Abstract
Background
Hepatitis C virus (HCV) coinfection among people with human immunodeficiency virus (HIV) might perturb immune function and HIV persistence. We aimed to evaluate the impact of HCV clearance with direct-acting antivirals (DAAs) on immune activation and HIV persistence in HIV/HCV-coinfected individuals on antiretroviral therapy (ART).
Methods
In a prospective observational study, ART-treated participants with HIV/HCV coinfection received sofosbuvir/daclatasvir ± ribavirin (n = 19). Blood samples were collected before DAA therapy, at the end of treatment, and 12 months after DAA termination (12MPT). T- and natural killer (NK)-cell phenotype, soluble plasma factors, cell-associated (CA)-HIV deoxyribonucleic acid (DNA) forms (total, integrated, 2LTR), CA-unspliced (US) and multiple-spliced ribonucleic acid (RNA), and plasma HIV RNA were evaluated.
Results
Hepatitis C virus clearance was associated with (1) a downmodulation of activation and exhaustion markers in CD4+, CD8+ T, and NK cells together with (2) decreased plasma levels of Interferon gamma-induced protein 10 (IP10), interleukin-8 (IL-8), soluble (s)CD163 and soluble intercellular adhesion molecule (sICAM). Cell-associated US HIV RNA was significantly higher at 12MPT compared to baseline, with no change in HIV DNA or plasma RNA.
Conclusions
Elimination of HCV in HIV/HCV-coinfected individuals alters immune function and the transcriptional activity of latently infected cells. This report provides insights into the effects of HCV coinfection in HIV persistence and regards coinfected subjects as a population in which HIV remission might prove to be more challenging.
Keywords: direct antiviral agents, hepatitis C, HIV reservoir, immune activation
This article addresses the immediate and long-term effects of direct-acting antiviral-mediated HCV clearance on the HIV reservoir dynamics and immune function, in a cohort of HIV/HCV-coinfected individuals under antiretroviral treatment.
Antiretroviral therapy (ART) quickly and persistently suppresses viral replication resulting in improved quality of life for people with human immunodeficiency virus/acquired immune deficiency syndrome (PWHA), reduced AIDS-associated death rates, reduced morbidity events, and increased life expectancy [1]. However, treatment is life-long with several limitations [2]. If treatment is interrupted, plasma viral load (VL) rapidly rebounds, due to the persistence of long-lived and proliferating latently infected cells that persist on ART [3, 4].
Because of overlapping pathways of transmission between HIV and hepatitis C virus (HCV), approximately 2 to 5 million individuals worldwide are estimated to be coinfected [5, 6]. In Argentina, coinfected individuals represent approximately 20% of PWHA [7]. Hepatitis C virus direct antiviral agents (DAAs), which target specific steps of HCV replication cycle, represent a major development in the treatment of HCV, with the possibility of >95% of cure and low rate of adverse events, even with advanced or decompensated cirrhosis [8]. The DAA-mediated clearance of HCV is associated with loss of intrahepatic immune activation by interferon (IFN)-α, recovery of T-cell proliferation, normalization of natural killer (NK)-cell phenotype and function [9], and restoration of type I IFN response both in acute [10] and chronic [11] HCV infection, as well as enhanced HCV-specific CD8+ T-cell responses [12].
Understanding the interaction between HCV coinfection before and after clearance of HCV on the HIV reservoir is important, because HIV/HCV coinfection is common and the high cure rate after DAAs allows for the opportunity to assess the impact of HCV on HIV persistence. Hepatitis C virus/HIV coinfection has been associated with increased levels of immune activation compared with HIV monoinfection including higher levels of microbial products [13, 14], low levels of detectable plasma HIV in PWHA on ART [15, 16], and increased risk of HIV virological failure [17]. Early reports showed that cell-associated (CA) HIV ribonucleic acid (RNA) levels decreased after HCV treatment with IFN-α plus ribavirin (RBV) [18], whereas no effect [18] or a decrease [19, 20] was observed in HIV DNA. More recent studies have reported an increase in CD4+ T cells harboring integrated HIV deoxyribonucleic acid (DNA) in HIV/HCV even after spontaneous HCV clearance [21], whereas DAA-mediated HCV clearance was associated with stable or increased levels of HIV CA DNA [22, 23].
We hypothesized that the elimination of HCV coinfection with DAAs would modulate the size and/or transcriptional activity of the HIV reservoir due to the restoration of HCV-driven immune dysregulation. We quantified the immediate and long-term effects of DAA-mediated HCV clearance in HIV/HCV-coinfected participants on multiple markers of immune function and on HIV persistence in blood. Overall, we observed a downmodulation of NK- and T-cell activation markers as well as of soluble plasma activation markers after treatment with DAAs. This was accompanied by an increase in peripheral CA HIV unspliced (US)-RNA with no change in HIV DNA. These results suggest a relationship between HCV and transcriptional activity of the HIV reservoir.
METHODS
This was a longitudinal, single-center study approved by the local Ethics Committee of the Huésped Foundation (Buenos Aires, Argentina). All participants were included after signing the informed consent from March 17, 2016 to May 12, 2016, and samples were analyzed during 2017, 2018, and 2019. Sample sizes were determined using the Harris, Horvitz, and Mood method to provide 80% power, at the 5% level of significance. Peripheral blood from 19 ART-treated participants with HIV/HCV coinfection were collected at baseline (BSL) before the start of DAA, at the end of treatment (EOT) (either at completion of 12 or 24 weeks of DAA), and after 12 months of DAA termination (12MPT). At 12MPT, samples from 2 participants were not available due to lung cancer diagnosis and loss of follow up, respectively. All individuals received HCV treatment with sofosbuvir (SOF) and daclatasvir (DCV), and 12 participants also received ribavirin (RBV). Diagnosis of liver cirrhosis was made by liver biopsy or hepatic transient elastography (>14 kPa). No participants had signs of hepatic decompensation at the time of enrollment. Sustained virological response (SVR) to DAA was defined by nondetectable HCV RNA (lower limit of detection, 12 IU/mL) at 12 weeks after EOT. Hepatitis C virus RNA was also evaluated at 48 weeks after EOT and it was nondetectable in all cases. Inclusion criteria were successful ART with HIV VL <40 copies/mL for more than 24 months. Peripheral blood mononuclear cells (PBMCs) were obtained from 60 mL whole blood by Ficoll-Hypaque density gradient centrifugation (GE Healthcare, UK) and cryopreserved in liquid nitrogen.
Cell-associated HIV RNA (US and multiple-spliced [MS]) and DNA forms (total HIV, HIV-integrated, and 2LTR circles) were evaluated in sorted CD4+ T cells by quantitative real-time polymerase chain reaction (PCR), as described previously [24]. Ultrasensitive HIV plasma VL was evaluated in all samples by replicate testing using the Aptima HIV-1 quant assay (Hologic). T-cell and NK-cell phenotyping was performed by flow cytometry. Soluble plasma factors were quantified by enzyme-linked immunosorbent assay. Detailed methods are described in Supplementary Material S1.
