Abstract
Due to limits on specificity and purity to allow for in-depth protein profiling, a standardized method for exosome isolation has yet to be established. In this study, we describe a novel, in-house microfluidic-based device to isolate exosomes from culture media and patient samples. This technology overcomes contamination issues because sample separation is based on the expression of highly specific surface markers CD63 and EpCAM. Top exosome proteins were identified based on their fold change and statistical significance between groups. Mass spectrometry revealed over 25 exosome proteins that are differentially expressed in high-grade serous ovarian cancer (HGSOC) cell lines compared to normal cells - ovarian surface epithelia cells (OSE) and fallopian tube secretory epithelial cells (FTSEC). Ingenuity Pathway Analysis identified STAT3 and HGF as top regulator proteins. We further validated exosome proteins of interest (pSTAT3, HGF, and IL-6) in HGSOC samples of origin-based cell lines (OVCAR-8, FTSEC) and in early stage HGSOC patient serum exosome samples using LC-MS/MS and Proximity Extension Assay (PEA). Our microfluidic device will allow us to make new discoveries for exosome-based biomarkers for the early detection of HGSOC and contribute to the development of new targeted therapies based on signaling pathways that are unique to HGSOC, both of which could improve the outcome for women with HGSOC.
Keywords: Exosomes, Microfluidics chip, proteomic profiling, ovarian cancer, STAT3
Introduction:
High-grade serous ovarian carcinoma (HGSOC) is an aggressive disease with a poor prognosis due to lack of early detection and the development of chemotherapy resistance (1–3). So far, no known specific and sensitive biomarkers, including CA125, have proven to be effective for early detection of ovarian cancer. Existing markers are not successful for early detection of ovarian cancer because: (I) they are not origin-specific for the disease; (II) they lack a clinically-relevant animal model for HGSOC; (III) and there is a lack of robust and reproducible technologies and data analysis for validation of protein biomarkers. Although early detection of high grade serous ovarian cancer is needed to improve overall-survival, currently, only 25% of ovarian cancers are detected at an early stage (4, 5).
Exosomes are extracellular vesicles, ~20–120 nm in diameter, that originate from multi-vesicular endosomes (MVEs) and play a key role in intercellular communication in cancers. They are highly stable, can be obtained from any biological fluid (6–9), and reflect the cell of origin, thus improving the sensitivity of biomarker amplification and reducing the number of false negative results. The protein and microRNA content of exosomes may serve as important biomarkers for various cancers, including ovarian cancer (10, 11). There is currently a lack of a standardized method that is reproducible, practical, and feasible for most labs for the isolation and purification of exosomes from cell culture medium. Comprehensive exploration of the proteome of biofluid-derived extracellular vesicles, such as exosomes, has been limited due to the difficulty in isolating circulating vesicles from clinical specimens, especially serum and plasma, with adequate yield and purity to allow for in-depth protein profiling (12–14). To address this challenge, we developed a microfluidic device to allow for efficient capture of exosomes based on their specific membrane markers from ovarian cancer cells and patient serum samples.
There is a growing body of evidence suggesting that the secretory epithelial cells in the distal fallopian tube (FT) serve as the point of origin for the majority of HGSOCs (15–17). Proteins secreted by tumors are found within the shed vesicles released from the tumor cells (18, 19). Detecting shed vesicle proteins with a high throughput technique such as mass spectrometry can improve specificity and sensitivity as compared to whole serum proteomics, as the presence of highly abundant proteins influences the dynamic range of detection (20–22). To circumvent this issue, we mined the exosomes from the conditioned media of epithelial ovarian cancer (EOC) cell lines and their normal counterparts, making a more specific biologic specimen to work with. The biologic contents within the exosomes reflect the physiologic state of the cell from which it is derived, making them a reliable and relevant source of the information they carry (10, 23, 24). We hypothesize that proteins shed or secreted by EOC cell lines within the exosomes are similar to those secreted or shed by EOC tumors and that these proteins can serve as biomarkers. In addition, analyzing the biological characteristics of ovarian cancer-derived exosomes can further help improve our understanding of tumor progression, and potentially help in monitoring the therapeutic effect(s) of treatment.
Recent studies highlight the comparative proteome profile of ovarian cancer cell lines with their precursor cell lines (25). However, to our knowledge there are no studies comparing the proteome profile of exosomes in HGSOC to the exosomes derived from their site of disease origin. Since HGSOC is commonly classified as epithelial in origin, we sought to isolate and characterize the proteome profile of exosomes released from a HGSOC cell line (OVCAR-8) and compare it with the exosomes derived from their precursor epithelial cells originated from normal fallopian tube secretory epithelial cells (FTSEC) and ovarian surface epithelial cells (OSE). By understanding signaling pathways that are significantly elevated in HGSOC cells, as compared to their normal counterparts, we aimed to identify possible targets for chemotherapy. We further evaluated key signaling molecules, along with downstream effector molecules, in normal and HGSOC patient samples to determine the clinical correlation and potential use of these molecules as biomarkers in the early detection and prognostication of ovarian cancer.
Material and Methods:
Exosome isolation by Microfluidic Device:
Immortalized FT33 cell lines were obtained from Dr. Ronny Drapkin (University of Pennsylvania). FT33 cells are very well characterized and commercially available from ABM or BioCat (15, 26, 27). Upon thawing, each vial of cells was passaged for a short time (n = 5) and tested for mycoplasma every 2 months. Ovarian cancer cells were grown to 70%–80% confluency in RPMI medium with 10% FBS. Media was removed and cells were rinsed three times with phosphate-buffered saline (PBS) and then grown in serum-free media. After 48 hrs. of incubation at 37 °C at 5% CO2, the conditioned media was collected, centrifuged once at 500 x g for 10 min., and then 2000 x g for 20 min. at 4 °C to eliminate cellular debris [10]. The cell-free supernatant was further concentrated using 100KD amicon filters (Millipore Sigma). For patient samples, cell-free serum was thawed on ice, diluted in PBS (final dilution dependent on the number of replicates in the experiment), filtered through a 0.2 μm syringe filter and concentrated for separation of exosomes by the microfluidic device.
