in total cell extract and supernatant (lanes 2 & 3), as well as in the contents of the mitochondrial and the proteinase K-treated mitochondria fractions (lanes 4 & 5), consistent with TRIT1 presence in the mitochondrial matrix, whereas TRIT1(R17E/R21E)-GFP was relatively lacking in the mitochondrial and proteinase K-treated mitochondria samples (lanes 8 & 9). This is consistent with localization of TRIT1 to nuclei and cytoplasm including in the mitochondrial matrix where its mt-tRNA substrates reside as previously determined by subcellular fractionation [14]. Additional analysis of the same fractions revealed the expected results for NDUFA9, an inner mitochondrial membrane protein, evident by its comparable detection in the mitochondrial and proteinase K treated mitochondria fractions in both wild-type TRIT1 and the MTS (R17E/R21E) (RR>EE) mutant TRIT1 extracts (NDUFA9, lanes 4 & 5, and 8 & 9). Likewise, the expected fractionation was also observed for Tom20, a mitochondrial outer membrane protein, in both sample sets, in the mitochondrial (Tom20, lanes 4 & 8) but not the proteinase K treated mitochondria fractions (lanes 5 & 9), and for La, an abundant soluble nuclear and cytoplasmic protein (Fig 2D, as indicated). We note that the slight mobility differences observed for the TRIT1-GFP bands in lanes 3 and 7 and their neighboring lanes is likely due to perturbation from BSA added to those samples as per the protocol (asterisks in Fig 2D bottom panel).
Our collective data support the idea that TRIT1 contains multiple trafficking elements that distribute it to nuclei, cytoplasm and inner mitochondrial matrix, similar to Mod5 [34–36, 38, 55, 56]. The MTS R17E/R21E mutations shifts distribution away from mitochondria, mostly to nuclei (Fig 2B). Importantly, as will be shown below, the MTS mutations impair i6A37 modification of mt-tRNAs specifically, without impairing modification of the nuclear-encoded cy-tRNAs by TRIT1.
TRIT1 MTS-dependent i6A37 modification of mt-tRNAs in S. pombe
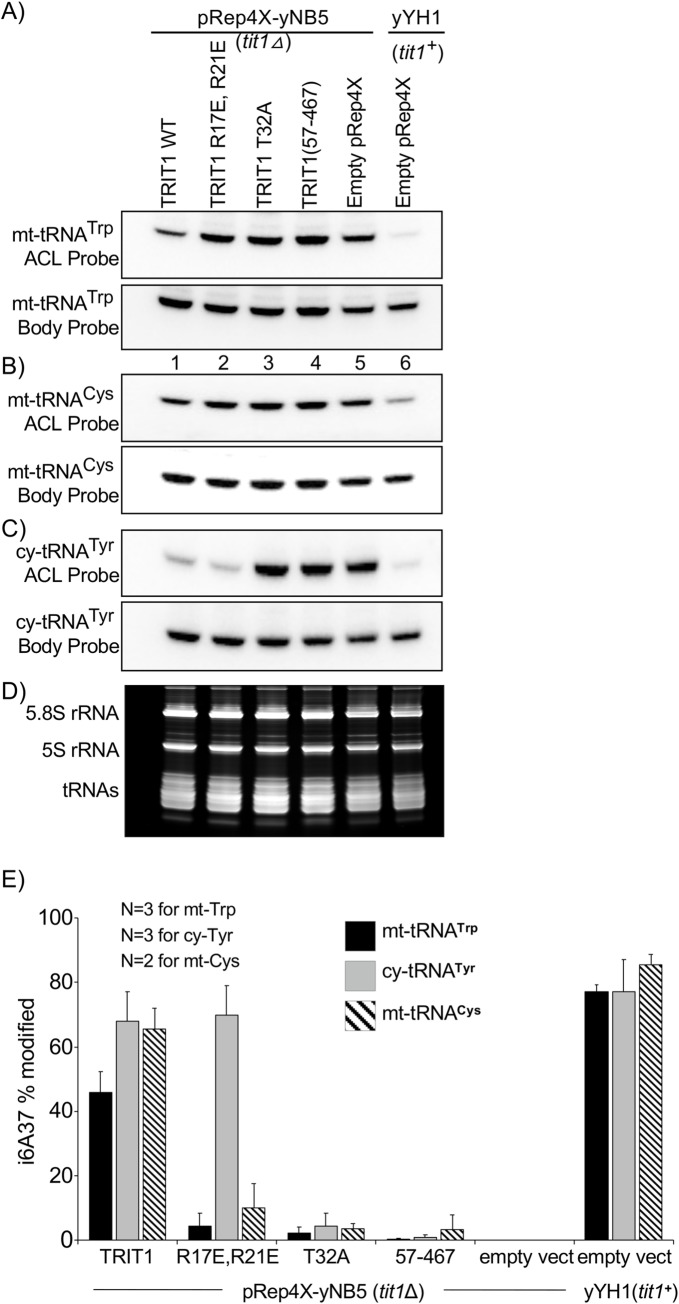
Mitochondrial protein import pathways have been functionally conserved in eukaryotes including mammals and yeasts [57, 58]; S. cerevisiae, S. pombe, D. melanogaster and H. Sapiens share Tom20, five other TOM homologs, and twelve of the thirteen TIM (translocase of the inner mitochondrial membrane) system components [58]. Analysis of S. pombe mt-DNA by tRNAscan-SE predicts 25 mt-tRNAs (including 3 for Met), of which two, mt-tRNATrpCCA and mt-tRNACysGCA contain A36-A37-A38. We examined the R17E/R21E point mutations that impaired mitochondrial localization in human cells for effects on mt-tRNA and cy-tRNA modification in S. pombe (Fig 3A–3E). For this, we used the unaltered nmt1+ promoter in the pRep4X expression vector as was done previously to examine mt-tRNATrpCCA modification by Mod5 in S. pombe [6]. MOD5 uses alternative translation start sites, M1 to target the MTS-containing protein to mitochondria and cytoplasm, and M12 which produces the MTS-lacking protein that accumulates in S. cerevisiae cytoplasm and nuclei [34, 35]. Fig 3A shows results of an i6A37-sensitive PHA6 blot assay for mt-tRNATrpCCA, mt-tRNACysGCA and cy-tRNATyrGUA. The PHA6 (Positive hybridization in Absence of i6A37) assay is based on loss of oligo-DNA probe hybridization to the ACL due to presence of i6A37, and is calibrated for quantitation by a body probe to the T stem loop on the same tRNA (Fig 3A upper and lower panels) [6, 43, 59, 60]. Here, pRep4X-TRIT1 led to ~45% i6A37 modification of mt-tRNATrpCCA and ~65% modification of mt-tRNACysGCA (Fig 3A, 3B & 3E). By contrast, TRIT1(R17E/R21E) led to ~6% i6A37 modification of mt-tRNATrpCCA and ~10% modification of mt-tRNACysGCA, a 7.5 and 6.5 fold reduction for these mt-tRNAs respectively, but showed high modification (~70%) of cy-tRNATyrGUA in the same cells, no reduction compared to TRIT1 (Fig 3A, 3C and 3E). Thus, the R17E/R21E mutations that impaired TRIT1 MTS-mediated mitochondrial localization in HEK293 cells, also impaired modification of mt-tRNATrpCCA and mt-tRNACysGCA in S. pombe, while they had no effect on modification of cy-tRNATyrGUA (Fig 3C and 3E), the latter of which is evidence that the R17E/R21E mutations do not impair enzymatic activity. Furthermore, as the mtDNA-encoded tRNAs reside in the mitochondrial matrix and are modified in a TRIT1 MTS-dependent manner, these data provide evidence of functional localization of the TRIT1 enzyme to the mitochondrial matrix of S. pombe cells.
Fig 3. TRIT1 MTS-dependent modification of mt-tRNAs in S. pombe.
A-C) The PHA6 northern blot assay (Positive Hybridization in Absence of i6A37) uses i6A37-sensitive ACL probes. This assay is based on loss of oligo-DNA probe hybridization to the ACL probe due to presence of the bulky i6A modification, and is calibrated for quantitation by a body probe to the T stem loop on the same tRNA. The panels show sequential probing (after stripping, Methods) for mt-tRNATrpCCA, mt-tRNACysGCA, and cy-tRNATyrGUA with ACL and body probes as indicated; lanes are numbered below panels A. Samples in lanes 1–4 are from tit1△ cells (yNB5) transformed with IPTases indicated above the lanes or empty vector (lane 5); lane 6 is from positive control, tit1+ (yYH1) cells. D) shows the small RNA region of the gel after staining with EtBr. E) Quantitation of % i6A37 modification of the tRNAs in A-C from biological duplicate, or triplicate PHA6 blots as indicated; error bars indicate standard deviation (SD).
Substitution of invariant Thr-12 in the P-loop of Tit1 inactivates it for cy-tRNASerUCA TMS [43]. Tit1 T12A is homologous to T19A of E. coli IPTase which resulted in the largest decrease in kcat (600-fold) of all mutations tested [47]. Indeed, the homologous T23 in Mod5 is positioned for catalysis [24]. We show here that as expected, the corresponding TRIT1-T32A mutant was inactive on both the mt- and cy- tRNA substrates (Fig 3A–3E). We also examined a variant isoform, TRIT1(57–467) [61], missing the P-loop entirely as well as D55 (see Fig 1), which in Mod5 acts as a general base for catalysis [24]. This variant represents an isoform from which translation was proposed to start at the second methionine, M57, and could complement cy-tRNATyrUUA TMS in S. cerevisiae [61]. Our TRIT1(57–467) construct was inactive on the mt-tRNA and cy-tRNA substrates (Fig 3A–3E).
TRIT1 MTS-dependent i6A37 modification of mt-tRNAs in S. cerevisiae
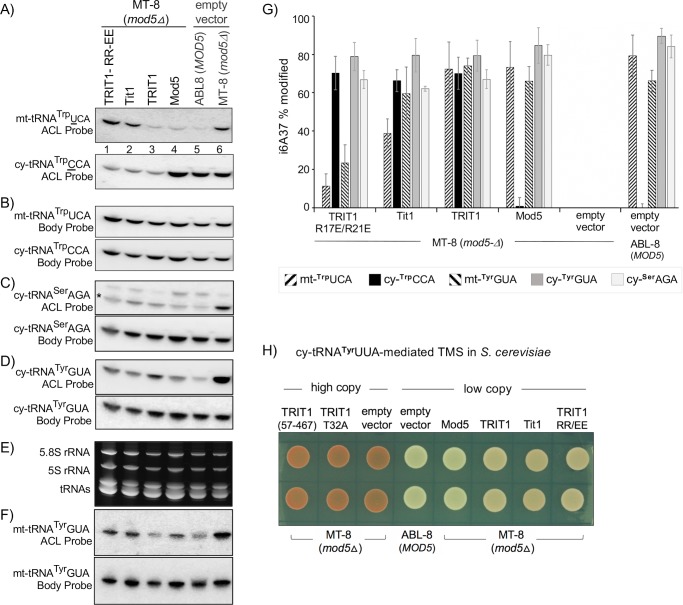
We examined TRIT1 and other IPTases in the mod5Δ, S. cerevisiae strain, MT-8 [34]; ABL8 is a MOD5 replete strain. PHA6 blot analysis for multiple tRNAs are shown in Fig 4, including quantifications in replicate. Fig 4A shows the ACL probings for mt-tRNATrpUCA and cy-tRNATrpCCA, with the body probings for these tRNAs in Fig 4B. Results for cy-tRNASerAGA, cy-tRNATyrGUA and mt-tRNATyrGUA are shown in Fig 4C–4G. TRIT1 modified the three cy-tRNAs and both mt-tRNAs in S. cerevisiae whereas TRIT1-R17E/R21E was deficient only for the mt-tRNAs. TRIT1-R17E/R21E-mediated modification of cy-tRNAs but not mt-tRNAs supports the conclusion that the TRIT1 MTS is functional for delivery of TRIT1 IPTase activity to the inner mitochondrial matrix in S. cerevisiae.
Fig 4. TRIT1 MTS-dependent modification of mt-tRNAs in S. cerevisiae.