Statistical analyses were performed using GraphPad Prism 7 (GraphPad Software) and InfoStat (UNC, Cordoba, Argentina) and R project (R Foundation for Statistical Computing, Vienna, Austria) softwares. Data were analyzed using nonparametric methods. Longitudinal association between CA US-HIV RNA and plasma RNA was estimated by using a generalized linear mixed-effects regression model. Correlation analyses were performed using Spearman’s rank test. For the phenotypic analyses, SPICE 6.0 software (https://niaid.github.io/spice/) was used following the experimental and technical considerations published by the software developers [25]. All tests were considered significant when P < .05.
RESULTS
Study Group
A total of 19 HIV/HCV-coinfected individuals were enrolled. Most participants were male (74%), and the median age was 49 years (interquartile range, 46–53). All participants were receiving ART with at least 2 years of documented undetectable HIV VL. During DAA, all individuals received integrase inhibitor-based ART. Sofosbuvir/DCV with RBV was prescribed for 12 weeks in 9 participants and 24 weeks in 3 participants, and 7 participants received SOF/DCV for 24 weeks without RBV. The use of RBV depended on HCV genotype and drug tolerance. All participants achieved SVR. Further clinical details are summarized in Table 1.
Table 1.
Subjects Characteristics
| Characteristics | BSL n = 19 | EOT n = 19 | 12MPT n = 17 | P Valuea,b |
|---|---|---|---|---|
| Age (years)c | 49 (46–53) | -- | 49 (45–52) | -- |
| Male sex (n, %)d | 14 (73.4) | -- | 13 (76.4) | -- |
| CD4 count (cells/μL)c | 291 (231–776) | 460 (205–692) | 506 (233–1058) | >.999/.455 |
| CD8 count (cells/μL)c | 849 (498–1263) | 917 (407–1293) | 1041 (503–1389) | .851/.216 |
| NK cells (%)c | 8.9 (5.6–16.2) | 8.1 (5.7–39.6) | 9.5 (4.2–28.2) | .903/.845 |
| CD4/CD8 ratioc | 0.56 (0.33–0.78) | 0.52 (0.39–0.78) | 0.51 (0.39–0.71) | .025/.397 |
| Time of HCV infection (years)c | 13 (11–22) | -- | -- | -- |
| Time of HIV infection (years)c | 19 (12–21) | -- | -- | -- |
| HCV viral load (log10 copies)c | 5.89 (5.56–6.13) | All <1.3 | All <1.3 | <.001/<.001 |
| Time of ARV (years)c | 10.5 (4–16.5) | -- | -- | -- |
| Routes of Transmission (n, %)d | ||||
| IDU | 14 (73.7) | -- | -- | -- |
| Heterosexual | 5 (21.1) | -- | -- | -- |
| MSM | 1 (5.3) | -- | -- | -- |
| HCV Genotype (n, %)d | ||||
| 1a | 12 (63.2) | -- | -- | -- |
| 1b | 1 (5.3) | -- | -- | -- |
| 1 | 3 (15.8) | -- | -- | -- |
| 3 | 3 (15.8) | -- | -- | -- |
| Liver stiffness (kPa)c | 22.2 (17.9–32.2) | ND | ND | -- |
| APRI scorec | 1.16 (0.58–2.09) | 0.76 (0.29–0.84) | 0.55 (0.29–1.03) | .0084/.0004 |
| ALT (IU/L)c | 67.5 (46–82) | 27 (17–49) | 33 (24–49) | <.001/<.001 |
| AST (IU/L)c | 78.5 (68.2–93.7) | 33 (26–45) | 38 (32–51) | <.001/<.001 |
| Albumin (g/dL)c | 4.2 (3.6–4.5) | 4.2 (3.9–4.4) | 4.4 (4.1–4.5) | .382/.020 |
| Platelets (×103/mm3)c | 111 (98–213) | 121 (87–229) | 121 (77–193) | .922/.130 |
| Total bilirubin (μg/dL)c | 0.85 (0.72–1.1) | 0.80 (0.55–1.42) | 0.80 (0.70–1.0) | .183/.450 |
| Prothrombin time (%)c | 74 (63–93) | 71 (63–81) | 76 (69–85) | .531/.867 |
Abbreviations: 12MPT, 12 month post treatment; ALT, alanine aminotransferase; APRI, aspartate aminotransferase to platelet ratio; ARV, antiretroviral therapy; AST, aspartate transaminase; BSL, baseline; EOT, end of treatment; HCV, hepatitis C virus; HIV, human immunodeficiency virus; IDU, injecting drug user; MSM, men who have sex with men; ND, not determined; NK, natural killer.
aBSL vs EOT, Wilcoxon test.
bBSL vs 12MPT, Wilcoxon test.
cMedian (interquartile range).
dNumber of cases (number/total in %).
Human Immunodeficiency Virus Persistence After Direct-Acting Antiviral Treatment
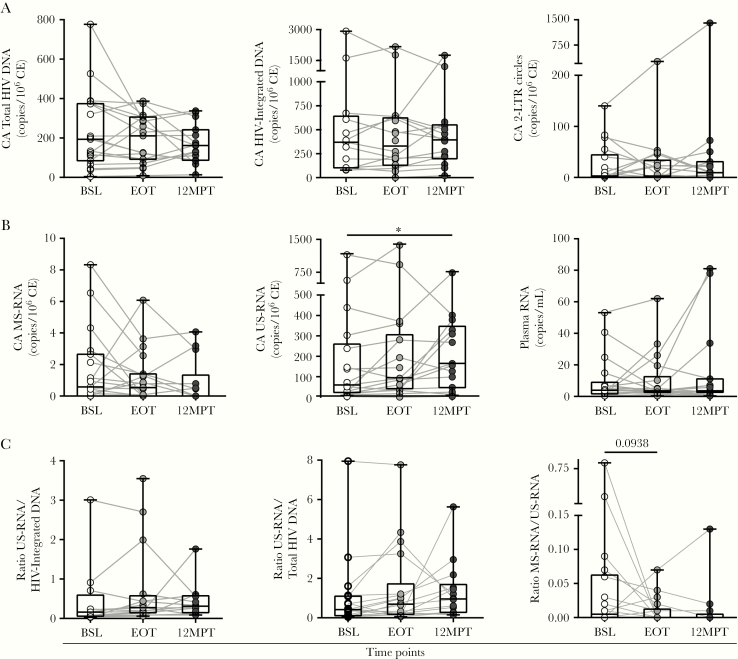
We quantified the frequency and transcriptional activity of HIV-infected cells before (BSL sample) and after DAA treatment (EOT and 12MPT samples) as an indicator of HIV persistence. No statistically significant differences were observed in CA viral DNA (total, integrated, and 2LTR circles) (Figure 1A), ie, overall levels remained stable from BSL to 12MPT. No differences in plasma HIV RNA, measured by ultrasensitive single-copy assay, were found among time points (Figure 1B).
