Abstract
Fowl cholera, caused by Pasteurella multocida, continues to be a challenge in meat-chicken-breeder operations and has emerged as a problem for free-range meat chickens. Here, using whole-genome sequencing (WGS) and phylogenomic analysis, we investigate isolate relatedness during outbreaks of fowl cholera on a free-range meat chicken farm over a 5-year period. Our genomic analysis revealed that while all outbreak isolates were sequence type (ST) 20, they could be separated into two distinct clades (clade 1 and clade 2) consistent with difference in their lipopolysaccharide (LPS) type. The isolates from the earlier outbreaks (clade 1) were carrying LPS type L3 while those from the more recent outbreaks (clade 2) were LPS type L1. Additionally, WGS data indicated high inter- and intra-chicken genetic diversity during a single outbreak. Furthermore, we demonstrate that while a killed autogenous vaccine carrying LPS type L3 had been successful in protecting against challenge from L3 isolates it might have driven the emergence of the closely related clade 2, against which the vaccine was ineffective. The genomic results also revealed a 14 bp deletion in the galactosyltransferase gene gatG in LPS type L3 isolates, which would result in producing a semi-truncated LPS in those isolates. In conclusion, our study clearly demonstrates the advantages of genomic analysis over the conventional PCR-based approaches in providing clear insights in terms of linkage of isolate within and between outbreaks. More importantly, it provides more detailed information than the multiplex PCR on the possible structure of outer LPS, which is very important in the case of strain selection for killed autogenous vaccines.
Keywords: fowl cholera, whole-genome sequencing, phylogeny, lipopolysaccharides, Pasteurella multocida
Data Summary
Genome sequence data generated in this study have been deposited to the National Center for Biotechnology Information (NCBI) under BioProject accession number PRJNA590306. Raw Illumina sequence read data have been deposited to the sequence read archive (SRA) under the accession numbers SRR10485145 to SRR10485219. The programs used to analyse raw sequence reads, for polymorphism discovery, and whole-genome-sequence-based phylogenies are available as described in Methods. The authors confirm all supporting data, code and protocols have been provided within the article or through supplementary data files.
Impact Statement.
Our data indicated high inter- and intra-chicken genetic diversity in P. multocida isolates during a single outbreak for the first time. Importantly, it demonstrates the benefit of genomics analysis in providing more detailed information on the possible structure of outer LPS, which is very important in the case of strain selection for killed autogenous vaccine. Using genomics data, we could demonstrate that although all the isolates belonged to the same clone, they were carrying two different LPS outer core biosynthesis loci type. In addition to that, our data revealed a major genetic mutation carried by all the LPS type 3 isolates, which would cause them to produce a truncated LPS outer core biosynthesis loci.
Introduction
The members of the family Pasteurellaceae consists of Gram-negative, obligate parasites that have adapted to living on the mucosal surfaces of many animal species [1]. Pasteurella multocida, a heterogeneous species from this family, is found in multiple hosts and causes a range of markedly different diseases that include haemorrhagic septicaemia in cattle and buffaloes, snuffles in rabbits, fowl cholera in poultry and atrophic rhinitis in pigs [2]. The organism can also cause septicaemia and sudden death in a range of captive wildlife such as fur seals [3] and squirrel gliders [4], thus confirming the capacity of this organism to cause similar disease in a variety of host species. Fowl cholera is a re-emerging problem in the free-range layer [5] and meat-chicken industries in Australia [6]. The disease manifestation can vary from per-acute to acute to chronic, with the recovered birds becoming a reservoir of the organism [7].
P. multocida strains are currently classified into 16 somatic (Heddleston) serovars (1 to 16) based on their lipopolysaccharide (LPS) antigens [8, 9]. An LPS multiplex PCR (LPS-mPCR) is currently in use to assign the 16 Heddleston serovars to one of the eight LPS genotypes (L1 to L8) based on their LPS outer core biosynthesis locus [10]. LPS type 3 (consisting of Heddleston serovars 3 and 4) has been recognized as the most prevalent LPS type associated with fowl cholera in Australia, followed by LPS types L1 (shared between Heddleston serovars 1 and 4) and L6 (represented by Heddleston serovars 10, 11, 12 and 15) [10, 11].
Recent work has demonstrated that killed whole cell vaccines give protection only against strains with identical or nearly identical LPS structures [12]. In Australia, multi-locus sequence typing (MLST), Heddleston serotyping, LPS genotyping and repetitive element PCR finger-printing (rep-PCR) are used for epidemiological investigations and also guide autogenous vaccinations in the poultry industry [11]. However, none of these techniques are discriminatory enough to provide the required insight into the LPS structure of isolates. Many of the fowl cholera associated isolates carrying LPS type L3 produce a truncated LPS, which suggests that the full-length LPS is not necessary to cause disease [13].
