Abstract
A new human coronavirus named SARS-CoV-2 was identified in several cases of acute respiratory syndrome in Wuhan, China in December 2019. On March 11 2020, WHO declared the SARS-CoV-2 infection to be a pandemic, based on the involvement of 169 nations. Specific drugs for SARS-CoV-2 are obviously not available. Currently, drugs originally developed for other viruses or parasites are currently in clinical trials based on empiric data. In the quest of an effective antiviral drug, the most specific target for an RNA virus is the RNA-dependent RNA-polymerase (RdRp) which shows significant differences between positive-sense and negative-sense RNA viruses. An accurate evaluation of RdRps from different viruses may guide the development of new drugs or the repositioning of already approved antiviral drugs as treatment of SARS-CoV-2. This can accelerate the containment of the SARS-CoV-2 pandemic and, hopefully, of future pandemics due to other emerging zoonotic RNA viruses.
SARS-CoV-2 is a new human coronavirus identified in patients with acute respiratory syndrome in Wuhan, China in December 2019 [1, 2]. Since then, the SARS-CoV-2 infection has become a pandemic, reaching almost every Country in all Continents with more than 3 million positive and 217.000 deaths, globally (on 29/04/2020 https://gisanddata.maps.arcgis.com/apps/opsdashboard/index.html#/bda7594740fd40299423467b48e9ecf6).
Similarly to the two new human CoV emerged in the past 20 years, namely the severe acute respiratory syndrome CoV (SARS-CoV) and the Middle East respiratory syndrome CoV (MERS-CoV) [3], also the current SARS-CoV-2 is suggested to be originated as a zoonosis from bats [4]. Moreover, evidences show that in bats there is a continuous circulation of additional SARS-like and MERS-like coronaviruses able to replicate efficiently in primary human lung cells [5]. It is therefore predictable that new epidemics/pandemics due to zoonotic coronaviruses will emerge in humans in the future. Consequently, together with a preventive vaccine, effective antivirals are needed to be developed within a broader program of Global preparedness.
Inter-human transmission of coronaviruses is mediated by saliva droplets reaching the new host through coughing and sneezing, both in symptomatic and asymptomatic positive subjects [6, 7]. Moreover, active virus replication in the upper respiratory tract is observed in patients also after the peak of respiratory symptoms, which may result in prolonged viral spreading of the infection [8]. Therefore, reducing the viral titer represents a major goal in order to slow/block the disease progression as well as to significantly limit the viral shedding. The latter objective would result in the lowering of R0 from 2.5 (2.5 people infected by a positive subject) to a value < 1 (less than one person infected by a positive subject). In this way the spread of the virus would be drastically contained and lastly blocked. Finally, the reduced viral replication would give the opportunity to the adaptive humoral immune system (with the production of antibodies, hopefully neutralizing) enough time to mount a sufficient primary response capable of containing further viral replication and eradicating the infection.
SARS-CoV-2 is a positive-sense RNA virus belonging to the Orthocoronavirinae (coronavirus, CoV) family and, in particular, to the genus beta (group 2) together with the other two new human coronaviruses SARS-CoV and MERS-CoV. Similarly to all RNA viruses, the viral genome replication and transcription processes of SARS-CoV-2 depend on an RNA-dependent RNA polymerase (RdRp), which is encoded by the RNA virus to catalyze the RNA synthesis from RNA templates. Consequently, RdRp is the key enzyme in the viral biological cycle of all RNA viruses, regardless the polarity of the viral RNA genome [9, 10]. Although different for each RNA viruses, all viral RdRps are characterized by a 500–600 residue catalytic module with palm, fingers, and thumb domains forming an encircled human right hand architecture. Seven catalytic motifs are located in the RdRp palm and fingers domains, comprising the most conserved parts of the RdRp and are responsible for the RNA-only specificity in catalysis. Nevertheless, RdRps from positive-sense and negative-sense RNA viruses show differences with significant implications in the enzymatic mode of action [11]. In particular, the palm catalytic subdomain (Motif C) is the most conserved region of all monomeric viral RNA polymerases with two aspartic acid residues that coordinate the two metal ions necessary for the phosphoryl transfer reaction [12, 13]. However, while positive-sense RNA viruses show a glycine preceding the two aspartates, negative-sense RNA viruses show a serine, resulting in the alternative interactions with the metal ions [14]. Mutation of the first aspartic residue results in a complete loss of RNA polymerase activity, whereas mutations of the second aspartate diminish the polymerase activity or modify the metal cofactor requirements, but do not inactivate the enzyme [15, 16]. Negative-sense RNA viruses, have an asparagine instead of the second aspartic acid in Motif C which has been shown to enable viral RNA polymerases to use manganese (Mn2+) instead of magnesium (Mg2+) as cofactor [17].
