Abstract
Surface functionalization with targeting ligands confers to nanomaterials the ability of selectively recognize a biological target. Therefore, a quantitative characterization of surface functional molecules is critical for the rational development of nanomaterials-based applications, especially in nanomedicine research. Single-molecule localization microscopy can provide visualization of surface molecules at the level of individual particles, preserving the integrity of the material and overcoming the limitations of analytical methods based on ensemble averaging. Here we provide single-molecule localization data obtained on streptavidin-coated polystyrene particles, which can be exploited as a model system for surface-functionalized materials. After loading of the active sites of streptavidin molecules with a biotin-conjugated probe, they were imaged with a DNA-PAINT imaging approach, which can provide single-molecule imaging at subdiffraction resolution and molecule counting. Both raw records and analysed data, consisting in a list of space-time single-molecule coordinates, are shared. Additionally, Matlab functions are provided that analyse the single-molecule coordinates in order to quantify features of individual particles. These data might constitute a valuable reference for applications of similar quantitative imaging methodologies to other types of functionalized nanomaterials.
Keywords: Single-molecule localization microscopy, Super-resolution microscopy, DNA-PAINT, Nanoparticles, Functional materials
Specifications table
| Subject | Nanotechnology |
| Specific subject area | Super-resolution microscopy |
| Type of data | Time-lapse records |
| Lists of single-molecule localization space-time coordinates | |
| Matlab functions | |
| How data were acquired | Total internal reflection fluorescence microscope |
| Instrument: Nikon N-STORM microscope. | |
| Softwares: NIS-elements (Nikon) | |
| Data format | Raw |
| analyzed | |
| Parameters for data collection | Particles on glass coverslip were imaged with continuous laser excitation (647 nm) under total internal reflection conditions. Fluorescence time-lapse records of 10,000–50,000 frames were acquired at 10–20 Hz onto a region of the camera of 40 µm in size. |
| Description of data collection | Binding sites exposed on streptavidin-coated polystyrene particles were loaded with biotin-conjugated short single strands of DNA and seeded on glass coverslip. A complementary free single strand of DNA, conjugated with a fluorophore, was introduced at low concentration (0.1 – 5 nM) in solution and time-lapse fluorescence imaging acquisitions were recorded. |
| Data source location | Institution: Institute for Bioengineering of Catalonia (IBEC) |
| City: Barcelona | |
| Country: Spain | |
| Data accessibility | In a public repository |
| Repository name: 4TU-center for Research Data | |
| Data identification number: uuid:50bd0ad4-52d8-4138-8eb1-40b4d7fc2286 | |
| Direct URL to data: https://doi.org/10.4121/uuid:50bd0ad4-52d8-4138-8eb1-40b4d7fc2286 | |
| Related research article | Delcanale et al., Nanoscale Mapping Functional Sites on Nanoparticles by Points Accumulation for Imaging in Nanoscale Topography (PAINT), ACS Nano, 10.1021/acsnano.7b09063 [1] |
Value of the Data
-
•
The data presented are acquired using a single-molecule localization methodology called DNA-PAINT [2]. These data contain information about the spatial distribution of binding sites on the surface of individual particles. The number of available binding sites can be obtained at single particle level from these data, through a quantitative PAINT procedure [3]. This allows an accurate characterization of heterogeneities within a nanoparticle population.
-
•
Rational design and quantitative characterization of nanomaterials for biomedical applications are often lacking [4], [5], [6], [7]. Single-molecule localization imaging is a promising approach to address this issue [8]. These data might result interesting for researchers who want to apply DNA-PAINT for a quantitative characterization of functional nanomaterials.
-
•
The reported data were obtained with a standard total internal reflection microscope, using commercially available products and relatively simple analysis process. Therefore, they represent an easy-to-replicate reference model. The same methodology might be employed to a variety of bio-medically relevant materials, differing for type, shape, size and surface functionalization.