Whole-genome sequencing (WGS) and single nucleotide polymorphism (SNP) analysis of the core genome is now becoming the preferred method for investigations into isolate relatedness in nosocomial [14, 15] and food-borne outbreaks [16] as well as within host diversity of bacterial pathogens [17]. Here we report on our analysis of WGS data to examine the relatedness of P. multocida isolates associated with fowl cholera outbreaks on a free-range meat-chicken farm.
Methods
Farm and isolates
A total of 78 isolates from outbreaks of fowl cholera from 2009 to 2013 on a free-range chicken-meat farm were included in this study. Isolates were mostly obtained from the pericardial sac, heart blood, liver and bone marrow with multiple single colonies taken from the primary isolation plates of the bone marrow (Table S1, available in the online version of this article). The farm was a large complex containing a hatchery, processing plant, fertilizer composting facilities as well as several growing sheds. Each individual shed was operated on an all-in-all-out basis, while the farm was multi-age. Chickens were kept in a fully enclosed shed till day 19, when they were transferred to an open-access shed till processing.
An additional single isolate associated with fowl cholera (PM1439) for which its LPS outer structure has been fully analysed previously [18], was also included in the study. This isolate was obtained in 2010 from a chicken-meat breeder farm located in northeast New South Wales.
Genotyping
For initial genotyping, the repetitive extragenic palindromic PCR (rep-PCR) was performed as previously described [19], with the exception that the genomic DNA was prepared using the PrepMan Ultra Sample Preparation reagent.
Whole-genome sequencing
The bacterial isolates were revived and cultured aerobically overnight on 5 % sheep blood agar. DNeasy UltraClean Microbial Kit (Qiagen GmbH, Hilden, Germany) was used for DNA extractions according to the manufacturer’s instructions and sequenced on the Illumina NextSeq 500 platform (150 bp paired ends) by the Australian Centre for Ecogenomics at the University of Queensland (St Lucia QLD 4072, Australia). Raw Illumina sequencing data was quality filtered using Trimmomatic v0.36 [20], quality assessed using FastQC v0.11.5 [21] and de novo assembled using SPAdes v3.10.1 [22]. quast v2.3 was used to check the final assemblies’ metrics [23]. In silico MLST profiling was performed using mlst v2.8 (https://githib.com/tseemann/mlst) and the P. multocida RIRDC-MLST scheme [24, 25]. blastn v2.2.31+ was used to extract the relevant contigs containing the LPS outer core biosynthesis loci from the draft assemblies. Geneious R11 (https://www.geneious.com) was used for annotation of the gene in the LPS region and ClustalW v2.1 [26] for alignments and comparisons to the corresponding loci in P. multocida type strain PM70 (NC_002663.1) in the case of LPS type 3 carrying isolates and the P. multocida Heddleston serovar 1 reference strain X73 (HQ873311) for those carrying LPS type 1.
As well, the amino acid sequence of the recently identified phosphoethanolamine (PEtn) transferase (PetG), responsible for the addition of the two PEtn molecules to the outer core galactose in strain X73 [27], was used as the query protein sequence using blastx against the draft assemblies of the LPS type 1 carrying isolates.
Nesoni v0.132 (https://github.com/Victorian-Bioinformatics-Consortium/nesoni) was used for read mapping of the trimmed paired end reads against PM70 and X73 for LPS type 3 and LPS type 1 carrying isolates, respectively. Artemis v16.0.0 [28] was used to check the read pile ups.
Parsnp v1.2 [29] was used for core genome alignment and SNP calling in the draft genome assemblies. Phylogenetic trees were then constructed with RAxML x8.2.9 using the core genome SNP alignment after removal of predicted recombination sites by Gubbins v2.1.0 [30]. A general-time reversible nucleotide substitution model with a GAMMA correction for site variation was used for tree construction (bootstrap 1000 with Lewis ascertainment correction). Phylogenetic trees were visualized using FigTree v1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) and Phandango [31]. PHYLOViZ v2.0 was used to build a minimum spanning tree from core genome SNPs of the 2009 isolates [32].
Results
Outbreaks
The details of the farm, the 2009 to 2012 samplings and the association of the 2011 and 2012 outbreaks with feral cats have been previously reported [6]. The first outbreak of fowl cholera occurred in 2009 with high mortality happening as early as 30 days of age. In the 2009 outbreak, five dead birds were sent to the University of Queensland from which 29 P . multocida isolates were obtained and included in the current study (isolates PM1330 to PM1358). At the same time, six dead birds from three different sheds (sheds 4, 11 and 15) were sent to the vaccine production company (two birds from each shed), from which isolates PM1766, PM1767 and PM1768 were kept (one isolate per shed). The farm has been using a single strain autogenous alum hydroxide gel vaccine subcutaneously at day of hatch (PM1766) since 2009. However, while initially successful, the vaccination program did not fully protect. Fowl cholera control was achieved after improvements in biosecurity, improved management and a change of the coccidiosis vaccination program [6]. However, flocks again suffered from fowl cholera outbreaks in 2011 and 2012 with an average of 55 % mortality to processing [6]. In 2013, fowl cholera was still affecting some sheds with chickens as young as 24 days of age being affected. The observed mortality in the 2013 outbreak ranged from 6.7 % in 27-day-old chickens to 15.9 % in 41-day-old chickens. A total of 18 isolates from the 2013 outbreaks were included in this study – an original five isolates from the 24-week-old chickens (PM1707 to PM1711) together with 13 isolates from the reoccurrence of the disease in the same flocks (PM1754 to PM1765 and PM1779) (Table 1).