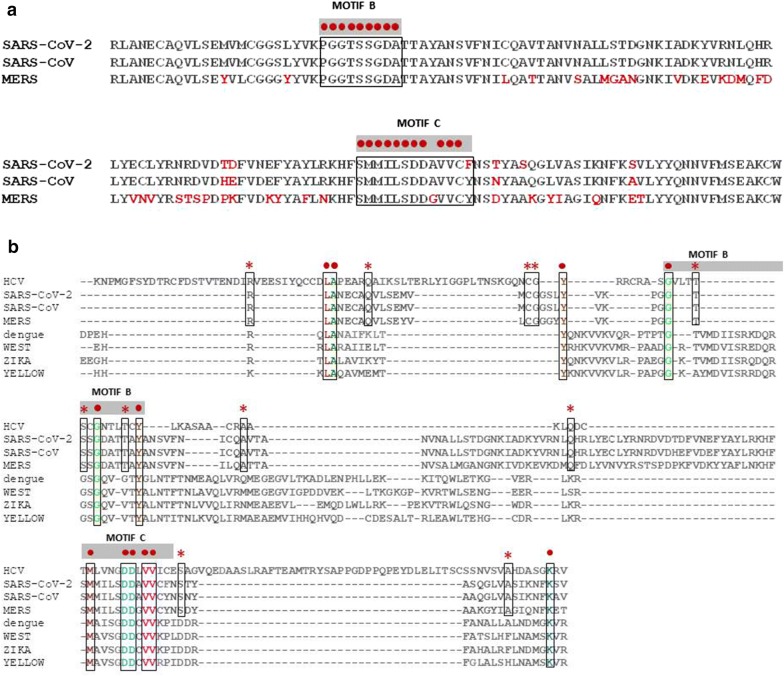
The alignment of RdRp sequences from the three epidemic/pandemic coronaviruses, confirms the high homology and conservation along the sequence (Fig. 1a). Such homology and conservation is strongly retained when the analysis includes other positive-sense RNA virus, namely HCV, Dengue, West Nile, Zika and Yellow Fever viruses (Fig. 1b). In particular, HCV shows the greatest number of identical residues with the coronaviruses. In Motif C, the DDxVV pattern has a 100% conservation but the three coronavirus are the only ones to show a serine instead of a glycine preceding the two aspartate residues.
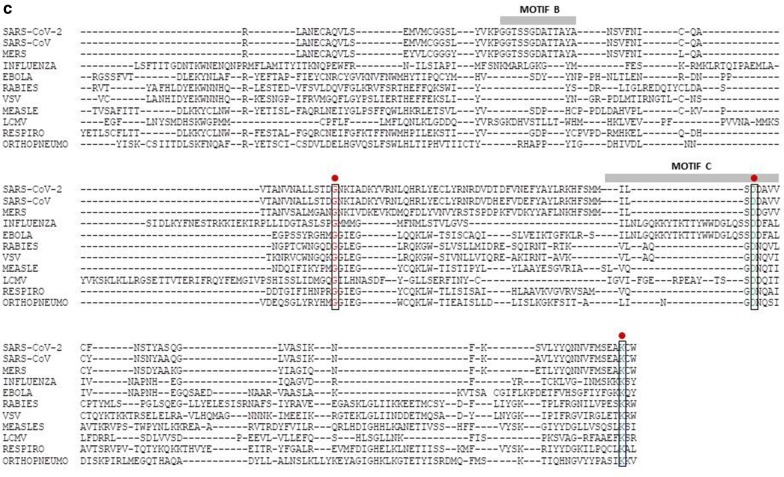
Fig. 1.