A) A PHA6 i6A37-sensitive northern blot after transformation of the strains indicated above the lanes, showing sequential probing for mt-tRNATrpUCA and cy-tRNATrpCCA with the respective ACL-specific probes. MT-8 is mod5Δ and ABL-8 is a MOD5 positive control. B) The same blot showing reprobing (after stripping, Methods) with the body probes for mt-tRNATrpUCA and cy-tRNATrpCCA as indicated. C) The same blot showing ACL and body probings for cy-tRNASerAGA; asterisk indicates mature tRNA species, upper band is likely a cy-tRNASerAGA gene-specific precursor. D) The same blot showing ACL and body probings for cy-tRNATyrGUA with ACL and body probes as indicated. E) The small RNA region of the gel after staining with EtBr. F) Shows the ACL and body probe results for mt-tRNATyrGUA from a different blot. G) Quantitation of % i6A37 modification of the tRNAs in A-D, F from biological duplicate, or triplicate PHA6 blots as indicated; error bars indicate standard deviation (SD). H) Cy-tRNATyrUUA-mediated TMS in S. cerevisiae MT-8 (mod5Δ) transformed with empty vector or the IPTases indicated.
Three additional points are relevant. First, substrate-specific restriction against cy-tRNATrpCCA-A37-A38 by Mod5 relative to the efficient modification of the mt-tRNATrpUCA-A37-A38, mt-tRNATyrGUA-A37-A38, cy-tRNASerAGA, and cy-tRNATyrGUA, was demonstrated for the endogenous and ectopic Mod5 enzyme (compare both panels of Fig 4A, and upper panels of C, D, F, and panel G). Second, TRIT1 modified S. cerevisiae cy-tRNATrpCCA-A37-A38 with efficiency similar to Tit1 and with efficiency similar to Mod5 for the other tRNAs (Fig 4G). Third, Tit1 showed somewhat low activity on the mt-tRNAs (Fig 4G). We suspect this may reflect a mitochondrial import mechanism that isn't fully efficient for Tit1 in S. cerevisiae, consistent with the alignment in Fig 1 although other explanations are possible.
We also examined the IPTases for cy-tRNATyrUUA-mediated TMS in S. cerevisiae which is dependent on efficient i6A37 modification. This revealed that while TRIT1, Tit1 and TRT1-R17E/R21E (TRIT1-RR/EE) were active, TRIT1-T32A and TRIT1(57–467) were inactive (Fig 4H). We note that for TMS, Mod5, Tit1, TRIT1 and TRIT1-TRT1-R17E/R21E were expressed from a low copy plasmid whereas TRT1-T32A and TRT1(57–467) were from a high copy plasmid (Fig 4H).
Complementation of cy-tRNASerUCA-mediated suppression in S. pombe
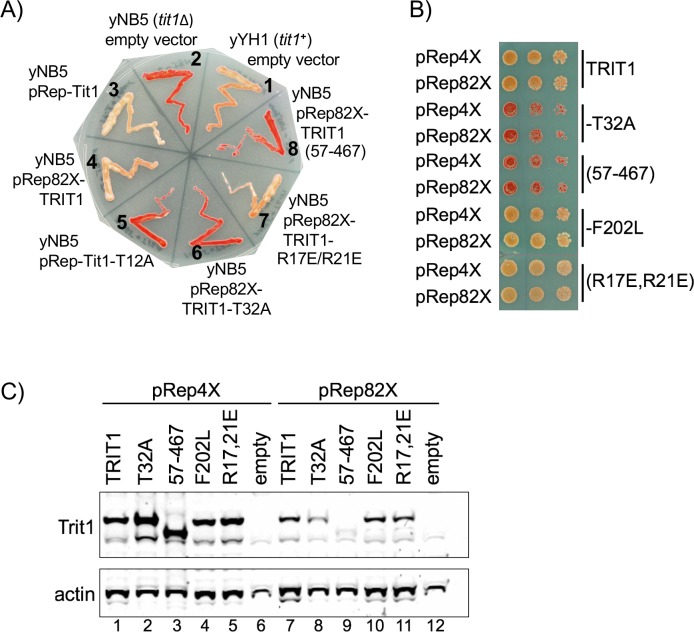
The monitoring of codon-specific insertion of an amino acid in the β-galactosidase active site was used to show that i6A37 increases tRNA decoding activity of the two different tRNAs tested by 3 to 4-fold in S. pombe, consistent with i6A37 leading to general increased tRNA affinity for ribosome decoding [60]. TMS makes use of a codon-specific reporter to examine TRIT1 mutants for modification activity of cy-tRNASerUCA in S. pombe deleted of tit1+ (yNB5 tit1Δ, Fig 5A). Cy-tRNASerUCA is dependent on efficient i6A37 modification for serine insertion at codon 215 of ade6-704 mRNA [62] with consequent decrease in cellular red pigment during growth in limiting adenine [33]. The pathogenic TRIT1-R323Q variant exhibits substantially reduced modification activity relative to TRIT wild type in the S. pombe TMS assay using the pRep82X expression vector [14]. In this assay, the positive control strain yYH1 (tit1+) is pink (moderately suppressed) whereas yNB5(tit1Δ) transformed with empty vector is unsuppressed, red (Fig 5A, sectors 1, 2). Transformation of yNB5 with pRep-Tit1 under control of the tit1+ promoter, and pRep82X-TRIT1, led to suppression (sectors 3, 4). The catalytic mutants, pRep-Tit1-T12A and pRep82X-TRIT1-T32A, were inactive, as was pRep82X-TRIT1(57–467) (sectors 5, 6, 8), consistent with the PHA6 results in Fig 3.
Fig 5. Determinants of TRIT1 critical for cy-tRNASerUCA-mediated TMS in S. pombe.
A) Cy-tRNASerUCA-mediated TMS assay in S. pombe yNB5 (tit1Δ) transformed with the empty plasmid pRep82X (sector 2) or plasmids expressing the proteins indicated (sectors 3–8), and the tit1+ control strain, yYH1 (sector 1). B) TMS assay as in A but displayed as dilution spot assay, for plasmids expressing TRIT1 and variants from pRep82X and from the stronger nmt1+ promoter (pRep4X) as indicated on the left. C) Western blot of extracts of cells in B detected with anti-TRIT1 (top panel), and anti-actin antibody; lanes numbers are indicated below actin panel.
Importantly, pRep82X-TRIT1(R17E/R21E) was active for TMS (Fig 5A, sector 7) comparable to pRep82X-TRIT1. This provided additional evidence that the R17E/R21E mutations that impair mitochondrial targeting do not impair enzymatic functional activity for the cy-tRNA.
Moderate over-expression does not rescue inactive TRIT1 alleles for cy-tRNASerUCA TMS
Human TRIT1 was identified as a candidate tumor suppressor [61]. An allele that encodes TRIT1-F202L was later found to be associated with modulation of lung cancer survival [63] (also see [64], [65]) and was reported to be active for cy-tRNATyrUAA-TMS in S. cerevisiae using a high level expression plasmid system [61]. Therefore, we wanted to examine TMS using both a weakened and a stronger promoter. The pRep4X promoter fused to chloramphenicol acetyltransferase (CAT) or LacZ, showed 45–75 fold more output than pRep82X, the latter of which was created by mutating the TATA box of the natural nmt1+ promoter [66, 67]. pRep4X-TRIT1(57–467) indeed produced more TRIT1(57–467) than did pRep82X-TRIT1(57–467) (Fig 5C), although without a hint of increased TMS activity relative to pRep82X-TRIT1(57–467) (Fig 5B). TRIT1-T32A also remained inactive in pRep4X (Fig 5B and 5C). TRIT1-F202L was as active as TRIT1 and not limited by expression levels, and this was also true for TRIT1-(R17E/R21E) (Fig 5B and 5C).
TRIT1 exhibits substrate-specific restriction against modification of S. pombe cy-tRNATrpCCA
We note that while comparison of different IPTases in a heterologous 'in vivo' system is not physiological, it provides an experimental model with which to gauge activity of an ectopic IPTase for a particular tRNA relative to other tRNA substrates in the cells and compare the overall tRNA substrate activity pattern to that of a different ectopic IPTase in the same system. We did that for Fig 3 by using S. cerevisiae as the heterologous system and in Fig 6 using S. pombe as the heterologous system. Unexpectedly, the cy-tRNATrpCCA-A37-A38 in S. pombe was a very poor substrate of TRIT1 as compared to cy-tRNASerAGA and cy-tRNATyrGUA, and Tit1 (Fig 6). Unexpected because TRIT1 efficiently modified the S. cerevisiae cy-tRNATrpCCA-A37-A38 in S. cerevisiae. Similarly, it was unexpected that Mod5 would modify the S. pombe cy-tRNATrpCCA-A37-A38 to as high as 60% (Fig 6E) based on its restriction against S. cerevisiae cy-tRNATrpCCA-A37-A38.
Fig 6. TRIT1 exhibits substrate-specific restriction against cy-tRNATrpCCA in S. pombe.
A) Schematics showing constructs used for analysis of tRNA i6A37 modification in S. pombe. Left: amino acid sequence representation of N-termini only, similar to Fig 1; the invariant Thr demarcated by yellow rectangle. The second, alternative translation initiation sites, where present are shown in red. Right: translation initiation contexts of the various constructs used. Lines 1, 2 and 8 represent translation start site consensuses as indicated [101–103], the former of which was derived for S. cerevisiae but applies to other yeasts including S. pombe, at the -3 position. B) The i6A37-sensitive PHA6 northern blot assay for three tRNAs from S. pombe expressing IPTases from pRep82X plasmids indicated above the lanes. The blot was sequentially probed with the i6A37-sensitive ACL probes to the cy-tRNAs indicated to the left of the panels. C) The body probes for each of the tRNAs in B. D) The same blot as in B-C, probed for mt-tRNATrpCCA using the ACL and body probes indicated. E) Quantitation of % i6A37 modification of the cy-tRNAs from biological replicate PHA6 northern blots as in panels B-C; n = 5 for cy-tRNATrpCCA and all others except n = 1 for cy-tRNATyr in Mod5 and Mod5M1A. F) Quantitation; % i6A37 modification of mt-tRNATrpCCA from biological duplicate PHA6 northern blots; error bars indicate standard deviation (SD).
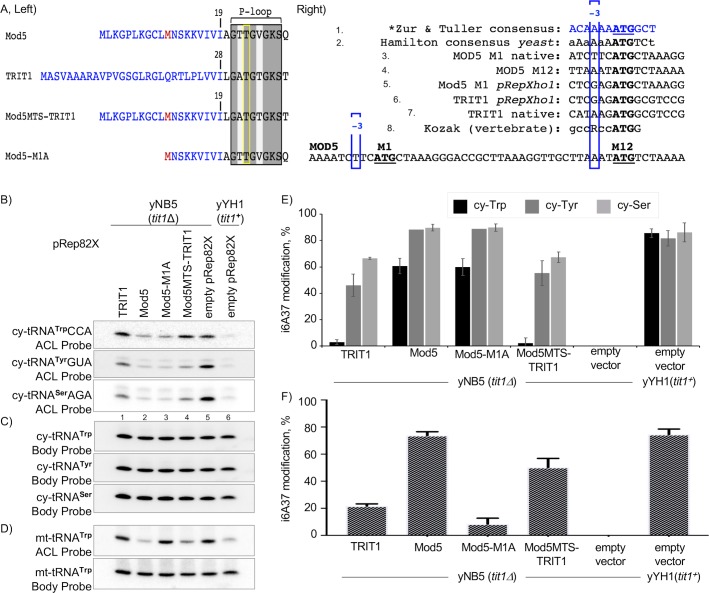
Effects of increasing TRIT1 levels on tRNA modification were also examined. Two additional IPTase constructs were included in this analysis, Mod5-M1A and Mod5MTS-TRIT1, the latter of which was created by replacing the first twenty-eight codons of TRIT1 with the first nineteen codons of MOD5 which includes sequences in its MTS and the second translation initiation site at M12, as schematized in the left and right panels of Fig 6A. In addition to usefulness for assessing mt-tRNA modification, the MOD5 leader sequence also produces increased levels of cytoplasmic isoforms of the IPTases because of the known high favorability of the translation initiation context of the second methionine, M12 relative to MOD5 M1 [68] and TRIT1 M1 (Fig 6A, S1A–S1D Fig). Mod5 and Mod5-M1A were only modestly deficient for cy-tRNATrpCCA modification, whereas TRIT1 and Mod5MTS-TRIT1 were severely deficient for cy-tRNATrpCCA modification relative to cy-tRNAs TyrGUA and SerAGA (Fig 6B, 6C & 6E). Hypomodification of cy-tRNATrpCCA is reflected by strong hybridization signals in lanes 1 and 4 relative to lanes 2 and 6 of the PHA6 ACL probe blots in Fig 6B, upper panel, and by the biological replicate data in Fig 6E. The data indicate that while Mod5 sees the S. pombe cy-tRNATrpCCA as the least preferred of the cy-tRNA substrates, this is modest compared to the more severe substrate-specific restriction against this tRNA by TRIT1. The cy-tRNATrpCCA is efficiently modified by Tit1 consistent with previous data [43].