Table 1.
Meat-chicken farm outbreaks and isolates
|
Year |
Outbreak isolate(s) |
No. of isolates |
No. of chickens |
Sequence type |
LPS type |
Note |
|---|---|---|---|---|---|---|
|
2009 |
PM1330 to PM1358, PM1766*, PM1767, PM1768 |
29 |
5 |
20 |
L3 |
gatG 14 bp deletion |
|
August 2010 |
PM1422 to PM1430 |
9 |
6 |
20 |
L3 |
gatG 14 bp deletion |
|
June 2012 |
PM1614, PM1615 |
2 |
na |
20 |
L1 |
None |
|
July 2012 |
PM1634 to PM1651† |
17(19)‡ |
4 |
20 |
L1 |
None |
|
August 2013 |
PM1707 to PM1711 |
5 |
na |
20 |
L1 |
None |
|
October 2013 |
PM1754 to PM1765† PM1799* |
11(12)‡ |
|
20 |
L1 |
None |
*Used in autogenous vaccine. na, data not available.
†Reads from isolates PM1642, PM1648 and PM1755 were removed from the analysis due to contamination.
‡Numbers in the bracket indicate the original number of isolates.
Genotyping and phylogenomic analysis of P. multocida isolates
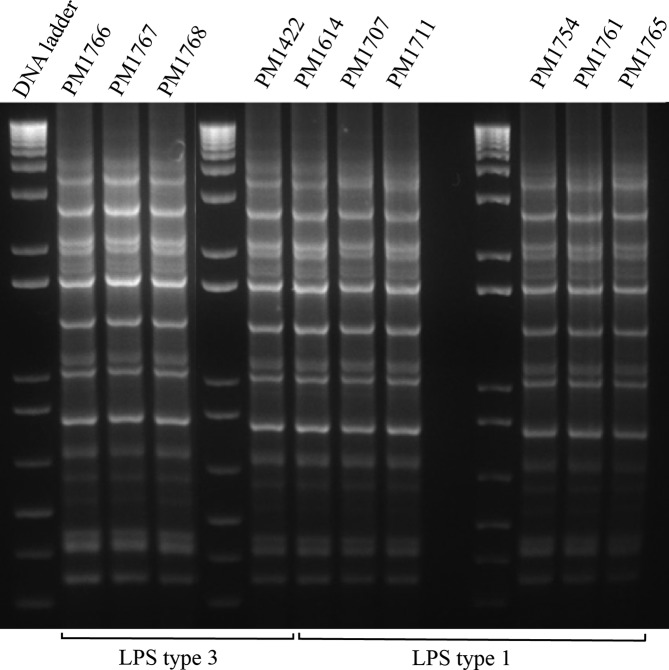
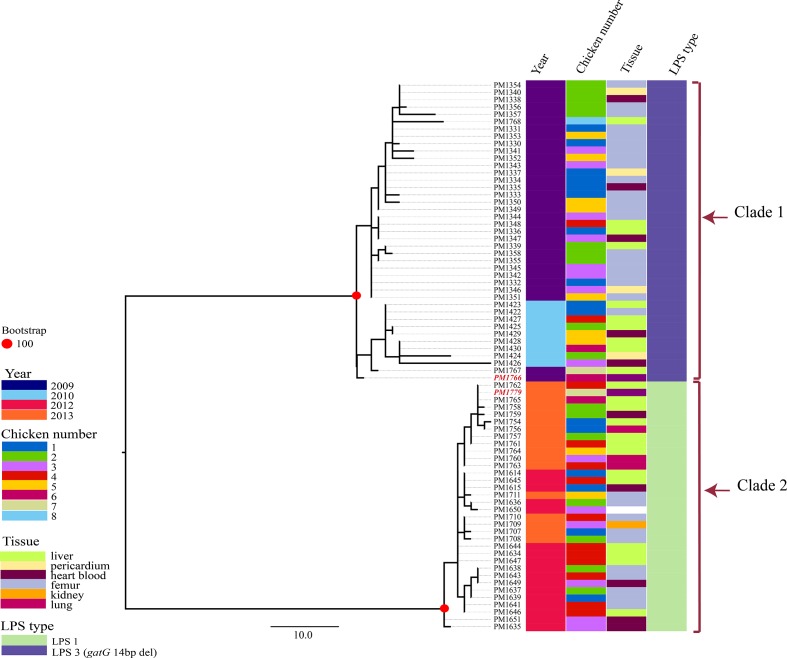
The in silico MLST revealed all isolates belonged to ST20 (Table 1). However, the in silico LPS typing resulted in recognition of two different LPS types, L1 and L3. While the 2009 and 2010 isolates together with the vaccine strain PM1766 carried LPS type L3, isolates from the 2012–2013 outbreaks carried LPS type L1. The rep-PCR typing, however, agreed with the MLST results in recognition of only one genetic type across the years in this farm (Fig. 1). Between zero to 78 non-recombinant SNPs were identified between the core genomes of all isolates. The maximum likelihood analysis separated the isolates into two clades (clade 1 and clade 2), in agreement with their LPS types (L3 and L1, respectively) (Fig. 2).