Microfluidic Device Design and Fabrication:
Channels with herringbone patterns were fabricated using a multilayer photolithography process (EVG 620 Contact Aligner) (28). Following treatment with hexamethyldisilazane (HMDS), Sylgard 184 polydimethylsiloxane (PDMS) was used to create molds of the SU-8 patterns (Fig. 1A–B). Inlet and effluent ports were punched, channels were bonded to thin PDMS bases using air plasma at 1000 mTorr for 3 min. (Harrick Plasma Cleaner) and baked for 15 min. at 60 °C before beginning surface chemistry modification. The functionalization of the PDMS was performed to covalently bond antibodies (CD63) to the inner channel surface, as explained previously (29). Cell-free serum was thawed on ice, diluted in PBS, and filtered as before. Serum was flowed through the device and then flushed with filtered PBS. Effluent tubing was replaced and the endings of the tubing were placed in 2 ml microcentrifuge tubes preloaded with Tris-HCl buffer (1M, pH 8.5). Captured exosomes were eluted from the device with Glycine-HCl buffer (1M, pH 2.2) and stored at −20 °C until characterization.
Fig. 1:

A) Fabrication schematic showing photolithography (I-VI). B) Assembled devices with both parallel and in series connections in the microfluidic chip. C) Representative nanoparticle tracking analysis (NTA) showing a snap shot of the video recordings of the particles along with a plot of the particle size and concentration. D) Graph representing the vesicle concentration and size in nm for FTSEC, OSE and OVCAR-8. E) Bar graph showing the concentration of the exosomes derived from three biological replicates of FTSEC, OSE and OVCAR-8 cells (n=3±SEM; p≤ 0.05, 0.01) normalized to their cell counts. F) Representative classical TEM images of the vesicles isolated from FTSEC, OSE and OVCAR-8 cells (scale bar-100 nm). G) Protein expression of exosome specific markers – CD-63, TSG-101 and epithelial cell specific marker EpCAM confirmed by Western blots.
RESULTS:
Comparison of Microfluidic chip (MFD) based exosome isolation with conventional technologies from HGSOC cells
We isolated exosomes from conditioned media in OSE, FTSEC and OVCAR-8 cell lines using our microfluidic device (Fig. 1A & B) that separates the vesicles from culture medium and serum based on the presence of exosome-based surface marker CD63 and epithelial cell specific marker EpCAM, as shown in the schematic of the work flow (Sup. Fig. 1A). To validate our technique, we measured the concentration and size of the isolated molecules by Nanoparticle Tracking Analyzer (NTA) (Fig. 1C), which measures the hydrodynamic diameter of the vesicles based on the Brownian movement of the particles. We found a significant increase in vesicle concentration released from OVCAR-8 cancer cells when compared to the normal FTSEC and OSE cells (Fig. 1D & E). Transmission electron microscopy was performed to measure the size of vesicles isolated by our microfluidic device (Fig. 1F), and ranged from 30–150nm. In addition, when evaluating the protein expression of the epithelial cell marker EpCAM, we found an increase in EpCAM-positive exosomes in those derived from OVCAR-8, compared to the FTSEC and OSE cells, as shown in western blot with control markers TSG101 and CD63 (Fig. 1G).
Given that the most common form of ovarian cancer, HGSOC, is derived from epithelial cells, we further tested for the epithelial specificity of the isolated vesicles by labeling them with FITC and measuring the fluorescence intensity using Image stream analysis (ISA). ISA showed a dramatic increase in the capture of the EpCAM-positive exosomes by the microfluidics device (MFD), as compared to the other two methods as gated in Sup. Fig. 1B & Fig 2A. We selected this as the best option among the three techniques in moving forward with exosome isolation, as it offered quicker processing, lower cost, higher specificity, and better yield – traits attractive clinical translation. Next, we used NTA to compare the total vesicle concentration isolated from TR127 cell culture media with the three different techniques. A higher vesicle concentration was observed in the commercial exosome isolation kit method compared to ultracentrifugation (UC) and MFD (Sup. Fig. 1C). The total concentration of the vesicles isolated from the MFD using CD9 antibody for the immune-affinity capture was similar to the concentration in the UC method. The peaks were observed for smaller sized vesicles (<100nm) in both techniques, reflecting a consistency in the size of the vesicles isolated. However, the kit method isolated a greater number of larger vesicles (>100nm), as shown in the graph (Fig. 2B). Though a significant difference in the total concentration of the vesicles was observed using MFD with CD9 immune-affinity capture, EpCAM-based capture more specifically targeted vesicles derived from epithelial cells. This suggests that EpCAM is a better target for epithelial-cell derived vesicles. We also observed a lower mean co-efficient of variation (CV) across the biological replicates in Mass Spectrometry proteomic profiling in our MFD, suggesting that this method is more reproducible than commercial kit isolation (n=5) (Fig. 2C). Further the spectral count distribution of the peptides and a pairwise comparison for the biological replicates of exosomes isolated by MFD showed greater consistency and reproducibility than the conventional UC and commercial Kit methods. (Fig. 2D&E). MFD shows a very strong positive correlation with replicates for the peptide spectral matches (PSMs) for each protein listed (Pearson correlation = 0.95 – 0.98) while the kit method shows less precision and larger variability (Pearson correlation ranges from 0.22 to 0.81) (Fig. 2F). The above results favor the reliability and predict the translational opportunity of the microfluidics device in the use for the isolation of exosomes.