We confirmed that Mod5-M1A, which initiates at M12 and therefore lacks a MTS [34, 35], is deficient for mt-tRNATrpCCA modification [6] (Fig 6D and 6F). Notably, pRep82X-TRIT1 reproducibly modified mt-tRNATrpCCA to 20% in the same cells in which it modified cy-tRNATrpCCA to only 5% (Fig 6E). Mod5MTS-TRIT1 exhibited 50% modification of mt-tRNATrpCCA, ~2.5-fold higher than TRIT1 (Fig 6F). Analysis of protein expression levels indicate that levels of cytoplasmic isoform of TRIT1 accumulate to >10-fold higher from pRep82X-Mod5MTS-TRIT1 relative to pRep82X-TRIT1 (S1C and S1D Fig). However, this had little if any effect on the modification efficiency of S. pombe cy-tRNATrpCCA (Fig 6E). S. pombe cy-tRNATrpCCA modification can be increased by increasing TRIT1 protein levels, but this requires even higher expression, from pRep4X-Mod5MTS-TRIT1 (S1G and S1H Fig).
Position 32 influences tRNATrpCCA substrate-specific restriction by Mod5 and TRIT1 IPTases
Our data indicate that Mod5 and TRIT1 exhibit differential restriction against the cy-tRNAsTrpCCA of S. cerevisiae and S. pombe which differ at C32 vs. U32 as well as other positions more distant to their ACLs (S2 Fig). Given the modification circuit relationship between position 32 and i6A37 among tRNAsSer [12, 13] and other potential effects of position 32 on ACL structure (and function [69, 70]), we suspected position 32 as a possible determinant of the differential activities of TRIT1 and Mod5 on the S. cerevisiae and S. pombe cy-tRNAsTrpCCA.
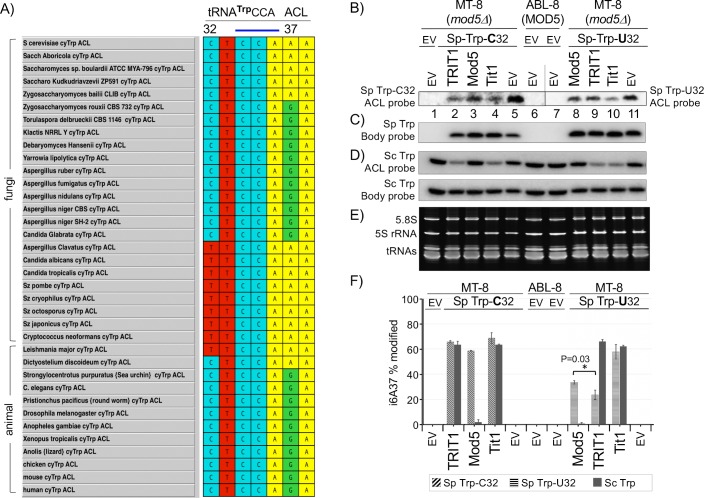
To attempt to gain broader perspective we collected tRNA gene sequences encoding the ACLs of cy-tRNAsTrpCCA in representative related yeasts as well as animal cells available in an appropriate database [71] and subjected them to multiple sequence alignment (Fig 7A). Three patterns emerged. The related Saccharomyces and Zygoaccharyomyces bailii species, at the top of the alignment, revealed a C32-A37 pattern different from the other fungi. The remaining nineteen fungal species exhibited two different covariations; eight with T32-G37 and eleven with C32-A37 (Fig 7A). The C32-G37 pattern was predominant in the animal cell species, with the exceptions being the primitive Leishmania and Dictyostelium. For comparison, ACL alignments of cy-tRNACysGCA was also done which revealed a less clear modal 32–37 covariation among the fungi than for cy-tRNAsTrpCCA (S3 Fig). For all fungal cy-tRNAsTyrGUA examined, the C32-A37 pattern was set, whereas the C32-G37 pattern was set for all eukaryotes examined (S3 Fig). The data are consistent with the idea that potential for i6A37 modification of cy-tRNAsTrpCCA was relatively variable in yeasts due to A37 vs. G37 identity and became progressively less variable and then fixed by G37 identity in higher eukaryotes.
Fig 7. Position 32 as a determinant of differential IPTase activity for cy-tRNATrpCCA in eukaryotes.
A) Sequence alignment of the cy-tRNATrpCCA ACLs in the eukaryotes indicated; the 32 and 37 positions are numbered above; a horizontal blue bar indicates the AC. B) Results of PHA6 i6A37-sensitive PHA6 northern blot after expressing a S. pombe cy-tRNATrpCCA gene in MT-8 (mod5Δ) S. cerevisiae containing the native U32 (right panel) or this position mutated to C32 (left panel), or empty vector (EV) as indicated above the lanes, in the presence the IPTases or empty vector (EV) also indicated above the lanes; ABL-8 is a MOD5 positive strain. The right and left panel blots show separate hybridizations with ACL probes. Note that the order of the IPTases in the left and right panels differs, and carries through to panel F. C) The body probe result for S. pombe cy-tRNATrpCCA. D) ACL probe (upper) and body probe (lower) results for endogenous S. cerevisiae cy-tRNATrpCCA as indicated. E) shows the EtBr-stained gel from which the blot was made. F) Quantification of % i6A37 modification of the three different cy-tRNAsTrpCCA as indicated; error bars indicate standard deviation (SD) from biological duplicates. P value was obtained by the paired t-test.
The observed differences in relative modification of S. cerevisiae and S. pombe cy-tRNAsTrpCCA by TRIT1 and Mod5 in the two yeast systems might be due to differences in the tRNA sequences themselves, posttranslational modifications of the IPTases, or subcellular distributions. Considering the wide range of functional transferability of IPTases from multiple distant phylogenetic groups to another (Introduction), we suspected that a basis of the modification differences may be in the cy-tRNAsTrpCCA themselves. We set out to look into this by examining IPTase activity for the S. pombe cy-tRNATrpCCA carrying its native encoded U32 or with this position mutated to C32, expressed in mod5Δ MT-8 S. cerevisiae. We chose this approach because S. pombe tRNA genes can be readily expressed in S. cerevisiae, but not vice versa [72], and because this tRNA can be distinguished from the S. cerevisiae cy-tRNATrpCCA [43]. A PHA6 ACL probed blot for the S. pombe cy-tRNATrpCCA with U32 and C32 is shown in the Right and Left panels of Fig 7B, and the body probe blot is shown in Fig 7C. The ACL probe for the endogenous S. cerevisiae cy-tRNATrpCCA is shown in Fig 7D, upper panel, with the body probe in the lower panel. Fig 7E shows the lower part of the gel from which the blot was made. Panel F shows quantitation of the PHA6 data for all three tRNAs. The PHA6 analysis of the S. pombe cy-tRNATrpCCA with U32 and C32 was consistent with Figs 4 and 6 in so far as TRIT1 was less active than Mod5 on the cy-tRNATrpCCA with its native U32 and more active than Mod5 on cy-tRNATrpCCA substituted with C32 (Fig 7B and 7C) which better matches the S. cerevisiae cy-tRNATrpCCA ACL. The data showed that substrate-specific restriction by TRIT1 against S. pombe cy-tRNATrpCCA could be demonstrated in S. cerevisiae, while maintaining efficient modification of endogenous S. cerevisiae cy-tRNATrpCCA (Fig 7F), with a trend of general relative activities similar to observed for TRIT1 in the two yeasts. Moreover, the single point mutation, U32C, converted S. pombe cy-tRNATrpCCA from a poor to better substrate of TRIT1, while Tit1 modified both substrates to comparable efficiencies (Fig 7B and 7F).
However, this experiment also produced unexpected results that raised a new hypothesis. Mod5 was more active on the C32 tRNATrp than expected, which bears an encoded ACL that matchs that of the endogenous S. cerevisiae cy-tRNATrpCCA, while it remained unable to modify the latter (Fig 7D and 7F). This suggests that another difference(s) in the cy-tRNAsTrpCCA of the two yeasts may be responsible, for example, G26 vs. U26 respectively (see S2 Fig).
Evidence against a hyper-restriction model for TRIT1
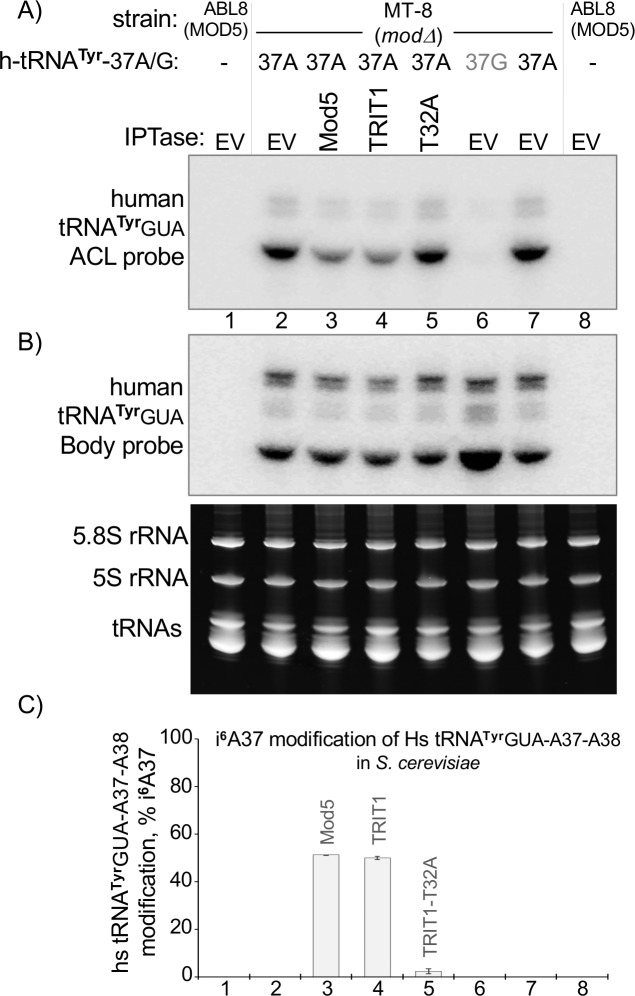
Finally, we designed an experiment to address if a hypothetical hyper-restriction IPTase model may apply to TRIT1 or if the alternative simple model of A36-A37-A38 as the principal positive determinant of activity applies. As noted in the Introduction, the hyper-restriction model would predict that TRIT1 would not modify human cy-tRNATyrGUA-G37-A38 even if it had A37 because other negative determinants would prevail. Whereas in the simple model, cy-tRNATyrGUA-A37-A38 would be a ready substrate of TRIT1. We therefore made a point mutation, G37A, in a human cy-tRNATyrGUA gene and examined it in MT-8 cells. This converted the tRNATyrGUA into a substrate that was modified with comparable efficiency by both enzymes (Fig 8). This was as expected of the simple model of A36-A37-A38 recognition in substrates that do not have a YYA anticodon.
Fig 8. Evidence against a hyper-restriction model for TRIT1.
A) Results of PHA6 i6A37-sensitive PHA6 northern blot assay after expressing a human nuclear gene for tRNATyrGUA-G37-A38 with a G37A point mutant in mod5Δ MT-8 S. cerevisiae cells (lanes 2–5, 7) and with native G37 (lane 6), in the presence of empty vector (EV) or the IPTases indicated above the lanes (T32A = TRIT1-T32A); lanes 1 and 8 show ABL-8, the MOD5 positive strain. B) Body probe of the blot in A (upper panel), and EtBr stained gel (lower panel). C) Quantitation of % i6A37 modification of the cy-tRNATyrGUA from biological duplicate PHA6 northern blots as in panels A-B; error bars indicate standard deviation (SD).
