Abstract
Coniothyrium minitans is a mycoparasite of the notorious plant pathogen Sclerotinia sclerotiorum. To further understand the parasitism of C. minitans, we assembled and analysed its genome and performed transcriptome analyses. The genome of C. minitans strain ZS-1 was assembled into 350 scaffolds and had a size of 39.8 Mb. A total of 11 437 predicted genes and proteins were annotated, and 30.8 % of the blast hits matched proteins encoded by another member of the Pleosporales, Paraphaeosphaeria sporulosa, a worldwide soilborne fungus with biocontrol ability. The transcriptome of strain ZS-1 during the early interaction with S. sclerotiorum at 0, 4 and 12 h was analysed. The detected expressed genes were involved in responses to host defenses, including cell-wall-degrading enzymes, transporters, secretory proteins and secondary metabolite productions. Seventeen differentially expressed genes (DEGs) of fungal cell-wall-degrading enzymes (FCWDs) were up-regulated during parasitism, with only one down-regulated. Most of the monocarboxylate transporter genes of the major facilitator superfamily and all the detected ABC transporters, especially the heavy metal transporters, were significantly up-regulated. Approximately 8 % of the 11 437 proteins in C. minitans were predicted to be secretory proteins with catalytic activity. In the molecular function category, hydrolase activity, peptidase activity and serine hydrolase activity were enriched. Most genes involved in serine hydrolase activity were significantly up-regulated. This genomic analysis and genome-wide expression study demonstrates that the mycoparasitism process of C. minitans is complex and a broad range of proteins are deployed by C. minitans to successfully invade its host. Our study provides insights into the mechanisms of the mycoparasitism between C. minitans and S. sclerotiorum and identifies potential secondary metabolites from C. minitans for application as a biocontrol agent.
Keywords: Coniothyrium minitans, Sclerotinia sclerotiorum, mycoparasitism, biological control, genome, differentially expressed genes (DEGs)
Data Summary
1. The assembled genome of Coniothyrium minitans (Taxonomy ID: 565426) strain ZS-1 has been deposited at DDBJ/ENA/GenBank under the accession VFEO00000000. The version described in this paper is version VFEO01000000.
2. The sequencing raw data for genome assembly of C. minitans is available via the NCBI Sequence Read Archive with BioProject: PRJNA548470, BioSample: SAMN12034817. The SRA accession number of SRR9290694 is for the 500-bp-long sequencing library and the number SRR9290695 is for the 6-Kbp-long mate-pair library on Illumina platforms; the SRA accession of SRR9290696 is for 20-Kbp-long sequencing library on PacBio RS II platform.
3. The mapped RNA-seq data files on the assembled genome of C. minitans from the mixed interaction RNA-seq of C. minitans with Sclerotinia sclerotiorum at 0, 4 and 12 h p.i. were uploaded into SRA with accessions of SRR9301108, SRR9301109, and SRR9301107 under biosample accession with SAMN12046022, respectively.
4. The 15 RNA-seq data files corresponding to five stages of the growth and development of C. minitans ZS-1 were uploaded into SRA with accessions of SRR9988701 to SRR9988715 under biosample accession with SAMN12588607.
Impact Statement.
Coniothyrium minitans is a specific and excellent biocontrol mycoparasite of the worldwideplant pathogen, Sclerotinia sclerotiorum. We used a combination of Illumina and single-molecule real-time (SMRT) sequencing to generate the sequences for the de novo assembly of the C. minitans genome. Comparative analysis of this genome with those of other plant pathogens (i.e. Sclerotinia spp. and Botrytis spp.) and biocontrol fungi (i.e. Trichoderma spp.) showed shared and unique characteristics with respect to genes for cell-wall-degrading enzymes, major facilitator superfamily (MFS), ABC transporter proteins, secretory proteins and secondary metabolite biosynthesis gene clusters. Dual RNA-seq analysis of C. minitans and co-culturing with its host fungus S. sclerotiorum demonstrated that cell-wall-degrading enzymes, transporters, secretory proteins (i.e. effectors) and secondary metabolites participate in the process of mycoparasitism of S. sclerotiorum at the early stage. The results of these genome analyses and RNA-seq surveys during mycoparasitism by C. minitans provide an understanding of the mycoparasitic mechanism and can further promote research on this antifungal agent.
INTRODUCTION
Coniothyrium minitans is a homotypic synonym of Paraconiothyrium minitans or Paraphaeosphaeria minitans, belongs to the phylum Ascomycota, the class Dothideomycetes, the order Pleosporales, the family Didymosphaeriaceae, and is more properly placed in the genus Paraphaeosphaeria (according to Taxbrowser on NCBI website). It is congeneric with Paraphaeosphaeria sporulosa, a worldwide soilborne fungus with biocontrol abililty. C. minitans is a sclerotial mycoparasite distributed worldwide in soil [1–3]. Its remarkable biocontrol ability has attracted great attention since it was first isolated and identified in 1947 by Campbell [4]. C. minitans shares a similar optimal growth temperature (20 °C) with its host S. sclerotiorum [2, 4, 5]. It can parasitize the sclerotia of Sclerotinia spp. and produce antifungal substances that inhibit host growth [5–10], and hence successfully control crop diseases caused by S. sclerotiorum in the field [11, 12]. For example, C. minitans destroyed the sclerotia, blocked the germination of apothecia by approximate 90 %, and reduced disease incidence in a bean crop by 50 % [5, 13]. As the main effective component in commercial biological control agents, C. minitans was first registered in Europe and was used to control the diseases caused by S. sclerotiorum and S. minor on bean, lettuce, rape, peanut, sunflower, witloof chicory, alfalfa, etc. in the glasshouse or in the field [11–19].
Mycoparasitism is an important mechanism by which C. minitans acts against S. sclerotiorum. C. minitans can penetrate both the hyphae and sclerotia of the host [7, 20, 21]. No appressorium-like structures are formed during the mycoparasitism [8]. Mechanical pressure and enzymatic hydrolysis are the two main invasion methods for C. minitans to parasitize the host [21–24]. To date, several mycoparasitism-related genes have been identified and verified in C. minitans. The fermentation broth of C. minitans showed β−1,3-glucanase and chitinase activity and could decompose the cell wall of hyphae and the sclerotial parenchyma cells of S. sclerotiorum in vitro [22, 25]. The gene cmg1 encoding a β−1,3-glucanase in C. minitans, was highly up-regulated, and high chitinase activity was also detected during mycoparasitism of the sclerotia and hyphae of S. sclerotiorum [23, 26, 27]. Genes encoding the components of the MAP kinase cascade [28], NADPH oxidase [29], oxalate decarboxylase [30], transcription factors [31–33], the peroxisome [34] and heat shock factors [35] have been shown to be involved in mycoparasitism by C. minitans.
C. minitans may synthesize several secondary substances to inhibit the growth of other fungi or bacteria. The antifungal substances (AFS), mainly macrosphelide A, produced by C. minitans inhibit the hyphal growth of S. sclerotiorum and control Sclerotinia diseases [6, 10, 36, 37]. The production of AFS is regulated by the ambient pH, which is strongly affected by the oxalic acid secreted by S. sclerotiorum. C. minitans synthesizes antibacterial substances (ABS) to inhibit the growth of bacteria such as Clavibacter and Xanthomonas [38, 39]. These ABS may alter the balance of microflora on the plant surface and to facilitate colonization by C. minitans [40].
Although it has been developed as a successful biocontrol agent commercially, further investigation of C. minitans is still needed to enhance biological control efficiency and to uncover novel genes and active secondary substances. Thus, we sequenced the entire genome of C. minitans and performed transcriptomic studies at the early stage of interaction with its host, S. sclerotiorum. The purpose of this research was to investigate the possible mechanisms of mycoparasitism, potential active secondary substances (antifungal or antibacterial substances) and gene resources for resistance breeding against fungal diseases using genomic sequencing and transcriptomic analyses.
METHODS
Strain and growth conditions
C. minitans strain ZS-1, originally isolated from a single conidium of C. minitans in China, was used for genome sequencing. S. sclerotiorum 1980 was used for mycoparasitism ‘virulence test’. All fungi were grown on PDA (potato dextrose agar, BD, Sparks, MD, USA) at 20 °C.
Nucleic acid preparation and sequencing
Genomic DNA of C. minitans was extracted from 4 day-old mycelia of strain ZS-1 following the protocol described in the handbook of the DNeasy Plant Kit (Cat. 69104, Qiagen, Germany). The genomic DNA was further purified using the magnetic beads method at Beijing Genomics Institute (BGI, Shenzhen, PR China) for preparing the DNA libraries.
The genomic DNA was sequenced by Illumina HiSeq 2500 sequencing (Illumina, CA, USA) and PacBio RS II sequencing (Pacific Biosciences, CA, USA) platforms at BGI. A total of three libraries (one sequenced by our lab in 2008) were used for genome analysis: a 500 bp paired-end shotgun library and one 6 Kbp mate-pair library for Illumina platform, and one 20 Kbp library for PacBio RS II (Table S1, available in the online version of this article).
In addition, 15 RNA-seq libraries corresponding to the growth and development stages of C. minitans ZS-1 were used to generate transcripts for helping the prediction of gene models. Conidia were collected from PDA slants and washed with sterilized water twice to remove culture residues. A total of 400 µl of conidial suspension at 106 ml−1 was evenly spread on each PDA plate covered with cellophane. To provide evidence for the genes predicted in C. minitans ZS-1, samples of C. minitans at different growth stages, namely, mature conidia (0 h), conidial germination (24 h), early hyphal growth (36 h), hyphal growth (48 h), and pycnidia formation (72 h), were collected, quickly frozen in liquid nitrogen and stored at −80 °C for RNA extraction, with three biological replicates for each sample. All 15 RNA samples were used to make the sequencing libraries and analysed via an Illumina HiSeq 2500 sequencer with a read length of PE150 (150 bp paired-end reads) at BGI.
To extract RNA of C. minitans interacting with S. sclerotiorum, the conidia of strain ZS-1 were shaken at 20 °C in PDB (final concentration: 106 conidia ml−1) at 100 r.p.m. for 36 h, washed with sterilized water for three times and resuspended in an equal volume of water. The germinated conidial suspension was spread on a 24-h-old colony of S. sclerotiorum growing on PDA covered with cellophane. The mixed mycelial samples were collected immediately [as 0 h post-inoculation (h p.i.)], 4 h p.i. and 12 h p.i., and RNA was extracted using RNA reagent (NewBio Industry, Tianjin, PR China) following the manufacturer's instructions. Each mixed RNA sample of the three interaction points were sequenced on an Illumina HiSeq 2000 with 49 bp of single-end reads at BGI, respectively. Complementary DNA (cDNA) was synthesized and 23 genes were detected to verify the transcriptome using quantitative reverse transcription PCR (qRT-PCR) with primer pairs listed in Table S7.
Genome assembly
The sequencing reads of C. minitans ZS-1 were assembled using SOAPdenovo2 (SOAPdenovo2, RRID: SCR_014986) [41] genome assembler with the 6 Kbp mate-pair library data. The most complete preassembly with the largest N50 contig size and the lowest number of the contigs was assembled as the initial assembly (Version1.0). The GapCloser v1.12 (GapCloser, RRID: SCR 015026) was used to fill gaps into Version1.0 with the 500 bp paired-end library data; then the contigs were linked to the assembled genome scaffolds using the software SSPACE (SSPACE, RRID: SCR_005056) with the PacBio library data [42, 43]. The scaffolds of the initial assembly were compared against the NCBI NT database (17 August 2016) (ftp://ftp.ncbi.nlm.nih.gov/blast/db/) for removing the contamination by bacteria. Omission of assembly scaffolds less than 1 Kbp, the genome assembly of C. minitans ZS-1 was obtained.
Genome annotation
Gene predictions on the masked genome were performed using both transcript mapping-based and ab initio methods. Transcript mapping-based methods: Transcripts were generated by first mapping the RNA-seq reads to the assembled genome using Hisat2 (Hisat2, RRID: SCR_015530) [44], and SAMtools (Samtools, RRID: SCR_002105) [45] was used to convert the sam files to bam files. A set of transcripts was generated by Cufflinks (Cufflinks, RRID: SCR_014597) with the mapping data [46]. Unmapped reads from the 15 RNA libraries were de novo assembled using Trinity (Trinity, RRID: SCR_013048) [47]. Then, the Cufflinks transcripts and Trinity-assembled transcripts were combined to get an initial gene model (GeneModel v1.1). The second gene model (GeneModel v1.2) was performed based on the RNA-seq using the program to assemble spliced alignments (PASA) (PASA, RRID: SCR_014656) [48]. Ab initio methods: GeneMark (GeneMark, RRID: SCR_011930) [49, 50] was used to generate a set of gene models (GeneModel v2.1). The bam mapping file generated by TopHat v2.1.1 (TopHat, RRID: SCR_013035) [51] was used to generate an intron/exon hint file. The file was used to predict another gene model (GeneModel v2.2) using Augustus v3.2.3 (Augustus, RRID: SCR_008417). Gene model evidence from RNA-seq-based (GeneModel v1.1 and GeneModel v1.2) and ab initio methods (GeneModel v2.1 and GeneModel v2.2) were imported into the EVidenceModeler package (EVidenceModeler, RRID: SCR_014659) [52] for consensus gene model predictions. Higher weight was given to the RNA-seq alignment predictions than to the ab initio predictions. The RNA-seq data were mapped to the repeat-masked genome and used to update the set of gene models using PASA. Genes with 150 nt or more were retained for subsequent analysis. After removing duplicated sequences using blast2GO (blast2GO, RRID: SCR_005828), artificial intelligence error correction of the final predicted genes was performed using WebApollo (WebApollo: a web-based sequence annotation editor for community annotation, RRID: SCR_005321) [53] with evidence of gene prediction.