Alignment of amino acid sequences from RdRp of RNA viruses. a Sequences from positive-sense SARS-CoV-2, SARS-CoV and MERS viruses; b Sequences as in (a) with the addition of positive-sense viruses HCV, Dengue, West Nile, Zika and Yellow Fever viruses; c Sequences from positive-sense coronaviruses and negative-sense viruses Influenza, Ebola, Rabies, Vesicular Stomatitis virus, Measle, LCMV, Respirovirus and Orthopneumovirus. Red dots indicate 100% conservation of the indicated aa residues. Red asterisks indicate 100% conservation among HCV and human coronaviruses. Motif B and C of the RdRp are indicated
On the contrary, such homology is very poor when RdRp sequences from the three epidemic/pandemic coronaviruses are aligned with negative-sense RNA viruses, namely Ebola, Influenza, Rabies, Vesicular Stomatitis virus, Measle, LCMV, Respirovirus and Orthopneumovirus (Fig. 1c). As predicted, the DDxVV pattern in Motif C is not present in the negative-sense RNA viruses. The Influenza virus is the only one showing the two aspartates (DDxYY), while the other three show an asparagine substituting the second aspartate (DNxYY), suggesting the use manganese (Mn2+) instead of magnesium (Mg2+) as cofactor.
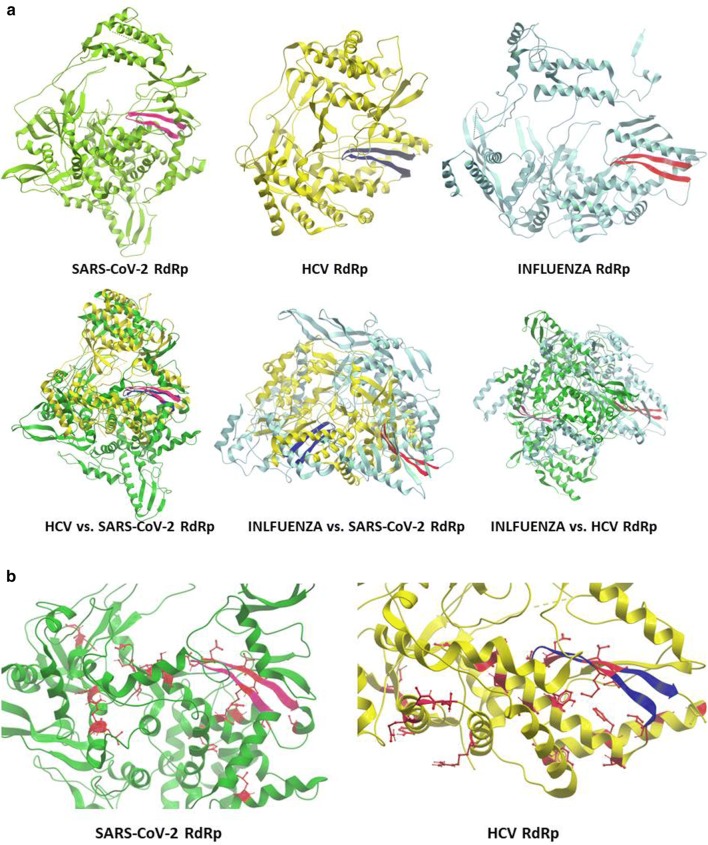
The structure modeling confirms the differences between the RdRps from positive-sense (HCV and SARS-CoV-2) and negative-sense (i.e. Influenza) RNA viruses. Indeed, considering the Motif C β-strand structure, only the alignment of RdRp structures from the two positive-sense RNA viruses results in the superimposition of the two Motifs (Fig. 2a). Furthermore, the highly conserved residues reside all in the inner part of the molecules which are in direct contact with the target genomic viral RNA and are responsible of the elongation of the nascent RNA molecule (Fig. 2b).
Fig. 2.