Discussion
In this paper, two related topics concerning i6A37 modification of cytoplasmic and mitochondrial tRNAs were advanced. First, a newly predicted MTS in the amino-terminal sequence of TRIT1 comprised of a TOM20 receptor binding site followed by an amphipathic helix was shown to be functional by the mutation of two key basic residues in the amphipathic helix. The mutations disabled targeting of TRIT1-GFP to the inner mitochondria of human cells, and impaired TRIT1-mediated i6A37 modification of mt-tRNAs in S. pombe and S. cerevisiae without affecting i6A37 modification of cy-tRNAs in the cells. Use of these yeast in vivo systems was critical for demonstrating function of the TRIT1 MTS in mt-tRNA modification. Understanding TRIT1 mitochondrial function is important because human TRIT1-deficiency leads to i6A37-hypomodification of mt-tRNAs and consequent impaired translation of mt-mRNAs that encode respiratory complex components [14].
That TRIT1 can access and modify cy- and mt- tRNAs in two evolutionarily diverged yeasts suggests that no specialized posttranslational modification of TRIT1 is required for function, or if so, it is highly conserved [73]. A model of IPTase action of direct recognition of the tRNAs as observed in cocrystal structures is consistent with this and the ability of phylogenetically divergent species' IPTases to complement tRNA-mediated TMS in yeast (Introduction).
This study also advanced insight into determinants of tRNA-i6A37 identity variation. It should be noted that tRNA-i6A37 variation in Table 1 is in contrast to that for threonylcarbamoyl-A37 (t6A37) which occurs nearly without exception on all tRNAs for ANN codons in all species [74, 75, reviewed in 76].
Although biological relevance of tRNA-i6A37 variation is yet unknown, that i6A37 is a functionally important modification is clear. I6A37 tRNA hypomodification that results from pathogenic mutations in TRIT1 causes debilitating childhood neurodevelopmental disorders [14, 15]. Deletion of TRIT1 is embryonic lethal in mice [77] and RNAi-mediated IPTase knockdown in silkworms leads to death [28]. Studies in S. pombe support the idea that changes in i6A37 levels can affect translation of specific mRNAs rich in i6A37-cognate codons [6, 60]. Because i6A37 formation is linked to basal metabolism, it is plausible that tRNA-i6A37 levels could vary under physiologic conditions and regulate the translation of specific mRNAs [78, see 79]. As i6A37 is a unique modification that increases tRNA affinity for the ribosome and specific activity for decoding (Introduction), it may serve its translation systems distinctively and plastically [80]. Full appreciation of i6A37 function will require better understanding of individual IPTase-tRNA systems. With the R17E,R21E mutations, researchers can attempt to specifically rescue cy-tRNA i6A37 modification in patient-derived cells while maintaining mt-tRNA hypomodification to begin to dissect the potential contribution of the cy-tRNAs to the phenotype.
This study represents comparative analyses of three IPTases and multiple tRNA substrates in two heterologous 'in vivo' model organisms, S. pombe and S. cerevisiae deleted of their endogenous IPTases. Quantitative modification analysis used the PHA6 assay, i6A37-sensitive northern blot probing of more than ten individual tRNAs followed by tRNA-specific internal calibration for each tRNA species, five endogenous substrates in S. cerevisiae, four endogenous substrates in S. pombe, and multiple heterologous tRNAs. We showed for the first time, the large quantitative extent to which cy-tRNATrpCCA is a poor substrate of Mod5 (endogenous and ectopic) in S. cerevisiae, while other substrates, including mt-tRNATrpUCA, are efficiently modified in the same cells (Fig 4A–4G). IPTase activities were also examined in S. pombe (Fig 6). A conclusion is that the three divergent IPTases exhibit largely unrestrained i6A37 modification activity on a range of tRNAs-A36-A37-A38 with the exception of tRNATrpCCA which is a least favored substrate for Mod5 and TRIT1, albeit conditional on position 32, whereas Tit1 does not discriminate against this type of substrate. Thus, although restrictive type IPTase discrimination against recognition of certain anticodons does exists in some eukaryotes, this appears to have been limited to cy-tRNATrpCCA. This discriminatory feature was retained by TRIT1 even though its cy-tRNATrpCCA contains G37 and is not a substrate for i6A modification. Data in Fig 7 suggest that elements beyond the ACL of S. cerevisiae cy-tRNATrpCCA-A37-A38 contribute to the remarkable extent to which this tRNA is a specifically poor substrate of its cognate IPTase (Fig 4A and 4G).
The N-terminal MTS of TRIT1
The mechanisms by which Mod5 localizes to mitochondria, nuclei and cytoplasm in S. cerevisiae have been well investigated [34–36, 68, 81, 82]. Mod5 and GRO-1 of the round worm, C. elegans use alternative translation initiation to target to mitochondria or cytoplasm [34, 35, 44]. Our data indicate that TRIT1 uses a single translation start site to produce mitochondrial, cytoplasmic and nuclear protein. Use of alternative translation initiation start sites is attractive because it could theoretically provide opportunity for regulation of the relative amount of the isoforms. The second methionine of the conserved metazoan IPTase is M57 of human TRIT1, part of a DSMQ motif involved in A37 recognition (Fig 1). A cDNA representing TRIT1(57–467) was isolated from human lung cDNA (referred to as Tr1 in ref [61]). This variant was found to be inactive for TMS in S. pombe and S. cerevisiae when compared to full length TRIT1 (Figs 4 and 5). It should be noted that although TRIT1(57–467)/Tr1 appeared active for TMS relative to other variants Tr2-8, it was not compared to full length TRIT1 [61] as was done here. Based on data here including TRIT1-T32A, we suggest that TRIT1 variant activities should be compared to TRIT1.
Because mt-tRNA i6A37 modification is critical to TRIT1 function we wanted to examine its mitochondrial import mechanism. As noted, MitoProt II predicted post-import cleavage at position 47, yet such a protein would lack a P-loop and the invariant T32 whose homologous mutations in TRIT1, Tit1-T12A, and MiaA inactivated them. MitoFates [53] is a newer and improved algorithm that predicted a new N-terminal MTS for TRIT1 but with low probability of cleavage. Prior subcellular fractionation showed TRIT1 localizes to the mitochondrial matrix [14]. Our data confirmed and extended this by point mutagenesis of two residues of the MTS which debilitated localization in human cells as well as i6A37 modification of mt-tRNAs, but not cy-tRNAs, in S. pombe and S. cerevisiae. In summary, our data do not support a model in which translation initiation at the second methionine, M57, nor cleavage after mitochondrial import at position 47, can produce efficient tRNA i6A37 modification activity.
Interestingly, it was concluded that the N-terminal 11 amino acids of Mod5 is required as part of a longer MTS that is probably not cleaved after import [34, 35]. This is supported by MitoFates analysis that predicts positively charged amphiphilicity in the Mod5 N-terminal region but with a very low probability of cleavage, 0.026. The earlier studies also indicate that Mod5 MTS is inefficient at import, consistent with a functional cytoplasmic pool of N-terminal extended protein [34]. Our data with Mod5MTS-M12A-TRIT1 are consistent with this as it retained significant i6A37 modification of cy-tRNAs. Perhaps related, biochemical and subcellular fractionation suggest a similar feature of TRIT1 [14]. Yarham et al. could not exclude the possibility that a significant fraction of cytoplasmic TRIT1 may be associated with the outer mitochondrial membrane [14]. This is consistent with microscopy that showed a substantial amount of the total cytoplasmic TRIT1-GFP and TRIT1(1–51)-GFP appeared to be mitochondria-associated (Fig 2B).
Future experiments may address the possibility that TRIT1 and Mod5 localize on the outer mitochondrial surface. Such localization has recently been noted for other tRNA ACL modification enzymes as well as processing factors for tRNA and other RNAs in yeast and metazoa [reviewed in 2]. Specifically, the enzyme responsible for formation of cyclic t6A37 [74] localizes to S. cerevisiae mitochondrial surface [see 2]. Similar IPTase localization might augment tRNA i6A37 formation via ready access to DMAPP, a product of the HMG-CoA reductase pathway, as it exits mitochondria, and enhance the link between i6A and basal metabolism [78, see 79].
Different TRIT1 modification efficiencies for cy- vs. mt- tRNATrpCCA-A37-A38 in S. pombe
At different TRIT1 expression levels, mt-tRNATrpCCA-A37-A38 was modified more efficiently than cy-tRNATrpCCA-A37-A38 (Fig 6, S1F and S1G Fig). Although the data support that the differences are significant yet do not provide mechanistic resolution, some points can nonetheless be considered. While the encoded ACL nucleotides of these cy- and mt- tRNAsTrp are identical (S1G Fig), the difference in i6A37 efficiency might reflect differential modification at another site(s) or some other feature(s). Related to this we note that all eukaryotic cy-tRNAsTrpCCA in the database representing mammals, chicken, plants and S. cerevisiae, contain 2'-O-methyl-cytidine (Cm34) at position 34 [83]. An encoded C34 in mt-tRNAsTrp is found only in plants [83] and Schizosaccharomyces species [84]. The U34 of mt-tRNATrpUCA in metazoan and many yeasts is hypermodified to decode numerous UGA (Trp) codons in their mt-mRNAs [83, 85, 86]. It was suggested that the wobble C34 of S. pombe mt-tRNATrpCCA is modified to facilitate recognition of the UGA (Trp) codon found in mt rps3 [84]. However, the only known modified C that can decode A is lysidine, which has so far been found in tRNAsIleCUA [87]. We did not detect a difference at C34 of S. pombe cy- and mt- tRNAsTrpCCA by primer extension using low (0.01 mM) dNTPs [88].
A Mod5-type restriction against CCA anticodon recognition appears inherent to TRIT1
Biological relevance of the exceptional absence of i6A37 modification of S. cerevisiae tRNATrpCCA-A37-A38 had been enigmatic. Because human mt-tRNATrpUCA is one of several mt-tRNAs with i6A37, it might have been unexpected that TRIT1 would exhibit strong substrate-specific restriction against S. pombe tRNATrpCCA-A37-A38 (Fig 6) [43]. This is important because it is evidence that a restrictive type IPTase is not a peculiarity of Mod5. Further, it indicates that restriction toward a tRNATrpCCA ACL is an IPTase characteristic in extant higher eukaryotes even though their cy-tRNAsTrpCCA contain G37. Yet, additional findings suggest that TRIT1 exhibits a restrictive type recognition that is conditional on features of the YYA ASL that is distinct from Mod5 type recognition. This raises possibilities for future studies of potentially new modes of ASL recognition.
Insight into tRNA-i6A37 identity profiles of eukaryotes
Phylogenetic analysis of gene sequences encoding cy-tRNAs for UNN codons other than Ser in representative advanced animal species shows that G is uniformly present at position 37 as noted for Table 1. By contrast, primitive animals, Dictyostelium discoidium and Strongylocentrotus purpuratus, as well as yeasts S. pombe, S. cerevisiae, Aspergillus and Candida subspecies, have G or A at positions 37 of tRNAsTrp and tRNAsCys [71, 89], also noted for Table 1, Fig 7 and S3 Fig. These data suggest that tRNAs-i6A37 identity was relatively variable in yeasts and primitive animal cells but became progressively less variable and fixed in higher eukaryotes, by patterns predominantly set by base identity of A or G at position 37.
We set an experiment to compare the IPTases from three divergent species, on S. pombe cy-tRNATrpCCA-A37-A38 substrates that differ at one encoded nucleotide, C32 vs. U32, one of the differences between the S. cerevisiae and S. pombe cy-tRNAsTrpCCA-A37-A38. When the S. pombe cy-tRNATrpCCA with either C32 or U32 was expressed in S. cerevisiae the results were consistent with our prior experiments in which the IPTases were compared in each of the two yeasts. The substrate-specific restriction observed for TRIT1 against S. pombe cy-tRNATrpCCA -U32, was demonstrated in S. cerevisiae while this IPTase maintained efficient modification of the endogenous S. cerevisiae cy-tRNATrpCCA. Moreover, the point mutation, U32C, converted the S. pombe cy-tRNATrpCCA from a poor to a better substrate for TRIT1, while Tit1 modified both substrates to comparable efficiencies. The results also raised a new hypothesis, that another difference in the cy-tRNAsTrpCCA of the two yeasts, G26, may be responsible for the higher activity of Mod5 for the S. pombe cy-tRNATrpCCA -C32, while remaining relatively inactive for the endogenous cy-tRNATrpCCA which encodes U26 (S2 Fig, Fig 7B–7D). The hypothesis that modification of G26 to m22G26 may promote i6A37 modification of certain tRNAs, including S. pombe cy-tRNATrpCCA which carries both modifications [90] is plausible because m22G26 can influence ASL structure [91]. This is a testable hypothesis fit for future study.