Functional annotation
Non-redundant (Nr) annotation, gene ontology, GO-Slim (the high-level subset of GO) and InterPro classification were performed using blast2GO (version 5.1) with default parameter settings [54]. Protein sequences of C. minitans were annotated against databases of UniProtKB/Swiss-Prot database (RRID: SCR 004426) [55], KOG (KOG: Phylogenetic Clusters of Orthologous Groups Ranking, RRID: SCR_008223) [56] using NCBI+blastp (blastp, RRID: SCR_001010) with an e-value cutoff of 1e−5. All the predicted proteins were searched in profile hidden Markov models of CDD (updated 28 March 2017) (CDD: Conserved Domain Database, RRID: SCR_002077) [57] and Pfam domains (protein families and domains; Pfam 31, 16 712 families) (Pfam, RRID: SCR_004726) [58] under HMMER v3.2.1 (Hmmer, RRID: SCR_005305). All matches with e-value cutoff of 1e−10 and coverage ≥0.4 were accepted. KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analyses were conducted using the KAAS online webserver with the BBH (bi-directional best hit) method (https://www.genome.jp/kegg/kaas/) [59].
Analysis of differentially expressed genes (DEGs)
Raw RNA data from the three interactional libraries were generated by Illumina HiSeq 2000 sequencing. Adaptors, reads with more than 10 % unknown bases and low-quality reads (quality value ≤5 of a read) were removed from the raw reads to obtain the clean reads (Table S2). The clean reads were mapped onto the C. minitans genome using Tophat with a bowtie2 index [46]. Mismatches of no more than two bases were allowed in the alignment. The gene-expression level was calculated using the RPKM method (reads per kb per million reads) [60].
A method described in ‘The significance of digital gene expression profiles’ [61] was used to screen for DEGs. We used false discovery rate (FDR)≤0.001 and the absolute value of log2 ratio≥1.5 as the threshold to judge the significance of gene-expression differences.
Three groups of RPKM-based gene-expression data were obtained from the three mixed RNA-seq libraries involving C. minitans in this research. The three libraries, CmSs0h, CmSs4h and CmSs12h, represented the gene expression of C. minitans at the 0, 4 and 12 h stages of mycoparasitism with S. sclerotiorum, respectively. Gene-expression data of C. minitans in CmSs0h, CmSs4h and CmSs12h with clean reads ≥10 at any one of the time points during the mycoparasitism stage of C. minitans with S. sclerotiorum were retained. Based on the gene RPKM, a series of comparison groups were generated to analyse the mycoparasitism genes at the primary interaction stage. We examined the DEGs in C. minitans at different interaction stages and three comparisons were conducted: CmSs0-4, DEGs at the stage of 4 h (CmSs4h) versus 0 h (CmSs0h); CmSs0-12, DEGs at the stage of 12 h (CmSs12h) versus CmSs0h; and CmSs4-12, DEGs in CmSs12h versus CmSs0h. The significant DEGs of different interaction stages are listed in Table S3.
The RNA-seq clean reads from the 15 RNA libraries (described in Nucleic acid preparation and sequencing) at five different growth stages of C. minitans were obtained and mapped to the C. minitans genome using the same methods described above. The expression levels of transcripts were calculated based on transcripts per million (t.p.m.). The DEGs between samples were analysed using DESeq (Version 1.12.4). The DEGs with a |log2ratio|≥2 and FDR≤0.001 were considered statistically significant. Compared to the mature conidia (0 h), the significant DEGs during the spore production process of the pycnidial formation (72 h) are listed in Table S4.
Orthologous protein analysis
All the proteins of the detected species were downloaded from NCBI or JGI. We used the OrthoMCL DB v2.0.9 (OrthoMCL DB: Ortholog Groups of Protein Sequences, RRID: SCR_007839) [62] to identify orthologous protein clusters between C. minitans and 38 genomes of other ascomycete fungi (genomes detail information in Table S5).
Orthologous protein groups were picked from the above results of the Cm group (including C. minitans ZS-1 and P. sporulosa), Tr group (including three biocontrol Trichoderma spp. of T. atroviride, T. harzianum and T. virens), and Pa group (including the host fungi S. sclerotiorum and S. borealis, three non-host fungi of plant pathogens Botrytis cinerea BcDW1/B05.10/T4, and two necrotrophic plant pathogenic fungi (Phaeosphaeria nodorum N15 and Pyrenophora tritici-repentis Pt-1C-BFP) (identified orthologous clusters are listed in Table S6).
Identification of carbohydrate-active enzymes (CAZymes)
We took into account all presented CAZyme classes: glycoside hydrolases (GH), glycoside transferase (GT), polysaccharide lyases (PL), carbohydrate esterases (CE), auxiliary redox enzymes (AA) and carbohydrate-binding modules (CBM). Genomes of C. minitans and the other 38 fungi were screened for CAZymes. NCBI blastp and dbCAN server (dbCAN, RRID: SCR_013208) [63] were conducted against the sequence libraries and profiles in the CAZy database (http://www.cazy.org/) (CAZy-Carbohydrate Active Enzyme, RRID: SCR_012909). All positive matched hits with e-value≥1e−15 and coverage less than 0.5 were examined manually for final prediction. Fungal and plant cell-wall-degrading enzymes (CWDEs) were classified based on the active domains in the CAZy database following Zhao et al. and Lyu et al. [64, 65]
MFS and ABC transporter proteins of C. minitans
The transporter classification database (TCDB) (TCDB, RRID: SCR_004490) [66, 67] is a class database of transporters and in the current version, the transporter proteins were classified into 1345 families based on phylogenetic, sequence, structural and functional analyses in TCDB using the Transporter Classification (TC) system. All the predicted proteins in C. minitans were blasted against the TCDB with the diamond blastp program (Diamond, RRID: SCR_009457) [68] with a high scoring pair (HSP) with an expectation value (e-value) cutoff of 1e−5. The query coverage was set as ≥50 %, and the identity level was set at ≥20 %. The blast results were scanned with TMHMM server v.2.0 (TMHMM server, RRID: SCR_014935) [69] with ≥10 transmembrane helices (TMHs).
Identification of secondary metabolite biosynthesis gene clusters
Identification of secondary metabolite biosynthesis gene clusters was carried out using the genome assemblies and annotation files of C. minitans and 11 other genomes: P. sporulosa, P. tritici-repentis, P. nodorum, S. sclerotiorum, S. borealis, B. cinerea B05.10, B. cinerea BcT4, B. cinerea BcDW1, T. atroviride, T. harzianum and T. virens. The ClusterFinder algorithm of antiSMASH fungal (version 4.0) with the default parameters was used to predict the secondary metabolite-encoding gene clusters [70]. The gene clusters of PKS and NRPS were investigated based on the prediction results. Combination with the functional annotation, the identified putative clusters of unknown type (cf_putative) were predicted and confirmed with amino acid similarity using NCBI+blastp program.
Secretome prediction
Protein sequences with extracellular secretory signals were predicted using SignalP 4.1 (SignalP, RRID: SCR_015644) [71, 72] with the default settings for eukaryotic organisms. Proteins were considered to be secreted if the signal peptide probability was greater than or equal to 0.9 and a cleavage site was within the first approximately 25 amino acids. These predictions were further refined using TargetP v1.1 with a non-plant program and default cutoff parameters [73, 74], and candidate secreted proteins predicted to target the mitochondrion were discarded. Subsequently, these candidate secreted proteins were checked for transmembrane domains using TMHMM v.2.0 [69] and compared by WoLF PSORT (WoLF PSORT, RRID: SCR_002472) [75] with the weighted k-nearest neighbour classifier >15. Putative small secreted proteins (SSPs), which were also considered effector-like proteins, were predicted as no more than 300 amino acids long and no less than 4 % cysteine content.
Construction of effector-like encoding gene vectors and transformation of S. sclerotiorum
In order to examine the influence of these effector-like proteins, full-length sequences of four effector genes, CMZSB_03902, CMZSB_06513, CMZSB_08537 and CMZSB_09504, with or without signal peptides, were transformed into S. sclerotiorum strain 1980 mediated by Agrobacterium as described by Yu et al. [76]. Primer sequences are listed in Table S7. The colony morphology of the transformants was observed after culturing on PDA at 20 °C, and the radial growth was measured using the cross method.
Quantitative reverse transcription PCR (qRT-PCR)
RNA of C. minitans parasitizing S. sclerotiorum at 0, 4 or 12 h p.i. was prepared and cDNA synthesis was conducted according to protocol (EasyScript cDNA synthesis kits, TransGen Biotech, Beijing, PR China). qRT-PCR experiments were performed with gene-specific primers (Table S7) using iTaq Universal SYBR Green Supermix (Bio-Rad, CA, USA) and a CFX96 Real-Time PCR detection system (Bio-Rad, CA, USA). The actin gene sequence of C. minitans was chosen as the internal control. The qRT-PCR was carried out in triplicate with a reaction mixture of total volume 20 µl containing 10 µl of 2×SYBR Green Supermix, 1 µl of 1 : 10 diluted cDNA, 0.5 µl each of the forward and reverse primer, and 8 µl of DEPC-H2O. The PCR involved three steps: 95 °C for 3 min, followed by 42 cycles at 95 °C for 10 s, 56 °C for 10 s and 72 °C for 25 s. The analysis was based on the Cq values of the PCR products. Melting curve analysis of the products was performed at the end of each PCR amplification. The Cq values for the comparison between the amplified genes and actin gene (ΔCq) were calculated. The expression level of the target genes was then calculated as the normalized expression (ΔΔCq).
RESULTS
General features of the C. minitans genome assembly
The DNA of C. minitans strain ZS-1 was sequenced on the Illumina and PacBio platforms at BGI, and the genome sequence was assembled using SOAPdenovo2 [41]. In all, the data provided an average of 80-fold sequence coverage of the genome (Table S1). The resulting assembly length was 38.7 Mb across 2575 contigs, and the N50 length was 64 002 bp. The contigs were assembled into 350 scaffolds that were longer than 1 kb with a total length of 39.8 Mb (including gaps between contigs) using SSPACE [42], and the final N50 length was 653 176 bps (Table 1).
Table 1.
Features of the C. minitans genome assembly
|
Genome feature |
Coniothyrium minitans ZS-1 |
|---|---|
|
Size (bp, draft) |
39,772,940 |
|
No. of scaffolds |
350 |
|
Scaffold N50 length (bp)* |
653,176 |
|
G+C content |
51.70% |
|
Scaffold minimum length (bp) |
1007 |
|
Scaffold maximum length/bp |
1,862,540 |
|
Total gene size (bp) |
22,430,464 |
|
Protein-coding genes |
11 437 |
|
Average coded protein length (AA) |
481 |
|
Predicted proteins with signal peptide |
1171 |
*Scaffold N50, scaffold length at which 50 % of total bases in the assembly are in scaffolds of that length or greater.