Fig. 2:
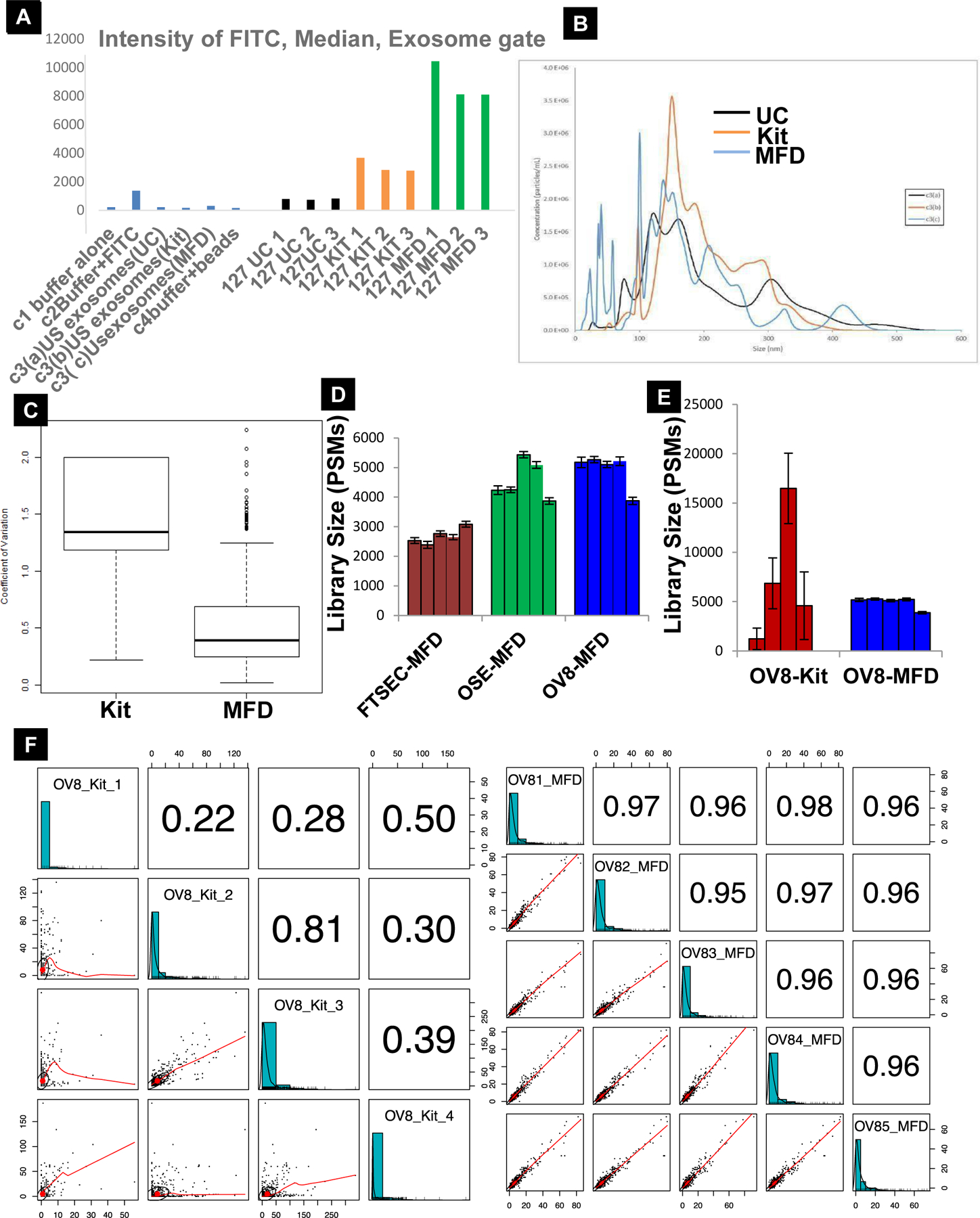
A) Intensity of the FITC median exosomes captured in different techniques (n=3), unstained (US) exosomes controls for different techniques. B) NTA analysis showing the average concentration and size of the vesicles captured via UC, Kit and MFD (n=3). C) Reproducibility and consistency of our MFD is demonstrated by the lower mean coefficient of variation over the Kit method (n=5, p≤0.05). D & E) Count distribution of the exosomes isolated by MFD across ovarian cell lines and within the same cell line using kit and MFD for technology validation. Error bars represent SE associated within each sample set (n = 4 for kit, n = 5 for all others). F) Pairwise comparisons depicting the reproducibility of MFD (OV81-OV85, right panel) over the commercial kit method (OV8_Kit_1-OV8_Kit_4, left panel). MFD shows a very strong positive correlation with replicates for peptide spectral matches (PSMs) for each protein listed (Pearson correlation = 0.95 – 0.98) while the kit method shows less precision and larger variability (Pearson correlation ranges from 0.22 to 0.81). Text size of the Pearson correlation correspond to how highly correlated the samples are.
Exosomes derived from HGSOC cells reveal common deregulated molecules
Mass spectrometry of exosomes isolated from FTSEC, OSE and OVCAR-8 cells (n=5) led to the identification of 988 total protein groups across all samples: 355 protein groups in FTSEC, 473 protein groups in OSE and 870 in OVCAR-8, with a common overlap of 195 protein groups (FDR = 2.6%, Sup. Fig. 2A). Prior to EdgeR analysis, we performed principal component analysis (PCA) and visualized count distribution with boxplots on the raw counts and TMM normalized counts. We saw clear separation of the three cell types with a 95% CI, and it appears that the exosome proteome of OVCAR-8 cancer cells has been highly diversified from the exosomes of normal FTSEC and OSE cells (Sup. Fig. 2B & C). The EdgeR Mean variance plot shows a positive correlation with significant overlaps across the proteins in all three comparisons (Sup Fig. 3A–C). The MA plots following filtering, TMM normalization, and exact test demonstrate the relationship between protein abundance (reported as log counts per million or logCPM) and expression (log fold change or logFC) of each protein group across the pairwise comparisons. This analysis resulted in a total (up- and down-regulated) DE of 335 protein groups in FTSEC/OVCAR-8, 463 protein groups in OSE/OVCAR-8 and 176 protein groups in FTSEC/OSE with an absolute FC greater than four (Fig. 3A–i, 2A–ii and Sup. Fig. 4A & B). Since we are interested in identifying up-regulated proteins and their associated pathways in HGSOC cells for further validation in biomarker studies and targeted therapies, we chose to focus on the up-regulated DE protein groups moving forward. The number of DE protein groups in the OSE/OVCAR-8 comparison was 407, while 306 were up-regulated in FTSEC/OVCAR-8 (Fig. 3B). The top differentially expressed proteins significantly up-regulated in FTSEC/OVCAR-8 and OSE /OVCAR-8 (Fig. 3C–i, 3C–ii; Sup. Fig. 5A–C) included hepatocyte growth factor (HGF) and slit homolog 2 protein (SLIT2), log 2 FC > 8, p-value < 0.0001.