We note that our findings also have additional implications. Human mt-tRNATrpUCA-A37-A38 carries U32 and U34. This and other observations suggest the possibility that the cmnm5U34 and tm5U34 modifications on S. cerevisiae and human mt-tRNAsTrpUCA [4, 83, 92] enhance activities of their substrate tRNAs for the respective IPTases. It is tempting to speculate that bulky modified uridines would also enhance other YYA AC IPTase substrates, the suppressors tRNATyrUUA, tRNASerUCA, and the tRNASer[Sec]UCA, which contain mcm5U (or hypermodified versions) [93–96].
Materials and methods
S. pombe and S. cerevisiae yeast transformations were by standard methods. All strains were published previously as noted. MT-8 (mod5Δ) lacks codons 1–250 of Mod5 and 35 bp upstream [34].
DNA constructs
The different TRIT1 mutants and wild-type for mammalian transfection were cloned in the HindIII and AgeI sites of the pEGFP-N1 vector. A linker of 5 glycine residues was introduced between the TRIT1 sequences and GFP.
All primers and oligos were obtained from Eurofins Genomics (Tables 2 and 3). For S. pombe, Tit1 and Tit1-T12A, both with a C-terminal HA tag were expressed from the tit1+ promoter in a pRep vector in which the nmt1+ promoter was excised [43]. TRIT1 and Mod5 were expressed from the nmt1+ promoters in pRep82X or 4X, cloned such that their ATG immediately followed the Xho I site. Mutants were made using site-directed mutagenesis or PCR. The Mod5MTS-M12A-TRIT1 construct was generated by site directed mutagenesis using the Q5 Site-Directed Mutagenesis kit (New England Biolabs). All constructs were sequence verified.
Table 2. Oligo-DNAs used for PHA6 assay; ACL (anticodon loop), BP (body probe).
| cy-tRNA-Tyr-GUA-BP (HS) | CGAGCCGGAATCGAACCAGCG |
| cy-tRNA-TrpCCA-A37-ACL (HS) | CCTAAGGATTTACAGTCCTCCGC |
| cy-tRNA-TrpCCA-G37-ACL (HS) | CCTAAGGATCTACAGTCCTCCGC |
| mt-TrpCCA-ACL (SC) | CCTAATGATTGATTTGAAATCAACTG |
| mt-TyrGUA-ACL (SC) | AAGTCATTGAGTTTACAGCTCAACT |
| cy-SerAGA-ACL (SC) | GCCCAAAAGATTTCTAATCTTTCG |
| cy-tRNA-Trp-T32 ACL (SP) | CTTTGAGATTTGGAATCTCACAC |
| cy-tRNA-Trp-C32 ACL (SP) | CTTTGAGATTTGGAGTCTCACAC |
| mt-tRNATrp-UCA-ACL (SC) | CCTAATGATTGATTTGAAATCAACTG |
| mt-tRNATyr-GUA-ACL (SC) | AAGTCATTGAGTTTACAGCTCAACT |
| cy-tRNATrp-CCA-ACL (SC) | CAACCCTTCGATTTGGAGTCGAA |
| cy-tRNASer-AGA-ACL (SC) | GCCCAAAAGATTTCTAATCTTTCG |
| cy-tRNATyr-GUA-ACL (SC) | GATCTCAAGATTTACAGTCTTGC |
| mt-tRNATrp-UCA-BP (SC) | TAAAGAGATTCGAACTCCTAATG |
| mt-tRNATyr-GUA-BP (SC) | GGAATAGGAATTGAACCTATGAAG |
| cy-tRNATrp-CCA-BP (SC) | GACAGGAATTGAACCTGCAAC |
| cy-tRNASer-AGA-BP (SC) | CTGCAGGACTCGAACCTGC |
| cy-tRNATyr-GUA-BP (SC) | GGCGAGTCGAACGCCCGAT |
Table 3. PCR Primers used in this study.
| TRIT1F202L_Fwd | TCATAGTGAAcTTCTCCATCG |
| TRIT1F202L_Rev | GAGATTCCTGTTTCTTCAAAAAC |
| Mod5MTS-TRIT1_M12A_F | TTGCTTAAATgctTCTAAAAAAGTTATAGTGATTCTCGGG |
| Mod5MTS-TRIT1_M12A_R | CCTTTAAGCGGTCCCTTTAG |
| TRIT1_29-467F | CTCGGGGCCACGGGCAC |
| TRIT1_Xma1R | ATTACCCGGGTTAAACGCTGCATTTCAGCTC |
| TRIT1_Rev | ATATGGATCCTTAAACGCTGCATTTCAGCTCTTG |
| TRIT1_Fwd | ATTACTCGAGATGGCGTCCGTGGCGGCT |
| Xma1_Tit1_R | ATTACCCGGGTCATTTTAAAATTCCAATGTTTTTTAAACG |
| Xho1-TRIT1_R | ATATCTCGAGTTAAACGCTGCATTTCAGCTCTTG |
| Xho1-Tit11_R | ATATCTCGAGTCATTTTAAAATTCCAATGT |
| Xho1-Mod5_R | ATATCTCGAGTCATTCCACAGTCTCCTTCTTGTT |
| Nco1-TRIT1_F | ATTACCATGGTGGCGTCCGTGGCGGCT |
| Nco1_57-467TRIT1_F | ATATCCATGGTGCAGGTCTATGAAGGCCTAG |
| Xho1_57-467TRIT1_R | ATTACTCGAGTTAAACGCTGCATTTCAGCTCT |
| Nco1_TRIT1_Fwd | ATTACCATGGTGGCGTCCGTGGCGGCT |
| Xho1_TRIT1_Rev | ATATCTCGAGTTAAACGCTGCATTTCAGCTCTTG |
| Nco1_TRIT1 R17E/R21E_F | ATTACCATGGTGGCGTCCGTGGCGGCT |
| Xho1_TRIT1 R17E/R21E_R | ATATCTCGAGTTAAACGCTGCATTTCAGCTCTTG |
| HindIII_Tit1_F | GCAATCAAGCTTatcttataaaatgttaaagcccctttgcgttgta |
| XhoI_Tit1_R | AATTCCCTCGAGTCATTTTAAAATTCCAATGTTTTTTAAACGAATTTG |
| HindIII_Mod5_F | AGGATCAAGCTTAAAATCTTCATGCTAAAGGGACCGCTTAAAGG |
| Xho1_Mod5_R | AATTCCCTCGAGTCATTCCACAGTCTCCTTCTTGTTTAT |
| Nco1_TRIT1 T32A_F | ATTACCATGGTGGCGTCCGTGGCGGCT |
| Xho1_TRIT1 T32A_R | ATATCTCGAGTTAAACGCTGCATTTCAGCTCTTG |
| Nco1_TRIT1 57–467_F | ATATCCATGGTGCAGGTCTATGAAGGCCTAG |
| Xho1_TRIT1 57–467_R | ATTACTCGAGTTAAACGCTGCATTTCAGCTCT |
Cloning of IPTases in S. cerevisiae expression plasmids. TRIT1, Tit1 and Mod5 ORF nucleotide sequences were PCR amplified using gene specific forward and reverse primers containing HindIII/Xho1 restriction sites. For Tit1 and Mod5, an existing ATG in pYX141 was disrupted prior to insertion of the ORF; pYX141 (low copy, 786 promoter) was digested with EcoRI and BamHI, followed by treatment with Klenow fragment and ligation. Sequencing confirmed disruption of the ATG. The HindIII/Xho1 restriction sites were then used for ORF insertion. For TRIT1, TRIT1-R17E/R21E, TRIT1-T32A, and TRIT1(57–467), the sequences were PCR amplified from the corresponding pREPx plasmid constructs using primers that contained Nco1/Xho1 sites, and these were cloned into the corresponding sites of pYX141 or pYX242 (high copy, TP1 promoter).
tRNA gene cloning. HindIII fragments containing the human nuclear-encoded tRNATyrGUA gene 2–1 (TRA-GUA2-1) [71] with G37 or A37 and including 150 bp upstream and 90 bp downstream of the mature tRNA sequence [97] were synthesized by Genewiz (https://www.genewiz.com/), then cloned into the HindIII site of YCplac33 (CEN4). All DNA clones were confirmed by sequencing.
Cell culture and transfection
HEK293 cells were maintained in DMEM plus Glutamax (Gibco) supplemented with 10% heat-inactivated FBS in humidified 37°C, 5% CO2 incubator. The cells were transfected using Lipofectamine 2000 according to manufacturer’s instructions using 2.5 ug plasmid DNA and 6.25 ul lipofectamine on cells that were 80% confluent in 6-well plates. The next day, cells were seeded onto coverslips for confocal microscopy (see below), or divided over multiple 10 cm plates for mitochondrial preparation (see below) and western blot analysis.
Mitochondrial preparation and cell subfractionation
Subcellular fractions were isolated as described previously [98], with a few modifications. Forty-eight hours post transfection, HEK293 cells (8 x 10 cm plates at 80% confluency per condition) were washed with 10 ml warm PBS per plate and scraped in 5 ml ice-cold PBS per plate and combined in a 50 ml Falcon tube. Cells were spun down for 3 mins at 1200 rpm and the pellets were washed twice in 10 ml ice-cold PBS. An aliquot of cell pellet was kept for isolation of total protein in RIPA buffer (Pierce) containing protease inhibitors (Roche). Pellets were suspended in 2 ml of homogenization buffer (HB, 0.6 M Mannitol, 1 mM EGTA, 10 mM Tris-HCl pH 7.4) +0.1% BSA) and transferred to a hand-held 10 ml glass-Teflon homogenizer. After 25 strokes, the suspension was transferred to a 2 ml Eppendorf tube and centrifuged at 400g for 10 min at 4C. The supernatant, containing crude mitochondria was kept. The pellet was resuspended in 2 ml HB and homogenization was repeated using 15 strokes. Suspension was transferred to a 2 ml Eppendorf tube and centrifuged at 400g for 10 min at 4C. Again, the supernatant, containing crude mitochondria was kept. This process was repeated one more time with 15 strokes. All three supernatants were centrifuged again at 1000g for 10 mins at 4C. Supernatants were transferred into fresh Eppendorf tubes without disturbing the pellets and spun at 11,000g for 10 mins at 4C to pellet the mitochondria. The supernatant of the first homogenization was kept as the soluble cytoplasmic fraction. Mitochondria were washed twice with 1ml HB without BSA and PMSF and combined into one tube and centrifuged again at 11,000g for 5 mins at 4C to generate a single pellet. This was taken up in 500 ul of HB without BSA and PMSF and split into 2 tubes (200 ul/tube). One tube was put on ice (ProtK fraction) and the other one (mitochondrial fraction) was spun down and the pellet taken up in 40 ul RIPA buffer (Pierce) containing protease inhibitors (Roche). Protein concentrations of the total protein- and mitochondrial fractions were determined using BCA (Pierce). 40 ug of the protK fraction in a total of 190 ul HB without BSA and PMSF was treated with 1.6 ug proteinase K (Viagen) by incubating on ice for 15 mins and subsequently at room temperature for 15 mins. 5 mM PMSF was added to inactivate the Proteinase K and the mitochondria were spun down at 11,000g for 5 mins at 4C. Mitochondria were washed with 1 ml HB including PMSF, but without BSA and taken up in 40 ul RIPA buffer (Pierce) containing protease inhibitors (Roche). For the cytosolic fraction, equal protein concentration between wild-type and mutant was determined using nanodrop using HB as blank.17.5 ug of each fraction was separated on SDS-PAGE and analyzed on western blot.
Western blots
Total protein was extracted from transfected HEK293 cells lysed in RIPA buffer (Pierce) containing protease inhibitors (Roche) 48 hours post transfection and size separated using SDS-PAGE. Proteins were then transferred to a nitrocellulose membrane. Antibodies used were anti-GFP (Santa Cruz, sc-8334), anti-Tom20 (Santa Cruz, sc-11415), anti-NDUFA9 (Abcam, ab14713) and polyclonal human anti-La (Go) was a gift of J. D. Keene and D. J. Kenan (Duke Medical Center) [99]. Anti-TRIT1 Ab was generated in rabbits using the peptide KLHPHDKRKVARSLQVFEE as an antigen and was affinity purified using the same peptide (Pierce protein research). Primary antibodies were detected by secondary antibodies from LI-COR Biosciences, which were conjugated to either IRDye 800CW or 680RD and western blot were scanned and quantified using the Odyssey CLx imaging system (LI-COR Biosciences).