Structure modelling of the RdRps. a RdRp structures were derived from PDB databank: SARS-CoV-2 (6M71), Influenza virus (6QCT); HCV (3MWV). The whole molecules are presented independently or superimposed. b Zoomin of the SARS-CoV-2 and HCV core molecules highlighting in red color the conserved residues. Modelling was performed with Molsoft Browser
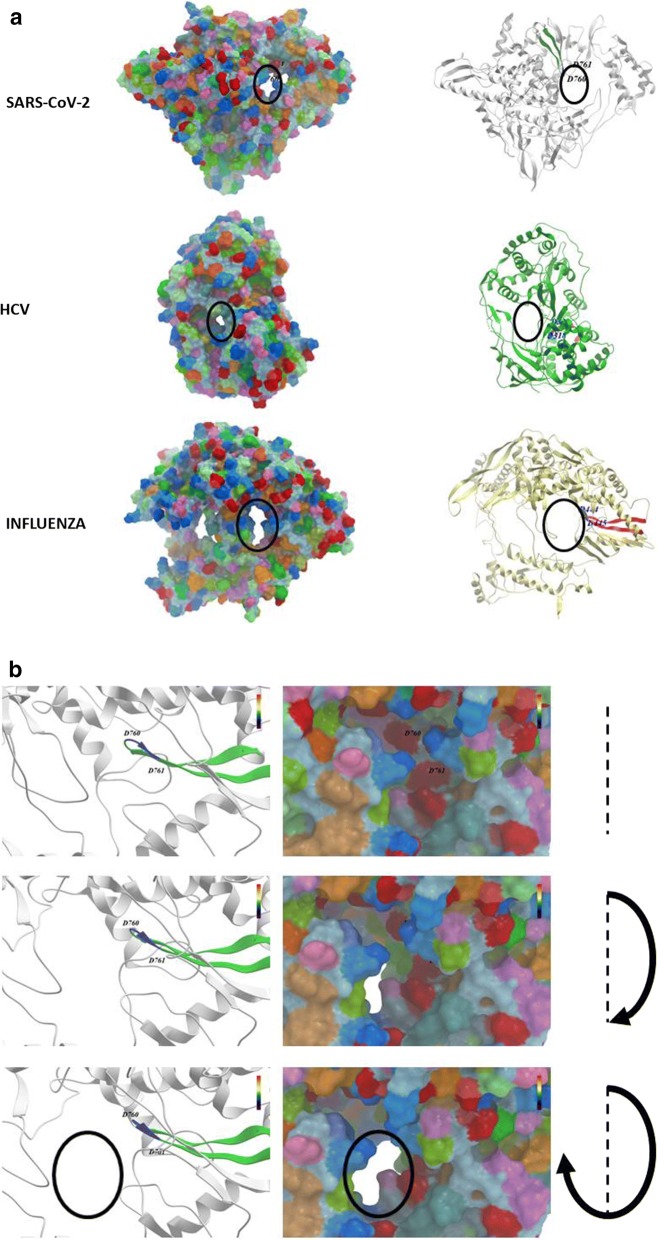
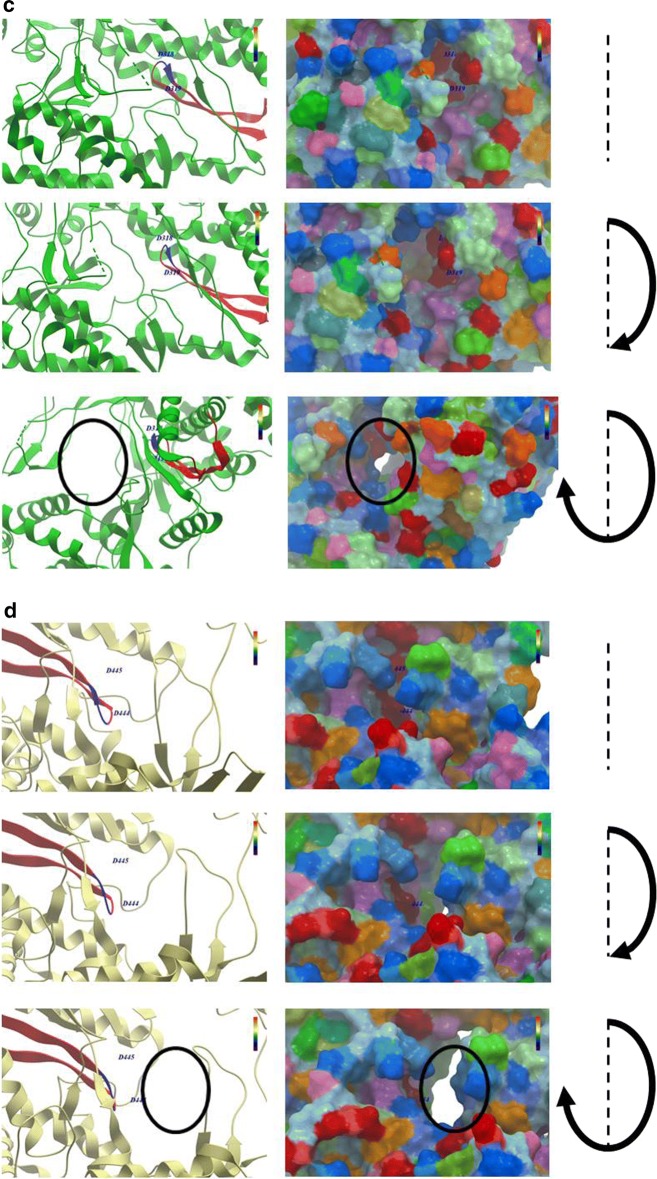
The molecular modelling which includes also the surface of molecules is a clearer representation of the overall structural difference between RdRps of positive-sense and negative-sense RNA viruses. In particular, this is clearly evidenced looking at the conformation of the core channel in which the Motif C β-strand-loop-β-strand structure protrudes (Fig. 3a–d).
Fig. 3.
Structure modelling of the RdRps. RdRp structures were derived from PDB databank and modelling was performed as described in Fig. 2. a External surface and internal structures of RdRps were compared. b SARS-CoV-2 RdRp; c HCV RdRp; d Influenza RdRp, zoomingin the channel in which the Motif C protrudes (black empty circle). Each of the latter three panels shows three different snapshots in a clockwise rotation
Specific drugs for SARS-CoV-2 should be developed to target the RdRp regions directly involved in the viral genome replication and transcription. Nucleoside analogs would be the most obvious class of drugs to be repositioned or de novo developed. In particular, few analogs are already available originally developed to target RdRps of other RNA viruses, such as Remdesivir (Ebola virus) [18], Favipiravir (Influenza virus) [19], NHC EIDD-1931 (broad spectrum) [20] and Sofosbuvir (Hepatitis C