Fig. 1.
Rep-PCR analysis of the P. multocida isolates obtained across the year. The finger printing shows the isolates carrying LPS type 1 and 3 share the same DNA finger print.
Fig. 2.
Maximum likelihood analysis of the core genome SNPs of the isolates from the studied meat-chicken farm. Phylogenomic analysis of all the 75 isolates obtained during the years in this farm. A total of 116 core genome SNPs was identified between the isolates. The tree is rooted from midpoint. Scale bar shows the number of SNPs. Isolates used in the killed autogenous vaccine are highlighted by red bold italic font.
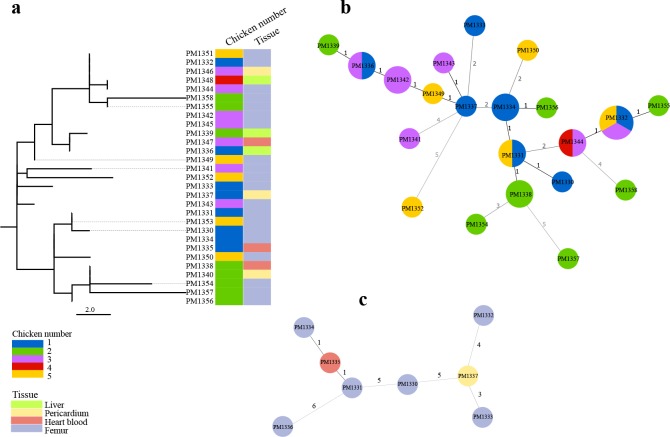
The 2009 outbreak was further investigated for both inter-host and intra-host genetic diversity of the P. multocida isolates. For the inter-host diversity investigation, a total of 29 isolates including five single colony picks from the original femur cultures of each chicken and also single colonies from internal organs such as liver, heart blood and pericardium were included. The pairwise comparisons between these isolates identified 0 to 12 SNPs, with the highest number found between isolates PM1352 and PM1358 obtained from femurs of chicken numbers 5 and 2, respectively (Fig. 3a). Some isolates obtained from different chickens appeared to be identical at the core genome level, e.g. isolates PM1332 and PM1351 and PM1346, with the first two isolates obtained from femur, and the last one from pericardium of three different chickens (Fig. 3b). This investigation also showed that within chicken diversity in chickens 1, 2, 3 and 5 was comparable or greater than between chicken diversity. For example, the core genomes from the eight isolates obtained from chicken one differed by between 1 and 11 SNPs (mean 6.1±2.5) (Fig. 3c).
Fig. 3.
Isolate relatedness in the 2009 outbreak. (a) Maximum likelihood tree of the core genome SNPs for the 29 isolates obtained from five chickens. A total of 30 core genome SNPs between these 29 isolates was identified. The tree is rooted from midpoint. Scale bar shows the number of SNPs. (b) Minimum spanning tree constructed from the total of 210 SNPs identified between the core genomes of the 29 isolates. Isolates with identical core genome: PM1336 and PM1347, PM1331 and PM1353, PM1344 and PM1348, PM1332 with PM1346 and PM1351. (c) Minimum spanning tree when isolates from bird one were compared to each other only.
No SNP was identified in the LPS outer core biosynthesis loci between the LPS type 3 isolates or between the LPS type 1 isolates. Two of the isolates from each group (PM1333 and PM1352 in the LPS type 3 isolates and PM1760 and PM17644 in the LPS type 1 isolates) had their LPS outer core biosynthesis loci over 2 contigs (broken natC gene and broken pcgD gene, respectively). A review of the read pile ups of these four isolates as well as from those with unbroken LPS confirmed a very low read coverage in those regions in general.