Western blots for S. pombe protein: For protein isolation from S. pombe, cells were transferred to a 1.5 mL microcentrifuge tube, an equal volume of 0.7 N NaOH solution was added, mixed, then incubated at room temperature for 3 minutes. The tube was centrifuged at 5,000 rpm for one min and the supernatant discarded. SDS-PAGE sample buffer was added, mixed well and the sample was heated at 95°C for 5 min centrifuged again and the resulting supernatant recovered for analysis. Proteins were then transferred to a nitrocellulose membrane and subsequently analyzed. Anti-β-actin was from Abcam (mAbcam 8224) and used at a 1:5000 dilution. Blots were scanned as described above using the Odyssey CLx imaging system (LI-COR Biosciences).
Confocal microscopy
24 hrs. after transfection cells were seeded onto coverslips. The next day the live cells were treated with MitoTracker (ThermoFisher) at 200 nM in PBS for 20 min. at 37°C. Cells were then fixed with 4% paraformaldehyde for 15 min. at room temperature and washed with PBS. PBS containing 1 ug/ml Hoechst 33342 (Sigma) was added and allowed to sit for 10 min, followed by a final PBS wash. Imaging was with a Leica DM IRE2 confocal microscope using a HCX PL APO CS 63.0x1.32 oil UV objective. Images were scanned in sequential mode in Hoechst, GFP and MitoTracker channels.
PHA6 northern blotting
We recently found that increasing the post-hybridization wash temperature, to as high as 15°C above the Ti (hybridization incubation temperature, below) increased the difference in signal between IPTase-positive and IPTase-negative sample tRNAs, especially, mt-tRNAs. We note that the optimal wash temperature for ACL probes varies among individual tRNAs possibly due to other ACL modifications and other characteristics. We found that subjecting each tRNA-ACL probing to sequential washes at progressively higher temperatures yielded more reliable and higher apparent % modification efficiencies than subjecting all tRNA-ACL probe combinations to a final wash at the same temperature. After the maximum difference in IPTase-positive and -negative sample tRNAs is recorded, the next high temperature signal from all samples may substantially or drastically decrease. The results reported and quantified in this paper are from blot washings that reveal the greatest difference in ACL probe signal between IPTase positive and IPTase negative sample tRNAs that were obtained using this approach. After the maximum difference in IPTase-positive and -negative sample tRNAs occurs, the next high temperature wash may drastically decrease signal from all samples and may not be useful for quantification.
Novex 10% TBE-urea polyacrylamide gels and transferred to GeneScreen plus nylon membranes using Invitrogen iBlot transfer apparatus. Blots are UV crosslinked then baked in a vacuum oven at 80°C for 1–2 hrs. The blot is incubated in 10–15 ml of PHA6 hybridization buffer contains 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 0.2% SDS, 1× Denhardt's solution [Life Technologies], 100 μg/ml sheared salmon sperm DNA (supplier) at the oligonucleotide DNA probe hybridization temperature for 1 to 2 h. A 32P-end-labeled antisense DNA oligonucleotide at >108 cpm/μg DNA was added to the hybridization buffer (final concentration, 105 cpm/ml) and left overnight for hybridization at the Ti. The Ti is calculated as follows: Ti = Tm—15°C where Tm is equal to 16.6 log[M] + 0.41[Pgc] + 81.5 − Pm − (B/L) − 0.65, where M is the molar salt concentration, Pgc is the percent G+C content in the oligonucleotide DNA probe, Pm is the percentage of mismatched bases, if any, B is 675, and L is the oligonucleotide DNA probe length [100]. Blots were washed with 2× SSC, 0.2% SDS two times at room temperature (10 min each) followed by 30 min at Ti +5°C, +7.5°C, +10°C, +12°C or +15°C (determined empirically, described above). Optimal wash temperatures for different tRNAs has ranged from Ti to T1+15°C.
Before the next hybridization, blots were stripped with 0.1 × SSC, 0.1% SDS at 85–90°C, and the nearly complete removal (≥90%) of 32P was confirmed by Phosphor Imager analysis. In general, multiple ACL probings are done followed by the body probes. Table 2 provides sequences of the oligo-DNAs used as ACL and body probes.
The formula used to calculate % i6A37 modification is [1-(ACLtit1-Δ (transformed with IPTase constructs)/BPtit1-Δ (transformed IPTase constructs))/(ACLtit1Δ (empty vector)/BPtit1Δ (empty vector))] x 100, where % modification in yNB5 (tit1-Δ) = 0, “ACL” indicates the ACL probe and “BP” indicates the body probe. In cases where MT-8 cells were used rather than yNB5, mod5Δ would substitute for tit1Δ. As described, the formula is internally normalized by the tit1Δ or mod5Δ samples.
Supporting information
A) Translation initiation contexts of ATG codons; the first two lines show consensuses derived for S. cerevisiae [highly expressed genes, refs 101, 102] and the last line shows a consensus derived for vertebrate [103]. Lines 3 and 4 are for the alternative ATGs that encode MOD5 M1 and M12 in their native context; lines 5 and 6 are for the MOD5 M1 and TRIT1 M1 after cloning into the Xho1 site of S. pombe expression vectors pRep4X or pRep82X (pRepXho1). Line 7 shows the native TRIT1 M1 ATG context. B) Nucleotide sequence of MOD5 including its M1 and M12 ATG sites and the M12A substitution mutation in the Mod5MTS-M12A-TRIT1 construct. C) Western blot of TRIT1 protein developed using anti-TRIT1 Ab, from cells with constructs containing the strong (pRep4X) or weak (pRep82X) nmt1+ promoter (see text), indicated above the lanes as 4X or 82X. Lanes are numbered below. MW markers are indicated in kDa. The shared band below 25 kDa is an internal control. Lower panel shows β-actin as loading control used for quantitative normalization. D) Determination of TRIT1 levels in C by quantitative Odyssey CLx imaging (Methods); numbers above bars indicate TRIT1/β-actin levels in each sample; The pRep vectors used are indicated as 4X and 82X are indicated along the X-axis. E) Northern blot of 2 cy- and 2 mt- tRNAs by TRIT1 and Mod5MTS-TRIT1 each from the strong promoter, pRep4X and weak promoter pRep82X, as indicated above the lanes, as 4X and 82X. The top four panels show the ACL probings as indicated to the left, and the bottom four panels show the corresponding body probings. F) Quantitation of % i6A37 modification of the mt-tRNAs and the cy-tRNAs; the pRep vectors used are indicated as 4X and 82X along the X-axis. G) Clover leaf representations of S. pombe cy-tRNATrpCCA and mt-tRNATrpCCA as encoded by the nuclear and mitochondrial DNA and folded by tRNAscan-SE [112] (Table 1, Discussion).
(TIF)
(TIF)
Sequence alignments of the ACLs of cy-tRNATrpCCA (A), cy-tRNACysGCA (B), and cy-tRNATyrGUA (C), in the eukaryotes indicated; the 32 and 37 positions are numbered and the horizontal bar indicates the AC. The empty boxes reflect that no genes for this tRNA were indicated for this species [ref 112].
(TIF)
Acknowledgments
We thank Anup Dey (NICHD) for assistance with microscopy, Monika Olahova (Wellcome Centre for Mitochondrial Research, UK) for the mitochondrial isolation protocol, Rima Sakhawala for technical assistance, and Tom Dever and Alan Hinnebusch for discussion.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH. The Grant number is HD000412-30 to RJM. The URL of the funders website is https://data.nidb.nih.gov/reports/ The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sokolowski M, Klassen R, Bruch A, Schaffrath R, Glatt S. Cooperativity between different tRNA modifications and their modification pathways. Biochim Biophys Acta. 2017 [DOI] [PubMed] [Google Scholar]
- 2.Chatterjee K, Nostramo RT, Wan Y, Hopper AK. tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location. Biochim Biophys Acta Gene Regul Mech. 2018;1861(4):373–86 10.1016/j.bbagrm.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vare VY, Eruysal ER, Narendran A, Sarachan KL, Agris PF. Chemical and Conformational Diversity of Modified Nucleosides Affects tRNA Structure and Function. Biomolecules. 2017;7(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014;42(11):7346–57 10.1093/nar/gku390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei FY, Zhou B, Suzuki T, Miyata K, Ujihara Y, Horiguchi H, et al. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab. 2015;21(3):428–42 10.1016/j.cmet.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 6.Lamichhane TN, Arimbasseri AG, Rijal K, Iben JR, Wei FY, Tomizawa K, et al. Lack of tRNA-i6A modification causes mitochondrial-like metabolic deficiency in S. pombe by limiting activity of cytosolic tRNATyr, not mito-tRNA. RNA. 2016;22(4):583–96 10.1261/rna.054064.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schweizer U, Bohleber S, Fradejas-Villar N. The modified base isopentenyladenosine and its derivatives in tRNA. RNA biology. 2017:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou HJ, Donnard E, Gustafsson HT, Garber M, Rando OJ. Transcriptome-wide Analysis of Roles for tRNA Modifications in Translational Regulation. Mol Cell. 2017;68(5):978–92 e4 10.1016/j.molcel.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han L, Phizicky EM. A rationale for tRNA modification circuits in the anticodon loop. RNA. 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guy MP, Shaw M, Weiner CL, Hobson L, Stark Z, Rose K, et al. Defects in tRNA Anticodon Loop 2'-O-Methylation Are Implicated in Nonsyndromic X-Linked Intellectual Disability due to Mutations in FTSJ1. Hum Mutat. 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guy MP, Phizicky EM. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA. 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arimbasseri AG, Iben J, Wei FY, Rijal K, Tomizawa K, Hafner M, et al. Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: a subset of tRNAsSer requires N6-isopentenylation of A37. RNA. 2016;22(9):1400–10 10.1261/rna.056259.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han L, Marcus E, D'Silva S, Phizicky EM. S. cerevisiae Trm140 has two recognition modes for 3-methylcytidine modification of the anticodon loop of tRNA substrates. RNA. 2016;23:406–19 10.1261/rna.059667.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yarham JW, Lamichhane T., Mattijssen S., Bruni F., McFarland R., Maraia R.J., Taylor R.W. Defective i6A37 Modification of Mitochondrial and Cytosolic tRNAs Results from Pathogenic Mutations in TRIT1 and its Substrate tRNA. PLoS Genetics. 2014;June 5;10(6):e1004424 10.1371/journal.pgen.1004424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kernohan KD, Dyment DA, Pupavac M, Cramer Z, McBride A, Bernard G, et al. Matchmaking facilitates the diagnosis of an autosomal-recessive mitochondrial disease caused by biallelic mutation of the tRNA isopentenyltransferase (TRIT1) gene. Hum Mutat. 2017;38:511–6 10.1002/humu.23196 [DOI] [PubMed] [Google Scholar]
- 16.Edvardson S, Elbaz-Alon Y, Jalas C, Matlock A, Patel K, Labbe K, et al. A mutation in the THG1L gene in a family with cerebellar ataxia and developmental delay. Neurogenetics. 2016;17(4):219–25 10.1007/s10048-016-0487-z [DOI] [PubMed] [Google Scholar]