virus) [21]. Remdesivir and Favipiravir are currently evaluated in clinical trials to assess the efficacy in SARS-CoV-2 infected subjects (Remdesivir: NCT04292899; NCT04257656) (Favipiravir: NCT04310228). NHC EIDD-1931 has been shown to inhibit SARS-CoV-2 replication in vitro and in a pre-clinical animal model [22]. However, all three of them have been developed for negative-sense RNA viruses which show a significant difference in the RdRp sequence and structure compared to the positive-sense SARS-CoV-2 RNA virus. In this respect, Sofosbuvir could represent the optimal nucleoside analog to be repositioned to treatment of SARS-CoV-2 infection. Indeed, it has been developed for the positive-sense HCV RNA virus which shares high sequence and structural homology with SARS-CoV-2. Moreover, Sofosbuvir has been already shown to be effective for other positive-sense RNA viruses, namely Yellow Fever and Hepatitis A virus [23, 24].
In conclusion, as for all RNA viruses, the RdRp of the newly identified positive-sense human SARS-CoV-2 RNA virus represents the most optimal target for an antiviral drug. Linear amino acid sequence as well as molecule structure show the highest homology to RdRps of other positive-sense RNA viruses. Therefore, it is highly predictable that an antiviral developed for an RNA virus with a genome of the same polarity (i.e. Sofosbuvir for HCV) could have a higher inhibitory efficacy against the SARS-CoV-2, compared to those developed for negative-sense RNA viruses. Overall, the possibility of repositioning already available drugs will significantly accelerate the containment of the SARS-CoV-2 pandemic and, hopefully, of future pandemics due to other emerging zoonotic RNA viruses.