The LPS type 3 isolates carried 36 nucleotide differences in the 6176 bp region of LPS outer core biosynthesis loci when compared with that from PM70, including a 14 bp deletion in their gatG gene from position 491 to 505, as well as a one bp insertion in their natC gene (313_314insA). The LPS type 1 carrying isolates were also identical to each other in their 5573 bp LPS outer core biosynthesis loci with all carrying only two synonymous substitutions compared to that from Heddleston serovar 1 type strain (G666T in gatA and C525T in the pcgC gene).
In order to investigate if the LPS type 1 carrying isolates had the potential of a decorated outer core galactose with PEtn, the draft genome assemblies were subjected to bioinformatic examination. The 525 amino acid long PetG sequence was used to search against the draft genome assemblies of LPS type 1 isolates using blastx. However, only one region with similar amino acid size was identified where it shared only 67 % amino acid identity with the PEtn transferase responsible for the addition of PEtn to the outer core glucose of the Heddleston serovar 1 reference strain.
Discussion
P. multocida is a rare example of a multi-host pathogen within the family Pasteurellaceae [1]. The aim of the current study was to provide some insight into the natural genetic diversity occurring within an outbreak of fowl cholera. Repeated outbreaks of fowl cholera on one farm can be simply the re-emergence of the very same strain (from carrier birds or a stable environmental source) or the introduction of new strains (replacement stock or a new environmental source) [5, 6].
MLST has been used to understand the epidemiology of fowl cholera outbreaks in Australia [6] and overseas [33] allowing a recognition of when strains have been associated with repeated outbreaks across time and when new strains have been introduced. Our results in this study, however, showed that while all the isolates in this farm belonged to ST20, they belonged to two separate, albeit closely related, clades when comparing their whole-genome SNPs. A relationship was also found between the clades and their capacity to carry LPS type L3 or L1 (clade 1 and 2, respectively) highlighting the role of horizontal gene transfer in the evolution of the LPS outer core biosynthesis loci, as reported previously [13]. In addition, the results of our study suggest that vaccination with isolates from clade 1 carrying LPS type L3 has been successful in halting the initial outbreak. However, the vaccine has not been sufficient to halt subsequent outbreaks. As even minor changes of the LPS structure has been shown to cause a loss of vaccine cross-protection provided by killed vaccines [12], the use of the killed vaccine with LPS type 3 might even have driven the selection for P. multocida with LPS type 1.
Our results also show that the currently in-use DNA finger-printing technique, rep-PCR, can fail to identify the variations in the LPS outer core biosynthesis loci as isolates with LPS type L1 or L3 produced the same rep-PCR pattern. Hence, rep-PCR finger-printing can be misleading in guiding vaccine selection if used as the only tool, as it is not always discriminatory enough to pick the difference in the LPS outer core biosynthesis loci of isolates. This lack of discrimination capacity by the typing schemes in-use, meant that on-going outbreaks of fowl cholera were not understood to be due to an antigenically very different challenge strain.
Within host diversity of bacterial pathogens has been reported in a number of important bacterial pathogens such as Staphylococcus aureus and Pseudomonas aeruginosa [17, 34]. Our laboratory has previously reported within-host variations in the core genome of P. multocida isolates obtained from cases of pasteurellosis in captive pinnipeds [3]. In our current work, five single colony picks from the primary isolation plates of femur bone marrow of several diseased chickens across different years revealed population diversity even in a single organ. Our analysis suggests that WGS from a single colony would not be sufficient to accurately determine transmission pathways between different chickens. Despite high diversity within individual chickens, some isolates from different chickens were nearly identical at the core genome level, indicating transmission from a common source, which carried a mixed population or repeated cross transmission between infected chickens. Microbiome analysis of tissues has the potential to offer greater insight into the diversity of co-infecting organisms within a fowl cholera outbreak. Although most standard microbiome analyses do not discriminate between closely related strains of the same species, it is feasible to resolve strain heterogeneity within metagenome data [35, 36]. These methods have the potential to capture, from a single sample, the P. multicide diversity defined by our conventional multiple isolate examination.