- 17.Tarnopolsky MA, Brady L, Tetreault M, Care4Rare Canada C. TRMT5 mutations are associated with features of complex hereditary spastic paraparesis. Neurology. 2017;89(21):2210–1 10.1212/WNL.0000000000004657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Powell CA, Kopajtich R, D'Souza AR, Rorbach J, Kremer LS, Husain RA, et al. TRMT5 Mutations Cause a Defect in Post-transcriptional Modification of Mitochondrial tRNA Associated with Multiple Respiratory-Chain Deficiencies. Am J Hum Genet. 2015;97(2):319–28 10.1016/j.ajhg.2015.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bohnsack MT, Sloan KE. The mitochondrial epitranscriptome: the roles of RNA modifications in mitochondrial translation and human disease. Cell Mol Life Sci. 2018;75(2):241–60 10.1007/s00018-017-2598-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Crecy-Lagard V, Boccaletto P, Mangleburg CG, Sharma P, Lowe TM, Leidel SA, et al. Matching tRNA modifications in humans to their known and predicted enzymes. Nucleic Acids Res. 2019;47(5):2143–59 10.1093/nar/gkz011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackman JE, Phizicky EM. tRNAHis guanylyltransferase adds G-1 to the 5' end of tRNAHis by recognition of the anticodon, one of several features unexpectedly shared with tRNA synthetases. RNA. 2006;12(6):1007–14 10.1261/rna.54706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christian T, Gamper H, Hou YM. Conservation of structure and mechanism by Trm5 enzymes. RNA. 2013;19(9):1192–9 10.1261/rna.039503.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goto-Ito S, Ito T, M K, Bessho Y, Yokoyama S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat Struct Mol Biol. 2009;16:1109–15 10.1038/nsmb.1653 [DOI] [PubMed] [Google Scholar]
- 24.Zhou C, Huang RH. Crystallographic snapshots of eukaryotic dimethylallyltransferase acting on tRNA: insight into tRNA recognition and reaction mechanism. Proc Natl Acad Sci U S A. 2008;105(42):16142–7 10.1073/pnas.0805680105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Golovko A, Hjalm G, Sitbon F, Nicander B. Cloning of a human tRNA isopentenyl transferase. Gene. 2000;258(1–2):85–93 10.1016/s0378-1119(00)00421-2 [DOI] [PubMed] [Google Scholar]
- 26.Yevdakova NA, von Schwartzenberg K. Characterisation of a prokaryote-type tRNA-isopentenyltransferase gene from the moss Physcomitrella patens. Planta. 2007;226(3):683–95 10.1007/s00425-007-0516-0 [DOI] [PubMed] [Google Scholar]
- 27.Patil G, Nicander B. Identification of two additional members of the tRNA isopentenyltransferase family in Physcomitrella patens. Plant Mol Biol. 2013;82(4–5):417–26 10.1007/s11103-013-0072-x [DOI] [PubMed] [Google Scholar]
- 28.Chen Y, Bai B, Yan H, Wen F, Qin D, Jander G, et al. Systemic disruption of the homeostasis of transfer RNA isopentenyltransferase causes growth and development abnormalities in Bombyx mori. Insect Mol Biol 2019;28(3):380–91 10.1111/imb.12561 [DOI] [PubMed] [Google Scholar]
- 29.Golovko A, Sitbon F, Tillberg E, Nicander B. Identification of a tRNA isopentenyltransferase gene from Arabidopsis thaliana. Plant Mol Biol. 2002;49(2):161–9 10.1023/a:1014958816241 [DOI] [PubMed] [Google Scholar]
- 30.Janner F, Vogeli G, Fluri R. The antisuppressor strain sin1 of Schizosaccharomyces pombe lacks the modification isopentenyladenosine in transfer RNA. J Mol Biol. 1980;139(2):207–19 10.1016/0022-2836(80)90305-8 [DOI] [PubMed] [Google Scholar]
- 31.Laten HM. Antisuppression of class I suppressors in an isopentenylated-transfer RNA deficient mutant of Saccharomyces cerevisiae. Curr Genet. 1984;8(1):29–32 10.1007/BF00405428 [DOI] [PubMed] [Google Scholar]
- 32.Dihanich ME, Najarian D, Clark R, Gillman EC, Martin NC, Hopper AK. Isolation and characterization of MOD5, a gene required for isopentenylation of cytoplasmic and mitochondrial tRNAs of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7(1):177–84 10.1128/mcb.7.1.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rijal K, Maraia RJ, Arimbasseri AG. A methods review on use of nonsense suppression to study 3' end formation and other aspects of tRNA biogenesis. Gene. 2015;556(1):35–50 10.1016/j.gene.2014.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gillman EC, Slusher LB, Martin NC, Hopper AK. MOD5 translation initiation sites determine N6-isopentenyladenosine modification of mitochondrial and cytoplasmic tRNA. Mol Cell Biol. 1991;11(5):2382–90 10.1128/mcb.11.5.2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boguta M, Hunter LA, Shen WC, Gillman EC, Martin NC, Hopper AK. Subcellular locations of MOD5 proteins: mapping of sequences sufficient for targeting to mitochondria and demonstration that mitochondrial and nuclear isoforms commingle in the cytosol. Mol Cell Biol. 1994;14(4):2298–306 10.1128/mcb.14.4.2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tolerico LH, Benko AL, Aris JP, Stanford DR, Martin NC, Hopper AK. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics. 1999;151(1):57–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pratt-Hyatt M, Pai DA, Haeusler RA, Wozniak GG, Good PD, Miller EL, et al. Mod5 protein binds to tRNA gene complexes and affects local transcriptional silencing. Proc Natl Acad Sci U S A. 2013;110(33):E3081–9 10.1073/pnas.1219946110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smaldino PJ, Read DF, Pratt-Hyatt M, Hopper AK, Engelke DR. The cytoplasmic and nuclear populations of the eukaryote tRNA-isopentenyl transferase have distinct functions with implications in human cancer. Gene. 2015;556(1):13–8 10.1016/j.gene.2014.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12(16):2463–8 10.1101/gad.12.16.2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302(5649):1399–401. 10.1126/science.1089814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kessler AC, d'Almeida GS, Alfonzo JD. The role of intracellular compartmentalization on tRNA processing and modification. RNA biology. 2017:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cherkasova V, Bahler J, Bacikova D, Pridham K, Maraia RJ. Altered nuclear tRNA metabolism in La-deleted S. pombe is accompanied by a nutritional stress response involving Atf1p and Pcr1p that is suppressible by Xpo-t/Los1p. Mol Biol Cell. 2012;23:480–91 10.1091/mbc.E11-08-0732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lamichhane TN, Blewett NH, Maraia RJ. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA. 2011;17:1846–57 10.1261/rna.2628611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lemieux J, Lakowski B, Webb A, Meng Y, Ubach A, Bussiere F, et al. Regulation of physiological rates in Caenorhabditis elegans by a tRNA-modifying enzyme in the mitochondria. Genetics. 2001;159(1):147–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leung HC, Chen Y, Winkler ME. Regulation of substrate recognition by the MiaA tRNA prenyltransferase modification enzyme of Escherichia coli K-12. J Biol Chem. 1997;272(20):13073–83 10.1074/jbc.272.20.13073 [DOI] [PubMed] [Google Scholar]
- 46.Soderberg T, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: essential elements for recognition of tRNA substrates within the anticodon stem-loop. Biochemistry. 2000;39(21):6546–53 10.1021/bi992775u [DOI] [PubMed] [Google Scholar]
- 47.Soderberg T, Poulter CD. Escherichia coli dimethylallyl diphosphate:tRNA dimethylallyltransferase: site-directed mutagenesis of highly conserved residues. Biochemistry. 2001;40(6):1734–40 10.1021/bi002149t [DOI] [PubMed] [Google Scholar]
- 48.Chimnaronk S, Forouhar F, Sakai J, Yao M, Tron CM, Atta M, et al. Snapshots of dynamics in synthesizing N(6)-isopentenyladenosine at the tRNA anticodon. Biochemistry. 2009;48(23):5057–65 10.1021/bi900337d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Motorin Y, Bec G, Tewari R, Grosjean H. Transfer RNA recognition by the Escherichia coli delta2-isopentenyl-pyrophosphate:tRNA delta2-isopentenyl transferase: dependence on the anticodon arm structure. RNA. 1997;3(7):721–33 [PMC free article] [PubMed] [Google Scholar]
- 50.Sibler AP, Bordonne R, Dirheimer G, Martin R. [Primary structure of yeast mitochondrial tryptophan-tRNA capable of translating the termination U-G-A codon]. C R Seances Acad Sci D. 1980;290(11):695–8 [PubMed] [Google Scholar]
- 51.Sprinzl M, Gauss DH. Compilation of tRNA sequences. Nucleic Acids Res. 1982;10(2):r1–55 [PMC free article] [PubMed] [Google Scholar]
- 52.Claros MG. MitoProt, a Macintosh application for studying mitochondrial proteins. Comput Appl Biosci. 1995;11(4):441–7 10.1093/bioinformatics/11.4.441 [DOI] [PubMed] [Google Scholar]
- 53.Fukasawa Y, Tsuji J, Fu SC, Tomii K, Horton P, Imai K. MitoFates: improved prediction of mitochondrial targeting sequences and their cleavage sites. Mol Cell Proteomics. 2015;14(4):1113–26 10.1074/mcp.M114.043083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A. 2009;106(25):10171–6 10.1073/pnas.0900604106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin NC, Hopper AK. How single genes provide tRNA processing enzymes to mitochondria, nuclei and the cytosol. Biochimie. 1994;76(12):1161–7 10.1016/0300-9084(94)90045-0 [DOI] [PubMed] [Google Scholar]
- 56.Murawski M, Szczesniak B, Zoladek T, Hopper AK, Martin NC, Boguta M. maf1 mutation alters the subcellular localization of the Mod5 protein in yeast. Acta Biochim Pol. 1994;41(4):441–8 [PubMed] [Google Scholar]
- 57.Eckers E, Cyrklaff M, Simpson L, Deponte M. Mitochondrial protein import pathways are functionally conserved among eukaryotes despite compositional diversity of the import machineries. Biol Chem. 2012;393(6):513–24 10.1515/hsz-2011-0255 [DOI] [PubMed] [Google Scholar]
- 58.Fukasawa Y, Oda T, Tomii K, Imai K. Origin and Evolutionary Alteration of the Mitochondrial Import System in Eukaryotic Lineages. Mol Biol Evol. 2017;34(7):1574–86 10.1093/molbev/msx096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamichhane TN, Mattijssen S, Maraia RJ. Human cells have a limited set of tRNA anticodon loop substrates of the tRNA isopentenyltransferase TRIT1 tumor suppressor. Mol Cell Biol. 2013;33:4900–8 10.1128/MCB.01041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lamichhane TN, Blewett NH, Cherkasova VA, Crawford AK, Iben JR, Farabaugh PJ, et al. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol Cell Biol. 2013;33 10.1128/MCB.00278-13 :2918–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spinola M, Galvan A, Pignatiello C, Conti B, Pastorino U, Nicander B, et al. Identification and functional characterization of the candidate tumor suppressor gene TRIT1 in human lung cancer. Oncogene. 2005;24:5502–9 10.1038/sj.onc.1208687 [DOI] [PubMed] [Google Scholar]
- 62.Park JM, Intine RV, Maraia RJ. Mouse and Human La Proteins Differ in Kinase Substrate Activity and Activation Mechanism for tRNA Processing. Gene Expression. 2007;14:71–81 10.3727/105221607783417619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spinola M, Falvella FS, Galvan A, Pignatiello C, Leoni VP, Pastorino U, et al. Ethnic differences in frequencies of gene polymorphisms in the MYCL1 region and modulation of lung cancer patients' survival. Lung Cancer. 2007;55(3):271–7 10.1016/j.lungcan.2006.10.023 [DOI] [PubMed] [Google Scholar]
- 64.Yue Z, Li HT, Yang Y, Hussain S, Zheng CH, Xia J, et al. Identification of breast cancer candidate genes using gene co-expression and protein-protein interaction information. Oncotarget. 2016;7(24):36092–100 10.18632/oncotarget.9132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen S, Zheng Z, Tang J, Lin X, Wang X, Lin J. Association of polymorphisms and haplotype in the region of TRIT1, MYCL1 and MFSD2A with the risk and clinicopathological features of gastric cancer in a southeast Chinese population. Carcinogenesis. 2013;34(5):1018–24 10.1093/carcin/bgt010 [DOI] [PubMed] [Google Scholar]
- 66.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123(1):131–6 10.1016/0378-1119(93)90552-e [DOI] [PubMed] [Google Scholar]
- 67.Forsburg SL. Comparison of Schizosaccharomyces pombe expression systems. Nucleic Acids Res. 1993;21(12):2955–6 10.1093/nar/21.12.2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Slusher LB, Gillman EC, Martin NC, Hopper AK. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc Natl Acad Sci U S A. 1991;88(21):9789–93 10.1073/pnas.88.21.9789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Auffinger P, Westhof E. Singly and bifurcated hydrogen-bonded base-pairs in tRNA anticodon hairpins and ribozymes. J Mol Biol. 1999;292(3):467–83 10.1006/jmbi.1999.3080 [DOI] [PubMed] [Google Scholar]