Fig. 3:
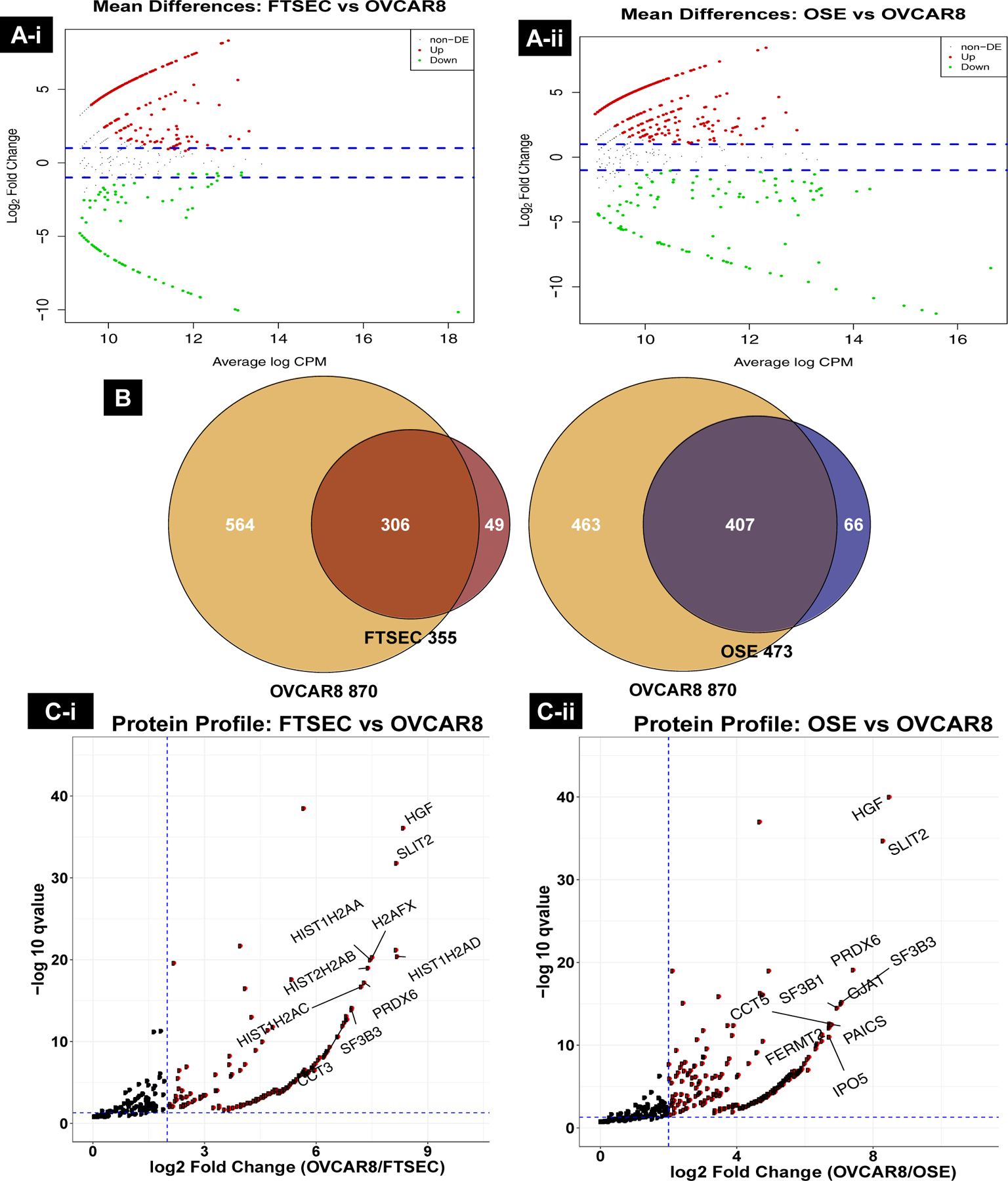
A) (i & ii): Differential protein expression resulting from EdgeR analysis of FTSEC (i) and OSE (ii) vs OVCAR-8 comparisons. The blue ablines represent log 2 fold change (FC) values greater than 2-fold difference in expression. Proteins in red are up-regulated in OVCAR-8 and those in green are up-regulated in FTSEC and OSE. B) Proteomic profiling of exosomes identified 306 and 407 up-regulated proteins in OVCAR-8 over FTSEC (left) and OSE (right), respectively. C) (i & ii) Top differentially up-regulated proteins in OVCAR-8 exosomes. Vertical abline is a log 2 FC of 2 or four-fold difference in expression while horizontal abline is a qvalue threshold of < 0.05. Proteins in red are up-regulated in OVCAR-8.
Prediction of signaling pathways in HGSOC cells
Hierarchical clustering analysis performed on all the candidate protein lists identified across the three sample sets (Fig. 4A) shows individual clusters of proteins that were differentially upregulated in the OVCAR-8 exosomes (Fig. 4B). Their functional significance, as identified by PANTHER Classification System, is nested primarily in signaling pathways and molecules related to integrin signaling and inflammation mediated by chemokines. (Table 1). The Gene Ontology (GO) term analysis associated with the molecular functions, biological process and cellular components of those differentially upregulated proteins in OVCAR-8 exosomes is shown in Sup. Table 1, 2 & 3. In this study, we validated the presence of HGF, STAT3 and IL-6 by western blot in the parental cell lysates from which the exosomes were isolated to confirm their origin from FTSEC, OSE, OVCAR-8 (Fig. 4C). Additionally, we also validated the expression of these proteins within the exosomes of other HGSOC cell lines and patient-derived primary cells (Fig. 4D). IPA core analysis (http://www.ingenuity.com) performed on the differentially expressed proteins predicted the upregulation of integrin and PI3K/AKT signaling along the HGF-mediated signaling pathway, and a downregulation of the HIPPO and PTEN signaling mechanisms in OVCAR8 cells, based on their exosome protein dataset (expressed as Log-p-values, Fig. 5A & B) when compared to FTSEC/OSE cells (Sup. Fig. 6A). Furthermore, these proteins were shown to be involved in cancer, reproductive system disease, cellular movement, inflammatory disease, cellular growth, and proliferation, as shown by IPA (Sup. Fig. 6B & C). The HGF network pathways predicted, along with its downstream effector proteins, are shown in Sup. Fig. 7A & B. The most significant proteins in this pathway that were found in our exosome data is shown in Sup. Table 4.
Fig. 4:

A)A cluster analysis of exosomal proteins identified in OVCAR-8, FTSEC and OSE cell lines. Filtering criteria required at least 3 observations in each replicate with a minimum of 10 peptide spectral matches (PSMs). B) Cluster showing differential upregulated proteins in OVCAR-8 exosomes like HGF, FAS, PCNA, SLIT2, NID1 & CD81. C) Validation of HGF, IL6 and STAT3 in the parental cell lysates from FTSEC, OSE, OVCAR-8. D) Validation of HGF, pSTAT3 and IL6 expression within exosomes isolated from patient derived primary cells and HGSOC cell lines - OVCAR-4, TR-127, TR-182 and PEOC1. The protein expressions were normalized to GAPDH as a loading control.
Table: 1.