Prediction of the coding proteins and functional analysis of C. minitans
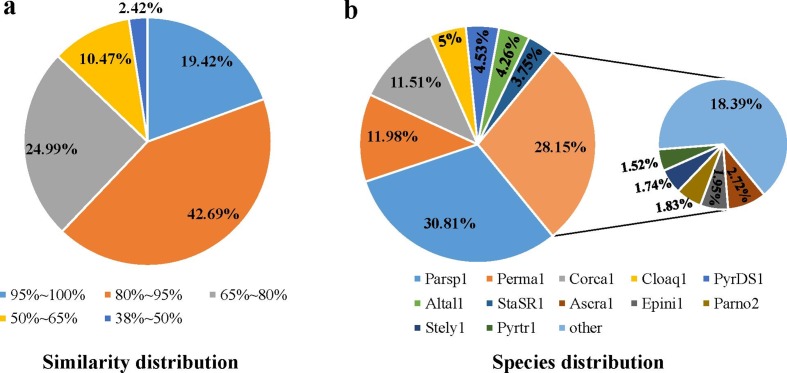
Based on further analysis and RNA-seq expression data, 11 437 protein-coding genes (11 146 longer than 100 amino acids) were predicted, and all proteins had an average length of 481 amino acids (Table 1). Similarity analysis of protein sequence or structure was performed against Nr, InterPro, KOG, CDD, KEGG, Pfam, GO-Slim, GO ontology and UniProtKB/Swiss-Prot, and significant differences were found (Fig. S1). A total of 3193 proteins were annotated against CDD, while 10 884 (95.0 %) of the predicted proteins matched the local Nr database (updated on 20180717) with an e-value cutoff of 1e−10, and 32 364 blast hits were returned with no more than three hits mapped for each protein. The distributions of the blast hits with Nr annotation were determined, and the similarity distribution indicated that 61.1 % of predicted genes of C. minitans had over 80.0 % similarity to proteins of other fungal species (Fig. 1a). A total of 30.8 % of the protein blast hits mapped to protein sequences from the closely related P. sporulosa AP3s5-JAC2a, which is saprophytic on wood and exhibits biocontrol ability by nutrient and niche competition [77, 78]. Altogether 12.0 % mapped to proteins from Periconia macrospinosa, a ‘dark septate endophytic’ fungus [79], and 11.5 % matched the proteins of Corynespora cassiicola, an endophyte, saprobe or necrotrophic plant pathogen [80]. Five percent matched proteins of Clohesyomyces aquaticus, a saprotrophic fungus originally isolated from submerged wood [81], while 4.5 % matched proteins similar to sequences of Pyrenochaeta sp. DS3sAY3a, which plays a role in the bioremediation of metal-polluted environments due to the oxidation of manganese (Mn) compounds [77]. Overall, 4.3 % matched proteins of Alternaria alternata, a worldwide plant pathogen known to perform lignocellulose degradation and Mn(II) oxidation [77] (Fig. 1b, Table S8).
Fig. 1.
Non-redundant (Nr) classification analysis of the encoded proteins in the genome of C. minitans ZS-1. (a) and (b) represent the similarity distribution and species distribution of the Nr annotation based on the blast hit number, respectively. Parsp1, P. sporulosa; Perma1, P. macrospinosa; Corca1, C. cassiicola Philippines; Cloaq1, C. aquaticus; PyrDS1, Pyrenochaeta sp. DS3sAY3a; Altal1, Alternaria alternate; StaSR1, Stagonospora sp. SRC1lsM3a; Ascra1, Ascochyta rabiei; Epini1, Epicoccum nigrum; Parno2, Parastagonospora nodorum; Stely1, Stemphylium lycopersici, Pyrtr1, Pyrenophora tritici-repentis; other, the rest of the 185 species matched five or more blast hits (Table S8).
Mycoparasitic DEG identification of C. minitans and qRT-PCR verification
Three RNA libraries from different stages of C. minitans induced by S. sclerotiorum at 0, 4 and 12 h p.i. resulted sequences approximately 26.4, 42.3 and 37.7 % were mapped on the genome of C. minitans ZS-1, respectively (detail information presented in Table S2).
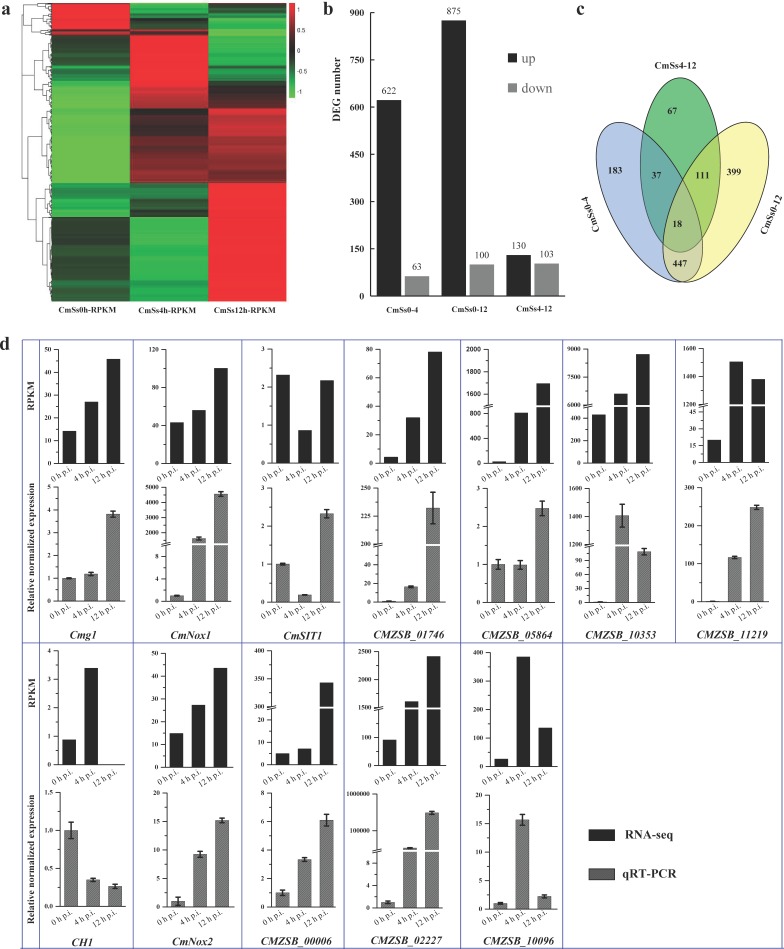
The number of DEGs increased along with the early mycoparasitic process on S. sclerotiorum. Compared to 0 h p.i., 685 and 974 DEGs were identified at 4 h p.i. (CmSs0-4) and 12 h p.i. (CmSs0-12) at level of |log2Ratio|≥1.50 and FDR≤0.001 (Fig. 2a, b). More DEGs were up-regulated than down-regulated in each comparison group (Fig. 2b). Among the DEGs of 12 h p.i. compared to 4 h p.i. (CmSs4-12), 18 DEGs were common in all the three comparison groups (Fig. 2c). The up-regulated DEGs were enriched in the functions of binding (including ion binding, drug binding, small molecular binding, and heterocyclic compound binding) and hydrolase activity (GO:0016787), and also mapped to the biological processes of response to stimulus (GO:0050896), biological regulation (GO:0065007), cell communication (GO:0007154), establishment of localization (GO:0051234), heterocycle biosynthesis (GO:0018130) and transport processes (including transport, ion transport, and lipid transport); while no enrichment was observed for the down-regulated DEGs.
Fig. 2.
Statistical analysis of DEGs in C. minitans induced by hyphae of S. sclerotiorum and confirmation of the gene-expression pattern of the DEGs using qRT-PCR. (a) Based on the RPKM value, the heatmap of all the DEGs detected of C. minitans at the early stages with 0, 4 and 12 h induced by S. sclerotiorum. The detailed DEG list is presented in Table S3. (b) The statistical number of the up-regulated and down-regulated DEGs in the comparison pairwise of 4 h p.i. (CmSs0-4) and 12 h p.i. (CmSs0-12) to the control set of 0 h p.i. and 12 h p.i. to 4 h p.i. (CmSs4-12). (c) The common and particular DEGs in comparison groups of CmSs0-4, CmSs0-12 and CmSs4-12. (d) Confirmation of gene-expression pattern in C. minitans. Mycelial RNA of C. minitans parasitizing on S. sclerotiorum at 0, 4 or 12 h p.i. was used. The black bars represent the RPKM value of the genes in RNA-seq and the bars in grey shades show the qRT-PCR results of the DEGs. Error bars indicate the standard deviation of three replicates.
Five known mycoparasitism-related genes and seven abundantly expressed genes were selected for confirmation using qRT-PCR (Fig. 2d). The expression of genes Cmg1, CMZSB_00006, CMZSB_01746 and CMZSB_10353 increased successively throughout the early parasitic process on hyphae of S. sclerotiorum. While the genes CMZSB_10096 and CMZSB_11219 were up-regulated at 4 h p.i. but decreased at 12 h p.i. In our analysis, the chitinase encoding gene CH1 showed decreased expression during the early mycoparasitism stages and low abundance detected in RNA-seq (Fig. 2d). The gene-expression patterns by qRT-PCR were consistent with the RNA transcriptome.
Carbohydrate-active enzymes (CAZymes) in C. minitans
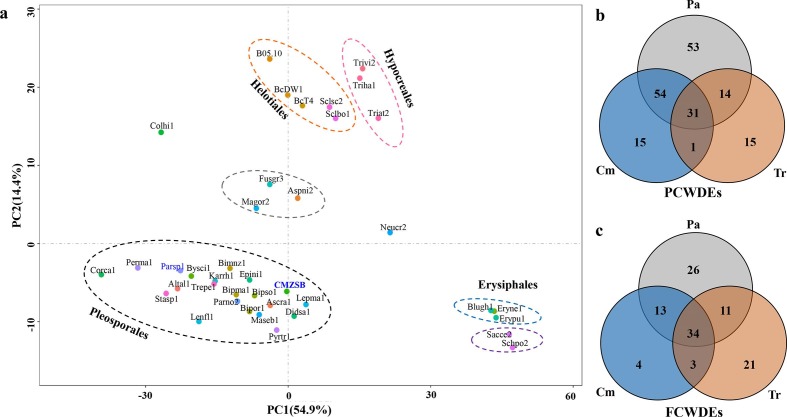
The distribution of each predicted CAZyme family was conserved in the analysed fungal genomes from the same order or with similar nutrition types. The CAZyme families predicted in C. minitans, along with other fungal genomes from the order Pleosporales, were grouped into a cluster using principal component analysis (PCA), and the fungi from Hypocreales, Helotiales and Erysiphales were also placed into corresponding groups (Fig. 3a and Table S9). Magnaporthe grisea, Fusarium graminearum and Aspergillus nidulans from different fungal orders were grouped into a cluster. Not considering the number of carbohydrate-binding modules (CBMs), C. minitans encoded 434 catalytic protein modules in CAZymes, which is a moderate number compared with the other 38 tested species in our analysis (Table S10). The number of modules was lower than those in the two phylogenetically nearest fungi, P. sporulosa (556) and Karstenula rhodostoma (507) and higher than the average level (380) of the three species in Trichoderma and two species in Sclerotinia. GO analysis showed that the CAZyme protein-coding genes were annotated with functions of catalytic activity (GO: 0003824), carbohydrate-binding (GO: 0030246), transferase activity (GO: 0016757 and GO: 0016758), hydrolase activity (GO: 0016787) (P≤0.05), cell-wall biosynthesis (GO: 0071554) (P≤0.05) and metabolic process (GO: 0008152) (P≤0.05) (Table S11).
Fig. 3.
Comparison of CAZymes encoded in C. minitans ZS-1 and other 38 ascomycetes. (a) Multivariate grouping of ascomycete fungal genomes based on the protein number profiles of predicted CAZyme families using PCA. The CAZyme proteins from the fungal genomes were identified against the CAZy database. A number list of all CAZyme proteins in each genome is shown in Table S10. (b) Orthologous analysis of PCWDE-related protein comparison of C. minitans (Cm) with Trichoderma spp. (Tr) and seven plant pathogenic fungi (Pa); (c) PCWDE-related protein comparison of Cm with Tr and Pa. Orthologous groups were determined among the CAZymes encoded in the genomes of C. minitans, three Trichoderma spp. (T. atroviride, T. harzianum and T. virens) and seven plant pathogenic fungi (S. sclerotiorum strain 1980, S. borealis, B. cinerea strain DW1, B. cinerea strain B05.10, B. cinerea strain BcT4, P. nodorum strain SN15, and P. tritici-repentis strain Pt-1C-BFP) (details in Table S6). P(F)CWDEs, plant (fungus) cell-wall-degrading enzymes. Cm, CAZyme orthologous groups related to the cell wall in C. minitans; Tr, CAZyme groups related to cell wall encoded in Trichoderma spp.; Pa, CAZyme orthologous groups encoded by the seven plant pathogenic fungi genomes.
C. minitans encodes 118 PCWDEs distributed in 26 families of CAZymes, which is less than its phylogenetically related fungus P. sporulosa (170 PCWDEs in the same 26 families) and more than Trichoderma spp. (average of 55 in 22 families), Sclerotinia spp. (78 in 21 families) and Botrytis spp. (101 in 22 families). Ten glycoside hydrolases and five polysaccharide lyases were specifically present in C. minitans, while 13 and 46 unique orthologous groups were present in Trichoderma spp. and plant pathogenic fungi, respectively. A total of 54 unique orthologous groups (including 58 proteins of C. minitans) were shared by C. minitans and the other plant pathogenic fungi analysed. One orthologous group protein encoding a predicted endo-1,4-beta-xylanase (in the GH43 family) was identified only in C. minitans and Trichoderma spp., while 13 specific orthologous groups were commonly present in Trichoderma spp. and the other pathogens (Fig. 3b). These results suggest that C. minitans also has the potential to break down complex components of plant cell walls.