- 70.Olejniczak M, Uhlenbeck OC. tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie. 2006;88(8):943–50 10.1016/j.biochi.2006.06.005 [DOI] [PubMed] [Google Scholar]
- 71.Chan PP, Lowe TM. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res. 2015;44(D1) 10.1093/nar/gkv1309 :44(D1):D184–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hamada M, Huang Y, Lowe TM, Maraia RJ. Widespread Use of TATA Elements in the Core Promoters for RNA Polymerases III, II, and I in Fission Yeast. Mol Cell Biol. 2001;21(20):6870–81 10.1128/MCB.21.20.6870-6881.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Forsburg SL. The best yeast? Trends Genet. 1999;15(9):340–4 10.1016/s0168-9525(99)01798-9 [DOI] [PubMed] [Google Scholar]
- 74.Miyauchi K, Kimura S, Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol. 2013;9(2):105–11 10.1038/nchembio.1137 [DOI] [PubMed] [Google Scholar]
- 75.Rojas-Benitez D, Thiaville PC, de Crecy-Lagard V, Glavic A. The Levels of a Universally Conserved tRNA Modification Regulate Cell Growth. J Biol Chem. 2015;290(30):18699–707 10.1074/jbc.M115.665406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maraia RJ, Arimbasseri AG. Factors That Shape Eukaryotic tRNAomes: Processing, Modification and Anticodon-Codon Use. Biomolecules. 2017;7(1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mattijssen S, Gaidamakov S, Maraia RJ. A mouse model of human tRNA isopentenyltransferase-1 deficiency. Forthcoming. 2020:Forthcoming
- 78.Benko AL, Vaduva G, Martin NC, Hopper AK. Competition between a sterol biosynthetic enzyme and tRNA modification in addition to changes in the protein synthesis machinery causes altered nonsense suppression. Proc Natl Acad Sci U S A. 2000;97(1):61–6 10.1073/pnas.97.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Helm M, Alfonzo JD. Posttranscriptional RNA Modifications: Playing Metabolic Games in a Cell’s Chemical Legoland. Chem Biol. 2014;21(2):174–85 10.1016/j.chembiol.2013.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maraia RJ, Iben JR. Different types of secondary information in the genetic code. RNA. 2014;20(7):977–84 10.1261/rna.044115.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li JM, Hopper AK, Martin NC. N2,N2-dimethylguanosine-specific tRNA methyltransferase contains both nuclear and mitochondrial targeting signals in Saccharomyces cerevisiae. J Cell Biol. 1989;109(4 Pt 1):1411–9 10.1083/jcb.109.4.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ellis SR, Hopper AK, Martin NC. Amino-terminal extension generated from an upstream AUG codon increases the efficiency of mitochondrial import of yeast N2,N2-dimethylguanosine-specific tRNA methyltransferases. Mol Cell Biol. 1989;9(4):1611–20 10.1128/mcb.9.4.1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Machnicka MA, Olchowik A, Grosjean H, Bujnicki JM. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA biology. 2014;11(12):1619–29 10.4161/15476286.2014.992273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bullerwell CE, Leigh J, Forget L, Lang BF. A comparison of three fission yeast mitochondrial genomes. Nucleic Acids Res. 2003;31(2):759–68 10.1093/nar/gkg134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, et al. MODOMICS: a database of RNA modification pathways—2013 update. Nucleic Acids Res. 2013;41(Database issue):D262–7 10.1093/nar/gks1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jia W, Higgs PG. Codon usage in mitochondrial genomes: distinguishing context-dependent mutation from translational selection. Mol Biol Evol. 2008;25(2):339–51 10.1093/molbev/msm259 [DOI] [PubMed] [Google Scholar]
- 87.Yokoyama S, Nishimura S. Modified Nucleosides and Codon Recognition. tRNA: Structure, Biosynthesis and Function. Wash., D.C: ASM Press; 1995. p. 207–23. [Google Scholar]
- 88.Jackman JE, Montange RK, Malik HS, Phizicky EM. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA. 2003;9(5):574–85 10.1261/rna.5070303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hofmann J, Schumann G, Borschet G, Gosseringer R, Bach M, Bertling WM, et al. Transfer RNA genes from Dictyostelium discoideum are frequently associated with repetitive elements and contain consensus boxes in their 5' and 3'-flanking regions. J Mol Biol. 1991;222(3):537–52 10.1016/0022-2836(91)90495-r [DOI] [PubMed] [Google Scholar]
- 90.Arimbasseri AG, Blewett NH, Iben JR, Lamichhane TN, Cherkasova V, Hafner M, et al. RNA polymerase III output is functionally linked to tRNA dimethyl-G26 modification. PLoS Genetics. 2015; 10.1371/journal.pgen.1005671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Steinberg S, Cedergren R. A correlation between N2-dimethylguanosine presence and alternate tRNA conformers. RNA. 1995;1(9):886–91 [PMC free article] [PubMed] [Google Scholar]
- 92.Martin RP, Sibler AP, Gehrke CW, Kuo K, Edmonds CG, McCloskey JA, et al. 5-[[(carboxymethyl)amino]methyl]uridine is found in the anticodon of yeast mitochondrial tRNAs recognizing two-codon families ending in a purine. Biochemistry. 1990;29(4):956–9 10.1021/bi00456a016 [DOI] [PubMed] [Google Scholar]
- 93.Johnson PF, Abelson J. The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature. 1983;302(5910):681–7 10.1038/302681a0 [DOI] [PubMed] [Google Scholar]
- 94.Heyer WD, Thuriaux P, Kohli J, Ebert P, Kersten H, Gehrke C, et al. An antisuppressor mutation of Schizosaccharomyces pombe affects the post-transcriptional modification of the "wobble" base in the anticodon of tRNAs. J Biol Chem. 1984;259(5):2856–62 [PubMed] [Google Scholar]
- 95.Chittum HS, Baek HJ, Diamond AM, Fernandez-Salguero P, Gonzalez F, Ohama T, et al. Selenocysteine tRNA[Ser]Sec levels and selenium-dependent glutathione peroxidase activity in mouse embryonic stem cells heterozygous for a targeted mutation in the tRNA[Ser]Sec gene. Biochemistry. 1997;36(28):8634–9 10.1021/bi970608t [DOI] [PubMed] [Google Scholar]
- 96.Johansson MJ, Esberg A, Huang B, Bjork GR, Bystrom AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol. 2008;28(10):3301–12 10.1128/MCB.01542-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mattijssen S, Arimbasseri AG, Iben JR, Gaidamakov S, Lee J, Hafner M, et al. LARP4 mRNA codon-tRNA match contributes to LARP4 activity for ribosomal protein mRNA poly(A) tail length protection. Elife. 2017;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bruni F, Gramegna P, Oliveira JM, Lightowlers RN, Chrzanowska-Lightowlers ZM. REXO2 is an oligoribonuclease active in human mitochondria. PLoS One. 2013;8(5):e64670 10.1371/journal.pone.0064670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Intine RV, Dundr M, Vassilev A, Schwartz E, Zhao Y, Depamphilis ML, et al. Nonphosphorylated human La antigen interacts with nucleolin at nucleolar sites involved in rRNA biogenesis. Mol Cell Biol. 2004;24:10894–904. 10.1128/MCB.24.24.10894-10904.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leonard G. Davis MDDaJFB. Basic Methods in Molecular Biology: Elsevier; 1986. 388 p. [Google Scholar]
- 101.Zur H, Tuller T. New universal rules of eukaryotic translation initiation fidelity. PLoS computational biology. 2013;9(7):e1003136 10.1371/journal.pcbi.1003136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hamilton R, Watanabe CK, de Boer HA. Compilation and comparison of the sequence context around the AUG startcodons in Saccharomyces cerevisiae mRNAs. Nucleic Acids Res. 1987;15(8):3581–93 10.1093/nar/15.8.3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15(20):8125–48 10.1093/nar/15.20.8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McIlwain SJ, Peris D, Sardi M, Moskvin OV, Zhan F, Myers KS, et al. Genome Sequence and Analysis of a Stress-Tolerant, Wild-Derived Strain of Saccharomyces cerevisiae Used in Biofuels Research. G3 (Bethesda). 2016;6(6):1757–66 10.1534/g3.116.029389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147 10.1038/13779 [DOI] [PubMed] [Google Scholar]
- 106.Kohli J, Munz P, Soll D. Informational suppression, transfer RNA, and intergenic conversion. In: Nasim A, Young P, Johnson BF, editors. Molecular Biology of the Fission Yeast. Cell Biology San Diego: Academic Press, Inc.; 1989. p. 75–96. [Google Scholar]
- 107.Hamada M, Sakulich AL, Koduru SB, Maraia R. Transcription termination by RNA polymerase III in fission yeast: A genetic and biochemical model system. J Biol Chem. 2000;275:29076–81 10.1074/jbc.M003980200 [DOI] [PubMed] [Google Scholar]
- 108.Schon A, Bock A, Ott G, Sprinzl M, Soll D. The selenocysteine-inserting opal suppressor serine tRNA from E. coli is highly unusual in structure and modification. Nucleic Acids Res. 1989;17(18):7159–65 10.1093/nar/17.18.7159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bonitz SG, Berlani R, Coruzzi G, Li M, Macino G, Nobrega FG, et al. Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci U S A. 1980;77(6):3167–70 10.1073/pnas.77.6.3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kerscher S, Durstewitz G, Casaregola S, Gaillardin C, Brandt U. The complete mitochondrial genome of yarrowia lipolytica. Comp Funct Genomics. 2001;2(2):80–90 10.1002/cfg.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zivanovic Y, Wincker P, Vacherie B, Bolotin-Fukuhara M, Fukuhara H. Complete nucleotide sequence of the mitochondrial DNA from Kluyveromyces lactis. FEMS Yeast Res. 2005;5(4–5):315–22 10.1016/j.femsyr.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 112.Chan PP & Lowe TM. tRNAscan-SE: Searching for tRNA Genes in Genomic Sequences. Methods Mol Biol. 2019; 1962:1–14 10.1007/978-1-4939-9173-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Translation initiation contexts of ATG codons; the first two lines show consensuses derived for S. cerevisiae [highly expressed genes, refs 101, 102] and the last line shows a consensus derived for vertebrate [103]. Lines 3 and 4 are for the alternative ATGs that encode MOD5 M1 and M12 in their native context; lines 5 and 6 are for the MOD5 M1 and TRIT1 M1 after cloning into the Xho1 site of S. pombe expression vectors pRep4X or pRep82X (pRepXho1). Line 7 shows the native TRIT1 M1 ATG context. B) Nucleotide sequence of MOD5 including its M1 and M12 ATG sites and the M12A substitution mutation in the Mod5MTS-M12A-TRIT1 construct. C) Western blot of TRIT1 protein developed using anti-TRIT1 Ab, from cells with constructs containing the strong (pRep4X) or weak (pRep82X) nmt1+ promoter (see text), indicated above the lanes as 4X or 82X. Lanes are numbered below. MW markers are indicated in kDa. The shared band below 25 kDa is an internal control. Lower panel shows β-actin as loading control used for quantitative normalization. D) Determination of TRIT1 levels in C by quantitative Odyssey CLx imaging (Methods); numbers above bars indicate TRIT1/β-actin levels in each sample; The pRep vectors used are indicated as 4X and 82X are indicated along the X-axis. E) Northern blot of 2 cy- and 2 mt- tRNAs by TRIT1 and Mod5MTS-TRIT1 each from the strong promoter, pRep4X and weak promoter pRep82X, as indicated above the lanes, as 4X and 82X. The top four panels show the ACL probings as indicated to the left, and the bottom four panels show the corresponding body probings. F) Quantitation of % i6A37 modification of the mt-tRNAs and the cy-tRNAs; the pRep vectors used are indicated as 4X and 82X along the X-axis. G) Clover leaf representations of S. pombe cy-tRNATrpCCA and mt-tRNATrpCCA as encoded by the nuclear and mitochondrial DNA and folded by tRNAscan-SE [112] (Table 1, Discussion).
(TIF)
(TIF)
Sequence alignments of the ACLs of cy-tRNATrpCCA (A), cy-tRNACysGCA (B), and cy-tRNATyrGUA (C), in the eukaryotes indicated; the 32 and 37 positions are numbered and the horizontal bar indicates the AC. The empty boxes reflect that no genes for this tRNA were indicated for this species [ref 112].
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.