1. Data description
The shared data consist in a set of single-molecule localization imaging acquisitions of binding sites exposed on the surface of streptavidin coated polystyrene particles. Data were obtained with DNA Points-Accumulation for Imaging in Nanoscale Topography (DNA-PAINT) procedure [2], where localization of single-molecules is associated to the transient annealing between short single strands of DNA: a docking strand loaded on particles binding sites and a dye-conjugated imager strand freely diffusing in solution. Raw experimental records are diffraction limited time-lapses of several thousand frames, therefore very large, and are shared as TIF files. Time-lapse acquisitions were analysed using a NIS-elements software from Nikon in order to identify single-molecule positions with sub-diffraction resolution. Data generated after this analysis are shared as text files (TXT format) comprising all the values retrieved by the analysis, including the space-time coordinates of single molecules (localizations). The shared data were obtained from four different types of experiments, carried out using the same instrumentation and methodology, and were grouped accordingly in four subsets: one colour data; three-dimensional data; multi-colour data; titration data. Additionally, Matlab functions are provided that process text files in order to extract specific parameters of individual particles. A detailed description of the different data subsets is offered below, while a complete list of all the file names together with the corresponding list of exact parameters employed for sample preparation, acquisition and analysis are provided as metadata in the repository.
1.1. One colour data
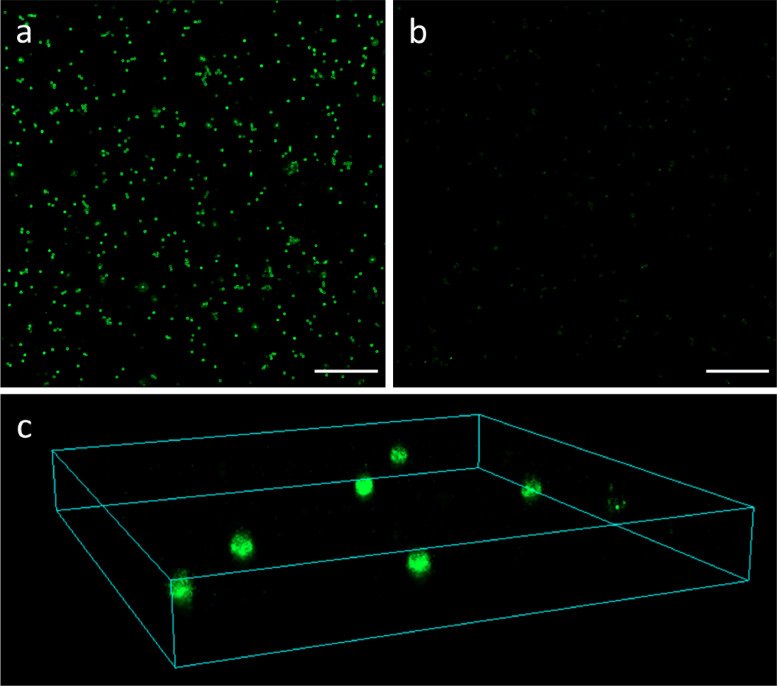
Two time-lapse acquisitions for particles imaged with complementary docking-imager pair and one acquisition obtained with wrong docking-imager are shared as TIF files. The last one constitutes a control experiment to assess the specificity of this imaging methodology, which relies on the selective recognition between DNA sequences. Additionally, the corresponding text files obtained from the analysis of these time-lapse acquisitions with the NIS-elements software are provided. Fig. 1a and b shows to two rendered DNA-PAINT images obtained from these data, for particles imaged with a complementary (Fig. 1a) and non-complementary (Fig. 1b) docking-imager pair.
Fig. 1.
Representative examples of DNA-PAINT images of particles reconstructed from provided data. (a-b): Two-dimensional DNA-PAINT images obtained using a complementary (a) and a non-complementary (b) docking-imager strands pair. Scale bars 5 µM. (c): Three-dimensional DNA-PAINT image of a selected region of interest, obtained with complementary docking-imager pair. Size 8 × 8 × 1 µm.
1.2. Three-dimensional data
An additional time-lapse acquisition obtained for particles imaged with complementary docking-imager pair is shared. This was acquired inserting a cylindrical lens in the light path so that, besides x-y coordinates, an additional z-coordinate can be retrieved by the analysis, leading to three-dimensional single-molecule localization. The corresponding text file obtained with the NIS-elements software and containing the results of the analysis is shared. A selected region of interest from a rendered DNA-PAINT image obtained from this data is showed in Fig. 1c.