Figure 1.
Human immunodeficiency virus (HIV) reservoir dynamics in HIV/hepatitis C virus-coinfected individuals treated with direct-acting antivirals (DAA). Cell-associated (CA) HIV deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), as well as plasma HIV RNA were evaluated in coinfected individuals before treatment initiation (baseline [BSL]), at the end of treatment (EOT), and 12 months after finalizing DAA therapy (12MPT). The CA total HIV DNA, integrated DNA and 2LTR (A), CA multiple-spliced (MS)-RNA, unspliced (US)-RNA, and plasma RNA (B), and US-RNA/integrated DNA, US-RNA/total HIV DNA, and MS/US-RNA ratios (C) are shown. Viral DNA and RNA copies were calculated relative to 106 cell equivalents (CE). Individual values, median and 25th and 75th percentiles, are indicated. Statistical comparisons were performed using Wilcoxon test, P < 0.05.
Levels of CA MS-RNA remained low and stable along all the studied time points (Figure 1B). However, a statistically significant increase in CA US-RNA was observed between BSL and 12MPT (P = 0.0203) (Figure 1B). Of note, the higher increments in US-RNA were observed in those participants who did not receive RBV within their regimens (Supplementary Figure S1); US-RNA was higher at BSL in the RBV group, nevertheless no differences in any of the clinical or laboratory parameters were observed between both groups that could account for that disparity.
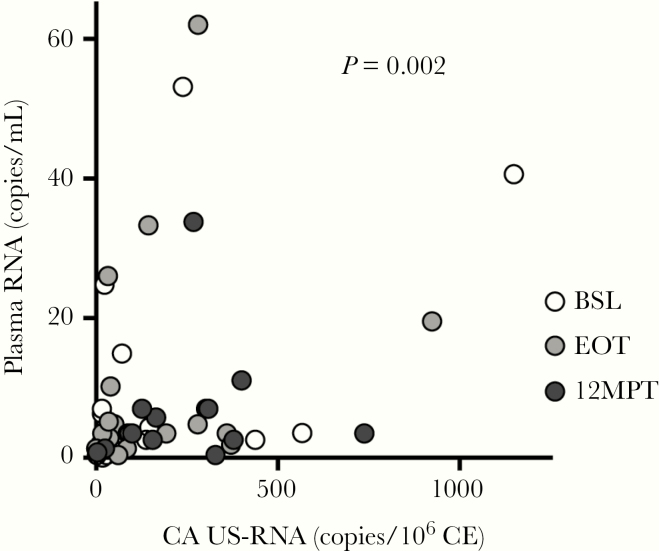
To provide further insight into the transcriptional activity of the infected cells, the ratio of CA US-RNA and both the integrated and total HIV DNA were calculated. In addition, the ratio between CA MS- and US-RNA was obtained to estimate the relative efficiency of transcriptional elongation and splicing. No differences were found among BSL, EOT, and 12MPT (Figure 1C). Finally, we hypothesized that increased US-RNA, if associated with increased viral transcription, would be correlated with plasma RNA. The longitudinal study of CA US-HIV RNA and plasma RNA across all 3 time points evaluated depicted a potential association between both variables (P = 0.002) (Figure 2).
Figure 2.
Association analysis between cell-associated (CA) unspliced (US) and plasma human immunodeficiency virus (HIV) ribonucleic acid (RNA) in HIV/hepatitis C virus-coinfected individuals treated with direct-acting antivirals (DAA). The CA US-RNA as well as plasma HIV RNA were evaluated in coinfected individuals before treatment initiation (baseline [BSL]), at the end of treatment (EOT), and 12 months after finalizing DAA therapy (12MPT). Relationship between variables was measured by applying a generalized linear mixed-effects model with plasma RNA as the independent variable and CA US-RNA and time as fixed-effect predictors; P value for the CA US-RNA coefficient is shown. White, gray, and black filled dots represent individual measures belonging to BSL, EOT, and 12MPT subgroups, respectively.
T-Cell Immune Phenotyping
The profile of different memory subsets and the expression of different markers were studied on CD8+ and CD4+ T cells (gating strategy is shown in Supplementary Figure S2). We defined 6 CD8+ or CD4+ T-cell subpopulations: naive (TNaive, CCR7+CD45RO−CD28+CD95−), stem memory (TSCM, CCR7+CD45RO−CD28+CD95+), central memory (TCM, CCR7+CD45RO+CD28+CD95+), transitional memory (TTM, CCR7−CD45RO+CD28+CD95+), effector memory (TEM, CCR7−CD45RO+CD28−CD95+), and terminal effector (TTE, CCR7−CD45RO−CD28−CD95+) cells. The distribution of these subsets was analyzed as previously described [26]. The memory profile within the CD4+ T-cell compartment showed the following hierarchical distribution: TTM cells represented the highest proportions, followed by TCM, TNaive, TEM, TSCM, and TTE cells (Supplementary Figure S3A). Regarding the CD8+ T-cell compartment, the distribution was as follows: TEM cells comprised the highest proportions, followed by TTE, TTM, TNaive, TCM, and TSCM cells (Supplementary Figure S3B). Memory distribution observed on both T-cell compartments were conserved across the 3 time points analyzed (BSL, EOT, and 12MPT), and no significant differences were observed between them.
We next quantified the expression of immune surface markers (such as CD38, HLA-DR, CD127, PD-1, and CD25) in both T-cell compartments. At BSL, elevated proportions of CD4+/HLA-DR+, CD4+/PD-1+, CD4+/CD38+/HLA-DR+, and CD4+/CD38+/HLA-DR+/PD-1+ T cells were observed. These proportions were significantly reduced by 12MPT (Supplementary Figure S4A). The decay in activation markers observed from BSL to 12MPT was observed in all CD4+ T-cell subpopulations, with CD4+ TTM and TEM having the largest and most significant changes.
A similar scenario was observed on CD8+ T cells. At BSL, higher proportions of CD8+ T cells expressing CD38 and HLA-DR were recorded. After DAA treatment, the expression of these activation markers was significantly reduced. The proportions of double-positive CD8+/CD38+/HLA-DR+ and triple-positive CD8+/CD38+/HLA-DR+/PD-1+ T cells were significantly reduced at EOT and 12MPT, respectively (Supplementary Figure S5A). Similar to the CD4+ T-cell compartment, the decay in CD38, HLA-DR, and PD-1 expression was observed in all CD8+ T-cell subpopulations (Supplementary Figure S5B). In this compartment, the subpopulations that exhibited more pronounced modifications after HCV clearance were the more differentiated phenotypes (CD8+ TTM, TEM, and TTE).