Both of the LPS outer core biosynthesis loci L1 and L3 start with a heptosyltransferase coding gene for the addition of the first outer core sugar, being a heptose, followed by a gene coding for a bifunctional glycosyltransferase [13]. The LPS type L3 consists of a total of six glycosyltransferase coding genes named htpE, gctC, gatF, natB, gatG and natC [18]. Previous studies on the field isolates of P. multocida , have revealed that a total of six different LPS outer core structures could be produced by the L3 bearing isolates with some being the result of major genetic mutation in the linked transferase gene [18]. Our analysis of the genomes of LPS type L3 carrying isolates revealed that all carried a 14 bp deletion in their gatG gene in contrast to the P. multocida type strain PM70, which has a fully functional gatG. This genetic mutation has been reported in previously studied P. multocida ST20 isolate PM1439 [18]. Previous studies have revealed that the gatG gene codes for a galactosyltransferase, which adds the second galactose (Gal II) to the fourth position of the first galactose (Gal I) [18]. The structural analysis of the LPS from isolate PM1439 as well as one of the strains from this farm (PM1422) has previously revealed their outer core finishes at the first galactose and consists of Hep.[Glu].Glu.Gal [12, 18]. This confirms the role of the 14 bp deletion in gatG, in the LPS truncation beyond Gal I. Since the isolates carrying LPS type L3 in the current study had LPS outer core biosynthesis loci that were identical to that of previously studied isolate PM1422 and PM1439, it is highly likely that the LPS type L3 carrying isolates in this farm all had an LPS outer structure finishing at Gal I. Our laboratory has previously reported an insertion in the position 777 of the gatG gene of P. multocida isolates associated with pasteurellosis in captive fur seals resulting in a CDS frame shift [3], which most likely results in the same LPS outer structure. These results indicate that there may be a strong selective advantage in truncating the LPS at Gal I.
The LPS outer core biosynthesis locus L1 consists of six genes including four genes involved in biosynthesis of phosphocoline (PCho) (pcgD, pcgA, pcgB and pcgC) as well as two glycosyltransferases htpE and gatA (for the addition of the outer core heptose and a bifunctional galactosyltransferase which adds Gal I and Gal II to the heptose, respectively) [37]. This LPS type is shared by the Heddleston serovars 1 and 14 serovar reference strains. The structure analysis of the Heddleston serovar 1 reference strain X-73 has revealed that the LPS outer core of this strain consists of one heptose, with two galactoses attached to its positions 4 and 6 [37]. One PCho and one PEtn molecule are also attached to the positions 3 and 6 of both galactose molecules, respectively [13]. It has been suggested previously that fowl cholera isolates carrying LPS type L1, mostly have an outer structure similar to that of VP161 [2PCho, 2Hex, Hep], which lacks the two PEtn found in X_73 [10]. A number of PEtn transferases has been identified in P. multocida with some responsible for adding PEtn to the inner core, lipid A, as well as outer core biosynthesis loci galactose [27]. The gene coding for the addition of the PEtn molecule has been recognized recently and named petG [27]. However, no protein sequence with high similarity to that of PetG could be identified in the LPS type 1 carrying P. multocida in our study. The fact that both strains VP161 and X_73 are overseas isolates, this finding might suggest a difference between Australian isolates carrying LPS type L1 and overseas strains in terms of the presence of particular PEtn transferase genes. The extracted PEtn transferase region from the LPS type 1 isolates in our study showed high degree of amino acid similarity with that from lipooligosaccharide phosphoethanolamine transferase A (LptA) (accession number WP_064968839.1) using blastp function in NCBI. However, full understanding of the diversity in the phosphoethanolamine transferase genes of P. multocida was beyond the scope of the current project.
In conclusion, whole-genome sequencing and core-genome SNP analysis is a suitable method for replacing PCR-based techniques in examining outbreaks of fowl cholera. As well, genomic analysis provides clearer insights into the associations of the P. multocida isolates within and between outbreaks as well as within a single chicken during an outbreak. The occurrence of within-host, as well as within-outbreak, diversity of fowl cholera-associated P. multocida isolates is quite a novel finding that requires further studies to fully understand the implications for prevention and control programs. WGS data is routinely used now in human bacterial pathogen research to investigate strain relatedness, monitor the emergence of bacterial antibiotic resistance and/or bacterial virulence factors [38], e.g. in food-borne disease outbreaks [39]. Since infectious disease outbreaks in animals can follow different epidemiology scenarios, the issue of flexible WGS-based case definitions of a ‘clonal outbreak’ that has been suggested for food-borne outbreaks [39] needs to be explored for veterinary disease outbreaks investigated by WGS. Our data strongly supports and expands the original observations of Singh et al. [6] – effective fowl cholera vaccination programs require effective strain selection for killed vaccines as well as other management and biosecurity-related strategies. Future efforts are needed to characterize P. multicida diversity by WGS of isolates across a larger geographical area. Importantly, the application of microbiome analysis to both chicken tissues and the surrounding environment are likely to be relevant and practical strategies in the near future for investigation of strain diversity and particular genes such as LPS outer core biosynthesis loci.
Data Bibliography
1. Omaleki L, Blackall PJ, Cuddihy T, Beatson SA, Forde BM, Turni C. All the sequencing data used in this study were generated within the study, NCBI BioProject number PRJNA590306 (2019).
Supplementary Data
Funding information
This project was co-funded by AgriFutures Australia and Australian Eggs.
Acknowledgements
We thank the farm management and the veterinary consultants linked to the study farm for their assistance and expertise.