1.3. Multi-colour data
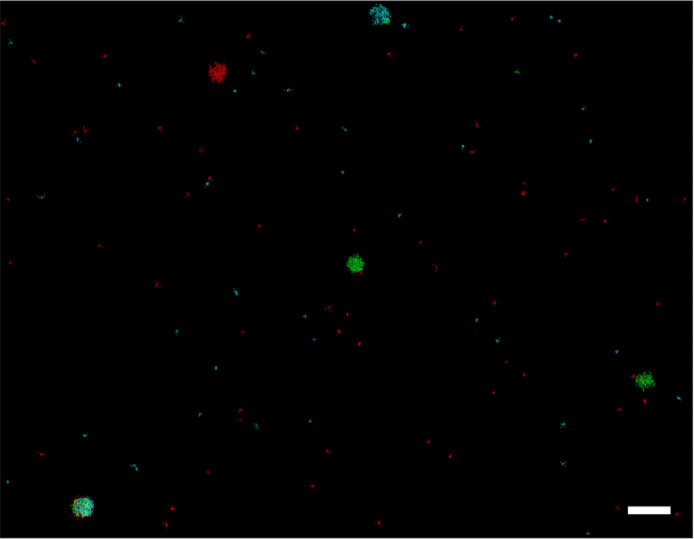
Two-dimensional DNA-PAINT multi-colour data are also shared. In this case, the sample consists of a mix of streptavidin-coated particles that expose different docking strands on their surface. Using three types of biotin-conjugated docking strands (docking1, docking2 and docking3), four populations of particles were prepared exposing: only docking1, only docking2, only docking3, or all of them simultaneously. Three separate time lapse records (TIF format) acquired on the same field of view are shared, together with the corresponding text files obtained from analysis with the NIS-elements software. Each of these acquisitions was obtained introducing sequentially three imager strands (imager1, imager2 and imager3), bearing the same dye but having an oligonucleotide sequence complementary to docking1, docking2 and docking3, respectively. Fig. 2 shows a selected region of interest obtained from a multi-colour rendered DNA-PAINT image that was reconstructed from these data. In this figure, an arbitrary false colour (cyan, green, red) was assigned to the three DNA-PAINT images of the same field of view obtained with the three imager strands (imager1, imager2 and imager3), that were subsequently merged.
Fig. 2.
Representative example of a multi-colour DNA-PAINT image of particles loaded with different docking strands, reconstructed from provided data. Different false colours are assigned to results obtained with imager strand 1 (cyan), 2 (green) and 3 (red), complementary to docking1, docking2 and docking3, respectively. Scale bar 0.6 µM. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
1.4. Titration data
Additional two-dimensional single-color DNA-PAINT data are shared. These were obtained on six different samples of the same streptavidin-coated particles, that were loaded with six increasing concentrations of docking strand. Multiple acquisitions (1 to 4) were collected for each of these samples, forming a statistically significant dataset. Due to the large size, only one representative raw time-lapse acquisition (TIF format) is shared for each of these six conditions. Conversely, all the text files (1–4 for each condition, 16 in total) obtained from the analysis of all the time-lapse acquisitions with the NIS-elements software are provided. Additional time-lapse acquisitions can be made available by the authors upon reasonable request.
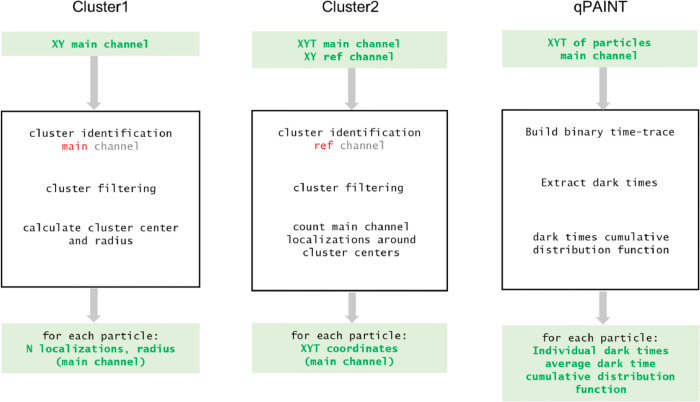
Finally, four Matlab functions are provided for the analysis of the text files generated with the NIS-elements software. ReadCoords.m reads the space-time coordinates (x-y-t) of single-molecules in the text files obtained from the NIS-elements software and store them. Cluster1.m and Cluster2.m identify particles in the DNA-PAINT data as clusters of localizations and calculate relevant parameters of individual particles such as size and number of localizations. Cluster1.m requires one set of coordinates as input (main channel) while Cluster2.m requires two set of coordinates: a reference channel used to identify the particles centers, and a main channel. Finally, qPAINT.m provides further processing, reconstructing a time-trace for each of the previously identified particle. An overview of these functions is offered in Fig. 4 and a more detailed description of the analysis methodology is given in the following section, while examples of the commands required for setting parameters and running the functions in the Matlab environment are provided as metadata.