Natural Killer Cell Immune Phenotyping
Compared with BSL, CD38 expression on NK cells was significantly reduced at EOT, and it continued at this lower level throughout the study (Supplementary Figure S6A). The frequency of HLA-DR-expressing NK cells was significantly reduced at EOT (Supplementary Figure S5B), whereas there was a slight increase by 12MPT levels, but it still remained at lower levels than those from BSL (Supplementary Figure S6B). When analyzing the distribution of NK-cell subsets defined by the expression of both HLA-DR and CD38, we found a significant reduction in the frequency of HLA-DR+/CD38+ NK cells, with a concomitant increase in the frequency of cells negative for both markers at EOT and 12MPT (Supplementary Figure S6C). In addition, the apoptosis-inducing receptor CD95 was studied. Percentages of CD95-expressing cells at EOT were lower than those from BSL; however, CD95 levels rebounded at 12MPT, reaching similar levels observed at BSL (Supplementary Figure S6D). Moreover, a trend towards a decreased frequency of both CD25+ and CD69+ NK cells at EOT and 12MPT was observed; however, neither CD25 nor CD69 expression differed among time points (Supplementary Figure S6E and F). It is interesting that, similar to our previous results, the frequency of CD25+/CD69+/CD95+ NK cells was reduced at EOT and 12MPT, whereas percentages of triple-negative NK cells were significantly augmented (Supplementary Figure S6G). Finally, expression of NK cell-activating receptor NKG2D and natural cytotoxic receptors (NCRs) NKp30 and NKp46 were evaluated. As shown in Supplementary Figure S5H, the percentage of NKG2D-expressing NK cells was significantly decreased at EOT, compared with BSL. When relative fluorescence intensities (RFIs) were examined, a significant reduction in NKG2D expression was also obtained at 12MPT (Supplementary Figure S6H, right panel). Regarding NCRs, although NKp46 expression was not differentially modulated, we observed a trend towards a reduction in the expression of NKp30 at EOT and 12MPT. No differences were found when analyzing RFI (Figure 6H).
Soluble Factors
Then, plasma concentrations of different soluble factors, frequently associated with markers of immune activation and inflammation, were evaluated. Interleukin (IL)-17 and IL-1β were below the limit of detection so they could not be quantified. For IL-6, IL-2, soluble (s)CD14, IFN-γ, tumor necrosis factor-α, and sCD23, no differences were observed along the different time points evaluated (Supplementary Figure S7A−F). In contrast, IFN-inducible protein 10 (IP-10), IL-8 soluble intercellular adhesion molecule 1 (sICAM-1), and sCD163 were elevated at BSL but were significantly reduced at EOT (P < 0.0001, P = 0.0015, P < 0.0001, and P = 0.0002, respectively) and remained low (always compared with BSL) at 12MPT (P = 0.0107, P = 0.0426, P = 0.0001, and P = 0.002, respectively) (Supplementary Figure S7G−J).
Association Between Immune Status and Human Immunodeficiency Virus Persistence
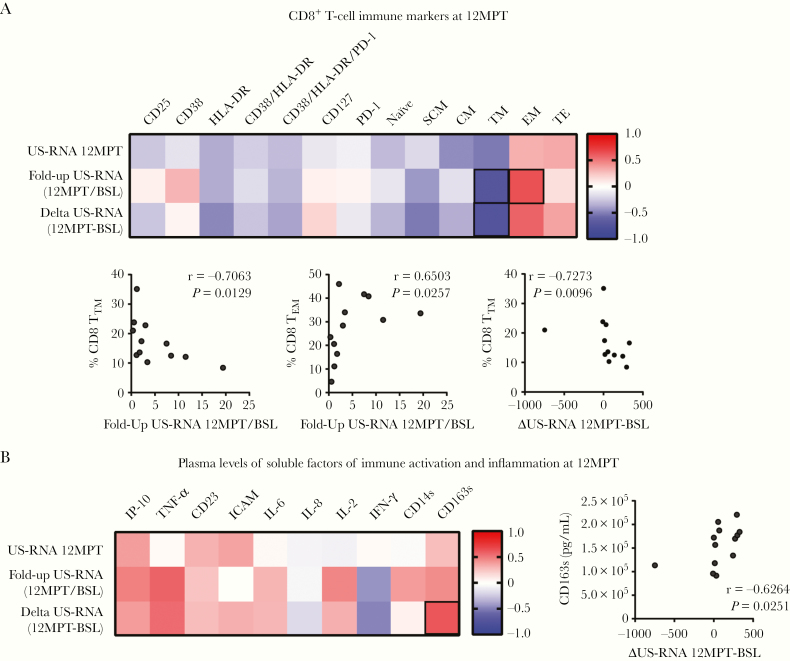
Finally, we assessed the relationship between the different immune parameters and HIV viral persistence. All immune measurements (T and NK phenotype and plasma soluble factors) evaluated at 12MPT were compared with US-RNA levels at 12MPT, the US-RNA fold-up between 12MPT and BSL (12MPT/BSL), and the US-RNA change (delta) between 12MPT and BSL (12MPT-BSL). For simplicity, results are shown in heat maps denoting r values. Significant correlations are highlighted, and the corresponding y versus x plot is shown. No statistically significant correlations were found between US-RNA at 12MPT, US-RNA fold-increase, or delta US-RNA with parameters evaluated on CD4+ or NK cells (data not shown). In contrast, US-RNA fold-increase was negatively associated with the proportions of CD8 TTM cells and positively associated with %CD8 TEM cells (Figure 3A). Levels of most cytokines at 12MPT showed positive r values with the 3 virological parameters evaluated; however, only sCD163 showed a statistically significant correlation with delta US-RNA (Figure 3B).
Figure 3.
Correlation analyses between cellular and soluble markers of immune activation and inflammation, and human immunodeficiency virus (HIV) reservoirs in HIV/hepatitis C virus-coinfected individuals treated with direct-acting antivirals (DAA). Heat map representation of Spearman rank correlation coefficients computed for the expression of CD8+ T-cell immune markers at 12 months after finalizing DAA therapy (12MPT) (A) and plasma levels of soluble factors of immune activation and inflammation at 12MPT (B) versus HIV unspliced (US)-ribonucleic acid (RNA) at 12MPT (US-RNA 12 MPT), US-RNA fold up between 12MPT and baseline (BSL) (Fold-up US-RNA 12MPT/BSL), and differences between US-RNA at 12MPT minus levels at BSL (Delta US-RNA 12MPT-BSL). The colors denote both the correlation direction and strength of association, ranging from −1 (blue) to 1 (red). Statistical significant associations are further shown below each panel in individual × vs y plots. Spermans’s r and P values are shown. ICAM, intercellular adhesion molecule; IL, interleukin; IP-10, interferon-inducible protein 10; TNF, tumor necrosis factor.