C. minitans encodes 64 FCWDEs belonging to 14 GH families in CAZymes. C. minitans has thus fewer FCWDEs than T. atroviride (89), T. harzianum (97) and T. virens (97). However, the plant pathogenic fungi also have a similar number of FCWDE families to C. minitans. Comparing the FCWDE families of C. minitans with those of the selected plant pathogenic fungi (seven fungi) and Trichoderma spp. (three species), most orthologous groups of C. minitans were shared with either Trichoderma spp. or plant pathogenic fungi, while four unique orthologous groups (four proteins) were found only in C. minitans (Table S6). These four orthologous groups encode glycoside hydrolase family 16 protein (GH16), concanavalin A-like lectin/glucanase (GH16), GPI-anchored cell wall beta-1,3-endoglucanase (GH17) and glycoside hydrolase (GH20). Interestingly, there were three orthologous groups shared by C. minitans and Trichoderma spp., while not existing in the tested plant pathogenic fungi (Fig. 3c). These three orthologous groups encoded four proteins: two hexosaminidases (GH20), a glycoside hydrolase (GH25) and a putative exo-beta-1,3-glucanase (GH55). Interestingly, C. minitans and plant pathogenic fungi shared 47 orthologous groups of FCWDEs (57 proteins) (Fig. 3c).
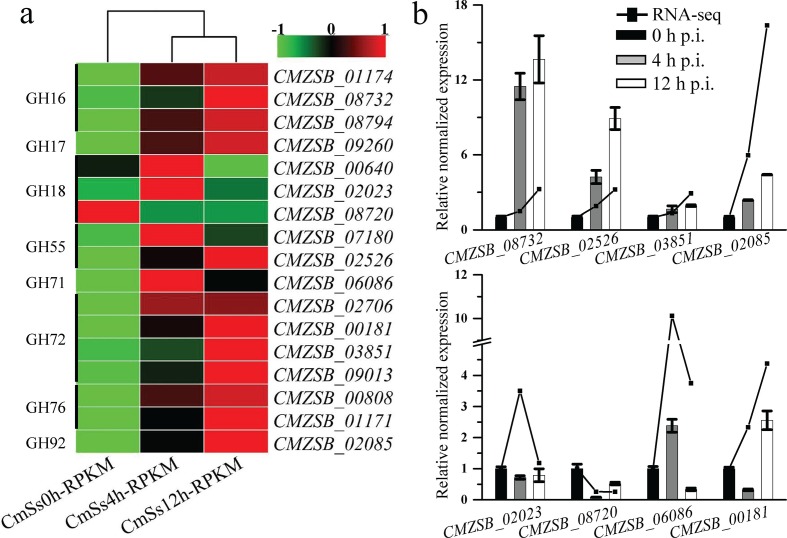
Out of 64 genes encoding FCWDE, 45 were expressed and the expression of 17 genes were significantly changed at 4 or 12 h p.i. as compared to 0 h p.i. Most genes predicted encoding GH families were up-regulated (Fig. 4a), while only one gene CMZSB_08720 distributed in all the fungi tested, was significantly down-regulated. However, the expression of the four unique FCWDEs and three Trichoderma-shared FCWDEs was not significantly altered. Expression of eight FCWDE encoding genes were examined by qRT-PCR and the patterns were consistent with the interactional transcriptome (Fig. 4b). Interestingly, PCWDE expression in C. minitans was also significantly changed (Fig. S2). These results suggest that cell-wall-degrading enzymes may play important roles during the early interaction of C. minitans and its host.
Fig. 4.
Expression of FCWDEs in the CAZymes detected during mycoparasitism. (a) The FCWDE-encoding genes among the DEGs of C. minitans induced by the hyphae of S. sclerotiorum at the early mycoparasitism time points of 0, 4 and 12 h. (b) Quantitative RT-PCR amplification of FCWDE-encoding DEGs of C. minitans. The black lines represent the RPKM value of the genes in RNA-seq and the bars show the qRT-PCR results of the DEGs. Error bars indicate the standard deviation of three replicates.
MFS and ABC transporters in C. minitans
The major facilitator superfamily (MFS) and the ATP-binding cassette (ABC) primary transporter superfamily are the most ubiquitous transporters in fungi and other organisms [82]. Based on the transporter classification databases (TCDB), the transporters of the MFS and ABC superfamilies were identified in C. minitans and 11 other fungal genomes.
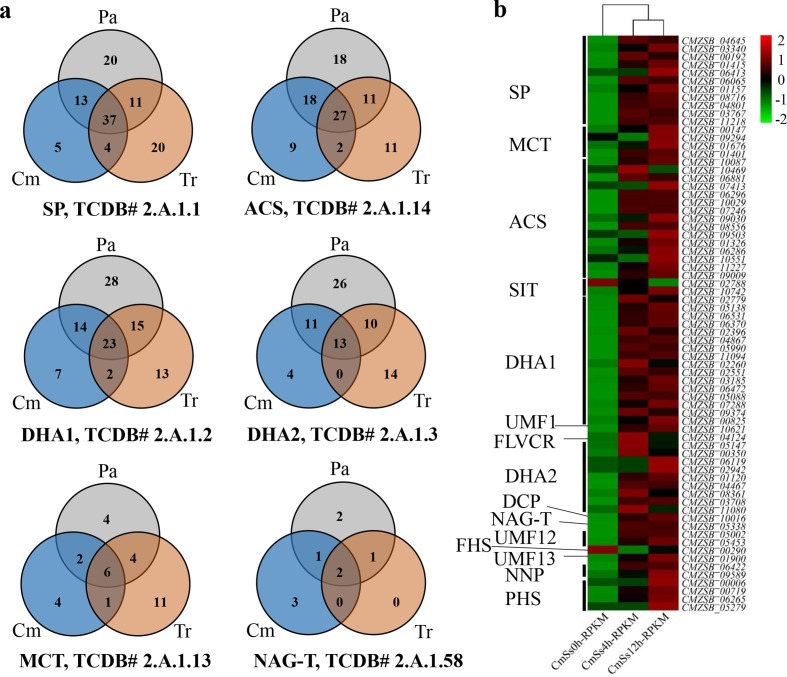
A total of 284 predicted MFS transporters of 25 classes were identified in the genome of C. minitans. Sugar Porter (SP), Drug: H+Antiporter-1 (12 Spanner) (DHA1), Anion: Cation Symporter (ACS), and Drug: H+Antiporter-2 (14 Spanner) (DHA2) were the most abundant MFSs in all the analysed genomes, including the plant pathogens Sclerotinia spp., B. cinerea, Parastagonospora nodorum and Pyrenophora tritici-repentis, and the biocontrol fungi Trichoderma spp. Ten orthologous MFS transporters were present in C. minitans and Trichoderma spp. and lacking in plant pathogenic fungi (Pa). In addition, 32 orthologous transporters, including five sugar porters, four monocarboxylate transporters (MCT), nine Anion: Cation symporters, seven Drug: H+antiporter-1 (DHA-1), four Drug: H+antiporter-2 (DHA-2) and three N-acetylglucosamine transporters (NAG-T), were unique to C. minitans (Fig. 5a, Table S12).
Fig. 5.
Distribution of the MFS transporters in the C. minitans ZS-1 genome and the DEGs involved in mycoparasitism. (a) Orthologous proteins groups distribution of SP, ACS, DHA1, DHA2, MCT and NAG-T in MFS transporters in three groups of Cm, Tr and Pa. (b) Gene expression of MFS transporters, which detected significantly changed during the mycoparasitism stages of 0, 4 and 12 h, respectively, in C. minitans. SP: Sugar Porter Family, ACS: Anion:Cation Symporter Family, DHA1: Drug:H+Antiporter-1 (12 Spanner) Family, DHA2: Drug:H+Antiporter-2 (14 Spanner) Family, MCT: Monocarboxylate Transporter Family, NAG-T: N-Acetylglucosamine Transporter Family, FHS: Fucose: H+Symporter Family, PHS: Phosphate: H+Symporter Family, OCT, Organic Cation Transporter Family, V-BAAT, Vacuolar Basic Amino Acid Transporter Family, SIT: Siderophore-Iron Transporter Family, LplT: Lysophospholipid Transporter Family, NNP: Nitrate/Nitrite Porter family, UMF1: Unidentified Major Facilitator-1 Family, LAT3: l-Amino Acid Transporter-3 Family, UMF12: Unidentified Major Facilitator-12 Family, UMF23: Unidentified Major Facilitator-23 Family, MHS: Metabolite:H+Symporter Family, SHS: Sialate:H+Symporter Family, FLVCR: Feline Leukemia Virus Subgroup C Receptor/Heme Importer Family, DCP: Major Facilitator Superfamily Domain-containing Protein 5 Family, Pl-Cu-UP: Plant Copper Uptake Porter, UMF30: Unidentified Major Facilitator-30 Family, YnfM: Acriflavin-sensitivity Family, PAT: Peptide/Acetyl-Coenzyme A/Drug Transporter Family, OFA: Oxalate:Formate Antiporter Family, ACDE: Aromatic Compound/Drug Exporter Family, PCFT/HCP: Proton Coupled Folate Transporter/Heme Carrier Protein Family, Pht: Proteobacterial Intraphagosomal Amino Acid Transporter Family. Sclsc2, S. sclerotiorum strain 1980; Sclbo1, S. borealis; BcDW1, B. cinerea strain DW1; B05.10, B. cinerea strain B05.10; BcT4, B. cinerea strain T4; Parno2, P. nodorum strain SN15; Pyrtr1, P. tritici-repentis strain Pt-1C-BFP; Triat2, T. atroviride; Triha1, T. harzianum; Trivi2, T. virens; CMZSB, C. minitans strain ZS-1 (this research); Parsp1, P. sporulosa. Cm, orthologous groups of MFS transporters in C. minitans; Tr, orthologous groups of MFS transporters in Trichoderma spp.; and Pa, orthologous groups of MFS transporters in plant pathogenic fungi.
A total of 185 MFS transporter genes were detected during mycoparasitism at 4 h p.i. and 12 h p.i. to 0 h p.i., and 71 MFS transporters showed significant changes in expression (Fig. 5b). Among the 71 genes, 69 were up-regulated, while only a predicted siderophore-iron transporter (Sit) gene (CMZSB_02788) and a putative Fucose: H+Symporter (FHS) gene (CMZSB_00290) were down-regulated. These results indicate that MFS transporters likely play an important role during the early stage of mycoparasitism. Nine of the 15 MCT protein-encoding genes in C. minitans were detected as expressed during the early stages of mycoparasitism. Among these nine genes, four were significantly up-regulated, and one was down-regulated. The results suggest the involvement of MCT in the biocontrol event.
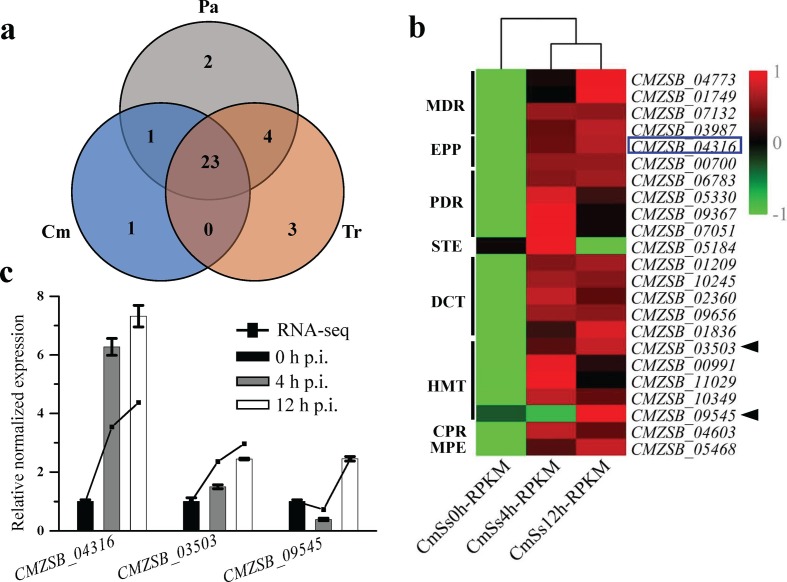
Fifty-one ABC transporters in 13 subfamilies were predicted in C. minitans, which is similar to P. sporulosa (52), S. sclerotiorum (49) and T. atroviride (51), while the numbers were 44, 47 and 43 in S. borealis, P. nodorum and P. tritici-repentis, respectively. The numbers of ABC transporters in B. cinerea B05.10, B. cinerea BcDW1, T. virens and T. harzianum were 65, 56, 66 and 64, respectively (Table S13). No orthologous ABC transporters were only shared between C. minitans and Trichoderma spp., while 23 transporters were present in all the fungi tested (Fig. 6a). This suggested that C. minitans, Trichoderma spp. and the plant pathogenic fungi analysed share a similar transporter distribution of the ABC superfamily, or the function of ABC transporters is conserved in different fungi. C. minitans has seven HMTs (heavy metal transporters), which is almost twice as many as the other 11 fungi analysed. Group 2983 (TC 3.A.1.210.1), group 494 (TC 3.A.1.210.2) and group 242 (TC 3.A.1.210.2) were present in all fungi analysed, while CMZSB_03503 (TC 3.A.1.210.2) of group 12899, CMZSB_09545 (TC 3.A.1.210.2) and CMZSB_11035 (TC 3.A.1.210.7) were specific to C. minitans (Table S6).