Author contributions
The study was conceptualized by L.O., P.J.B., S.A.B and C.T. Whole-genome analysis was carried out by L.O., T.C. and B.M.F. The manuscript was drafted by L.O., and reviewed and edited by P.J.B., T.C., S.A.B., B.M.F. and C.T. All authors have read and approved the final version of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This work was undertaken on stored bacterial cultures and no samples were collected from any birds during this study
Footnotes
Abbreviations: Gal, galactose; Glu, glucose; Hep, heptose; LPS, lipopolysaccharide; LptA, lipopolysaccharide phosphoethanolamine transferase A; MLST, multi locus sequence typing; mPCR, multiplex PCR; NCBI, National Center for Biotechnology Information; PCho, phosphocoline; PCR, polymerase chain reaction; PEtn, phosphoethanolamine; Rep-PCR, repetitive extragenic palindromic PCR; SNP, single nucleotide polymorphism; SRA, sequence read archive; ST, sequence type; WGS, whole genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. A supplementary table is available with the online version of this article.
References
- 1.Blackall PJ, Norskov-Lauritsen N. Pasteurellaceae - the view from the diagnostic laboratory. In: Kuhnerts P, Christensen H, editors. Pasteurellaceae: Biology, Genomics and Molecular Aspects. Norwich, UK: Horizon Scientific Press; 2008. pp. 227–260. [Google Scholar]
- 2.Hunt ML, Adler B, Townsend KM. The molecular biology of Pasteurella multocida . Vet Microbiol. 2000;72:3–25. doi: 10.1016/S0378-1135(99)00183-2. [DOI] [PubMed] [Google Scholar]
- 3.Crawford RL, Blyde D, Blackall PJ, Forde BM, Beatson SA, et al. Novel insights into pasteurellosis in captive pinnipeds. Vet Microbiol. 2019;231:232–237. doi: 10.1016/j.vetmic.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Omaleki L, Beatson SA, Thomrongsuwannakij T, Blackall PJ, Buller N, et al. Phase variation in latB gene associated with a fatal Pasteurella multocida outbreak in captive squirrel gliders . 2019 doi: 10.1016/j.vetmic.2020.108612. [DOI] [PubMed] [Google Scholar]
- 5.Singh R, Blackall PJ, Remington B, Turni C. Studies on the presence and persistence of Pasteurella multocida serovars and genotypes in fowl cholera outbreaks. Avian Pathol. 2013;42:581–585. doi: 10.1080/03079457.2013.854861. [DOI] [PubMed] [Google Scholar]
- 6.Singh R, Remington B, Blackall P, Turni C. Epidemiology of fowl cholera in free range broilers. Avian Dis. 2014;58:124–128. doi: 10.1637/10656-090313-Reg.1. [DOI] [PubMed] [Google Scholar]
- 7.Christensen JP, Dietz HH, Bisgaard M. Phenotypic and genotypic characters of isolates of Pasteurella multocida obtained from back-yard poultry and from two outbreaks of avian cholera in avifauna in Denmark. Avian Pathol. 1998;27:373–381. doi: 10.1080/03079459808419354. [DOI] [PubMed] [Google Scholar]
- 8.Brogden KA, Rhoades KR, Heddleston KL. A new serotype of Pasteurella multocida associated with fowl cholera. Avian Dis. 1978;22:185–-90. doi: 10.2307/1589525. [DOI] [PubMed] [Google Scholar]
- 9.Heddleston KL, Gallagher JE, Rebers PA. Fowl cholera: gel diffusion precipitin test for serotyping Pasteruella multocida from avian species. Avian Dis. 1972;16:925–936. doi: 10.2307/1588773. [DOI] [PubMed] [Google Scholar]
- 10.Harper M, John M, Turni C, Edmunds M, St Michael F, et al. Development of a rapid multiplex PCR assay to genotype Pasteurella multocida strains by use of the lipopolysaccharide outer core biosynthesis locus. J Clin Microbiol. 2015;53:477–485. doi: 10.1128/JCM.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turni C, Singh R, Blackall PJ. Genotypic diversity of Pasteurella multocida isolates from pigs and poultry in Australia. Aust Vet J. 2018;96:390–394. doi: 10.1111/avj.12748. [DOI] [PubMed] [Google Scholar]
- 12.Harper M, John M, Edmunds M, Wright A, Ford M, et al. Protective efficacy afforded by live Pasteurella multocida vaccines in chickens is independent of lipopolysaccharide outer core structure. Vaccine. 2016;34:1696–1703. doi: 10.1016/j.vaccine.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 13.Harper M, Boyce JD. The properties of Pasteurella multocida lipopolysaccharide. Toxins. 2017;9:E254. doi: 10.3390/toxins9080254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartley PB, Ben Zakour NL, Stanton-Cook M, Muguli R, Prado L, et al. Hospital-wide eradication of a nosocomial Legionella pneumophila serogroup 1 outbreak. Clin Infect Dis. 2016;62:273–279. doi: 10.1093/cid/civ870. [DOI] [PubMed] [Google Scholar]