DISCUSSION
Human immunodeficiency virus remission and cure clinical research has largely occurred in high-income settings where (1) most participants are men who have sex with men, (2) subtype B is the most prevalent viral variant, (3) malnutrition is not a regular finding, and (4) there is a low burden of coinfections. All of these factors may be important determinants of the size and transcriptional activity of the HIV reservoir, and cure strategies may need to be different for high- and low-middle income settings, especially if the intervention is dependent on a change in immune function [27]. In this study, we found that DAA-mediated HCV clearance in HIV/HCV-coinfected subjects is associated with the following: a clear improvement in liver functionality and a tendency to improved CD4+ T cells counts; a decrease in the surface expression of activation and exhaustion markers in CD4+ and CD8+ T cells and in NK cells; lower plasma levels of IP-10 and IL-8 and indicators of macrophage and monocyte activation, such as sCD163 and sICAM; and higher levels of US-RNA at 12 months post-DAA treatment.
Therapies with DAAs are very effective oral and short-term treatments, with more than 90% of SVR, even in HIV-coinfected individuals [28]. This has been reflected in our study, where all participants achieved viral clearance and improved liver function parameters, despite the presence of advanced hepatic disease. In addition, a recovery in both CD4+ and CD8+ T-cell phenotype (in terms of the expression of activation and exhaustion markers) was registered after DAA treatment. These findings are in line with previous reports after DAA for HIV/HCV coinfection describing improved HCV-specific CD8+ T-cell functionality and lower PD-1 expression, recovery of the CD4+ T-cell compartment, and a replenishment of T cells with memory/effector phenotype [29–31]. Likewise, a decrease in the proportion of activated NK cells was found after DAA treatment, in agreement with previous publications describing reduced expression of activation markers and cytolytic activity [9, 29, 31, 32]. Although others have observed an increase in NK cell frequency after DAA treatment [33–37], this was not found in our study. It might be because in this study, all individuals presented with end-stage liver fibrosis, which is linked to low NK cell frequencies [34]. In these individuals, liver damage could be a stronger factor modifying NK-cell population than HCV clearance. Finally, lower levels of soluble inflammatory factors, such as IP-10, were previously reported [38] and recapitulated in our study.
The first reports regarding the impact of HCV on HIV persistence on ART, described that IFN-α/RBV treatment was associated with decreased levels of total and integrated HIV DNA in CD4+ T cells [20] and 2LTR circles from PBMCs [19]—as assessed by reverse-transcription PCR—and also CD4+ T-cell HIV RNA reduction [18]. In this latter report, no differences were found regarding CD4+ T-cell proviral HIV DNA, 2LTR circles, and replication-competent reservoirs measured with quantitative viral outgrowth assay (qVOA). Nevertheless, it is important to highlight that those therapies were based on 2 drugs (IFN-α and RBV) that could also have a direct effect on HIV replication [39, 40]. More recently, 3 studies have evaluated the levels of CA HIV DNA in PBMCs before and after DAAs for HCV treatment. Parisi et al [22] described that there was an increase or decrease in total HIV DNA after DAA treatment depending on the magnitude of HIV viremia (low-level versus undetectable) before starting DAAs. In other works, HIV DNA remained stable before and after DAA treatment [23, 41]. All 3 forms of HIV DNA measured here (total, integrated, and 2LTR) remained stable during follow up. The differential findings might be explained by the use of purified CD4+ T cells instead of total PBMCs to quantify HIV DNA and a more rigorous criteria regarding HIV undetectable VL before enrollment. It is also worth noting that, although extensively used, PCR-based techniques for measuring both total and integrated HIV DNA face considerable limitations, because they tend to overestimate the size of the reservoir due to the high prevalence of defective proviruses. Thus, these results should not be interpreted as if there was no effect on the size of the competent reservoir [42].
We also studied the transcriptional activity of infected cells by measuring US-RNA and MS-RNA and plasma RNA. A significant increase in US-RNA was found 12 months after HCV clearance with no increase in CA MS-RNA or plasma RNA levels. Correlation analysis indicated that the elevation of US-RNA was accompanied by diminished proportions of CD8+ TTM cells and higher proportions of CD8+ TEM cells. This could be an indicator that higher transcription might be accompanied by production of at least some viral proteins that might be priming memory CD8+ T-cell responses. On the other hand, the steady state of MS-RNA and plasma RNA levels could be reflecting a block in viral cycle termination. This raises different hypotheses. Higher levels of US-RNA might represent higher rates of genuine HIV transcripts but also host-HIV read-through transcripts [43, 44]. However, these latter transcripts have been shown to contribute poorly to the bulk of HIV RNA, so this hypothesis seems unlikely [44]. Yukl et al [43] showed that nonactivated latently infected CD4+ T cells show substantial transcription initiation that is subsequently blocked at the elongation, polyadenylation, and splicing steps. In addition, evidence indicates that unstimulated naive and memory CD4+ T-cell subsets support transcription initiation and elongation with different capacity; however, upon stimulation, TEM cells are the cell subset that more efficiently achieves transcript elongation [45]. Although we did not observe differences in the bulk distribution of CD4+ T-cell subsets from BSL to 12MPT, we cannot exclude modifications within particular T helper subsets that might justify the higher frequency of US-RNA-positive cells in periphery. The potential variations in T helper subsets could contribute to our findings as a consequence of increased transcription initiation, enhanced blockade, or even reduced trafficking of these cells to the tissues secondary to the changes of chemokines and chemokine receptors expression after HCV clearance. Moreover, HCV has been shown to replicate in lymphocytes [46], thus HCV elimination might modify signaling pathways leading to increased HIV transcription in these cells. Finally, it has been demonstrated that DAA-mediated viral clearance was accompanied by a lower activity of type I IFN (α, γ, β) receptors, meaning a downregulation of IFN-stimulated genes, in HCV-monoinfected individuals [11]. This might impact the transcriptional activity of HIV latently infected cells. A recent study showed that, in vitro, once latency is stablished, IFN-α could act as a reversal agent promoting viral replication [47]. A priori, this result is in contrast with our findings. However, it should be noted that the establishment and maintenance of latency is a complex multifactorial phenomenon governed by mixed, sometimes opposed, mechanisms, and the net result of this is what is observed ex vivo.