Fig. 6.
Orthologous proteins analysis of ABC family transporters in C. minitans and related fungal genomes. (a) Analysis of orthologous groups of ABC transporters in C. minitans, Trichoderma spp. and plant pathogenic fungi. (b) Identified DEGs of transporters during the early stages of mycoparasitism. PDR: Pleiotropic drug resistance, EPP: Eye pigment precursor transporter, Gld: Gliding motility ABC transporter, CPR: Cholesterol/phospholipid/retinal flippase, TauT: Taurine Uptake Transporter, U-ABC1: Unknown-ABC1, DrugE1: Drug exporter-1, BIT: Brachyspira Iron Transporter, B12-P: Cobalamine Precursor, PepT: Peptide/Opine/Nickel Uptake Transporter, PAAT: Polar Amino Acid Uptake Transporter, EVE: Ethyl Viologen Exporter, STE: a-Factor sex pheromone exporter, MPE: Mitochondrial peptide exporter, P-FAT: Peroxysomal fatty acyl CoA transporter, HMT: Heavy metal transporter, DCT: Drug conjugate transporter, MDR: Multidrug resistance exporter, DrugRA1: Drug resistance ATPase-1, DrugRA2: Drug resistance ATPase-2. The species-specific proteins encoding genes expression in C. minitans was represented of the black triangle; the blue box represented the gene of ABC transporters of the shared orthologous groups of C. minitans with Trichoderma spp. (c) qRT-PCR valued the DEGs of three ABC transporters encoding genes. 0, 4 h and 12 h p.i. represent different mycoparasitic time points of C. minitans parasitizing on the hyphae of S. sclerotiorum. The black lines represent the RPKM value of the genes in RNA-seq and the bars show the qRT-PCR results of the DEGs. Error bars indicate the standard deviation of three replicates.
Thirty-six of the 51 predicted ABC transporter genes were up-regulated during the early mycoparasitic stages of C. minitans and 23 were significantly altered (Fig. 6b). Five of the seven HMT coding genes, including CMZSB_03503 (orthologous groups 12899) and CMZSB_09545 (specific to C. minitans), were significantly up-regulated at the early stage of infection (Fig. 6b) and all seven genes were mapped to the iron-binding (GO: 0005506) molecular function based on GO enrichment analysis.
Secretory proteins enriched in the mycoparasitism process in C. minitans
A total of 908 secretory proteins were predicted in C. minitans, which accounts for 8.3 % of the total proteins. The proportion of secretory proteins was 5.5 % in Sclerotinia spp., 6.5 % in Botrytis spp., 6.7 % in Trichoderma spp. and 9.4 % in P. sporulosa. Excluding CAZymes, the 695 predicted secretory proteins were classified into 675 orthologous groups, and 45 were unique to C. minitans without orthologs in Trichoderma spp. or the seven selected plant pathogenic fungi. In addition, 307, 499 and 1041 specific orthologs were predicted in C. minitans, Trichoderma spp. and the plant pathogenic fungi, respectively. Twenty-four orthologous groups were specific to C. minitans and Trichoderma spp. and absent in the plant pathogenic fungi. The 45 unique secretory proteins in C. minitans were annotated. Five of these secretory proteins were predicted as a PR-1-like protein (CMZSB_08830), an Acyl-CoA N-acyltransferase (CMZSB_04745), an amino acid transporter (CMZSB_05748), a protein with the DUF605 multi-domain (CMZSB_00858), and a FAD/NAD(P)-binding domain-containing protein (CMZSB_06166), while the rest 40 proteins were unknown proteins or hypothetical proteins with no specifically putative functions reported.
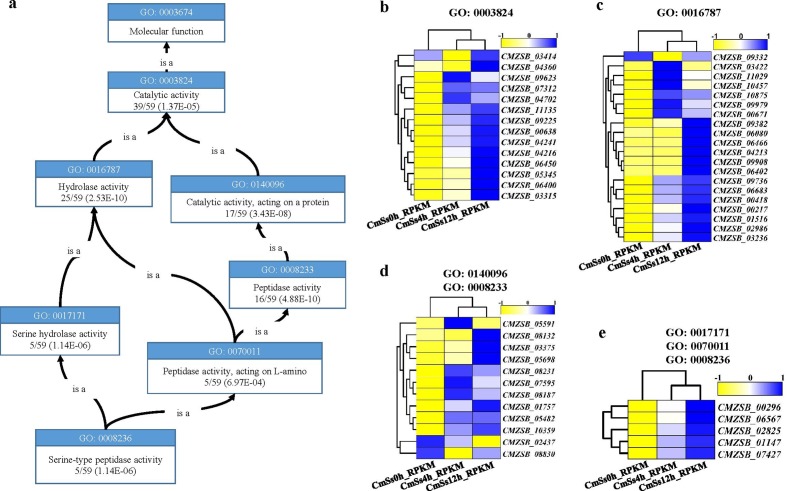
A total of 526 secretory protein-coding genes were detected during the early mycoparasitism stages of C. minitans, and 132 DEGs were predicted via CAZymes. The CAZymes DEGs were enriched in catalytic activity (GO: 0003824), hydrolase activity (GO: 0016787), peptidase activity (GO: 0008233) and serine hydrolase activity (GO: 0017171) in molecular function (P≤0.01) (Fig. 7a). Almost all the genes involved in GO terms were significantly up-regulated at the early stages of mycoparasitism in C. minitans (Fig. 7c, d, e). The only exception of CMZSB_02437, encoding a putative Asp-domain-containing protein, was suppressed all the way (Fig. 7d).
Fig. 7.
Serine peptidase secreted proteins in the mycoparasitism process in C. minitans. (a) Enriched GO terms' graph of serine peptidase of the DEG-encoding secreted proteins. (b) Expression of the unique DEGs involved in GO: 0003824. (c) Expression of the unique DEGs involved in hydrolase activity in GO molecular function. (d) Expression of the unique DEGs related to GO: 0140096 and GO: 0008233, with CMZSB_08830 in GO: 0140096. (e) Expression of the five DEGs related to each of GO: 0017171, GO: 0070011 and GO: 0008236. The graph of GO terms was downloaded and modified from https://www.ebi.ac.uk/QuickGO.
Eighty effector-like proteins were identified in C. minitans strain ZS-1, and for 50 of them, expression of their encoding genes was detected during mycoparasitism. Four effector-like encoding genes (CMZSB_03902, CMZSB_06513, CMZSB_08537 and CMZSB_09504) from the C. minitans were transformed into the host fungus (S. sclerotiorum strain 1980) with the help of Agrobacterium . All of them had an influence on the phenotype of the S. sclerotiorum transformants, including partial inhibition of the hyphal growth rates (Fig. S3). More analysis should be conducted on these four effector-like genes to elucidate their functions during mycoparasitism.
NRPS- and PKS-encoding genes and their expression in C. minitans
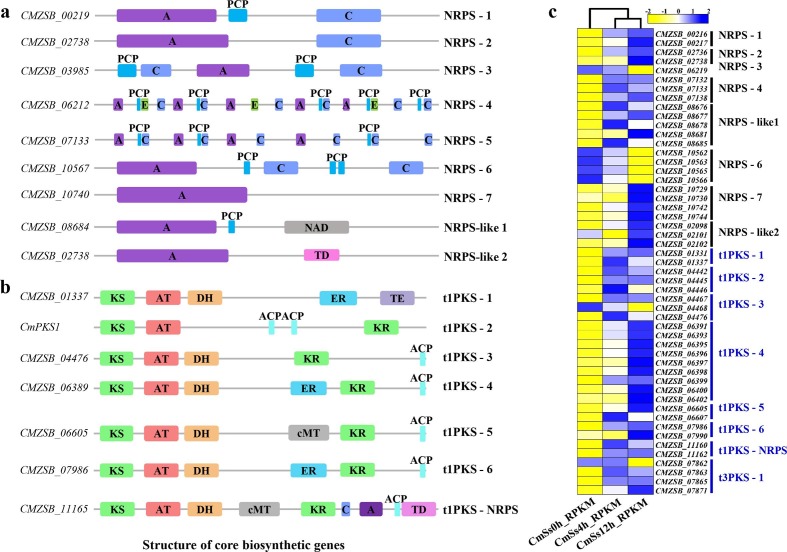
The C. minitans genome encoded seven non-ribosomal peptide synthase (NRPS) clusters and two NRPS-like clusters. Each NRPS cluster was identified with different domains (Fig. 8a). Besides the basic AMP-binding domain, NRPS-like 1 cluster carried a nicotinamide adenine dinucleotide (NAD) (NRPS-like 1) domain and NRPS-like 2 cluster with a thioesterase domain (TD) (Fig. 8a). Six type I polyketide synthase (PKS) (t1PKS) clusters, one type III PKS (t3PKS) cluster and one hybrid t1PKS-NRPS gene cluster were identified in the C. minitans genome (Fig. 8b). During the early stages of mycoparasitism by C. minitans, 53 DEGs were identified among the genes detected in the NRPS and PKS biosynthesis gene clusters. Of these, compared to 0 h p.i., 31 were up-regulated at 4 h p.i., while 40 were up-regulated and seven were down-regulated at 12 h p.i. At the mycoparasitism stage (12 h p.i. versus 4 h p.i.), 13 genes were up-regulated, and eight were down-regulated (Fig. 8c). Most of the core biosynthetic genes of NRPS involved in mycoparasitism were up-regulated, but the NRPS-3-related core gene CMZSB_06212 and the NRPS-6-related gene CMZSB_10567 were down-regulated significantly (Fig. 8c). The expression of all the core biosynthetic genes of t1PKS, t3PKS, and hybrid t1PKS-NRPS was significantly induced when in contact with S. sclerotiorum (Fig. 8c).
Fig. 8.
Gene expression and core biosynthetic genes related to secondary metabolism in C. minitans during the early stages of mycoparasitism. (a) Structures of core NRPS biosynthetic proteins predicted to be involved in mycoparasitism in C. minitans strain ZS-1. A: adenylation (AMP) domain; C: condensation domain; PCP: peptidyl-carrier protein domain; cMT: cycle methyltransferase; NAD: nicotinamide adenine dinucleotide; E: epimerization domain. (b) Structures of core PKS or hybrid PKS-NRPS biosynthetic proteins predicted to be involved in mycoparasitism in C. minitans strain ZS-1. KS: beta-ketoacyl-synthase domain; AT: acyl transferase domain; DH: polyketide synthase dehydrogenase; ER: enoyl reductase domain; KR: ketoreductase domain; TE: thioesterase; ACP: acyl carrier protein; TD: thioesterase domain. (c) Heatmap created with MeV (version 4.9.0) based on the RPKM values of the detected genes in C. minitans strain ZS-1; the RPKM values were normalized within the row for each gene.
Nine genes in five domains related to gliotoxin were predicted into an unknown type of gene cluster in C. minitans: one GliA (gliotoxin A) (CMZSB_02816), two GliI (gliotoxin I) (CMZSB_02903 and CMZSB_03980), two GliK (gliotoxin K) (CMZSB_03977 and CMZSB_08616), two GliC (gliotoxin C) (CMZSB_03976 and CMZSB_03979), two GliT (gliotoxin T) (CMZSB_03988) and one GliP (gliotoxin P) (CMZSB_03975) involved in gliotoxin biosynthesis. The expression of these genes could not be detected at 4 h p.i., and only two were expressed at 12 h p.i. (Table S3).
DISCUSSION
In this study, the genome of C. minitans was sequenced, assembled and annotated, and the potential mechanism of parasitism was analysed based on RNA-seq data. The size of the genome of C. minitans was approximately 39.8 Mb, which is moderate among ascomycetes. C. minitans has a large number of genes encoding enzymes for both plant and fungal cell-wall degradation; it also has many genes for MFS and ABC transporters, PKS and NRPS; a high proportion of genes coding for secretory proteins were also predicted.