Up-regulated protein pathways associated with the exosome proteome identified by Pantherdb analysis in OVCAR-8 exosomes when compared to their precursor cells’ exosomes.
| S.NO | Signaling Pathways | Total No: genes | % gene hit against total # genes | % gene hit against total # process hits |
|---|---|---|---|---|
| 1 | Integrin signaling (P00034) | 44 | 8.20% | 10.10% |
| 2 | Inflammation mediated by chemokine and cytokine signaling pathway(P00031) | 19 | 3.50% | 4.40% |
| 3 | Cytoskeletal regulation by Rho GTPase (P00016) | 28 | 5.20% | 6.40% |
| 4 | EGF receptor signaling pathway(P00018) | 11 | 2.00% | 2.50% |
| 5 | Cadherin signaling pathway(P00012) | 10 | 1.90% | 2.30% |
| 6 | FGF signaling pathway(P00021) | 13 | 2.40% | 3.00% |
| 7 | Wnt signaling pathway(P00057) | 12 | 2.20% | 2.80% |
| 8 | Glycolysis(P00024) | 11 | 2.00% | 2.50% |
| 9 | Heteromeric G-protein signaling pathway-Giα and GSα mediated pathway(P00026) | 10 | 1.90% | 2.30% |
| 10 | Gonadotropin–releasing hormone receptor pathway(P06664) | 10 | 1.90% | 2.30% |
| 11 | Heteromeric G-protein signaling pathway-Gqα and Goα mediated pathway(P00027) | 9 | 1.70% | 2.10% |
| 12 | Cell cycle(P00013) | 7 | 1.30% | 1.60% |
| 13 | Plasminogen activating cascade(P00050) | 4 | 0.70% | 0.90% |
| 14 | P53 pathway(P00059) | 6 | 1.10% | 1.40% |
| 15 | PI3 kinase pathway(P00048) | 3 | 0.60% | 0.70% |
Fig. 5:
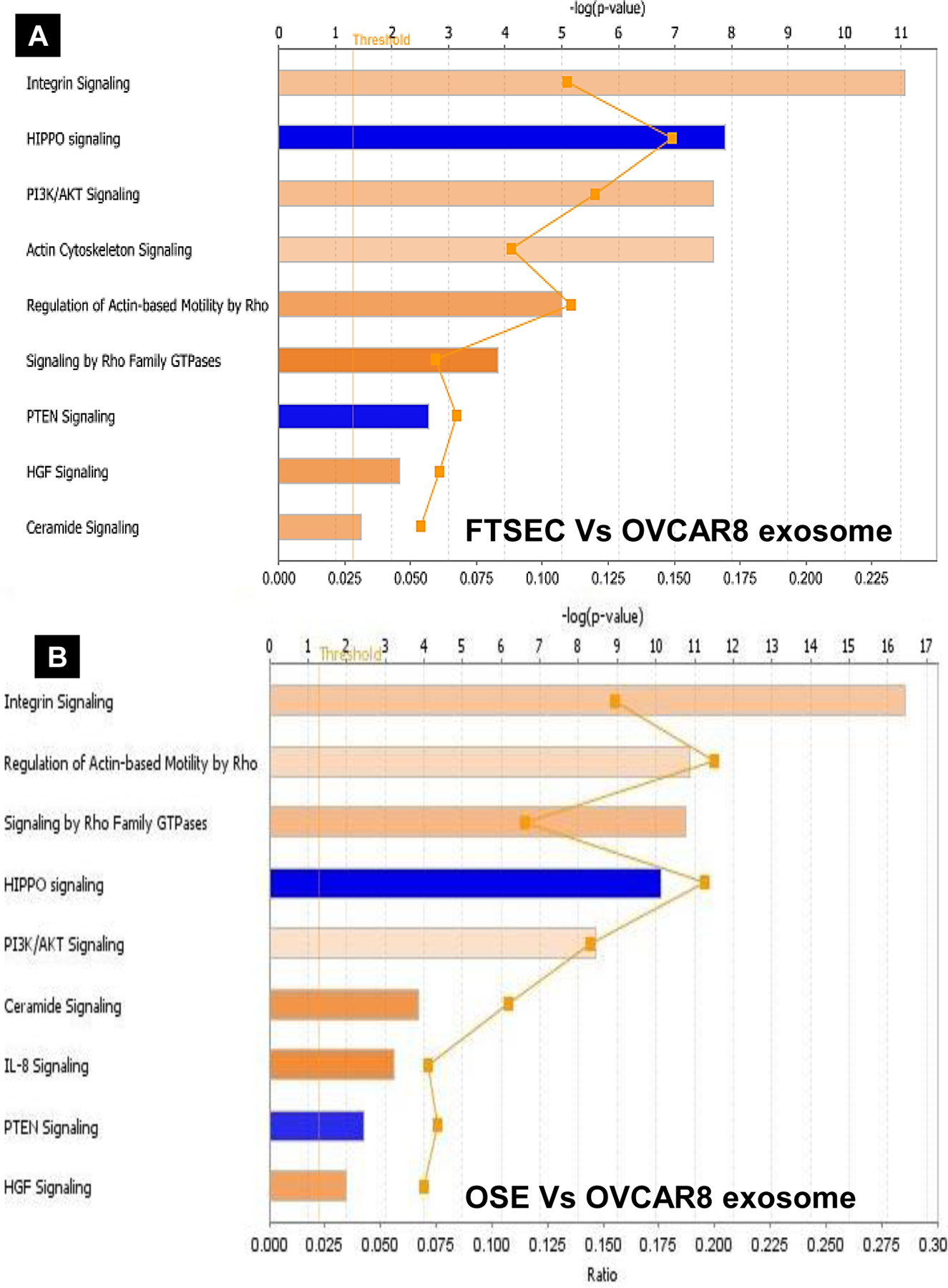
IPA core analysis revealing the top canonical pathways associated with the exosome proteins in A) FTSEC vs OVCAR-8 and B) OSE vs OVCAR-8. Values are expressed as -log(p-value) above a significance threshold based on Fisher’s exact test (p < 0.05).
Evaluation of HGF, IL-6 and STAT3 expression as early indicators of HGSOC
HGF was predicted to be the top upregulated molecule in both FTSEC/OVCAR-8 and OSE/OVCAR-8 comparisons, showing a log FC of 8.3 and 8.5, respectively (Sup. Table 5 & 6). Our lab has previously reported that cancer exosomes carry the activated form of STAT3 (10, 30) and that STAT3 is a known downstream effector of HGF activation (31). Furthermore, we analyzed the proportion of epithelial cell specific EpCAM positive vesicles (Exo-Red labeled) relative to the total CD9-positive vesicle populations (FITC labeled) within each sample by Image stream analysis. The results confirmed a higher proportion of EpCAM positive vesicles in HGSOC patient serum exosome sample as compared to benign sample (Fig. 6A). Protein expression of HGF, STAT3 and IL6 in serum exosomes of benign and HGSOC patient samples, analyzed by Western blot and ELISA were significantly elevated in early stage samples (Fig. 6B, C & Sup. Fig. 7A & B) as represented in Heat map and boxplot (Fig. 6D). Also, the Immunohistochemistry performed on patient tissues showed increased expression of the same proteins in stage I, and stage IV human ovarian tissues, which helped us determine the origin of these proteins. Additionally, stage I tissue samples showed a higher percentage of expression when compared to benign and stage IV; therefore, suggesting that HGF, STAT3 and IL-6 represent valuable candidates as early indicators of ovarian cancer for future validation studies.