CONCLUSIONS
This work opens new perspectives that should be addressed. First, we used PCR-based techniques to measure viral reservoirs. These assays tend to overestimate the frequency of replication competent virus, because both defective and nondefective viral strains are detected. The aim of future studies should include assays aimed at identifying the translation-competent reservoir [48] as well as qVOA, TILDA (Tat/rev Induced Limiting Dilution Assay), and IPDA (intact proviral DNA assay) assays to address this limitation [49, 50]. Second, longer time of follow up would provide more information regarding the long-term effect of HCV clearance in HIV transcriptional activity. In addition, it would be relevant to evaluate the magnitude and quality of HIV-specific immune response after HCV clearance and its association with HIV persistence, because it could be hypothesized that the increase in HIV transcriptional activity might be associated with a boosting of HIV-specific T cells. Third, all of the individuals included in the present study presented advanced liver fibrosis; it will be important to evaluate whether the results found are reproducible in a cohort of participants with low to mild liver fibrosis. Finally, results could not be extended to the intrahepatic CD4+ T cells, but a similar or higher increase in HIV transcription is expected. There is certain evidence to suggest that hepatocytes and hepatic stellate cells support HIV infection [51]. Thus, the modification in the liver environment, including decreased HCV-specific immune surveillance, could lead to a relapse in HIV transcription in these cells. Nevertheless, results presented here provide an important insight into the effects of chronic HCV infection on virus persistence, confirming HCV coinfection as a relevant factor imposing an extra challenge in HIV remission studies, even after HCV clearance.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary File 1. Detailed description of methods used in this manuscript.
Supplementary Figure S1. Cell-associated (CA) HIV US-RNA dynamics in HIV/HCV-coinfected individuals treated with DAA with or without ribavirin. Cell-associated HIV unspliced (US)-RNA was evaluated in coinfected individuals receiving DAA or DAA plus ribavirin (RBV), before treatment initiation (baseline [BSL]), at the end of treatment (EOT), and 12 months after finalizing therapy (12MPT). Viral DNA and RNA copies were calculated relative to 106 cell equivalents (CE). Individual values, median and 25th and 75th percentiles, are indicated. Statistical comparisons were performed using Wilcoxon test; *, P ˂ 0.05.
Supplementary Figure S2. Gating strategy used for the identification of the studied cellular populations, by flow cytometry. To study immune phenotype of both NK and T cells, initial gating was performed on a forward scatter area (FSC-A) versus FSC-height (FSC-H) plot to remove doublets. Dead cells were then excluded on the bases of Zombie NIR fluorescence. Then, lymphocytes were selected in FSC versus side scatter (SSC) plot. Subsequently, CD3 expression was analyzed in a CD3 versus SSC-H dot plot (A). For NK analysis, CD3 and CD56 expression was analyzed on total lymphocytes. CD3−/CD56+ cells were selected. Then, expression of cellular markers of activation was studied. Representative flow cytometry plots for Nkp46, NKp30, NKG2D, CD69, and CD95 are shown (B). For identification of CD4+ and CD8+ T-cells, a CD8 versus CD4 dot plot was constructed. Expression of CD25, CD38, HLA-DR, CD127, and PD-1 on CD8+ cells is shown from 1 representative subject (C, top panel). To analyze the distribution of the different phenotype subsets on CD8+ T cells, CD45RO versus CCR7 dot plots were constructed and shown in C (bottom panel). Subsequently, CD28 and CD95 were used to define on (1) CCR7+/CD45RO− gate, naive and stem cell memory (SCM) T-cells, (2) CCR7+/CD45RO+ gate, central memory T cells, (3) CCR7−/CD45RO+ gate, transitional memory (TM) and effector memory T cells (EM), and (4) CCR7−/CD45RO− gate, terminal effector T cells (TE). Identical analysis was performed on the CD4+ T-cell subset (data not shown ).
Supplementary Figure S3. Distribution of memory subpopulations within CD4+ and CD8+ T-cells from HIV/HCV-coinfected individuals treated with DAA. Proportion of naive cells; stem cell memory (SCM), central memory (CM), transitional memory (TM), effector memory (EM), and terminal effector (TE) T cells were evaluated in CD4+ (A) or CD8+ T-cells (B) from coinfected individuals at baseline (BSL), end of treatment (EOT), and 12 months after finalizing DAA therapy (12MPT). The analyses were performed with SPICE software. In bar graphs, the color code shown at the bottom mirrors the one shown in the pies. Each dot represent individual values, boxes extend from 25th to 75th percentiles, and lines within boxes represent the median value.
Supplementary Figure S4. Immune marker expression in CD4+ T cells from HIV/HCV-coinfected individuals treated with DAA. Modulation of immune cell markers was evaluated before DAA therapy initiation (baseline [BSL]), at the end of treatment (EOT), and 12 months after finalizing DAA therapy (12MPT). Frequencies of CD38, HLA-DR, PD-1, CD38/HLA-DR, and CD38/HLA-DR/PD-1 positive CD4+ T cells are shown. CD25 and CD127 expression were also monitored, but no differences were found and thus no results are shown. (A). Analysis of distribution of CD4+ T-cell populations in terms of memory subsets was performed with SPICE software (B). Individual values are indicated. Boxes represent the IQR of 25–75, and horizontal lines within boxes represent the medians. P values were calculated using the Wilcoxon test. Asterisks denote different P values: *, P ˂ 0.05; **, P ˂ 0.01.
Supplementary Figure S5. Immune marker expression in CD8+ T cells from HIV/HCV-coinfected individuals treated with DAA. Modulation of immune cell markers was evaluated before DAA therapy initiation (baseline [BSL]), end of treatment (EOT), and 12 months after finalizing DAA therapy (12MPT). Frequencies of CD38, HLA-DR, CD38/HLA-DR, and CD38/HLA-DR/PD-1 positive CD8+ T cells are shown. CD25 and CD127 expression were also monitored, but no differences were found and thus no results are shown. (A). Analysis of distribution of CD8+ T-cell populations in terms of memory subsets was performed with SPICE software (B). Individual values are indicated. Boxes represent the IQR of 25–75, and horizontal lines within boxes represent the medians. P values were calculated using the Wilcoxon test. Asterisks denote different P values: *, P ˂ 0.05; **, P ˂ 0.01; ***, P ˂ 0.001.
Supplementary Figure S6. Study of NK cell activation in HIV/HCV-coinfected individuals treated with DAA therapy. NK cell immune profile was assessed in coinfected individuals at BSL, EOT, and 12MPT. Frequencies of CD38 (A), HLA-DR (B), CD95 (D), CD69 (E), CD25 (F), and NKG2D, Nkp30, and NKp46 positive NK cells (H) are shown. Analysis of distribution of NK cells subsets in terms of HLA-DR/CD38 (C) and CD25/CD69/CD95 expression (G) was performed with SPICE software. Relative fluorescence intensity (RFI) for NKG2D, Nkp30, and NKp46 stains on NK cells (H, right panel) are shown. Individual values are indicated. Boxes represent the IQR of 25–75, and horizontal lines within boxes represent the medians. P values were calculated using Wilcoxon test. Asterisks denote different P values: *, P ˂ 0.05; ** P ˂ 0.01; ***, P ˂ 0.001.