Fig. 4.
Schematic workflow of the operations implemented in the provided codes for analysis.
2. Experimental design, materials, and methods
These data were acquired using a commercial N-STORM super-resolution microscope from Nikon, equipped for total internal reflection and endowed with Perfect Focus system to prevent z-drift. Fluorescence was detected by a Hamamatsu ORCA Flash 4.0 CMOS camera. Three dimensional acquisitions (subset of data named Three-dimensional data) were obtained with the astigmatism method by introducing a cylindrical lens in the light path [9]. Extensive description of the materials, experimental setup and sample preparation can be found elsewhere [1].
Briefly, streptavidin-coated polystyrene particles (320 nm diameter) were incubated with biotin-conjugated DNA docking strand. For the subsets of data named one-colour data and three-dimensional data, particles were exposed to a large excess (10 μM) of docking strand in order to saturate the surface. Conversely, for the subset of data named titration data, the particles in the six samples were exposed to six docking strand concentrations (0.5, 5, 20, 50, 500, 6250 nM) in order to obtain an increasing amount of strands exposed on the particle surface in the different samples.
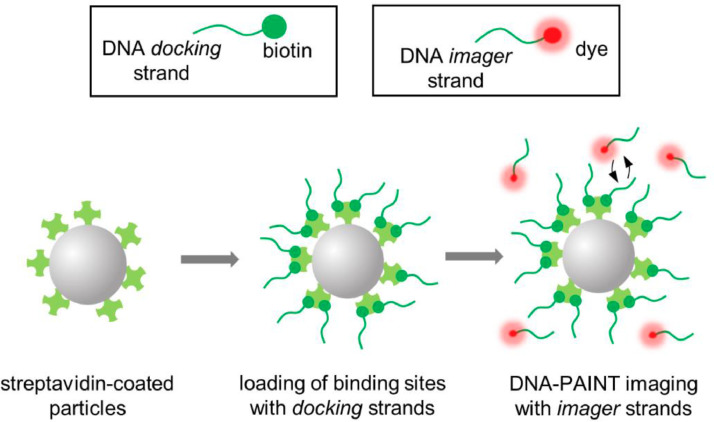
After loading of the DNA docking strands on available surface binding sites, the particles were adsorbed on a pre-cleaned glass coverslip. Then, Atto647N-conjugated DNA imager strands were introduced at low concentrations (0.1 to 5 nM) for DNA-PAINT imaging. A specific imaging buffer was employed to reduce non-specific interactions of the imager strand onto glass coverslip surface. A schematic representation of the sample preparation and DNA-PAINT imaging procedure is displayed in Fig. 3.
Fig. 3.
Schematic representation of the procedure followed for sample preparation and DNA-PAINT imaging of sites exposed on streptavidin coated particles.
In addition, before loading the binding sites with docking strands, some streptavidin molecules (approximately 30 per particle) exposed on the particle surface were covalently labelled with Cy3 dye, emitting in a spectrally distinguishable acquisition channel than the imager strand. This procedure was useful to localize the particles center through the signal of Cy3. This signal was employed as fiducial marker during data analysis for correction of the mechanical drift occurring in x-y along the acquisition. A detailed description of this procedure is found in Ref. [1].