Fig. 6:
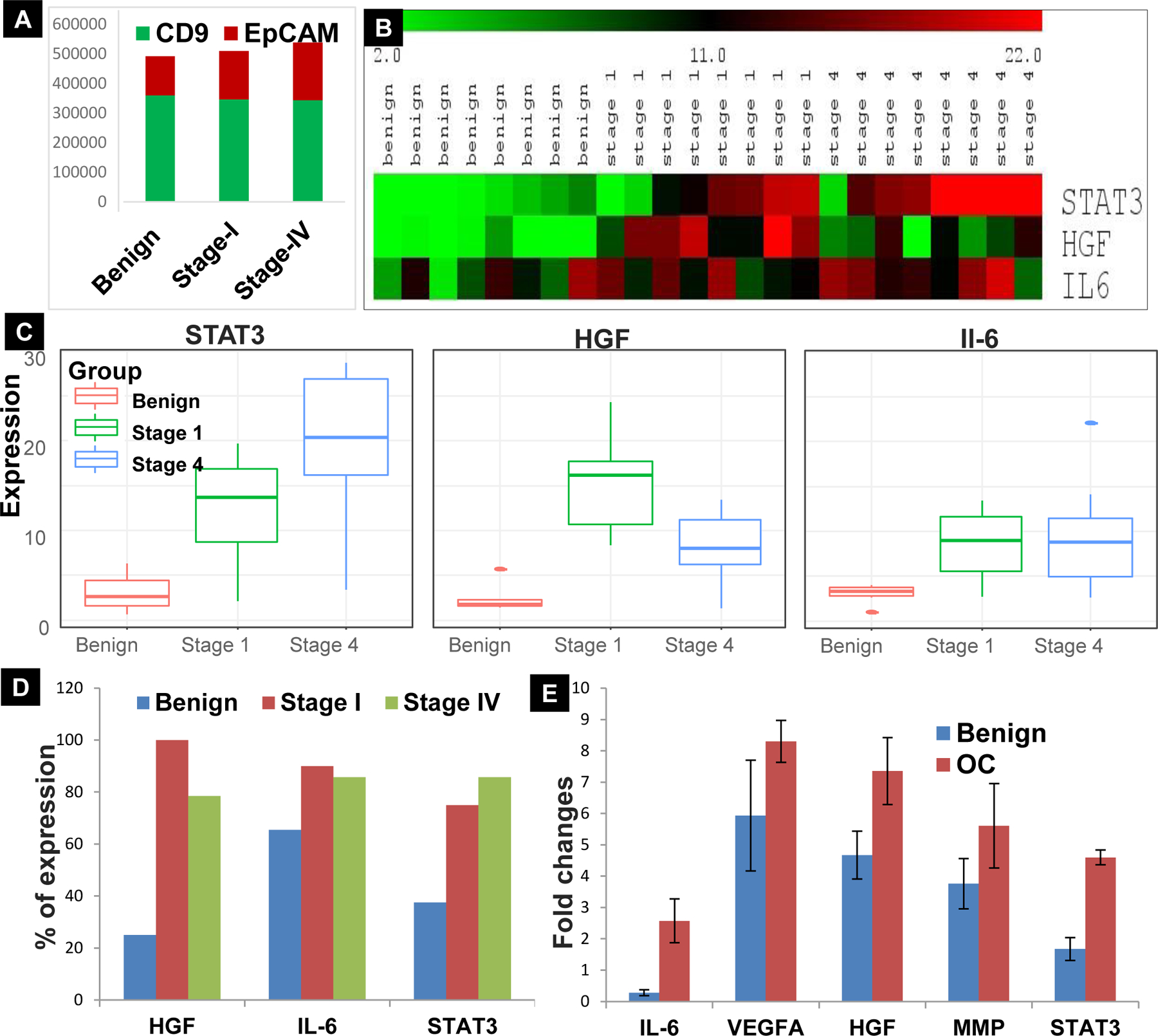
A) Stage wise comparison showing relative proportion of EpCAM positive (Exo-red labeled) vesicles to the total population of CD9 positive (FITC–green labeled) in Benign, Stage-I and IV serum exome sample measured by Fluorescence intensity on Image stream analysis (ISA). B) Expression levels of STAT3, HGF and IL-6 in Benign, Stage-1 and Stage-IV patient serum exosome samples (n=8). C) Levels of HGF (n=13), STAT3 (n=9) and IL-6 (n=9) in benign, Stage-1 and IV serum exosomes (p < 0.05); D) Immunohistochemistry analysis for HGF, IL-6 and STAT3 expression performed in patient tissues show increased percentage of expression in Stage-1 when compared to Benign and Stage-IV tissues (n=4). E) Proximity extension assay in patient serum exosome samples of Benign and stage-IV HGSOC patients (n=5) showing alterations in the levels of STAT3 related proteins–IL-6, VEGF-A, HGF, MMP2.
To account for the reproducibility of the proteins identified through mass spectrometry, we also analyzed a total of 12 exosome lysates from five benign and seven HGSOC patient serum samples for a panel of 92 immuno-oncology proteins by proximity extension assay (PEA). With a FC of 1.5 and P<0.02 we identified the top proteins as given in Sup. Fig. 8 & Sup. Table 7. We observed reproducible results for a few proteins in PEA that were initially identified in Mass spectrometry as well (Fig. 6E), along with other proteins that were found to be significantly elevated in HSGOC exosomes when compared to benign samples in PEA alone. These proteins can be validated in future studies to elucidate their significance in the context of biomarkers.
Discussion:
Currently, there is no exosome-based biomarker known for early detection of ovarian cancer. Using a novel technology with our in-house developed MFD, we present a new method to isolate and purify exosomes to aid in identification of differentially expressed proteins for biomarker candidates in HGSOC. In the present study, we have shown the following important findings: (a) Successful development of a microfluidic-based device for intact exosome isolation necessary for downstream processing with high purity and quality compared with conventional standard technology; (b) Identification of a protein signature for HGSOC by utilizing disease origin cell lines FTSEC and OSE; (c) HGF, STAT3, and IL-6 are highly elevated in early stage HGSOC patient serum exosomes, compared to benign and late stage HGSOC.