Supplementary Figure S7. Evaluation of soluble markers of immune activation and inflammation in HIV/HCV-coinfected individuals treated with DAA therapy. Plasma from coinfected individuals was evaluated before DAA therapy initiation (baseline [BSL]), at the end of treatment (EOT), and after 12 months of DAA therapy finalization (12MPT). Plasma IL-6 (A), IL-2 (B), sCD14 (C), TNF-α (D), IFN-γ (E), sCD23 (F), IP-10 (G), IL-8 (H), sICAM (I), and sCD163 (J) levels are indicated. Boxes represent the IQR of 25–75, whiskers extend from minimum to maximum, and horizontal lines within boxes represent the medians. P values were calculated using Wilcoxon test. Asterisks denote different P values: *, P ˂ 0.05; **, P ˂ 0.01; ***, P ˂ 0.001.
Acknowledgments
We acknowledge study participants for agreeing to collaborate in this study and to provide blood samples. We thank Drs. Michael Busch and Mars Stone from Vitalant Research Institute for scientific input; Sabrina Azzolina for technical help during sample processing; Drs. Carla Pascuale and Virginia Polo for technical assistance in multiparametric flow cytometry; and Andrea Peña Malavera for statistical advice.
Financial support. S. R. L. is funded by the National Health and Medical Research Council of Australia and the National Institutes for Health Delaney AIDS Research Enterprise (DARE) Collaboratory (U19 A1096109, UM1AI126611]). This work was funded by grants from Consejo Nacional de Investigaciones Científicas y Técnicas (Grant Number PUE2016-INBIRS), the Agencia Nacional de Promoción Científica y Tecnológica (PICT2016, Grant Number 930), ViiV Healthcare Investigator Sponsored Studies (reference number 209424), and Fundación Florencio Fiorini.
Potential conflicts of interest. S. R. L. has received financial support for investigator-initiated, industry-funded research from Merck, Viiv Healthcare, Gilead Sciences, and Leidos. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Hull M, Lange J, Montaner JS. Treatment as prevention–where next? Curr HIV/AIDS Rep 2014; 11:496–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Deeks SG, Tracy R, Douek DC. Systemic effects of inflammation on health during chronic HIV infection. Immunity 2013; 39:633–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siliciano JD, Kajdas J, Finzi D, et al. . Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med 2003; 9:727–8. [DOI] [PubMed] [Google Scholar]
- 4. Strain MC, Little SJ, Daar ES, et al. . Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J Infect Dis 2005; 191:1410–8. [DOI] [PubMed] [Google Scholar]
- 5. Alter MJ. Epidemiology of viral hepatitis and HIV co-infection. J Hepatol 2006; 44:S6–9. [DOI] [PubMed] [Google Scholar]
- 6. Platt L, Easterbrook P, Gower E, et al. . Prevalence and burden of HCV co-infection in people living with HIV: a global systematic review and meta-analysis. Lancet Infect Dis 2016; 16:797–808. [DOI] [PubMed] [Google Scholar]
- 7. Laufer N, Quarleri J, Bouzas MB, et al. . Hepatitis B virus, hepatitis C virus and HIV coinfection among people living with HIV/AIDS in Buenos Aires, Argentina. Sex Transm Dis 2010; 37:342–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sikavi C, Chen PH, Lee AD, et al. . Hepatitis C and human immunodeficiency virus coinfection in the era of direct-acting antiviral agents: no longer a difficult-to-treat population. Hepatology 2018; 67:847–57. [DOI] [PubMed] [Google Scholar]
- 9. Serti E, Chepa-Lotrea X, Kim YJ, et al. . Successful interferon-free therapy of chronic hepatitis C virus infection normalizes natural killer cell function. Gastroenterology 2015; 149:190–200.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlton-Smith C, Holmes JA, Naggie S, et al. ; of the ACTG A5327 study group IFN-free therapy is associated with restoration of type I IFN response in HIV-1 patients with acute HCV infection who achieve SVR. J Viral Hepat 2018; 25:465–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meissner EG, Wu D, Osinusi A, et al. . Endogenous intrahepatic IFNs and association with IFN-free HCV treatment outcome. J Clin Invest 2014; 124:3352–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Debes JD, de Knegt RJ, Boonstra A. The path to cancer and back: immune modulation during hepatitis C virus infection, progression to fibrosis and cancer, and unexpected roles of new antivirals. Transplantation 2017; 101:910–5. [DOI] [PubMed] [Google Scholar]
- 13. Gonzalez VD, Landay AL, Sandberg JK. Innate immunity and chronic immune activation in HCV/HIV-1 co-infection. Clin Immunol 2010; 135:12–25. [DOI] [PubMed] [Google Scholar]
- 14. Shmagel KV, Saidakova EV, Shmagel NG, et al. . Systemic inflammation and liver damage in HIV/hepatitis C virus coinfection. HIV Med 2016; 17:581–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baroncelli S, Pirillo MF, Galluzzo CM, et al. . Rate and determinants of residual viremia in multidrug-experienced patients successfully treated with raltegravir-based regimens. AIDS Res Hum Retroviruses 2015; 31:71–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pugliese P, Delpierre C, Cuzin L, et al. ; Dat AIDS Study Group An undetectable polymerase chain reaction signal in routine HIV plasma viral load monitoring is associated with better virological outcomes in patients receiving highly active antiretroviral therapy. HIV Med 2013; 14:509–15. [DOI] [PubMed] [Google Scholar]
- 17. Calcagno A, Motta I, Ghisetti V, et al. . HIV-1 very low level viremia is associated with virological failure in highly active antiretroviral treatment-treated patients. AIDS Res Hum Retroviruses 2015; 31:999–1008. [DOI] [PubMed] [Google Scholar]
- 18. Morón-López S, Gómez-Mora E, Salgado M, et al. . Short-term treatment with interferon alfa diminishes expression of HIV-1 and reduces CD4+ T-cell activation in patients coinfected with HIV and hepatitis C virus and receiving antiretroviral therapy. J Infect Dis 2016; 213:1008–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiao YM, Weng WJ, Gao QS, et al. . Hepatitis C therapy with interferon-α and ribavirin reduces the CD4 cell count and the total, 2LTR circular and integrated HIV-1 DNA in HIV/HCV co-infected patients. Antiviral Res 2015; 118:118–22. [DOI] [PubMed] [Google Scholar]
- 20. Sun H, Buzon MJ, Shaw A, et al. . Hepatitis C therapy with interferon-α and ribavirin reduces CD4 T-cell-associated HIV-1 DNA in HIV-1/hepatitis C virus-coinfected patients. J Infect Dis 2014; 209:1315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. López-Huertas MR, Palladino C, Garrido-Arquero M, et al. ; Multidisciplinary Group of viral coinfection HIV/Hepatitis (COVIHEP) HCV-coinfection is related to an increased HIV-1 reservoir size in cART-treated HIV patients: a cross-sectional study. Sci Rep 2019; 9:5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parisi SG, Andreis S, Basso M, et al. . Time course of cellular HIV-DNA and low-level HIV viremia in HIV-HCV co-infected patients whose HCV infection had been successfully treated with directly acting antivirals. Med Microbiol Immunol 2017; 206:419–28. [DOI] [PubMed] [Google Scholar]