Plant and fungal cell-wall-degrading enzymes in C. minitans
C. minitans has 434 putative catalytic protein modules in CAZymes, and this number is moderate compared with the other 39 fungal species in this study. Many of these proteins were predicted to be enzymes for degrading either plant or fungal cell walls. The number of genes coding for PCWDEs is not smaller than those of necrotrophic plant pathogenic fungi such as M. oryzae, B. cinerea and its host, S. sclerotiorum, and is significantly larger than those of biotrophic fungal pathogens. Compared to plant pathogenic fungi, C. minitans has 17 unique PCWDEs (distributed in 15 orthologous proteins). These genes may enable C. minitans to live on dead plant tissues. Interestingly, we found that the expression of many PCWDE genes was highly induced at the early stage of C. minitans interaction with S. sclerotiorum, suggesting that these PCWDEs may also play an important role in parasitism. Further experiments are needed to confirm this hypothesis.
FCWDEs may play essential roles in fungal development [83–85]; thus, no fungi without FCWDEs could survive in nature. As a mycoparasite, C. minitans might reasonably have a powerful fungal cell-wall-degrading enzyme system to attack S. sclerotiorum. However, the number of FCWDE genes in C. minitans is not significantly different from that in plant pathogenic fungi. Furthermore, C. minitans shared 47 orthologous FCWDEs with the plant pathogenic fungi tested. Interestingly, the mycoparasitic Trichoderma spp. has many more FCWDEs than C. minitans, and only three orthologues in the GH18 family that were not detected during the early stage of interaction between C. minitans and S. sclerotiorum, were shared by C. minitans and Trichoderma spp. The GH18 family is considered to play an important role during mycoparasitism [86–89]. These genes may suggest some reasons for the difference between C. minitans and Trichoderma spp. in regarding host range. The chitinase gene CH1 (CMZSB_00640) and β−1,3-endoglucanase (CMZSB_02526) of C. minitans are usually used as two marker genes for mycoparasitism [90], while their orthologues can also be found in plant pathogenic fungi. Furthermore, these two genes and CMZSB_02023 (also predicted encoding chitinase) were up-regulated significantly (log2 ratio=2.6, FDR≤0.001) during conidiation (Table S4). This suggests that the FCWDEs may play a role not only in parasitizing S. sclerotiorum, but also in shaping the cell wall of C. minitans, as reported for S. sclerotiorum [64].
Secondary metabolites and their functions in C. minitans
Fungi are known to produce a large number of secondary metabolites, and genes for secondary metabolism are often distributed in clusters on genomes. Many biocontrol agents can produce antagonistic secondary metabolites to suppress the growth of pathogens or to occupy advantageous niches [91–93]. Early experiments found that C. minitans could produce both antifungal [10] and antibacterial substances [38], among which macrosphelide A has antimicrobial activity against some ascomycetes, such as Sclerotinia spp., basidiomycetes, oomycetes and Gram-positive bacteria [37, 94]. Macrosphelide A, benzenediol and 5-aminopentanoate were the three most strongly accumulated compounds when C. minitans was co-cultured with S. sclerotiorum for 2 days [36].
PKS and NRPS are the main synthases of polyketides and peptides. As in S. sclerotiorum, B. cinerea, P. nodorum and mycoparasitic Trichoderma spp., C. minitans contains gene clusters related to secondary metabolism, including seven NRPS clusters, six t1PKS clusters, one t3PKS cluster and one hybrid t1PKS-NRPS gene cluster. The number of related genes was counted within each cluster, and no significant differences when compared to all the tested fungal genomes were found. Previously, we found that disruption of a type I PKS gene CmPKS1 and a transcription factor gene CmMR1 led to an absence of melanin, but did not significantly affect the mycoparasitism of S. sclerotiorum [31, 95].
Gliotoxin is a secondary metabolite produced by many fungi. It can suppress immunity, promote apoptosis of mammalian cells [96] and inhibit the growth of micro-organisms by disrupting NADPH oxidase activity [97]. In A. fumigatus, the non-ribosomal peptide synthetase GliP catalyses the first biosynthetic step in the synthesis of gliotoxin and was reported as the determinant of host-specific virulence [98]. When gliP was disrupted, T. virens lost the ability to produce gliotoxin and to parasitize S. sclerotiorum [99]. T. virens and T. harzianum have six and nine NRPS-encoding genes [96, 100] respectively, but non-mycoparasitic T. reesei has nine genes in the biosynthetic cluster. Although gliotoxin has not yet been identified in C. minitans, an NRPS cluster with nine genes related to gliotoxin synthesis was predicted on Scaffold 12. The homologous gene cluster of gliotoxin biosynthesis in C. minitans displays similarity of 47.0 % with that of T. reesei and much lower than that of the biocontrol Trichoderma species. We speculate that C. minitans encodes a type of gliotoxin biosynthetic gene cluster different from those in biocontrol Trichoderma spp., but similar to that in T. reesei. Two of the annotated gliotoxin-related genes were up-regulated at 12 h p.i. comparing to 0 h p.i. in C. minitans induced by S. sclerotiorum. This evidence suggests that gliotoxin or gliotoxin-like substances may be synthesized and contribute to the parasitism of C. minitans.
MFS and ABC transporters in C. minitans
The MFS and ABC transporter superfamilies are the most ubiquitous transporters in fungi and other organisms [82]. MFS and ABC transporters play important roles in resistance against drug and plant toxic components for pathogens and are also important in beneficial micro-organisms [101–104]. The expression of MFS and ABC transporters was induced in C. minitans during interaction with S. sclerotiorum, suggesting that these two classes of transporters may play important roles in the mycoparasitism process.
Siderophores are responsible for the acquisition of iron and the protection of cells from oxidative stress [105, 106]. In T. virens, deletion of TvTex10, an intracellular siderophore biosynthesis-related gene, increased the fungal growth rate and sensitivity to oxidative stress and simultaneously decreased conidia and gliotoxin production [107]. Fifty-four ABC transporters in 14 subfamilies were predicted in C. minitans. Interestingly, seven HMTs were predicted in C. minitans, while only three–four HMTs were found in the other fungi tested. Six of these HMTs have putative iron-binding functions and were expressed during the early interaction between C. minitans and S. sclerotiorum. CmSIT1, a siderophore iron transporter gene (CMZSB_03548) annotated in Scaffold 10, was only expressed during the interaction with S. sclerotiorum [108]. The results suggest that siderophore iron transporters and related HMTs are important for the acquisition of iron by mycoparasites in a competitive environment and are involved in mycoparasitism.
Effector-like proteins in C. minitans
Typical effectors are small cysteine-rich secretory proteins (usually less than 150 aa) and are released by pathogens into host cells to regulate the host's resistance system. Cerato-platanin proteins are effectors for many plant pathogenic fungi [109] and were also identified in Trichoderma spp. to induce plant defense against pathogens [110]. SM1, an elicitor of induced systemic resistance (ISR), was identified as an effector-like protein involved in the colonization of maize roots by T. virens [111]. In Trichoderma spp., 233 effector-like proteins have been annotated, and the class II hydrophobin family gene tvhyiil, could help T. virens colonize plant roots and participate in antagonistic activity against Rhizoctonia solani [112]. In this study, 80 genes encoding effector-like proteins were identified in C. minitans, 50 of which were expressed during the early stage of interaction between C. minitans and S. sclerotiorum. We further found that some of these effector-like protein genes could induce a hypersensitive reaction when transiently expressed in the leaves of Nicotiana benthamiana and four other effector-like protein genes led to a decrease in hyphal growth when expressed in S. sclerotiorum, suggesting that some of these effector-like proteins may potentially function in plants, and some may function in the parasitism of C. minitans. It may not be surprising that Trichoderma spp. release effector-like proteins into plant cells, since they can live endophytically in plant roots. However, C. minitans is a soil-borne fungus and has not been found to live in plants. Considering that C. minitans can parasitize only fungi in the genus Sclerotinia, we hypothesize that some effector-like proteins of C. minitans may specifically function in the recognition of Sclerotinia spp. Further investigation is necessary to confirm the functions of effector-like proteins in the interaction between the host fungus and mycoparasite.
Mycoparasitism of C. minitans is a two-way process
Previously, a model for the interaction between C. minitans and S. sclerotiorum was established. C. minitans senses oxalic acid produced by S. sclerotiorum, or the low pH caused by oxalic acid, and produces antifungal substances to inhibit the growth of S. sclerotiorum and degrade the oxalic acid with oxalate decarboxylase. After the ambient pH rises, C. minitans secretes fungal cell-wall-degrading enzymes and enters the parasitic life stage [30, 32]. However, in this interaction system, although C. minitans could produce antifungal substances to weaken S. sclerotiorum and destroy the cell wall of S. sclerotiorum using FCWDEs, counteraction by S. sclerotiorum should be considered. As shown in this study, many genes for MFS transporters, ABC transporters, effector-like proteins and secondary metabolites were significantly up-regulated during the early stage of interaction, suggesting that these substances play essential roles in parasitism of S. sclerotiorum. ABC transporters, especially HMTs, may be used by C. minitans to compete with S. sclerotiorum for heavy metal elements such as iron; MFS transporters may play roles in releasing antifungal substances and in the efflux of toxic compounds produced by S. sclerotiorum. There is a high possibility that the effector-like proteins enter the hyphae of S. sclerotiorum and specifically inhibit the counteraction system of S. sclerotiorum. Once it subdues S. sclerotiorum, C. minitans carries on a parasitic life in its host.
In this study, we just surveyed the response of C. minitans during the early mycoparasitic stages. The interaction of C. minitans with S. sclerotiorum is a two-way process. Meanwhile, a considerable number of reads mapped to the host fungus S. sclerotiorum genome were also included in the interactive RNA_seq libraries. In order to elucidate the two-way mechanisms during the interaction between C. minitans and S. sclerotiorum, the responses of S. sclerotiorum challenged by C. minitans need further study and a great opportunity would be provided to understand the mycoparasitism mechanism.
Data Bibliography
1. The UniProt C. UniProt: the universal protein knowledgebase. Nucleic Acids Res 45(D1): D158-D169 (2017).
2. Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43(Database issue): D261-269 (2015).
3. Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1): D200-D203 (2017).
4. Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44(D1): D279-285 (2016). Pfam database HMM file (release version Pfam32.0) was downloaded from the website http://pfam.xfam.org/.
5. Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35(Web Server issue): W182-185 (2007).
6. Zhang H, Huang L, Wu P, Yang Z, Entwistle S et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46(W1): W95-W101 (2018). The database file CAZyDB.07312018.fa (2018-08-24) was downloaded from http://bcb.unl.edu/dbCAN2/download/.
7. Lyu X, Shen C, Fu Y, Xie J, Jiang D et al. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci Rep 5:15565 (2015).
8. Zhao Z, Liu H, Wang C, Xu J-R. Erratum to: Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics 15(1):6 (2014).
9. Saier MH, Jr., Reddy VS, Tsu BV, Ahmed MS, Li C et al. The Transporter Classification Database (TCDB): recent advances. Nucleic Acids Res 44(D1): D372-D379 (2016).
10. Blin K, Medema MH, Kottmann R, Lee SY, Weber T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res 45(D1): D555-D559 (2017).
11. Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods, Correspondence 8:785 (2011).
12. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden markov model: application to complete genomes. J Mol Biol 305(3):567-580 (2001).
13. Horton P, Park KJ, Obayashi T, Fujita N, Harada H et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res 35(Web Server issue): W585-587 (2007).
14. The genome sequence and proteins of Parsp1, Perma1, Coral1, Ascra1, Stasp1, Epini1, Lepma1, Parno2, Altal1, Pyrtr1, BcDW1, B05.10, BcT4, Sclbo1, Sclsc2, Triat2, Triha1, Trivi2, Fusgr3, Magor2, Colhi1, Neucr2, Aspni2, Bipma1, Bipor1, Bipso1, Eryne1, Blugh1, Sacce2 and Schpo2 are available from the NCBI database and details of samples, BioProject accession codes and the download sites are given in Table S5.
15. The genome sequence of Karrh1, Bimnz1, Didsa1, Bysci1, Maseb1, Lenfl1, and Trepe1 are available from the JGI database; details of samples, BioProject accession codes and the download sites are given in Table S5.
Supplementary Data
Funding information
This research was supported by the National Key R and D Program of China (2017YFD0200400), the National Natural Science Foundation of China (Grant 31572048), and the earmarked fund for China Agriculture Research System (CARS-13).
Acknowledgements
We would like to acknowledge the anonymous reviewers for their work on this paper.
Author contributions
Y.F. and D.J. designed and managed the project. H.Z. and T.Z. collected materials, prepared and purified DNA and RNA samples for the genome sequencing and RNA-seq. H.Z., T.Z., J.X., T.C. and J.C., performed the genome assemblies and genome annotations. H.Z., Y.F. and D.J. wrote the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
No ethical issues are associated with this work as all sequence data analysed were acquired either from publicly accessible databases or were generated as part of this work.