Conventional methods to isolate exosomes in research labs are technically challenging, involve laborious ultracentrifugation, require a large sample volume, and are time consuming (13, 14). Commercial kits are costly and are non-specific. These drawbacks can be overcome through a microfluidic approach that can expedite exosomal isolation in a small sample volume with great precision and detection sensitivity. Microfluidic devices play an important role in many applications involving Biology, Chemistry, and Engineering, as there are many ways to fabricate the channel and customize the dimensions to specific needs. We successfully developed and fabricated a microfluidic chip using soft lithography with PDMS, which is a relatively inexpensive material. The total cost of the single chip is ~ $2-$5, as compared to the $1,070 kit used for 20 samples. Because of its relative cost and benefits, with special consideration to the commercialization prospects, microfluidics technology will be of great use to mankind if it is developed wisely. These advancements have allowed microfluidic devices to be fabricated using a wide range of materials and geometries, enabling new and advantageous physical behaviors and qualities in microfluidic devices.
We have demonstrated in our prior publications, as well as these current results, that a microfluidic device can directly isolate and fairly pure population of the exosomes using a specific surface marker and release them fully intact for further characterization and analysis (29). We have compared our technology to conventional isolation methods, including ultracentrifugation and a commercial exosome isolation kit. Our microfluidic chip showed higher yield, higher specificity, and comparatively rapid processing (<20 minutes for capture and release). In addition, it requires minimal sample volume, providing potential to be utilized as a rapid-screening tool in clinical samples. Also, previous reports showed that the conventional centrifuge method for exosome isolation contains contaminating proteins, and can even lead to vesicle damage. For these reasons, we adopted our MFD in this current study to isolate exosomes from cell lines and clinical samples (Fig. 6 and Table 1), which allowed for EpCAM- positive epithelial cell based selective isolation of exosomes in HGSOC and minimized damage to vesicles.
Using different techniques, we identified more than 20 unique exosomal proteins as high-priority candidate protein biomarkers based on their statistical significance and fold-change. Mass spectrometry of patient serum exosomes is a novel approach for diagnosis and disease monitoring. Pantherdb and IPA were used to understand the biological pathways associated with the exosome proteomes identified in the different cell groups. The integrin signaling pathway and cytokine-mediated inflammation pathways were the most prominent pathways elevated in the OVCAR-8 cells, suggesting that cytokines play an important role in cancer development and disease progression. The IPA analysis also showed that HGF was at the top of the canonical pathways that predicted the activation of specific cytokine signaling pathways. This further convinced us to consider HGF as a target for an in-depth analysis of the activation of its receptor cMet, a proto-oncogene. Since binding of HGF to the c-MET receptor has mitogenic, motogenic, and morphogenic effects on cells, we strongly believe that the HGF/cMet axis plays an important role in cancer development and progression (32–35).
Based on our pathway analysis by IPA and evaluation of HGF and its downstream effectors such as IL-6 and STAT3, we found that the HGF/STAT3 axis might be a potential therapeutic target in HGSOC. Clustering analysis of the FTSEC and OSE exosome proteome with the OVCAR-8 exosome proteome provides support that FTSEC and OSE are closely related and formed a single cluster, and that OVCAR-8 was highly diversified from the precursor cells FTSEC and OSE. The underlying differences between the exosome proteome in the two different clusters were also reflected in the biological pathways associated with their respective proteins. However, we cannot arrive at conclusions using a single HGSOC cell line, and these results need to be further elaborated using exosomes from different ovarian cancer cell lines in comparison to the exosomes from their precursor cells in order to substantiate or reject this concept. In addition, given that some of the proteins identified have been shown to also be elevated in non-ovarian cancers, the specificity of these exosomal proteins for ovarian cancer needs to be further delineated. We believe this could be done through further modification of the MFD to specifically identify ovarian epithelial exosomes, as well as expanding these studies to include non-HGSOC epithelial ovarian cancers and through larger cohort studies evaluating the differential expression of these proteins among different epithelial cancers in general.
The use of exosome proteomic profiling across different cell lines provides researchers in the field of ovarian cancer with additional resources for their research. Our current study showed that we can successfully isolate intact, label-free exosomes from cell lines and clinical samples using our microfluidic device, which saves time, cost and can be clinically translated at ease. We have also identified origin-based exosome proteins signatures (HGF, STAT3, IL-6, MMP7, and VEGF-A) in clinical specimens. Many previously identified biomarkers such as HGF, IL-6 and STAT3 were not confirmed in subsequent studies (36–38). The lack of confirmation is due to several issues, perhaps the most notable being that, for serum markers, the complexity or the serum proteome is very high and has broad dynamic range. Serum proteins that may serve as biomarkers are usually low abundance proteins that are very difficult to detect and tend to have higher variations. In exosomes, the proteome is less complex (due to the smaller number of proteins present) and has a reduced dynamic range. In addition, there is evidence that cancer patients have increased serum exosome content, presumably from cancer tissues. Therefore, exosomes (HGF, IL-6 and STAT3) are more likely to contain proteins (HGF, IL-6, STAT3) that can serve as ovarian cancer biomarkers, thus making biomarker discovery and confirmation in exosomes an easier task.
The identification of these proteins provides the opportunity for translational research investigating the use of these proteins for the early detection of and targeted therapies for HGSOC, which could ultimately lead to improved prognosis and survival in patients with HGSOC. Further exploration of the integrin signaling pathways could help to understand the role of exosomes in disease metastasis, especially given that the initial step in ovarian carcinoma dissemination occurs by the attachment of carcinoma cells onto the peritoneal surface via integrins.
Supplementary Material
Significance:
A unique platform utilizing a microfluidic device enables the discovery of new exosome-based biomarkers in ovarian cancer.
Acknowledgements:
The authors are thankful to the undergraduate students Puneet Modgil, Roshan Sivakumar, Marissa Werner and Audrey Whitaker for the cell culture and basic assay. We thank Dr. Zhang Liwen for proteomic support and Alex Cornwell for Image Stream Analysis.