Acknowledgements
Not applicable.
Authors’ contributions
LB and FMB elaborated the concept; MT and MLT provided critical reading of the manuscript; LB drafted the manuscript and coordinated the whole project. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hui DS, Azhar I, Madani TA, Ntoumi F, Kock R, Dar O, et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogoch II, Watts A, Thomas-Bachli A, Huber C, Kraemer MUG, Khan K. Potential for global spread of a novel coronavirus from China. J Travel Med. 2020;27:taaa011. doi: 10.1093/jtm/taaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu B, Ge X, Wang LF, Shi Z. Bat origin of human coronaviruses. Virol J. 2015;12:221. doi: 10.1186/s12985-015-0422-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menachery VD, Yount BL, Jr, Debbink K, Agnihothram S, Gralinski LE, Plante JA, et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med. 2015;21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.To KK, Tsang OT, Chik-Yan YC, Chan KH, Wu TC, Chan JMC, et al. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 9.Reich E, Franklin RM, Shatkin AJ, Tatum EL. Effect of actinomycin D on cellular nucleic acid synthesis and virus production. Science. 1961;134:556–557. doi: 10.1126/science.134.3478.556. [DOI] [PubMed] [Google Scholar]
- 10.Harwig A, Landick R, Berkhout B. The battle of RNA synthesis: virus versus host. Viruses. 2017;9:309. doi: 10.3390/v9100309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jia H, Gong P. A structure–function diversity survey of the RNA-dependent RNA polymerases from the positive-strand RNA viruses. Front Microbiol. 2019;10:1945. doi: 10.3389/fmicb.2019.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacome R, Becerra A, de Ponce LS, Lazcano A. Structural analysis of monomeric RNA-dependent polymerases: evolutionary and therapeutic implications. PLoS ONE. 2015;10:e0139001. doi: 10.1371/journal.pone.0139001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ng KK, Arnold JJ, Cameron CE. Structure–function relationships among RNA-dependent RNA polymerases. Curr Top Microbiol Immunol. 2008;320:137–156. doi: 10.1007/978-3-540-75157-1_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lohmann V, Korner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/JVI.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vazquez AL, Alonso JM, Parra F. Mutation analysis of the GDD sequence motif of a calicivirus RNA-dependent RNA polymerase. J Virol. 2000;74:3888–3891. doi: 10.1128/JVI.74.8.3888-3891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan J, Vakharia VN, Tao YJ. The structure of a birnavirus polymerase reveals a distinct active site topology. Proc Natl Acad Sci USA. 2007;104:7385–7390. doi: 10.1073/pnas.0611599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warren TK, Jordan R, Lo MK, Ray AS, Mackman RL, Soloveva V, et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furuta Y, Komeno T, Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci. 2017;93:449–463. doi: 10.2183/pjab.93.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reynard O, Nguyen XN, Alazard-Dany N, Barateau V, Cimarelli A, Volchkov VE. Identification of a new ribonucleoside inhibitor of Ebola virus replication. Viruses. 2015;7:6233–6240. doi: 10.3390/v7122934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bourliere M, Pietri O. Hepatitis C virus therapy: no one will be left behind. Int J Antimicrob Agents. 2019;53:755–760. doi: 10.1016/j.ijantimicag.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 22.Sheahan TP, Sims AC, Zhou S, Graham RL, Pruijssers AJ, Agostini ML, et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci Transl Med. 2020 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendes EA, Pilger DRB, Santos Nastri ACS, Malta FM, Pascoalino BDS, Carneiro D’Albuquerque LA, et al. Sofosbuvir inhibits yellow fever virus in vitro and in patients with acute liver failure. Ann Hepatol. 2019;18:816–824. doi: 10.1016/j.aohep.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 24.Jiang W, Muhammad F, Ma P, Liu X, Long G. Sofosbuvir inhibits hepatitis A virus replication in vitro assessed by a cell-based fluorescent reporter system. Antivir Res. 2018;154:51–57. doi: 10.1016/j.antiviral.2018.04.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.