Footnotes
Abbreviations: blast, basic local alignment search tool; bp, base pairs; CAZymes, carbohydrate-active enzymes; CBMs, carbohydrate-binding modules; CDD, conserved domain databases; DEG, different expression gene; FCWDEs, fungal cell wall-degrading enzymes; Gb, giga of base pairs; GliA, gliotoxin A; GliP, gliotoxin P; GO, gene ontology; GO-Slim, the high-level subset of GO; hpi, hour post-inoculation; JGI, DOE, Joint Genome Institute; Kbp, thousands of base pairs; KEGG, Kyoto Encyclopedia of Genes and Genomes; KOG, eukaryotic orthologous groups; Mb, millions of base pairs; MFS, major facilitator superfamily; NCBI, National Center for Biotechnology Information; Nr database, non-redundant protein sequences database; NRPS, non-ribosomal peptide synthases; PacBio, Pacific Bioscience; PCA, principal component analysis; PCWDEs, plant cell wall-degrading enzymes; Pfam domains, protein families and domains; PKS, polyketide synthases; RNA-seq, RNA sequencing; RRID, research resource identifier; Swiss-Prot, the section of the UniProtKB (universal protein knowledgebase); TCDB, the transporter classification databases.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Three supplementary figures and 13 supplementary tables are available with the online version of this article.
References
- 1.Sandys-Winsch C, Whipps JM, Gerlagh M, Kruse M. World distribution of the sclerotial mycoparasite Coniothyrium minitans . Mycol Res. 1993;97:1175–1178. doi: 10.1016/S0953-7562(09)81280-X. [DOI] [Google Scholar]
- 2.Li G, Wang D, Zhang S, Dan H. Characterization of the sclerotial parasite Coniothyrium minitans I: biological characteristics and the natural distribution in Hubei Province. J Huazhong Agr Univ. 1995;14:125–130. [Google Scholar]
- 3.Verkley GJM, da Silva M, Wicklow DT, Crous PW. Paraconiothyrium, a new genus to accommodate the mycoparasite Coniothyrium minitans, anamorphs of Paraphaeosphaeria, and four new species. Stud Mycol. 2004;50:323–335. [Google Scholar]
- 4.Campbell WA. A new species of Coniothyrium parasitic on sclerotia. Mycologia. 1947;39:190–195. doi: 10.1080/00275514.1947.12017603. [DOI] [Google Scholar]
- 5.Whipps JM, Gerlagh M. Biology of Coniothyrium minitans and its potential for use in disease biocontrol. Mycol Res. 1992;96:897–907. doi: 10.1016/S0953-7562(09)80588-1. [DOI] [Google Scholar]
- 6.Yang R, Han YC, Li GQ, Jiang DH, Huang HC. Suppression of Sclerotinia sclerotiorum by antifungal substances produced by the mycoparasite Coniothyrium minitans . Eur J Plant Pathol. 2007;119:411–420. doi: 10.1007/s10658-007-9174-0. [DOI] [Google Scholar]
- 7.JC Tu. Mycoparasitism by Coniothyrium minitans on Sclerotinia sclerotiorum and its effect on sclerotial germination. J Phytopathol. 1984;109:261–268. doi: 10.1111/j.1439-0434.1984.tb00716.x. [DOI] [Google Scholar]
- 8.Huang HC, Hoes JA. Penetration and infection of Sclerotinia sclerotiorum by Coniothyrium minitans . Can J Bot. 1976;54:406–410. doi: 10.1139/b76-039. [DOI] [Google Scholar]
- 9.Turner GJ, Tribe HT. On Coniothyrium minitans and its parasitism of sclerotinia species. Transactions Brit Mycol Soc. 1976;66:97–105. doi: 10.1016/S0007-1536(76)80098-8. [DOI] [Google Scholar]
- 10.McQuilken MP, Gemmell J, Whipps JM. Some nutritional factors affecting production of biomass and antifungal metabolites of Coniothyrium minitans . Biocontrol Sci Techn. 2002;12:443–454. doi: 10.1080/09583150220146022. [DOI] [Google Scholar]
- 11.Budge SP, Whipps JM. Potential for integrated control of Sclerotinia sclerotiorum in glasshouse Lettuce using Coniothyrium minitans and reduced fungicide application. Phytopathology. 2001;91:221–227. doi: 10.1094/PHYTO.2001.91.2.221. [DOI] [PubMed] [Google Scholar]
- 12.Elsheshtawi M, Elkhaky MT, Sayed SR, Bahkali AH, Mohammed AA, et al. Integrated control of white rot disease on beans caused by Sclerotinia sclerotiorum using Contans and reduced fungicides application. Saudi J Biol Sci. 2017;24:405–409. doi: 10.1016/j.sjbs.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerlagh M, Goossen-van de Geijn HM, Fokkema NJ, Vereijken PFG. Long-Term biosanitation by application of Coniothyrium minitans on Sclerotinia sclerotiorum infected crops. Phytopathology. 1999;89:141–147. doi: 10.1094/PHYTO.1999.89.2.141. [DOI] [PubMed] [Google Scholar]
- 14.Kamal MM, Savocchia S, Lindbeck KD, Ash GJ. Biology and biocontrol of Sclerotinia sclerotiorum (Lib.) de Bary in oilseed Brassicas. Australasian Plant Pathol. 2016;45:1–14. doi: 10.1007/s13313-015-0391-2. [DOI] [Google Scholar]
- 15.Benigni M, Bompeix G. Chemical and biological control of Sclerotinia sclerotiorum in witloof chicory culture. Pest Manag Sci. 2010;66:1332–1336. doi: 10.1002/ps.2019. [DOI] [PubMed] [Google Scholar]
- 16.Chitrampalam P, Figuli PJ, Matheron ME, Subbarao KV, Pryor BM. Biocontrol of lettuce drop caused by Sclerotinia sclerotiorum and S. minor in desert agroecosystems. Plant Disease. 2008;92:1625–1634. doi: 10.1094/PDIS-92-12-1625. [DOI] [PubMed] [Google Scholar]
- 17.Partridge DE, Sutton TB, Jordan DL, Curtis VL, Bailey JE. Management of sclerotinia blight of peanut with the biological control agent Coniothyrium minitans . Plant Disease. 2006;90:957–963. doi: 10.1094/PD-90-0957. [DOI] [PubMed] [Google Scholar]
- 18.GQ L, Huang HC, Acharya SN, Erickson RS. Effectiveness of Coniothyrium minitans and Trichoderma atroviride in suppression of sclerotinia blossom blight of alfalfa. Plant Pathol. 2005;54:204–211. [Google Scholar]
- 19.McLaren DL, Huang HC, Kozub GC, Rimmer SR. Biological control of sclerotinia wilt of sunflower with Talaromyces flavus and Coniothyrium minitans . Plant Dis. 1994;78:231–235. doi: 10.1094/PD-78-0231. [DOI] [Google Scholar]
- 20.Jones D, Johnson RPC. Ultrastructure of frozen, fractured and etched pycnidiospores of Coniothyrium minitans . Transactions Brit Mycol Soc. 1970;55:83–IN9. doi: 10.1016/S0007-1536(70)80098-5. [DOI] [Google Scholar]
- 21.Huang HC, Kokko EG. Ultrastructure of hyperparasitism of Coniothyrium minitans on sclerotia of Sclerotinia sclerotiorum . Can. J. Bot. 1987;65:2483–2489. doi: 10.1139/b87-337. [DOI] [Google Scholar]
- 22.Huang HC, Kokko EG. Penetration of hyphae of Sclerotinia sclerotiorum by Coniothyrium minitans without the formation of Appressoria. J Phytopathol. 1988;123:133–139. doi: 10.1111/j.1439-0434.1988.tb04460.x. [DOI] [Google Scholar]
- 23.Jones D, Gordon AH, Bacon JSD. Co-operative action by endo- and exo-β-(1→3)-glucanases from parasitic fungi in the degradation of cell-wall glucans of Sclerotinia sclerotiorum (Lib.) de Bary. Biochem J. 1974;140:47–55. doi: 10.1042/bj1400047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips AJL, Price K. Structural aspects of the parasitism of sclerotia of Sclerotinia sclerotiorum (Lib) de Bary by Coniothyrium minitans Campb. J Phytopathol. 1983;107:193–203. doi: 10.1111/j.1439-0434.1983.tb00537.x. [DOI] [Google Scholar]
- 25.Hu Y, Li G, Yang L. Characterization of factors affecting enzymatic activity of chitinase produced by mycoparasite Coniothyrium minitans . Chin J Appl Environ Biol. 2009;2010:226–229. [Google Scholar]
- 26.Giczey G, Kerenyi Z, Fulop L, Hornok L. Expression of cmg1, an Exo- -1,3-glucanase gene from Coniothyrium minitans, increases during sclerotial parasitism. Appl Environ Microbiol. 2001;67:865–871. doi: 10.1128/AEM.67.2.865-871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren L, Li G, Han YC, Jiang DH, Huang H-C. Degradation of oxalic acid by Coniothyrium minitans and its effects on production and activity of β-1,3-glucanase of this mycoparasite. Biol Control. 2007;43:1–11. doi: 10.1016/j.biocontrol.2007.06.006. [DOI] [Google Scholar]
- 28.Zeng F, Gong X, Hamid MI, Fu Y, Jiatao X, et al. A fungal cell wall integrity-associated MAP kinase cascade in Coniothyrium minitans is required for conidiation and mycoparasitism. Fungal Genet Biol. 2012;49:347–357. doi: 10.1016/j.fgb.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Wei W, Zhu W, Cheng J, Xie J, Jiang D, et al. Nox complex signal and MAPK cascade pathway are cross-linked and essential for pathogenicity and conidiation of mycoparasite Coniothyrium minitans . Sci Rep. 2016;6:24325. doi: 10.1038/srep24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng L-M, Zhang J, Han Y-C, Yang L, Wu M-de, et al. Degradation of oxalic acid by the mycoparasite Coniothyrium minitans plays an important role in interacting with Sclerotinia sclerotiorum . Environ Microbiol. 2014;16:2591–2610. doi: 10.1111/1462-2920.12409. [DOI] [PubMed] [Google Scholar]
- 31.Luo C, Zhao H, Yang X, Qiang C, Cheng J, et al. Functional analysis of the melanin-associated gene CmMR1 in Coniothyrium minitans . Front Microbiol. 2018;9:2658. doi: 10.3389/fmicb.2018.02658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lou Y, Han Y, Yang L, Wu M, Zhang J, et al. CmpacC regulates mycoparasitism, oxalate degradation and antifungal activity in the mycoparasitic fungus C oniothyrium minitans . Environ Microbiol. 2015;17:4711–4729. doi: 10.1111/1462-2920.13018. [DOI] [PubMed] [Google Scholar]
- 33.Han Y-C, Li G-Q, Yang L, Jiang D-H. Molecular cloning, characterization and expression analysis of a pacC homolog in the mycoparasite Coniothyrium minitans . World J Microb Biot. 2011;27:381–391. doi: 10.1007/s11274-010-0469-5. [DOI] [Google Scholar]
- 34.Wei W, Zhu W, Cheng J, Xie J, Li B, et al. CmPEX6, a gene involved in peroxisome biogenesis, Is essential for parasitism and conidiation by the sclerotial parasite Coniothyrium minitans . Appl Environ Microbiol. 2013;79:3658–3666. doi: 10.1128/AEM.00375-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hamid MI, Zeng F, Cheng J, Jiang D, Fu Y. Disruption of heat shock factor 1 reduces the formation of conidia and thermotolerance in the mycoparasitic fungus Coniothyrium minitans . Fungal Genet Biol. 2013;53:42–49. doi: 10.1016/j.fgb.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 36.He Z, Hu X, Fu Y, Jiang D. Metabolic profile of Coniothyrium minitans co-cultured with Sclerotinia sclerotiorum . J Huazhong Agr Univ. 2017;36:35–41. [Google Scholar]
- 37.McQuilken MP, Gemmell J, Hill RA, Whipps JM. Production of macrosphelide A by the mycoparasite Coniothyrium minitans . FEMS Microbiol Lett. 2003;219:27–31. doi: 10.1016/S0378-1097(02)01180-1. [DOI] [PubMed] [Google Scholar]
- 38.Jiang D, Li G, Yi X, Fu Y, Wang D. Studies on the properties of antibacterial substance produced by Coniothyrium minitans . Acta Phytopathol Sin. 1998;28:29–32. [Google Scholar]
- 39.Wang H, Hu X, Jiang D. Separation of the metabolic product of Coniothyrium minitans against Xanthomonas oryzae pv. oryzae. J Huazhong Agr Univ. 2009;28:148–150. [Google Scholar]
- 40.Yang L, Miao HJ, Li GQ, Yin LM, Huang H-C. Survival of the mycoparasite Coniothyrium minitans on flower petals of oilseed rape under field conditions in central China. Biological Control. 2007;40:179–186. doi: 10.1016/j.biocontrol.2006.10.002. [DOI] [Google Scholar]