- 23. Rozera G, Fabbri G, Lorenzini P, et al. . Peripheral blood HIV-1 DNA dynamics in antiretroviral-treated HIV/HCV co-infected patients receiving directly-acting antivirals. PLoS One 2017; 12:e0187095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghiglione Y, Trifone C, Salido J, et al. . PD-1 expression in HIV-specific CD8+ T cells before antiretroviral therapy is associated With HIV persistence. J Acquir Immune Defic Syndr 2019; 80:1–6. [DOI] [PubMed] [Google Scholar]
- 25. Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A 2011; 79:167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Salido J, Ruiz MJ, Trifone C, et al. . Phenotype, polyfunctionality, and antiviral activity of in vitro stimulated CD8+ T-cells from HIV+ subjects who initiated cart at different time-points after acute infection. Front Immunol 2018; 9:2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rossouw T, Tucker JD, van Zyl GU, et al. . Barriers to HIV remission research in low- and middle-income countries. J Int AIDS Soc 2017; 20:21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salmon D, Mondelli MU, Maticic M, Arends JE; ESCMID Study Group for Viral Hepatitis The benefits of hepatitis C virus cure: every rose has thorns. J Viral Hepat 2018; 25:320–8. [DOI] [PubMed] [Google Scholar]
- 29. Burchill MA, Golden-Mason L, Wind-Rotolo M, Rosen HR. Memory re-differentiation and reduced lymphocyte activation in chronic HCV-infected patients receiving direct-acting antivirals. J Viral Hepat 2015; 22:983–91. [DOI] [PubMed] [Google Scholar]
- 30. Martin B, Hennecke N, Lohmann V, et al. . Restoration of HCV-specific CD8+ T cell function by interferon-free therapy. J Hepatol 2014; 61:538–43. [DOI] [PubMed] [Google Scholar]
- 31. Urbanowicz A, Zagożdżon R, Ciszek M. Modulation of the immune system in chronic hepatitis C and during antiviral interferon-free therapy. Arch Immunol Ther Exp (Warsz) 2019; 67:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mondelli MU. Direct-acting antivirals cure innate immunity in chronic hepatitis C. Gastroenterology 2015; 149:25–8. [DOI] [PubMed] [Google Scholar]
- 33. Li Y, Zeng Y, Zeng G, et al. . The effects of direct-acting antiviral agents on the frequency of myeloid-derived suppressor cells and natural killer cells in patients with chronic hepatitis C. J Med Virol 2019; 91:278–86. [DOI] [PubMed] [Google Scholar]
- 34. Nakamura I, Furuichi Y, Sugimoto K. Restoration of natural killer cell activity by interferon-free direct-acting antiviral combination therapy in chronic hepatitis C patients. Hepatol Res 2018; 48:855–61. [DOI] [PubMed] [Google Scholar]
- 35. Ning G, Li YT, Chen YM, et al. . Dynamic changes of the frequency of classic and inflammatory monocytes subsets and natural killer cells in chronic hepatitis C patients treated by direct-acting antiviral agents. Can J Gastroenterol Hepatol 2017; 2017:3612403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spaan M, van Oord G, Kreefft K, et al. . Immunological analysis during interferon-free therapy for chronic hepatitis C virus infection reveals modulation of the natural killer cell compartment. J Infect Dis 2016; 213:216–23. [DOI] [PubMed] [Google Scholar]
- 37. Stevenson TJ, Barbour Y, McMahon BJ, et al. . observed changes in natural killer and T cell phenotypes with evaluation of immune outcome in a longitudinal cohort following sofosbuvir-based therapy for chronic hepatitis C infection. Open Forum Infect Dis 2019; 6:ofz223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Carlin AF, Aristizabal P, Song Q, et al. . Temporal dynamics of inflammatory cytokines/chemokines during sofosbuvir and ribavirin therapy for genotype 2 and 3 hepatitis C infection. Hepatology 2015; 62:1047–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Snell NJC. The activity of ribavirin against the human immunodeficiency virus: a review of laboratory and clinical experience. Antivir Chem Chemother 1991; 2:257–63. [Google Scholar]
- 40. Rivero-Juárez A, Frias M, Rivero A. Current views on interferon therapy for HIV. Expert Opin Biol Ther 2016; 16:1135–42. [DOI] [PubMed] [Google Scholar]
- 41. Parisi SG, Andreis S, Mengoli C, et al. . Soluble CD163 and soluble CD14 plasma levels but not cellular HIV-DNA decrease during successful interferon-free anti-HCV therapy in HIV-1-HCV co-infected patients on effective combined anti-HIV treatment. Med Microbiol Immunol 2018; 207:183–94. [DOI] [PubMed] [Google Scholar]
- 42. Bruner KM, Murray AJ, Pollack RA, et al. . Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 2016; 22:1043–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yukl SA, Kaiser P, Kim P, et al. . HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med 2018; 10:eaap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pasternak AO, DeMaster LK, Kootstra NA, et al. . Minor contribution of chimeric host-HIV readthrough transcripts to the level of HIV cell-associated gag RNA. J Virol 2016; 90:1148–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ruiz MJ, Plantin J, Pagliuzza A, et al. . Different HIV transcriptional profiles in memory CD4+ T cells subsets during ART. Conference on HIV Science IAS2019 (Mexico City). July 21–24, 2019. [Google Scholar]
- 46. Blackard JT, Sherman KE. HCV/ HIV co-infection: time to re-evaluate the role of HIV in the liver? J Viral Hepat 2008; 15:323–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Van der Sluis RM, Zerbato JM, Rhodes JW, et al. . Diverse effects of interferon alpha on the establishment and reversal of HIV latency. PLoS Pathog 2020; 16:e1008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Baxter AE, O’Doherty U, Kaufmann DE. Beyond the replication-competent HIV reservoir: transcription and translation-competent reservoirs. Retrovirology 2018; 15:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bruner KM, Wang Z, Simonetti FR, et al. . A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 2019; 566:120–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Horsburgh BA, Palmer S. Measuring HIV persistence on antiretroviral therapy. Adv Exp Med Biol 2018; 1075:265–84. [DOI] [PubMed] [Google Scholar]
- 51. Ganesan M, Poluektova LY, Kharbanda KK, Osna NA. Liver as a target of human immunodeficiency virus infection. World J Gastroenterol 2018; 24: 4728–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.