For the subset of data named multi-colour data, three biotin-conjugated DNA docking strands (docking1, docking2 and docking3), only differing by the order of the bases in the oligonucleotide sequence, were employed. Four populations of the same streptavidin-coated particles loaded with different docking strands (docking1, docking2, docking3 and docking1-2-3) were prepared, mixed and adsorbed on the surface of a microfluidic chamber. The edges of the chamber were connected to two syringes, through micro-tubes, to allow injection and removal of the solutions. Thanks to this microfluidic system, three sequential acquisitions of the very same field of view were obtained exchanging the imaging solution, without removing the sample from microscope holder. A measurement cycle consisted in: (I) introduction of a solution containing the imager strand, (II) acquisition of a time-lapse record and (III) washing of the imager strand solution using buffer. The cycle was repeated three times introducing sequentially three Atto647N-conjugated imager strands (imager1, imager2 and imager3), having an oligonucleotide sequence complementary to docking1, docking2 and docking3 respectively. More detailed description of the multi-colour imaging with DNA-PAINT can be found elsewhere [1,2].
All the diffraction-limited movies were analysed using a tool in the NIS-elements software, from Nikon. This software generates a list of single-molecule localizations by Gaussian fitting diffraction-limited fluorescent spots in the acquired movies [10,11]. No additional filters are applied and all the information retrieved by the analysis is reported. Finally, the data obtained by the analysis with NIS-elements and provided as text file can be further processed using different codes, developed with Matlab, in order to obtain quantitative information at the level of individual particles. Here we provide four main different Matlab functions, obtained by the modification of previously developed codes [12], for analysis of two-dimensional DNA-PAINT localizations obtained on particles with spherical shape. First, a preliminary function ReadCoords.m scan the text file in the format generated by NIS-elements, extract the x-y-t coordinates for two different acquisition channels (a main channel and a reference channel) and store these coordinates in separate files of reduced size. These coordinates can be further analysed with other provided Matlab functions.
Cluster1.m reads two-dimensional spatial coordinates (x-y, in nm) of the DNA-PAINT main channel (imager strand signal) and performs a mean-shift clustering algorithm, based on a user-introduced bandwidth parameter (in nm), to identify clusters of localizations corresponding to particles. In order to remove clusters with unrealistic size and shape, they are then filtered according to user defined parameters: minimum number of localizations, maximum elongation and size, and minimum distance between clusters. Finally, refined mass center positions, size and number of localizations is returned for each selected particle. In addition, the function produces four graphical outputs for a visual evaluation of the results: (I) A plot of the localizations with the results of cluster filtering: clusters having a number of localization above the threshold value are fitted with an ellipse model and discarded if too elongated (cyan ellipses) or too close (yellow ellipses), while ellipses corresponding to selected clusters are reported in magenta. The indexes assigned to the selected clusters are also displayed. (II) A plot displaying in red the localizations within selected clusters, together with circles corresponding to the optimized radius retrieved by the analysis. Other localizations are in black. (III) Histogram with the distribution of localization number per selected cluster. (IV) Histogram with the diameter distribution. For testing Cluster1.m, the following values can be employed as an initial guess for the required input parameters, even though more appropriate values should be selected depending on the data: bandwidth = 250 nm; minimum number of localizations in cluster = 80; maximum diameter = 400 nm. Additional optional input parameters can be set to default values or modified for further refinement of the analysis.
Cluster2.m reads two-dimensional space-time coordinates (x-y-t, in nm and frame number) of both main channel (imager strand signal) and space coordinates (x-y, in nm) of reference channel (in these data, associated to Cy3-conjugated particles). A clustering analysis followed by a cluster filtering process are then performed on the localizations in the reference channel. This procedure aims to identify the center positions of particles, as more extensively described in [1]. Finally, the function identifies and returns, for each particle, all the space-time coordinates (x-y-t) of localizations of the main channel that are located within a user-defined distance from the particle center. The function produces three graphical outputs for a visual evaluation of the results: (I) A plot of the localizations with the results of cluster filtering on the reference channel: the clusters having a number of localization above the threshold value are fitted with an ellipse model and discarded if too elongated (cyan ellipses) or too close (yellow ellipses), while ellipses corresponding to selected clusters are reported in magenta. The indexes assigned to the selected clusters are also displayed. (II) A plot displaying in red the localizations of the main channel associated to selected clusters. Localizations in reference channel are in green and non-selected localizations of the main channel are black. Red circles define the areas in which main-channel localizations are considered associated to the particle. Black circles indicate the retrieved radius of the cluster of localizations in reference channel. Indexes assigned to selected clusters are reported. (III) Histogram with the distribution of main-channel localizations within selected clusters. For testing Cluster2.m, the following values can be employed as an initial guess for the required input parameters, even though more appropriate values should be selected depending on the data: bandwidth = 50 nm; minimum number of localizations in cluster = 30; maximum cluster diameter = 200 nm; maximum distance of main-channel localizations from center = 400 nm. Additional optional input parameters can be set to default values or modified for further refinement of the analysis. The output generated by Cluster2, can be read by the function qPAINT.m that performs an analysis of the time-trace on the main channel signal, for each particle detected. This function generates a binary time-trace for each particle, assigning a zero-value to acquisition frames without localization detected on the particle and a one-value to frames with one (or more) localization detected. Then, dark times are extracted for each time-trace, corresponding to consecutive dark frames between non-zero frames. Individual dark times are stored for each particle and their cumulative distribution function and average value are calculated and returned.