- 41.Luo R, Liu B, Xie Y, Li Z, Huang W, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1 doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 43.Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15:211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, et al. Differential gene and transcript expression analysis of RNA-Seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haas BJ, Zeng Q, Pearson MD, Cuomo CA, Wortman JR. Approaches to fungal genome annotation. Mycology. 2011;2:118–141. doi: 10.1080/21501203.2011.606851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Besemer J, Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 2005;33:W451–W454. doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoff KJ, Lange S, Lomsadze A, Borodovsky M, Stanke M. BRAKER1: unsupervised RNA-Seq-Based genome annotation with GeneMark-ET and AUGUSTUS: table 1. Bioinformatics. 2016;32:767–769. doi: 10.1093/bioinformatics/btv661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9:R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee E, Helt GA, Reese JT, Munoz-Torres MC, Childers CP, et al. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 2013;14:R93. doi: 10.1186/gb-2013-14-8-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 55.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat Methods. 2008;5:621. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 61.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 62.Fischer S, Brunk BP, Chen F, Gao X, Harb OS, et al. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr Protoc Bioinformatics. 2011;Chapter 6:6.12.11–6.12.16. doi: 10.1002/0471250953.bi0612s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, et al. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018;46:W95–W101. doi: 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lyu X, Shen C, Fu Y, Xie J, Jiang D, et al. Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci Rep. 2015;5:15565. doi: 10.1038/srep15565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao Z, Liu H, Wang C, Xu J-R. Correction: comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2014;15:6. doi: 10.1186/1471-2164-15-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saier MH, Reddy VS, Tsu BV, Ahmed MS, Li C, et al. The transporter classification database (tcdB): recent advances. Nucleic Acids Res. 2016;44:D372–D379. doi: 10.1093/nar/gkv1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saier MH, Tran CV, Barabote RD. Tcdb: the transporter classification database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using diamond. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 69.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 70.Blin K, Medema MH, Kottmann R, Lee SY, Weber T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2017;45:D555–D559. doi: 10.1093/nar/gkw960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nielsen H. Predicting secretory proteins with SignalP. In: Kihara D, editor. Methods in Molecular Biology. Vol. 1611. New York, NY: Humana Press; 2017. pp. 59–73. editor. [DOI] [PubMed] [Google Scholar]
- 72.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 73.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 74.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 75.Horton P, Park K-J, Obayashi T, Fujita N, Harada H, et al. Wolf PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu Y, Jiang D, Xie J, Cheng J, Li G, et al. Ss-Sl2, a novel cell wall protein with PAN modules, is essential for sclerotial development and cellular integrity of Sclerotinia sclerotiorum . PLoS One. 2012;7:e34962. doi: 10.1371/journal.pone.0034962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeiner CA, Purvine SO, Zink EM, Paša-Tolić L, Chaput DL, et al. Comparative analysis of secretome profiles of Manganese(II)-oxidizing ascomycete fungi. PLoS One. 2016;11:e0157844. doi: 10.1371/journal.pone.0157844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turhan G. Further hyperparasites of Rhizoctonia solarni Kühn as promising candidates for biological control. J Plant Dis Protect. 1990;97:208–215. [Google Scholar]
- 79.Knapp DG, Németh JB, Barry K, Hainaut M, Henrissat B, et al. Comparative genomics provides insights into the lifestyle and reveals functional heterogeneity of dark septate endophytic fungi. Sci Rep. 2018;8:6321. doi: 10.1038/s41598-018-24686-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lopez D, Ribeiro S, Label P, Fumanal B, Venisse J-S, et al. Genome-wide analysis of Corynespora cassiicola leaf fall disease putative effectors. Front Microbiol. 2018;9:276. doi: 10.3389/fmicb.2018.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang H, Hyde KD, Mckenzie EHC, Bahkali AH, Zhou D. Sequence data reveals phylogenetic affinities of Acrocalymma aquatica sp. nov., Aquasubmersa mircensis gen. et sp. nov. and Clohesyomyces aquaticus (Freshwater Coelomycetes) Cryptogamie Mycol. 2012;33:333–346. doi: 10.7872/crym.v33.iss3.2012.333. [DOI] [Google Scholar]
- 82.Perlin MH, Andrews J, Toh SS. Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity. Adv Genet. 2014;85:201–253. doi: 10.1016/B978-0-12-800271-1.00004-4. [DOI] [PubMed] [Google Scholar]
- 83.Donzelli BGG, Harman GE. Interaction of ammonium, glucose, and chitin regulates the expression of cell wall-degrading enzymes in Trichoderma atroviride strain P1. Appl Environ Microbiol. 2001;67:5643–5647. doi: 10.1128/AEM.67.12.5643-5647.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Gruber S, Seidl-Seiboth V. Self versus non-self: fungal cell wall degradation in Trichoderma . Microbiology. 2012;158:26–34. doi: 10.1099/mic.0.052613-0. [DOI] [PubMed] [Google Scholar]
- 85.NAR G, Latge J-P, Munro CA. The fungal cell wall: structure, biosynthesis, and function. Microbiol Spectr. 2017;5:FUNK-0035-2016. doi: 10.1128/microbiolspec.funk-0035-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kubicek CP, Herrera-Estrella A, Seidl-Seiboth V, Martinez DA, Druzhinina IS, et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma . Genome Biol. 2011;12:R40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seidl V, Huemer B, Seiboth B, Kubicek CP. A complete survey of Trichoderma chitinases reveals three distinct subgroups of family 18 chitinases. Febs J. 2005;272:5923–5939. doi: 10.1111/j.1742-4658.2005.04994.x. [DOI] [PubMed] [Google Scholar]
- 88.Carsolio C, Benhamou N, Haran S, Cortés C, Gutiérrez A, et al. Role of the Trichoderma harzianum endochitinase gene, ech42, in mycoparasitism. Appl Environ Microbiol. 1999;65:929–935. doi: 10.1128/AEM.65.3.929-935.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 90.Yang X, Cui H, Cheng J, Xie J, Jiang D, et al. A HOPS protein, CmVps39, is required for vacuolar morphology, autophagy, growth, conidiogenesis and mycoparasitic functions of Coniothyrium minitans . Environ Microbiol. 2016;18:3785–3797. doi: 10.1111/1462-2920.13334. [DOI] [PubMed] [Google Scholar]
- 91.Keller NP. Fungal secondary metabolism: regulation, function and drug discovery. Nat Rev Microbiol. 2019;17:167–180. doi: 10.1038/s41579-018-0121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vinale F, Sivasithamparam K, Ghisalberti EL, Marra R, Barbetti MJ, et al. A novel role for Trichoderma secondary metabolites in the interactions with plants. Physiol Mol Plant Pathol. 2008;72:80–86. doi: 10.1016/j.pmpp.2008.05.005. [DOI] [Google Scholar]
- 93.Mukherjee PK, Horwitz BA, Kenerley CM. Secondary metabolism in Trichoderma – a genomic perspective. Microbiology. 2012;158:35–45. doi: 10.1099/mic.0.053629-0. [DOI] [PubMed] [Google Scholar]
- 94.Tomprefa N, McQuilken MP, Hill RA, Whipps JM. Antimicrobial activity of Coniothyrium minitans and its macrolide antibiotic macrosphelide A. J Appl Microbiol. 2009;106:2048–2056. doi: 10.1111/j.1365-2672.2009.04174.x. [DOI] [PubMed] [Google Scholar]
- 95.Xiang Y. Cloning and Functional Analysis of Melanins Associated Gene CmPKS1 in Coniothyrium minitans. Master Diss: Huazhong Agricultural University; 2011. [Google Scholar]
- 96.Scharf DH, Brakhage AA, Mukherjee PK. Gliotoxin - bane or boon? Environ Microbiol. 2016;18:1096–1109. doi: 10.1111/1462-2920.13080. [DOI] [PubMed] [Google Scholar]
- 97.Tsunawaki S, Yoshida LS, Nishida S, Kobayashi T, Shimoyama T. Fungal metabolite gliotoxin inhibits assembly of the human respiratory burst NADPH oxidase. Infect Immun. 2004;72:3373. doi: 10.1128/IAI.72.6.3373-3382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Spikes S, Xu R, Nguyen CK, Chamilos G, Kontoyiannis DP, et al. Gliotoxin production in Aspergillus fumigatus contributes to host-specific differences in virulence. J Infect Dis. 2008;197:479–486. doi: 10.1086/525044. [DOI] [PubMed] [Google Scholar]
- 99.Vargas WA, Mukherjee PK, Laughlin D, Wiest A, Moran-Diez ME, et al. Role of gliotoxin in the symbiotic and pathogenic interactions of Trichoderma virens . Microbiology. 2014;160:2319–2330. doi: 10.1099/mic.0.079210-0. [DOI] [PubMed] [Google Scholar]
- 100.Atanasova L, Crom SL, Gruber S, Coulpier F, Seidl-Seiboth V, et al. Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism. BMC Genomics. 2013;14:121. doi: 10.1186/1471-2164-14-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Woo SL, Scala F, Ruocco M, Lorito M. The molecular biology of the interactions between Trichoderma spp., phytopathogenic fungi, and plants. Phytopathology. 2006;96:181–185. doi: 10.1094/PHYTO-96-0181. [DOI] [PubMed] [Google Scholar]
- 102.Ruocco M, Lanzuise S, Vinale F, Marra R, Turrà D, et al. Identification of a new biocontrol gene in Trichoderma atroviride: the role of an ABC transporter membrane pump in the interaction with different plant-pathogenic fungi. Mol Plant Microbe Interact. 2009;22:291–301. doi: 10.1094/MPMI-22-3-0291. [DOI] [PubMed] [Google Scholar]
- 103.Karlsson M, Durling MB, Choi J, Kosawang C, Lackner G, et al. Insights on the evolution of mycoparasitism from the genome of Clonostachys rosea . Genome Biol Evol. 2015;7:465–480. doi: 10.1093/gbe/evu292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dubey MK, Jensen DF, Karlsson M. An ATP-binding cassette pleiotropic drug transporter protein is required for xenobiotic tolerance and antagonism in the fungal biocontrol agent Clonostachys rosea . Mol Plant Microbe Interact. 2014;27:725–732. doi: 10.1094/MPMI-12-13-0365-R. [DOI] [PubMed] [Google Scholar]
- 105.Oide S, Krasnoff SB, Gibson DM, Turgeon BG. Intracellular siderophores are essential for ascomycete sexual development in heterothallic Cochliobolus heterostrophus and homothallic Gibberella zeae . Eukaryot Cell. 2007;6:1339–1353. doi: 10.1128/EC.00111-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wallner A, Blatzer M, Schrettl M, Sarg B, Lindner H, et al. Ferricrocin, a siderophore involved in intra- and transcellular iron distribution in Aspergillus fumigatus . Appl Environ Microbiol. 2009;75:4194–4196. doi: 10.1128/AEM.00479-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mukherjee PK, Hurley JF, Taylor JT, Puckhaber L, Lehner S, et al. Ferricrocin, the intracellular siderophore of Trichoderma virens, is involved in growth, conidiation, gliotoxin biosynthesis and induction of systemic resistance in maize. Biochem Biophys Res Commun. 2018;505:606–611. doi: 10.1016/j.bbrc.2018.09.170. [DOI] [PubMed] [Google Scholar]
- 108.Sun X, Zhao Y, Jia J, Xie J, Cheng J, et al. Uninterrupted expression of CmSIT1 in a sclerotial parasite Coniothyrium minitans leads to reduced growth and enhanced antifungal ability. Front Microbiol. 2017;8:2208. doi: 10.3389/fmicb.2017.02208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Pazzagli L, Seidl-Seiboth V, Barsottini M, Vargas WA, Scala A, et al. Cerato-platanins: elicitors and effectors. Plant Sci. 2014;228:79–87. doi: 10.1016/j.plantsci.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 110.Gomes EV, Costa MdoN, de Paula RG, de Azevedo RR, da Silva FL, et al. The cerato-platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection. Sci Rep. 2015;5:17998. doi: 10.1038/srep17998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Crutcher FK, Moran-Diez ME, Ding S, Liu J, Horwitz BA, et al. A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by Trichoderma virens . Fungal Biol. 2015;119:476–486. doi: 10.1016/j.funbio.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 112.Guzmán-Guzmán P, Alemán-Duarte MI, Delaye L, Herrera-Estrella A, Olmedo-Monfil V. Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 2017;18:16. doi: 10.1186/s12863-017-0481-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.