- 15.Quainoo S, Coolen JPM, van Hijum SAFT, Huynen MA, Melchers WJG, et al. Whole-Genome sequencing of bacterial pathogens: the future of nosocomial outbreak analysis. Clin Microbiol Rev. 2017;30:1015–1063. doi: 10.1128/CMR.00016-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pijnacker R, Dallman TJ, Tijsma ASL, Hawkins G, Larkin L, et al. An international outbreak of Salmonella enterica serotype Enteritidis linked to eggs from Poland: a microbiological and epidemiological study. Lancet Infect Dis. 2019;19:778–786. doi: 10.1016/S1473-3099(19)30047-7. [DOI] [PubMed] [Google Scholar]
- 17.Stanczak-Mrozek KI, Manne A, Knight GM, Gould K, Witney AA, et al. Within-Host diversity of MRSA antimicrobial resistances. J Antimicrob Chemother. 2015;70:2191–2198. doi: 10.1093/jac/dkv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harper M, St Michael F, John M, Vinogradov E, Steen JA, et al. Pasteurella multocida Heddleston serovar 3 and 4 strains share a common lipopolysaccharide biosynthesis locus but display both inter- and intrastrain lipopolysaccharide heterogeneity. J Bacteriol. 2013;195:4854–4864. doi: 10.1128/JB.00779-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gunawardana GA, Townsend KM, Frost AJ. Molecular characterisation of avian Pasteurella multocida isolates from Australia and Vietnam by REP-PCR and PFGE. Vet Microbiol. 2000;72:97–109. doi: 10.1016/S0378-1135(99)00191-1. [DOI] [PubMed] [Google Scholar]
- 20.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andrews S. FastQC: a quality control tool for high throughput sequence data 2010. [Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/]
- 22.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolley KA, Chan M-S, Maiden MCJ. mlstdbNet - distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subaaharan S, Blackall LL, Blackall PJ. Development of a multi-locus sequence typing scheme for avian isolates of Pasteurella multocida . Vet Microbiol. 2010;141:354–361. doi: 10.1016/j.vetmic.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 26.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 27.Harper M, Wright A, St Michael F, Li J, Deveson Lucas D, et al. Characterization of two novel lipopolysaccharide phosphoethanolamine transferases in Pasteurella multocida and their role in resistance to Cathelicidin-2. Infect Immun. 2017;85 doi: 10.1128/IAI.00557-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hadfield J, Croucher NJ, Goater RJ, Abudahab K, Aanensen DM, et al. Phandango: an interactive viewer for bacterial population genomics. Bioinformatics. 2018;34:292–293. doi: 10.1093/bioinformatics/btx610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nascimento M, Sousa A, Ramirez M, Francisco AP, Carriço JA, et al. PHYLOViZ 2.0: providing scalable data integration and visualization for multiple phylogenetic inference methods. Bioinformatics. 2017;33:128–129. doi: 10.1093/bioinformatics/btw582. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Zhu J, Lu C, Wu B, Liu D, et al. Evidence of circulation of an epidemic strain of Pasteurella multocida in Jiangsu, China by multi-locus sequence typing (MLST) Infect Genet Evol. 2013;20:34–38. doi: 10.1016/j.meegid.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 34.Sherrard LJ, Tai AS, Wee BA, Ramsay KA, Kidd TJ, et al. Within-host whole genome analysis of an antibiotic resistant Pseudomonas aeruginosa strain sub-type in cystic fibrosis. PLoS One. 2017;12:e0172179. doi: 10.1371/journal.pone.0172179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo C, Knight R, Siljander H, Knip M, Xavier RJ, et al. Constrains identifies microbial strains in metagenomic datasets. Nat Biotechnol. 2015;33:1045–1052. doi: 10.1038/nbt.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oh J, Byrd AL, Deming C, Conlan S, Kong HH, Barnabas B, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harper M, St Michael F, John M, Vinogradov E, Adler B, et al. Pasteurella multocida Heddleston serovars 1 and 14 express different lipopolysaccharide structures but share the same lipopolysaccharide biosynthesis outer core locus. Vet Microbiol. 2011;150:289–296. doi: 10.1016/j.vetmic.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 38.Walker MJ, Beatson SA, Outbreaks O. Epidemiology outsmarting outbreaks. Science. 2012;338:1161–1162. doi: 10.1126/science.1232327. [DOI] [PubMed] [Google Scholar]
- 39.Brown E, Dessai U, McGarry S, Gerner-Smidt P. Use of whole-genome sequencing for food safety and public health in the United States. Foodborne Pathog Dis. 2019;16:441–450. doi: 10.1089/fpd.2019.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.