Acknowledgments
Acknowledgments
The authors The authors thank the Spanish Ministry of Economy, Industry and Competitiveness through the project SAF2016-75241-R, the Generalitat de Catalunya through the Centres de Recerca de Catalunya (CERCA) programme, the EuroNanoMed II platform through the NanoVax project, the Obra Social La Caixa foundation and the European Research Council (ERC- StG-757397).
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.105468.
Appendix. Supplementary materials
References
- 1.Delcanale P., Miret-Ontiveros B., Arista-Romero M., Pujals S., Albertazzi L. Nanoscale mapping functional sites on nanoparticles by points accumulation for imaging in nanoscale topography (PAINT) ACS Nano. 2018;12:7629–7637. doi: 10.1021/acsnano.7b09063. [DOI] [PubMed] [Google Scholar]
- 2.Schnitzbauer J., Strauss M.T., Schlichthaerle T., Schueder F., Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 2017;12:1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- 3.Jungmann R., Avendaño M.S., Dai M., Woehrstein J.B., Agasti S.S., Feiger Z., Rodal A., Yin P. Quantitative super-resolution imaging with qPAINT. Nat. Methods. 2016;13:439–442. doi: 10.1038/nmeth.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Meel R., Sulheim E., Shi Y., Kiessling F., Mulder W.J.M., Lammers T. Smart cancer nanomedicine. Nat. Nanotechnol. 2019;14:1007–1017. doi: 10.1038/s41565-019-0567-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabanel J.-.M., Adibnia V., Tehrani S.F., Sanche S., Hildgen P., Banquy X., Ramassamy C. Nanoparticle heterogeneity: an emerging structural parameter influencing particle fate in biological media? Nanoscale. 2019;11:383–406. doi: 10.1039/C8NR04916E. [DOI] [PubMed] [Google Scholar]
- 6.Pujals S., Albertazzi L. Super-resolution microscopy for nanomedicine research. ACS Nano. 2019;13:9707–9712. doi: 10.1021/acsnano.9b05289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faria M., Björnmalm M., Thurecht K.J., Kent S.J., Parton R.G., Kavallaris M., Johnston A.P.R., Gooding J.J., Corrie S.R., Boyd B.J., Thordarson P., Whittaker A.K., Stevens M.M., Prestidge C.A., Porter C.J.H., Parak W.J., Davis T.P., Crampin E.J., Caruso F. Minimum information reporting in bio–nano experimental literature. Nat. Nanotechnol. 2018;13:777–785. doi: 10.1038/s41565-018-0246-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pujals S., Feiner-Gracia N., Delcanale P., Voets I., Albertazzi L. Super-resolution microscopy as a powerful tool to study complex synthetic materials. Nat. Rev. Chem. 2019;3:68–84. doi: 10.1038/s41570-018-0070-2. [DOI] [Google Scholar]
- 9.Huang B., Wang W., Bates M., Zhuang X. Three-Dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Betzig E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 11.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Zwaag D., Vanparijs N., Wijnands S., De Rycke R., De Geest B.G., Albertazzi L. Super resolution imaging of nanoparticles cellular uptake and trafficking. ACS Appl. Mater. Interfaces. 2016;8:6391–6399. doi: 10.1021/acsami.6b00811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.