Financial Support: This work was funded by NCI RO1-CA176078 grant (K.S. and D.E.C) and KOH ovarian cancer foundation grant to DKDP, the Beucler Family Fund (D.E. Cohn), NCI R01GM122436 (Michael Freitas) and Pelotonia fellowship (Miranda Gardner).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References:
- 1.Bowtell DD, Bohm S, Ahmed AA, Aspuria PJ, Bast RC Jr., Beral V, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nature reviews Cancer. 2015;15:668–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis A, Tinker AV, Friedlander M. “Platinum resistant” ovarian cancer: what is it, who to treat and how to measure benefit? Gynecologic oncology. 2014;133:624–31. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA: a cancer journal for clinicians. 2019;69:7–34. [DOI] [PubMed] [Google Scholar]
- 4.Terry KL, Schock H, Fortner RT, Husing A, Fichorova RN, Yamamoto HS, et al. A Prospective Evaluation of Early Detection Biomarkers for Ovarian Cancer in the European EPIC Cohort. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:4664–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIntosh MW, Drescher C, Fitzgibbon MM. Ovarian Cancer Early Detection Needs Better Imaging, Not Better Algorithms or Biomarkers. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016;34:199–200. [DOI] [PubMed] [Google Scholar]
- 6.Shender VO, Pavlyukov MS, Ziganshin RH, Arapidi GP, Kovalchuk SI, Anikanov NA, et al. Proteome-metabolome profiling of ovarian cancer ascites reveals novel components involved in intercellular communication. Molecular & cellular proteomics : MCP. 2014;13:3558–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalluri R. The biology and function of exosomes in cancer. The Journal of clinical investigation. 2016;126:1208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. Journal of molecular medicine. 2013;91:431–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorayappan KD, Wallbillich JJ, Cohn DE, Selvendiran K. The biological significance and clinical applications of exosomes in ovarian cancer. Gynecologic oncology. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecologic oncology. 2008;110:13–21. [DOI] [PubMed] [Google Scholar]
- 12.Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H. New Technologies for Analysis of Extracellular Vesicles. Chemical reviews. 2018;118:1917–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Momen-Heravi F, Balaj L, Alian S, Mantel PY, Halleck AE, Trachtenberg AJ, et al. Current methods for the isolation of extracellular vesicles. Biological chemistry. 2013;394:1253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liga A, Vliegenthart AD, Oosthuyzen W, Dear JW, Kersaudy-Kerhoas M. Exosome isolation: a microfluidic road-map. Lab on a chip. 2015;15:2388–94. [DOI] [PubMed] [Google Scholar]
- 15.Perets R, Wyant GA, Muto KW, Bijron JG, Poole BB, Chin KT, et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer cell. 2013;24:751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karst AM, Levanon K, Drapkin R. Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proc Natl Acad Sci U S A. 2011;108:7547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perets R, Drapkin R. It’s Totally Tubular….Riding The New Wave of Ovarian Cancer Research. Cancer research. 2016;76:10–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dutta DK, Dutta I. Origin of ovarian cancer: molecular profiling. J Obstet Gynaecol India. 2013;63:152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eckert MA, Pan S, Hernandez KM, Loth RM, Andrade J, Volchenboum SL, et al. Genomics of Ovarian Cancer Progression Reveals Diverse Metastatic Trajectories Including Intraepithelial Metastasis to the Fallopian Tube. Cancer discovery. 2016;6:1342–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Li C, Zhou T, Liu X, Liu X, Li X, et al. Role of exosomal proteins in cancer diagnosis. Molecular cancer. 2017;16:145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He M, Zeng Y. Microfluidic Exosome Analysis toward Liquid Biopsy for Cancer. Journal of laboratory automation. 2016;21:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elzek MA, Rodland KD. Proteomics of ovarian cancer: functional insights and clinical applications. Cancer metastasis reviews. 2015;34:83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li A, Zhang T, Zheng M, Liu Y, Chen Z. Exosomal proteins as potential markers of tumor diagnosis. Journal of hematology & oncology. 2017;10:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J, Li J, Huang B, Liu J, Chen X, Chen XM, et al. Exosomes: novel biomarkers for clinical diagnosis. TheScientificWorldJournal. 2015;2015:657086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coscia F, Watters KM, Curtis M, Eckert MA, Chiang CY, Tyanova S, et al. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nature communications. 2016;7:12645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karst AM, Drapkin R. Primary culture and immortalization of human fallopian tube secretory epithelial cells. Nature protocols. 2012;7:1755–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levanon K, Ng V, Piao HY, Zhang Y, Chang MC, Roh MH, et al. Primary ex vivo cultures of human fallopian tube epithelium as a model for serous ovarian carcinogenesis. Oncogene. 2010;29:1103–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hisey CL, Dorayappan KDP, Cohn DE, Selvendiran K and Hansford DJ. Microfluidic Affinity Separation Chip for Selective Capture and Release of Label-free Ovarian Cancer Exosomes. Lab on a Chip. 2018;In Press. [DOI] [PubMed] [Google Scholar]
- 29.Hisey CL, Dorayappan KDP, Cohn DE, Selvendiran K, Hansford DJ. Microfluidic affinity separation chip for selective capture and release of label-free ovarian cancer exosomes. Lab on a chip. 2018;18:3144–53. [DOI] [PubMed] [Google Scholar]
- 30.Dorayappan KDP, Wanner R, Wallbillich JJ, Saini U, Zingarelli R, Suarez AA, et al. Hypoxia-induced exosomes contribute to a more aggressive and chemoresistant ovarian cancer phenotype: a novel mechanism linking STAT3/Rab proteins. Oncogene. 2018;37:3806–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomida M, Saito T. The human hepatocyte growth factor (HGF) gene is transcriptionally activated by leukemia inhibitory factor through the Stat binding element. Oncogene. 2004;23:679–86. [DOI] [PubMed] [Google Scholar]
- 32.Zhou HY, Pon YL, Wong AS. HGF/MET signaling in ovarian cancer. Current molecular medicine. 2008;8:469–80. [DOI] [PubMed] [Google Scholar]
- 33.Mariani M, McHugh M, Petrillo M, Sieber S, He S, Andreoli M, et al. HGF/c-Met axis drives cancer aggressiveness in the neo-adjuvant setting of ovarian cancer. Oncotarget. 2014;5:4855–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan C, et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Molecular cancer. 2018;17:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell. 2016;166:755–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goode EL, Chenevix-Trench G, Hartmann LC, Fridley BL, Kalli KR, Vierkant RA, et al. Assessment of hepatocyte growth factor in ovarian cancer mortality. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2011;20:1638–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han C, Bellone S, Siegel ER, Altwerger G, Menderes G, Bonazzoli E, et al. A novel multiple biomarker panel for the early detection of high-grade serous ovarian carcinoma. Gynecologic oncology. 2018;149:585–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saini U, Suarez AA, Naidu S, Wallbillich JJ, Bixel K, Wanner RA, et al. STAT3/PIAS3 Levels Serve as “Early Signature” Genes in the Development of High-Grade Serous Carcinoma from the Fallopian Tube. Cancer research. 2018;78:1739–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


